ABSTRACT
Bacterial cell division is a complex process that relies on a multiprotein complex composed of a core of widely conserved and generally essential proteins and on accessory proteins that vary in number and identity in different bacteria. The assembly of this complex and, particularly, the initiation of constriction are regulated processes that have come under intensive study. In this work, we characterize the function of DipI, a protein conserved in Alphaproteobacteria and Betaproteobacteria that is essential in Caulobacter crescentus. Our results show that DipI is a periplasmic protein that is recruited late to the division site and that it is required for the initiation of constriction. The recruitment of the conserved cell division proteins is not affected by the absence of DipI, but localization of DipI to the division site occurs only after a mature divisome has formed. Yeast two-hybrid analysis showed that DipI strongly interacts with the FtsQLB complex, which has been recently implicated in regulating constriction initiation. A possible role of DipI in this process is discussed.
IMPORTANCE Bacterial cell division is a complex process for which most bacterial cells assemble a multiprotein complex that consists of conserved proteins and of accessory proteins that differ among bacterial groups. In this work, we describe a new cell division protein (DipI) present only in a group of bacteria but essential in Caulobacter crescentus. Cells devoid of DipI cannot constrict. Although a mature divisome is required for DipI recruitment, DipI is not needed for recruiting other division proteins. These results, together with the interaction of DipI with a protein complex that has been suggested to regulate cell wall synthesis during division, suggest that DipI may be part of the regulatory mechanism that controls constriction initiation.
KEYWORDS: Caulobacter crescentus, SH3 domain, bacterial cell division, constriction initiation, divisome
INTRODUCTION
Bacterial cell division is a complex process involving the action of a multiprotein complex known as the divisome (1, 2). In Escherichia coli, the divisome is composed of approximately 30 proteins that assemble into a complex spanning from the cytoplasm to the outer membrane (OM). Of these proteins, only 12 are essential in E. coli and 11 are widely conserved in other bacteria, suggesting that they constitute the core of the divisome (3, 4). The divisome complex starts assembling when a ring formed by the FtsZ protein is stabilized at the site where division will occur (5, 6). FtsZ is a tubulin-related cytoplasmic protein that is brought near the cytoplasmic membrane through its interactions with the membrane-associated protein FtsA and, in E. coli, the transmembrane protein ZipA (7, 8). The FtsZ ring generates a force sufficient to deform lipid membranes and probably drives the constriction of the inner membrane (9, 10). After establishment of the FtsZ ring, the FtsEX complex is recruited through the interaction of FtsE with FtsZ (11). The divisome then expands to the periplasmic space by the addition of several transmembrane proteins. The first of these is FtsK. The cytoplasmic domain of this protein is involved in chromosome resolution and is not essential for cell division, whereas the transmembrane and periplasmic domains are essential but their only known function is in the recruitment of other division proteins (2, 12, 13). After a time delay, the FtsQ, FtsL, FtsB, FtsW, FtsI, and FtsN proteins are rapidly recruited (14). These proteins probably do not accumulate at the division site simultaneously, since FtsQLB forms a complex that is required for divisome stabilization and the recruitment of other division proteins (2, 15–18). The FtsQLB complex has also been proposed to activate the synthesis of septal peptidoglycan in response to the state of the divisome (19, 20). It was initially thought that FtsN was the last essential protein to be recruited to the division site and that its function was to stabilize the divisome (21–23). However, this protein interacts with FtsA, enabling the early recruitment of a small amount of FtsN that interacts with the cell wall-synthesizing proteins FtsI (PBP3) and PBP1B, thereby promoting the synthesis of murein at the division site (24–29). This new cell wall is then processed by amidases, enabling the C-terminal domain of FtsN to bind to the cell wall, which results in further accumulation of FtsN through a self-stimulating mechanism (30). The amount of FtsN present at the division site may also change the oligomerization state of the FtsA filaments, changing the activity of FtsZ and triggering constriction (19).
Caulobacter crescentus is a Gram-negative bacterium that has been studied as a bacterial cell differentiation model, resulting in an extensive description of the control of its cell cycle in which the differential localization of key cell cycle regulatory proteins has a central role (31). In C. crescentus, the initiation of divisome assembly is coordinated with chromosome replication and segregation through the action of the MipZ protein (32). The divisome is present during a longer fraction of the cell cycle in C. crescentus than in E. coli, and the time needed for its maturation and the initiation of constriction is also longer (32, 33), suggesting differences in the assembly and regulation of the divisome. Probably as a consequence of the long presence of the divisome, the zonal growth of the cell wall is more relevant in this bacterium (33). In addition, the study of the cell division process in C. crescentus has revealed a different order of protein recruitment to the divisome, as well as robustness in the absence of a division protein (34). In line with this, although FtsA is essential in C. crescentus, it is not necessary for the initiation of constriction (35) and is recruited late to the division site (4, 34). There is no homolog of ZipA in C. crescentus; instead, the interaction of the Z-ring with the inner membrane in the first stages of divisome assembly seems to be mediated by other proteins, such as the membrane-associated protein FzlC (36, 37). It has been recently proposed that the activation of the divisome also occurs in C. crescentus, since point mutations in FtsW, FtsN, and FtsI cause premature division (38, 39). In this work, we describe a new conserved protein essential for cell division in C. crescentus. DipI is recruited late to the divisome, and in its absence, cell constriction is not observed. The interactions between DipI and the FtsQ and FtsB proteins suggest that it might have a regulatory role in the activity of the divisome.
RESULTS
Depletion of DipI causes cell filamentation.
To identify new proteins that might be involved in cell division, the hypothetical or poorly annotated proteins from a list of homologs present in rhizobia and Caulobacter crescentus (40) were selected. From this list, we decided to examine the function of the CC3721 protein, as the gene coding for this protein was identified as essential in a recent high-throughput mutagenesis study (41). Moreover, the homolog of CC3721 in Sinorhizobium meliloti (SMc02848) is under direct control of CtrA (42). An analysis of the primary structure of CC3721 revealed the presence of two SH3_4 domains and an N-terminal transmembrane domain or a possible signal peptide (Fig. 1A), suggesting that it is a membrane or a soluble periplasmic protein. Examination of the coding sequence of CC3721 revealed an alternative start codon 12 codons downstream of the annotated protein start. The score of the predicted signal peptide improves when the protein starts from this alternative start codon. A search in Pfam showed that the SH3_4 domain is present mainly in predicted periplasmic proteins and is widely distributed in Alphaproteobacteria and Betaproteobacteria but can also be found in some firmicutes, bacteroidetes, actinobacteria, and cyanobacteria. However, proteins with a domain composition that is the same as CC3721 (two SH3_4 domains) are found only in Alphaproteobacteria and Betaproteobacteria and in some cyanobacteria. The role of bacterial SH3 domains is just beginning to be investigated. In Gram-positive bacteria, the SH3-like domains are frequently associated with cell wall-hydrolyzing proteins that contain a peptidase NlpC/P60 domain (43) and have been proposed to work as auxiliary domains that increase the activity of the enzyme by binding peptidoglycan (44). Interestingly, SH3 domains have also been shown to determine the substrate specificity of the catalytic domain (45–47).
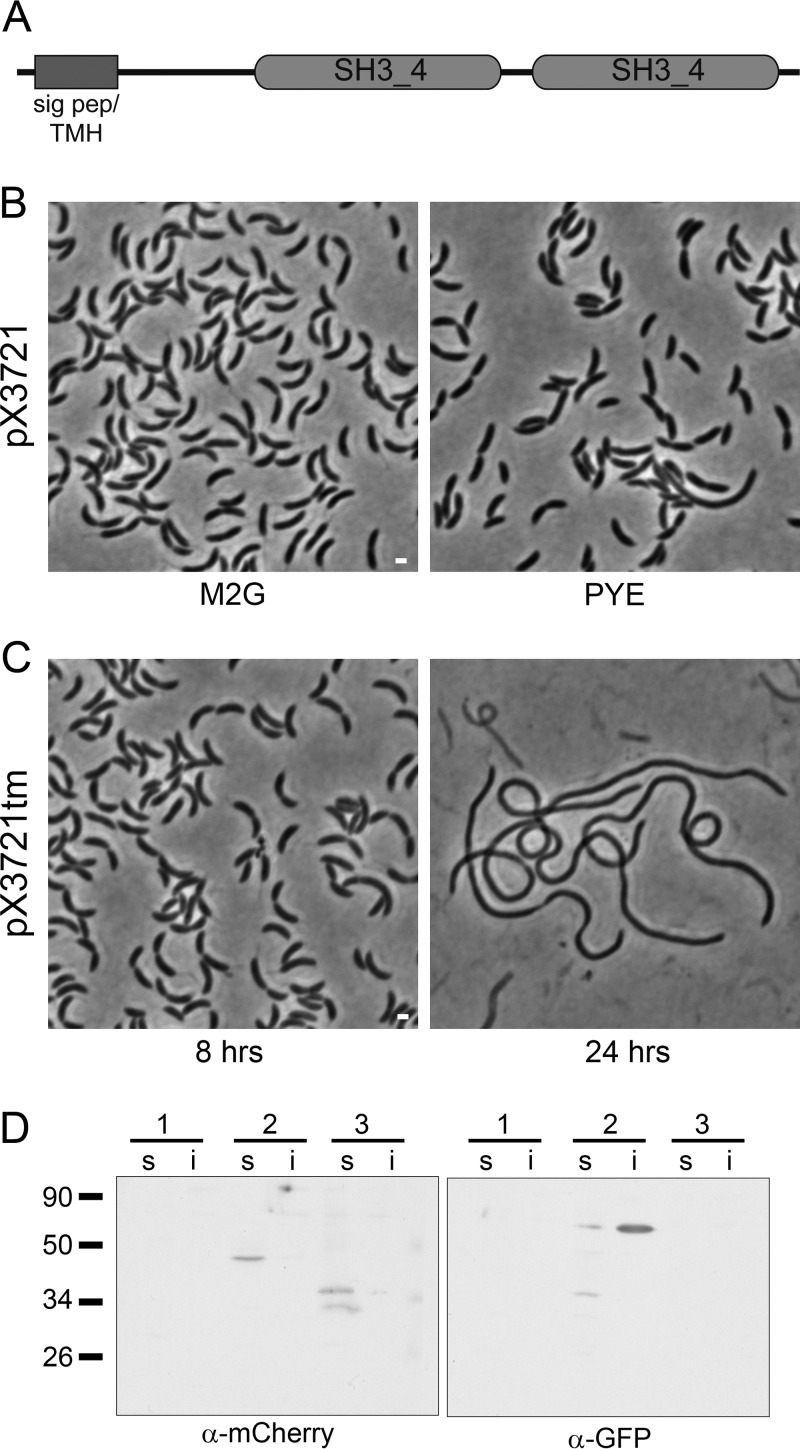
FIG 1.
Depletion of DipI causes cell filamentation. (A) The domain structure of DipI is shown. (B) A strain (SP2) with a single chromosomal copy of dipI under the control of a xylose-inducible promoter (pX3721) was grown in minimal medium (M2G) or rich medium (PYE) in the absence of xylose. (C) A strain (SP3) with a single chromosomal copy of dipI fused with the sequence for the degradation signal encoded by tmRNA under the control of a xylose-inducible promoter (pX3721tm) was grown from an overnight culture in M2G medium without xylose for 8 or 24 h. (D) Presence of DipI-mCherry in the soluble fraction. Western blots of soluble (s) and insoluble (i) fractions of the following strains were probed with the indicated antibodies: 1, CB15N (wild type); 2, SP22 (expressing DipI-mCherry and Venus-FtsN protein fusions); and 3, CJW2959 (expressing periplasmic mCherry). Expected molecular masses of the proteins in kilodaltons were as follows: periplasmic mCherry, 32.8; Venus-FtsN, 55; and periplasmic DipI-mCherry, 44.2. Bars, 1 μm.
We unsuccessfully tried to obtain a CC3721 null mutant by selecting double recombinants that had the target gene replaced by the Ω-spc cassette, corroborating its previous classification as an essential gene. To determine the phenotype of cells lacking the product of CC3721, we deleted CC3721 in a strain carrying a second copy of the gene under the control of the xylX promoter (xylXp). In the absence of xylose, the resultant strain grew normally in M2G minimal medium, and only a moderate filamentation was observed in cells grown in peptone-yeast extract (PYE) rich medium (Fig. 1B). An explanation for this result may be that a small amount of the CC3721 protein produced from leaky activity of xylXp is sufficient to fulfill its function. Alternatively, CC3721 might be essential only in PYE. For this reason, we tried to obtain the mutant in M2G liquid and solid media, but as before, we obtained only sucrose-resistant mutants. To further reduce the amount of CC3721, we added at the end of the gene a region of the transfer-messenger RNA (tmRNA) present in C. crescentus that codes for a peptide that marks the protein for degradation (48). A similar approach was used previously with cytoplasmic proteins (49, 50); however, this is to our knowledge the first time that this approach has been used with a periplasmic protein. In the absence of xylose, the resultant strain (SP3) reached stationary phase (optical density at 600 nm [OD660] of 1.2) in the first overnight (ON) culture. Cells from this culture showed a normal or slightly filamented morphology, but a reinoculated culture did not reach saturation, obtaining a maximal OD660 of ∼0.3. Cells from this second culture showed strong filamentation, growing as smooth filaments with occasional constrictions (Fig. 1C), indicating that this protein is involved in cell division. A similar result was obtained when this strain was grown in M2G minimal medium, supporting the idea that CC3721 is an essential protein (see Fig. S1 in the supplemental material). Filamentation was not due to a combination of the absence of CC3721 and allowing the culture to reach stationary phase, since the cells from a culture that was maintained in exponential phase also filamented after 24 h (Fig. S1). Following the nomenclature proposed for the soluble periplasmic protein DipM (51–53), from here on, we will refer to CC3721 as DipI (division-involved protein I).
The primary structure analysis indicated that DipI is either a transmembrane or a soluble periplasmic protein. To distinguish between these possibilities, we tagged DipI with the fluorescent mCherry protein and tested whether the fusion protein was found in the soluble or insoluble fraction of cell extracts. For this, we obtained a strain that expresses a carboxy-terminal fusion of DipI with mCherry and an N-terminal fusion of FtsN with the Venus fluorescent protein (strain SP22). The dipI-mCherry fusion is expressed from the native promoter and the venus-ftsN fusion is expressed from a vanillic acid-inducible promoter. The Venus-FtsN fusion and the periplasmic mCherry protein expressed in strain CJW2959 were used as controls for membrane and soluble proteins, respectively. Detection with an anti-green fluorescent protein (anti-GFP) antibody showed the presence of Venus-FtsN mainly in the insoluble fraction (Fig. 1D, right panel). An identical blot probed with an anti-mCherry antibody showed that the periplasmic mCherry and DipI-mCherry were only in the soluble fraction (Fig. 1D, left panel), indicating that DipI is a soluble periplasmic protein. This result also suggests that the previously mentioned alternative translation start site is more likely to be the correct one.
DipI localizes to the division site.
To determine if DipI is directly involved in cell division, we investigated whether this protein is recruited to the division site. However, cells expressing DipI-mCherry (strain SP15) showed mild filamentation and a reduced growth rate when grown in PYE rich medium but not in M2G minimal medium (see Table S1), indicating that the DipI-mCherry fusion is partially functional. Fusion proteins are frequently partially functional or nonfunctional, and the localization of these proteins can be determined by coexpressing them with the wild-type protein. Inducible N-terminal and C-terminal fusions of DipI with mCherry were introduced into the wild-type strain as second chromosomal copies, and the stabilities of these fusion proteins were verified by Western blotting (see Fig. S2A). These proteins were not localized at induction levels above or below those of DipI-mCherry under the control of the native promoter (Fig. S2), suggesting that the wild-type protein outcompetes the fusion versions of DipI. As we were unable to obtain a fluorescent fusion of DipI capable of localizing in the presence of the wild-type protein, we decided to determine the localization of DipI-mCherry when expressed as the single version of this protein (strain SP15). Except for the depletion experiments (see below), the localization of DipI-mCherry was determined from cells grown in M2G medium.
In an unsynchronized cell population, DipI-mCherry was observed mainly in cells that were already constricting but also in a few cells that did now show any constriction (Fig. 2A, cells marked with an arrow), indicating that DipI probably arrives to the divisome near the time that constriction is initiated. To determine the time of DipI-mCherry localization in the cell cycle, we monitored the localization of DipI in a synchronized culture (Fig. 2B). As has been reported previously for several division proteins of C. crescentus (4, 34, 35, 54, 55), localization of DipI-mCherry was observed at the new pole of the swarmer cells (Fig. 2B, first panel). This result was verified by the polar colocalization of DipI-mCherry with other cell division proteins (Fig. 3). Localization of DipI at midcell was observed in the last third of the cell cycle but only in a maximum of 30% of the cells (Fig. 2B and C). This percentage was similar to that of cells with localized DipI observed in an unsynchronized population (localized DipI, 28% ± 4%; constricting cells, 50% ± 2%), suggesting that the synchronization procedure may be affecting the localization of DipI-mCherry. However, the time at which DipI starts localizing is similar to the time of constriction appearance, supporting the idea that DipI is recruited near the time that constriction is initiated. In C. crescentus, the expression and stabilities of many of the proteins involved in cell division are coordinated with the cell cycle. The expression of dipI shows a 2-fold increase after the swarmer-to-stalked transition, and it has been suggested to be a cell cycle-regulated gene (56, 57). Immunodetection of DipI-mCherry showed that the protein is present during the entire cell cycle (Fig. 2D), indicating that the localization of DipI is not restricted by its expression pattern.
FIG 2.
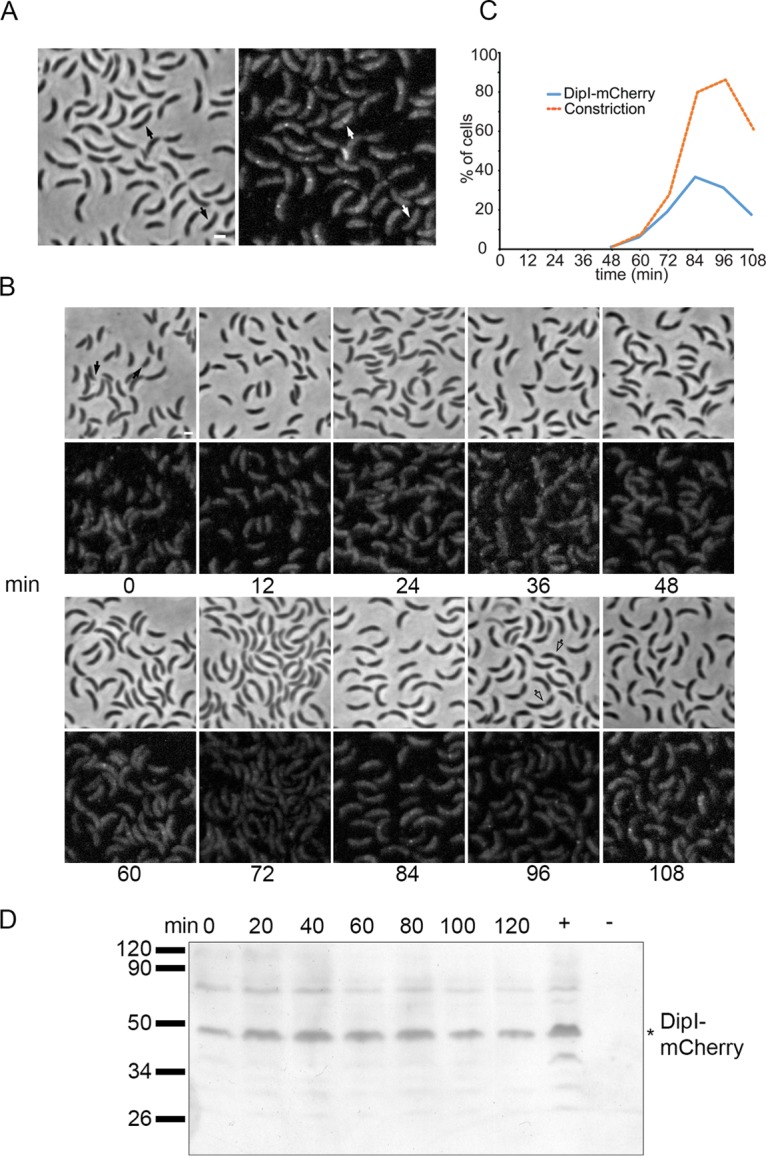
DipI is recruited late to the division site. (A) Localization of DipI-mCherry in an unsynchronized cell population. Cells from a culture of strain SP15 grown in minimal medium (M2G) were observed when the culture reached an OD660 of 0.3. Arrows indicate cells with no visible constriction in which DipI-mCherry was already localized at midcell. (B) Time-lapse images for localization of DipI-mCherry in a synchronized cell population. A culture with an OD660 of 0.3 was synchronized, and aliquots were taken for observation every 12 min. Empty arrows indicate cells with deep constriction that did not show localization of DipI. (C) Quantification of the localization of DipI to the division site. The percentage of cells that showed constriction or localization of DipI was determined in 300 cells for each time point after synchronization. (D) Presence of DipI-mCherry during the cell cycle. Total cell extracts from a synchronized culture of the SP15 strain were obtained every 15 min and the presence of DipI-mCherry was determined by Western blotting. Total cell extracts from unsynchronized SP15 and CB15N cultures were used as positive and negative controls, respectively. Migration of molecular weight markers is shown at the left. The asterisk indicates the migration of the DipI-mCherry protein. Bars, 1 μm.
FIG 3.
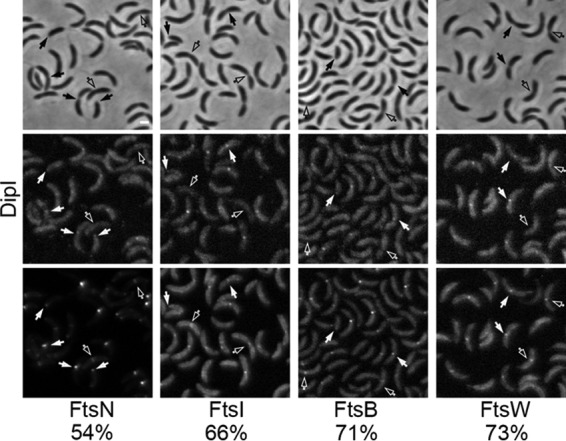
DipI is recruited late to the division site. The colocalization of DipI-mCherry with other inducible cell division protein fusions was quantified in exponential cultures. Top panels, phase-contrast images; middle panels, DipI-mCherry fluorescence images; bottom panels, fluorescence images of Venus fusions with the division protein indicated at the bottom of the image (strains from left to right: SP22, SP26, SP21, and SP23). Empty arrows indicate dividing cells where localization of DipI-mCherry was expected but not observed; solid arrows indicate division sites or cell poles were colocalization was observed. The percentages of colocalization are shown at the bottom. Percentages were calculated only from cells that showed localization of the indicated division protein at the division site (n ≈ 300). Bar, 1 μm.
The low percentage of cells showing localized DipI-mCherry (Fig. 2C) caught our attention, as in a synchronized population the percentage of cells showing localization of a division protein is usually approximately 80% (34). This is in part due to the absence of a signal in constricting cells that should have DipI-mCherry at the division site (Fig. 2B, empty arrows) but is probably also due to the combination of high background caused by nonlocalized protein and a weak localized signal in early dividing cells. Both problems likely result from the partial functionality of the DipI-mCherry protein but may also be caused by partial stability of the fusion protein; however, no significant proteolysis was detected in a Western blot (Fig. 1D and 2D; see also Fig. S2), suggesting that the mCherry moiety interferes with the recruitment of the DipI fusion protein to the division site.
To establish the time of DipI recruitment to the divisome, the localization of DipI-mCherry was compared with that of coexpressed inducible fluorescent fusions of the late cell division proteins FtsB, FtsI, FtsN, and FtsW (Fig. 3). As expected from the time course experiments, DipI did not arrive before any of the cell division proteins in any of the cells observed. To determine at what point during the assembly of the divisome DipI is recruited, we calculated the percentage of DipI-mCherry colocalization with the other cell division proteins in an unsynchronized population. Colocalization increased accordingly with the previously reported order of assembly from 54% for FtsN/DipI and 66% for FtsI/DipI up to 71 and 73% for FtsB/DipI and FtsW/DipI, respectively. These results suggest that DipI is recruited to the division site at the same time as FtsB or FtsW, both of which are recruited at the end of divisome assembly in C. crescentus. Although this conclusion is in agreement with the DipI localization dependency of other division proteins (see below), a functional DipI fluorescent fusion would provide more conclusive evidence of the time at which DipI is recruited to the divisome.
DipI is not required for the recruitment of other division proteins.
To determine if DipI is required for the formation of a mature divisome (i.e., a divisome to which all the essential division proteins have been recruited), the localization of fluorescently tagged division proteins was determined in DipI-depleted cells. Fluorescent fusion proteins were expressed as second copies from inducible promoters or, in the case of MurG-mCherry, as a single copy from its native promoter; all strains were grown in PYE medium. The assembly order of the conserved divisome proteins, as well as their recruitment dependencies, has been previously reported (34), enabling us to select proteins from different stages of divisome assembly and with different recruitment dependencies. In the previous work, recruitment dependency was established only if no localization was observed in the absence of another protein. In the present study, the same standard was followed. As expected, FtsZ localization was not affected by the absence of DipI, and a similar result was obtained with the early-localizing MurG protein (Fig. 4). The localization of the early-localizing MreB and DipM was then tested. Both proteins showed weak localization and extensive signal that was not localized, indicating that their recruitment was affected. It should be noted that a partially functional internal fusion of MreB with mCherry (MreBswmCherry) was used; this fusion was obtained using a strategy previously described (58). We then tested the localization of FtsI and FtsN, both of which are recruited just before cell constriction starts. For this reason, they are less frequently found localized in unconstricted cells in a nonsynchronized population than the previously used proteins. However, in the DipI-depleted cells, we easily observed localization of FtsI and FtsN in sites where no constriction was visible (Fig. 4, black arrows). In contrast to that in E. coli, the FtsB protein is not essential in C. crescentus, and the FtsL protein is required for the localization of FtsQ and FtsB (34). The expression of fluorescent fusions of FtsL and FtsQ in cells depleted of DipI showed that both proteins were recruited to division sites that did not show any constriction, as was observed for the FtsI and FtsN fusions. The last essential division protein to be recruited to the C. crescentus divisome is FtsW. Recruitment of FtsW partially depends on the presence of the FtsQLB complex (34). We observed a clear localization of FtsW in the absence of DipI, indicating that DipI is not required for the maturation of the divisome. As we frequently observed recruitment of all the division proteins tested to division sites that showed no constriction, we presume that in the absence of DipI a mature divisome assembles and that DipI is required to initiate constriction.
FIG 4.
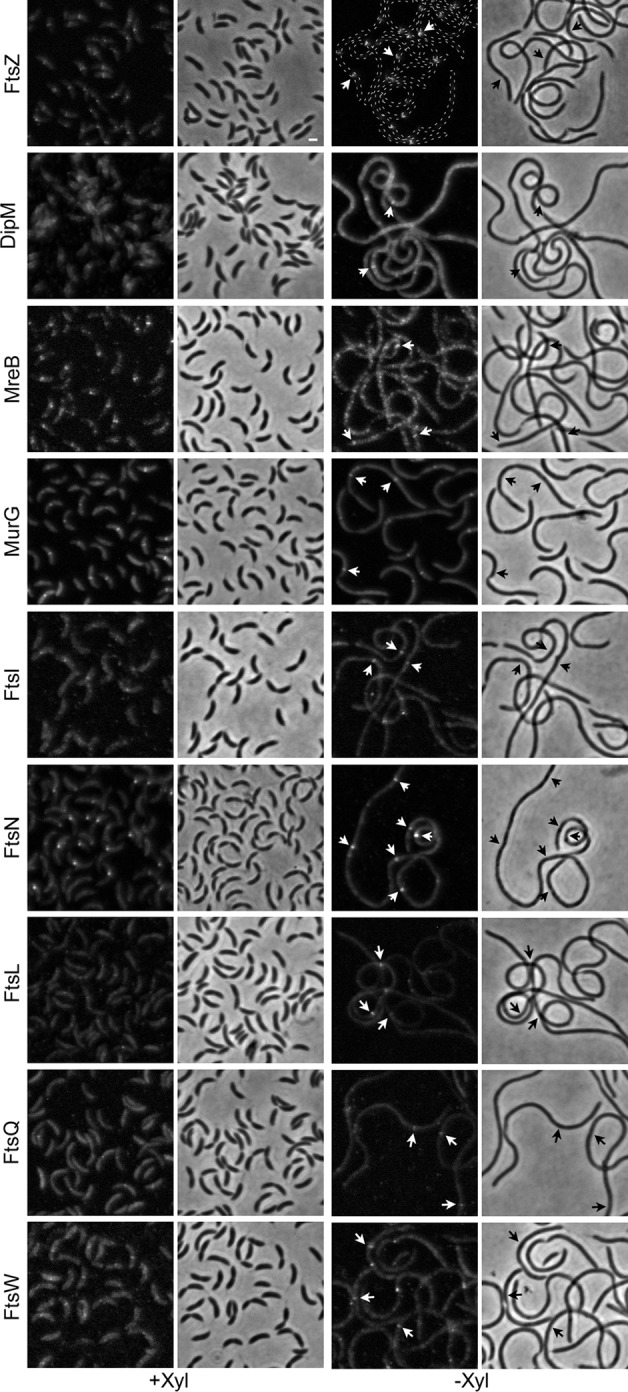
Mature divisomes form in the absence of DipI. The localization of different cell division proteins was determined in cells expressing DipI (+Xyl) or depleted of DipI (−Xyl). Fluorescent protein fusions of the different cell division proteins (indicated at the left of each row) were introduced as second copies and expressed from a vanillic acid-inducible promoter. The murG gene was substituted with the allele coding for the fluorescent fusion. All the strains were grown in rich medium (PYE) in the presence or absence of xylose. Depletion was carried out as described in Materials and Methods. When required, vanillic acid was added 3 h before observation. Arrows indicate localization of the division proteins in sites where no constriction was visible. From top to bottom, the strains used are as follows: SP4, SP6, SP7, SP8, SP9, SP5, SP27, SP28, and SP10. Bar, 1 μm.
DipI interacts with the FtsQLB complex.
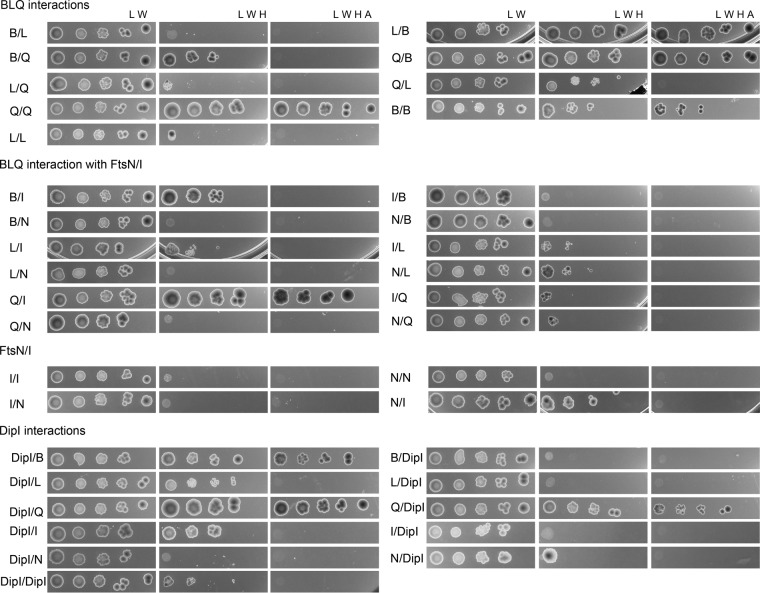
To investigate whether the recruitment of DipI to the division site depends on a late division protein, we tested whether DipI interacts with the periplasmic domains of different division proteins required for cell constriction. For this, the coding sequences of the periplasmic domains of FtsI, FtsN, FtsQ, FtsL, and FtsB, as well as the mature DipI protein, were fused to the DNA-binding and activator domains of the GAL4 transcriptional activator. The interactions between these proteins were tested in a yeast two-hybrid assay. In these experiments, a positive interaction results in the restoration of histidine prototrophy if the interaction is weak and the restoration of adenine if it is strong. Spurious activation of the reporter promoters was discarded by recording the growth phenotype of the strains carrying only one of the plasmids to be tested together with the complementary plasmid expressing a nonrelated protein fusion (see Fig. S3).
To verify that the yeast two-hybrid assay could be used with division proteins, we started by testing the interactions between the conserved division proteins. It was shown by in vitro and in vivo methods that the FtsQLB proteins from other bacteria interact with each other and form a complex (15, 16, 29, 59). The same seems to be true for the C. crescentus FtsQLB proteins, as we detected strong interactions between FtsQ and FtsB and between FtsL and FtsB and a weak interaction between FtsQ and FtsL (Fig. 5). These interactions are in agreement with the interactions reported for the periplasmic domains of these proteins (16). In addition, as has been shown in two-hybrid assays for FtsQ (28, 29), our results show that FtsQ from C. crescentus interacts with itself; however, we also detected self-interaction of FtsB. The FtsQLB complex has been shown to interact with the FtsI and FtsN proteins through FtsQ (28, 29). When we tested these interactions, we detected a strong interaction only between FtsQ and FtsI and a weak interaction between FtsB and FtsI. In agreement with previous reports, we detected a positive interaction between FtsI and FtsN (28, 29).
FIG 5.
DipI interacts with the FtsQLB complex. Interactions of DipI with the periplasmic domains of different cell division proteins were tested in a yeast two-hybrid assay. The mature DipI protein and the periplasmic domains of FtsQ, FtsL, FtsB, FtsI, and FtsN were fused to the activator and DNA-binding domains (AD and DBD, respectively) of the Gal4 protein, and their abilities to interact were determined by the loss of histidine auxotrophy (if the interaction was weak) and histidine and adenine auxotrophy (if it was strong). Serial dilutions of the yeast strain carrying the plasmids being tested were spotted on agar plates lacking Leu and Trp (growth control), Leu, Trp, and His (weak interaction), and Trp, Leu, His, and adenine (strong interaction). The missing amino acids or nucleotide bases are indicated at the top of each column. Protein fusions are indicated in the following order: DBD fusion/AD fusion. B, FtsB; L, FtsL; Q, FtsQ; I, FtsI; and N, FtsN.
A summary of these interactions is shown in Fig. 6. Some differences from what has been observed for the E. coli proteins are the homodimerization or multimerization of FtsB and the possible interaction between FtsB and FtsI. We did not detect self-interactions of FtsN and FtsI. Some of these discrepancies may be caused by the absence of the cytoplasmic and transmembrane regions that have been reported to be important for FtsI dimerization (28, 29, 60).
FIG 6.
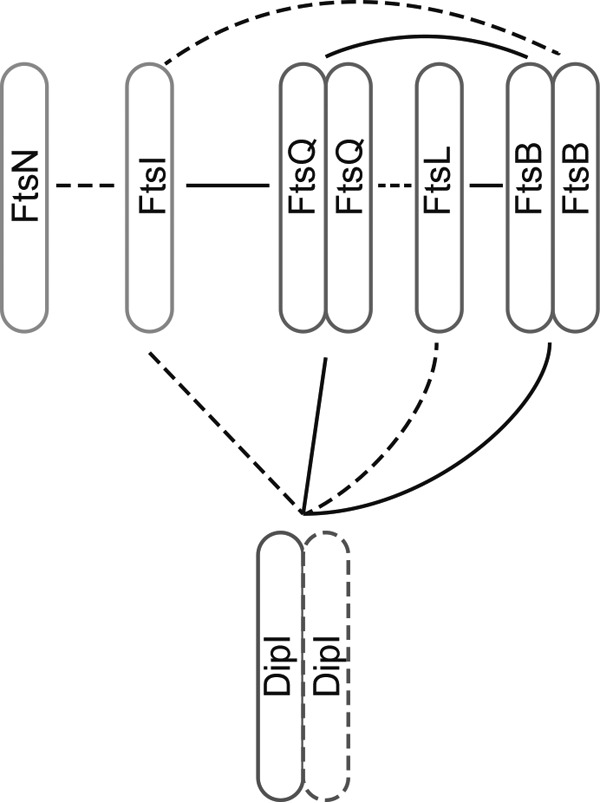
Interaction of the cell division proteins. A summary of the results obtained in the yeast two-hybrid experiments is shown. Strong interactions (homo- or heteromeric) are shown as solid lines and weak interactions are shown as broken lines.
As most of the previously reported interactions between the conserved division proteins were also observed in our assay, we tested the interaction of these proteins with DipI. The strongest DipI interactions were observed with the FtsQ and FtsB proteins, indicating that DipI may be part of the FtsQLB complex or interact with it at some point during the assembly of the divisome. We observed that DipI also interacts weakly with FtsI, as has been shown for FtsQ (29).
Recruitment of DipI depends on the presence of a mature divisome.
To establish which of the interactions detected in the two-hybrid assay were important for the recruitment of DipI to the divisome and to obtain additional evidence for the time at which DipI is recruited to the divisome, we tested the localization of DipI-mCherry in cells depleted of different division proteins. As a positive control, we used an FtsA depletion strain, as in the absence of FtsA C. crescentus cells cannot divide but are still able to initiate constriction (34, 35), indicating the presence of mature divisomes. DipI-mCherry showed strong localization in constriction sites of FtsA-depleted cells and infrequently colocalized in sites where no constriction was visible (Fig. 7). In contrast, no localization of DipI was observed in cells depleted of FtsZ, FtsI, FtsN, or FtsL. The depletion of FtsN and the presence of DipI-mCherry seem to have a synthetic negative effect, because unlike the parental strains, these cells showed extensive lysis and outer membrane blebbing. We verified the presence of DipI-mCherry in the depleted cells to rule out the possibility that the absence of DipI-mCherry localization was due to the degradation of the protein (see Fig. S4). The last protein to be recruited to the division site in C. crescentus is FtsW (34), and the arrival of this protein likely marks the completion of the division machinery. To determine if FtsW was also required for the localization of DipI, we introduced the dipI-mCherry allele into an FtsW depletion strain. However, the cells of the resultant strain grew as filaments even in the presence of the inducer and had no fluorescence. As the synthetic phenotypes observed in the depletion strains may have been due to the partial functionality of the DipI-mCherry fusion, we grew the previous strains in M2G minimal medium in the absence and presence of xylose. Depletion of the division proteins took longer in this medium than in PYE, but except for the FtsW depletion strain, which still showed extensive filamentation even in the presence of xylose, no synthetic phenotypes were observed. Confirming our previous result, no localization of DipI-mCherry was detected in the cells depleted of FtsZ, FtsI, FtsN, or FtsL (see Fig. S5).
FIG 7.
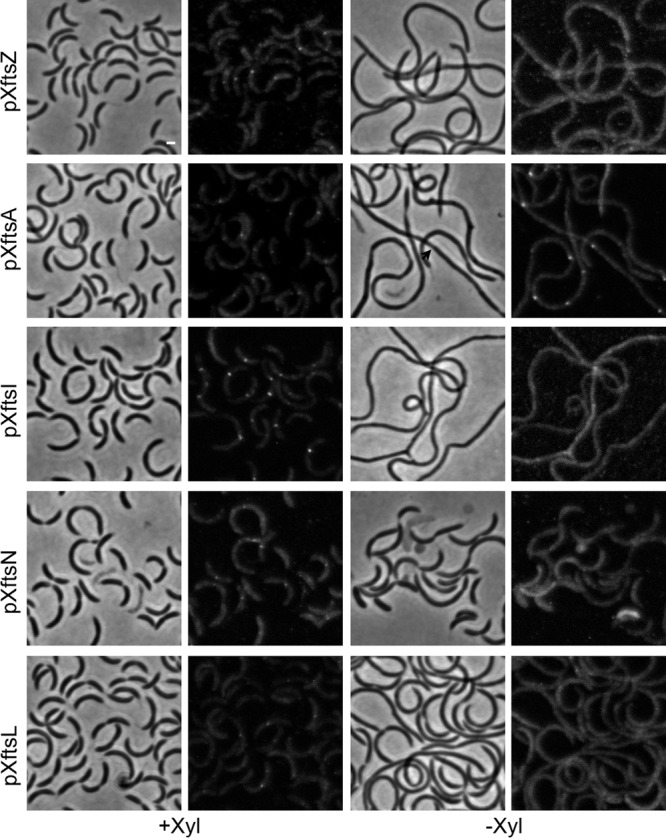
Maturation of the divisome is required for DipI recruitment. The localization of DipI-mCherry was determined in cells depleted of different cell division proteins (indicated at the left of each row). Cells were grown in PYE and depletion was carried out as described in Materials and Methods. Strains used from top to bottom are as follows: SP29, SP16, SP18, SP19, and SP17. Bar, 1 μm.
DISCUSSION
In this work, we describe a new essential cell division protein in C. crescentus that is conserved in Alphaproteobacteria and Betaproteobacteria. The involvement of DipI in cell division was first suggested by the filamentation of the depletion mutant and further supported by the localization of the fluorescent fusion to the division sites. Cells depleted of DipI grew as smooth filaments with infrequent constrictions, suggesting that DipI is required for the initiation of cell division. The presence of the few constrictions observed in the DipI-depleted cells might be explained by a small amount of DipI in some cells of the population, as the degradation signal present in the inducible DipI copy is only functional while the protein is in the cytoplasm; however, constrictions have been observed in cells depleted of other essential division proteins (34). An alternative reason for the filamentation phenotype observed in the cells depleted of DipI might be that the absence of DipI reduces the constriction speed. This enables a new round a chromosome replication, which leads to the movement of the MipZ gradient, resulting in the disassembly of stalled or slowly constricting divisomes. This effect has been shown to be at least partially responsible for the filamentation phenotype of a strain mutated in the accessory division protein DipM (52, 53). However, in contrast to that of DipM, the absence of DipI is lethal to the cells, supporting the idea that DipI is essential for cell division.
A possible reason why a protein that has no apparent enzymatic activity and that is not part of the conserved division proteins is essential for cell constriction is that it is required for recruiting a conserved essential division protein. This possibility is appealing, as the assembly order of the C. crescentus divisome is different from that of E. coli and the recruitment is less hierarchical (34), suggesting a different interaction network between these proteins that might be facilitated by an additional protein. However, in the absence of DipI, a divisome to which all the essential division proteins are recruited still forms but constriction does not start. In a yeast two-hybrid assay, DipI interacts strongly with the FtsQ and FtsB proteins. In C. crescentus, the recruitment of these two proteins depends on FtsL and, as expected, the localization of DipI-mCherry is lost in FtsL-depleted cells. Surprisingly, the localization of DipI was also lost in the absence of FtsN and FtsI, even though in a two-hybrid assay DipI interacts feebly with FtsI and not at all with FtsN. Since in the absence of FtsN the FtsQLB complex and the rest of the division proteins are still recruited (34), the delocalization of DipI in the FtsN depletion strain does not seem to be due to the absence of other division proteins. DipI's dependence on these many proteins for localization is not a common characteristic among the C. crescentus divisional proteins, which do not depend as much as those in E. coli on the presence of other proteins for their localization (34, 38). In general, DipI did not localize in the absence of proteins essential for constriction, but it localized in cells depleted of FtsA that were still able to initiate constriction. These results suggest that recruitment of DipI depends on the presence of a divisome that is capable of initiating constriction and not just on the presence of a single protein.
In E. coli, the initiation of constriction has been proposed to be regulated on the cytoplasmic and periplasmic sides of the inner membrane by FtsA, FtsN, and the FtsQLB complex. On the cytoplasmic side, the interaction between FtsA and FtsN changes the multimerization state of FtsA, triggering the constriction of the FtsZ ring (30). On the periplasmic side, FtsN seems to allosterically change the FtsQLB complex from an off to an on state, which would enable stronger activation of septal peptidoglycan synthesis (19, 20, 29, 61–63). Possible interactions between FtsA and the FtsQLB complex may also be relevant in this process. In. C. crescentus, FtsI, FtsN, and FtsW seem to be relevant in the activation of the divisome (38, 39). A weak interaction between DipI and FtsI was detected in the two-hybrid analysis, and a strong synthetic phenotype was obtained when the DipI-mCherry fusion was introduced into the FtsW depletion strain. However, because DipI strongly interacts with the FtsQLB complex, it is possible that the state of this complex is the signal enabling the localization of DipI, and the recruitment of DipI might be required for activating peptidoglycan synthesis and constriction. This activation might be mediated exclusively by the FtsQLB complex after DipI is recruited, or DipI might be directly involved in this process through its possible interaction with FtsI.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains of E. coli were grown in LB medium with the appropriate antibiotic at 37°C. Plasmids were maintained and purified from E. coli TOP10 strains. Strains of C. crescentus were grown at 30°C unless otherwise specified, in PYE rich medium or M2G medium (64). Antibiotics were used at the following concentrations for E. coli: kanamycin, 50 μg ml−1; spectinomycin, 50 μg ml−1; tetracycline, 10 μg ml−1; gentamicin, 20 μg ml−1; and nalidixic acid, 20 μg ml−1. For C. crescentus, antibiotics were used at the following concentrations for liquid and solid media, respectively: kanamycin, 5 and 20 μg ml−1; spectinomycin, 25 and 100 μg ml−1; tetracycline, 3 and 3.3 μg ml−1; and gentamicin, 2 and 5 μg ml−1. Strains and plasmids are listed in Table 1 and their construction is explained in the supplemental material.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| AH109 | Saccharomyces cerevisiae strain for two-hybrid analysis | Clontech |
| CB15N | Synchronizable derivative of CB15 | 66 |
| CJW3186 | CB15N ftsA::pXftsA | C. Jacobs-Wagner, unpublished data |
| CJW2959 | CB15N vanA::pVsigpepCHYN-4 | 53 |
| SP2 | CB15N ΔdipI::ΩSpc xylR::pXdipI5 | This study |
| SP3 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5 | This study |
| SP4 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pMT383 | This study |
| SP5 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVmCHYftsN6 | This study |
| SP6 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVdipMCHYC2 | This study |
| SP7 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVmreBswCHY4 | This study |
| SP8 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-murG::pmurGCHY4 | This study |
| SP9 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVmCHYftsI4 | This study |
| SP10 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVmCHYftsW6 | This study |
| SP12 | CB15N ΔftsN xylR::pXftsN5 | This study |
| SP13 | CB15N ΔftsI xylR::pXftsI5 | This study |
| SP14 | CB15N ΔftsL xylR::pXftsL5 | This study |
| SP15 | CB15N dipI::pdipICHY4 | This study |
| SP16 | CB15N ftsA::pXftsA2-dipI::pdipICHY4 | This study |
| SP17 | CB15N ΔftsL xylR::pXftsL5-dipI::pdipICHY4 | This study |
| SP18 | CB15N ΔftsI xylR::pXftsI5-dipI::pdipICHY4 | This study |
| SP19 | CB15N ΔftsN xylR::pXftsN5-dipI::pdipICHY4 | This study |
| SP20 | CB15N vanR::pVmCHYftsN6 | This study |
| SP21 | CB15N dipI::pdipICHY4-xylR::pXVENftsB2 | This study |
| SP22 | CB15N dipI::pdipICHY4-xylR::pXVENftsN2 | This study |
| SP23 | CB15N dipI::pdipICHY4-xylR::pXVENftsW2 | This study |
| SP24 | CB15N xylR::pXDipICHY5 | This study |
| SP25 | CB15N vanR::pVsp2DipICHY4 | This study |
| SP26 | CB15N dipI::pdipICHY4-xylR::pXVENftsI2 | This study |
| SP27 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVftsLCHY4 | This study |
| SP28 | CB15N ΔdipI::ΩSpc xylR::pXdipItm5-vanR::pVftsQCHY4 | This study |
| SP29 | CB15N ftsZ::pBJM1-dipI::pdipICHY4 | This study |
| XL1-Blue | Cloning strain | Invitrogen |
| Plasmids | ||
| pBGKT7 | Matchmaker system plasmid | Clontech |
| pADdipI | pGADT7 carrying dipI | This study |
| pADftsB | pGADT7 carrying ftsB | This study |
| pADftsI | pGADT7 carrying ftsI | This study |
| pADftsL | pGADT7 carrying ftsL | This study |
| pADftsN | pGADT7 carrying ftsN | This study |
| pADftsQ | pGADT7 carrying ftsQ | This study |
| pBDdipI | pBGKT7 carrying dipI | This study |
| pBDftsB | pBGKT7 carrying ftsB | This study |
| pBDftsI | pBGKT7 carrying ftsI | This study |
| pBDftsL | pBGKT7 carrying ftsL | This study |
| pBDftsN | pBGKT7 carrying ftsN | This study |
| pBDftsQ | pBGKT7 carrying ftsQ | This study |
| pBOR | pBluescript carrying a 2-kb EcoRI fragment from pHP45Ω | C. Stevens, unpublished data |
| pBJM1 | pBGST18 carrying 5′-terminal region of ftsZ fused to the xylXp promoter | 73 |
| pCHYC-2 | pMB1 replicon carrying mCherry | 74 |
| pdipICHY4 | pCHYC-4 carrying the 3′-terminal region of dipI fused to mCherry | This study |
| pGADT7 | Matchmaker system plasmid | Clontech |
| pMT383 | Integration vector with ftsZ-eyfp under the control of vanAp | 32 |
| pmurGCHY4 | pCHYC-4 carrying the 3′-terminal region of murG fused to mCherry | This study |
| pNPTDdipIW | pNPTS138 carrying ΔdipI::ΩSpc | This study |
| pNPTDftsI | pNPTS138 carrying ΔftsI | This study |
| pNPTDftsL | pNPTS138 carrying ΔftsL | This study |
| pNPTDftsN | pNPTS138 carrying ΔftsN | This study |
| pNPTS138 | pLitmus derivative carrying oriT and sacB | MRK Alley |
| pVCHYC-2 | pMB1 replicon carrying mCherry under the control of vanAp and vanR | 74 |
| pVCHYftsI4 | pVCHYN-4 carrying ftsI fused to mCherry | This study |
| pVCHYftsN6 | pVCHYN-6 carrying ftsN fused to mCherry | This study |
| pVchyftsW6 | pVCHYN-6 carrying ftsW fused to mCherry | This study |
| pVCHYN-6 | pMB1 replicon carrying vanR, vanAp, and mCherry | 74 |
| pVdipMCHY2 | pVCHYC-2 carrying dipM fused to mCherry | This study |
| pVmreBswCHY4 | pVCHYC-4 derivative carrying MreBswmCherry | This study |
| pXdipI5 | pXTCYC-5 derivative carrying dipI | This study |
| pXdipItm5 | pXTCYC-5 derivative carrying dipI fused with tmRNA degradation sequence | This study |
| pXftsI5 | pXTCYC-5 derivative carrying ftsI | This study |
| pXftsL5 | pXTCYC-5 derivative carrying ftsL | This study |
| pXftsN5 | pXTCYC-5 derivative carrying ftsN | This study |
| pXVENftsB2 | pXVENN-2 derivative carrying ftsB | This study |
| pXVENftsN2 | pXVENN-2 derivative carrying ftsN | This study |
| pXVENftsW2 | pXVENN-2 derivative carrying ftsW | This study |
| pXVENN-2 | pMB1 replicon carrying Venus under the control of xylXp | 74 |
| pXTCYC-5 | pMB1 replicon carrying xylR, xylXp, and tetracysteine tag | 74 |
| pVsp2CHYN-4 | pVCHYN-4 carrying dipMΔ109–609 | This work |
| pVsp2dipICHY4 | pVsp2CHYN-4 carrying dipIΔ1–33 | This work |
| pXdipICHY5 | pXCHYC-5 carrying dipI | This work |
Genetic and molecular biology techniques and Western blotting.
Restriction enzymes and T4 DNA ligase were bought from New England BioLabs or Invitrogen. PCRs were performed with TaKaRa PrimeSTAR HS enzyme with high-GC buffer. Transductions were carried out as described previously (64). Strains and plasmid constructions are described in the supplemental material. Plasmid sequences were confirmed and the genotypes of the strains were verified by PCR and phenotype or only by phenotype when the strain was obtained from a transduction. For Western blots, SDS-PAGE-resolved proteins were transferred to nitrocellulose membranes and were incubated with a mouse polyclonal anti-red fluorescent protein (anti-RFP) antibody raised against 6×His-tagged mouse RFP (65) or with a commercial anti-GFP monoclonal mouse antibody (Clontech). For detection, an alkaline phosphatase-conjugated anti-mouse antibody (Sigma) was used together with Tropix CDP-Star/Nitro-block substrate. All samples were collected at an OD600 of 0.3, and the protein amount in each extract was quantified by a Bradford assay. Equal amounts of protein (3 μg) were loaded into every well.
Cell fractionation.
Exponential cultures (OD660 of 0.3; 60 ml) of strains SP22 and CJW2959 induced for 2 h with 75 and 300 μM vanillic acid, respectively, and a culture of the wild-type strain at the same optical density were centrifuged and resuspended in 1/60 of the original volume of phosphate-buffered saline (PBS). These samples were sonicated for 10 s three times with 1-min rest intervals in an ice bath. Cell lysates were centrifuged at low speed (8,000 × g) for 5 min. The supernatants were recovered, total protein was determined by the Bradford assay (Bio-Rad), and the protein concentrations were adjusted to that of the sample with the lowest concentration in a final volume of 5 ml. Samples were centrifuged at 35,000 × g for 2 h at room temperature in an SW50 rotor, and the supernatants were recovered and labeled as the soluble fractions. The pellets were resuspended in the same volumes as those recovered from the supernatants with PBS.
Growth curves and synchronization.
Aliquots of 100 μl from overnight cultures of the strains of interest grown in PYE were used to inoculate 125-ml flasks containing 20 ml of PYE, and the cultures were incubated at 30°C in a water bath with shaking at 200 rpm. Samples were taken every 2 h for 12 h, and the OD660 values were measured. Data obtained from three independent experiments were processed to obtain the average generation time.
Cell synchronization was carried out as previously described (66, 67).
Fluorescence microscopy.
For fluorescence microscopy, 1.5 ml of culture was concentrated to approximately 150 μl, and a 2 μl sample was placed on a microscope slide covered with a 1.5% agarose pad freshly made with M2G medium. Images were taken with a Nikon E600 microscope, a Hamamatsu ORCA-ER camera, and an X-Cite 120 as the light source for fluorescence images. Fluorescence pictures of mCherry- and Venus-labeled proteins were taken using Chroma filters 39010 and 39003, respectively. Images were processed with ImageJ (68). The background was subtracted using a rolling ball radius of 50 pixels, and contrast and brightness were adjusted before copying the relevant selections to Photoshop, where labels were added and the final image size was adjusted. Image analysis to obtain different cell statistics was carried out with microbeTracker (69) either automatically or manually. All experiments were repeated at least two independent times, but the results of single representative experiments are shown.
Depletion of cell division proteins and induction of fluorescent fusions.
To deplete unstable cell division proteins (FtsZ and FtsA), a culture with or without inducer was inoculated with an aliquot of an overnight (ON) culture grown in the presence of the inducer and depletion was allowed to proceed for 8 to 12 h, until cells showed extensive filamentation. For slowly depleting proteins, an ON culture without inducer was inoculated with a cell colony taken from a solid medium plate. The next day, a culture without inducer was inoculated with an aliquot of the ON cultures and incubated for 4 to 6 h until the cells showed extensive filamentation. To induce the expression of the fluorescent fusions, vanillic acid or xylose was added to a growing culture between 2 and 3 h before observation. The following final micromolar concentrations of vanillic acid were used: FtsZ-yellow fluorescent protein (YFP), 150; MreBswmCherry, 200; DipM-mCherry, 200; mCherry-FtsI, 50; mCherry-FtsN, 10; and mCherry-FtsW, 50. For xylose-inducible fusions, the following concentrations were used: Venus-FtsN, 0.025%; Venus-FtsI, 0.075%; Venus-FtsB, 0.05%; and Venus-FtsW, 0.025%.
Yeast two-hybrid assays.
Matchmaker GAL4 two-hybrid system 3 (Clontech) was used to test the interactions between FtsB, FtsL, FtsI, FtsN, FtsQ, and DipI. The region encoding the mature polypeptide (amino acids 35 to 181 of the DipI original annotation) or the periplasmic domains corresponding to amino acids 24 to 100 of FtsB, 36 to 147 of FtsL, 62 to 298 of FtsQ, 55 to 266 of FtsN, and 70 to 589 of FtsI were amplified by PCR using the primers described in supplemental material. The products of these reactions were cloned into pGBKT7 and pGADT7, which encode the DNA-binding domain (BD) and the activation domain (AD) of GAL4, respectively. Interactions were examined by introducing the plasmids that express the proteins to be tested into the reporter strain, AH109. The double transformants were selected as tryptophan (Trp) and leucine (Leu) prototrophs. Transformants were grown ON in synthetic defined (SD) minimal medium without Leu and Trp but supplemented with histidine (His) and adenine (Ade). Aliquots of the cultures were washed once with SD minimal medium without supplements and then normalized to an OD600 of 0.5. Immediately, 10-fold serial dilutions were made in the same medium. From these dilutions, 10-μl aliquots were seeded onto selection plates lacking Trp, Leu, and His or lacking Trp, Leu, His, and Ade.
Protein primary analysis.
Domain composition and domain distribution in different species were analyzed in Pfam and NCBI Conserved Domains websites (70, 71). Signal peptide prediction was done using SignalP (72) and transmembrane helix prediction was done using the TMHMM server.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Consejo Nacional de Ciencia y Tecnologia (SEP-CONACYT 178685) and DGAPA/UNAM (IA201314).
We thank the Unidad de Biologia Molecular IFC UNAM.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00811-16.
REFERENCES
- 1.Nanninga N. 1991. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol 5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 2.Egan AJ, Vollmer W. 2013. The physiology of bacterial cell division. Ann N Y Acad Sci 1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 3.Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J. 2006. Septum enlightenment: assembly of bacterial division proteins. J Bacteriol 188:19–27. doi: 10.1128/JB.188.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moll A, Thanbichler M. 2009. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol Microbiol 72:1037–1053. doi: 10.1111/j.1365-2958.2009.06706.x. [DOI] [PubMed] [Google Scholar]
- 5.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 6.Addinall SG, Bi E, Lutkenhaus J. 1996. FtsZ ring formation in fts mutants. J Bacteriol 178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogales E, Downing KH, Amos LA, Lowe J. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 8.Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osawa M, Anderson DE, Erickson HP. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Hsin J, Zhao L, Cheng Y, Shang W, Huang KC, Wang HW, Ye S. 2013. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science 341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbin BD, Wang Y, Beuria TK, Margolin W. 2007. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol 189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenga L, Luzi G, Paolozzi L, Ghelardini P. 2008. The Escherichia coli FtsK functional domains involved in its interaction with its divisome protein partners. FEMS Microbiol Lett 287:163–167. doi: 10.1111/j.1574-6968.2008.01317.x. [DOI] [PubMed] [Google Scholar]
- 13.Yu XC, Tran AH, Sun Q, Margolin W. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol 180:1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 15.Buddelmeijer N, Beckwith J. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol Microbiol 52:1315–1327. doi: 10.1111/j.1365-2958.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- 16.Glas M, van den Berg van Saparoea HB, McLaughlin SH, Roseboom W, Liu F, Koningstein GM, Fish A, den Blaauwen T, Heck AJ, de Jong L, Bitter W, de Esch IJ, Luirink J. 2015. The soluble periplasmic domains of Escherichia coli cell division proteins FtsQ/FtsB/FtsL form a trimeric complex with submicromolar affinity. J Biol Chem 290:21498–21509. doi: 10.1074/jbc.M115.654756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez MD, Akbay EA, Boyd D, Beckwith J. 2010. Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. J Bacteriol 192:2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez MD, Beckwith J. 2009. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J Bacteriol 191:2815–2825. doi: 10.1128/JB.01597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Persons L, Lee L, de Boer PA. 2015. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol 95:945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsang MJ, Bernhardt TG. 2015. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol 95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goehring NW, Robichon C, Beckwith J. 2007. Role for the nonessential N terminus of FtsN in divisome assembly. J Bacteriol 189:646–649. doi: 10.1128/JB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Addinall SG, Cao C, Lutkenhaus J. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol 25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 23.Rico AI, Garcia-Ovalle M, Palacios P, Casanova M, Vicente M. 2010. Role of Escherichia coli FtsN protein in the assembly and stability of the cell division ring. Mol Microbiol 76:760–771. doi: 10.1111/j.1365-2958.2010.07134.x. [DOI] [PubMed] [Google Scholar]
- 24.Busiek KK, Margolin W. 2014. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol Microbiol 92:1212–1226. doi: 10.1111/mmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busiek KK, Eraso JM, Wang Y, Margolin W. 2012. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol 194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Disteche M, Vollmer W. 2007. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem 282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 27.Corbin BD, Geissler B, Sadasivam M, Margolin W. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol 186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Lallo G, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353–3359. doi: 10.1099/mic.0.26580-0. [DOI] [PubMed] [Google Scholar]
- 29.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol 191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkpatrick CL, Viollier PH. 2012. Decoding Caulobacter development. FEMS Microbiol Rev 36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 32.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 33.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 34.Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. 2011. Assembly of the Caulobacter cell division machine. Mol Microbiol 80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin ME, Trimble MJ, Brun YV. 2004. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol 54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- 36.Goley ED, Dye NA, Werner JN, Gitai Z, Shapiro L. 2010. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol Cell 39:975–987. doi: 10.1016/j.molcel.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meier EL, Razavi S, Inoue T, Goley ED. 2016. A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol Microbiol 101:265–280. doi: 10.1111/mmi.13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modell JW, Hopkins AC, Laub MT. 2011. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev 25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modell JW, Kambara TK, Perchuk BS, Laub MT. 2014. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol 12:e1001977. doi: 10.1371/journal.pbio.1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castillo-Ramirez S, Gonzalez V. 2008. Factors affecting the concordance between orthologous gene trees and species tree in bacteria. BMC Evol Biol 8:300. doi: 10.1186/1471-2148-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. 2011. The essential genome of a bacterium. Mol Syst Biol 7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pini F, De Nisco NJ, Ferri L, Penterman J, Fioravanti A, Brilli M, Mengoni A, Bazzicalupo M, Viollier PH, Walker GC, Biondi EG. 2015. Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet 11:e1005232. doi: 10.1371/journal.pgen.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anantharaman V, Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol 4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eldholm V, Johnsborg O, Straume D, Ohnstad HS, Berg KH, Hermoso JA, Havarstein LS. 2010. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol Microbiol 76:905–917. doi: 10.1111/j.1365-2958.2010.07143.x. [DOI] [PubMed] [Google Scholar]
- 45.Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Chiu M, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Lam WW, Marciano D, Miller MD, Morse AT, Nigoghossian E, Nopakun A, Okach L, Puckett C, Reyes R, Tien HJ, Trame CB, van den Bedem H, Weekes D, Wooten T, Yeh A, Hodgson KO, Wooley J, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA. 2010. Structure of the gamma-d-glutamyl-l-diamino acid endopeptidase YkfC from Bacillus cereus in complex with l-Ala-gamma-d-Glu: insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1354–1364. doi: 10.1107/S1744309110021214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Q, Mengin-Lecreulx D, Liu XW, Patin D, Farr CL, Grant JC, Chiu HJ, Jaroszewski L, Knuth MW, Godzik A, Lesley SA, Elsliger MA, Deacon AM, Wilson IA. 2015. Insights into substrate specificity of NlpC/P60 cell wall hydrolases containing bacterial SH3 domains. mBio 6:e02327-14. doi: 10.1128/mBio.02327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Q, Sudek S, McMullan D, Miller MD, Geierstanger B, Jones DH, Krishna SS, Spraggon G, Bursalay B, Abdubek P, Acosta C, Ambing E, Astakhova T, Axelrod HL, Carlton D, Caruthers J, Chiu HJ, Clayton T, Deller MC, Duan L, Elias Y, Elsliger MA, Feuerhelm J, Grzechnik SK, Hale J, Han GW, Haugen J, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Kumar A, Marciano D, Morse AT, Nigoghossian E, Okach L, Oommachen S, Paulsen J, Reyes R, Rife CL, Trout CV, van den Bedem H, Weekes D, White A, Wolf G, Zubieta C, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. 2009. Structural basis of murein peptide specificity of a gamma-d-glutamyl-l-diamino acid endopeptidase. Structure 17:303–313. doi: 10.1016/j.str.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Himeno H, Nameki N, Kurita D, Muto A, Abo T. 2015. Ribosome rescue systems in bacteria. Biochimie 114:102–112. doi: 10.1016/j.biochi.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 49.Kim JH, Wei JR, Wallach JB, Robbins RS, Rubin EJ, Schnappinger D. 2011. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic Acids Res 39:2210–2220. doi: 10.1093/nar/gkq1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shapland EB, Reisinger SJ, Bajwa AK, Ryan KR. 2011. An essential tyrosine phosphatase homolog regulates cell separation, outer membrane integrity, and morphology in Caulobacter crescentus. J Bacteriol 193:4361–4370. doi: 10.1128/JB.00185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. 2010. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol Microbiol 77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. 2010. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol Microbiol 77:90–107. doi: 10.1111/j.1365-2958.2010.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poggio S, Takacs CN, Vollmer W, Jacobs-Wagner C. 2010. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Mol Microbiol 77:74–89. doi: 10.1111/j.1365-2958.2010.07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa T, Priyadarshini R, Jacobs-Wagner C. 2008. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol Microbiol 70:634–651. doi: 10.1111/j.1365-2958.2008.06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quardokus EM, Din N, Brun YV. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol Microbiol 39:949–959. doi: 10.1046/j.1365-2958.2001.02287.x. [DOI] [PubMed] [Google Scholar]
- 56.Fang G, Passalacqua KD, Hocking J, Llopis PM, Gerstein M, Bergman NH, Jacobs-Wagner C. 2013. Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene coexpression and evolution. BMC Genomics 14:450. doi: 10.1186/1471-2164-14-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. 2007. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol 25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- 58.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khadria AS, Senes A. 2013. The transmembrane domains of the bacterial cell division proteins FtsB and FtsL form a stable high-order oligomer. Biochemistry 52:7542–7550. doi: 10.1021/bi4009837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraipont C, Alexeeva S, Wolf B, van der Ploeg R, Schloesser M, den Blaauwen T, Nguyen-Disteche M. 2011. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157:251–259. doi: 10.1099/mic.0.040071-0. [DOI] [PubMed] [Google Scholar]
- 61.Daniel RA, Noirot-Gros MF, Noirot P, Errington J. 2006. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J Bacteriol 188:7396–7404. doi: 10.1128/JB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robichon C, King GF, Goehring NW, Beckwith J. 2008. Artificial septal targeting of Bacillus subtilis cell division proteins in Escherichia coli: an interspecies approach to the study of protein-protein interactions in multiprotein complexes. J Bacteriol 190:6048–6059. doi: 10.1128/JB.00462-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rowland SL, Wadsworth KD, Robson SA, Robichon C, Beckwith J, King GF. 2010. Evidence from artificial septal targeting and site-directed mutagenesis that residues in the extracytoplasmic beta domain of DivIB mediate its interaction with the divisomal transpeptidase PBP 2B. J Bacteriol 192:6116–6125. doi: 10.1128/JB.00783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. doi: 10.1016/0076-6879(91)04019-K. [DOI] [PubMed] [Google Scholar]
- 65.Ginez LD, Osorio A, Poggio S. 2014. Localization of the outer membrane protein OmpA2 in Caulobacter crescentus depends on the position of the gene in the chromosome. J Bacteriol 196:2889–2900. doi: 10.1128/JB.01516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schrader JM, Shapiro L. 2015. Synchronization of Caulobacter crescentus for investigation of the bacterial cell cycle. J Vis Exp 98. doi: 10.3791/52633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Jones BD, Brun YV. 2001. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol 40:347–360. doi: 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 74.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.