ABSTRACT
Geobacter sulfurreducens, an anaerobic metal-reducing bacterium, possesses type IV pili. These pili are intrinsic structural elements in biofilm formation and, together with a number of c-type cytochromes, are thought to serve as conductive nanowires enabling long-range electron transfer (ET) to metal oxides and graphite anodes. Here, we report that a posttranslational modification of a nonconserved amino acid residue within the PilA protein, the structural subunit of the type IV pili, is crucial for growth on insoluble extracellular electron acceptors. Matrix-assisted laser desorption ionization (MALDI) mass spectrometry of the secreted PilA protein revealed a posttranslational modification of tyrosine-32 with a moiety of a mass consistent with a glycerophosphate group. Mutating this tyrosine into a phenylalanine inhibited cell growth with Fe(III) oxides as the sole electron acceptor. In addition, this amino acid substitution severely diminished biofilm formation on graphite surfaces and impaired current output in microbial fuel cells. These results demonstrate that the capability to attach to insoluble electron acceptors plays a crucial role for the cells' ability to utilize them. The work suggests that glycerophosphate modification of Y32 is a key factor contributing to the surface charge of type IV pili, influencing the adhesion of Geobacter to specific surfaces.
IMPORTANCE Type IV pili are bacterial appendages that function in cell adhesion, virulence, twitching motility, and long-range electron transfer (ET) from bacterial cells to insoluble extracellular electron acceptors. The mechanism and role of type IV pili for ET in Geobacter sulfurreducens is still a subject of research. In this study, we identified a posttranslational modification of the major G. sulfurreducens type IV pilin, suggested to be a glycerophosphate moiety. We show that a mutant in which the glycerophosphate-modified tyrosine-32 is replaced with a phenylalanine has reduced abilities for ET and biofilm formation compared with those of the wild type. The results show the importance of the glycerophosphate-modified tyrosine for surface attachment and electron transfer in electrode- or Fe(III)-respiring G. sulfurreducens cells.
KEYWORDS: filaments, fimbriae, type IV pili, glycerophosphate, attachment, microbial fuel cells
INTRODUCTION
Long-range electron transfer (ET) within microbial biofilms is of great interest because of its importance in biogeochemical reactions and bioelectrical systems. Data support the view that type IV pili from Geobacter sulfurreducens are extracellular filamentous appendages that play an intrinsic role in long-range ET in biofilms, mediating contacts among cells within a biofilm and between biofilms and terminal electron acceptors, such as insoluble metal oxides or graphite electrodes (1, 2).
G. sulfurreducens is a member of the Geobacteraceae family. This group of dissimilatory metal-reducing microorganisms is highly abundant in metal-rich subsurface environments. Geobacteraceae predominate during subsurface bioremediation as they can couple the oxidation of a wide range of organic substrates with the reduction of soluble metals and insoluble metal oxides via anaerobic respiration (3–6). In G. sulfurreducens, electrons derived from central metabolism are transferred to external insoluble Fe(III) oxides, a process that involves a number of different c-type cytochromes in addition to microbial nanowires (1, 7–9). The nanowires of G. sulfurreducens are thin appendages originating from the cell surface and are composed principally, although not exclusively, of thousands of PilA subunits. They have been reported to be electrically conductive, facilitating electron transfer to insoluble electron acceptors, and are an important structural element of the current-producing biofilm (1, 10–13).
The mechanism of electron transfer through a G. sulfurreducens biofilm to an insoluble extracellular electron acceptor is an area of intense investigation. Two major model mechanisms for conductivity have been proposed: π-orbital electron-mediated metal-like conductivity and the super exchange, or electron hopping, model. Malvankar et al. (14) observed temperature- and pH-dependent conductivities of a dried crude extract of pili and of a bacterial biofilm and inferred metal-like properties for the nanowires of type IV pili. These nanowires were proposed to conduct electrons along the length of the pilus, wiring cells within the biofilm and channeling electrons to the graphite electrode in microbial fuel cells. The authors attributed the conductivity of type IV pili to p-type charge carriers in the PilA protein and proposed a stacked arrangement of aromatic amino acid residues, resulting in overlapping π electron systems in the PilA tertiary structure and enabling intermolecular electron delocalization among pilin subunits (14). To test this model, five point mutations were introduced in the pilA gene of G. sulfurreducens, replacing three tyrosine and two phenylalanine residues with alanines (F24A, Y27A, Y32A, F51A, and Y57A in Fig. 1A). The mutant strain Aro-5 was incapable of producing current in microbial fuel cells and was deficient in Fe(III) oxide reduction (15). However, the five mutations were not tested separately, leaving it unclear which aromatic amino acid residues are necessary or sufficient for electron transfer.
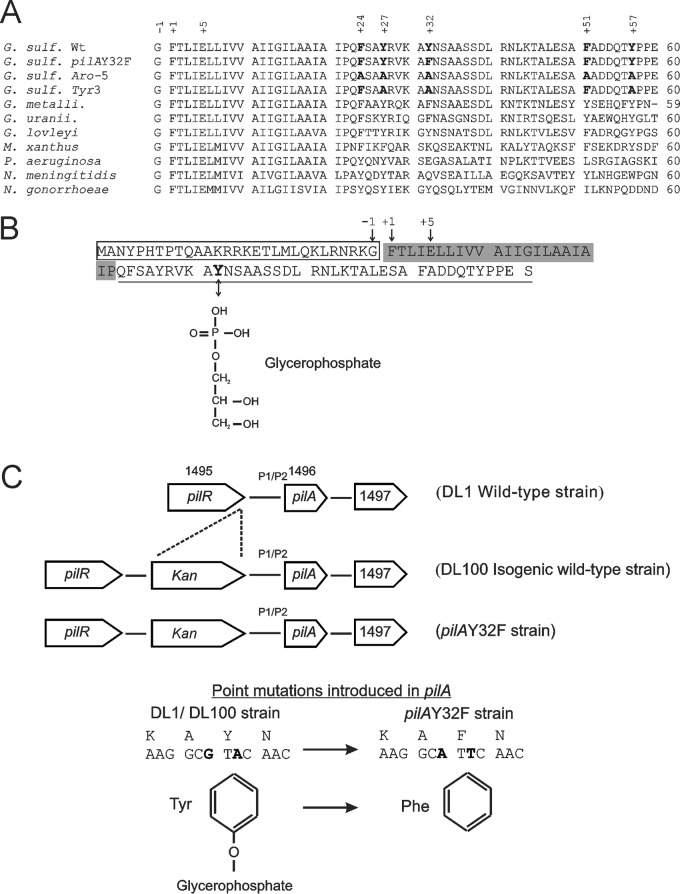
FIG 1.
(A) Sequence alignment of the first 60 amino acid residues of the cleaved PilA homologs from the Geobacter sulfurreducens wild type (G. sulf. Wt; gi 39996596), the G. sulfurreducens pilAY32F mutant produced in this work, G. sulfurreducens Aro-5 (15), G. sulfurreducens Tyr3 (16), G. metallireducens (G. metalli.; gi 78222611), G. uraniireducens (G. uranii.; gi 148264718), G. lovleyi (gi 189425155), Myxococcus xanthus (gi 108761074), Pseudomonas aeruginosa PAO1 (gi 15599721), Neisseria meningitidis (gi 45180), and Neisseria gonorrhoeae (gi 59717790). The conserved glycine at position −1, N-methylphenylalanine at +1, and glutamate at +5 are indicated. Amino acids occupying positions 24, 27, 32, 51, and 57 are in bold in G. sulfurreducens strains (wild type and mutants). (B) Amino acid sequence of the full-length (uncleaved) PilA protein (GSU1496). The conserved cleavage site is glycine at position −1, so that the mature protein starts with N-methylphenylalanine at position +1. The consensus hydrophobic segment of the mature protein is shaded, including the conserved N-methylphenylalanine at +1 and glutamate at +5. Tyrosine-32 is in boldface font and the chemical structure of the posttranslational modification of this tyrosine, a glycerophosphate moiety, is displayed. (C) Genomic organization of pilA and the surrounding genes in the wild-type and pilAY32F strains. A kanamycin resistance cassette was inserted upstream of the P1 and P2 promoter regions in the isogenic wild-type and pilAY32F strains. The base changes leading to the tyrosine-32-phenylalanine amino acid replacement are in boldface font.
Feliciano et al. constructed a Tyr3 mutant strain in which the three tyrosine residues of PilA (Y27, Y32, and Y57 in Fig. 1A) were replaced by alanines (16). The mutations severely inhibited cellular respiration when utilizing an insoluble electron acceptor. The Tyr3 strain was defective in Fe(III) oxide reduction and, although Tyr3 cells were capable of colonizing graphite anodes and forming a thick biofilm, their current production decreased compared with that of wild-type (Wt) biofilms. Feliciano et al. proposed a model of the G. sulfurreducens pilus fiber structure using molecular dynamic simulation and the structure of the type IV pilus of Neisseria gonorrhoeae as a template (16). The model suggested a right-handed helical clustering of aromatic amino acid residues of PilA with interaromatic distances optimal for a multistep electron hopping movement but not for stacking π orbitals as required for metallic conductivity.
On the other hand, Strycharz-Glaven et al. proposed that the super exchange model could explain the conductivity of G. sulfurreducens biofilms (17). Using an electron source and an electron drain, they measured electron diffusion from low- to high-redox-potential areas across the anode biofilm (18). According to their findings, electron transfer reactions proceeded through a mechanism involving a redox gradient within the biofilm that transferred electrons from redox cofactors located far from the anode to those in proximity to the anode surface (19). Heme-containing c-type cytochromes located at the bacterial outer membrane (10, 20), along the pili (21, 22), and within the extracellular matrix (12, 23) were proposed as redox-active factors facilitating electron hopping within the biofilm and to the anode surface.
Bonanni et al. modeled the G. sulfurreducens pilus structure based on the structure of the homologous pilus of Pseudomonas aeruginosa and proposed a stepping stone mechanism for long-range electron transfer (24). The stepping stone model attributes the electron transfer across the thick biofilm and onto graphite electrodes in microbial fuel cells to a synergistic effect of the aromatic amino acids in the PilA structure and the cytochromes bound to the pilus filament.
The PilA protein of G. sulfurreducens shares many of the structure-defining characteristics seen in other type IV pilin proteins. It is expressed as a prepilin with a signal peptide that is cleaved after the glycine at position −1 by a peptidase, PilD (Fig. 1B) (25, 26). Interestingly, the pilA gene has two functional translational start codons leading to two PilA preprotein isoforms, namely, short and long. The two isoforms appear to be processed into a single mature form (7 kDa) (26). The mature PilA sequence contains N-methylphenylalanine and glutamate residues at positions 1 and 5, respectively, known to be essential for the attraction between pilin subunits (Fig. 1B) (27, 28). PilA has a highly conserved hydrophobic region that is known to fold into a helical structure and is required for retaining the pilin subunits in a membrane-anchored state prior to pilus assembly (Fig. 1A and B) (29–32). Moreover, the N-terminal domain contains other structurally important amino acids, including conserved tyrosine/phenylalanine residues at positions 24 and 27 (Fig. 1A and B). The phenol/phenyl rings of tyrosine/phenylalanine-24 and tyrosine-27 from one subunit stack with the phenyl ring of the conserved N-methylphenylalanine at position 1 of an adjoining pilin subunit, thereby conferring stability to the pilus structure (33). The C-terminal domain of type IV pilin proteins is mainly hydrophilic and is exposed to the external environment. The sequences of this region vary across bacterial species in a manner that correlates with variation in pilus function. The mature PilA protein of G. sulfurreducens is smaller than other studied type IV pilins, having only 61 amino acids in total. It lacks the ∼80 amino acids that usually fold into the globular C-terminal domain (29, 32, 34).
Type IV pilin proteins undergo posttranslational modifications on the N- and C-terminal domains (31). The modifications identified on residues in the solvent-exposed region include glycosylation, as seen in Neisseria gonorrhoeae (35), Neisseria meningitidis (36), and Pseudomonas aeruginosa (37, 38), and a modification with glycerophosphate, as in Neisseria meningitidis (39). These modifications were reported to modulate pilus function (40–42). The type IV pilin protein of N. meningitidis was shown to be modified with glycerophosphate at Ser69 and Ser93 (43). The phosphorylation level of Ser69 appeared to be constant, while that of Ser93 was triggered by direct interaction of N. meningitidis with the host cells. Bacterial contact with the host surface induced the expression of pptB, encoding pilin phosphotransferase B that mediates the addition of glycerophosphate to Ser93. The solvent-exposed glycerophosphate moiety of Ser93 introduces a negative charge to the pilus surface, causing a reduction in pilus bundling and bacterial aggregation (43).
In this work, we demonstrate through mass spectrometric and mutational analyses that a glycerophosphate moiety attached to Tyr32 in the PilA protein of G. sulfurreducens is crucial for surface attachment and, consequently, for reducing Fe(III) oxides and for current production in microbial fuel cells.
RESULTS
Tyrosine-32 of the mature PilA protein of Geobacter sulfurreducens is posttranslationally modified with a glycerophosphate moiety.
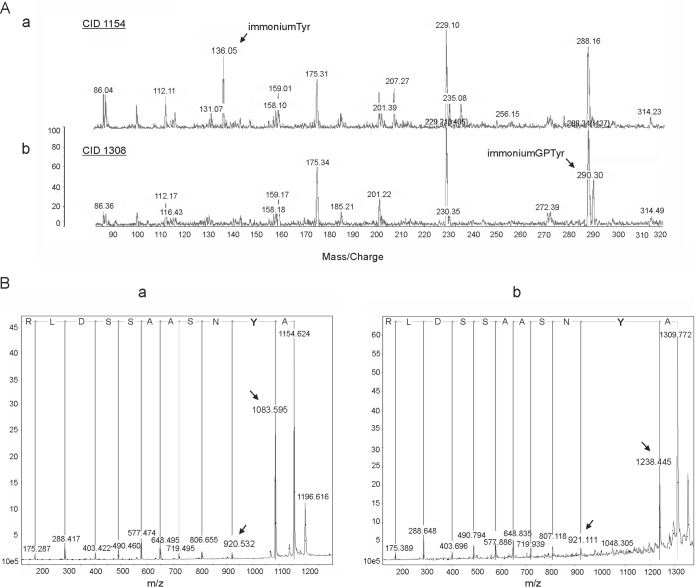
To test for any posttranslational modifications to the mature secreted form of wild-type PilA (GSU1496) protein, a concentrated secreted protein fraction was electrophoresed and the gel band corresponding to 7 kDa was excised and analyzed by matrix-assisted laser desorption ionization tandem mass spectrometry (MALDI MS/MS). The peptides detected matched the sequence predicted for the G. sulfurreducens PilA protein (44) and are as follows: KAYNSAASSDLRN, KAYNSAASSDLRN with glycerophosphate Y, RVKAYNSAASSDLRN, RVKAYNSAASSDLRN with glycerophosphate Y, KTALESAFADDQTYPPES, and KTALESAFADDQTYPPES with a C-terminal Na cation, where the underlined amino acids denote trypsin cleavage sites. MS/MS spectra obtained from collision-induced dissociation (CID) and post-source decay (PSD) techniques gave nearly complete coverage of the PilA C-terminal domain but not the amino-terminal sequence. This was presumably due to the high hydrophobicity of the amino terminus (Fig. 1B). The mass spectrometric data revealed a shift in mass in two tryptic digested peptides: AYNSAASSDLR and VKAYNSAASSDLR. Both peptides appeared to be modified with a moiety of 154 Da (Fig. 2A). This is consistent with the mass of a glycerophosphate according to the protein modification database (45). To verify the site of modification, tryptic peptides were derivatized with 4-sulfophenylisothiocyanate to increase the efficiency of the PSD fragmentation (46) and were sequenced (Fig. 2B). The site of the posttranslational modification with glycerophosphate was determined to be the tyrosine residue at position 32 (Fig. 2B).
FIG 2.
Mass spectrometry analyses of the mature PilA protein of the G. sulfurreducens wild-type strain. (A) Matrix-assisted laser desorption ionization (MALDI) mass spectrometric data for the tryptic digested peptide AYNSAASSDLR of the PilA protein secreted by the wild-type G. sulfurreducens strain. Comparison of the MALDI spectra of the unmodified (a) and modified (b) AYNSAASSDLR peptide obtained by collision-induced dissociation (CID). The 1,154-Da peptide displays the unmodified tyrosine residue at 136 Da, which is not visible in the 1,308-Da peptide. The new peak emerging at 290 Da (in the 1,308-Da peptide) corresponds to a mass difference of 154 Da, suggesting a glycerophosphate-modified tyrosine. (B) MALDI-PSD (post-source decay) mass spectrometric spectra of the unmodified (a) and modified (b) AYNSAASSDLR peptide. The amino acid sequence was deduced from the spectrum on the basis of the mass difference between adjacent peaks (indicated by arrows). The mass difference of 317 Da between the peaks at 1,238 and 921 Da (b) corresponds to the combined mass of a 4-sulfophenyl-modified tyrosine residue (163 Da) (a) and a glycerophosphate group (154 Da).
Characterization of the pilAY32F mutant strain.
To understand whether tyrosine-32 and its posttranslational modification have roles in the ability of G. sulfurreducens to reduce iron (via the nanowire/pilus model), site-directed mutagenesis was performed to substitute tyrosine-32 with phenylalanine and the mutation was introduced into the chromosomal copy of pilA (Fig. 1C). We chose the Tyr-Phe substitution because it is the minimal change necessary for testing the effect of removing the glycerophosphate while conserving as much as possible of the structure of PilA. The two aromatic amino acids share similar conformational preferences (47) and only differ by the hydroxyl group to which the glycerophosphate can attach (Fig. 1C). In addition, the phenylalanine substitution was chosen over an alanine or some other nonaromatic substitution to preserve the aromatic character of position 32 so as not to interrupt potential conductivity along the pilus filament, as predicted by the PilA metal-like conductance model (14, 15). The Y32F mutation was transferred to the chromosome as described in Materials and Methods. It was confirmed by DNA sequencing. To confirm that the mutation removed the glycerophosphate at the phenylalanine at position 32, a concentrated secreted protein fraction prepared from the pilAY32F mutant strain was electrophoresed and the band corresponding to 7 kDa was prepared for MALDI MS/MS analysis in a fashion similar to what was done for the wild-type PilA. While the Y32F mutant spectrum reveals the presence of other Geobacter proteins with molecular masses similar to that of PilA, the presence of a peak at 1,138 kDa and the lack of a peak at 1,308 kDa indicate that the sample contains the PilA peptide with phenylalanine and no glycerophosphate modification (AFNSAASSDLR) while lacking a PilA peptide with glycerophosphate-modified tyrosine-32 (see Fig. S1 in the supplemental material).
It has been established that type IV pili of G. sulfurreducens are not required for cellular growth on soluble electron acceptors, as the pilA deletion mutant strain is capable of growth on fumarate and Fe(III) citrate (1). We tested the ability of the pilAY32F mutant strain to grow on soluble electron acceptors. Figure 3A and B show that it displayed no deficiency in growth on fumarate or Fe(III) citrate as assessed by measuring the culture optical density or production of Fe(II), respectively. The Y32F mutation did not affect the expression or subcellular distribution of the PilA protein at 25°C as measured by Western blotting. All three fractions of the PilA(Y32F) mutant protein, namely, the secreted, soluble nonsecreted cellular, and membrane-associated fractions, were detected by Western blotting at levels comparable to those in the wild type DL1 (Fig. 3C).
FIG 3.
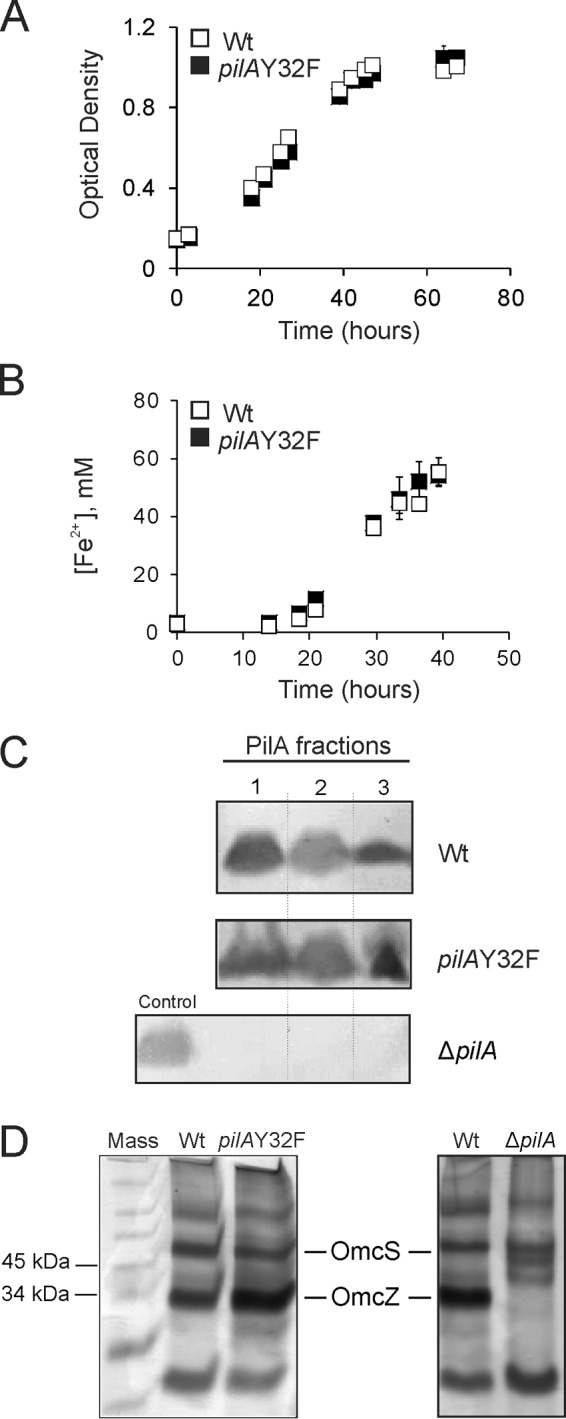
Characterization of the pilAY32F mutant strain. (A) Growth curves of the wild type DL1 (Wt) and the pilAY32F mutant. Cells were grown under conditions suitable for pilus expression (NBAF medium, 25°C). The optical densities (600 nm) are averages from six biological replicates from two independent experiments. Error bars are standard errors of the means. (B) Reduction of soluble Fe(III) citrate. Wild-type DL1 (Wt) and pilAY32F mutant cells were grown in freshwater medium with acetate and Fe(III) citrate under strict anaerobic conditions. The Fe(II) concentrations are average measurements from six biological replicates from two independent experiments. Error bars are standard errors of the means. (C) Immunoblots of the wild-type DL1 (Wt), pilAY32F, and ΔpilA in-frame deletion (26) strains using a PilA-specific antibody. Cells were grown at 25°C. PilA fractions: 1, secreted; 2, nonsecreted soluble; and 3, membrane associated. PilA protein migrated at 7 kDa. Each lane contained 10 μg of total protein. Membranes were equally contrast-enhanced to better visualize the immunoreactive bands. (D) Heme-stained SDS-PAGE of loosely bound outer surface c-type cytochromes prepared from the wild-type DL1, pilAY32F, and ΔpilA strains. The heme-positive bands that migrated at the molecular masses of OmcS and OmcZ are indicated. Each lane contained 3 μg protein.
The PilA protein was demonstrated to modulate proper secretion and localization of OmcZ, an outer membrane c-type cytochrome essential for biofilm conductivity in microbial fuel cells (12). Mutations that eliminated the expression of the pilA gene, the secretion of pili (26), or pilus assembly by targeting amino acids involved in pilin-pilin interaction (48) resulted in the absence of OmcZ on the cell's outer surface. The Y32F mutation did not interfere with its role in the proper secretion of OmcZ as indicated by the pattern of loosely bound outer surface cytochromes on heme-stained gels (Fig. 3D).
Substitution of tyrosine-32 with phenylalanine modulates attachment to graphite surfaces regardless of their utilization as electron acceptors.
Type IV pili are known to be a structural biofilm component that mediate cell-cell interactions as well as surface colonization (49). It has been established that in G. sulfurreducens, type IV pili are essential for achieving a maximal biomass, even on surfaces that do not serve as an electron acceptor (50). Therefore, we performed attachment assays to evaluate the effect of the Y32F mutation on cell adhesion to glass and graphite. The ΔpilA in-frame deletion mutant strain was used as a negative control (26). The isogenic wild-type DL100, pilAY32F, and ΔpilA strains were grown anaerobically in the presence of the soluble electron acceptor fumarate at 25°C. It is known that these conditions induce pilus expression (1). Confocal laser scanning microscopy (CLSM) revealed no defect in the ability of the pilAY32F strain to attach to glass surfaces in comparison with the wild type (Fig. 4A and B), whereas the defect of ΔpilA was confirmed (Fig. 4C). However, attachment to graphite surfaces appeared to be severely inhibited for the pilAY32F mutant compared with the wild type (Fig. 4D and E). Interestingly, the Y32F substitution diminished the attachment to graphite surfaces to a greater degree than the deletion of the entire pilA gene (Fig. 4F). We additionally measured the cell density of surface-attached cells. The cell density of the pilAY32F strain was about 25-fold less than that of the wild type on graphite surfaces (Fig. 4G). Altogether, these data strongly support the idea that the presence of the glycerophosphate moiety is critical for attachment to graphite.
FIG 4.
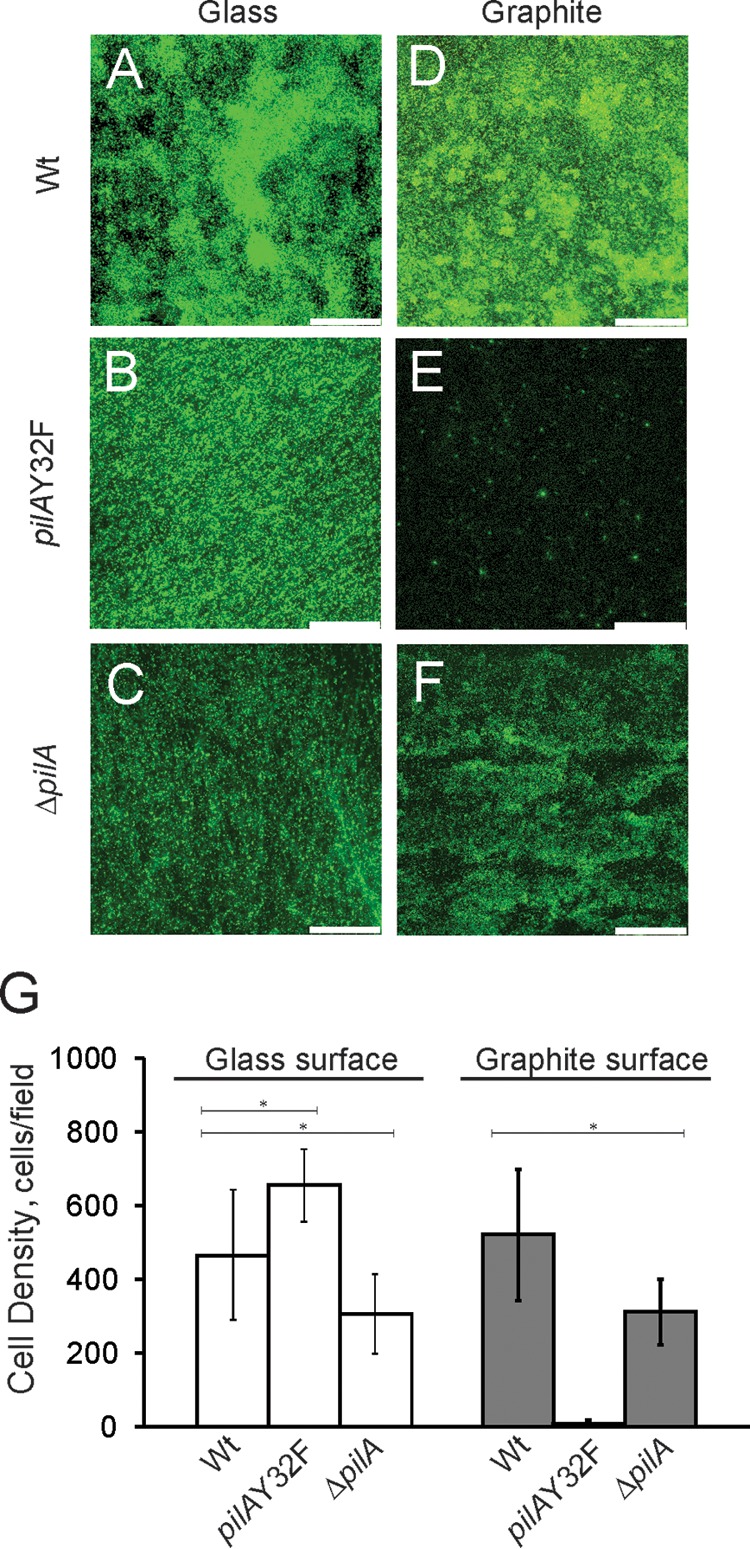
(A to F) Confocal microscopic images of biofilms formed with the isogenic wild-type DL100, pilAY32F, and ΔpilA in-frame deletion strains on glass and graphite surfaces. Cells were incubated with soluble electron donor and acceptor (10 mM acetate and 40 mM fumarate, respectively) on glass/graphite slips for 4 days under anaerobic conditions. Bars, 75 μm. (G) Average numbers of cells of the wild-type DL100, pilAY32F, and ΔpilA strains attached to glass and graphite surfaces after 4 days of incubation. The results are the averages and standard errors of the means from six biological replicates in two independent experiments. Statistical analysis was performed by Student's t test; *, P < 0.001.
The pilAY32F strain is impaired in current production in microbial fuel cells.
Type IV pili of G. sulfurreducens are a crucial element in the conductive anode biofilm (11–13). They are reported to have metal-like electric conductivity (14) and are required for achieving maximal current production in microbial fuel cells (26).
To study the impact of the Y32F mutation on the ability of G. sulfurreducens to generate electricity, cells were grown on graphite electrodes in microbial fuel cells with acetate as the electron donor and the poised graphite electrode as the sole electron acceptor (51). The wild-type strain produced current shortly after incubation and reached maximal current density around day 4 (Fig. 5). The pilAY32F strain exhibited a significant delay (9 days) before utilizing the graphite electrode as an electron acceptor (Fig. 5), and a maximal current density of 14.9 ± 0.4 mA was achieved only by day 16 (Fig. 5). CLSM of biofilms after maintaining maximal current production for 3 days (day 7 for the wild type [Wt] and day 19 for pilAY32F) indicated structural variations in substratum surface coverage (67% and 52%) and the maximum pillar heights (55.00 μm and 50.00 μm) for the wild-type and mutant strains, respectively (see Fig. S2).
FIG 5.
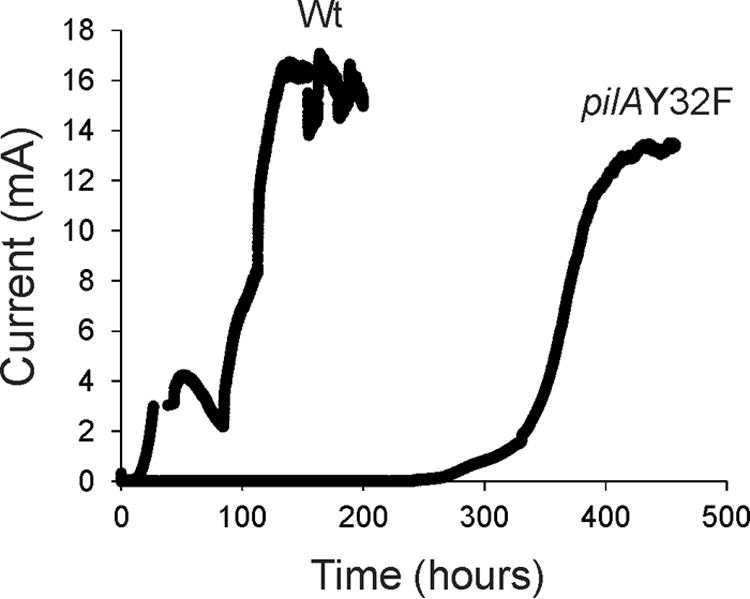
Current production by the DL100 wild-type (Wt) and pilAY32F mutant strains in microbial fuel cells. The data are representative of three biological replicates. Anodes were poised at +300 mV versus an Ag/AgCl reference electrode.
Together, our data clearly support the previous observations that the glycerophosphate moiety is essential for initial attachment to graphite and initial current production. The delayed current production by the pilAY32F mutant strain can be explained by at least two different mechanisms, namely, by adaptation and by the growth of a suppressor (see Discussion).
Tyrosine-32 and its glycerophosphate moiety are important for growth on insoluble Fe(III) oxides.
To grow, G. sulfurreducens can utilize insoluble Fe(III) oxides as an electron acceptor (2, 52). It is believed that the critical function of the type IV pilus in this process is its metal-like conductivity by which electrons generated in the central metabolism are transferred from the cells to distant extracellular electron acceptors (14). Furthermore, evidence suggests that there are other proteins and/or mechanisms for accessing or attaching to Fe(III) oxide particles and Fe(III) oxide-coated surfaces; however, this is not sufficient for growth when Fe(III) oxides are the sole electron acceptor (1, 2, 50).
To evaluate the contribution of Y32 and its glycerophosphate modification to cell growth on Fe(III) oxides, pilAY32F cells were tested for anaerobic growth with a soluble electron donor, acetate, and an insoluble electron acceptor, Fe(III) oxides (Fig. 6A). The wild-type DL100 strain reduced insoluble Fe(III) oxide particles, and the concentrations of Fe(II) produced from Fe(III) reduction increased gradually, whereas no significant change in Fe(II) concentration was detected for the samples taken from pilAY32F cultures, even after 6 months of incubation. Similarly, strains lacking pilA (1) or extracellular pili (26) were deficient in growth on insoluble Fe(III) oxides.
FIG 6.
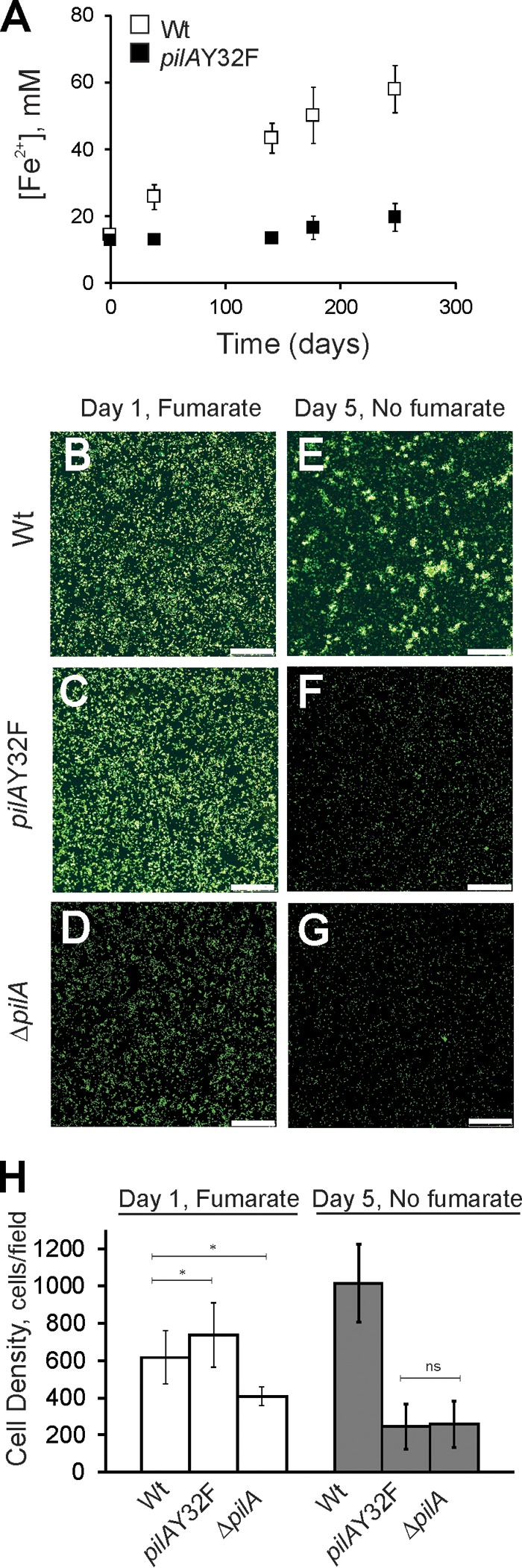
(A) Growth curves of the DL100 (Wt) and pilAY32F strains in insoluble Fe(III) oxide medium. Cells were provided with 100 mM poorly crystalline Fe(III) oxides and 15 mM acetate. Ferrozine assays were applied to determine the concentration of Fe(II) produced by bacterial reduction of Fe(III). The results are averages from eight biological replicates from two independent experiments. The error bars are standard errors of the means. (B to G) Confocal microscopic analysis of the isogenic wild-type DL100, pilAY32F, and ΔpilA biofilms formed on insoluble Fe(III) oxide-coated glass. Images were taken after incubation for 24 h in medium with 40 mM fumarate, and 4 days later, after the fumarate was removed. Bars, 70 μm. (H) Average cell growth of the wild-type DL100, pilAY32F, and ΔpilA strains in biofilms attached to insoluble Fe(III) oxide-coated glass. Cell densities were measured after incubation for 24 h in the presence of 40 mM fumarate and 4 days later after fumarate was removed. The results are the averages from six biological replicates from two independent experiments. The error bars are standard errors of the means. Statistical analysis was performed by Student's t test; *, P < 0.008; ns, not significant.
Attachment to glass surfaces coated with Fe(III) oxides and respiration of this electron acceptor were assayed. For all of the strains tested (wild type DL100, the pilAY32F substitution mutant, and the ΔpilA in-frame deletion mutant), cells were capable of attachment to Fe(III) oxide-coated glass surfaces after 24 h of incubation in the presence of a soluble electron acceptor, fumarate (Fig. 6B, C, and D). After the removal of fumarate and incubation of cells for an additional 4 days with insoluble Fe(III) oxides as the sole electron acceptor, significant growth was observed only for the wild-type cells, which demonstrated an average increase of 395 cells/field (Student's t test, P < 0.0001) (Fig. 6E and H). On the contrary, no biofilm growth was observed for the pilAY32F or ΔpilA mutant strain (Fig. 6F and G). The average biomasses of pilAY32F and ΔpilA biofilms decreased by 490 and 150 cells/field, respectively (Student's t test, P < 0.0001) (Fig. 6H). The results clearly demonstrate a growth deficiency for the pilAY32F strain with insoluble Fe(III) oxides as the sole electron acceptor, and the phenotype of pilAY32F is comparable to that observed with the ΔpilA deletion mutant strain.
DISCUSSION
Type IV pili of G. sulfurreducens have been shown to be an important structural element in current-producing biofilms attached to graphite electrodes and are believed to be conductive nanowires serving as an electron conduit for insoluble electron acceptors (1, 13, 14, 17). This work highlights the significance of a glycerophosphate-modified tyrosine residue in the mature PilA protein for attachment to surfaces that can be utilized as extracellular electron acceptors by G. sulfurreducens.
Mass spectrometry of the PilA protein (GSU1496) secreted by G. sulfurreducens confirmed the amino acid sequence expected for the mature cleaved protein. It also indicated a posttranslational modification consistent with the mass of glycerophosphate of a nonconserved tyrosine (Y32) in the C-terminal domain. The phosphate group of glycerophosphate is a likely attachment point for the hydroxyl group of tyrosine. Glycerophosphate forms a phosphodiester bond with the hydroxyl group of serine-93 in the type IV pilin of N. meningitidis (39). However, the G. sulfurreducens genome does not contain a homolog to the pilin phosphotransferase B gene (pptB) of N. meningitidis, identified by Chamot-Rooke et al. to modify Ser93 (43). Yet, a possible candidate for catalyzing the attachment of glycerophosphate to Tyr32 is a phosphoglycosyl-diphosphate-polyprenyl-phosphate phosphoglycosyl transferase encoded by the GSU1502 gene. GSU1502 is part of the xap operon (GSU1498 to GSU1505) located downstream in close proximity to the pilA gene (GSU1496). In addition to GSU1502, the xap operon encodes ATP-dependent transporters, membrane proteins, a cell wall glycosylation enzyme, and other proteins of unknown function. It is important to note that coexpression of pilA and the xap operon is required for the production and assembly of PilA (26).
We show in Fig. 3C that the PilA fractions (secreted, soluble nonsecreted, and membrane associated) recovered from the pilAY32F strain were comparable to those from the wild type, indicating proper expression and secretion of the PilA protein. Detection of G. sulfurreducens type IV pili using electron microscopy was unfortunately not feasible. G. sulfurreducens cells possess several filamentous appendages with diameters and lengths comparable to those expected for type IV pili (53), and consequently, microscopic imaging would not yield a definitive answer for whether the Y32F mutation affected pilus structure. In addition, the available anti-PilA antibody was generated against a peptide in the PilA C-terminal domain and does not bind to the native PilA protein; therefore, immunoelectron microscopy could not be used for detecting the PilA filaments. Therefore, we relied on the available resources of Western blotting and known molecular features of type IV pilin proteins for interaction and assembly for assessing the expression of PilA and its presence in subcellular fractions.
This study varies significantly from the two previously published studies of the Aro-5 and Tyr3 mutants in that those studies mutated five and three aromatic amino acid residues to alanine, respectively, whereas this study mutated a tyrosine to a phenylalanine residue, removing only a single hydroxyl group. While this single mutant did not have the entire spectrum of phenotypes that the other two mutants had, it is clear that removal of the single hydroxyl, the site of the glycerophosphate modification, has a significant phenotypic effect. The Y32F mutation in PilA is unlikely to affect pilus assembly because Y32 is located in the C-terminal domain that is not reported to participate in pilin-pilin intersubunit interactions or pilus assembly (28, 29, 32) and, in models of G. sulfurreducens pilus structure, is positioned on the pilus surface with its side chain protruding into the solvent (16, 24). In addition, a pilus model for the Tyr3 strain (with Y27A, Y32A, and Y57A) indicated that there was proper pilus assembly (16). A mutation of the type IV pilin gene of N. meningitidis to substitute the glycerophosphate-attached Ser93 with alanine had no effect on the level of piliation (43). Similar observations were reported for N. gonorrhoeae, where a Ser63Ala mutation in the pilE gene that eliminated glycosylation at position 63 resulted in a mutant strain with levels of expressed pilin and piliated cells comparable to those of the wild type (54).
The pilAY32F strain was impaired in attachment to graphite surfaces. The attachment deficiency is a consequence of a single amino acid change, which suggests that the glycerophosphate moiety attached to Y32 is exposed to the external environment and contributes to the pilus surface charge (43, 54). The bacterial cell envelope generally has a negatively charged surface that is known to influence the bacterium-surface physical/chemical interactions (55). Attachment occurs in two stages, reversible adhesion followed by irreversible adhesion (56). Reversible adhesion is initiated via long-range forces, including electrostatic attraction and van der Waals interactions. Irreversible adhesion is mediated by short-range forces, such as covalent and hydrogen bonding, and results in strong cell-surface attachment (56). Bacterial appendages, such as flagella and pili, are among the biomolecular factors involved in irreversible adhesion (49, 57, 58). Wild-type G. sulfurreducens cells favor positively charged and hydrophilic anodes for attachment and biofilm formation (59). The mature PilA protein carries a negative overall net charge with six negatively and two positively charged amino acids within the entire cleaved sequence (Fig. 1B). The glycerophosphate moiety contributes to the negative charge of the PilA protein (Fig. 1B). A pilus filament is a polymer of numerous PilA protein molecules. A point mutation that eliminates the glycerophosphate modification would result in a shift to a cell with an overall less-negative charge. This, in turn, might impact bacterial surface attachment. Moreover, a change in the pilus surface charge can cause electrostatic attraction or repulsion among pilus fibers. This influences their morphology (bundling) and, in turn, modulates their function (43, 60). Interestingly, the Y32F mutation resulted in less cell attachment to graphite than the complete deletion of pilA when a soluble electron acceptor was provided (Fig. 4E and F). Deletion of pilA may have enabled other structures of the cell's outer surface to replace the pilus filaments and maintain some affinity of the cell's outer surface toward graphite.
The pilAY32F mutant strain did not grow on insoluble Fe(III) oxides even after an extended period of 6 months (Fig. 6A). Yet, it grew as well as the wild type on soluble electron acceptors [fumarate or Fe(III) citrate] (Fig. 3A and B). The pilAY32F mutant formed a biofilm on Fe(III) oxide-coated glass but did not utilize Fe(III) oxides as an electron acceptor (Fig. 6C and F). Similarly, a pilA-deficient strain accessed Fe(III) particles, as shown by electron microscopy, but did not respire Fe(III) oxides (1). Given the fact that G. sulfurreducens does not utilize an electron shuttle (9), a physical interaction between pili and the Fe(III) particles is required for electron transfer. As the pilAY32F strain still produced and secreted the mutant PilA protein under the applied experimental conditions, its lack of growth on Fe(III) oxides may be attributed to the absence of the Fe(III)-binding site in the mutant pili. Therefore, it is plausible that the glycerophosphate moiety binds Fe(III) and/or aids in bringing Fe(III) oxide particles in proximity to the pilus surface, thereby facilitating electron transfer to Fe(III) oxides.
Transcripts for the outer membrane hexaheme c-type cytochrome OmcS were reported to be highly upregulated in cells grown on insoluble Fe(III) oxides (61) and observed to be localized along filaments that were not conclusively shown to be type IV pili (21). Our data show that OmcS is still properly secreted (Fig. 3D). Therefore, we believe that the Y32F mutation does not affect the binding of OmcS to filaments. Furthermore, this is supported by the fact that OmcS was associated with filaments in the Aro-5 mutant (15). Thus, our data are consistent with the model in which Y32F pili do not bind Fe(III) oxide particles.
The pilAY32F cells did not produce current in microbial fuel cells even after an extended incubation of >200 h (Fig. 5). However, they produced current after an ∼300-h delay. Two possible explanations for this behavior are that the population somehow adapted to the situation or that a suppressor mutant arose in the population (62–64). While we cannot choose between these two with the data presented, we favor the possibility that a suppressor mutant arose because it was shown elsewhere that pilA deletion mutants readily acquire suppressors enabling their growth under similar selective conditions (64). Therefore, it is possible that the lack of current production observed for the pilAY32F strain can be attributed to a deficiency in pilus-mediated attachment or to a combination of impairments in attachment and electron transfer. According to the metal-like conductivity model, electron transfer along the pilus structure is facilitated by π-π stacking of aromatic amino acid side chains (14). On the basis of this model, replacing tyrosine-32 with phenylalanine, another aromatic amino acid, should not interrupt electron transfer to graphite anodes. Therefore, to test whether the modified tyrosine at position 32 is required for attachment only or for attachment and electron transfer, further experiments are needed. The circumstance that a similar adaptation was not noted in the pilAY32F cells incubated for 6 months on solid Fe(III) oxides can be explained by the constant requirement for quick and efficient attachment to new iron particles during growth, as opposed to a requirement for one-time attachment only on fuel cell graphite electrodes. Once the first layer of biofilm is initiated on the anode, the biofilm can mature and a three-dimensional structure may be established via exopolysaccharides (65).
In comparing our work to previously reported studies on Aro-5 (14) and Tyr3 (16), the majority of phenotypes are observed from a single mutation (Y32F), suggesting that the other 2 to 4 amino acids have small contributions to the noted phenotypes. Our results provide further insight into the respiration of G. sulfurreducens cells on insoluble electron acceptors and show that tyrosine-32, with its posttranslational modification in the PilA protein, is essential for these cells to attach to graphite and for growth on an insoluble extracellular electron acceptor.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Wild-type and mutant strains of G. sulfurreducens and the plasmids used in this work are listed in Table 1. Escherichia coli strain TOP10 was purchased from Invitrogen Co. (Carlsbad, CA) and was used for subcloning PCR products and for DNA manipulations.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmida | Descriptionb | Source or reference |
|---|---|---|
| Strains | ||
| DL1 (Wt) | Wild type | 66 |
| DL100 (isogenic Wt) | Wild type; pilA Kanr | 26 |
| pilAY32F mutant | pilA contains the Tyr-32-Phe mutation (GCG TAC → GCA TTC)c; Kanr | This work |
| ΔpilA (in-frame deletion) mutant | In-frame deletion mutation; Kanr | 26 |
| Plasmids | ||
| pCD341 | Source of Kanr cassette | 74 |
| pLC3 | pilA (wild type) Specr | 26 |
| pLC9 | pilA with Tyr-32-Phe mutation; Specr | This work |
All strains were G. sulfurreducens and were derived from DL1 (66).
Kanr, kanamycin resistance; Specr, spectinomycin resistance.
Boldface font denotes mutated bases.
DNA manipulations and plasmid construction.
Genomic DNA of the G. sulfurreducens wild-type strain DL1 (66) was purified using the MasterPure complete DNA purification kit (Epicentre Technologies, Madison, WI). Plasmid DNA purification, PCR product purification, and gel extraction were performed using the QIAprep spin mini plasmid purification, QIAquick PCR purification, and QIAquickgel extraction kits, respectively (Qiagen, Inc., Valencia, CA). Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs, Inc. (Beverly, MA). Primers for PCR and for site-directed mutagenesis were purchased from Operon Biotechnologies, Inc. (Huntsville, AL). All PCRs were performed with high-fidelity Phusion polymerase using reaction conditions specified by the manufacturer (Finnzymes, Inc., Woburn, MA).
Plasmid pLC3, expressing the wild-type pilA (GSU1496) under the control of native promoters (26), served as the template for replacing the tyrosine codon (TAC) with a phenylalanine codon (TTC). A standard site-directed mutagenesis technique (QuikChange II XL site-directed mutagenesis kit; Stratagene, Inc., La Jolla, CA) was performed with the Y32F_Fwd primer and the complementary primer (Y32F_Rev) to introduce a BsmI restriction site polymorphism upon replacing tyrosine-32 with phenylalanine (Table 2). The restriction site was introduced for screening to verify that the plasmid carried the phenylalanine mutation. DNA sequencing confirmed that the plasmid carried only the desired mutation. The resulting plasmid, pLC9, was used to introduce the Tyr-32-Phe point mutation into the chromosomal copy of pilA as described below.
TABLE 2.
Primers used in this work
| Purpose | Name | Sequence,a 5′ → 3′ | Description |
|---|---|---|---|
| pilAY32F construction | rLC43F | AGAGCAGGTGAAGGAAGGGAGTTT | Amplifies the last 500 bp of GSU1495 (pilR) with the kanamycin cassette |
| rLC46R | tgacgattttcgtcactggctcctctTGGATCCCCCGGGCTGCAGGAATTCG | ||
| rLC47F | cgaattcctgcagcccgggggatccaAGAGGAGCCAGTGACGAAAATCGTCA | Amplifies pilA | |
| rLC51R | CTATTGCGCACAATGGCTATTCCCTGCATTGCGA | ||
| Plasmid pLC9 construction | Y32F_Fwd | CAGTTCTCGGCGTATCGTGTCAAGGCATTCAACAGCGCGGCGTCAAGCG | Introduces the Tyr-32-Phe mutation |
| Y32F_Rev | CGCTTGACGCCGCGCTGTTGAATGCCTTGACACGATACGCCGAGAACTG |
Annealing nucleotides are in lowercase, BsmI restriction sites are italicized and underlined, and base pairs replaced by site-directed mutagenesis are in boldface font.
Construction of G. sulfurreducens mutant strain.
The pilAY32F mutant strain was constructed by recombinant PCR as previously described (67). The pLC9 plasmid carrying the Tyr-32-Phe mutation in pilA was the template for amplification of pilA, to generate a linear mutagenic fragment. Using the chromosomal DNA of strain DL100 as the template, a segment extending from within pilR (GSU1495) to the pilA P1/P2 promoter region (Fig. 1C), with a kanamycin resistance marker inserted, was amplified. The two amplicons were joined by crossover PCR. The point mutation in pilA was introduced into the chromosome of DL1 by homologous recombination using the kanamycin resistance marker to select for recombinants. The Tyr-32-Phe mutation was confirmed by PCR using Phusion polymerase and different sets of primers to map the pilA region, by digestion at the introduced BsmI restriction site in pilA, and by DNA sequencing. Two purified and sequenced colonies of the pilAY32F strain were independently characterized. The isogenic strain DL100, which has the wild-type version of pilA but the same kanamycin resistance cassette insertion as pilAY32F (Fig. 1C), was used as a second control (in addition to the original wild type).
Culturing conditions and growth media.
G. sulfurreducens wild-type and mutant strains were routinely cultured under anaerobic conditions (N2/CO2: 80/20) (5, 67). Plating and incubation on solid NBAF medium (NB medium with 15 mM acetate, 40 mM fumarate, 1.5% agar, 0.1% yeast extract, and 1 mM cysteine) were performed in an anaerobic chamber at 30°C as previously described (67).
For liquid cultures, 15 mM acetate served as the electron donor and either fumarate (40 mM), Fe(III) citrate (60 mM), or poorly crystalline Fe(III) oxide (100 mM) was the electron acceptor. The growth temperature was 30°C, except when fumarate was the electron acceptor, in which case cells were grown at 25°C, the pilus expression-inducing condition (1). Cells were adapted for growth on Fe(III) citrate for three consecutive transfers before their growth was monitored on poorly crystalline Fe(III) oxides.
Microbial fuel cell experiments were conducted as previously described (51). The source of carbon/electrons/energy was 10 mM acetate, and the graphite electrode serving as electron acceptor was poised with a potentiostat at +300 mV versus an Ag/AgCl reference electrode. The system was switched from batch mode into continuous flow of 10 mM acetate (30 ml h−1) upon current initiation (11).
Immunoblotting and heme-staining analyses.
PilA protein fractions were prepared from G. sulfurreducens cultures according to published protocols (26, 68) to separate the secreted and nonsecreted cellular PilA. Briefly, 100-ml batch bacterial cultures were grown under strictly anaerobic conditions in NBAF medium at 25°C. Cells were harvested at late exponential phase and subjected to a 15-min centrifugation at 5,000 × g and 4°C to separate the culture supernatant from the sedimented cells without shearing the cells. Supernatants were concentrated using a 5,000-molecular-weight-cutoff (MWCO) membrane. Further filtration was achieved using 3,000-MWCO ultrafiltration tubes (Millipore Corp., Billerica, MA) to a final volume of 500 μl. This fraction was referred to as the secreted protein fraction. Cell pellets were resuspended in 50 mM Tris-HCl buffer (pH 7.5) and vigorously sonicated and then subjected to 15-min centrifugation at 5,000 × g and 4°C. The collected supernatant was referred to as the soluble nonsecreted protein fraction, whereas the pellet was referred to as the membrane-associated protein fraction. Protein fractions were separated by electrophoresis on 15% acrylamide–Tris-Tricine–SDS gels, and the protein bands were transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA) using a semidry transfer unit (Trans-Blot SD, Bio-Rad Laboratories). The membranes were probed with a PilA-specific antibody (69), and the immunoreactive bands were visualized with the One-Step Western kit (Genscript Corp., Piscataway, NJ) according to the manufacturer's instructions.
For detection of loosely bound outer surface c-type cytochromes, protein samples were prepared according to the published protocol (8) and were separated by electrophoresis using 12% Next gels (Amresco, Inc., Solon, OH). N,N,N′,N′-Tetramethylbenzidine was used for heme staining as previously described (70).
SeeBlue Plus2 prestained standard (Invitrogen Corp., Carlsbad, CA) was used as the molecular weight marker in all electrophoresis gels.
All protein concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Rockford, IL) with bovine serum albumin as a standard (71).
Mass spectrometry.
For mass spectrometry of the PilA wild type and the Y32F mutant, the secreted protein fractions prepared from wild-type DL1 and pilAY32F strains as described above were separated by electrophoresis in a 15% Tris-Tricine gel and the PilA protein band corresponding to 7 kDa was excised. The in-gel sample digestion, purification, and matrix-assisted laser desorption ionization tandem mass spectrometry (MALDI MS/MS) were carried out at the University of Massachusetts Medical School, Laboratory for Mass Spectrometry, Worcester, MA.
Biofilm characterization and analysis.
Bacterial biofilms were stained with a nucleic acid stain, Syto 9 L7012 component A (Invitrogen Corp., Carlsbad, CA), and examined by CLSM using a Leica TCS SP5 microscope with a HCX PL APO 100× objective (numerical aperture, 1.4). CLSM images of the anode biofilms were processed using Leica LAS AF software (Leica Microsystems GmbH, Wetzlar, Germany) to create three-dimensional projections and cross sections of the biofilms (14, 26). Thicknesses and percent coverage of biofilms were statistically determined using the biofilm analysis software PHLIP (72) from a minimum of 15 image stacks per condition.
Cell attachment assays.
Wild-type and mutant cells grown in freshwater medium (5) with 10 mM acetate and 40 mM fumarate were exposed to graphite or glass surfaces for 4 days under anaerobic conditions. In the case of Fe(III) oxide attachment, cells were exposed to Fe(III) oxide-coated glass, prepared as described previously (73), in the presence of soluble electron donor and acceptor (10 mM acetate and 40 mM fumarate) for 24 h. Thereafter, the medium was swapped and cells were incubated for an additional 4 days with only acetate, with the Fe(III) oxide-coated glass as the sole electron acceptor. Two independent sets of experiments were conducted with three biological replicates per strain per condition for all attachment assays. Biofilms were examined under CLSM and three images for each of the six biological replicates were generated. Cells were manually counted at ×1,000 magnification at three random locations per image. Statistical analyses were conducted using Student's t test, and the P values are reported in Results and in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Hanno Richter and Muktak Aklujkar for critically reading the manuscript.
The U.S. Department of Energy Office of Science (BER) supported this research under Cooperative Agreement no. DEFC02-02ER63446.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00716-16.
REFERENCES
- 1.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 2.Lovley DR. 2011. Live wires: direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energy Environ Sci 4:4896–4906. doi: 10.1039/C1EE02229F. [DOI] [Google Scholar]
- 3.Cologgi DL, Lampa-Pastirk S, Speers AM, Kelly SD, Reguera G. 2011. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc Natl Acad Sci U S A 108:15248–15252. doi: 10.1073/pnas.1108616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovley DR, Baedecker MJ, Lonergan DJ, Cozzarelli IM, Phillips EJP, Siegel DI. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297–300. doi: 10.1038/339297a0. [DOI] [Google Scholar]
- 5.Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes DE, Finneran KT, O'Neil RA, Lovley DR. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl Environ Microbiol 68:2300–2306. doi: 10.1128/AEM.68.5.2300-2306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leang C, Coppi MV, Lovley DR. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol 185:2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol 71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nevin KP, Lovley DR. 2000. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl Environ Microbiol 66:2248–2251. doi: 10.1128/AEM.66.5.2248-2251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methe BA, Didonato RJ, Covalla SF, Franks AE, Liu A, Lovley DR. 2009. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628. doi: 10.1371/journal.pone.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol 72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter H, Nevin KP, Jia H, Lowy DA, Lovley DR, Tender LM. 2009. Cyclic voltammetry of biofilms of wild type and mutant Geobacter sulfurreducens on fuel cell anodes indicates possible roles of OmcB, OmcZ, type IV pili, and protons in extracellular electron transfer. Energy Environ Sci 2:506–516. doi: 10.1039/B816647A. [DOI] [Google Scholar]
- 13.Franks AE, Nevin KP, Glaven RH, Lovley DR. 2010. Microtoming coupled to microarray analysis to evaluate the spatial metabolic status of Geobacter sulfurreducens biofilms. ISME J 4:509–519. doi: 10.1038/ismej.2009.137. [DOI] [PubMed] [Google Scholar]
- 14.Malvankar NS, Vargas M, Nevin KP, Franks AE, Leang C, Kim BC, Inoue K, Mester T, Covalla SF, Johnson JP, Rotello VM, Tuominen MT, Lovley DR. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat Nanotechnol 6:573–579. doi: 10.1038/nnano.2011.119. [DOI] [PubMed] [Google Scholar]
- 15.Vargas M, Malvankar NS, Tremblay PL, Leang C, Smith JA, Patel P, Snoeyenbos-West O, Nevin KP, Lovley DR. 2013. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio 4:e00105-13. doi: 10.1128/mBio.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feliciano GT, Steidl RJ, Reguera G. 2015. Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys Chem Chem Phys 17:22217–22226. doi: 10.1039/c5cp03432a. [DOI] [PubMed] [Google Scholar]
- 17.Strycharz-Glaven SM, Snider RM, Guiseppi-Elie A, Tender LM. 2011. On the electrical conductivity of microbial nanowires and biofilms. Energy Environ Sci 4:4366–4379. doi: 10.1039/C1EE01753E. [DOI] [Google Scholar]
- 18.Snider RM, Strycharz-Glaven SM, Tsoi SD, Erickson JS, Tender LM. 2012. Long-range electron transport in Geobacter sulfurreducens biofilms is redox gradient-driven. Proc Natl Acad Sci U S A 109:15467–15472. doi: 10.1073/pnas.1209829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bond DR, Strycharz-Glaven SM, Tender LM, Torres CI. 2012. On electron transport through Geobacter biofilms. ChemSusChem 5:1099–1105. doi: 10.1002/cssc.201100748. [DOI] [PubMed] [Google Scholar]
- 20.Ding YH, Hixson KK, Giometti CS, Stanley A, Esteve-Nunez A, Khare T, Tollaksen SL, Zhu W, Adkins JN, Lipton MS, Smith RD, Mester T, Lovley DR. 2006. The proteome of dissimilatory metal-reducing microorganism Geobacter sulfurreducens under various growth conditions. Biochim Biophys Acta 1764:1198–1206. doi: 10.1016/j.bbapap.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Leang C, Qian X, Mester T, Lovley DR. 2010. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl Environ Microbiol 76:4080–4084. doi: 10.1128/AEM.00023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian X, Mester T, Morgado L, Arakawa T, Sharma ML, Inoue K, Joseph C, Salgueiro CA, Maroney MJ, Lovley DR. 2011. Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim Biophys Acta 1807:404–412. doi: 10.1016/j.bbabio.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Inoue K, Leang C, Franks AE, Woodard TL, Nevin KP, Lovley DR. 2011. Specific localization of the c-type cytochrome OmcZ at the anode surface in current-producing biofilms of Geobacter sulfurreducens. Environ Microbiol Rep 3:211–217. doi: 10.1111/j.1758-2229.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 24.Bonanni PS, Massazza D, Busalmen JP. 2013. Stepping stones in the electron transport from cells to electrodes in Geobacter sulfurreducens biofilms. Phys Chem Chem Phys 15:10300–10306. doi: 10.1039/c3cp50411e. [DOI] [PubMed] [Google Scholar]
- 25.Strom MS, Lory S. 1992. Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J Bacteriol 174:7345–7351. doi: 10.1128/jb.174.22.7345-7351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter LV, Sandler SJ, Weis RM. 2012. Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. J Bacteriol 194:2551–2563. doi: 10.1128/JB.06366-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aas FE, Winther-Larsen HC, Wolfgang M, Frye S, Lovold C, Roos N, van Putten JP, Koomey M. 2007. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect Type IV pilus assembly, dynamics and associated functions. Mol Microbiol 63:69–85. doi: 10.1111/j.1365-2958.2006.05482.x. [DOI] [PubMed] [Google Scholar]
- 28.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT, Tainer JA. 2003. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell 11:1139–1150. doi: 10.1016/S1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 29.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 30.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L, Tainer JA. 2011. Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J Biol Chem 286:44254–44265. doi: 10.1074/jbc.M111.297242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Egelman EH, Craig L. 2012. Structure of the Vibrio cholerae type IVb pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J Mol Biol 418:47–64. doi: 10.1016/j.jmb.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reardon PN, Mueller KT. 2013. Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J Biol Chem 288:29260–29266. doi: 10.1074/jbc.M113.498527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aas FE, Vik A, Vedde J, Koomey M, Egge-Jacobsen W. 2007. Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol 65:607–624. doi: 10.1111/j.1365-2958.2007.05806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Power PM, Seib KL, Jennings MP. 2006. Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem Biophys Res Commun 347:904–908. doi: 10.1016/j.bbrc.2006.06.182. [DOI] [PubMed] [Google Scholar]
- 37.Voisin S, Kus JV, Houliston S, St-Michael F, Watson D, Cvitkovitch DG, Kelly J, Brisson JR, Burrows LL. 2007. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J Bacteriol 189:151–159. doi: 10.1128/JB.01224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kus JV, Kelly J, Tessier L, Harvey H, Cvitkovitch DG, Burrows LL. 2008. Modification of Pseudomonas aeruginosa Pa5196 type IV pilins at multiple sites with d-Araf by a novel GT-C family arabinosyltransferase, TfpW. J Bacteriol 190:7464–7478. doi: 10.1128/JB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stimson E, Virji M, Barker S, Panico M, Blench I, Saunders J, Payne G, Moxon ER, Dell A, Morris HR. 1996. Discovery of a novel protein modification: alpha-glycerophosphate is a substituent of meningococcal pilin. Biochem J 316 (Pt 1):29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smedley JG III, Jewell E, Roguskie J, Horzempa J, Syboldt A, Stolz DB, Castric P. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect Immun 73:7922–7931. doi: 10.1128/IAI.73.12.7922-7931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jennings MP, Jen FE, Roddam LF, Apicella MA, Edwards JL. 2011. Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell Microbiol 13:885–896. doi: 10.1111/j.1462-5822.2011.01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen LC, Taguchi F, Tran QM, Naito K, Yamamoto M, Ohnishi-Kameyama M, Ono H, Yoshida M, Chiku K, Ishii T, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2012. Type IV pilin is glycosylated in Pseudomonas syringae pv. tabaci 6605 and is required for surface motility and virulence. Mol Plant Pathol 13:764–774. doi: 10.1111/j.1364-3703.2012.00789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, Imhaus AF, Martin P, Trellet M, Clary G, Chafey P, Camoin L, Nilges M, Nassif X, Dumenil G. 2011. Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331:778–782. doi: 10.1126/science.1200729. [DOI] [PubMed] [Google Scholar]
- 44.Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 45.Creasy DM, Cottrell JS. 2004. Unimod: protein modifications for mass spectrometry. Proteomics 4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 46.Chen P, Nie S, Mi W, Wang XC, Liang SP. 2004. De novo sequencing of tryptic peptides sulfonated by 4-sulfophenyl isothiocyanate for unambiguous protein identification using post-source decay matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom 18:191–198. doi: 10.1002/rcm.1280. [DOI] [PubMed] [Google Scholar]
- 47.Williams RW, Chang A, Juretic D, Loughran S. 1987. Secondary structure predictions and medium range interactions. Biochim Biophysica Acta 916:200–204. doi: 10.1016/0167-4838(87)90109-9. [DOI] [PubMed] [Google Scholar]
- 48.Steidl RJ, Lampa-Pastirk S, Reguera G. 2016. Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat Commun 7:12217. doi: 10.1038/ncomms12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Toole GA, Wong GC. 2016. Sensational biofilms: surface sensing in bacteria. Curr Opin Microbiol 30:139–146. doi: 10.1016/j.mib.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reguera G, Pollina RB, Nicoll JS, Lovley DR. 2007. Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J Bacteriol 189:2125–2127. doi: 10.1128/JB.01284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bond DR, Lovley DR. 2003. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl Environ Microbiol 69:1548–1555. doi: 10.1128/AEM.69.3.1548-1555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovley DR. 1995. Microbial reduction of iron, manganese, and other metals. Adv Agron 54:175–231. doi: 10.1016/S0065-2113(08)60900-1. [DOI] [Google Scholar]
- 53.Klimes A, Franks AE, Glaven RH, Tran H, Barrett CL, Qiu Y, Zengler K, Lovley DR. 2010. Production of pilus-like filaments in Geobacter sulfurreducens in the absence of the type IV pilin protein PilA. FEMS Microbiol Lett 310:62–68. doi: 10.1111/j.1574-6968.2010.02046.x. [DOI] [PubMed] [Google Scholar]
- 54.Marceau M, Forest K, Beretti JL, Tainer J, Nassif X. 1998. Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Mol Microbiol 27:705–715. doi: 10.1046/j.1365-2958.1998.00706.x. [DOI] [PubMed] [Google Scholar]
- 55.Li BK, Logan BE. 2004. Bacterial adhesion to glass and metal-oxide surfaces. Colloids Surf B Biointerfaces 36:81–90. doi: 10.1016/j.colsurfb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 56.Ploux L, Ponche A, Anselme K. 2010. Bacteria/material interfaces: role of the material and cell wall properties. J Adhes Sci Technol 24:2165–2201. doi: 10.1163/016942410X511079. [DOI] [Google Scholar]
- 57.Mattick JS. 2002. Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 58.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 59.Guo K, Freguia S, Dennis PG, Chen X, Donose BC, Keller J, Gooding JJ, Rabaey K. 2013. Effects of surface charge and hydrophobicity on anodic biofilm formation, community composition, and current generation in bioelectrochemical systems. Environ Sci Technol 47:7563–7570. doi: 10.1021/es400901u. [DOI] [PubMed] [Google Scholar]
- 60.Forest KT, Dunham SA, Koomey M, Tainer JA. 1999. Crystallographic structure reveals phosphorylated pilin from Neisseria: phosphoserine sites modify type IV pilus surface chemistry and fibre morphology. Mol Microbiol 31:743–752. doi: 10.1046/j.1365-2958.1999.01184.x. [DOI] [PubMed] [Google Scholar]
- 61.Aklujkar M, Coppi MV, Leang C, Kim BC, Chavan MA, Perpetua LA, Giloteaux L, Liu A, Holmes DE. 2013. Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159:515–535. doi: 10.1099/mic.0.064089-0. [DOI] [PubMed] [Google Scholar]
- 62.Kim BC, Leang C, Ding YH, Glaven RH, Coppi MV, Lovley DR. 2005. OmcF, a putative c-Type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J Bacteriol 187:4505–4513. doi: 10.1128/JB.187.13.4505-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krushkal J, Leang C, Barbe JF, Qu Y, Yan B, Puljic M, Adkins RM, Lovley DR. 2009. Diversity of promoter elements in a Geobacter sulfurreducens mutant adapted to disruption in electron transfer. Funct Integr Genomics 9:15–25. doi: 10.1007/s10142-008-0094-7. [DOI] [PubMed] [Google Scholar]
- 64.Smith JA, Tremblay PL, Shrestha PM, Snoeyenbos-West OL, Franks AE, Nevin KP, Lovley DR. 2014. Going wireless: Fe(III) oxide reduction without pili by Geobacter sulfurreducens strain JS-1. Appl Environ Microbiol 80:4331–4340. doi: 10.1128/AEM.01122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rollefson JB, Stephen CS, Tien M, Bond DR. 2011. Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J Bacteriol 193:1023–1033. doi: 10.1128/JB.01092-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caccavo F Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol 60:3752–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coppi MV, Leang C, Sandler SJ, Lovley DR. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol 67:3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu SS, Wu J, Cheng YL, Kaiser D. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol Microbiol 29:1249–1261. doi: 10.1046/j.1365-2958.1998.01013.x. [DOI] [PubMed] [Google Scholar]
- 69.Yi H, Nevin KP, Kim BC, Franks AE, Klimes A, Tender LM, Lovley DR. 2009. Selection of a variant of Geobacter sulfurreducens with enhanced capacity for current production in microbial fuel cells. Biosens Bioelectron 24:3498–3503. doi: 10.1016/j.bios.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Thomas PE, Ryan D, Levin W. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem 75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 71.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 72.Mueller LN, de Brouwer JF, Almeida JS, Stal LJ, Xavier JB. 2006. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol 6:1. doi: 10.1186/1472-6785-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Schie PM, Fletcher M. 1999. Adhesion of biodegradative anaerobic bacteria to solid surfaces. Appl Environ Microbiol 65:5082–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.