Abstract
Vesivirus 2117 is an adventitious agent that, in 2009, was identified as a contaminant of Chinese hamster ovary cells propagated in bioreactors at a pharmaceutical manufacturing plant belonging to Genzyme. The consequent interruption in supply of Fabrazyme and Cerezyme (drugs used to treat Fabry and Gaucher diseases, respectively) caused significant economic losses. Vesivirus 2117 is a member of the Caliciviridae, a family of small icosahedral viruses encoding a positive-sense RNA genome. We have used cryo-electron microscopy and three-dimensional image reconstruction to calculate a structure of vesivirus 2117 virus-like particles as well as feline calicivirus and a chimeric sapovirus. We present a structural comparison of several members of the Caliciviridae, showing that the distal P domain of vesivirus 2117 is morphologically distinct from that seen in other known vesivirus structures. Furthermore, at intermediate resolutions, we found a high level of structural similarity between vesivirus 2117 and Caliciviridae from other genera: sapovirus and rabbit hemorrhagic disease virus. Phylogenetic analysis confirms vesivirus 2117 as a vesivirus closely related to canine vesiviruses. We postulate that morphological differences in virion structure seen between vesivirus clades may reflect differences in receptor usage.
Keywords: calicivirus, capsid, virus structure, cryo-EM
Abbreviations
3D, three-dimensional; CHO, Chinese hamster ovary; cryo-EM, cryogenic electron microscopy; EM, electron microscopy; FCV, feline calicivirus; NoV, Norwalk virus; RHDV, rabbit hemorrhagic disease virus; SMSV, San Miguel sea lion virus; SV, sapporovirus; VESV, vesicular exanthema of swine virus; VLP, virus-like particle.
Introduction
Caliciviruses are non-enveloped icosahedral viruses that have single-stranded, positive-sense RNA genomes. The Caliciviridae are divided into five genera, namely, Norovirus, Sapovirus, Vesivirus, Lagovirus and Nebovirus. The representative viruses of these genera are Norwalk virus (NoV), sapporovirus (SV), feline calicivirus (FCV), rabbit hemorrhagic disease virus (RHDV) and Newbury-1 virus, respectively. The noroviruses and sapoviruses cause gastroenteritis in humans, while neboviruses have been shown to cause gastroenteritis in cattle. Vesiviruses and lagoviruses cause a range of symptoms in different animal species including stomatitis, conjunctivitis, respiratory illness and hemorrhagic disease [1–3].
Calicivirus genomes are around 7.5 kb in length, encode up to four ORFs and are polyadenylated. The first ORF encodes the non-structural proteins and in the lagoviruses and sapoviruses the major capsid protein, VP1. In the genera Norovirus and Vesivirus, the VP1 protein is encoded by a second ORF and the minor structural protein, VP2, is encoded by ORF3, which is translated following ribosomal termination–reinitiation [4]. The ORF1 polyprotein is post-translationally cleaved by the autocatalytic viral protease to yield several non-structural proteins [5]. The major capsid protein, VP1, is also post-translationally processed, resulting in the removal of the N-terminal 124 amino acids to produce the mature form of the protein (62 kDa) [6]. Murine norovirus (MuNV) is the only member of the Caliciviridae that encodes a fourth ORF, the product of which, referred to as VF1, is involved in the regulation of the innate response to infection [7].
Members of the Caliciviridae exhibit a characteristic virion morphology of 32 cup-shaped depressions on their outer surface. The T=3 capsid is composed of three quasi-equivalent forms of the VP1 protein, termed A, B and C. These give rise to two classes of dimeric capsomere: A/B and C/C. Ninety dimers/capsomeres assemble to form a 35–40 nm spherical capsid that encloses the viral genome. A/B and C/C capsomeres differ only slightly in their conformations. An atomic model of RHDV shows the A/B capsomeres (described as bent) arranged around the fivefold symmetry axis, while the C/C capsomere, which adopts a flatter conformation, is located at the twofold symmetry axis [8–11]. The VP1 protein has been divided into three domains: the N-terminal arm, the shell domain (S) and the protruding domain (P). The N-terminal arm is hypothesized to be involved in the switch between the A/B and C/C conformations. The S domain contains a β-barrel motif and forms the floor of the viral capsid. Sequences within the S domain have been shown to interact with the viral RNA-dependent RNA polymerase to promote de novo RNA synthesis [12]. The P domain is further divided into two subdomains, P1 and P2, with the P2 domain containing the antigenic and receptor-binding sites [13–15].
Recently, vesivirus 2117 was identified as a contaminant of bioreactors containing Chinese hamster ovary (CHO) cells at Genzyme in both their Allston Landing and Geel sites. This led to an interruption in production of Cerezyme and Fabrazyme biopharmaceuticals for the treatment of patients suffering from Gaucher disease and Fabry disease, respectively. Vesivirus 2117 was first described as an adventitious agent from an unknown source following observations of cytopathic changes in CHO cells grown in culture [16]. Electron microscopy (EM) showed the presence of viral particles measuring approximately 40 nm in diameter and exhibiting the typical morphology of caliciviruses. Upon infection of CHO cells with vesivirus 2117, cells become rounded and detach from cell culture plastic within 24 h, which accounts for the loss of cell viability seen at Genzyme. It is thought that the viral contamination at Genzyme was introduced from reagents used in the manufacturing process. It was estimated that approximately 5×109 virus particles were present in each millilitre of bioreactor fluid. Subsequently, the bioreactors were closed for decontamination, causing a significant delay in the production of Cerezyme and Fabrazyme, which affected around 8000 patients. Stock analysts estimate that the contamination and interruption in manufacturing may have cost Genzyme in the region of $200–300 million [17].
Adventitious agent testing is mandated for the prevention of contamination by viruses able to replicate in CHO cells and has led to the detection of five incidents in the past 20 years caused by murine minute virus, reovirus, Cache Valley virus, epizootic hemorrhagic disease virus and vesivirus 2117 [18–23]. Currently, three processes are adopted to minimize the risk of introducing contaminants into cell culture: control and testing of raw materials, testing at key stages during manufacturing and the use of virus inactivation techniques. These steps are necessary owing to the extent to which CHO cells in particular are used for the production of therapeutic agents. Although these cells are less permissive to infection than, for example, baby hamster kidney cells, episodes of contamination have been documented. In each case, the source of the contaminants was suspected to be input raw materials. Since 1998, the Food and Drug Administration has made it a regulatory requirement in the production of biopharmaceuticals to demonstrate the lack of adventitious agents [18, 20, 24, 25].
Another calicivirus, SV (the type member of the genus Sapovirus), was first identified by EM in 1976 from stool samples of infants presenting with gastroenteritis. Sapovirus strains are classified into five genogroups (GI–GV) based on their capsid protein sequences. Currently, only GIII viruses can be propagated in cell culture; however, members of this genogroup cause porcine infections, while the other genogroups cause gastroenteritis in humans. As is the case for many members of the Caliciviridae, expression of a recombinant form of the major capsid protein is sufficient for the assembly of virus-like particles (VLPs), which exhibit the same morphology as virions [26–29].
Here we describe the structures of VLPs formed by recombinant expression of vesivirus 2117 VP1 and a chimeric sapovirus VP1, alongside an improved structure for FCV strain F9, all solved by cryogenic EM (cryo-EM) and three-dimensional (3D) image reconstruction. These data reveal that the vesivirus 2117 capsid structure more closely resembles that of sapoviruses, and surprisingly a lagovirus, than it does other known vesivirus structures. Phylogenetic analysis of capsid protein sequences supports the classification of vesivirus 2117 as a member of the genus Vesivirus, although it resides in a clade distinct from FCV and the vesicular exanthema of swine virus (VESV) and San Miguel sea lion virus (SMSV) groups. It has recently been proposed that structural similarities may be used to infer common heritage of diverse virus groups in the absence of genetic similarity [30]. Here we see substantial differences in virion morphology between clades of the genus Vesivirus and, albeit at comparatively low resolution, similarities between vesivirus 2117 and structures for both sapoviruses and lagoviruses. The striking differences in P domain morphology within vesiviruses may indicate functional differences, perhaps related to receptor usage and entry mechanisms.
Results
Vesivirus 2117 VLPs are structurally distinct from known vesivirus capsid structures
Vesivirus 2117 VLPs were prepared by baculovirus expression of VP1 in Hi5 insect cells and purified by differential centrifugation. VLP preparations were vitrified by plunge freezing in liquid ethane and imaged in a frozen-hydrated state by cryogenic transmission electron microscopy (Fig. 1a). Nine hundred and sixty particle images were extracted from 242 micrographs and processed to calculate a 3D icosahedral reconstruction of the VLP at a resolution of 10 Å (Fig. 1b, c). Vesivirus 2117 VLPs were seen to exhibit the characteristic calicivirus morphology: a T=3 icosahedral capsid composed of VP1 dimers, giving rise to arch-like capsomeres. The VP1 dimers also appear to form intradimeric interactions between P2 domains and interdimeric interactions between P1 domains, similar to those seen in FCV and as previously described for SMSV [31]. Viewed perpendicular to the capsid floor, a side view of the VP1 dimeric capsomere (Fig. 1d) shows that the outer surface of the P2 domain has pronounced horn-like structures at either end. This is quite distinct from the morphology seen in previously solved vesivirus capsid structures [13, 14, 32]. This morphological divergence from known vesivirus capsid structures led us to hypothesize that 2117 might represent an intermediate between classical vesiviruses and other genera. We therefore set out to compare 2117 VLPs to other calicivirus capsid structures.
Fig. 1.
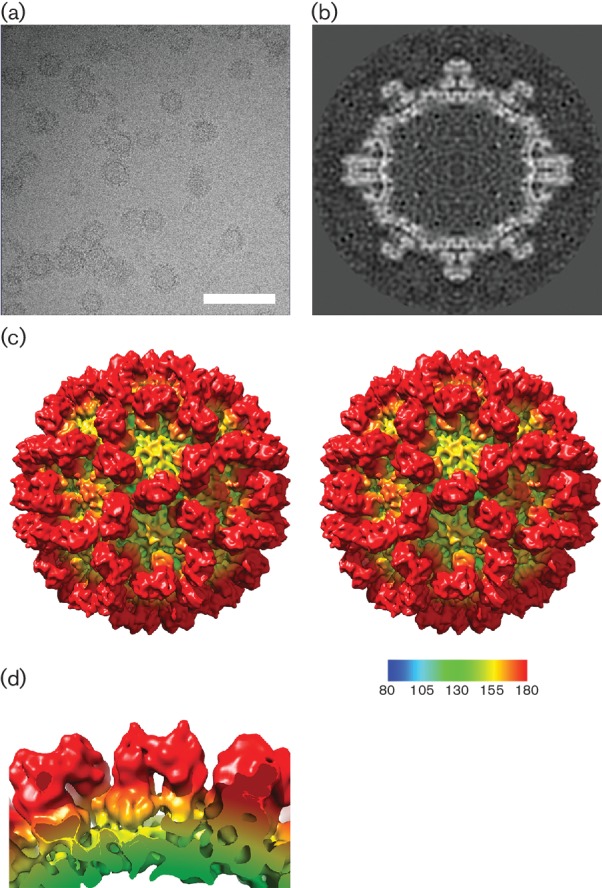
Cryo-EM structure of vesivirus 2117 at 10 Å resolution. (a) Cryo-electron micrograph of vesivirus 2117 VLPs imaged in a frozen-hydrated state. Bar, 100 nm. (b) A central slice through the 3D reconstruction of vesivirus 2117 shows the compact structure of the P domain. (c) Stereo pair images of the reconstruction, calculated at 10 Å resolution, viewed along the twofold symmetry axis. (d) A side view of the 2117 VP1 dimer viewed parallel to the capsid surface highlights the pronounced horn-shaped structures on the outer faces of the P domains.
Comparison of vesivirus 2117 VLP structure to that of a chimeric sapovirus
We observed that our 2117 structure closely resembled a published low-resolution structure of Parkville virus (a sapovirus) [31]. To draw a comparison between vesivirus 2117 capsids and those of the genus Sapovirus at higher resolution, we calculated a 3D structure from images of VLPs produced by a chimeric VP1 construct that has been shown to yield high levels of expression [28]. Briefly, a construct consisting of amino acids 1–289 of the Yokote strain VP1 and amino acids 290–560 of the Mc114 sapovirus strain was used to produce VLPs. The chimera consisted of the N-terminal arm, S domain and P1.1 domain of the Yokote strain and the P2 domain, P1.2 domain and VP2 of the Mc114 strain [28]. These particles were imaged in a frozen-hydrated state (Fig. 2a). A total of 2943 particles were picked from 222 micrographs and used to calculate a 3D reconstruction of the chimeric sapovirus VLP at 10 Å resolution (Fig. 2b, c). The sapovirus structure also exhibits typical calicivirus morphology, with notable similarities to that of vesivirus 2117. In particular, the P2 region of the dimeric capsomeres is compact and rounded. The outermost surface of the P2 domain of vesivirus 2117 shows horn-like protrusions at either end, whereas the P2 domains of the sapovirus VLPs appear to have a more curved structure without such large protuberances (Fig. 2d). The S and P1 domains of the two structures appear quite similar at this resolution.
Fig. 2.
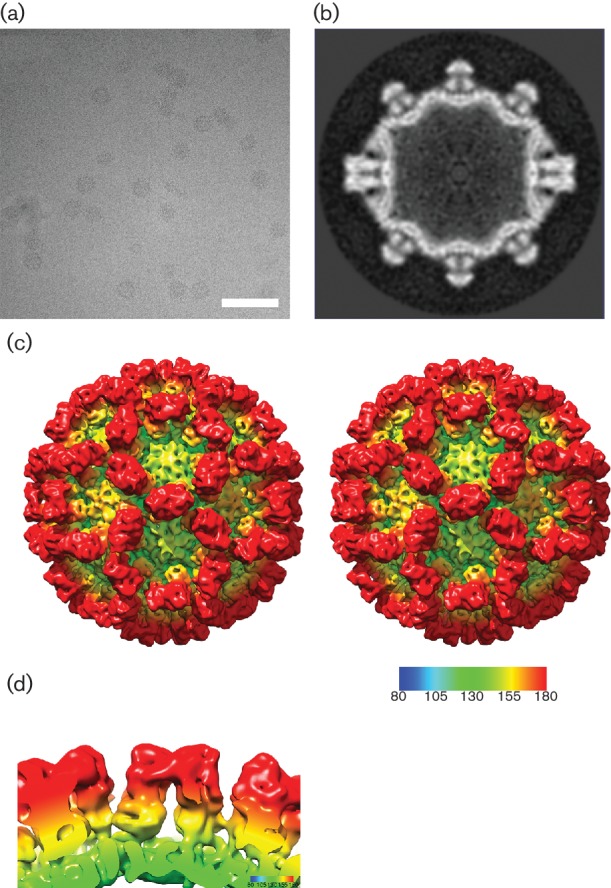
3D reconstruction of a chimeric sapovirus VLP. (a) Cryo-electron micrograph of chimeric sapovirus-like particles. Bar, 100 nm. (b) A central slice through the sapovirus VLP structure reveals a compact P domain structure, similar to that seen in 2117. (c) Stereo pair images of the reconstruction calculated at 10 Å resolution, viewed along the twofold symmetry axis. (d) Side view of the sapovirus VP1 dimer.
Structure of FCV at 7 Å resolution
We have extended the resolution of our structure for the vesivirus FCV strain F9 to 7 Å resolution using a new dataset. A total of 6965 particles were picked from 241 micrographs (Fig. 3a) and used to calculate the 3D reconstruction shown in Fig. 3(b, c). This structure exhibits striking differences from the vesivirus 2117 and sapovirus structures set out above; the FCV P2 region forms a flattened, rhombus-shaped outer face (Fig. 3c, d). Central sections through each reconstruction (Figs 1b, 2b and 3b) show that this region of the P domain of FCV has a broader conformation than that of either 2117 or sapovirus.
Fig. 3.
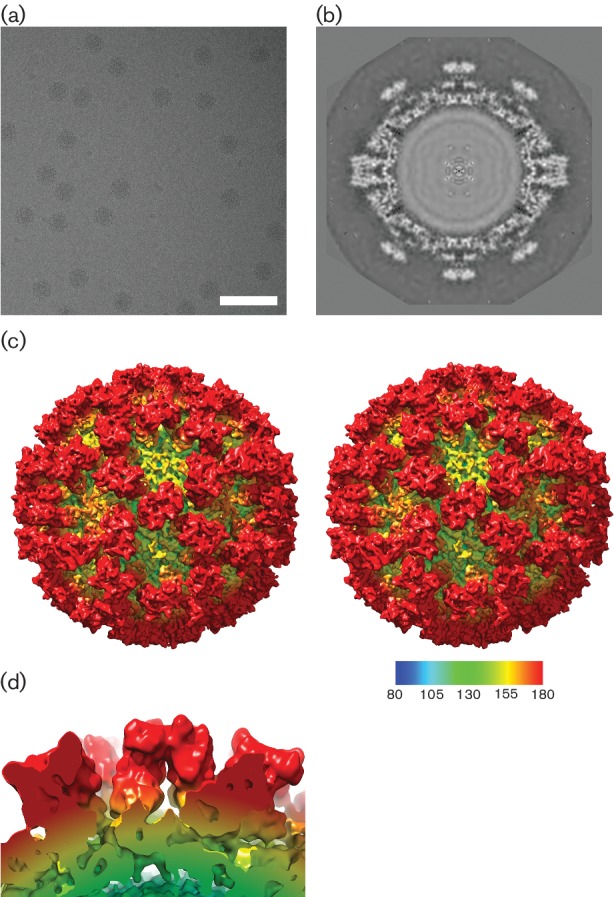
Structure of FCV. (a) Cryo-microscopy of native FCV virions. Bar, 100 nm. (b) The central section shows a broader P domain structure that presents a flatter outer face. (c) Stereo pair images of the reconstruction, calculated at 7 Å resolution, viewed along the twofold symmetry axis. (d) A side view of a dimeric FCV VP1 capsomere.
Structural comparison of members of the Caliciviridae
Given the clear differences in capsomere morphology between these three structures, we sought to extend our comparison to include a capsid structure from each genus of the Caliciviridae. The structures of a lagovirus (RHDV: EMDB 1933; Fig. 4c) and two noroviruses (EMDB 5374 and PDB 1IHM; Fig. 4e, f, respectively) were used to calculate density maps filtered to 10 Å resolution. Likewise, the sapovirus (Fig. 4b) and FCV (Fig. 4d) structures presented above were filtered to 10 Å (the resolution attained for vesivirus 2117; Fig. 4a). Fig. 4 shows a side-by-side comparison of the six 3D structures viewed along the twofold symmetry axis. The similarities (and differences) between the maps are clearly recognized at this resolution. Fig. 5 shows a side-by-side comparison of the P domains of each of the viruses compared in Fig. 4. The P domains of vesivirus 2117, sapovirus and RHDV (Fig. 5a–c, respectively) appear rather similar, showing a compact morphology. The capsomeres of FCV, norovirus GII.10 and NoV (Fig. 5d–f, respectively), however, appear to be quite distinct in conformation; indeed, there are notable differences between the two norovirus structures GII.10 and Norwalk. The P domains of vesivirus 2117, sapovirus and FCV appear to be raised off the S domain, as previously observed for RHDV and MuNV-1, in contrast to the P domains of NoV, which form a more collapsed conformation in relation to the S domain [33, 34].
Fig. 4.

Side-by-side comparison of (a) vesivirus 2117, (b) a chimeric sapovirus, (c) RHDV, (d) FCV, (e) norovirus GII.10 and (f) NoV at 10 Å resolution, viewed along the twofold symmetry axis.
Fig. 5.
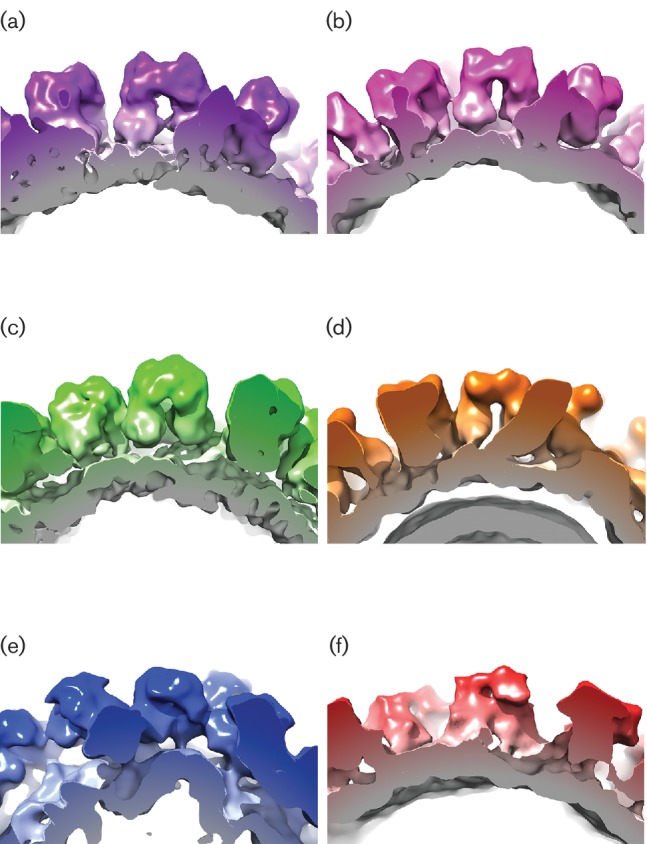
Side-by-side comparison of the P domains of (a) vesivirus 2117, (b) a chimeric sapovirus, (c) RHDV, (d) FCV, (e) norovirus GII.10 and (f) NoV at 10 Å resolution.
To provide a quantitative index of similarity between the maps under investigation, the maps were brought to a common alignment, resolution and scale. Cross-correlation coefficients between vesivirus 2117 and each of the other five structures were calculated (Table 1). In such analyses, identical maps would give a correlation of 1.0. The structure that produced the highest correlation value when aligned to the vesivirus 2117 structure was the chimeric sapovirus described here, giving a correlation value of 0.9214. This might be expected given the apparent similarities in the maps when visualized by isosurface rendering as in Fig. 4. It is immediately apparent that the sapovirus structure is the most comparable to that of vesivirus 2117, in both the S domains and the P domains of the VP1 proteins. When fitting the structure of RHDV into that of vesivirus 2117, a correlation value of 0.9007 was recorded, signifying a high degree of similarity between the two capsids, again in both the S and P domains of VP1. These results are unexpected given that all three viruses (vesivirus 2117, sapovirus and RHDV) are classified into different genera of the Caliciviridae. When the structure of FCV is fitted into the vesivirus 2117 structure, the correlation value obtained is comparatively low at 0.8478. While this value still indicates reasonable similarity, consistent with a common architecture, we might have expected these two viruses to exhibit a higher degree of correlation, as both FCV and vesivirus 2117 are classified in the same genus, Vesivirus. While the NoV structure produced a correlation value of 0.8117, the norovirus GII.10 structure produced a correlation value of 0.8498. This suggests that the structures of FCV and norovirus GII.10 are equally similar to that of vesivirus 2117. When observing the structures in Fig. 4, the P2 domains of norovirus GII.10 seem similar to those of vesivirus 2117, whereas the S domains and P1 domains of FCV seem to resemble those of vesivirus 2117. When comparing the structures side by side, it is likely that different domains of the capsid proteins of the two viruses have contributed to the close correlation values recorded. When the norovirus GII.10 structure is fitted into the FCV structure, a correlation value of only 0.6261 is produced, supporting this hypothesis of different components of the capsids contributing to the similar correlation values.
Table 1. Correlation values calculated by docking structures of several calicivirus capsids into that of vesivirus 2117.
A correlation value of 1.0 indicates perfect agreement.
| Virus structure fitted into vesivirus 2117 | Correlation value |
|---|---|
| A chimeric sapovirus | 0.9214 |
| RHDV | 0.9007 |
| FCV (F9) | 0.8478 |
| NoV | 0.8117 |
| Norovirus GII.10 | 0.8498 |
Phylogenetic analysis of vesivirus 2117
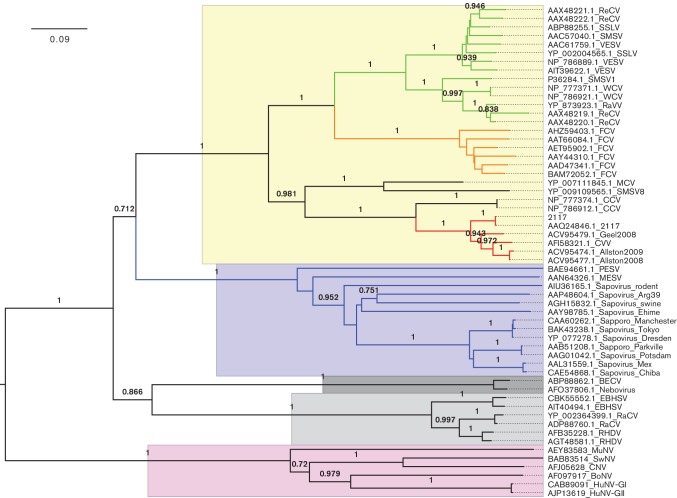
Fifty-one sequences of the VP1 capsid protein from the three genera highlighted as being structurally similar to that of vesivirus 2117 (Vesivirus, Lagovirus and Sapovirus) were aligned and analysed to compute a phylogenetic tree (Fig. 6). As expected, this analysis placed vesivirus 2117 firmly within the Vesivirus genus and showed that the sapoviruses are more closely related to the vesiviruses than the Lagovirus genus. Vesivirus 2117 isolates and canine vesivirus are clearly closely related and occupy their own clade. These viruses along with the canine caliciviruses, SMSV8 and mink calicivirus are distinct from the FCV and VESV clades. Cross-correlation data indicated a high degree of similarity between vesivirus 2117 and both sapovirus and RHDV but a lesser degree when compared with FCV. At 10 Å resolution, gross morphological similarities or differences may not indicate common tertiary structures; therefore, such comparisons should be interpreted with care. However, the P2 domain plays a critical role in the viral infection cycle as it contains both the major immunodominant epitopes and the receptor-binding site [32, 35]. Thus, differences in capsid morphology between the vesivirus clades and similarities between vesivirus 2117 and viruses in different genera within the Caliciviridae may allude to important functional differences and similarities, respectively.
Fig. 6.
Vesivirus 2117 neighbour-joining tree. The evolutionary history of the VP1 capsid of 2117 was inferred using the neighbour-joining method. The vesivirus 2117 cluster is highlighted in red; FCV cluster, in orange; VESV cluster (includes walrus, reptile and sea lion viruses), in green; and the sapovirus cluster, in blue. The proportions of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The genus Vesivirus is highlighted in yellow; the genus Sapovirus, in blue; the genus Nebovirus, in dark grey; the genus Lagovirus, in light grey; and the genus Norovirus, in pink. BECV, bovine enteric calicivirus; BoNV, bovine norovirus; CCV, canine calicivirus; CNV, canine norovirus; CVV, canine vesivirus; EBHSV, European brown hare syndrome virus; FCV, feline calicivirus; HuCV, human calicivirus; HuNV-GI, human norovirus GI; HuNV-GII, human norovirus GII; MCV, mink calicivirus; MuNV, murine norovirus; PESV, porcine enteric sapovirus; RaCV, rabbit calicivirus; RaVV, rabbit vesivirus; ReCV, reptile calicivirus; ReVV, reptile vesivirus; RHDV, rabbit hemorrhagic disease virus; SMSV, San Miguel sea lion virus; SMSV1, San Miguel sea lion virus 1; SSLV, Steller sea lion vesivirus; SwNV, swine norovirus; VESV, vesicular exanthema of swine virus; WCV, walrus calicivirus.
Discussion
Vesivirus 2117 has caused significant financial losses owing to the contamination of bioreactors at Genzyme (Allston Landing and Geel facilities) where CHO cells were used for the production of biopharmaceuticals. The contamination necessitated the shutdown and decontamination of bioreactors, causing a significant delay in the delivery of drugs to patients with Gaucher or Fabry disease (17). The substantial impact of this contamination highlights the need for effective adventitious agent testing.
We have determined the structure of vesivirus 2117 at 10 Å resolution by cryo-EM and 3D reconstruction. We also determined the structure of a chimeric sapovirus at 10 Å resolution and showed the high degree of morphological similarity between the two. Indeed, vesivirus 2117 appeared to be structurally closer to sapovirus and RHDV (a lagovirus) than other vesiviruses, including FCV. However, phylogenetic analysis based on VP1 sequences confirmed that vesivirus 2117 is correctly classified as a vesivirus, also showing that it resides in a clade distinct from FCV and VESV.
The dimeric capsomeres formed of vesivirus 2117 capsid protein present two horn-like structures on the outer surface of the P2 domains. Similar structures are present on the outer faces of P domains of sapovirus VP1, although they are less pronounced. MuNV also exhibits prominent horn-like protrusions on the outer surface of the P2 domain of VP1 [34]. Conversely, the P2 domains of FCV form a rhombus-shaped structure with a flatter outer surface (similar to that of SMSV and Tulane virus [36, 37]). Despite this obvious morphological difference, sequence analysis suggests that FCV is indeed the calicivirus most closely related to vesivirus 2117 for which we have a capsid structure.
The outer face of the FCV capsomere incorporates the receptor-binding site. Two molecules of feline junctional adhesion molecule-A bind to the flat surface in a head-to-tail arrangement [13, 32]. Receptor binding induces conformational changes in the capsid that are hypothesized to prime the virion for genome uncoating in the late endosome. FCV is the only calicivirus that has been shown to enter via attachment to a protein receptor. Sapoviruses and MuNV are thought to bind sialic acid moieties to mediate attachment [38, 39], while RHDV and human noroviruses bind histo-blood group antigens – complex carbohydrates linked to glycoproteins or glycolipids on the surface of erythrocytes and mucosal epithelial cells [40–42]. Our morphological comparison of vesivirus 2117 and sapovirus may therefore provide clues to entry pathways exploited by the 2117 and CCV clades of the genus Vesivirus.
There is a paucity of data concerning pathology caused by vesiviruses in the CCV and 2117 clades. Reports of enteritis associated with CCV suggest the possibility of sapovirus-like disease [43], whereas for 2117, the natural host and pathology remain unknown.
Interestingly, the first and second ORFs of vesivirus 2117 are separated by a stop codon, which is typical of the noroviruses and vesiviruses. However, the capsid protein is encoded within the first ORF along with the viral non-structural proteins, typical of the sapoviruses and lagoviruses [16]. This has, to date, only been described for canine calicivirus [44], which is possibly the closest related virus to vesivirus 2117.
While phylogenetic analysis indicated a close genetic relatedness between vesivirus 2117 and canine vesivirus, the striking morphological similarities with sapoviruses led us to question the taxonomic classification of vesivirus 2117 as a vesivirus. However, based on sequence data alone, vesivirus 2117 is undoubtedly more closely related to FCV than it is to the genus Sapovirus. Structural analysis has been proposed to provide insights into the common ancestry of distantly related viruses following the discovery of fold conservation between viruses that infect highly divergent branches on the tree of life [30]. Within the Caliciviridae, VP1 proteins share a common topology. The fact that the outer face of the P domain presents the receptor-binding site and major immunodominant epitopes means that it is subject to significant evolutionary pressure from immune surveillance. Typically, receptor-binding sites of viruses are embedded in hyper-variable regions of capsid proteins. Thus, morphological differences/similarities in the P domain may be uncoupled from genetic relatedness but may provide important clues concerning critical aspects of virus biology, such as entry pathway.
Methods
Virus culture and purification
Vesivirus 2117 VLPs were produced by baculovirus expression of the VP1 gene (lacking the predicted leader sequence) in Hi5 cells. Six days post-infection, cells were freeze–thawed and the lysate was clarified by centrifugation at 14 000 g for 30 min. The supernatant was filtered (0.45 µm vacuum filter) and protein was precipitated by addition of polyethylene glycol to a final concentration of 10 % followed by incubation at 4 °C. The precipitate was centrifuged at 8000 g and 4 °C for 30 min. The VLP-containing pellet was then resuspended in boric acid buffer (0.2 M boric acid, 0.5 M NaCl, pH 7.5). VLPs were then centrifuged through a sucrose cushion (30 %, 150 000 g at 4 °C). The VLP pellets were subsequently resuspended in boric acid buffer, centrifuged (8000 g, 4 °C for 10 min) and mixed with an equal volume of PBS containing 0.5 M NaCl and 4.51 M CsCl for ultracentrifugation using a Sw55 Ti rotor at 40 000 r.p.m. for 20 h at 4 °C. Purified VLPs were then collected and dialysed against PBS.
FCV strain F9 was propagated in Crandell Rees feline kidney cells for 8 h. The virus-infected cells were pelleted from the culture medium by centrifugation (1500 g, 10 min at 4 °C). The pellet was resuspended in TBS (250 mM NaCl and 85 mM Tris/HCl, pH 7.2) and freeze–thawed at −80 °C prior to sonication with an equal volume of Vertrel XF (Sigma-Aldrich). The cell suspension was then clarified by low-speed centrifugation (7000 g, 10 min at 4 °C), and the aqueous phase was subjected to repeated sonication. Virus was then purified from the aqueous phase by centrifugation through a caesium chloride gradient (1.31–1.45 g ml−1) using a SW-41 Ti rotor at 28 000 r.p.m. for 8 h at 12 °C. The purified virus was collected from the gradient and dialysed into virion buffer (10 mM Tris, 150 mM NaCl and 20 mM MgCl2, pH 7.2).
Sapovirus particles were made using a baculovirus expression system in Tn5 cells as previously described [27].
Electron microscopy
Vesivirus 2117 VLPs and FCV virions (4 µl) were loaded on-to glow discharged C-flat holey carbon support films (R2/2 ProtoChips), blotted at 4 °C for 4 s at 100 % humidity and plunged into liquid-nitrogen-cooled liquid ethane using an FEI Mark IV Vitrobot. Vitrified samples were imaged in a JEOL 2200 FS cryo-microscope equipped with a Gatan 626 cryo-stage. Energy-filtered images were recorded (with a 20 eV slit width) on a Gatan Ultrascan US4000 charge-coupled device camera at a magnification of ×100 000, corresponding to a pixel size of 1.05 Å per pixel (vesivirus 2117) or a Direct Electron DE20 direct detection device at a magnification of ×40 000 with a pixel size of 1.39 Å per pixel (FCV). Sapovirus VLPs were plunged using the same method but using R1.2/1.3 Mo 200 mesh holey carbon grids (Quantifoil), imaging was performed in a JEOL 2200FS at a magnification of ×80 000 and micrographs were recorded on a TVIPS 4k×4k CCD camera, giving a pixel size of 1.6 Å per pixel.
3D image reconstruction
Two hundred and forty-two micrographs of vesivirus 2117 VLPs, 241 micrographs of FCV and 222 micrographs of SV VLPs were processed to calculate 3D reconstructions. Images of particles were contrast transfer function corrected and extracted from micrographs using the BSoft program Bshow [45]. The vesivirus 2117 and SV particles were masked and sorted by size into five classes by cross-correlation against fuzzy ring models using spider [46]. The most populous class for each was selected for use in further processing. The origins and orientations of the particles were then determined using the polar Fourier transform method (PFT2), and the EM3DR2 program was used to create 3D reconstructions [47, 48]. Resolution estimates for each map were determined by dividing the data set into two and calculating 3D reconstructions from each half. The paired reconstructions were compared in Bresolve to compute a number of indices of similarity including the Fourier shell correlation (FSC). A FSC cut-off value of 0.5 was taken (Fig. S1, available in the online Supplementary Material). To draw comparisons between different calicivirus capsids, additional structures were downloaded from the Protein Data Bank and Electron Microscopy Data Bank public databases. Density maps were calculated from Protein Data Bank files using the eman program pdb2mrc. All density maps were then low pass filtered to a common resolution of 10 Å using eman [49]. UCSF Chimera was used for visualizing cryo-EM reconstructions (using an isosurface threshold of the mean plus 1 sd) and for calculation of correlation values between pairs of maps. Correlation values were calculated in UCSF Chimera using the ‘Fit in Map’ function [50].
Phylogenetic analysis
Our 2117 capsid amino acid sequence was used as a query sequence in a blastp search against the GenBank non-redundant database. The top hit was to an existing 2117 sequence (AAQ24846; 99 % identity), followed by three hits to other suspected 2117 sequences sampled in Geel, Belgium (ACV95479; 85 % identity) and Allston, USA (ACV95474, ACV95477; 84 % identity). The next most similar hits were canine vesivirus (AFI58321; 84 % identity) and canine calicivirus (NP_786912, NP_777374; 68 % identity) sequences. All of the above sequences had an E value of 0 and were selected for inclusion in the phylogenetic analysis. Representative sequences from other species within the genera Vesivirus, Sapovirus, Lagovirus and Norovirus of the family Caliciviridae were also selected from the blast hits for inclusion in the phylogenetic analysis. In total, 57 protein sequences were selected (including our 2117 capsid sequence) and aligned using clustalw within the mega software [51]. The evolutionary history of the 2117 capsid sequence was inferred using the neighbour-joining method (1000 bootstrap replicates) to generate a phylogenetic tree, evolutionary distances were computed using the JTT-matrix-based method and all positions in the alignment containing gaps and missing data were eliminated, leaving a total of 402 positions in the final dataset.
Funding information
M. C. was funded by a PhD studentship from the UK Biotechnology and Biological Sciences Research Council (BBSRC WestBIO DTP; grant number BB/J013854/1). D. G. C. was supported by an Erasmus scholarship. G. S. H. was funded by the Chica and Heinz Schaller Foundation, Heidelberg, Grant-in-Aid for Scientific Research, a grant from the Ministry of Health, Labor, and Welfare of Japan. K. M. was funded by a grant from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Collaborative Study Program of the National Institute for Physiological Sciences. D. T. performed this work in Japan as a Japan Society for the Promotion of Science/National Science Foundation East Asia and Pacific Summer Institute Fellow. I. G. G. was supported by a Wellcome Trust senior fellowship in basic biomedical science. R. O. and D. B. were supported by the UK Medical Research Council (MC_UU_12014/3 and MC_UU_12014/7 respectively).
Conflicts of interest
The authors declare there are no conflicts of interest.
Supplementary Data
References
- 1.Etherington GJ, Ring SM, Charleston MA, Dicks J, Rayward-Smith VJ, et al. Tracing the origin and co-phylogeny of the caliciviruses. J Gen Virol. 2006;87:1229–1235. doi: 10.1099/vir.0.81635-0. [DOI] [PubMed] [Google Scholar]
- 2.Green KY, Ando T, Balayan MS, Berke T, Clarke IN, et al. Taxonomy of the caliciviruses. J Infect Dis. 2000;181:S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- 3.Radford AD, Gaskell RM, Hart CA. Human norovirus infection and the lessons from animal caliciviruses. Curr Opin Infect Dis. 2004;17:471–478. doi: 10.1097/00001432-200410000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Sosnovtsev SV, Belliot G, Chang KO, Onwudiwe O, Green KY. Feline calicivirus VP2 is essential for the production of infectious virions. J Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oka T, Yamamoto M, Yokoyama M, Ogawa S, Hansman GS, et al. Highly conserved configuration of catalytic amino acid residues among calicivirus-encoded proteases. J Virol. 2007;81:6798–6806. doi: 10.1128/JVI.02840-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Molawi N, Beardmore VA, Carter MJ, Kass GE, Roberts LO. Caspase-mediated cleavage of the feline calicivirus capsid protein. J Gen Virol. 2003;84:1237–1244. doi: 10.1099/vir.0.18840-0. [DOI] [PubMed] [Google Scholar]
- 7.Mcfadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, et al. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 2011;7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/SQB.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 9.Harrison SC. The familiar and the unexpected in structures of icosahedral viruses. Curr Opin Struct Biol. 2001;11:195–199. doi: 10.1016/S0959-440X(00)00189-5. [DOI] [PubMed] [Google Scholar]
- 10.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–569. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Xu F, Liu J, Gao B, Liu Y, et al. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog. 2013;9:e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subba-Reddy CV, Yunus MA, Goodfellow IG, Kao CC. Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1. J Virol. 2012;86:10138–10149. doi: 10.1128/JVI.01208-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Bhella D, Gatherer D, Chaudhry Y, Pink R, Goodfellow IG. Structural insights into calicivirus attachment and uncoating. J Virol. 2008;82:8051–8058. doi: 10.1128/JVI.00550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Neill JD, Estes MK, Prasad BV. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc Natl Acad Sci USA. 2006;103:8048–8053. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad BV, Rothnagel R, Jiang X, Estes MK. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oehmig A, Büttner M, Weiland F, Werz W, Bergemann K, et al. Identification of a calicivirus isolate of unknown origin. J Gen Virol. 2003;84:2837–2845. doi: 10.1099/vir.0.19042-0. [DOI] [PubMed] [Google Scholar]
- 17.Qiu Y, Jones N, Busch M, Pan P, Keegan J, et al. Identification and quantitation of vesivirus 2117 particles in bioreactor fluids from infected Chinese hamster ovary cell cultures. Biotechnol Bioeng. 2013;110:1342–1353. doi: 10.1002/bit.24791. [DOI] [PubMed] [Google Scholar]
- 18.Berting A, Farcet MR, Kreil TR. Virus susceptibility of Chinese hamster ovary (CHO) cells and detection of viral contaminations by adventitious agent testing. Biotechnol Bioeng. 2010;106:598–607. doi: 10.1002/bit.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burstyn DG. Contamination of genetically engineered Chinese hamster ovary cells. Dev Biol Stand. 1995;88:199–203. [PubMed] [Google Scholar]
- 20.Garnick RL. Raw materials as a source of contamination in large-scale cell culture. Dev Biol Stand. 1998;93:21–29. [PubMed] [Google Scholar]
- 21.Kerr A, Nims R. Adventitious viruses detected in biopharmaceutical bulk harvest samples over a 10 year period. PDA J Pharm Sci Technol. 2010;64:481–485. [PubMed] [Google Scholar]
- 22.Plavsic M, Qiu Y, Jones N, Keegan J, Woodcock D, et al. Caliciviridae and vesivirus 2117. Bioprocessing (Williamsbg Va) 2011;9:6–12. doi: 10.12665/J92.Plavsic. [DOI] [Google Scholar]
- 23.Rabenau H, Ohlinger V, Anderson J, Selb B, Cinatl J, et al. Contamination of genetically engineered CHO-cells by epizootic haemorrhagic disease virus (EHDV) Biologicals. 1993;21:207–214. doi: 10.1006/biol.1993.1077. [DOI] [PubMed] [Google Scholar]
- 24.Onions D. Animal virus contaminants of biotechnology products. Dev Biol. 2004;118:155–163. [PubMed] [Google Scholar]
- 25.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 26.Han MG, Wang Q, Smiley JR, Chang KO, Saif LJ. Self-assembly of the recombinant capsid protein of a bovine norovirus (BoNV) into virus-like particles and evaluation of cross-reactivity of BoNV with human noroviruses. J Clin Microbiol. 2005;43:778–785. doi: 10.1128/JCM.43.2.778-785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansman GS, Oka T, Katayama K, Takeda N. Enhancement of sapovirus recombinant capsid protein expression in insect cells. FEBS Lett. 2006;580:4047–4050. doi: 10.1016/j.febslet.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki N, Taylor DW, Hansman GS, Murata K. Antigenic and cryo-electron microscopy structure analysis of a chimeric sapovirus capsid. J Virol. 2016;90:2664–2675. doi: 10.1128/JVI.02916-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka T, Hansman GS, Katayama K, Ogawa S, Nagata N, et al. Expression of sapovirus virus-like particles in mammalian cells. Arch Virol. 2006;151:399–404. doi: 10.1007/s00705-005-0613-y. [DOI] [PubMed] [Google Scholar]
- 30.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Chen R, Neill JD, Noel JS, Hutson AM, Glass RI, et al. Inter- and intragenus structural variations in caliciviruses and their functional implications. J Virol. 2004;78:6469–6479. doi: 10.1128/JVI.78.12.6469-6479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhella D, Goodfellow IG. The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J Virol. 2011;85:11381–11390. doi: 10.1128/JVI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katpally U, Wobus CE, Dryden K, Virgin HW, Smith TJ. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J Virol. 2008;82:2079–2088. doi: 10.1128/JVI.02200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katpally U, Voss NR, Cavazza T, Taube S, Rubin JR, et al. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J Virol. 2010;84:5836–5841. doi: 10.1128/JVI.00314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radford AD, Willoughby K, Dawson S, Mccracken C, Gaskell RM. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J Virol. 1999;73:8496–8502. doi: 10.1128/jvi.73.10.8496-8502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R, Neill JD, Prasad BV. Crystallization and preliminary crystallographic analysis of San Miguel sea lion virus: an animal calicivirus. J Struct Biol. 2003;141:143–148. doi: 10.1016/S1047-8477(02)00583-X. [DOI] [PubMed] [Google Scholar]
- 37.Yu G, Zhang D, Guo F, Tan M, Jiang X, et al. Cryo-EM structure of a novel calicivirus, Tulane virus. PLoS One. 2013;8:e59817. doi: 10.1371/journal.pone.0059817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim DS, Hosmillo M, Alfajaro MM, Kim JY, Park JG, et al. Both α2,3- and α2,6-linked sialic acids on O-linked glycoproteins act as functional receptors for porcine sapovirus. PLoS Pathog. 2014;10:e1004172. doi: 10.1371/journal.ppat.1004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taube S, Perry JW, Mcgreevy E, Yetming K, Perkins C, et al. Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J Virol. 2012;86:5584–5593. doi: 10.1128/JVI.06854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marionneau S, Ruvoën N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, et al. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyström K, Le Gall-Reculé G, Grassi P, Abrantes J, Ruvoën-Clouet N, et al. Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog. 2011;7:e1002188. doi: 10.1371/journal.ppat.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruvoën-Clouet N, Ganière JP, André-Fontaine G, Blanchard D, Le Pendu J. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol. 2000;74:11950–11954. doi: 10.1128/JVI.74.24.11950-11954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaffer FL, Soergel ME, Black JW, Skilling DE, Smith AW, et al. Characterization of a new calicivirus isolated from feces of a dog. Arch Virol. 1985;84:181–195. doi: 10.1007/BF01378971. [DOI] [PubMed] [Google Scholar]
- 44.Roerink F, Hashimoto M, Tohya Y, Mochizuki M. Genetic analysis of a canine calicivirus: evidence for a new clade of animal caliciviruses. Vet Microbiol. 1999;69:69–72. doi: 10.1016/S0378-1135(99)00091-7. [DOI] [PubMed] [Google Scholar]
- 45.Heymann JB. Bsoft: image and molecular processing in electron microscopy. J Struct Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 46.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 47.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 48.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, et al. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol. 2005;79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.