ABSTRACT
Dimethylarginine dimethylaminohydrolases (DDAHs) catalyze the hydrolysis of methylarginines to yield l-citrulline and methylamines as products. DDAHs and their central roles in methylarginine metabolism have been characterized for eukaryotic cells. While DDAHs are known to exist in some bacteria, including Streptomyces coelicolor and Pseudomonas aeruginosa, the physiological importance and genetic regulation of bacterial DDAHs remain poorly understood. To provide some insight into bacterial methylarginine metabolism, this study focused on identifying the key elements or factors regulating DDAH expression in P. aeruginosa PAO1. First, results revealed that P. aeruginosa can utilize NG,NG-dimethyl-l-arginine (ADMA) as a sole source of nitrogen but not carbon. Second, expression of the ddaH gene was observed to be induced in the presence of methylarginines, including NG-monomethyl-l-arginine (l-NMMA) and ADMA. Third, induction of the ddaH gene was shown to be achieved through a mechanism consisting of the putative enhancer-binding protein PA1196 and the alternative sigma factor RpoN. Both PA1196 and RpoN were essential for the expression of the ddaH gene in response to methylarginines. On the basis of the results of this study, PA1196 was given the name DdaR, for dimethylarginine dimethylaminohydrolase regulator. Interestingly, DdaR and its target ddaH gene are conserved only among P. aeruginosa strains, suggesting that this particular Pseudomonas species has evolved to utilize methylarginines from its environment.
IMPORTANCE Methylated arginine residues are common constituents of eukaryotic proteins. During proteolysis, methylarginines are released in their free forms and become accessible nutrients for bacteria to utilize as growth substrates. In order to have a clearer and better understanding of this process, we explored methylarginine utilization in the metabolically versatile bacterium Pseudomonas aeruginosa PAO1. Our results show that the transcriptional regulator DdaR (PA1196) and the sigma factor RpoN positively regulate expression of dimethylarginine dimethylaminohydrolases (DDAHs) in response to exogenous methylarginines. DDAH is the central enzyme of methylarginine degradation, and its transcriptional regulation by DdaR-RpoN is expected to be conserved among P. aeruginosa strains.
KEYWORDS: PA1196, DdaR, enhancer-binding protein, RpoN, dimethylarginine dimethylaminohydrolase, methylarginine, ADMA, Pseudomonas aeruginosa
INTRODUCTION
Many eukaryotic proteins are posttranslationally modified through arginine methylation. At the center of this posttranslational modification are enzymes known as protein arginine methyltransferases (PRMTs) that methylate the guanidino nitrogen atoms of arginine residues in S-adenosylmethionine-dependent reactions (1, 2). Because PRMTs have differences in specificity and prevalence, the methylation patterns vary among arginine residues. Consequently, a few methylarginines are known to exist, including NG-monomethyl-l-arginine (l-NMMA), asymmetric NG,NG-dimethyl-l-arginine (ADMA), and symmetric NG,NG′-dimethyl-l-arginine (SDMA). During protein turnover or proteolysis, ADMA and l-NMMA are released within the cell in their free forms (3, 4), which allows them to act as competitive inhibitors in arginine utilization pathways (5–7). To circumvent this problem, cells rely on an enzyme called dimethylarginine dimethylaminohydrolase (DDAH) that hydrolyzes l-NMMA and ADMA to generate l-citrulline and either mono- or dimethylamine as end products (3, 8). In contrast, SDMA is not an inhibitor of the arginine utilization pathways, nor is it a substrate for DDAH (8, 9). SDMA is removed from the body through excretion in the urine (4, 10).
The metabolism of methylarginines, especially ADMA, has been extensively studied as it pertains to eukaryotes. Interestingly, genes encoding DDAHs are found in some bacteria, including Mycobacterium tuberculosis, Pseudomonas aeruginosa, Sinorhizobium meliloti, and Streptomyces coelicolor (11). The DDAHs of M. tuberculosis, P. aeruginosa, and S. coelicolor have been biochemically investigated and were shown to catalyze the hydrolysis of both ADMA and l-NMMA (11). The presence of DDAH suggests that these bacteria might be able to utilize methylarginines as a source of carbon and/or nitrogen. As a first step in understanding such metabolism, we sought to identify the genes and regulatory mechanisms necessary for methylarginine utilization in P. aeruginosa PAO1.
DDAH of P. aeruginosa PAO1 has been biochemically characterized (12–15) and is encoded by the PA1195 (ddaH) gene. The ddaH gene has distinguishable features suggesting that it is under the regulation of the alternative sigma factor σ54 or RpoN (16, 17). First, located 50 bp upstream of the start codon of ddaH is a putative −24/−12 promoter (Sigma 54 Promoter Database [http://www.sigma54.ca]). The −24/−12 promoter is a highly conserved nucleotide sequence that is recognized by RpoN for transcriptional activation (17, 18). Second, the adjacent PA1196 gene encodes a putative enhancer-binding protein (EBP) with an unknown function (19, 20). EBPs are transcriptional regulators that interact specifically with RpoN to activate transcription from −24/−12 promoters (19, 21, 22). Upon stimulation, EBPs become active and hydrolyze ATP. This nucleotide hydrolysis provides the energy needed for the RpoN-RNA polymerase holoenzyme to transition from a closed to an open complex (21).
In the current study, PA1196 was hypothesized to be a key regulator of ddaH expression in P. aeruginosa PAO1. Specifically, it was proposed that PA1196 in concert with RpoN activates the transcription of ddaH in response to methylarginines, such as ADMA and l-NMMA. The PA1196 gene is in a predicted operon with PA1197, a gene encoding a putative Sir2 protein. Sir2 proteins often function as deacetylases and have regulatory roles in cellular processes, including transcriptional repression or posttranslational modifications (23, 24). It was therefore considered that PA1197 might also have some regulatory role in methylarginine utilization in P. aeruginosa PAO1.
RESULTS
PA1196 is required for utilization of ADMA as a nitrogen source for P. aeruginosa PAO1.
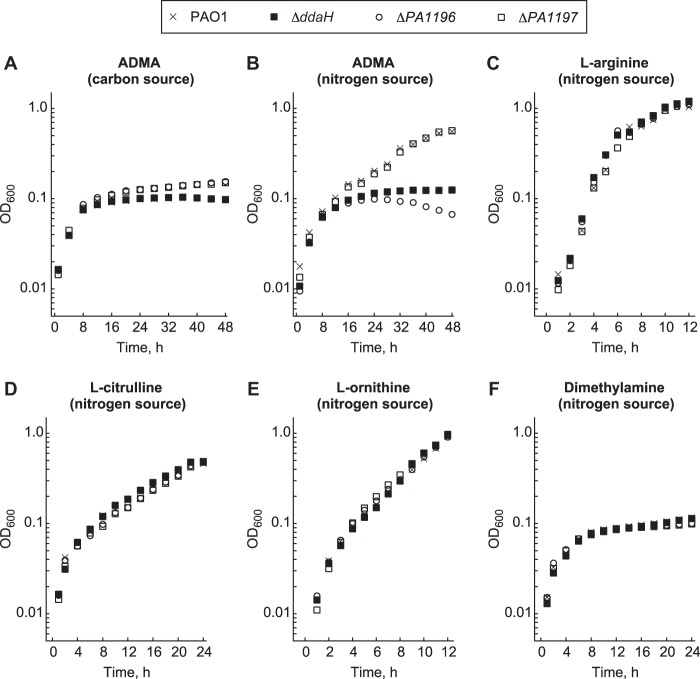
The ddaH, PA1196, and PA1197 genes were deleted from P. aeruginosa PAO1 to create a series of unmarked deletion mutants: the ΔddaH, ΔPA1196, and ΔPA1197 mutants. The mutants and wild-type PAO1 strain were grown in minimal medium supplemented with ADMA as a sole source of either carbon or nitrogen (Fig. 1). As a sole carbon source in the presence of NH4Cl, ADMA did not support the growth of any of the mutants or PAO1 (Fig. 1A). Cell densities reached a maximum optical density at 600 nm (OD600) of ∼0.1 at 48 h postinoculation on ADMA as a carbon source. However, when ADMA served as the sole nitrogen source in the presence of gluconate, slow growth was observed for the ΔPA1197 mutant and PAO1 (Fig. 1B). For these two strains, cell densities reached a maximum OD600 of ∼0.6 at 48 h postinoculation on ADMA as a nitrogen source. The ΔddaH and ΔPA1196 mutants exhibited no growth on ADMA as a nitrogen source; cell densities reached a maximum OD600 of ∼0.1 (Fig. 1B). Identical growth curves were observed for all strains on l-arginine, l-citrulline, and l-ornithine as nitrogen sources (Fig. 1C to E), suggesting that deletion of ddaH, PA1196, and PA1197 has no effect on the utilization of these compounds. Dimethylamine did not serve as a nitrogen source for any of the mutants or PAO1 (Fig. 1F).
FIG 1.
PA1196 is required for P. aeruginosa PAO1 to grow on ADMA as a nitrogen source. The growth of P. aeruginosa PAO1 and the ΔddaH, ΔPA1196, and ΔPA1197 mutants in minimal medium supplemented with the following nitrogen (10 mM) and carbon (15 mM) sources was measured: NH4Cl and ADMA (A), ADMA and gluconate (B), l-arginine and gluconate (C), l-citrulline and gluconate (D), l-ornithine and gluconate (E), and dimethylamine and gluconate (F). The strains were grown in 96-well polystyrene plates at 37°C for 24 to 48 h. Data points represent mean values (n = 4), with SDs being <5.0%. Error bars were omitted for clarity.
Expression of either the PA1196 or the ddaH gene from the lac promoter on plasmid pBBR1MCS-5 restored the growth of the ΔPA1196 mutant on ADMA as a nitrogen source (Fig. 2). The finding that plasmid-derived expression of ddaH compensated for the ADMA-related growth deficiency of the ΔPA1196 mutant was an indicator that expression of the ddaH gene is possibly deregulated or insufficient in the absence of the PA1196 gene. Collectively, the results of these growth assays demonstrate that ddaH (as expected) and PA1196 are essential for ADMA utilization in P. aeruginosa PAO1.
FIG 2.
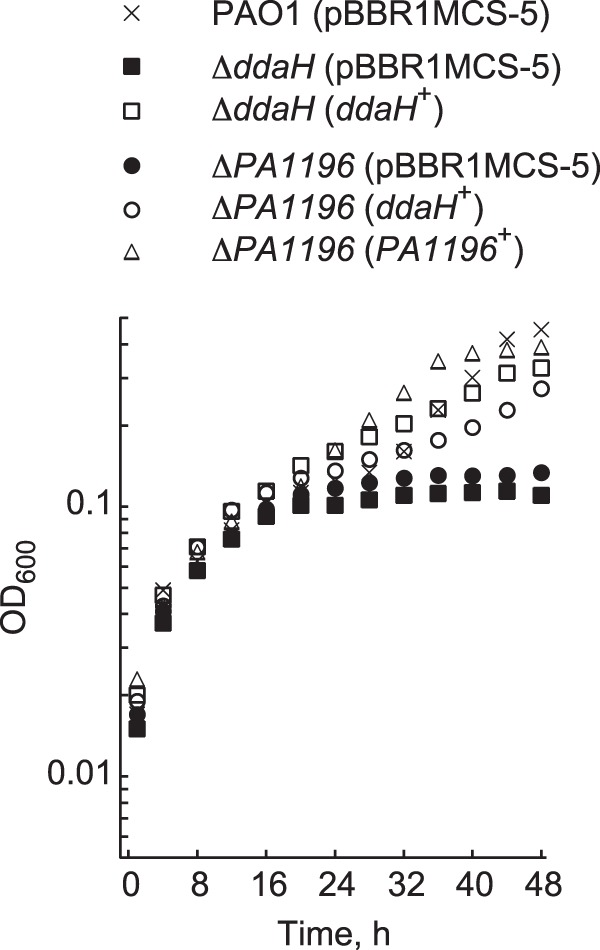
Plasmid-based expression of PA1196 or ddaH rescues the growth of the ΔPA1196 mutant on ADMA. Growth in minimal medium supplemented with 15 mM gluconate, 10 mM ADMA, and 30 μg ml−1 gentamicin was measured for P. aeruginosa PAO1 harboring an empty plasmid (pBBR1MCS-5), the ΔddaH mutant harboring either an empty plasmid (pBBR1MCS-5) or pBRL685 (ddaH+), and the ΔPA1196 mutant harboring either an empty plasmid (pBBR1MCS-5), pBRL685 (ddaH+), or pBRL686 (PA1196+). Note that the ddaH and PA1196 genes were cloned under the control of the lac promoter of pBBR1MCS-5 to generate pBRL685 and pBRL686, respectively. Recombinant strains were grown in 96-well polystyrene plates at 37°C for 48 h. Data points represent mean values (n = 4), with SDs being <5.0%. Error bars were omitted for clarity.
Expression of ddaH is induced by ADMA.
To explore the expression of ddaH, a LacZ reporter was constructed by fusing the ∼500-bp 5′ regulatory region of ddaH with the lacZ open reading frame (ORF) of Escherichia coli. The ddaH::lacZ fusion was cloned into the promoter-less, low-copy-number plasmid ΔPlac-pBBR1MCS-5. Afterwards, P. aeruginosa PAO1 harboring ddaH::lacZ was grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with various substrates, each at a final concentration of 0.1 mM. As shown in Fig. 3A, LacZ activities increased >2-fold at 1.0 h after the addition of ADMA. LacZ activities also increased >2-fold with the addition of l-NMMA, a monomethylated derivative of l-arginine. In contrast, LacZ activities did not change with the addition of l-arginine, l-citrulline, or no substrate.
FIG 3.
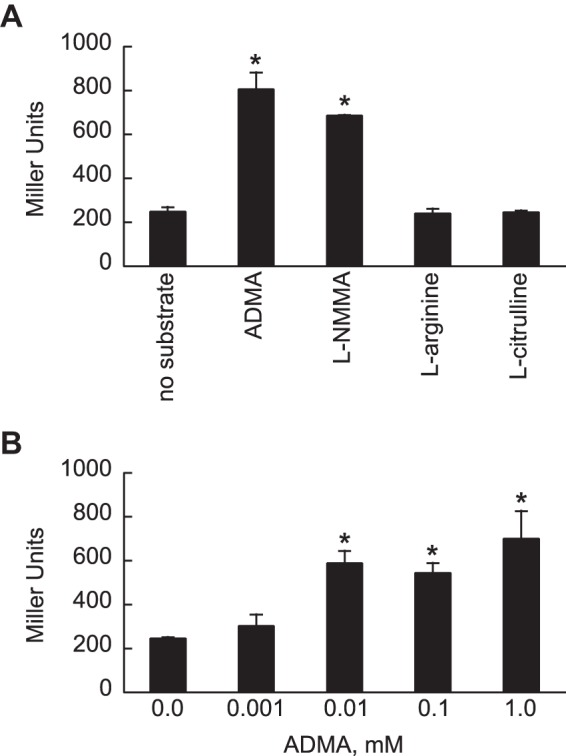
Methylarginines induce expression of ddaH::lacZ in P. aeruginosa PAO1. The ∼500-bp 5′ regulatory region of ddaH was fused to E. coli lacZ, and the resulting construct was cloned into a promoter-less plasmid to give ddaH::lacZ. P. aeruginosa PAO1 harboring ddaH::lacZ was grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with either 0.1 mM ADMA, l-NMMA, l-arginine, or l-citrulline (A) or ADMA at various concentrations, 0.001, 0.01, 0.1, or 1.0 mM (B). LacZ activities were measured 1.0 h after the addition of the substrate. Data points represent mean values (n = 3) ± SDs. ANOVA with a Dunnett's post hoc test (α value, 0.05) was done to identify significant differences (P < 0.0001), which are indicated with asterisks.
The effects of various concentrations of exogenous ADMA on the expression of ddaH::lacZ were measured. Final ADMA concentrations of 0.01, 0.1, and 1.0 mM caused LacZ activities to increase >2-fold when ddaH::lacZ was expressed in P. aeruginosa PAO1 (Fig. 3B). The addition of ADMA to 0.001 mM did not significantly affect ddaH::lacZ expression. These results indicate that expression of ddaH in P. aeruginosa PAO1 is inducible by ADMA at relatively low concentrations.
DDAH is regulated by PA1196.
It was next determined what role PA1196 and PA1197 might have on expression of the ddaH gene. Mutants (the ΔddaH, ΔPA1196, and ΔPA1197 mutants) and PAO1 harboring ddaH::lacZ were grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with either no substrate or 0.1 mM ADMA. LacZ activities were subsequently measured at 1.0 h after the addition of the substrate. It was found that the addition of ADMA induced the expression of ddaH::lacZ for both the ΔPA1197 mutant and PAO1; i.e., LacZ activities increased >2-fold for both strains (Fig. 4). For the ΔddaH mutant, LacZ activities increased >7-fold with the addition of ADMA. This elevated fold change compared to the activity observed in PAO1 was expected, because the ΔddaH mutant is unable to catabolize or enzymatically break down ADMA. For the ΔPA1196 mutant, there was no change in LacZ activity with the addition of ADMA. This result is consistent with PA1196 being a positive regulator of the ddaH gene.
FIG 4.
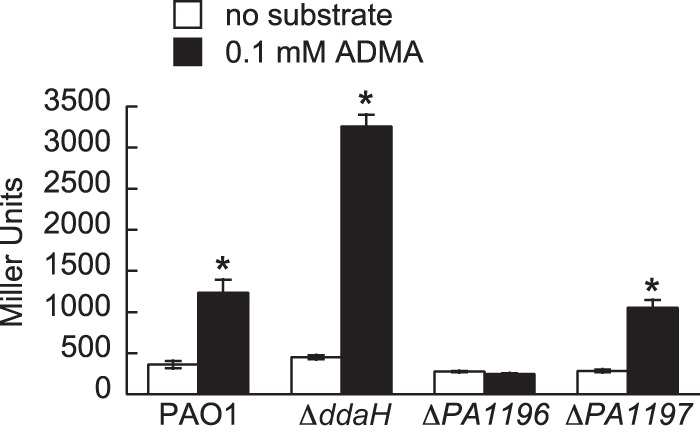
PA1196 is essential for induction of ddaH::lacZ in response to ADMA. Expression of ddaH::lacZ was determined in P. aeruginosa PAO1 and the ΔddaH, ΔPA1196, and ΔPA1197 mutants. Strains harboring ddaH::lacZ were grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with either no substrate or 0.1 mM ADMA. LacZ activities were measured at 1.0 h after the addition of the substrate. Data points represent mean values (n = 3) ± SDs. ANOVA with a Dunnett's post hoc test (α value, 0.05) was done to identify significant differences (P < 0.0001), which are indicated with asterisks.
Deletion of the PA1197 gene did not affect expression of ddaH::lacZ, indicating that PA1197 is not necessary for the transcription of ddaH in response to ADMA. Therefore, it was considered possible that PA1197 regulates the actual function or catalytic activity of DDAH. To address this possibility, cell extracts from mutant and PAO1 strains grown in gluconate-minimal medium were prepared in either the absence or the presence of ADMA. DDAH activity, i.e., the formation of l-citrulline from ADMA, was measured for each cell extract. For all mutant and PAO1 strains grown in the absence of ADMA, the resulting cell extracts yielded DDAH activities of ∼20 U (Fig. 5). However, when the bacteria were grown in the presence of ADMA, DDAH activities were ∼100 U in cell extracts prepared from the ΔPA1197 mutant and PAO1 (Fig. 5). The addition of ADMA caused an ∼5-fold increase in DDAH activity. In contrast, cell extracts prepared from the ΔddaH and ΔPA1196 mutants grown in the presence of ADMA generated DDAH activities of ∼20 U, which was similar to the value measured in the absence of ADMA. These findings confirm that PA1196 is essential for DDAH activity in P. aeruginosa PAO1.
FIG 5.
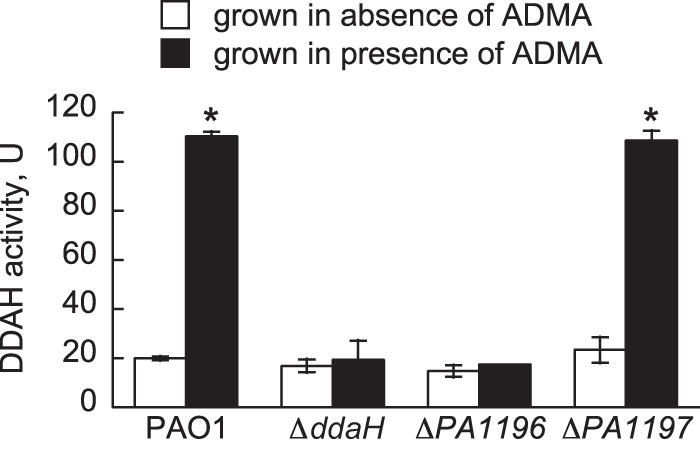
DDAH activity is dependent on PA1196. P. aeruginosa PAO1 and the ΔddaH, ΔPA1196, and ΔPA1197 mutants were grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with either no substrate or 1.0 mM ADMA. At 2.0 h after the addition of the substrate, cells were collected by centrifugation, washed, and subsequently lysed via sonication. The lysates were cleared of cellular debris, concentrated, and then assayed for DDAH activity, which is reported in units (U), defined as the formation of l-citrulline (nanomoles hour−1 milligram−1 total protein). Data points represent mean values (n = 2) ± SDs. ANOVA with a Dunnett's post hoc test (α value, 0.05) was done to identify significant differences (P < 0.0001), which are indicated with asterisks.
ADMA induces expression of ddaH::lacZ in Pseudomonas putida cells that express PA1196.
Because P. putida KT2440 does not possess any homologs of DDAH or PA1196, it was considered a suitable host to validate the central role of PA1196 in the expression of the ddaH gene. To this end, the PA1196 gene was cloned under the control of the lac promoter on plasmid pBBR1MCS-3, and the resulting plasmid-derived PA1196 was cotransformed with ddaH::lacZ into P. putida KT2440. Recombinant cells were grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with either no substrate or 0.1 mM ADMA. LacZ activities were measured 2.0 h after the addition of the substrate. As shown in Fig. 6, the addition of ADMA induced LacZ activity >20-fold for P. putida KT2440 cells harboring plasmid-derived PA1196 and ddaH::lacZ. PA1196 was essential for this induction, because cells that harbored ddaH::lacZ in combination with either an empty plasmid (pBBR1MCS-3) or a plasmid having PA1196 cloned backwards relative to the orientation of the lac promoter on pBBR1MCS-3 yielded only background levels of LacZ activity with the addition of ADMA.
FIG 6.
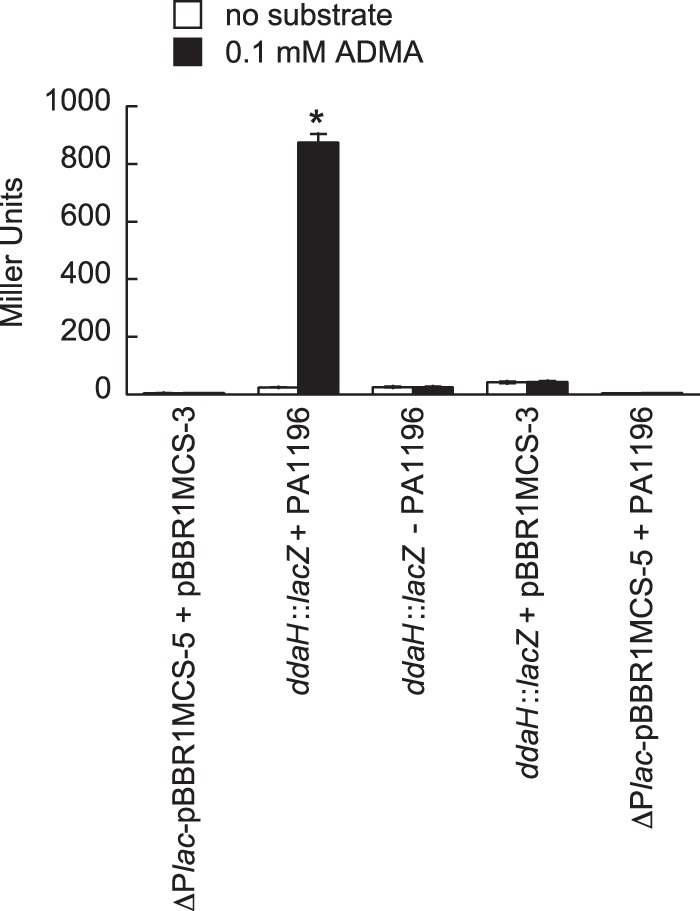
PA1196 is essential for induction of ddaH::lacZ in P. putida KT2440. P. putida KT2440 was cotransformed with several plasmid combinations, including (i) ddaH::lacZ and pBRL720 (+PA1196), (ii) ddaH::lacZ and an empty plasmid (pBBR1MCS-3), (iii) ddaH::lacZ and pBRL719 (−PA1196), (iv) ΔPlac-pBBR1MCS-5 and pBRL720 (+PA1196), and (v) empty plasmids (ΔPlac-pBBR1MCS-5 and pBBR1MCS-3). Recombinant strains were grown in gluconate-minimal medium to an OD600 of 0.3 and then challenged with either no substrate or 0.1 mM ADMA. LacZ activities were measured at 2.0 h after the addition of the substrate. Note that the PA1196 gene was cloned with either a forward (pBRL720) or a backward (pBRL719) orientation relative to the orientation of the lac promoter of pBBR1MCS-3. Data points represent mean values (n = 3) ± SDs. ANOVA with a Dunnett's post hoc test (α value, 0.05) was done to identify significant differences (P < 0.0001), which are indicated with an asterisk.
RpoN is required for induction of ddaH::lacZ.
A putative RpoN promoter with the sequence TGGCGCGTGGCTTGCA (Sigma 54 Promoter Database [http://www.sigma54.ca]) is located 50 bp upstream of the ddaH ORF. The GG and GC nucleotides (which appear in bold type in the promoter sequence) of the −24 and −12 elements, respectively, are crucial for promoter activity. In order to evaluate the role of this promoter in ddaH expression, the GG nucleotides of the −24 element were replaced with AA in the ddaH::lacZ reporter. As expected, this mutation (−PRpoN) rendered ddaH::lacZ unresponsive to ADMA in P. aeruginosa PAO1 (Fig. 7). Furthermore, the addition of ADMA did not induce expression of ddaH::lacZ in an rpoN mutant (the rpoN::Ω-Km mutant) of P. aeruginosa PAO1. These findings are in agreement and indicate that expression of ddaH is regulated through RpoN.
FIG 7.
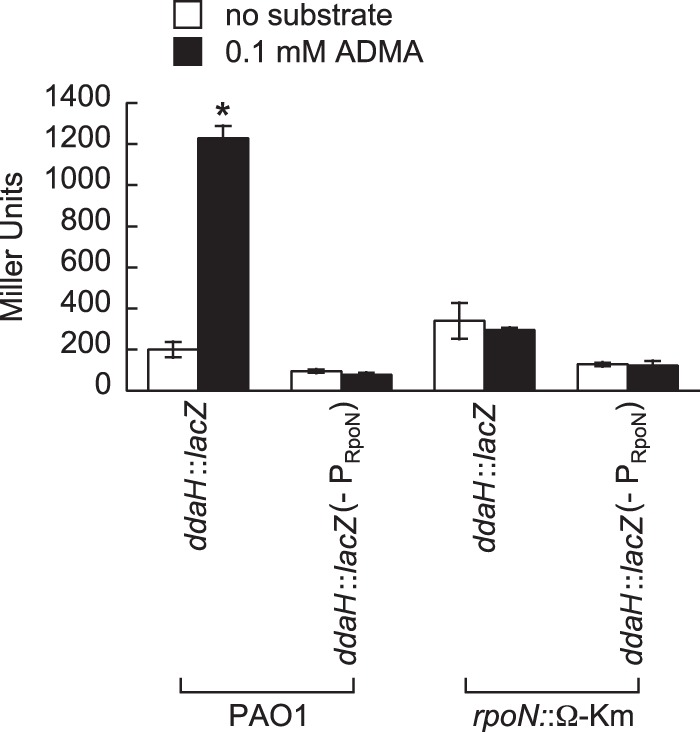
RpoN and its cognate promoter are required for induction of ddaH::lacZ in response to ADMA. Expression of ddaH::lacZ was measured in P. aeruginosa PAO1 and an rpoN mutant (the rpoN::Ω-Km mutant). In addition, expression was measured for a ddaH::lacZ construct that had a mutated RpoN promoter (−PRpoN); i.e., the conserved GG dinucleotide of the −24 element of the RpoN promoter was changed to AA. Strains harboring the ddaH::lacZ constructs were grown in gluconate-minimal medium supplemented with 1.0 mM l-glutamine to an OD600 of 0.3 and then challenged with either no substrate or 0.1 mM ADMA. LacZ activities were measured 1.0 h after the addition of the substrate. Data points represent mean values (n = 3) ± SDs. ANOVA with a Dunnett's post hoc test (α value, 0.05) was done to identify significant differences (P < 0.0001), which are indicated with an asterisk.
ADMA utilization in other strains of P. aeruginosa.
DDAHs and homologs of PA1196 are found in other P. aeruginosa strains (20), which suggests that methylarginine utilization is conserved among them. Indeed, growth on ADMA as a nitrogen source was observed for P. aeruginosa strains PA14 and PAK (Fig. 8A). In comparison, DDAHs are not predicted to occur in other Pseudomonas spp., and consistent with their absence, neither P. putida nor Pseudomonas fluorescens grew on ADMA as a nitrogen source (Fig. 8A). Lastly, we decided to evaluate the growth of P. aeruginosa PAO1 in gluconate-minimal medium supplemented with various concentrations of ADMA. As shown in Fig. 8B, identical growth curves were observed for P. aeruginosa PAO1 on 2.0, 5.0, and 10 mM ADMA. Supplementation with ADMA at 0.2 mM also resulted in detectable growth but at a reduced level compared to that observed for the higher ADMA concentrations.
FIG 8.
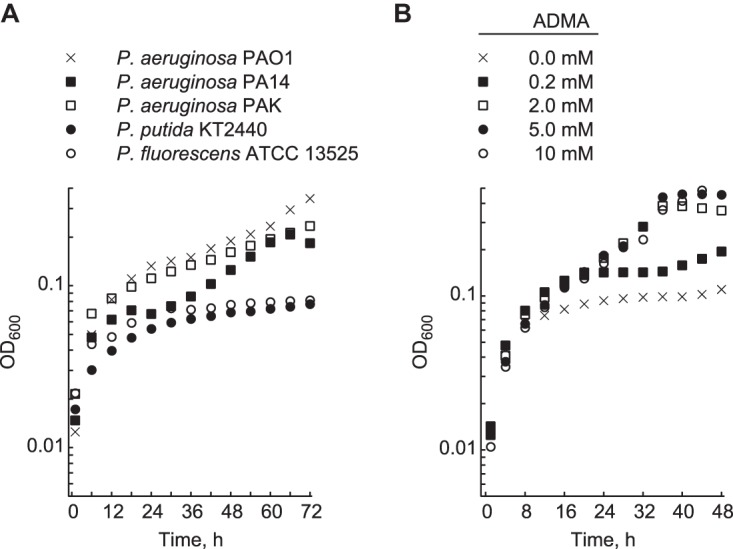
ADMA serves as a nitrogen source for other strains of P. aeruginosa. (A) Growth in minimal medium supplemented with 15 mM gluconate and 10 mM ADMA was measured for various P. aeruginosa strains (PAO1, PA14, and PAK), P. putida KT2440, and P. fluorescens ATCC 13525. Strains were grown in 96-well polystyrene plates at 30°C for 72 h. (B) Growth in minimal medium supplemented with 15 mM gluconate and various ADMA concentrations (0.0, 0.2, 2.0, 5.0, and 10 mM) was measured for P. aeruginosa PAO1 in 96-well polystyrene plates at 37°C for 48 h. Data points represent mean values (n = 4), with SDs being <5.0%. Error bars were omitted for clarity.
DISCUSSION
Methylarginine degradation in P. aeruginosa PAO1.
A proposed pathway for the breakdown of methylarginines in P. aeruginosa PAO1 is given in Fig. 9. Extracellular methylarginines are believed to be transported through PA1194 and/or ArcD (PA5170). The PA1194 gene encodes a transport protein exhibiting 60% similarity (49% identity) to ArcD, an l-arginine/l-ornithine antiporter (25, 26). Similar to DDAH and PA1196, homologs of PA1194 exist in other strains of P. aeruginosa. This suggests that PA1194 may have evolved to function as a methylarginine/l-ornithine antiporter. Interestingly, results from the LacZ assays involving P. putida KT2440 (Fig. 6) indicate that this bacterium is capable of the uptake of extracellular ADMA, even though it does not possess a homolog of PA1194. P. putida KT2440 does, however, harbor an l-arginine/l-ornithine antiporter, ArcD (PP_1002), which could be responsible for methylarginine transport in this bacterium. Although PA1194 and ArcD are potentially involved in the transport of methylarginines, further studies are needed to elucidate the actual mechanisms underlying the uptake of methylarginines in P. aeruginosa.
FIG 9.
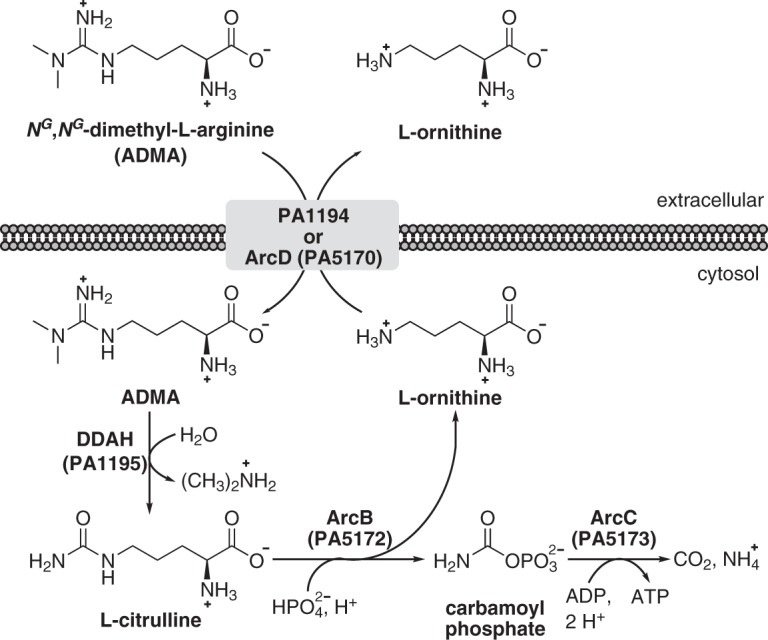
Proposed pathway for methylarginine degradation in P. aeruginosa PAO1. It is hypothesized that extracellular methylarginines are transported into the bacterium through PA1194 and/or ArcD (PA5170). PA1194 shares 60% similarity (49% identity) with ArcD (PA5170), an l-arginine/l-ornithine antiporter. This suggests that PA1194 might function as a dedicated methylarginine/l-ornithine antiporter. Upon entry, intracellular methylarginines are hydrolyzed into l-citrulline and either mono- or dimethylamine via DDAH (PA1195). Using phosphate as a substrate, ornithine carbamoyltransferase, ArcB (PA5172), catalyzes the conversion of l-citrulline into l-ornithine and carbamoyl phosphate. While the l-ornithine product can be used for the uptake of methylarginines, carbamoyl phosphate is broken down into ATP, CO2, and NH3 in a reaction catalyzed by carbamate kinase, ArcC (PA5173).
The central catabolic step in methylarginine utilization is catalyzed by DDAH, which hydrolyzes intracellular methylarginines to generate mono- or dimethylamine and l-citrulline as products. Mono- and dimethylamines have not been reported to be viable nutrients for P. aeruginosa, and in this study, dimethylamine did not serve as a nitrogen source for P. aeruginosa PAO1 (Fig. 1). However, l-citrulline is an intermediate of arginine degradation (the arginine deiminase or ArcABC pathway) and has been shown to be a poor nitrogen source for P. aeruginosa (27–29). On the basis of this information, it is predicted that the l-citrulline product of DDAH catalysis is converted into l-ornithine and carbamoyl phosphate via catabolic ornithine carbamoyltransferase (ArcB) (Fig. 9). Carbamate kinase (ArcC) then transfers the phosphorous group from carbamoyl phosphate onto ADP, thus yielding ATP, CO2, and NH3 as products. This series of reactions would account for the ability of P. aeruginosa PAO1 to use ADMA as a nitrogen source, albeit poorly (Fig. 1).
The significance or importance of methylarginine utilization by P. aeruginosa has yet to be established. Data from the literature indicate that there is ∼0.2 to 20 μmol of ADMA per kg of plant tissue (30) and ∼0.01 to 1.0 μmol of ADMA per g of animal protein (31), and plasma concentrations of ADMA are on the order of 0.3 to 0.7 μM in healthy humans (32, 33). These values in combination with the results from this study suggest that P. aeruginosa assimilates exogenous methylarginines to supplement its growth rather than using these compounds as a main source of nitrogen and/or energy. Additionally, l-NMMA and ADMA are inhibitors of nitric oxide (NO) production in eukaryotes (7), so it is possible that methylarginine degradation serves as a protective measure for NO signaling in P. aeruginosa.
DdaR (PA1196) as a regulator of DDAH in P. aeruginosa PAO1.
One of the more significant findings of this study was the identification of the putative EBP PA1196 as an essential regulator of the expression of the ddaH gene in response to methylarginines. Consequently, PA1196 was named DdaR, for dimethylarginine dimethylaminohydrolase regulator. DdaR has a predicted modular structure comprised of an N-terminal sensor domain, a central sigma 54 interaction domain, and a C-terminal DNA-binding domain that possesses a FIS-type helix-turn-helix motif (19, 20). DdaR is not a homolog of any previously characterized EBP and therefore represents a class of EBPs whose regulatory activities are mediated through methylarginines. Homologs of DdaR are found only in the genomes of other strains of P. aeruginosa. DdaR homologs do not occur in other Pseudomonas spp., nor are they found in other bacteria which possess DDAHs. This suggests that the DdaR-RpoN mechanism for regulating expression of ddaH is unique to P. aeruginosa. Despite DDAH and PA1196 being conserved among P. aeruginosa strains, the ability to grow on methylarginines as a sole source of nitrogen varies among them (Fig. 8). The reasons behind this growth variation is unclear but might be related to factors such as the experimental growth temperature, RpoN activity, l-citrulline degradation, and tolerance to dimethylamine.
Apart from ddaH, it is not clear what other genes are regulated by DdaR. The arcBC genes, which are central in l-citrulline degradation, do not possess any putative −24/−12 promoters (Sigma 54 Promoter Database [http://www.sigma54.ca]). Both PA1194 and ArcD (PA5170) are hypothesized to be involved in methylarginine transport, but neither corresponding gene possesses a −24/−12 promoter. The absence of −24/−12 promoters among these genes is a clear indicator that they are not direct targets of transcriptional activation by RpoN. More extensive analyses, e.g., transcriptomic studies, are needed to determine the scope and magnitude of genes whose expression is affected by DdaR.
The function of PA1197 remains unclear.
The ddaR gene is in a putative operon with PA1197, which encodes a Sir2-type deacetylase. While this operon structure would suggest that PA1197 has some role in methylarginine utilization, we found that a ΔPA1197 mutant exhibited wild-type growth on ADMA under the conditions tested in this study (Fig. 1). In addition, DDAH activity and expression of a ddaH::lacZ reporter were not affected by a ΔPA1197 mutation (Fig. 4 and 5). These findings argue that PA1197 is not essential for DDAH activity and methylarginine utilization in P. aeruginosa PAO1. It is possible that PA1197 affects methylarginine utilization under conditions not tested in this study, including under conditions of oxygen limitation and in biofilms. For example, in the presence of NO, the catalytic cysteine residue of DDAH undergoes S-nitrosylation (34, 35), a modification that inhibits enzymatic activity. PA1197 might participate in the alleviation of this inhibition through some unknown mechanism.
MATERIALS AND METHODS
Bacteria and media.
The bacteria used in the current study are listed in Table 1. Bacteria were grown in either Lennox broth (LB) or minimal medium (22 mM KH2PO4, 42 mM Na2HPO4, 8.6 mM NaCl, 1.0 mM MgSO4, 5.0 μM FeSO4, pH 7.0). Minimal medium was supplemented with 15 mM carbon source and 10 mM nitrogen source as specified for each set of experiments (see below). Solid bacteriological media were prepared with the addition of Difco Bacto agar at 15 g liter−1. For plasmid selection in E. coli, the media were supplemented with carbenicillin (100 μg ml−1), gentamicin (20 μg ml−1), or kanamycin (50 μg ml−1). For plasmid and marker selection in Pseudomonas spp., the media were supplemented with carbenicillin (200 μg ml−1), gentamicin (30 μg ml−1), or tetracycline (25 μg ml−1).
TABLE 1.
Bacteria used in the current study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type | 46 |
| PAO1 ΔddaH | ΔddaH (ΔPA1195) mutant derivative of PAO1 | This study |
| PAO1 ΔPA1196 | ΔPA1196 mutant derivative of PAO1 | This study |
| PAO1 ΔPA1197 | ΔPA1197 mutant derivative of PAO1 | This study |
| PAO6359 | rpoN::Ω-Km mutant derivative of PAO1 | 46 |
| PA14 | Wild type | 47 |
| PAK | Wild type | 48 |
| Pseudomonas putida KT2440 | Wild type | ATCC |
| Pseudomonas fluorescens ATCC 13525 | Wild type | ATCC |
| Escherichia coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1 λ− | Invitrogen |
General molecular biology procedures.
The plasmids and oligonucleotides (primers) used in the current study are listed in Table 2. The restriction endonucleases, T4 ligase, and Phusion DNA polymerase used for cloning purposes were purchased from New England BioLabs. Promega nucleic acid purification kits were used for DNA isolation. Genomic DNA from P. aeruginosa PAO1 served as the template in all PCRs.
TABLE 2.
Plasmids and oligonucleotides used in the current studya
| Plasmid or oligonucleotide | Relevant characteristics or sequence | Reference/source or purpose |
|---|---|---|
| Plasmids | ||
| pCR-Blunt | Cloning plasmid; Kmr | Invitrogen |
| pDONR221 | Gateway cloning plasmid; Kmr | Invitrogen |
| pET15b | Expression plasmid; Cbr | EMD Millipore |
| pBBR1MCS-3 | Broad-host-strain plasmid; Tcr | 39 |
| pBBR1MCS-5 | Broad-host-strain plasmid; Gmr | 39 |
| pEX18ApGW | Gene deletion plasmid for P. aeruginosa; Cbr | 36 |
| pFLP2 | Carries FLP recombinase; Cbr | 49 |
| pPS856 | Carries FRT-accC1-FRT marker; Gmr | 49 |
| ΔPlac-pBBR1MCS-5 | pBBR1MCS-5 minus the lac promoter; Gmr | 37 |
| pBRL631 | ddaH::FRT-aacC1-FRT in pDONR221; Kmr Gmr | This study |
| pBRL633 | PA1197::FRT-aacC1-FRT in pDONR221; Kmr Gmr | This study |
| pBRL635 | PA1196::FRT-aacC1-FRT in pDONR221; Kmr Gmr | This study |
| pBRL637 | ddaH::FRT-aacC1-FRT in pEX18ApGW; Cbr Gmr | This study |
| pBRL638 | PA1197::FRT-aacC1-FRT in pEX18ApGW; Cbr Gmr | This study |
| pBRL639 | PA1196::FRT-aacC1-FRT in pEX18ApGW; Cbr Gmr | This study |
| pBRL645 | ddaH::lacZ in pCR-Blunt; Kmr | This study |
| pBRL649 | ddaH::lacZ in ΔPlac-pBBR1MCS-5; Gmr | This study |
| pBRL671 | ddaH in pCR-Blunt; Kmr | This study |
| pBRL672 | PA1196 in pCR-Blunt; Kmr | This study |
| pBRL674 | ddaH in pET15b; Cbr | This study |
| pBRL675 | PA1196 in pET15b; Cbr | This study |
| pBRL685 | ddaH in pBBR1MCS-5; Gmr | This study |
| pBRL686 | PA1196 in pBBR1MCS-5; Gmr | This study |
| pBRL688 | ddaH::lacZ with mutated RpoN promoter; Gmr | This study |
| Oligonucleotides | ||
| BL342.f | ATGACCATGATTACGGATTCACT | E. coli lacZ |
| BL342.r | TTATTTTTGACACCAGACCAACTGG | E. coli lacZ |
| BL579.f | TACAAAAAAGCAGGCTCGAACGCATCCTGGTCATCAC | PA1197 UpF-GWL |
| BL579.r | TCAGAGCGCTTTTGAAGCTAATTCGTGCTTCTGCAACTCGGCGATG | PA1197 UpR-Gm |
| BL580.f | AGGAACTTCAAGATCCCCAATTCGGTACCGCGAATTGCGCAAAG | PA1197 DnF-Gm |
| BL580.r | TACAAGAAAGCTGGGTCGATAAATGTGACTTACCACCTG | PA1197 DnR-GWR |
| BL581.f | GGCCTGCAGCGTATCGATC | ddaH::lacZ |
| BL581.r | GAATCCGTAATCATGGTCATGACGATTTTCCTCGTGTGG | ddaH::lacZ |
| BL587.f | TACAAAAAAGCAGGCTCGCTTCGCGTGGAAATCGTC | ddaH UpF-GWL |
| BL587.r | TCAGAGCGCTTTTGAAGCTAATTCGCGAGCGATGATGTGCTTGAAC | ddaH UpR-Gm |
| BL588.f | AGGAACTTCAAGATCCCCAATTCGGTCAACGAAAGGGTGATCATGC | ddaH DnF-Gm |
| BL588.r | TACAAGAAAGCTGGGTCAGTTTTCCCTTGTCCGAATCG | ddaH DnR-GWR |
| BL614.f | GCCATATGTTCAAGCACATCATCGCTC | ddaH ORF |
| BL614.r | GCGGATCCGAGCTCTCAGAAGCGCAGCGACATGC | ddaH ORF |
| BL615.f | GCCATATGGCGCTTGTCGAGGAACC | PA1196 ORF |
| BL615.r | GCGGATCCGAGCTCTCAGGCTGGCGGATCGAG | PA1196 ORF |
| BL618.f | GGCGTTGGCTAACGCGTGGCTT | Mutagenesis of pBRL649 |
| BL618.r | CGGGAATTGCCTTGGAAATCC | Mutagenesis of pBRL649 |
| ZS412.f | TACAAAAAAGCAGGCTATGGCGCTTGTCGAGGAAC | PA1196 UpF-GWL |
| ZS412.r | TCAGAGCGCTTTTGAAGCTAATTCGGGCCCCCACGTAGGCCTG | PA1196 UpR-Gm |
| ZS413.f | AGGAACTTCAAGATCCCCAATTCGGTGATCCGGCGCAGCCTGTTGCTGC | PA1196 DnF-Gm |
| ZS413.r | TACAAGAAAGCTGGGTTCAGGCTGGCGGATCGAG | PA1196 DnR-GWR |
| Gm-F | CGAATTAGCTTCAAAAGCGCTCTGA | FRT-aacC1-FRT marker |
| Gm-R | CGAATTGGGGATCTTGAAGTTCCT | FRT-aacC1-FRT marker |
| GW-attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCT | Gateway cloning primer |
| GW-attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGT | Gateway cloning primer |
Plasmids carry the bla, aacC1, aph(3′)-II, and/or tetC gene encoding resistance to carbenicillin (Cbr), gentamicin (Gmr), kanamycin (Kmr), and/or tetracycline (Tcr), respectively. FRT, FLP recombination target. The following abbreviations referring to the target and purpose of gene deletion primers correspond to the gene deletion strategy for P. aeruginosa (49): UpF, up forward; UpR, up reverse; DnF, down forward; DnR, down reverse; GWL, Gateway left; GWR, Gateway right; and Gm, gentamicin marker.
Deletion of the PA1195 (ddaH), PA1196, and PA1197 genes in P. aeruginosa PAO1.
The PA1195 (ddaH), PA1196, and PA1197 genes were deleted from P. aeruginosa PAO1 using well-established procedures (36–38). The primers and plasmids pertaining to the gene deletions are given in Table 2.
Cloning of the PA1195 (ddaH) and PA1196 genes.
The ddaH and PA1196 ORFs were PCR amplified from the genomic DNA of P. aeruginosa PAO1 using primer pairs BL614.f/BL614.r and BL615.f/BL615.r, respectively. The desired ddaH and PA1196 PCR products were gel purified and cloned into pCR-Blunt (Invitrogen) to generate the plasmids pBRL671 and pBRL672, respectively. To install a 5′ ribosome-binding site (RBS), the ddaH and PA1196 ORFs were subcloned into the NdeI/BamHI sites of pET15b (EMD Millipore) to generate the plasmids pBRL674 and pBRL675, respectively. The RBS-ddaH and RBS-PA1196 fragments were then each subcloned into the XbaI/SacI sites of the expression plasmid pBBR1MCS-5 (39) to give plasmids pBRL685 and pBRL686, respectively. Lastly, the PA1196 ORF from pBRL672 was subcloned into the XbaI site of pBBR1MCS-3 (39) with either a backward (pBRL719) or a forward (pBRL720) orientation relative to the orientation of the lac promoter.
Cloning of the ddaH::lacZ reporter.
The ddaH::lacZ reporter was constructed through fusion PCR (40). Briefly, the primers BL581.f and BL581.r were used to PCR amplify the 5′ regulatory region (∼500 bp) of the ddaH gene. The primers BL342.f and BL342.r were used to PCR amplify the E. coli lacZ ORF (∼3000 bp). The ddaH and lacZ amplicons were next fused together in a PCR with the primers BL581.f and BL342.r. The desired ddaH::lacZ fusion was gel purified and cloned into pCR-Blunt (Invitrogen) to generate pBRL645. The ddaH::lacZ fusion was subcloned into the XbaI site of ΔPlac-pBBR1MCS-5 (37) to give pBRL649. In addition, Q5 site-directed mutagenesis (NEB) was performed using primers BL618.f and BL618.r to change the GG nucleotides of the −24 element of the RpoN promoter to AA in ddaH::lacZ of pBRL649. The resulting plasmid, pBRL688, was sequenced to verify the presence of the desired mutation.
Growth assays.
ADMA hydrochloride and l-NMMA acetate were purchased from Sigma-Aldrich. However, due to the presence of acetate in the l-NMMA preparation, this methylarginine was excluded from the growth assays. Furthermore, for the experiments to be cost-effective, growth was assayed using a microplate reader. Growth was measured in quadruplicate for each P. aeruginosa strain. Bacteria were initially grown on LB agar plates at 37°C for 24 h. Single colonies were inoculated into 2 ml of LB (in a 16- by 100-mm culture tube), and the inoculated or seed cultures were grown at 37°C at 200 rpm for 24 h. The wells of 96-well polystyrene plates were filled with 0.2 ml of minimal medium supplemented with 15 mM gluconate and 10 mM nitrogen source (ADMA, l-arginine, l-citrulline, dimethylamine, l-ornithine, or NH4Cl). The minimal medium-filled wells were inoculated with 1% (vol/vol) LB-grown seed culture. The inoculated plates were incubated at 37°C with continuous slow shaking in a Synergy HT BioTek microplate reader. The absorbance at 600 nm (OD600) of each well was measured at 1.0-h intervals for a total duration of 24 to 48 h.
Exceptions to this procedure included the following: (i) when ADMA was tested as a carbon source, the minimal media were supplemented with 15 mM ADMA and 10 mM NH4Cl, (ii) for complementation experiments, the media were supplemented 30 μg ml−1 gentamicin for plasmid selection, and (iii) a temperature of 30°C (instead of 37°C) was used when the growth of various Pseudomonas spp. on ADMA as a nitrogen source was tested.
β-Galactosidase (LacZ) assays.
LacZ activities were measured using the Miller assay (40, 41), and experiments were conducted in triplicate for each strain and/or condition. The ddaH::lacZ reporter (pBRL649) was electroporated into P. aeruginosa, and colonies were selected on LB agar plates supplemented with 30 μg ml−1 gentamicin. Individual colonies were inoculated into 2 ml of LB (in a 16- by 100-mm culture tube) supplemented with 30 μg ml−1 gentamicin, and the inoculated or seed cultures were grown at 37°C at 200 rpm for 24 h. Minimal medium (1 ml in a 16- by 100-mm culture tube) supplemented with 15 mM gluconate, 10 mM NH4Cl, and 30 μg ml−1 gentamicin was inoculated with 1% (vol/vol) LB-grown seed culture, and the inoculated cultures were grown at 37°C at 200 rpm. At an OD600 of 0.3, cultures were treated with a final concentration of 0.1 mM substrate, which included ADMA, l-NMMA, l-citrulline, or l-arginine. LacZ activities were measured at 1.0 h afer the addition of the substrate. Analysis of variance (ANOVA) was done using Dunnett's post hoc test (α value, 0.05) to identify significant differences (P < 0.0001) in LacZ activities.
P. putida KT2440 was cotransformed with the ddaH::lacZ reporter (pBRL649) and plasmid-derived PA1196 (pBRL720) via electroporation (42). Recombinant colonies were selected on LB supplemented with 30 μg ml−1 gentamicin and 25 μg ml−1 tetracycline. Individual colonies were inoculated into 2 ml of LB (in a 16- by 100-mm culture tube) supplemented with 30 μg ml−1 gentamicin and 25 μg ml−1 tetracycline, and the inoculated or seed cultures were grown at 30°C at 200 rpm for 24 h. Minimal medium (1 ml in a 16- by 100-mm culture tube) supplemented with 15 mM gluconate, 10 mM NH4Cl, 30 μg ml−1 gentamicin, and 25 μg ml−1 tetracycline was inoculated with 2% (vol/vol) LB-grown seed culture, and the inoculated cultures were grown at 30°C at 200 rpm. At an OD600 of 0.3, the cultures were treated with either no substrate or 0.1 mM ADMA. LacZ activities were measured at 2.0 h after the addition of the substrate. LacZ assays (controls) were also performed with P. putida KT2440 cotransformed with empty plasmids pBBR1MCS-3/ΔPlac-pBBR1MCS-5, pBBR1MCS-3/pBRL649, pBRL720/ΔPlac-pBBR1MCS-5, and pBRL719/pBRL649. ANOVA was done using Dunnett's post hoc test (α value, 0.05) to identify significant differences (P < 0.0001) in LacZ activities.
Measurement of DDAH activity.
DDAH activity was measured in duplicate for each P. aeruginosa strain per tested condition. Bacteria were initially grown on LB agar plates at 37°C for 24 h. Single colonies were used to inoculate 2 ml of LB (in a 16- by 100-mm culture tube), and the inoculated or seed cultures were grown at 37°C at 200 rpm for 24 h. Minimal medium (5 ml in a 50-ml conical centrifuge tube) supplemented with 15 mM gluconate and 10 NH4Cl was inoculated with 1% (vol/vol) LB-grown seed culture. The inoculated cultures were grown at 37°C at 200 rpm to an OD600 of 0.3 and subsequently treated with either no substrate or 1.0 mM ADMA. At 2.0 h after the addition of the substrate, bacteria were harvested via centrifugation. Extracts were prepared from the cells as previously described (43). Briefly, cells were lysed by sonication on ice, debris and unlysed cells were removed by centrifugation, and the cell-free supernatants were washed and concentrated in a Microcon centrifugal filter (Millipore). The protein content of cell extracts was quantified using the Bradford assay (Pierce). The DDAH activity in the cell extracts was measured using established procedures (44, 45) and is reported in units (U), defined as the amount of l-citrulline (nanomoles hour−1) formed per milligram of total protein. ANOVA was done using Dunnett's post hoc test (α value, 0.05) to identify significant differences (P < 0.0001) in DDAH activities.
ACKNOWLEDGMENTS
We gratefully acknowledge support from the Undergraduate Honors Program of SUNY-ESF, the Center for Applied Microbiology at SUNY-ESF, and CSTEP at SUNY-ESF.
REFERENCES
- 1.Gary JD, Clarke S. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol 61:65–131. doi: 10.1016/S0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 2.Morales Y, Càceres T, May K, Hevel JM. 2016. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch Biochem Biophys 590:138–152. doi: 10.1016/j.abb.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 3.Teerlink T. 2005. ADMA metabolism and clearance. Vasc Med 10(Suppl 1):S73–S81. doi: 10.1177/1358836X0501000111. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell S. 2010. The biochemistry, measurement and current clinical significance of asymmetric dimethylarginine. Ann Clin Biochem 47:17–28. doi: 10.1258/acb.2009.009196. [DOI] [PubMed] [Google Scholar]
- 5.Vallance P, Leone A, Calver A, Collier J, Moncada S. 1992. Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 339:572–575. doi: 10.1016/0140-6736(92)90865-Z. [DOI] [PubMed] [Google Scholar]
- 6.Palmer RM, Ashton DS, Moncada S. 1988. Vascular endothelial cells synthesize nitric oxide from l-arginine. Nature 333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- 7.Segarra G, Medina P, Vila JM, Chuan P, Domenech C, Torondel B, Lluch A. 2001. Inhibition of nitric oxide activity by arginine analogs in human renal arteries. Am J Hypertens 14:1142–1148. doi: 10.1016/S0895-7061(01)02208-7. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa T, Kimoto M, Sasaoka K. 1989. Purification and properties of a new enzyme, NG,NG-dimethylarginine dimethylaminohydrolase, from rat kidney. J Biol Chem 264:10205–10209. [PubMed] [Google Scholar]
- 9.Murray-Rust J, Leiper J, McAlister M, Phelan J, Tilley S, Santa Maria J, Vallance P, McDonald N. 2001. Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase. Nat Struct Biol 8:679–683. doi: 10.1038/90387. [DOI] [PubMed] [Google Scholar]
- 10.McDermott JR. 1976. Studies on the catabolism of NG-methylarginine, NG,NG-dimethylarginine and NG,NG-dimethylarginine in the rabbit. Biochem J 154:179–184. doi: 10.1042/bj1540179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santa Maria J, Vallance P, Charles IG, Leiper JM. 1999. Identification of microbial dimethylarginine dimethylaminohydrolase enzymes. Mol Microbiol 33:1278–1279. [DOI] [PubMed] [Google Scholar]
- 12.Stone EM, Person MD, Costello NJ, Fast W. 2005. Characterization of a transient covalent adduct formed during dimethylarginine dimethylaminohydrolase catalysis. Biochemistry 44:7069–7078. doi: 10.1021/bi047407r. [DOI] [PubMed] [Google Scholar]
- 13.Stone EM, Costello AL, Tierney DL, Fast W. 2006. Substrate-assisted cysteine deprotonation in the mechanism of dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry 45:5618–5630. doi: 10.1021/bi052595m. [DOI] [PubMed] [Google Scholar]
- 14.Plevin MJ, Magalhães BS, Harris R, Sankar A, Perkins SJ, Driscoll PC. 2004. Characterization and manipulation of the Pseudomonas aeruginosa dimethylarginine dimethylaminohydrolase monomer-dimer equilibrium. J Mol Biol 341:171–184. doi: 10.1016/j.jmb.2004.05.057. [DOI] [PubMed] [Google Scholar]
- 15.Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. 2014. Ligand-dependent dynamics of the active-site lid in bacterial dimethylarginine dimethylaminohydrolase. Biochemistry 53:1092–1104. doi: 10.1021/bi4015924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrick MJ. 1993. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol Microbiol 10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 17.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. 2000. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J Bacteriol 182:4129–4136. doi: 10.1128/JB.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrios H, Valderrama B, Morett E. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res 27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Studholme DJ, Dixon R. 2003. Domain architectures of sigma54-dependent transcriptional activators. J Bacteriol 185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morett E, Segovia L. 1993. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol 175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol Mol Biol Rev 76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blander G, Guarente L. 2004. The Sir2 family of protein deacetylases. Annu Rev Biochem 73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 24.Buck SW, Gallo CM, Smith JS. 2004. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol 75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 25.Verhoogt HJ, Smit H, Abee T, Gamper M, Driessen AJ, Haas D, Konings WN. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J Bacteriol 174:1568–1573. doi: 10.1128/jb.174.5.1568-1573.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourdineaud JP, Heierli D, Gamper M, Verhoogt HJ, Driessen AJ, Konings WN, Lazdunski C, Haas D. 1993. Characterization of the arcD arginine:ornithine exchanger of Pseudomonas aeruginosa. Localization in the cytoplasmic membrane and a topological model. J Biol Chem 268:5417–5424. [PubMed] [Google Scholar]
- 27.Stalon V, Vander Wauven C, Momin P, Legrain C. 1987. Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J Gen Microbiol 133:2487–2495. [DOI] [PubMed] [Google Scholar]
- 28.Shoesmith JH, Sherris JC. 1960. Studies on the mechanism of arginine-activated motility in a Pseudomonas strain. J Gen Microbiol 22:10–24. doi: 10.1099/00221287-22-1-10. [DOI] [PubMed] [Google Scholar]
- 29.Ramos F, Stalon V, Piérard A, Wiame JM. 1967. The specialization of the two ornithine carbamoyltransferases of Pseudomonas. Biochim Biophys Acta 139:98–106. doi: 10.1016/0005-2744(67)90116-7. [DOI] [PubMed] [Google Scholar]
- 30.Servillo L, Giovane A, Cautela D, Castaldo D, Balestrieri ML. 2013. The methylarginines NMMA, ADMA, and SDMA are ubiquitous constituents of the main vegetables of human nutrition. Nitric Oxide 30:43–48. doi: 10.1016/j.niox.2013.02.080. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima T, Matsuoka Y, Kakimoto Y. 1971. Isolation and identification of NG-monomethyl, NG,NG-dimethyl- and NG,NG-dimethylarginine from the hydrolysate of proteins of bovine brain. Biochim Biophys Acta 230:212–222. doi: 10.1016/0304-4165(71)90206-6. [DOI] [PubMed] [Google Scholar]
- 32.Thomazella MC, Góes MF, Andrade CR, Debbas V, Barbeiro DF, Correia RL, Marie SK, Cardounel AJ, daLuz PL, Laurindo FR. 2011. Effects of high adherence to Mediterranean or low-fat diets in medicated secondary prevention patients. Am J Cardiol 108:1523–1529. doi: 10.1016/j.amjcard.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goralczyk T, Tisonczyk J, Fijorek K, Undas A. 2012. High tea and vegetable consumption is associated with low ADMA generation in older healthy subjects. Metabolism 61:1171–1176. doi: 10.1016/j.metabol.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Leiper J, Murray-Rust J, McDonald N, Vallance P. 2002. S-Nitrosylation of dimethylarginine dimethylaminohydrolase regulates enzyme activity: further interactions between nitric oxide synthase and dimethylarginine dimethylaminohydrolase. Proc Natl Acad Sci U S A 99:13527–13532. doi: 10.1073/pnas.212269799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knipp M, Braun O, Gehrig PM, Sack R, Vasák M. 2003. Zn(II)-free dimethylargininase-1 (DDAH-1) is inhibited upon specific Cys-S-nitrosylation. J Biol Chem 278:3410–3416. doi: 10.1074/jbc.M209088200. [DOI] [PubMed] [Google Scholar]
- 36.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundgren BR, Villegas-Peñaranda LR, Harris JR, Mottern AM, Dunn DM, Boddy CN, Nomura CT. 2014. Genetic analysis of the assimilation of C5-dicarboxylic acids in Pseudomonas aeruginosa PAO1. J Bacteriol 196:2543–2551. doi: 10.1128/JB.01615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundgren BR, Sarwar Z, Pinto A, Ganley JG, Nomura CT. 2016. Ethanolamine catabolism in Pseudomonas aeruginosa PAO1 is regulated by the enhancer-binding protein EatR (PA4021) and the alternative sigma factor RpoN. J Bacteriol 198:2318–2329. doi: 10.1128/JB.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 40.Lundgren BR, Thornton W, Dornan MH, Villegas-Peñaranda LR, Boddy CN, Nomura CT. 2013. Gene PA2449 is essential for glycine metabolism and pyocyanin biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol 195:2087–2100. doi: 10.1128/JB.02205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Bremer H. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem 270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Mueller AP, Leong CR, Matsumoto K, Taguchi S, Nomura CT. 2010. Quick and efficient method for genetic transformation of biopolymer-producing bacteria. J Chem Technol Biotechnol 85:775–778. [Google Scholar]
- 43.Lundgren BR, Harris JR, Sarwar Z, Scheel RA, Nomura CT. 2015. The metabolism of (R)-3-hydroxybutyrate is regulated by the enhancer-binding protein PA2005 and the alternative sigma factor RpoN in Pseudomonas aeruginosa PAO1. Microbiology 161:2232–2242. doi: 10.1099/mic.0.000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyde TR, Rahmatullah M. 1980. Optimization of conditions for the colorimetric determination of citrulline, using diacetyl monoxime. Anal Biochem 107:424–431. doi: 10.1016/0003-2697(80)90404-2. [DOI] [PubMed] [Google Scholar]
- 45.Knipp M, Vasák M. 2000. A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal Biochem 286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 46.Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. 2003. Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J Bacteriol 185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price-Whelan A, Dietrich LE, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol 189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1→3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- 49.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range FLP-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]