ABSTRACT
Despite high pertussis vaccine coverage, reported cases of whooping cough (pertussis) have increased over the last decade in the United States and other developed countries. Although Bordetella pertussis is well known for its limited gene sequence variation, recent advances in long-read sequencing technology have begun to reveal genomic structural heterogeneity among otherwise indistinguishable isolates, even within geographically or temporally defined epidemics. We have compared rearrangements among complete genome assemblies from 257 B. pertussis isolates to examine the potential evolution of the chromosomal structure in a pathogen with minimal gene nucleotide sequence diversity. Discrete changes in gene order were identified that differentiated genomes from vaccine reference strains and clinical isolates of various genotypes, frequently along phylogenetic boundaries defined by single nucleotide polymorphisms. The observed rearrangements were primarily large inversions centered on the replication origin or terminus and flanked by IS481, a mobile genetic element with >240 copies per genome and previously suspected to mediate rearrangements and deletions by homologous recombination. These data illustrate that structural genome evolution in B. pertussis is not limited to reduction but also includes rearrangement. Therefore, although genomes of clinical isolates are structurally diverse, specific changes in gene order are conserved, perhaps due to positive selection, providing novel information for investigating disease resurgence and molecular epidemiology.
IMPORTANCE Whooping cough, primarily caused by Bordetella pertussis, has resurged in the United States even though the coverage with pertussis-containing vaccines remains high. The rise in reported cases has included increased disease rates among all vaccinated age groups, provoking questions about the pathogen's evolution. The chromosome of B. pertussis includes a large number of repetitive mobile genetic elements that obstruct genome analysis. However, these mobile elements facilitate large rearrangements that alter the order and orientation of essential protein-encoding genes, which otherwise exhibit little nucleotide sequence diversity. By comparing the complete genome assemblies from 257 isolates, we show that specific rearrangements have been conserved throughout recent evolutionary history, perhaps by eliciting changes in gene expression, which may also provide useful information for molecular epidemiology.
KEYWORDS: Bordetella pertussis, whooping cough, evolution, genomics, pertussis, rearrangement
INTRODUCTION
Bordetella pertussis is the causative agent of whooping cough (pertussis), a respiratory disease with the highest morbidity and mortality in young infants. The introduction of vaccines against pertussis during the 1940s dramatically reduced the disease incidence in the United States. However, despite high or increasing coverage with pertussis-containing vaccines, the number of reported pertussis cases in the United States and many other developed countries has increased over the last decade, with notable recent epidemics (1–3). Multiple factors likely contribute to increased disease reporting, including heightened awareness, expanded surveillance, improved laboratory diagnostic testing, and allelic mismatch between circulating and vaccine reference strains (2, 4, 5). Waning protection conferred by acellular vaccine formulations, which replaced whole-cell preparations in the United States during the 1990s, has also led to increased disease rates among vaccinated individuals (2, 6–8).
Often cited for its limited genetic variability, B. pertussis has earned a reputation as a monomorphic pathogen (9, 10). Previous genomic studies have identified only a few mutations within a limited number of genes, each of which have quickly spread throughout the circulating population with little evidence of geographic restriction (5, 11). These mutations have occurred in genes encoding immunogenic proteins, most notably those for antigens in currently used acellular pertussis vaccines, such as genes for pertussis toxin (ptxA and the promoter region ptxP) and fimbriae (fimH), leading many to conclude that they result from vaccine-driven selection (5, 12–15). Although sequence diversity has also been observed in the vaccine immunogen pertactin (Prn), the circulating isolates recovered in the United States have become predominantly Prn deficient in recent years by one of at least 16 different mutations to the prn gene (16). The global emergence of Prn deficiency is well documented (16–19), and deficiency may confer a fitness advantage during infection (20, 21). While Prn deficiency appears to be more prevalent in isolates recovered from fully vaccinated patients, it does not appear to impact the effectiveness of acellular pertussis vaccines (22, 23).
Such limited genetic variation appears contradictory to the restricted ecology of B. pertussis as an obligate human pathogen, where ongoing adaptation is expected for its continued survival in the host population. Low-resolution genomic approaches have previously identified chromosomal diversity among B. pertussis isolates within epidemics by marker gene mapping (24), hybridization (25), and pulsed-field gel electrophoresis (PFGE) (1). A genome sequence-level characterization of structural variation has only recently become possible through advances in long-read sequencing technology capable of spanning the many insertion sequence (IS) elements present in B. pertussis permitting circular assembly. While initial comparisons of complete genome assemblies have revealed considerable rearrangement plasticity among B. pertussis isolates, these studies have been limited to small sample sizes due to high sequencing costs (9, 26–28).
For these reasons, B. pertussis presents unique obstacles to comparative genomics and sequence-based molecular epidemiology that are not shared by many other bacterial pathogens. To address these challenges, we have reconstructed the rearrangement history of the genetic content in circulating B. pertussis by analyzing the complete genome assemblies from 257 isolates with varied chromosomal structures. The results from this study provide evidence of conservation within the observed patterns of rearrangement, some of which correlate with previously described allelic replacements, such as the switch from ptxP1 to ptxP3. Furthermore, the distribution of structures within a single nucleotide polymorphism (SNP) phylogeny uncovered specific inversions, as well as the local arrangement of functional genes nearby, that underlie the PFGE profiles predominant within the circulating population. These results broaden the understanding of ongoing B. pertussis evolution to include specific gene order changes.
RESULTS
Variability among B. pertussis genomic structures.
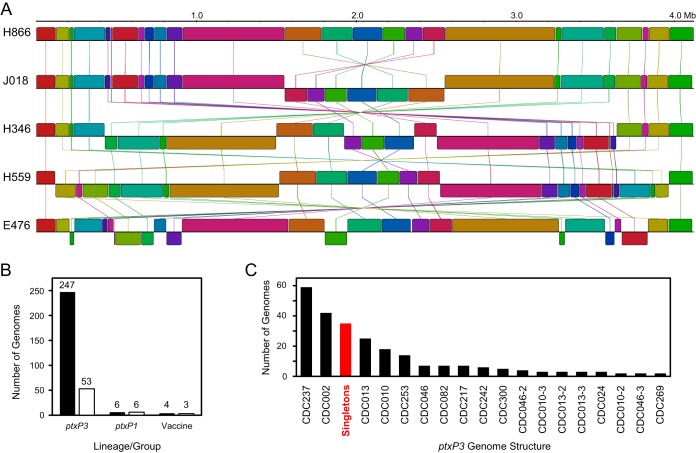
The current study compared 257 complete circular genome assemblies from isolates of B. pertussis, which captured the full complement of gene content and nucleotide sequence information (see Table S1 in the supplemental material). An alignment revealed considerable variation in genomic structures (Fig. 1). The assemblies presented here, most of which came from clinical isolates recovered in the United States over the past decade, exhibited 62 discrete genomic structures (Fig. 1B). Two hundred forty-seven of these genomes were recovered from isolates of the ptxP3 lineage and included 53 unique structures. These structures largely correlated with PFGE patterns such that genomes from isolates with the same PFGE profiles were frequently colinear. The abundances of the observed structures varied (Fig. 1C), and the most common were CDC237 (n = 59) and CDC002 (n = 42) (named according to the associated PFGE profile), reflecting their prevalences in the circulating population. Thirty-five structures were uniquely present in only one genome (“singletons”), suggesting that structural diversity remains undersampled. Likewise, the genomes of isolates from the ptxP1 lineage and the vaccine reference strains (10536 [B203], CS [C393], and Tohama I [E476 and J169]) all exhibited different structures (Fig. 1B). No differences in genomic structure were observed by restriction digest optical mapping or by genome sequencing following 11 serial passages of a clinical isolate. Although the genomes exhibited considerable rearrangement plasticity, structures appeared stable during routine laboratory manipulations and likely did not change between clinical isolation and genome sequencing.
FIG 1.
Genome structure variability among isolates of B. pertussis. (A) The order and orientation of genome content in recent circulating isolates (H866, J018, H346, and H559) varied, primarily due to large inversions, and differed greatly from those in vaccine reference strains, such as Tohama I (E476). (B) Genomes analyzed were derived primarily from isolates of the ptxP3 lineage, with only a few isolates from the ptxP1 lineage or vaccine reference strains, and comprised 62 unique chromosomal structures. Vaccine reference strains included 10536 (B203), CS (C393), and Tohama I (E476 and J169). Black bars indicate total numbers of genomes, and white bars indicate numbers of unique structures in each group. (C) The abundance distribution of structures observed in the 247 genomes of the ptxP3 lineage included 35 structures that were observed in only one isolate (singletons). Structures are named according to their associated PFGE profile. Minor variants with shared PGFE profiles are named CDC046-2 and CDC046-3, for example.
The observed rearrangements within genomes of different structures were primarily in the form of large inversions and were frequently flanked by insertions of IS481, a mobile genetic element with >240 copies per genome. All three copies of the rRNA operon were also observed flanking rearrangements. Minor variants of abundant structures were detected in smaller groups of isolates (e.g., CDC046-2) (Fig. 1C) and among singletons. For example, the genomes of some isolates with PFGE profile CDC010 differed by small unique inversions (see Fig. S1). Other minor variants resulted from small deletions, which averaged 5 kb, and were the only source of gene content variation detected among the ptxP3 genomes. These results are consistent with the prevailing view that B. pertussis gene content is relatively static and is occasionally altered through IS-mediated erosion but not horizontal acquisition.
Global relationships among genomic structures.
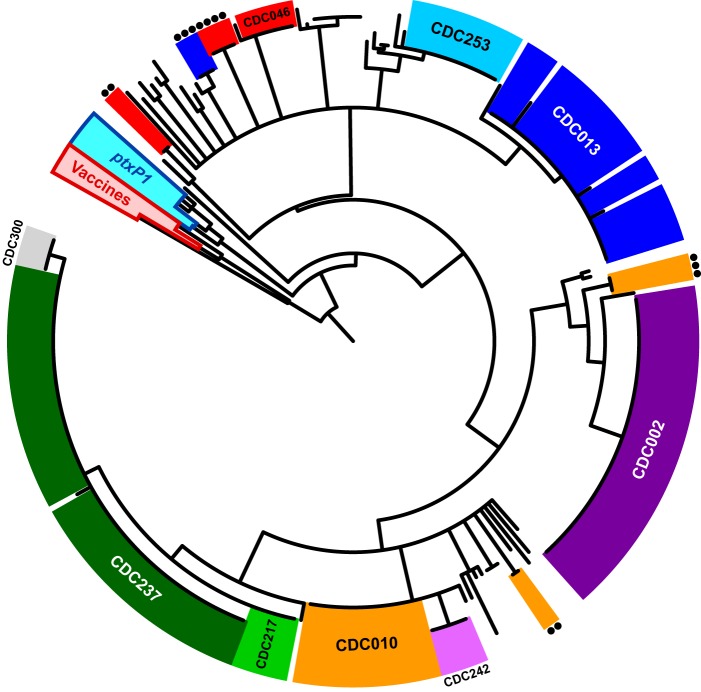
To illustrate the relationships between observed genomic structures, a subset representing each of the 62 unique structures was aligned and then clustered on the basis of relative differences in the order and orientation of shared homologous sequence blocks representing >94% of each genome. The resulting tree topology (Fig. 2) was largely concordant with a hierarchical clustering of PFGE profiles on the basis of shared genome fragment sizes (see Fig. S2). As with PFGE, the clustering according to global genomic structures discretely separated isolates of the ptxP1 and ptxP3 lineages from vaccine reference strains and from each other. Phylogenetic divergence of the ptxP1 and ptxP3 lineages is well documented (5), and these results suggest that the two groups are distinguishable by conserved genomic structural features, not just by nucleotide sequence polymorphisms. Despite the considerable structural heterogeneity observed among the 247 ptxP3 genomes, all were much more similar to each other than to the widely used reference strain Tohama I (E476).
FIG 2.
Global structure relationships among 257 B. pertussis genomes. Genome structure alignments were clustered according to relative positional changes in homologous sequence blocks. Genomes of the ptxP1 and ptxP3 lineages and vaccine reference strains, which have diverged through accumulation of SNPs, were structurally distinct. Common structures shared by colinear groups of ptxP3 isolates are color coded and labeled according to their associated PFGE profile. Colinear groups of minor variants that share a PFGE profile with a more common structure but differ by rearrangement (e.g., CDC046-2) are colored the same and indicated with circles.
The genome sequences of additional isolates, most recovered in Europe, were completed recently and are available in public databases (26, 27, 29, 30). An alignment of these 17 genomes with a representative subset of the data here revealed that their structures were largely different (see Fig. S3 and S4). Specifically, only two recent European isolates, B1865 (GenBank accession no. CP011441) and B3621 (accession no. CP011401), were colinear with the CDC013 structure. The structural differences between these data sets may simply reflect undersampling, particularly given the distribution of abundances observed within the data here (Fig. 1C).
Phylogenetic linkage of genomic structures.
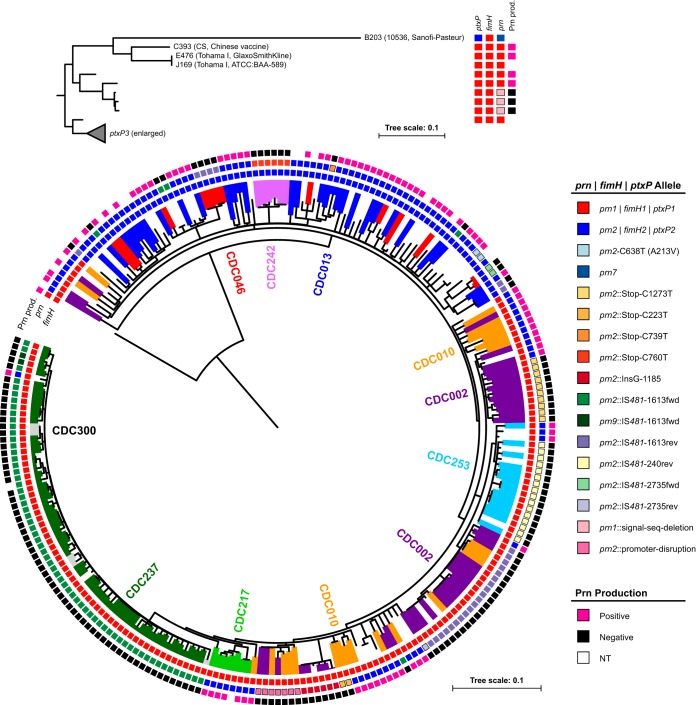
SNPs were predicted throughout the core genome for investigating the phylogenetic distribution of genomic structures. All IS elements, rRNA operons, and the highly variable prn gene were explicitly masked to minimize the influence of known sites of recombination and homoplasy. A total of 1,473 variable core positions were identified, representing 0.036% of the average B. pertussis genome, and were used to reconstruct the phylogeny with maximum parsimony (Fig. 3). The isolates separated into well-supported clades according to their ptxP and fimH (fim3) alleles, consistent with SNP phylogenies reported elsewhere (5, 11). Certain genomic structures appeared phylogenetically restricted, which is to say that strains with colinear genomes were also closely related according to their SNP patterns. For example, structures associated with PFGE profiles CDC217, CDC237, CDC242, and CDC253 each resided within a single clade in the phylogenetic tree (Fig. 3). Whether isolates with a common genomic structure were distributed across relatively small (e.g., CDC242) or large (e.g., CDC237) geographic distances, they shared an observable ancestral heritage.
FIG 3.
Phylogenetic reconstruction of 257 B. pertussis isolates using maximum parsimony. SNP profiles across 1,473 core variable positions discretely separated ptxP1 and ptxP3 lineages and vaccine reference strains (inset). The phylogenetic distribution of Prn production, prn alleles, and fimH alleles is color coded according to the key. Abundant genomic structures are color coded and labeled according to their associated PFGE profile. Scale bars indicate substitutions per site.
Other genomic structures did not coalesce within the phylogeny but instead comingled with other similar structures. Genomic structures represented by PFGE profiles CDC002 and CDC010 were intermingled, as were CDC013 and CDC046. Each of these pairs was confined to the SNP background defined by either fimH1 (CDC002 and CDC010) or fimH2 (CDC013 and CDC046). The distribution of structures within the phylogeny suggests that, despite the observed heterogeneity and possibly variable rearrangement rates, global genomic structures are surprisingly stable and thus potentially traceable.
Phylogenetic linkage of pertactin disruption.
The phylogeny of the genomes analyzed was reconstructed while excluding the nucleotide sequence of the highly variable gene prn, which encodes an immunogenic protein found in most current acellular vaccine formulations. Clinical isolates recovered in the United States have become increasingly Prn deficient in recent years by a variety of mutations to prn, including missense substitutions, insertions, deletions, and promoter disruption, but most frequently through IS481 insertion at one of three positions (16). Most mutations appeared to be restricted within the phylogeny of genomes here, with the exception of the IS481 insertions, even when closely related strains differed in genomic structure (Fig. 3). Homoplastic insertions likely resulted from independent events, and the observation of IS481 insertion in both forward and reverse orientations at positions 1,613 and 2,735 provides direct evidence of this. These results suggest that most prn-disrupting mutations occurred once in the circulating population but that prn-disrupting IS481 insertions have occurred repeatedly at the same positions.
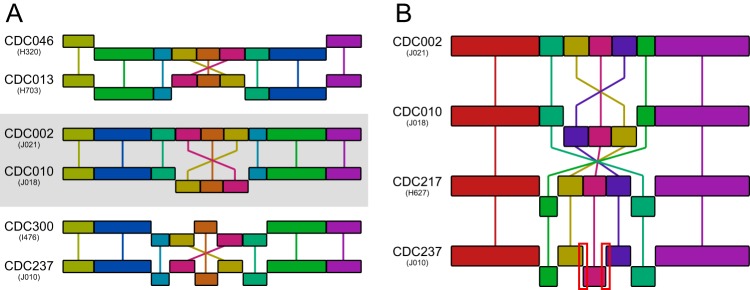
Repeated common inversion.
The phylogenetic distribution of genomic structures exposed pairs that appeared intermingled, namely, CDC002/CDC010 and CDC013/CDC046, within the SNP backgrounds defined by fimH1 and fimH2, respectively (Fig. 3). An alignment of representatives with these structures revealed that each pair differed by a common single inversion (Fig. 4A). The boundaries of this inversion contained a three-gene inverted repeat including IS481 and encoding a hypothetical protein and a predicted major facilitator superfamily (MFS) membrane protein. Nearby genes encoded proteins with various functions, including leucine biosynthesis and transport, fatty acid biosynthesis, organic acid transport, diguanylate cyclase activity, protein stability, and siderophore biosynthesis and transport (see Data Set S1). Structures CDC237 and CDC300 also differed by the same inversion (Fig. 4A), suggesting that this rearrangement had occurred repeatedly, and perhaps reversibly, within the ptxP3 lineage.
FIG 4.
Genomic rearrangement in a phylogenetic context. (A) A common inversion was detected in multiple genetic backgrounds within the ptxP3 lineage, between structures CDC046 and CDC013 (fimH2), CDC002 and CDC010 (fimH1), and CDC300 and CDC237 (fimH1). (B) Emergence of CDC237, the PFGE profile most commonly recovered in the United States every year since 2012, can be inferred from the phylogeny as a series of sequential inversions, which ultimately create novel, local structural conformations (red boxes) detailed in Fig. 5.
Emergence of CDC237 by sequential inversion.
The PFGE profile CDC237 was associated with the most common genomic structure in the data presented here (Fig. 1C). A phylogenetic reconstruction indicated that these 59 genomes shared a clonal SNP background and conserved IS481 disruption of prn at position 1613 (Fig. 3). The placement of the CDC237 clade shed light on its potential emergence from similar structures present in closely related isolates (Fig. 3), and a clear path of sequential inversion was inferred by an alignment of representative genome sequences (Fig. 4B). Whole-genome SNP patterns and structures together suggested that CDC237 descended from the progressive rearrangement of CDC002 to CDC010 to CDC217 to CDC237. The inversion from CDC217 to CDC237 produced novel gene order arrangements not observed elsewhere in the data set (Fig. 5). Annotation of nearby genes revealed the operon and dedicated regulator for the biosynthesis of the excreted polysaccharide Bps (Fig. 5; see also Data Set S1). Genomes in the clade containing CDC237 and CDC300 also included specific SNPs present in a few genes encoding central metabolic proteins (Data Set S1).
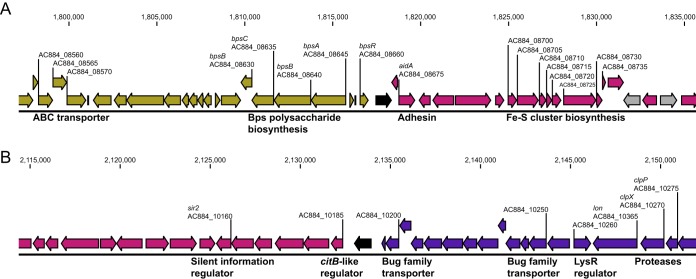
FIG 5.
Novel structural conformation in CDC237. A single inversion between IS481 insertions (black) differentiates structures CDC217 and CDC237, creating local conformations (highlighted in Fig. 4) not present in any other structures, except CDC300. Neighboring genes located at the boundaries encoded proteins with various functions, such as transporters, proteases, and proteins involved in Bps polysaccharide biosynthesis and Fe-S cluster assembly. Additional IS481 insertions are shaded gray. Indicated coordinates and locus tags correspond to positions in clinical isolate J010 (GenBank accession no. CP012085). A full list of annotated genes at these loci is available in Data Set S1.
Conserved local structure identification.
The clustering of genomes according to global structure discretely separated vaccine reference strains and ptxP1 and ptxP3 isolates, indicating that these groups had diverged in genomic structure (Fig. 2). Filtering of the structural differences observed in an alignment of representative genomes revealed a limited number of discrete deletions, inversions, and rearrangement boundaries specific to certain lineages (Table 1). Three conserved deletions were observed, including two in all clinical isolate genomes relative to vaccine reference strains and one specific to ptxP3 isolates only (Table 1; see also Data Set S2). Three inversions, all flanked by IS481 insertions, were also detected; one was present in all vaccine references, another was present in all ptxP3 genomes, and one differentiated ptxP3 fimH1 and ptxP3 fimH2 genomes (Table 1; see also Data Set S2). The vaccine- and ptxP3-specific inversions were asymmetric with respect to the predicted replication origin and terminus. Three individual rearrangement boundaries were specific to ptxP3 genomes, including two that were present and one that was absent compared with vaccine reference and ptxP1 genomes, and the local arrangement of genes at these boundaries differed between lineages (Table 1; see also Data Set S2). No specific local structures were identified among genomes of nonvaccine ptxP1 isolates or Prn-deficient isolates. These results indicated that structural features have become fixed throughout the phylogenetic history of B. pertussis and that such loci may contribute toward adaptive evolution.
TABLE 1.
Summary of conserved structural features
| Type | Description | Locationa |
|---|---|---|
| Deletionb | ∼25 kb absent in all nonvaccine strain genomes | E476: RD16_04530 to RD16_04650 |
| Deletionb | ∼10 kb absent in all nonvaccine strain genomes | E476: RD16_05650 to RD16_05695 |
| Deletionb | ∼25-kb ptxP3-specific deletion | E476: RD16_09730 to RD16_09830 |
| Inversion | ∼98-kb vaccine-specific inversion | E476: RD16_03925 to RD16_04345 |
| Inversion | ∼38-kb ptxP3-specific inversion | H866: ABC03_15465 to ABC03_15650 |
| Inversion | ∼1-Mb conserved inversion between all ptxP3 fimH1 and ptxP3 fimH2 isolates | H866: ABC03_02160 to ABC03_17235 |
| Rearrangement boundary | ptxP3-specific boundary | H866: ABC03_11025 |
| Rearrangement boundary | ptxP3-specific boundary | H866: ABC03_08550 |
| Rearrangement boundary | Boundary absent in all ptxP3 genomes | E476: RD16_08535 |
Example strain and locus tags within the structural feature. For deletions and inversions, locus tags indicate the total region, inclusively. For rearrangement boundaries, locus tags indicate the gene at the predicted rearrangement breakpoint. A full list of annotated genes at each locus is available in Data Set S2 in the supplemental material.
IS481 content variability.
Conserved insertion sites and IS element content variability were further tracked in two colinear groups, the phylogenetically linked CDC237 (n = 59) and phylogenetically diverse CDC002 (n = 42). In both groups, the majority of IS481 insertions (>90%) were present at conserved sites in all of the genomes, including among isolates recovered as much as 14 years apart (Fig. S5). In at least one genome, 23% of these conserved sites contained multiple neighboring (“duplicated”) insertions, in which a second or third IS481 had inserted adjacent to an existing copy, sharing a 6-bp target sequence (Fig. S5). Individual genomes in each group had an average of 17 sites with multiple insertions (range, 13 to 21). Variable insertion sites, i.e., those that lacked IS481 in one or more genomes, were also observed within both groups, but only three such insertions on average were present in each genome (range, 1 to 6). No changes in IS481 content were observed in either of two parallel replicates of a clinical isolate following serial passage. No variations in IS1002 (n = 5) or IS1663 (n = 16) content were observed in these two sets of colinear genomes. Therefore, IS481 copy number did not equal the number of unique loci disrupted by insertion, and the observed variation resulted primarily from multiple insertions at conserved sites rather than differential insertions at unique sites. The distribution patterns of some variable insertions within CDC002 genomes were phylogenetically linked but did not suggest that IS481 copy number had increased over time in this group (Fig. S5).
Unoccupied IS481 insertion sites.
IS481 is inserted at specific 6-bp target sequences, which become duplicated, flanking the element upon insertion (31). Forty-two unique sequences were observed flanking IS481 insertions, and the target sequence motif was calculated from the seven most common sequences in six phylogenetically disparate genomes (see Fig. S6). Using the resulting motif, all 257 genomes were queried for 6-bp target sequences to identify putative unoccupied sites susceptible to insertion. The genomes contained an average of 249 unoccupied sites (range, 242 to 255), suggesting that IS481 copies currently occupy approximately half of all possible insertion sites. On average, 146 available sites (range, 139 to 150) fell within predicted coding regions, with some genes containing multiple sites, and the remaining 103 (range, 99 to 107) were within intergenic regions. The 142 unique genes harboring IS481 target sequences encoded predicted proteins of various functions (Fig. S6; Data Set S3). The availability of these sites for future insertions implies that these genes either present opportunities for further genome erosion or are essential for B. pertussis growth.
DISCUSSION
In this study, we introduced a new comprehensive picture of B. pertussis genome variation through a comparative analysis of complete assemblies from 257 isolates. Just as the first closed sequence of Tohama I revealed much about B. pertussis speciation from a B. bronchiseptica-like ancestor (32), the data here further revise the understanding of B. pertussis evolution. Previous studies have indicated that the recent genetic history of B. pertussis has been punctuated by the successive accumulation of small mutations (5, 11). Advances in sequencing and optical mapping technologies make complete assemblies more accessible, and the results here reveal evidence of conservation in genomic structural changes, which may result from selective pressure in favor of specific gene arrangements.
Bacterial chromosome organization is not random but rather appears to favor a conserved core gene order, at least within species or lineages (33). Such organization is heavily influenced by bidirectional replication, which imprints strand biases for gene distribution (33, 34) and coordinates gene expression (35–37), imposing constraints on rearrangement. Comparative genomics across many species has suggested that selection operates to maintain replichore balance, such that rearrangements in the form of symmetric inversions centered on the replication origin or terminus appear to be a common feature of bacterial genome evolution (34, 38, 39). Indeed, many of the rearrangements reported here were symmetric inversions, and large uninterrupted regions were frequently observed, consistent with this understanding.
The observed linkage between SNPs and rearrangements in the population makes it challenging to infer which are under positive selection and which, if any, are hitchhiking. Although the ptxP3 allele is thought to increase pertussis toxin expression (40, 41), a recent comparison of isogenic mutants found that the ptxP3 allele and the genetic background, which includes genomic structure, independently enhance colonization in mice (42). Similarly, although alleles of fimH contain polymorphisms in the predicted surface epitope region, differences in immune recognition remain untested (43). If selective forces govern genomic architecture, then some rearrangements likely carry fitness effects, including possible benefits. Rearrangement-mediated adaptation has been observed during laboratory evolution experiments, including recombination between IS elements (44, 45). Perhaps recently observed population sweeps have actually been driven by selection in favor of genomic structure, not nucleotide sequence. Inversions specific to either isolates with the ptxP3 allele or vaccine reference strains appeared within a single replichore (i.e., they were asymmetric), suggesting that these rearrangements confer beneficial effects sufficiently large to offset the costs associated with disrupting coding strand bias. Further evidence of selection for gene order may exist in temporal fluctuations in PFGE profiles (Fig. 6B), a proxy for genomic structure, which generally occur in the absence of significant gene sequence variation in B. pertussis. However, until experiments can appropriately compare isogenic strains with varied genomic structures, the adaptive nature of B. pertussis rearrangement remains speculative.
FIG 6.
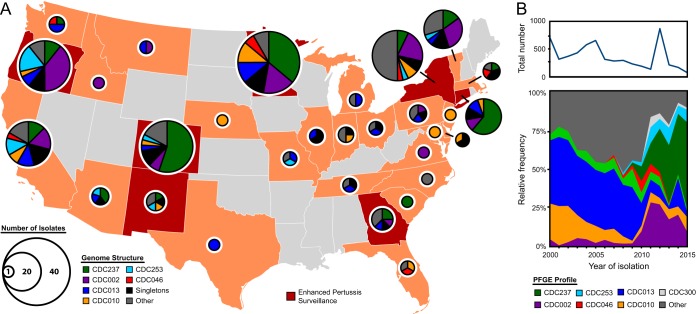
Geographic origin of U.S. B. pertussis isolates studied and their genomic structures. (A) Sequenced isolates analyzed in this study were recovered primarily in 30 states in 2000 to 2014, and some states contributed more isolates through participation in the Enhanced Pertussis Surveillance/Emerging Infection Program Network. The geographic distribution of these isolates does not reflect national patterns of disease incidence. See Materials and Methods for selection criteria and Table S1 for specific isolate information. Pie chart diameter represents numbers of isolates and colors indicate genomic structures, named according to their associated PFGE profile, as detailed in the key. (B) Relative frequencies of predominant PFGE profiles (bottom) and total numbers (top) of B. pertussis isolates recovered in the United States and submitted to the Centers for Disease Control and Prevention from 2000 to 2015.
A lack of geographical SNP clustering in the global B. pertussis population was reported previously (5, 11), and similarly, no such trends in genome structural variation were observed in the data here (Fig. 6). PFGE profile CDC237 has been the most abundant profile among U.S. clinical isolates since 2012 (46) and was reported for 55% of those received at the CDC in 2015 (Fig. 6B). Although isolates sequenced here with this profile were collected across 13 disparate states from 2010 to 2014, a phylogenetic reconstruction indicated that this group had a clonal origin. The data here highlighted a possible path of three successive inversions toward the emergence of CDC237, which produced novel local gene order conformations. Annotation of nearby genes revealed the operon and dedicated regulator for the biosynthesis of Bps, an excreted polysaccharide necessary for biofilm formation (47) that also confers resistance to complement-mediated killing (48). How this unique gene reorganization might impact B. pertussis fitness or virulence by altering Bps production remains unanswered, but the recent prevalence of CDC237 hints that this genotype has some putative advantage.
Contextualizing the genomic structural relatedness within the SNP phylogeny also revealed an identical inversion that transpired in the divergent fimH1 and fimH2 backgrounds, independently. Structures did not appear monophyletic within either the fimH1 or fimH2 background, suggesting that the inversion has occurred multiple times or perhaps reversibly in each. The switch from CDC237 to CDC300 also occurred by an inversion at the same boundaries in the fimH1 background, providing further evidence for reversibility. The biosynthesis and transport of alcaligin, an important siderophore produced by B. pertussis and B. bronchiseptica (49), were encoded nearby. Whether this inversion modulates alcaligin production or whether the observed parallelism reflects positive selection rather than neutral polymorphism has yet to be explored.
In contrast to that in many other bacterial pathogens, the genetic content in B. pertussis is largely invariable, even between epidemic and nonepidemic isolates (14). Although no evidence of gene gain has been reported, many previous studies have identified regions of difference resulting from apparent deletions conserved among circulating isolates in comparison with older references (12, 14, 50, 51). The conserved structural features identified here, specifically those which differentiated isolates with the ptxP3 allele and vaccine reference strains, included deletions flanked by IS481 insertions, each of which corresponds to regions of difference identified previously (12, 14, 25, 50–53). However, conserved rearrangements and inversions were also identified, illustrating that structural genome evolution in B. pertussis is not limited to reduction.
These new data expand the understanding of how extensively IS481 contributes to B. pertussis genome evolution, beyond previously reported gene disruption and recombination-mediated deletion. They also permit accurate accounting of content variation between related genomes, which has uncovered fluctuations in IS481 insertion numbers at conserved sites. Such neighboring insertions differ across short and long phylogenetic distances within the ptxP3 lineage and appear to be the primary determinants of total IS481 copy number variation between closely related isolates, rather than insertions at unique sites. The mechanism and effects of these fluctuations, perhaps by modulating downstream gene expression (54, 55), require further study as their detection is only now possible with such complete genomic data. Many coding regions harboring predicted, but unoccupied, IS481 insertion sites were conserved across phylogenetically disparate isolates. These loci may encode the proteins necessary for survival and thus might be useful targets for improved molecular typing or vaccine immunogens.
The structural plasticity reported here portrays a dynamic circulating population, replete with genome rearrangement as a source of variation for natural selection, independent of mutations to primary gene sequences, and further illustrates the dramatic divergence of circulating isolates away from common reference strains (56). How gene order influences fitness or virulence remains unanswered, and phenotypic comparisons between defined genomic structures are under way. Even if structural conservation simply reflects population bottlenecks experienced during each selective sweep, the data here clearly demonstrate signs of stability and suggest that limits to rearrangement or rearrangement rate exist within B. pertussis. Although the utility of genome structure for molecular epidemiology requires further exploration, application of this knowledge to routine surveillance is particularly challenging, as short-read benchtop sequencers, which are becoming common in public health laboratories, are currently incapable of detecting such diversity. The new depth of measurable variation among circulating B. pertussis isolates revealed here will strengthen investigations of recent disease resurgence.
MATERIALS AND METHODS
Strain selection.
U.S. B. pertussis isolates for sequencing were selected by various strategies from the CDC collection, including isolates collected through surveillance and outbreaks, and assembled genomes were included in the current study on the basis of their availability. One set was selected to capture potential regional diversity among 28 states by maximizing the variety of source states for isolates collected from 2000 to 2013, with an emphasis on those collected from 2010 to 2013 (n = 80). Another selection focused on isolates obtained through the Enhanced Pertussis Surveillance/Emerging Infection Program Network (57) in seven states from 2011 to 2014, which prioritized isolates from hospitalized cases and forced sampling of every possible combination of year, state, vaccination status, and age group (n = 130). Additionally, eight isolates selected prospectively from sporadic submission in 2014, 31 epidemic isolates sequenced previously (28), and eight others, such as vaccine reference strains, were also included.
Pulsed-field gel electrophoresis.
PFGE was performed using restriction enzyme XbaI according to the method developed by Gautom (58). PFGE patterns were compared with those in a database of B. pertussis isolate profiles maintained at the CDC, and profiles were assigned on the basis of bands in the 125- to 450-kb range using BioNumerics v5.01 (Applied Maths, Austin, TX).
Genomic DNA preparation.
Isolates were cultured on Regan-Lowe agar without cephalexin for 72 h at 37°C. Genomic DNA (gDNA) isolation and purification were performed using the Gentra Puregene yeast/bacteria kit (Qiagen, Valencia, CA) with slight modification. Briefly, two aliquots of approximately 109 bacterial cells were harvested and resuspended in 500 μl of 0.85% sterile saline and then pelleted by centrifugation for 1 min at 16,000 × g. Recovered genomic DNA was resuspended in 100 μl of DNA hydration solution. Aliquots were quantified using a Nanodrop 2000 (Thermo Fisher Scientific, Inc., Wilmington, DE).
Genome sequencing and assembly.
Whole-genome shotgun sequencing of isolates was performed using a combination of the PacBio RSII (Pacific Biosciences, Menlo Park, CA), Illumina HiSeq/MiSeq (Illumina, San Diego, CA), and Argus (OpGen, Gaithersburg, MA) platforms as described previously (28). Briefly, genomic DNA libraries were prepared for PacBio sequencing using SMRTbell template prep kit 1.0 and polymerase binding kit P4, while Illumina libraries were prepared using the NEB Ultra library prep kit (New England BioLabs, Ipswich, MA). De novo assembly was performed using the Hierarchical Genome Assembly Process (HGAP v3; Pacific Biosciences) (59). The resulting consensus sequences were manually checked for circularity using gepard (v1.30) (60) and then reordered to match the start of Tohama I (GenBank accession no. NC_002929). Assemblies were confirmed by comparison with restriction digest optical maps using the Argus system (OpGen) with MapSolver (v.2.1.1; OpGen) and further polished by mapping Illumina reads using the CLC genomics workbench (v8.5; CLC bio, Boston, MA). Assemblies were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP).
Genome structural variation and phylogenetic reconstruction.
Genomes were aligned in all pairwise combinations using progressiveMauve with default settings (61) and were determined to be colinear if alignments included no observable inversions or gaps of >1,500 bp. A nonredundant subset of genomes representing all unique structures was aligned using progressiveMauve with optimized parameters (seed-weight, 16; hmm-identity, 0.85). Permutation matrices of homologous sequence blocks of >1,500 bp were calculated from progressiveMauve output files with custom Perl scripts and then used to cluster genomes on the basis of structural similarity with the Maximum Likelihood for Gene Order (MLGO) pipeline (62). Phylogenetic reconstruction was calculated using kSNP3 (63), with k of 23, after masking the pertactin gene (prn) and all IS-element sequences with N's. A maximum parsimony tree was calculated from core variable positions in kSNP3. For all trees, internal nodes with <50% bootstrap support were collapsed into multifurcations in Archaeopteryx v0.9901 (64) and annotation was performed with iTOL v3.0 (65). Alleles for common molecular typing loci (ptxP, ptxA, ptxB, fimH, and prn) were assigned by a high-stringency BLASTn alignment to assembled genomes. Mutations to prn were assigned using a custom curated database.
Conserved structural changes were identified by searching the distribution of shared adjacencies from MLGO using Fisher's exact test with Benjamini-Hochberg correction for multiple testing in a subset of representative genomic structures, excluding all singleton ptxP3 genomes. The identified features were validated by a manual inspection of progressiveMauve and BLASTn alignments. The predicted protein sequences encoded by genes near specific rearrangement sites were functionally classified according to a betaproteobacterium-specific subset of EggNOG v4.1 (66) using HMMER v3.1b2 (http://hmmer.org) and further annotated by a manual query of Swiss-Prot (67) and the Conserved Domain Database (68) using DELTA-BLAST (69) via the NCBI web interface.
Genome structural stability.
A single glycerol stock bead of clinical isolate H866 was suspended in 100 μl saline and spread on Regan-Lowe agar without cephalexin at serial dilutions down to 10−6. A single colony (ancestor) was recovered from the most dilute plate, was suspended in 50 μl saline, and was spread onto a new plate. After incubation, bacterial growth was recovered with a cotton swab and suspended in saline. A small volume of the suspension was spread on a new plate and the remainder was used for gDNA extraction as described above. After 11 passages, multiple colonies were recovered for optical mapping and gDNA was extracted from the remaining culture for genome sequencing.
Variable IS481 insertion.
Positions of IS481 (GenBank accession no. M22031) insertions were identified by a BLASTn query of assembled genomes and then matched across colinear genomes according to exhaustive pairwise progressiveMauve alignments using the parameters described above in combination with custom Perl scripts. Insertions were classified as either “conserved” if their positions and orientations were shared in all genomes or “variable” if they were absent in one or more genomes. Sites of multiple insertion were identified if neighboring insertions were separated only by their 6-bp target sequence and were in the same orientation.
The insertion target consensus sequence was determined from the 6 bp flanking all IS481 insertions in six phylogenetically disparate strains (B203, E150, E476, E976, I344, and J090). A motif was calculated from the seven most abundant sequences in each, a total of 2,314 sequences representing >77% of sites in each genome, using MEME (v4.10.2) with a 5-order Markov frequency background model (70). Putative IS481 insertion target sequences were predicted with FIMO (v4.10.2) (71), followed by subtraction of sites with known insertions identified by BLASTn alignment. The predicted protein sequences from genes containing target sites were functionally classified as described above.
Source code.
The source code for custom scripts developed in the present study is available at https://github.com/mikeyweigand/Pertussis_n257.
Accession number(s).
The whole-genome shotgun sequences have been deposited at DDBJ/EMBL/GenBank under accession numbers CP011167 to CP011208, CP011234 to CP011244, CP011255, CP011687 to CP011768, CP012078 to CP012089, CP012129 to CP012134, CP013075 to CP013096, CP013863 to CP013866, CP013868 to CP013907, and CP013951 (see Table S1 in the supplemental material). The versions described in this paper are the first versions. Raw sequence data are available from the NCBI Sequence Read Archive, organized under a BioProject with accession number PRJNA279196.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leonard Mayer, Conrad Quinn, Stacy Martin, Tami Skoff, and Connie Lam (CDC) for insightful discussions that contributed to the development of this work, Christine Miner (CDC) for isolate metadata management, and the Enhanced Pertussis Surveillance/Emerging Infection Program Network sites and other state health departments for contributing isolates.
This work was supported by internal funds.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00806-16.
REFERENCES
- 1.Bowden KE, Williams MM, Cassiday PK, Milton A, Pawloski L, Harrison M, Martin SW, Meyer S, Qin X, DeBolt C, Tasslimi A, Syed N, Sorrell R, Tran M, Hiatt B, Tondella ML. 2014. Molecular epidemiology of the pertussis epidemic in Washington State in 2012. J Clin Microbiol 52:3549–3557. doi: 10.1128/JCM.01189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark TA. 2014. Changing pertussis epidemiology: everything old is new again. J Infect Dis 209:978–981. doi: 10.1093/infdis/jiu001. [DOI] [PubMed] [Google Scholar]
- 3.Winter K, Harriman K, Zipprich J, Schechter R, Talarico J, Watt J, Chavez G. 2012. California pertussis epidemic, 2010. J Pediatr 161:1091–1096. doi: 10.1016/j.jpeds.2012.05.041. [DOI] [PubMed] [Google Scholar]
- 4.Ausiello CM, Cassone A. 2014. Acellular pertussis vaccines and pertussis resurgence: revise or replace? mBio 5:e01339-14. doi: 10.1128/mBio.01339-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, Bouchez V, Cassiday PK, Chiang CS, Dalby T, Fry NK, Gaillard ME, van Gent M, Guiso N, Hallander HO, Harvill ET, He Q, van der Heide HG, Heuvelman K, Hozbor DF, Kamachi K, Karataev GI, Lan R, Lutynska A, Maharjan RP, Mertsola J, Miyamura T, Octavia S, Preston A, Quail MA, Sintchenko V, Stefanelli P, Tondella ML, Tsang RS, Xu Y, Yao SM, Zhang S, Parkhill J, Mooi FR. 2014. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio 5:e01074-14. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein NP, Bartlett J, Rowhani-Rahbar A, Fireman B, Baxter R. 2012. Waning protection after fifth dose of acellular pertussis vaccine in children. N Engl J Med 367:1012–1019. doi: 10.1056/NEJMoa1200850. [DOI] [PubMed] [Google Scholar]
- 7.Misegades LK, Winter K, Harriman K, Talarico J, Messonnier NE, Clark TA, Martin SW. 2012. Association of childhood pertussis with receipt of 5 doses of pertussis vaccine by time since last vaccine dose, California, 2010. JAMA 308:2126–2132. doi: 10.1001/jama.2012.14939. [DOI] [PubMed] [Google Scholar]
- 8.Warfel JM, Edwards KM. 2015. Pertussis vaccines and the challenge of inducing durable immunity. Curr Opin Immunol 35:48–54. doi: 10.1016/j.coi.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Belcher T, Preston A. 2015. Bordetella pertussis evolution in the (functional) genomics era. Pathog Dis 73:ftv064. doi: 10.1093/femspd/ftv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mooi FR. 2010. Bordetella pertussis and vaccination: the persistence of a genetically monomorphic pathogen. Infect Genet Evol 10:36–49. doi: 10.1016/j.meegid.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 11.van Gent M, Bart MJ, van der Heide HG, Heuvelman KJ, Mooi FR. 2012. Small mutations in Bordetella pertussis are associated with selective sweeps. PLoS One 7:e46407. doi: 10.1371/journal.pone.0046407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kallonen T, Grondahl-Yli-Hannuksela K, Elomaa A, Lutynska A, Fry NK, Mertsola J, He Q. 2011. Differences in the genomic content of Bordetella pertussis isolates before and after introduction of pertussis vaccines in four European countries. Infect Genet Evol 11:2034–2042. doi: 10.1016/j.meegid.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Octavia S, Maharjan RP, Sintchenko V, Stevenson G, Reeves PR, Gilbert GL, Lan R. 2011. Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection. Mol Biol Evol 28:707–715. doi: 10.1093/molbev/msq245. [DOI] [PubMed] [Google Scholar]
- 14.Sealey KL, Harris SR, Fry NK, Hurst LD, Gorringe AR, Parkhill J, Preston A. 2015. Genomic analysis of isolates from the United Kingdom 2012 pertussis outbreak reveals that vaccine antigen genes are unusually fast evolving. J Infect Dis 212:294–301. doi: 10.1093/infdis/jiu665. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, Liu B, Grondahl-Yli-Hannuksila K, Tan Y, Feng L, Kallonen T, Wang L, Peng D, He Q, Wang L, Zhang S. 2015. Whole-genome sequencing reveals the effect of vaccination on the evolution of Bordetella pertussis. Sci Rep 5:12888. doi: 10.1038/srep12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison MJ, Shang W, Williams MM, Bowden KE, Burgos-Rivera B, Qin X, Messonnier N, Tondella ML. 2014. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol 21:119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, He Q. 2012. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol 19:1703–1704. doi: 10.1128/CVI.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otsuka N, Han HJ, Toyoizumi-Ajisaka H, Nakamura Y, Arakawa Y, Shibayama K, Kamachi K. 2012. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One 7:e31985. doi: 10.1371/journal.pone.0031985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam C, Octavia S, Ricafort L, Sintchenko V, Gilbert GL, Wood N, McIntyre P, Marshall H, Guiso N, Keil AD, Lawrence A, Robson J, Hogg G, Lan R. 2014. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis 20:626–633. doi: 10.3201/eid2004.131478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegerle N, Dore G, Guiso N. 2014. Pertactin deficient Bordetella pertussis present a better fitness in mice immunized with an acellular pertussis vaccine. Vaccine 32:6597–6600. doi: 10.1016/j.vaccine.2014.09.068. [DOI] [PubMed] [Google Scholar]
- 21.Safarchi A, Octavia S, Luu LD, Tay CY, Sintchenko V, Wood N, Marshall H, McIntyre P, Lan R. 2015. Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model. Vaccine 33:6277–6281. doi: 10.1016/j.vaccine.2015.09.064. [DOI] [PubMed] [Google Scholar]
- 22.Martin SW, Pawloski L, Williams M, Weening K, DeBolt C, Qin X, Reynolds L, Kenyon C, Giambrone G, Kudish K, Miller L, Selvage D, Lee A, Skoff TH, Kamiya H, Cassiday PK, Tondella ML, Clark TA. 2015. Pertactin-negative Bordetella pertussis strains: evidence for a possible selective advantage. Clin Infect Dis 60:223–227. doi: 10.1093/cid/ciu788. [DOI] [PubMed] [Google Scholar]
- 23.Breakwell L, Kelso P, Finley C, Schoenfeld S, Goode B, Misegades LK, Martin SW, Acosta AM. 2016. Pertussis vaccine effectiveness in the setting of pertactin-deficient pertussis. Pediatrics 137:e20153973. doi: 10.1542/peds.2015-3973. [DOI] [PubMed] [Google Scholar]
- 24.Stibitz S, Yang MS. 1997. Genomic fluidity of Bordetella pertussis assessed by a new method for chromosomal mapping. J Bacteriol 179:5820–5826. doi: 10.1128/jb.179.18.5820-5826.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinig MM, Cummings CA, Sanden GN, Stefanelli P, Lawrence A, Relman DA. 2006. Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J Bacteriol 188:2375–2382. doi: 10.1128/JB.188.7.2375-2382.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bart MJ, van der Heide HG, Zeddeman A, Heuvelman K, van Gent M, Mooi FR. 2015. Complete genome sequences of 11 Bordetella pertussis strains representing the pandemic ptxP3 lineage. Genome Announc 3:e01394-15. doi: 10.1128/genomeA.01394-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bart MJ, Zeddeman A, van der Heide HG, Heuvelman K, van Gent M, Mooi FR. 2014. Complete genome sequences of Bordetella pertussis isolates B1917 and B1920, representing two predominant global lineages. Genome Announc 2:e01301-14. doi: 10.1128/genomeA.01301-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowden KE, Weigand MR, Peng Y, Cassiday PK, Sammons S, Knipe K, Rowe LA, Loparev V, Sheth M, Weening K, Tondella ML, Williams MM. 2016. Genome structural diversity among 31 Bordetella pertussis isolates from two recent U.S. whooping cough statewide epidemics. mSphere 1:e00036-16. doi: 10.1128/mSphere.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akamatsu MA, Nishiyama MY Jr, Morone M, Oliveira UC, Bezerra MF, Sakauchi MA, Raw I, Junqueira de Azevedo IL, Kitajima JP, Carvalho E, Ho PL. 2015. Whole-genome sequence of a Bordetella pertussis Brazilian vaccine strain. Genome Announc 3:e01570-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boinett CJ, Harris SR, Langridge GC, Trainor EA, Merkel TJ, Parkhill J. 2015. Complete genome sequence of Bordetella pertussis D420. Genome Announc 3:e00657-15. doi: 10.1128/genomeA.00657-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stibitz S. 1998. IS481 and IS1002 of Bordetella pertussis create a 6-base-pair duplication upon insertion at a consensus target site. J Bacteriol 180:4963–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeno-Tarraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, et al. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 33.Kang Y, Gu C, Yuan L, Wang Y, Zhu Y, Li X, Luo Q, Xiao J, Jiang D, Qian M, Ahmed Khan A, Chen F, Zhang Z, Yu J. 2014. Flexibility and symmetry of prokaryotic genome rearrangement reveal lineage-associated core-gene-defined genome organizational frameworks. mBio 5:e01867. doi: 10.1128/mBio.01867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esnault E, Valens M, Espeli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3:e226. doi: 10.1371/journal.pgen.0030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci U S A 109:E42–E50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. 2004. Macrodomain organization of the Escherichia coli chromosome. EMBO J 23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campo N, Dias MJ, Daveran-Mingot ML, Ritzenthaler P, Le Bourgeois P. 2004. Chromosomal constraints in Gram-positive bacteria revealed by artificial inversions. Mol Microbiol 51:511–522. doi: 10.1046/j.1365-2958.2003.03847.x. [DOI] [PubMed] [Google Scholar]
- 39.Eisen JA, Heidelberg JF, White O, Salzberg SL. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol 1:RESEARCH0011. doi: 10.1186/gb-2000-1-6-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, Heuvelman KJ, de Greeff SC, Diavatopoulos D, Teunis P, Nagelkerke N, Mertsola J. 2009. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15:1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Gouw D, Hermans PW, Bootsma HJ, Zomer A, Heuvelman K, Diavatopoulos DA, Mooi FR. 2014. Differentially expressed genes in Bordetella pertussis strains belonging to a lineage which recently spread globally. PLoS One 9:e84523. doi: 10.1371/journal.pone.0084523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King AJ, van der Lee S, Mohangoo A, van Gent M, van der Ark A, van de Waterbeemd B. 2013. Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains. PLoS One 8:e66150. doi: 10.1371/journal.pone.0066150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williamson P, Matthews R. 1996. Epitope mapping the Fim2 and Fim3 proteins of Bordetella pertussis with sera from patients infected with or vaccinated against whooping cough. FEMS Immunol Med Microbiol 13:169–178. doi: 10.1016/0928-8244(95)00123-9. [DOI] [PubMed] [Google Scholar]
- 44.Douillard FP, Ribbera A, Xiao K, Ritari J, Rasinkangas P, Paulin L, Palva A, Hao Y, de Vos WM. 2016. Polymorphisms, chromosomal rearrangements, and mutator phenotype development during experimental evolution of Lactobacillus rhamnosus GG. Appl Environ Microbiol 82:3783–3792. doi: 10.1128/AEM.00255-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raeside C, Gaffe J, Deatherage DE, Tenaillon O, Briska AM, Ptashkin RN, Cruveiller S, Medigue C, Lenski RE, Barrick JE, Schneider D. 2014. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. mBio 5:e01377-14. doi: 10.1128/mBio.01377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassiday PK, Skoff TH, Jawahir S, Tondella ML. 2016. Changes in predominance of pulsed-field gel electrophoresis profiles of Bordetella pertussis isolates, United States, 2000–2012. Emerg Infect Dis 22:442–448. doi: 10.3201/eid2203.151136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conover MS, Redfern CJ, Ganguly T, Sukumar N, Sloan G, Mishra M, Deora R. 2012. BpsR modulates Bordetella biofilm formation by negatively regulating the expression of the Bps polysaccharide. J Bacteriol 194:233–242. doi: 10.1128/JB.06020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganguly T, Johnson JB, Kock ND, Parks GD, Deora R. 2014. The Bordetella pertussis Bps polysaccharide enhances lung colonization by conferring protection from complement-mediated killing. Cell Microbiol 16:1105–1118. doi: 10.1111/cmi.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brickman TJ, Cummings CA, Liew SY, Relman DA, Armstrong SK. 2011. Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J Bacteriol 193:4798–4812. doi: 10.1128/JB.05136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King AJ, van Gorkom T, van der Heide HG, Advani A, van der Lee S. 2010. Changes in the genomic content of circulating Bordetella pertussis strains isolated from the Netherlands, Sweden, Japan and Australia: adaptive evolution or drift? BMC Genomics 11:64. doi: 10.1186/1471-2164-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam C, Octavia S, Sintchenko V, Gilbert GL, Lan R. 2014. Investigating genome reduction of Bordetella pertussis using a multiplex PCR-based reverse line blot assay (mPCR/RLB). BMC Res Notes 7:727. doi: 10.1186/1756-0500-7-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caro V, Hot D, Guigon G, Hubans C, Arrive M, Soubigou G, Renauld-Mongenie G, Antoine R, Locht C, Lemoine Y, Guiso N. 2006. Temporal analysis of French Bordetella pertussis isolates by comparative whole-genome hybridization. Microbes Infect 8:2228–2235. doi: 10.1016/j.micinf.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Heikkinen E, Kallonen T, Saarinen L, Sara R, King AJ, Mooi FR, Soini JT, Mertsola J, He Q. 2007. Comparative genomics of Bordetella pertussis reveals progressive gene loss in Finnish strains. PLoS One 2:e904. doi: 10.1371/journal.pone.0000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeShazer D, Wood GE, Friedman RL. 1994. Molecular characterization of catalase from Bordetella pertussis: identification of the katA promoter in an upstream insertion sequence. Mol Microbiol 14:123–130. doi: 10.1111/j.1365-2958.1994.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 55.Han HJ, Kuwae A, Abe A, Arakawa Y, Kamachi K. 2011. Differential expression of type III effector BteA protein due to IS481 insertion in Bordetella pertussis. PLoS One 6:e17797. doi: 10.1371/journal.pone.0017797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caro V, Bouchez V, Guiso N. 2008. Is the sequenced Bordetella pertussis strain Tohama I representative of the species? J Clin Microbiol 46:2125–2128. doi: 10.1128/JCM.02484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skoff TH, Baumbach J, Cieslak PR. 2015. Tracking pertussis and evaluating control measures through Enhanced Pertussis Surveillance, Emerging Infections Program, United States. Emerg Infect Dis 21:1568–1573. doi: 10.3201/eid2109.150023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gautom RK. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol 35:2977–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 60.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 61.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu F, Lin Y, Tang J. 2014. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics 15:354. doi: 10.1186/s12859-014-0354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 64.Han MV, Zmasek CM. 2009. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 71.Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.