Abstract
Background: MicroRNAs (miRNAs) are a class of endogenous, small non-coding RNAs which function as essential posttranscriptional modulators of gene expression tightly involved in a wide range of diseases, including the hepatocellular carcinoma (HCC). Here, the present study was designed to investigate the expression levels and cellular roles of miR-200a in HCC.
Methods: Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was used to detect the expression levels of miR-200a in serums and cell lines. Bioinformation analysis, the luciferase reporter assay, qRT-PCR and western blotting were employed to validate Foxa2 as a direct target gene of miR-200a. Cell proliferation, migration and invasion were assessed to identify whether miR-200a could regulate the biological behaviors of HCC cells by targeting Foxa2.
Results: In this study, a low level of miR-200a was observed in patients' serums and HCC cell lines. Overexpression of miR-200a in HCC cell lines reduced cell proliferation, migration and invasion. In addition, transcription factor forkhead box A2 (Foxa2) was identified as a novel target of miR-200a and downregulated at mRNA and protein levels in miR-200a overexpressed cells. Meanwhile, restoration of Foxa2 significantly reversed the tumor suppressive effects of miR-200a.
Conclusions: These findings indicate that miR-200a regulates the proliferation, migration and invasion of HCC cells by targeting Foxa2, suggesting that miR-200a may function as a potential therapeutic molecular for the diagnosis and treatment of the liver cancer.
Keywords: miR-200a, Foxa2, hepatocellular carcinoma, proliferation, migration, invasion.
Introduction
Hepatocellular carcinoma (HCC) is one of the most malignant tumors with high morbidity and mortality, which ranks as the third leading cause of cancer-related death worldwide at present1,2. In addition, most HCC patients have a poor prognosis due to late diagnosis and lack of effective treatment2,3. Though alpha fetal protein (AFP) is still the most widely used diagnostic marker, its positive rate is only about 70%4. Therefore, it is necessary to identify other specific biomarkers and clarify the molecular mechanism of HCC for the early diagnosis and timely treatment of the disease.
MicroRNAs (miRNAs) are small, noncoding, single-stranded RNAs known to regulate the expression of genes by binding to the 3'-untranslated region (3'-UTR) of target messenger RNA (mRNA) to participate in the regulation of cell growth, differentiation, apoptosis and metastasis of biological processes5, 6. Many studies have demonstrated that microRNAs have played important roles especially in the evolution and progression of HCC by regulating tumor oncogenes or suppressor genes7-10.
MicroRNA-200a (miR-200a), a member of the miR-200 family, has been reported to be dysregulated and function as a tumor inhibitor in the pathogenesis of various cancers, such as pancreatic carcinoma11, renal cell carcinoma12, nasopharyngeal carcinoma13, breast cancer14, ovarian cancer15and so on, including HCC. However, the exact role and molecular mechanism of miR-200a in liver cancer have not been elucidated yet.
In the study, we investigated in the expression and functions of miR-200a in the HCC. We found that miR-200a was decreased in HCC. Furthermore, miR-200a suppressed the growth and metastasis of HCC cells by directly forkhead box A2 (Foxa2) and supplement of Foxa2 significantly reversed the tumor suppressive role of miR-200a in HCC cells. Therefore, our study has revealed a novel target for miR-200a and may facilitate a novel treatment of HCC.
Methods
Patients and participants
Twenty patients with HCC who were treated at the Huashan Hospital affiliated of Fudan University (Shanghai, China) between July 2015 and September 2015 were enrolled into the present study. All the serum samples of HCC patients were collected before and after treatment. We also obtained serum samples from 20 healthy donors who were used as healthy controls. These controls, completely free from any known malignancy or active inflammatory condition, matched with the age, sex and ethnicity of the patients. Blood sample collection obtained informed consent of patients and healthy volunteers. All HCC patients were diagnosed clinically.
Cell culture
The human HCC cell lines (HepG2, Huh7, SMMC-7721 and MHCC-97H) and L02 cells were obtained from the Liver Cancer Institute, Zhongshan Hospital, Fudan University (Shanghai, China). The cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% Penicillin-Streptomycin (Gibco) and incubated at 37°C in a humidified incubator containing 5% CO2 following the manufacturer's instructions.
Lentiviral miR-200a upregulation assay
Lentiviruses expressing miR-200a, or negative control miRNA (miR-NC) were commercially obtained from Genechem (Shanghai, China). To transduce hepatocellular carcinoma cell line (Huh7 cell), lentiviruses were added into culture medium overnight with a multiplicity of infection (MOI) of 10. The next day, the fresh DMEM culture medium was replenished. After the cells were continuously cultured for another 4 to 5 days to stabilize the lentivirus transduction, puromycin was used to screen out the stable cell lines. Then, the transduction efficiency was verified by qRT-PCR.
Luciferase reporter assay
A luciferase reporter assay was performed in order to detect the direct binding of miR-200a to Foxa2. The entire 3′-UTR of human Foxa2 was cloned into the p-MIR-reporter vector (Ambion, Austin, TX, USA) and confirmed by sequencing to form a wild type luciferase reporter vector (Foxa2 WT 3'-UTR). To verify the binding specificity, the sequences of Foxa2 3'-UTR that bound with the miR-200a were mutated (from AGUGUU to UCACAA), and it was also inserted into p-MIR-reporter vector to generate a mutated luciferase reporter vector (Foxa2 Mut3'-UTR). For the luciferase reporter assays, HEK293 cells were plated in 96-well plates and then transiently cotransfected with luciferase reporter vectors with miR-200a-mimic or control microRNA using Lipofectamine 2000. After transfection for 48h, the relative luciferase activity was detected using a dual luciferase reporter assay system (Promega).
RNA extraction and quantitative real-time PCR
RNA was extracted from serum samples using miRNeasy Serum/Plasma kit (Qiagen, Germany) according to the manufacturer instructions. MiRNeasy Mini kit (Qiagen, Germany) was used for the extraction of total RNA from the cells. MiScript SYBR-Green PCR kit (Qiagen, Germany) was performed to detect the mature miR-200a expression in the cells. Expression of cel-miR-39 and RNU6B was used as an endogenous control in serum and cells testing respectively. The mRNA expression of Foxa2 was measured by SYBR-Green qPCR assay (Takara, Dalian, China). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control of Foxa2 mRNA and calculated using the 2-ΔΔCt formula, and then normalized to the internal control. The primers of miR-200a, cel-miR-39 and RNU6B (hsa-mir-200a, MS00003738; cel-miR-39, MS00019789; hsa-RNU6B, MS00033740) were supplied by Qiagen. PCR of Foxa2 was performed with specific primers: forward, 5'-CACCACCAGCCCCACAAA-3'; and reverse, 5'-GGGTAGTGCATCACCTGTTCGT-3'. Primers of GAPDH were as follows: 5'-CGTGGGCCGCCCTAGGCACCA-3' and 5'-TTGGCTTAGGGTTCAGGGGGG-3'.
Protein extraction and western blotting
All cells were washed with PBS for twice and lysed in IP lysis buffer (Beyotime, China) supplemented with protease and phosphatase inhibitor on ice for 30 min. After that, cell lysates were centrifuged for 30 min (12000 g, 4 °C). Then the protein sample was collected and the protein concentration was measured by using BCA protein concentration assay kit (Beyotime, China). Protein samples were diluted to a certain concentration (5 ug/ul) with the lysis buffer. The proteins (50 μg) were separated by 12%SDS-PAGE and transferred to PVDF membranes (Millipore). After being blocked with skimmed milk, the blots were incubated overnight at 4°C with rabbit anti-foxa2 and anti-β-actin. The membranes were then washed and incubated with goat anti-rabbit secondary antibodies for 60 min, and then developed with BeyoECL Plus kit (Beyotime). The antibodies were purchased from the following sources: Anti-Foxa2 (1:1000, Cell Signaling Technology #3143, USA), anti-β-actin (1:1200, Cell Signaling Technology #8457, USA) and goat anti-rabbit secondary antibodies (1:1200, Santa Cruz, sc2030, USA).
Cell proliferation assay
To assess cell proliferation, Huh7 cells were plated in 96-well plates at 5 × 103 cells per well in 100μL of culture medium. The cell proliferation index was measured using the Cell Counting Kit-8 (CCK-8; Diojindo Laboratories, Kumamoto, Japan) at 24, 48, 72, 96 and 120 h after transfection. According to the manufacturer's instructions, each well was treated with 10ul CCK8 solution and incubating for 4 h. The absorbance was measured at 450 nm on Synergy H1 Hybrid Microplate Reader. All experiments were repeated in triplicate.
Cell migration and invasion assay
The migration and invasion of cells were tested by using transwell chambers (Corning 3422, USA). In the invasion assay, filters were precoated with Matrigel (BD Biosciences). The cells (5×105 cells/200ul) were seeded in the top chamber and the bottom chamber was filled with 600ul DMEM containing 20%FBS. Cells that remained on the bottom surface of the membranes were fixed in methanol for 30 min and stained with 0.5 % crystal violet for 20 min after incubation for 24-48 h. Four random fields were counted for each group.
Statistical analysis
All of the western blotting images are representative of at least three independent experiments. Quantitative RT-PCR, the luciferase reporter, the cell proliferation, migration and invasion assays were performed in triplicate. The data are presented as the mean ± SD and all results are analyzed by using Stata 11.0 software. P value less than 0.05 is considered statistically significant.
Results
Clinical features of study participants
The characteristics of 20 HCC patients and 20 healthy volunteers were presented in Table 1. There was no significant difference in the distribution of age, sex and alanine aminotransferase (ALT) except viral infection and AFP between HCC patients and healthy people. In addition, Table 1 also shows that HCC was more prevalent in males than in females though few cases were studied in the group.
Table 1.
Clinicopathological characteristics of study participants. *** P<0.001.
| Pathology characteristics | HCC | Health | P value | χ2 | |
|---|---|---|---|---|---|
| Total | 20 | 20 | |||
| Gender | male | 15 | 12 | 0.3110 | 1.0256 |
| Female | 5 | 8 | |||
| Age (years old) | <55 | 12 | 10 | 0.4040 | 0.5250 |
| ≥55 | 8 | 10 | |||
| ALT (U/L) | <40 | 18 | 20 | 0.1470 | 2.1053 |
| ≥40 | 2 | 0 | |||
| Viral infection (HBV or HCV) |
Positive | 11 | 0 | <0.001*** | 15.1724 |
| Negative | 9 | 20 | |||
| AFP serum level (ng/ml) |
0-20 | 10 | 20 | <0.001*** | 13.3330 |
| >20 | 10 | 0 |
MiR-200a is downregulated in HCC serum and cell lines
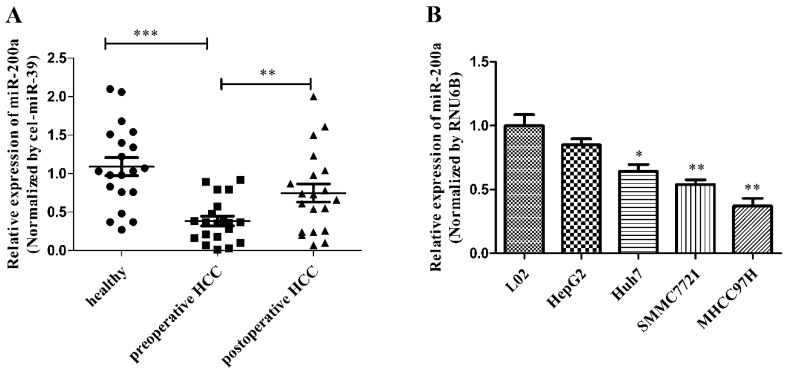
We measured the expression of circulating miR-200a in 20 HCC patients before and after resection and 20 healthy subjects using quantitative real-time polymerase chain reaction (qRT-PCR) with the endogenous control (cel-miR-39). The levels of circulating miR-200a expression were markedly decreased in the serum from preoperative patients vs healthy subjects. In addition, the expression level of miR-200a in the serum from the postoperative patients increased significantly compared with the preoperative serum (Fig.1A). We further analyzed whether the preoperative serum level of miR-200a was correlated with clinicopathological factors in 20 patients with HCC (Table 2). The results showed that large tumor sizes of more than 5 cm, TNM stage (III~IV), poor differentiation, the presence of metastasis and recurrences were significantly correlated with low levels of serum miR-200a (P=0.0327, 0.0463, 0.0190, 0.0190 and <0.001, respectively), whereas other clinical factors, such as gender, age, viral infection and liver cirrhosis were not correlated with serum miR-200a levels. The significant clinicopathological factors in univariate analysis were enrolled into a cox model for multivariate analysis, and metastasis and recurrence were identified as independent risk factors for the clinical relevance of miR-200a (Table 3). The above results indicate that miR-200a expression can be detected in the blood samples and may be used as a potential biomarker. To further determine the expression level of miR-200a in cell lines, we also investigated expression of miR-200a in 4 HCC cell lines (HepG2, Huh7, SMMC-7721 and MHCC-97H) and the hepatocyte cell line (L02). The expression of miR-200a in HCC cell lines was significantly downregulated compared with L02 cells (Fig.1B). Among them, Huh7 cell was selected for subsequent investigation. These data strongly suggest that miR-200a may be involved in the occurrence and development of HCC.
Figure 1.
MiR-200a was decreased in HCC serum and cell lines. (A) The levels of circulating miR-200a in serum were detected by qRT-PCR analysis. The relative expression levels of miR-200a in serum were qualified by using a spiked-in cel-miR-39 as an internal control. (B) Relative expression levels of miR-200a in 4 human HCC cell lines (HepG2, Huh7, SMMC-7721 and MHCC-97H), compared to the normal human hepatocyte (L02). Data represent Mean ± SD of 3 replicates. *P<0.05, **P<0.01, ***P<0.001, compared with the negative control (the negative control was preoperative HCC in figure 1A, the negative control was L02 cell in figure 1B).
Table 2.
Relationship between the relative expression of preoperative serum miR-200a and clinicopathological features. *P<0.05, ***P<0.001.
| Pathology characteristics |
Case | Preoperative serum miR-200a | ||
|---|---|---|---|---|
| Median | P value | |||
| Total | 20 | |||
| Gender | male | 15 | 0.350 | 0.1374 |
| Female | 5 | 0.480 | ||
| Age (years old) | <55 | 12 | 0.365 | 0.9378 |
| ≥55 | 8 | 0.325 | ||
| ALT (U/L) | <40 | 18 | 0.360 | 0.1225 |
| ≥40 | 2 | 0.625 | ||
| Viral infection (HBV or HCV) |
Positive | 11 | 0.370 | 0.9326 |
| Negative | 9 | 0.360 | ||
| AFP serum level (ng/ml) |
0-20 | 10 | 0.375 | 0.7245 |
| >20 | 10 | 0.265 | ||
| Liver cirrhosis | Positive | 11 | 0.370 | 0.8355 |
| Negative | 9 | 0.360 | ||
| Tumor size (cm) | <5 | 14 | 0.380 | 0.0327* |
| ≥5 | 6 | 0.140 | ||
| TNM stage | 0~II | 13 | 0.380 | 0.0463* |
| III~IV | 7 | 0.180 | ||
| Differentiation |
Middle or well | 13 | 0.380 | 0.0190* |
| Poor | 7 | 0.100 | ||
| Metastasis | Positive | 7 | 0.100 | 0.0190* |
| Negative | 13 | 0.380 | ||
| Recurrence | Positive | 3 | 0.070 | <0.001*** |
| Negative | 17 | 0.370 | ||
Table 3.
Multivariate analysis of clinical factors associated with preoperative serum miR-200a. *P<0.05.
| Factors | B | SE | Wald | df | P | EXP(B) | 95% CI | |
|---|---|---|---|---|---|---|---|---|
| Upper limit | Lower limit | |||||||
| Tumor size: >5 cm vs ≤5 cm | -1.643 | 0.078 | 1.004 | 1 | 0.063 | 0.193 | 0.518 | 0.212 |
| TNM stage: 0-II vs III-IV | -1.159 | 0.115 | 0.012 | 1 | 0.120 | 0.314 | 0.590 | 0.223 |
| Differentiation: middle vs poor | 0.715 | 0.018 | 2.054 | 1 | 0.051 | 2.044 | 0.401 | 0.035 |
| Metastasis: yes vs no | -0.536 | 0.072 | 4.351 | 1 | 0.032* | 0.029 | 0.506 | 0.141 |
| Recurrence: yes vs no | -2.285 | 0.034 | 3.008 | 1 | 0.028* | 0.001 | 0.432 | 0.067 |
Overexpression of miR-200a inhibits cell proliferation, migration and invasion in HCC cells
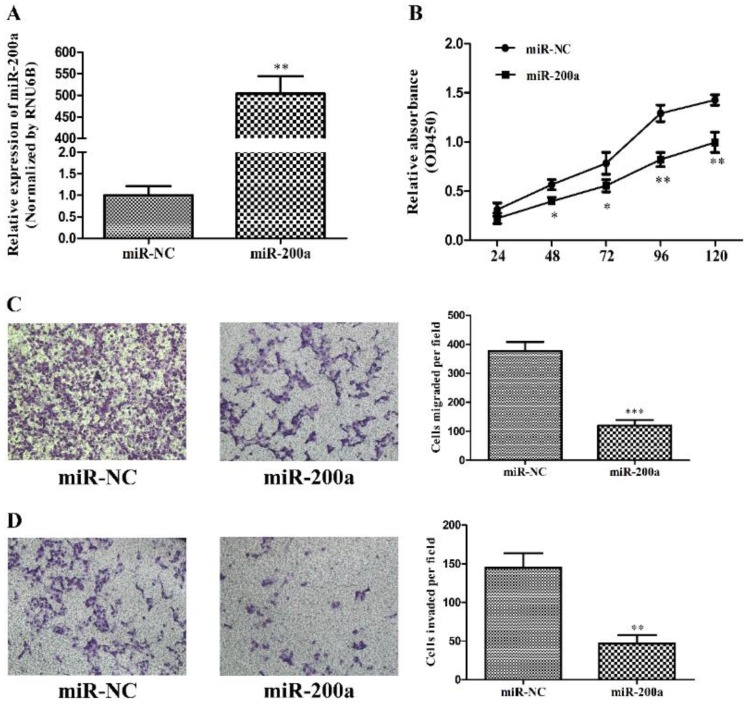
To investigate the effects of miR-200a on tumor biology, stable cell lines expressing miR-200a (Huh7-miR-200a) and its negative control (Huh7-miR-NC) were established by lentiviral transfection, which was validated by qRT-PCR (Fig.2A). Cell proliferation examined with CCK-8 assay demonstrated that overexpression of miR-200a markedly attenuated cell viability of the Huh7 cells as compared with those transfected with the negative control from 48h to 120h (Fig. 2B). The effects of miR-200a on the migration and invasion of HCC cells were analyzed initially in vitro using transwell assays. The results showed that both the migratory and invasive abilities of the HCC cells were suppressed by miR-200a overexpression when compared to the control (Fig. 2C and D). Collectively, the in vitro studies suggest the suppressive effects of miR-200a on tumor cell proliferation, migration and invasion in HCC.
Figure 2.
MiR-200a inhibited proliferation, migration and invasion of HCC cells. (A) Huh7 cells were transfected with miR-200a or negative control lentivirus, and qRT-PCR was performed to determine the expression of miR-200a. (B) Huh7 cells were transfected with miR-200a or negative control lentivirus, and proliferation ability of cells was detected by CCK8 testing kit. (C, D) Transwell assays of Huh7 cells transfected with miR-200a or negative control lentivirus were used to determine the migration and invasion (magnification×100, Nikon Te2000). Experiments were performed in triplicate. *P<0.05, **P<0.01, ***P<0.001, compared with the negative control (the negative control was Huh7 cells transfected with negative lentivirus in figure 2)
Identification of conserved miR-200a target sites within the 3'-UTR of Foxa2
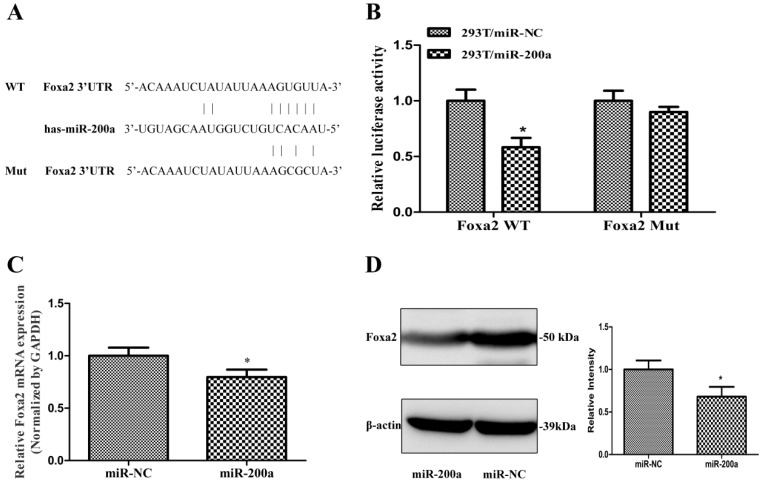
To further understanding the molecular mechanism of miR-200a in regulating HCC growth and metastasis, we used three publicly available algorithm (Targetscan, miRanda and PicTar), and found that miR-200a was identified as a candidate miRNA that could target Foxa2. The sequence of the predicted miR-200a binding sites and the Foxa2 3'UTR segments containing the miR-200a complementary sequence are shown in Fig. 3A. Luciferase activity assay demonstrated that miR-200a significantly inhibited the wild type (WT) but not the mutant (MUT) 3'-UTR of Foxa2 luciferase activity (Fig. 3B). Furthermore, qRT-PCR and western blotting assay were verified that overexpression of miR-200a markedly suppressed the endogenous mRNA and protein levels of Foxa2 respectively (P<0.05; Fig. 3C and 3D). These data indicate that miR-200a may negatively regulate the transcription of Foxa2 expression by directly targeting its 3'UTR.
Figure 3.
Foxa2 was predicted and confirmed as a potential target of miR-200a in HCC cells. (A)Schematic description of wild-type (WT) and mutant (MUT) 3′-UTR of the human Foxa2. (B) Analysis of luciferase activity results. HEK293 cells were cotransfected with wild-type or mutant 3′-UTR of Foxa2 and miR-200a or the control mimics. The luciferase activities were examined 48h after transfection. (C) Huh7 cells were transfected with miR-200a or the negative control lentivirus. QRT-PCR was performed to observe the mRNA change of Foxa2. (D)Western blotting was analyzed to detect the protein level of Foxa2. *P<0.05, compared with the negative control (the negative control was HEK293 cells transfected with the control mimics in Figure 3B, the negative control was Huh7 cells transfected with negative lentivirus in Figure 3C and 3D).
Overexpression of Foxa2 reverses the effects of miR-200a on HCC cells
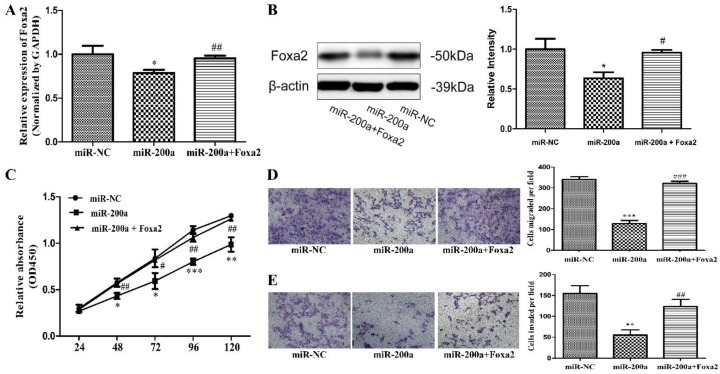
To further confirm Foxa2 was involved in the function of miR-200a, stable cell lines expressing miR-200a (Huh7/miR-200a) were transfected with Foxa2 overexpression plasmid. The expression efficiency of Foxa2 was examined by qRT-PCR and western blotting. The mRNA level and protein level of Foxa2 were both significantly restored in Foxa2 overexpression plasmid-transfected Huh7/miR-200a cells, compared with its negative control (Fig. 4A, 4B). Through analyzing the changes of cell proliferation, migration, and invasion, we found that overexpression of Foxa2 significantly attenuated the miR-200a-mediated tumor suppressive effects (Fig. 4C, D, E). The above results imply that miR-200a serves as a tumor inhibitor mediated by Foxa2.
Figure 4.
Overexpression of Foxa2 attenuated the effects of miR-200a. (A) stable cell lines expressing miR-200a (Huh7/miR-200a) were transfected with Foxa2 overexpression plasmid, and the mRNA level of Foxa2 was examined by qRT-PCR. (B) the protein level of Foxa2 was analyzed by western blotting. (C) up-regulation of Foxa2 in Huh7/miR-200a restored partly the ability of cell proliferation by CCK8 assay. (D, E) migration and invasion assays were performed (magnification×100, Nikon Te2000). The experiments were repeated three times. *P<0.05, **P<0.01, ***P<0.001, compared with the negative control (Huh7 cells transfected with negative lentivirus). #P<0.05, ##P<0.01, ###P<0.001, compared with the negative control (Huh7 cells transfected with miR-200a lentivirus).
Discussion
HCC is one of the most common causes of death from cancer. Its 5-year post-surgery survival rate is very low16. This poor prognosis is mainly due to the early metastasis and invasion of HCC, which result in an incurable stage most patients' condition entered 17, 18. The molecular mechanisms underlying the metastasis and invasion of HCC remain to be explored. Recently, a large number of studies have reported that many miRNAs could participate in the occurrence and development process of liver cancer19, 20. Among them, microRNA-200a has been found to be decreased in the liver cancer and play a critical role as tumor suppressor21-23. However, the specific mechanisms related with the role of miR-200a in HCC process are still not fully elucidated. In our study, we have for the first time demonstrated that Foxa2 gene was a direct downstream target gene of miR-200a in HCC.
To test the hypothesis that miR-200a could play an important role in suppressing the metastasis and invasion of HCC cells, we've first analyzed the level of miR-200a expression in the serums and cell lines. It was found that miR-200a was significantly decreased in the preoperative serum of HCC patients compared with the postoperative of the same patients and the healthy people. Furthermore, we investigated the relationship between preoperative serum miR-200a expression of HCC patients and their clinical characteristics. MiR-200a expression level in serum was closely associated with tumor size, tumor stage, differentiation, metastasis and recurrence. Multivariate analysis identified that metastasis and recurrence were independent risk factors for miR-200a. Because liver cancer tissues were difficult to be collected, we did not measure the expression level of miR-200a in tissues. But many literatures have reported that miR-200a is markedly decreased in the HCC21, 24, 25, consistent with the expected results. It shows that low expression of miR-200a in HCC tissues release less into the peripheral blood, resulting in the low expression of serum miR-200a correspondingly. Moreover, we investigated expression of mi-200a in HCC cell lines and the normal liver cell line (L02). The expression level of miR-200a in 4 kinds of HCC cell lines (HepG2, Huh7, SMMC-7721 and MHCC97H) was lower than the normal liver epithelial cell (LO2). In addition, we further researched the functional characteristics of one HCC cell line. Overexpression of miR-200a could suppress HCC cell proliferation, migration and invasion, which is consistent with prior researches 22. These data suggest that miR-200a may function as a potentially crucial tumor suppressor and the malignancy of liver cancer is restrained by high expression of miR-200a.
It has been well known that miRNAs regulate target gene expression at the posttranscriptional level through both translational inhibition and mRNA degradation. Foxa2 is a member of the fork head transcription factor family which is widely involved in the regulation of cell proliferation and differentiation26, 27. Prior studies have showed that Foxa2 may be abnormally expressed in a number of cancers, which sequently lead to the development of tumors24, 28, 29. A number of miRNAs have been shown to be able to regulate Foxa2 expression such as miR-1291, miR-29 and miR-124a27, 30, 31. But the potential relationship of miR-200a targeting Foxa2 in human hepatocellular carcinoma have not been studied before. In this study, we performed bioinformation software analysis, the luciferase reporter assay, qRT-PCR and western blotting to identify that miR-200a could combine to Foxa2 3'UTR sites and overexpression of miR-200a in Huh7 cell caused a reduction of Foxa2 mRNA and protein. In addition, introducing Foxa2 in miR-200a overexpressed cells could partially reverse miR-200a-mediated tumor suppressive effects. Therefore, it can be inferred that miR-200a may function as a potential therapeutic biomarker for HCC diagnosis.
This study has some potential limitations. In our research, we've just employed serums and the only one cell line to identify that miR-200a regulate the growth and metastasis of HCC by targeting the Foxa2 gene. It is felt that we should have confirmed the relationship between the expression status of miR-200a and the development of HCC on clinical tissues, more cell lines and in vivo experiments, which is one limitation of our study. Another limitation is that, though our results showed that miR-200a inhibited cell growth and metastasis through targeting Foxa2 expression, our study did not exclude specific downstream signal pathways that may modulate cell migration and invasion. Thus, in the following, we are going to overcome the attendant limitations of the present study.
Conclusions
Generally speaking, the results of our research demonstrate that miR-200a is frequently down-regulated in HCC patients' serum and HCC cells, which is likely to be a potential biomarker for diagnosis with an important role as a tumor suppressor in HCC. Decreased expression of miR-200a in HCC cells might lead to aberrant cell proliferation, migration and invasion by regulating the expression of Foxa2 directly, and subsequently contribute to the deterioration of liver cancer. All these results help to further clarifying the mechanisms in the pathogenesis of liver cancer. More importantly, miR-200a might be a novel molecular target for the therapy of HCC.
Acknowledgments
We thank Huashan Hospital, Fudan University, Shanghai, China for providing the HCC serum samples and Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai, China for providing cell lines.
Funding
This work was supported by the National Natural Science Foundation of China (81472224), the National Key Basic Research Program (973 project) from the Ministry of Science and Technology of China (2015CB554005), the Leading Investigator Program of Shanghai municipal government (2016), the National Major Scientific Research Program of China (201526) and the Youth Clinical Medical Technical Talents Training Program of Shanghai (201601).
Ethics approval and consent to participate
All subjects gave informed consent to the study (molecular characterization of liver tumors obtained after curative resection and clinicopathological data associated with the study). The study was approved by the ethics committee of Huashan Hospital, Fudan University.
Abbreviations
- HCC
hepatocellular carcinoma
- QRT-PCR
quantitative reverse-transcription polymerase chain reaction
- Foxa2
forkhead box A2
- AFP
alpha fetal protein
- miRNAs
microRNAs
- 3'-UTR
3'-untranslated region
- mRNA
messenger RNA
- DMEM
dulbecco's modified eagle's medium
- FBS
fetal bovine serum
- ALT
alanine aminotransferase
References
- 1.Lamarca A, Mendiola M, Barriuso J. Hepatocellular carcinoma: Exploring the impact of ethnicity on molecular biology. Critical reviews in oncology/hematology. 2016;105:65–72. doi: 10.1016/j.critrevonc.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nature reviews Cancer. 2006;6:674–87. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 3.Yin CQ, Yuan CH, Qu Z, Guan Q, Chen H, Wang FB. Liquid Biopsy of Hepatocellular Carcinoma: Circulating Tumor-Derived Biomarkers. Disease markers. 2016;2016:1427849. doi: 10.1155/2016/1427849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reichl P, Mikulits W. Accuracy of novel diagnostic biomarkers for hepatocellular carcinoma: An update for clinicians (Review) Oncology reports. 2016;36:613–25. doi: 10.3892/or.2016.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E. et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nature medicine. 2015;21:1280–9. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao S, Tian C, Ding Y, Ye Q, Gao Y, Yang N, Down-reguation of Kruppel-like factor-4 by microRNA-135a-5p promotes proliferation and metastasis in hepatocellular carcinoma by transforming growth factor-beta1. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW. et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q. et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. The Lancet Oncology. 2015;16:804–15. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhao N, Li S, Wang R, Xiao M, Meng Y, Zeng C. et al. Expression of microRNA-195 is transactivated by Sp1 but inhibited by histone deacetylase 3 in hepatocellular carcinoma cells. Biochimica et biophysica acta. 2016;1859:933–42. doi: 10.1016/j.bbagrm.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Wu G, Wu Z, Yao X, Li G. MiR-200a Suppresses the Proliferation and Metastasis in Pancreatic Ductal Adenocarcinoma through Downregulation of DEK Gene. Translational oncology. 2016;9:25–31. doi: 10.1016/j.tranon.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Jiang F, Song H, Li X, Xian J, Gu X. MicroRNA-200a-3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma. Biochemical and biophysical research communications. 2016;470:620–6. doi: 10.1016/j.bbrc.2016.01.095. [DOI] [PubMed] [Google Scholar]
- 13.Shi Z, Hu Z, Chen D, Huang J, Fan J, Zhou S. et al. MicroRNA-200a mediates nasopharyngeal carcinoma cell proliferation through the activation of nuclear factor-kappaB. Molecular medicine reports. 2016;13:1732–8. doi: 10.3892/mmr.2015.4738. [DOI] [PubMed] [Google Scholar]
- 14.Ming J, Zhou Y, Du J, Fan S, Pan B, Wang Y, Identification of miR-200a as a novel suppressor of connexin 43 in breast cancer cells. Bioscience reports; 2015. p. 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuberi M, Mir R, Das J, Ahmad I, Javid J, Yadav P. et al. Erratum to: Expression of serum miR-200a, miR-200b and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2015;17:840. doi: 10.1007/s12094-015-1355-2. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Wang S, Ma W, Lu Y. Transforming Growth Factor beta1 Promotes Migration and Invasion of Human Hepatocellular Carcinoma Cells Via Up-Regulation of Connective Tissue Growth Factor. Cell biochemistry and biophysics. 2015;73:775–81. doi: 10.1007/s12013-015-0693-6. [DOI] [PubMed] [Google Scholar]
- 17.Jiang YF, Yang ZH, Hu JQ. Recurrence or metastasis of HCC:predictors, early detection and experimental antiangiogenic therapy. World journal of gastroenterology. 2000;6:61–5. doi: 10.3748/wjg.v6.i1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Di C, Li W, Cai W, Tan X, Xu L. et al. Oncomirs miRNA-221/222 and Tumor Suppressors miRNA-199a/195 Are Crucial miRNAs in Liver Cancer: A Systematic Analysis. Digestive diseases and sciences. 2016;61:2315–27. doi: 10.1007/s10620-016-4156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han K, Li J, Zhao H, Liang P, Huang X, Zheng L. et al. Identification of the typical miRNAs and target genes in hepatocellular carcinoma. Molecular medicine reports. 2014;10:229–35. doi: 10.3892/mmr.2014.2194. [DOI] [PubMed] [Google Scholar]
- 20.Afonso MB, Rodrigues PM, Simao AL, Castro RE. Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. Journal of clinical medicine; 2016. p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng J, Wang J, Chen M, Chen G, Wu Z, Ying L. et al. miR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncology reports. 2015;33:713–20. doi: 10.3892/or.2014.3642. [DOI] [PubMed] [Google Scholar]
- 22.Zhong C, Li MY, Chen ZY, Cheng HK, Hu ML, Ruan YL. et al. MicroRNA-200a inhibits epithelial-mesenchymal transition in human hepatocellular carcinoma cell line. International journal of clinical and experimental pathology. 2015;8:9922–31. [PMC free article] [PubMed] [Google Scholar]
- 23.Hung CS, Liu HH, Liu JJ, Yeh CT, Chang TC, Wu CH. et al. MicroRNA-200a and -200b mediated hepatocellular carcinoma cell migration through the epithelial to mesenchymal transition markers. Annals of surgical oncology. 2013;20(Suppl 3):S360–8. doi: 10.1245/s10434-012-2482-4. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Wang J, Qu S, Zhang H, Ruan B, Gao Y. et al. MicroRNA-200a suppresses metastatic potential of side population cells in human hepatocellular carcinoma by decreasing ZEB2. Oncotarget. 2015;6:7918–29. doi: 10.18632/oncotarget.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhayat SA, Mardin WA, Kohler G, Bahde R, Vowinkel T, Wolters H. et al. The microRNA-200 family-a potential diagnostic marker in hepatocellular carcinoma? Journal of surgical oncology. 2014;110:430–8. doi: 10.1002/jso.23668. [DOI] [PubMed] [Google Scholar]
- 26.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer research. 2010;70:2115–25. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu MJ, Pan YZ, Qiu JX, Kim EJ, Yu AM. MicroRNA-1291 targets the FOXA2-AGR2 pathway to suppress pancreatic cancer cell proliferation and tumorigenesis. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton G, Hochmair M, Rath B, Klameth L, Zeillinger R. Small cell lung cancer: Circulating tumor cells of extended stage patients express a mesenchymal-epithelial transition phenotype. Cell adhesion & migration; 2016. pp. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li CM, Gocheva V, Oudin MJ, Bhutkar A, Wang SY, Date SR. et al. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes & development. 2015;29:1850–62. doi: 10.1101/gad.267393.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtz CL, Peck BC, Fannin EE, Beysen C, Miao J, Landstreet SR. et al. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes. 2014;63:3141–8. doi: 10.2337/db13-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing G, Westwell-Roper C, Chen J, Xu G, Verchere CB, Shalev A. Thioredoxin-interacting protein promotes islet amyloid polypeptide expression through miR-124a and FoxA2. The Journal of biological chemistry. 2014;289:11807–15. doi: 10.1074/jbc.M113.525022. [DOI] [PMC free article] [PubMed] [Google Scholar]