Abstract
Rheumatoid arthritis (RA) is characterized by synovial lining hyperplasia, which involves abnormal growth of fibroblast-like synoviocytes (FLSs). This study aimed to investigate the function and molecular mechanism of peptidylarginine deiminase type 4 (PADI4) in FLSs isolated from RA patients (RA-FLSs). FLSs were isolated from RA patients and transfected with small interfering RNAs (siRNAs) or PADI4 overexpression plasmid. FLSs were treated by Adriamycin (ADR) to induce apoptosis, and apoptotic cells were detected by flow cytometry. The expression of PADI4, p53 and p21 was detected by qRT-PCR and Western blot analysis. The recruitment of PADI4 and histone H3 arginine modifications to p21 promoter was measured by chromatin immunoprecipitation. The results showed that knockdown of PADI4 promoted the apoptosis of RA-FLSs and the expression of p53 and p21. Ectopic expression of PADI4 inhibited ADR-induced apoptosis of RA-FLSs, and down-regulated the expression of p53 and p21. In RA-FLSs, global H3 citrullination (CitH3) and H3 arginine 17 methylation levels were dynamically changed by PADI4 and ADR treatment. PADI4 and H3 could bind p21 promoter region to regulate p21 expression. In conclusion, PADI4 contributes to the pathogenesis of RA by protecting FLSs from apoptosis. PADI4 suppresses p21 transcription through altering histone H3 arginine modifications on p21 promoter region. Our study provides new insight into the anti-apoptotic role of PADI4 in RA development.
Keywords: PADI4, p21, histone arginine citrullination, cell apoptosis
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by chronic joint inflammation and variable degrees of bone and cartilage erosion. The etiology of RA is related to genetic and environmental factors, such as viral infection, hormone levels and mental state. The morbidity of RA in women is 2-3 times higher than in men, but the underlying mechanism remains unknown 1, 2. Recent evidence suggests that fibroblast-like synoviocytes (FLSs) are implicated in the pathogenesis of RA. The apoptosis and proliferation of FLSs play important role in the occurrence and development of RA, but the detailed mechanism remains to be explored 3.
Peptidylarginine deiminase (PAD) converts peptidylarginine into citrulline in the presence of Ca2+, in a process known as citrullination. The PAD family includes five distinct isozymes, PADI1, PADI2, PADI3, PADI4 and PADI6. Among them, PADI4 is expressed in a variety of blood cells 4, 5. However, overexpression of PADI4 has been observed in synovial organization and peripheral blood mononuclear cells (PBMCs) of RA patients 6, 7. Moreover, PADI4 expression was specifically induced by collagen in the inflammation period in RA rat model 8. These results suggest that abnormally high expression of PADI4 is a key factor in the development of RA.
Citrullination of the proteins leads to significant changes in their biological activity, such as the regulation of cell proliferation and apoptosis 9, 10. Histones H3 and H4 are the first substrate proteins identified which could be citrullinated by PADI4 at different Arginine (Arg, R) residues. In particular, PADI4 increases the citrullination at arginine 3 residue of histone H4 (H4R3cit) and arginine 17 residue of histone H3 (H3R17cit) 11. Recently, we demonstrated that non-histone protein vimentin is a direct substrate citrullinated by PADI4. The citrullinated vimentin effectively promotes the proliferation of FLSs isolated from RA patients 12. Given the fact that the major citrullination substrate of PADI4 is histone, we speculated that PADI4 might play a role in RA by regulating the citrullination on histone which modulates the transcription of key genes during the development of RA.
In the present study, we employed both gain and loss of function approaches to illustrate the anti-apoptosis function of PADI4 in RA-FLSs. PADI4 negatively regulated the apoptosis of RA-FLSs via inhibiting the expression of p53 and its target protein p21. Furthermore, chromatin immunoprecipitation (ChIP) assays indicated a decrease in histone H3 arginine citrullination and an increase in histone H3 arginine methylation at H3R17 on the p21 promoter.
Materials and methods
Patients
Primary synovial tissues were obtained from two female RA patients who underwent knee arthroscopic or replacement surgery (Table 1). The tissue samples were immediately suspended in 1640 medium and processed within 4 h for FLSs culture and histological and immunohistochemical analysis. The clinical diagnosis of RA was based on American College of Rheumatology (ACR) criteria. All patients were asked to consent before enrolling into this medical research. The Ethics Committee of Shanghai East Hospital approved the study protocol.
Table 1.
Clinical and serological characteristics of the patients
| Patient # | Diagnosis | Gender | Age (year) | Disease duration (year) |
Medications taken | ESR (mm/h) |
RF (IU/mL) |
CRP (mg/L) |
Anti-CCP (IU/L) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | RA | Female | 65 | 11.5 | MTX+Leflunomide | 45 | 136.5 | 34.8 | 25.6 |
| 2 | RA | Male | 59 | 8 | MTX+ SSZ | 39 | 76.5 | 27.6 | 41.3 |
RA, rheumatoid arthritis; MTX, methotrexate; SSZ, sulfasalazine; ESR, erythrocyte sedimentation rate; RF, rheumatoid factor; CRP, C-reactive protein; anti-CCP, anti-cyclic citrullinated peptide antibody.
Isolation and culture of FLSs
Synovial tissues were minced into pieces of 2-3 mm in size and spread on the bottom of cell culture flasks in 1640 medium at 37°C for 6 h. Next, the tissues were incubated with complete 1640 medium, supplemented with 10% fetal calf serum in a humidified atmosphere containing 5% CO2. The medium was changed every 3-5 days and non-adherent tissue pieces were carefully removed. FLSs were grown further over 4-6 passages. To characterize the cytological phenotype of synovial cultures, the third passage cells were stained with mouse mAb against human CD14 and CD90 (eBioscience, San Diego, CA, USA) and analyzed by flow cytometry.
RNA Interference and Plasmids Construction
Two small interfering RNAs (siRNAs) against human PADI4 (si-944: 5′-CAGGAGGTGTACGCGTGCAGTATTT-3′; si-1225: 5′-GGGTATCAGTGGACTGGACTCCTTT-3′) and negative control siRNA (si-NC: 5′-TTCTCCGAACGTGTCACGT-3′) were synthesized by Genepharma (Shanghai, China). Full-length cDNA encoding human PADI4 was amplified by using primers forward: GCTGAATTCATGGCCCAGGGGACATTGATC and reverse: AGTGGATCCGGGCACCATGTTCCACCA. pcDNA3.1-PAID4 was constructed by inserting the PADI4 PCR product. FLSs were transfected with siRNAs by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) at 30% confluency, while transfected with plasmids by using Lipo-LTX (Invitrogen, Carlsbad, CA, USA) at 80% confluency.
Quantitative Real-time-PCR (qRT-PCR)
Total RNA was extracted from FLSs by using TRIzol (Invitrogen, Carlsbad, CA, USA). The reverse transcription was performed by using a first strand cDNA synthesis kit (Takara, Dalian, China) according to the manufacturer's instructions. Real-time PCR was performed using Premix Ex Taq SYBR Green PCR (Takara, Dalian, China) according to the manufacturer's instructions on an ABI PRISM 7300 (Applied Biosystems, Foster City, CA, USA). The sequences of the primers were as follows: PADI4 forward: 5'-CACAGCTCTGGTTGGCTTCA-3', reverse: 5'-CTGCACGTCCTTCAGCATCA-3'; p21 forward: 5'-AGAACCCATGCGGCAGCAAG-3', reverse: 5'-TGGATGCAGCCCGCCATTAG-3'; p53 forward: 5'-GTGAGGGATGTTTGGGAGATG-3', reverse: 5'-CCTGGTTAGTACGGTGAAGTG-3'; β-actin forward: 5'-TGACTTCAACAGCGACACCCA-3', reverse: 5'-CACCCTGTTGCTGTAGCCAAA-3'. β-actin was used as internal control.
Western blot analysis
Whole cell lysates were prepared from about 2×105 cells by homogenization in lysis buffer and subsequent centrifugation at 14,000 rpm for 15 min. The protein concentration in the supernatant was determined using the Bradford method (Bio-Rad, Hercules, CA, USA). Protein samples were separated on 10% SDS-PAGE and then transferred onto nitrocellulose membranes (Amersham Pharmacia Biotech, Uppsala, Sweden). The membranes were blocked in 5% skimmed milk, and incubated overnight with primary antibodies anti-Flag (Sigma, #F3165), anti-Beta-actin (Santa Cruz, #sc-47778), anti-p53 (Abcam, # ab26), anti-PADI4 (Abcam, # ab38772), anti-p21 (Abcam, # ab7960), anti-citrullinated histone H3 (Abcam, # ab5103), anti-Arg-dimethyl histone H3 (Abcam, # ab8284) or anti-monomethyl histone H4 (Abcam, # ab17339), then washed 3 times with TBST every 5 min. The membrane was incubated with horseradish peroxidase-conjugated secondary antibody for one hour, and then washed with TBST 3 times every 10 min. The immunoreactive bands were visualized with SuperSignals west Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA).
Flow cytometry analysis of apoptosis
Adriamycin (ADR) was administered to FLSs at a dose of 1 μg/ml for 24 h. For flow cytometry analysis, the FLSs were trypsinized and collected for the detection of apoptosis by using an Annexin V-FITC Apoptosis Detection Kit (eBioscience, San Diego, CA, USA). Briefly, FLSs were washed twice with cold PBS and suspended in 500 μl binding buffer (10 mM HEPES-NaOH pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) at a concentration of 1×106 cells/ml. After the addition of 5 μl Annexin V-FITC solution and PI (1 μg/ ml), the cells were incubated for 15 min at room temperature and then analyzed by flow cytometer (Beckman Coulter, Fullerton, CA, USA).
Chromatin Immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) assay was performed by using an EZ-ChIP Chromatin Immunoprecipitation Kit (Millipore, MA, USA, #17-371) according to the manufacturer's instructions. Sonicated chromatin was incubated at 4°C overnight with antibodies against PADI4, CitH3 (citrullination of H3R2, R8 and R17), and H3R17me2, respectively. The bound DNA fragments were analyzed by PCR using HotStart Taq enzyme (Takara, Dalian, China). The primers designed to detect the -8 promoter region of p21 (forward: 5′-TATATCAGGGCCGCGCTG-3′ and reverse: 5′-GGCTCCACAAGGAACTGACTTC-3′) and the primers to detect GAPDH promoter region (forward: 5′-CAATTCCCCATCTCAGTCGT-3′ and reverse: 5′-TAGTAGCCGGGCCCTACTTT-3′) were synthesized by Invitrogen (Carlsbad, CA, USA). GAPDH was used as negative control 13.
Immunohistochemistry staining (IHC)
Synovial tissues were dissected and processed following standard IHC protocols. In brief, the tissue was fixed with 4% paraformaldehyde in phosphate buffered saline, dehydrated and embedded in paraffin wax. The sections were deparaffinized, rehydrated and incubated with PADI4 or p21 antibody (Abcam) followed with incubation with secondary rabbit antibody. The staining was performed following the manual of Vectastain ABC kit (Vector Laboratories). Stain images were captured using a Leica DM 4M light microscope (Leica).
Statistical analysis
Data were expressed as mean ± standard deviation (SD) and analyzed by using a SPSS16.0 program package (SPSS Inc., Chicago, IL, USA). Comparison was performed by an independent t-test. P <0.05 was considered statistically significant.
Results
Abundant PADI4 expression in RA is associated with less apoptosis
To explore the expression pattern of PADI4 in osteoarthritis (OA) and RA, we performed immunohistochemistry staining and quantitative RT-PCR on synovial tissues from RA and OA patients. PADI4 staining signal and mRNA level are both dramatically higher in RA compared to OA (Fig. 1A&B). In addition, we observed a negative correlation between PADI4 and p21 staining in synovial tissues from RA and OA patients (Table 2).
Figure 1.
PADI4 is highly expressed and inhibited the apoptosis of RA-FLSs. (A) Immunohistochemistry staining of PADI4 in arthritic synovial tissues from RA and OA patients. Original magnification: x200. Images represent PADI4 and p21 antibodies staining, IgG control staining (Negative) and HE staining. (B) Quantitative RT-PCR analysis of PADI4 mRNA level in RA and OA FLSs. Data are presented after normalization by GAPDH. (C) Left: flow cytometry analysis of apoptotic RA and OA FLSs after ADR treatment (Upper left, untreated OA; Upper right, ADR treated OA; Lower left, untreated RA; Lower right, ADR treated RA). Right: quantification of apoptotic cells. *, P<0.05, ** P<0.01.
Table 2.
Negative correlation between PADI4 and p21 staining in synovial tissues from RA and OA patients.
| Staining | p21 | Spearman r |
p | ||||
|---|---|---|---|---|---|---|---|
| -/+ | ++ | +++ | |||||
| PADI4 | -/+ | 2 | 2 | 6 | -0.518 | 0.003 | |
| ++ | 3 | 1 | 2 | ||||
| +++ | 10 | 3 | 1 | ||||
Furthermore, we treated RA and OA FLS cells with a well-known apoptosis-inducing agent Adriamycin (ADR), and measured the apoptotic cells by Annexin V/PI staining. Flow cytometry analysis showed that the percentage of apoptotic cells increased about 10-fold in RA-FLSs and almost 20-fold in OA-FLSs after ADR treatment (Fig. 1C). These data suggest that abundant expression of PADI4 in FLS cells is negatively associated with the induction of apoptosis.
PADI4 depletion promotes the apoptosis of RA-FLSs
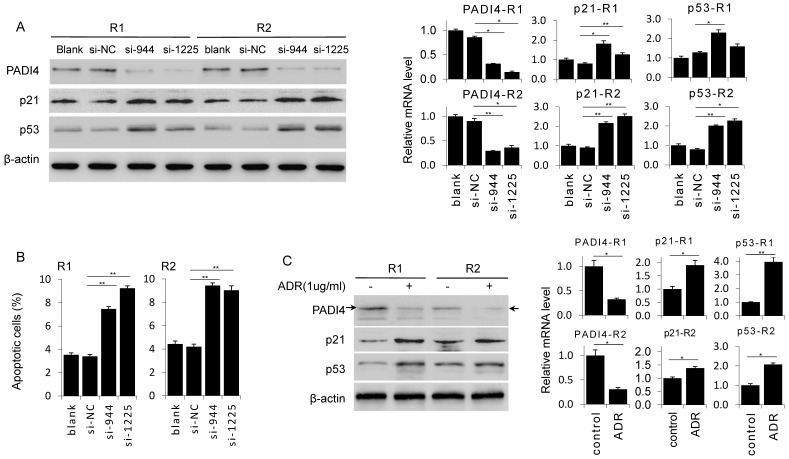
To confirm the anti-apoptosis function of PADI4, we depleted PADI4 expression in RA-FLSs by siRNAs. Western blot and RT-PCR analysis confirmed that two PADI4 specific siRNAs (si-944 and si-1225) both significantly decreased PADI4 expression at protein and mRNA levels in RA-FLSs derived from two RA patients (Fig. 2A). Next, we performed flow cytometry analysis and found that the ratio of apoptotic cells was significantly higher in PADI4 depleted RA-FLSs compared to RA-FLSs transfected with control siRNA (Fig. 2B).
Figure 2.
Knockdown of PADI4 promotes the apoptosis and increases the expression of p53 and p21 in RA-FLSs. (A) RA-FLSs were transfected with siRNAs and the levels of p21 and p53 were evaluated by Western blot analysis (left) and qRT-PCR (right). (B) Flow cytometry analysis of apoptotic RA-FLSs after PADI4 knockdown for 72 h. (C) Western blot and qRT-PCR analyses of PADI4, p53 and p21 in RA-FLSs (R1 and R2) after treatment with ADR (1 μg/ml) for 24 h. *, P<0.05, ** P<0.01.
To verify that PADI4 protects RA-FLSs from apoptosis, we detected the expression of two key pro-apoptotic proteins p53 and p21 in PADI4 depleted RA-FLSs. Western blot analysis showed that protein levels of p53 and p21 increased in RA-FLSs treated with PADI4 siRNAs (Fig. 2A). Furthermore, we found that PADI4 level was decreased in ADR treated RA-FLSs, while p53 and p21 levels were consistently increased (Fig. 2C).
PADI4 rescues the RA-FLSs from apoptosis through inactivation of p21 and p53
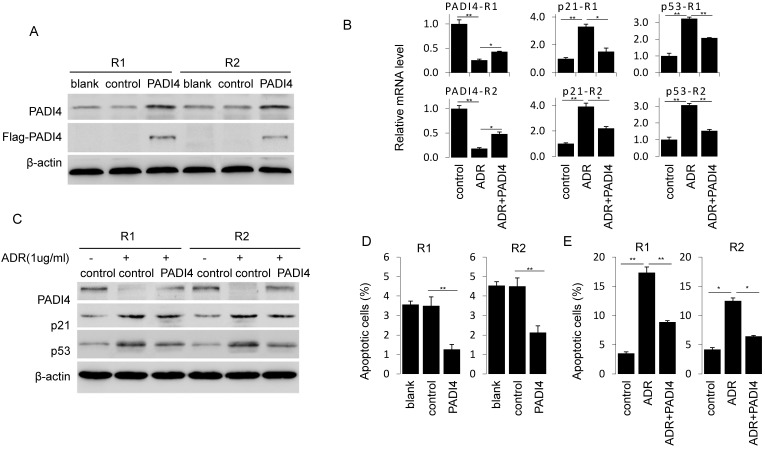
Next, we generated PADI4 overexpression plasmid that successfully achieve a strong PADI4 expression in RA-FLSs (Fig. 3A). PADI4 overexpression in RA-FLSs significantly decreased the ratio of apoptotic cells (Fig. 3D). Similarly, ADR induced apoptosis in RA-FLSs was reversed after ectopic expression of PADI4 (Fig. 3E).
Figure 3.
Ectopic expression of PADI4 inhibits the apoptosis and decreases p53 and p21 expression in RA-FLSs. (A) RA-FLSs were transfected with overexpression plasmids and PAID4 protein was evaluated by Western blot analysis. (B & C) RA-FLSs were transfected with PADI4 overexpression plasmid and 24 h later treated with ADR. mRNA and protein levels of PADI4, p53 and p21 were measured by qRT-PCR (B) and Western blot analysis (C). (D & E) Flow cytometry analysis of apoptotic RA-FLSs after PADI4 overexpression alone (D) and in combination with ADR treatment (E). *P<0.05, **P<0.01.
To explore the involvement of p21 and p53 in PADI4 mediated apoptosis inhibition, we compared the expression of p53 and p21 in ADR treated RA-FLSs with or without PADI4 overexpression. We found that PADI4 levels increased while p21 and p53 levels decreased in PADI4 overexpressing cells (Fig. 3B and 3C). Collectively, these results suggest that PADI4 inhibits the apoptosis of RA-FLSs through the inhibition of p21 and p53 transcription.
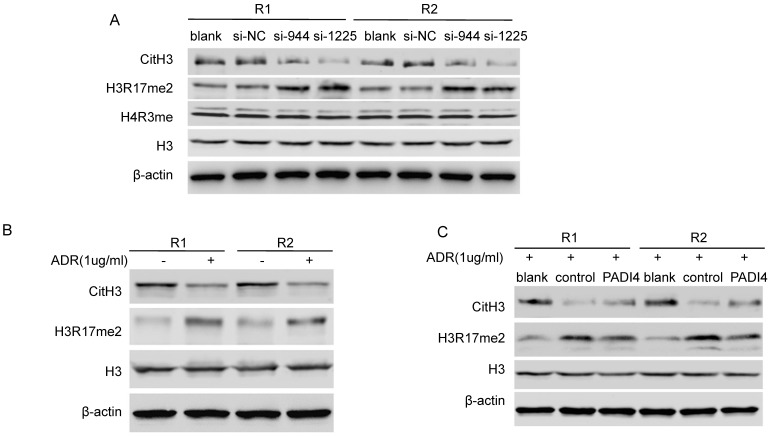
PADI4 reverses global H3 modification in RA-FLSs
Given that PADI4 citrullinates histone H3 and H4, we further hypothesized that PADI4 may inhibit p21 transcription through an epigenetic mechanism. We found that PADI4 depletion led to global repression of CitH3 (Citrullinated H3R2 + R8 + R17) and global deposition of H3R17me2 in whole cell lysate of RA-FLSs (Fig. 4A). Interestingly, we detected decreased CitH3 and increased H3R17me2 in ADR-treated RA-FLSs (Fig. 4B). However, the increase of H3R17me2 and decrease of CitH3 were both significantly reversed by PADI4 (Fig. 4C). These results suggest that PADI4 globally reverses the key histone arginine modifications.
Figure 4.
PADI4 modulates the citrullination/methylation of histone H3 in RA-FLSs. (A) Western blot analysis of the levels of CitH3 (citrullination of H3R2, R8 and R17), H3R17me2, H4R3me and total histone H3 in RA-FLSs after transfection with PADI4 siRNAs. (B) Western blot analysis of the levels of CitH3, H3R17me2 and total histone H3 in RA-FLSs with ADR treatment. (C) Western blot analysis of the levels of CitH3, H3R17me2 and total histone H3 in PADI4 overexpressing RA-FLSs after ADR treatment.
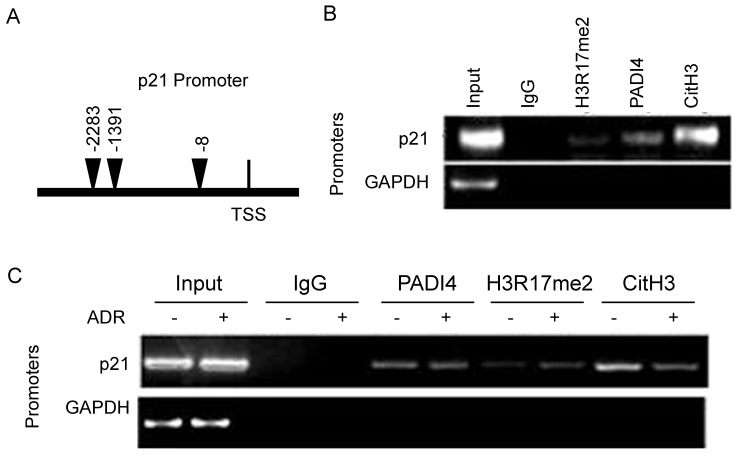
PADI4 enhances the recruitment of CitH3 to p21 promoter in RA-FLSs
To further determine the details of PADI4 inhibited p21 transcription, we conducted qChIP assays. We designed a pair of primers that specifically recognized the distal promoter region of p21 gene (Fig. 5A). Compared to IgG control, a decent PADI4 binding signal, a strong CitH3 binding signal and a weak H3R17me2 binding signal were detected on p21 promoter region (Fig. 5B). No PADI4 binding signal was detected with GAPDH promoter (data not shown).
Figure 5.
Recruitment of PADI4 and H3 to p21 promoter during apoptosis of RA-FLSs. (A) Schematic diagram of the primer l location on p21 distal promoter region. (B) ChIP assays to examine histone H3R17me2, PADI4 and CitH3 binding signals on the p21 promoter. (C) ChIP assays to examine histone H3R17me, PADI4 and CitH3 binding signals on the p21 promoter in RA-FLS with ADR treatment.
Compared to non-treated RA-FLSs, we observed weaker CitH3 occupancy and stronger H3R17me2 occupancy on p21 promoter region in ADR treated RA-FLSs (Fig. 5C). PADI4 occupancy on p21 promoter was not significantly affected after ADR treatment.
Discussion
Protein methylation, especially the methylation of lysine, has been regarded as one of the key epigenetic mechanisms that regulate gene expression. However, protein citrullination emerges as a new form of protein post-translational modification that plays a unique role in epigenetic gene regulation 14-17. PADI4 is a member of the PADI family that regulates protein citrullination. PADI4 was found to be highly expressed in RA synovial tissue but the biological function of PADI4 in RA development, especially in the apoptosis, remains controversial. Soem et al. showed that high expression of PADI4 induced cell apoptosis 18, 19, while others showed that down-regulation of PADI4 led to retroaction 20, 21. Our recent study reported that PADI4 was highly activated in RA-FLS by stimulating with citrullinated vimentin 12. In this study we provide further evidence that PADI4 protects RA-FLSs from apoptosis through suppressing p21 transcription via an epigenetic mechanism.
During RA development, the in vivo citrullinated proteins accumulate as a result of high expression of PADI4 22, 23. In addition, the locus of PADI4 gene SNP is highly correlated with RA susceptibility 6, 24, 25. It was reported that the positive rate of anti-PADI4 antibody and its titer in RA patients were higher than in patients with other rheumatic diseases or healthy people, indicating that PADI4 protein is likely to participate in RA autoimmune reaction 26, 27. However, the detailed mechanism by which PADI4 regulates RA development remains elusive.
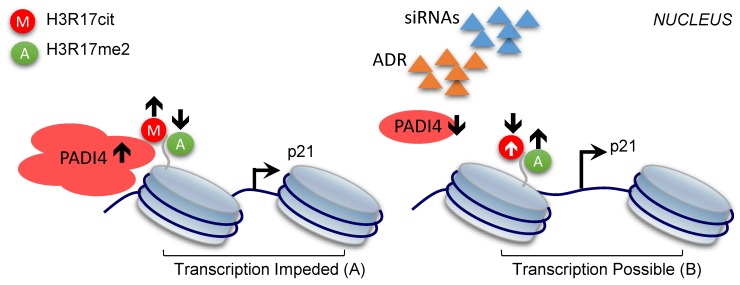
In the present study, we showed for the first time that PADI4 inhibits cell apoptosis of RA-FLSs, and the underlying mechanism involves the histone H3 modifications altered by PADI4. Based on our findings, we proposed a working model to explain the contribution of PADI4 to RA development. High level of PADI4 recruits repressive histone mark CitH3, including H3R17cit, to p21 promoter, which competitively removes the active histone mark (H3R17me2) on the same residue (Fig. 6A). As a result, p21 transcription was impeded and the cells were protected from apoptosis, which eventually leads to the occurrence of RA. On the other hand, when PADI4 is down-regulated, reduced CitH3 and enhanced H3R17me2 activate p21 transcription. Consequently, the apoptosis of RA-FLSs is promoted and RA development is inhibited (Fig. 6B).
Figure 6.
Model of the repressive role of PADI4 in p21 transcription. PADI4 suppresses p21 transcription through the deposition of repressive mark CitH3 (for example, H3R17cit) and inhibition of the active mark H3R17me2 on p21 promoter region (A). ADR induced PDK4 down-regulation or siRNAs mediated PADI4 depletion promotes p21 transcription (B). H3R17cit: repressive histone H3R17 citrullination; H3R17me2: active histone H3R17 di-methylation.
In addition, we found that PADI4 inhibited p53 expression in RA-FLSs. The downregulation of p53 may also explain the anti-apoptotic role of PADI4 in RA-FLSs. p21 is partly involved in the regulation of apoptosis by p53 28, 29. Moreover, several studies suggest that the function of p21 is dependent on p53 during the regulation of cell apoptosis by PADI4 12, 20. Intriguingly, PADI4 is also expressed at a high level in various types of adenocarcinomas, and one of the target genes of PADI4 is p53 30, 31. Further studies are needed to elucidate the mechanism by which PADI4 regulates p53 expression in RA-FLSs and investigate the role of p53 in the apoptotic regulation mediated by PADI4.
PADI4 catalyzes the citrullination of all proteins that contain arginine residues, such as H3, H4, vimentin, and fibronectin; and further catalytic substrates of PADI4 have been identified 17, 32-34. Our recent studies reported that citrullinated vimentin and citrullinated fibronectin elevated PADI4 expression and promoted the proliferation of RA-FLSs 11, 12. In this study we demonstrated that histone arginine citrullination in p21 promoter, as a poorly understood histone modification, could crosstalk with histone arginine methylation, which is one of the best-characterized modification that regulates gene transcription. More importantly, our data showed that this crosstalk plays a major role in the regulation of the apoptosis in RA-FLSs. Future studies are need to investigate how to interrupt PADI4 mediated citrullinations on histone proteins or other non-histone proteins as a potential therapy for RA. Additionally, our findings provide a clue for potential targeted therapeutic approaches to reverse the inflammation development of RA. One possibility is directly targeting histone arginine citrullination in p21 promoter for RA therapy. Another strategy is to develop small molecule inhibitors of PADI4. In vivo studies using animal RA model will be necessary to evaluate these approaches.
Conclusions
For the first time we showed that PADI4 contributes to the pathogenesis of RA by inhibiting the apoptosis of FLSs. Down-regulation of PADI4 could increase the apoptosis of RA-FLSs, while up-regulation of PADI4 could decrease the apoptosis of RA-FLSs. Mechanistically, PADI4 disrupts histone H3 arginine modifications and suppresses p21 transcription. Our study provides new insight into the anti-apoptotic role of PADI4 in RA development and illuminate the epigenetic mechanism.
Supplementary Material
Supplementary figures.
Acknowledgments
We thank the patients for their contribution to this study and thank all the members of the lab for stimulating discussion. This project was supported by the Natural Science Foundation of China (81373203, 81601407), Shanghai Municipal Health and Planning Commission (No. 20154Y0115), Pudong New Area Science and Technology Commission (No. PKJ2015-Y18).
Authors' contributions
Lieying Fan designed and directed the research and drafted the manuscript. Ming Zong, Dongyi He, Lishan Sun and Qin Ye collected clinical samples and data. Ming Zong, Ruhan Gong, Hui Zhang and Niu Li performed the experiments. All authors read and approved the final manuscript for publication.
Abbreviations
- RA
Rheumatoid arthritis
- FLSs
fibroblast-like synoviocytes
- PADI4
peptidylarginine deiminase type 4
- ACR
American College of Rheumatology
- SDS-PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- siRNAs
small interfering RNAs
- ChIP
Chromatin immunoprecipitation
- qRT-PCR
Quantitative Real-time-PCR.
References
- 1.Harris ED Jr. Rheumatoid arthritis, Pathophysiology and implications for therapy. N Engl J Med. 1990;322:1277–89. doi: 10.1056/NEJM199005033221805. [DOI] [PubMed] [Google Scholar]
- 2.Huber LC, Distler O, Tarner I. et al. Synovial fibroblasts: key players in rheumatoid arthritis. Rheumatology (Oxford) 2006;45:669–75. doi: 10.1093/rheumatology/kel065. [DOI] [PubMed] [Google Scholar]
- 3.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–55. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seri Y, Shoda H, Matsumoto I. et al. Peptidylarginine deiminase type4 (PADI4) role in immune system. Nihon Rinsho Meneki Gakkai Kaishi. 2014;37:154–9. doi: 10.2177/jsci.37.154. [DOI] [PubMed] [Google Scholar]
- 5.Arita K, Hashimoto H, Shimizu T. et al. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–83. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 6.Fan LY, Wang WJ, Wang Q. et al. A functional haplotype and expression of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Chinese. Tissue Antigens. 2008;72:469–73. doi: 10.1111/j.1399-0039.2008.01119.x. [DOI] [PubMed] [Google Scholar]
- 7.Chang X, Zhao Y, Sun S. et al. The expression of PADI4 in synovium of rheumatoid arthritis. Rheumatol Int. 2009;29:1411–6. doi: 10.1007/s00296-009-0870-2. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki A, Yamada R, Yamamoto K. Citrullination by peptidylarginine deiminase in rheumatoid arthritis. Ann N Y Acad Sci. 2007;1108:323–39. doi: 10.1196/annals.1422.034. [DOI] [PubMed] [Google Scholar]
- 9.Junta CM, Sandrin-Garcia P, Fachin-Saltoratto AL. et al. Differential gene expression of peripheral blood mononuclear cells from rheumatoid arthritis patients may discriminate immunogenetic, pathogenic and treatment features. Immunology. 2009;127:365–72. doi: 10.1111/j.1365-2567.2008.03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Helm-van Mil AH, Verpoort KN, Breedveld FC. et al. Antibodies to citrullinated proteins and differences in clinical progression of rheumatoid arthritis. Arthritis Res Ther. 2005;7:R949–58. doi: 10.1186/ar1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Wysocka J, Sayegh J. et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 12.Fan LY, He DY, Wang Q. et al. Citrullinated vimentin stimulates proliferation, pro-inflammatory cytokine secretion, and PADI4 and RANKL expression of fibroblast-like synoviocytes in rheumatoid arthritis. Scand J Rheumatol. 2012;41:354–8. doi: 10.3109/03009742.2012.670263. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Zhang L, Jones KA.Jones. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 2011;25:701–16. doi: 10.1101/gad.2002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Reinberg D.Transcription regulation by histone methylation. interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 15.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 16.McNee G, Eales KL, Wei W, Citrullination of histone H3 drives IL-6 production by bone marrow mesenchymal stem cells in MGUS and multiple myeloma. Leukemia; 2016. p. 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christophorou MA, Castelo-Branco G, Halley-Stott RP. et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–8. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu GY, Liao YF, Chang WH. et al. Overexpression of peptidylarginine deiminase IV features in apoptosis of haematopoietic cells. Apoptosis. 2006;11:183–96. doi: 10.1007/s10495-006-3715-4. [DOI] [PubMed] [Google Scholar]
- 19.Tanikawa C, Ueda K, Nakagawa H. et al. Regulation of protein Citrullination through p53/PADI4 network in DNA damage response. Cancer Res. 2009;69:8761–9. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 20.Yao H, Li P, Venters BJ. et al. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008;283:20060–8. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P, Yao H, Zhang Z. et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008;28:4745–58. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willemze A, Toes RE, Huizinga TW. et al. New biomarkers in rheumatoid arthritis. Neth J Med. 2012;70:392–9. [PubMed] [Google Scholar]
- 23.Yamada R, Suzuki A, Chang X. et al. Citrullinated proteins in rheumatoid arthritis. Front Biosci. 2005;10:54–64. doi: 10.2741/1506. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki A, Yamada R, Chang X. et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 25.Panati K, Pal S, Rao KV. et al. Association of single nucleotide polymorphisms (SNPs) of PADI4 gene with rheumatoid arthritis (RA) in Indian population. Genes Genet Syst. 2012;87:191–6. doi: 10.1266/ggs.87.191. [DOI] [PubMed] [Google Scholar]
- 26.Schellekens GA, de Jong BA, van den Hoogen FH. et al. Citrulline is an Essential Constituent of Antigenic Determinants Recognized by Rheumatoid Arthritis-specific Autoantibodies. J Immunol. 2015;195:8–16. [PubMed] [Google Scholar]
- 27.Guzmán-Guzmán IP, Reyes-Castillo Z, Muñoz-Barrios S. et al. Polymorphisms and functional haplotype in PADI4: further evidence for contribution on rheumatoid arthritis susceptibility and anti-cyclic citrullinated peptide antibodies in a western Mexican population. Immunol Lett. 2015;163:214–20. doi: 10.1016/j.imlet.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 28.Kuribayashi K, El-Deiry WS. Regulation of programmed cell death by the p53 pathway. Adv Exp Med Biol. 2008;615:201–21. doi: 10.1007/978-1-4020-6554-5_10. [DOI] [PubMed] [Google Scholar]
- 29.Garner E, Raj K. Protective mechanisms of p53-p21-pRb proteins against DNA damage-induced cell death. Cell Cycle. 2008;7:277–82. doi: 10.4161/cc.7.3.5328. [DOI] [PubMed] [Google Scholar]
- 30.Chang X, Hou X, Pan J. et al. Investigating the pathogenic role of PADI4 in oesophageal cancer. Int J Biol Sci. 2011;7:769–81. doi: 10.7150/ijbs.7.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang X, Han J, Pang L. et al. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang X, Yamada R, Suzuki A. et al. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 2005;44:40–50. doi: 10.1093/rheumatology/keh414. [DOI] [PubMed] [Google Scholar]
- 33.Sharma P, Azebi S, England P. et al. Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLoS Genet. 2012;8:e1002934. doi: 10.1371/journal.pgen.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seri Y, Shoda H, Suzuki A. et al. Peptidylarginine deiminase type 4 deficiency reduced arthritis severity in a glucose-6-phosphate isomerase-induced arthritis model. Sci Rep. 2015;5:13041. doi: 10.1038/srep13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.