Abstract
Purpose
Chemical-shift imaging (CSI) has long been considered the gold standard method for in-vivo hyperpolarized 13C metabolite imaging because of its high sensitivity. However, CSI requires a large number of excitations so it is desirable to reduce the number of RF excitations and the total acquisition time.
Methods
Centric phase encoding and 3D compressed sensing methods were adopted into a CSI acquisition to improve efficiency and reduce the number of excitations required for imaging hyperpolarized metabolites. The new method was implemented on a GE MR750W scanner for routine real time metabolic imaging experiments.
Results
Imaging results from phantoms and in-vivo animals using hyperpolarized 13C tracers demonstrate that when the entire CSI data set is treated as a single object, compressed sensing can be satisfactorily applied to spectroscopic CSI. Centric k-space trajectory data collection also greatly improves the acquisition efficiency. This combination of compressed sensing CSI and acquisition time reduction was used to perform a hyperpolarized 13C dynamic study.
Discussion
Compressed sensing can be satisfactorily applied to conventional CSI in hyperpolarized 13C metabolite MR imaging to reduce the number of RF excitations and accelerate the imaging speed to take advantage of conventional CSI in providing high sensitivity and a large spectral bandwidth.
INTRODUCTION
Imaging metabolism of hyperpolarized (HP) 13C-enriched substrates offers a new approach to molecular imaging in experimental animals and patients [1, 2]. Studies have shown the potential of hyperpolarized 13C metabolic imaging in cardiac, cancer and kidney diseases [3–5]. Chemical shift imaging (CSI) has been considered the gold standard method for in vivo metabolite imaging and has been widely applied for hyperpolarized 13C metabolic studies [1–5] because images can be easily generated from metabolite resonances that partially overlap with other resonances or have relatively low intensity by using common peak fitting or decomposition methods. Compared to accelerated CSI [12–14], conventional CSI has many advantages including a wide spectral bandwidth for detecting multiple metabolites with different chemical shifts and a relatively large number of acquisition points. One of the limitations of hyperpolarized 13C metabolite imaging is the nonrenewable 13C polarization and the relatively short T1 of the hyperpolarized materials. This is particularly true for CSI since it requires RF excitation for each phase encoding. For regular 2D especially 3D CSI, the number of RF excitations can become quite large. For these reasons, it is desirable to reduce the number of RF excitations for CSI of hyperpolarized 13C substrates as well as to reduce the imaging time. One possible method is to apply compressed sensing. Compressed Sensing (CS) was first developed for signal reconstruction from incomplete information [6] then further adapted by Lustig et al [7] to reduce the MRI scan time. In this study, we have applied the compressed sensing method to conventional 2D CSI for imaging of hyperpolarized 13C-enriched metabolites with exploratory phantoms and in vivo applications.
Compressed sensing requires that the MR image is compressible and the image artifact caused by k-space under-sampling is incoherent. When applying compressed sensing to conventional 2D CSI for example, the later can be realized with optimized random under-sampling pattern in both phase encoding directions; the first condition is not necessarily always achievable due to the nature of the structure of the spectra. To overcome this problem, Wang [8] demonstrated that if the entire 3-dimensional (3D) MRI set is treated as a single unit with all transform operators formed in intrinsic 3D, the sparse structure of the entire 3D subject can normally be satisfied. It was demonstrated that 3D CS provides better accuracy and optimization efficiency over slice-by-slice CS reconstruction for 3D MR images. In this work, we have adopted the principle of 3D compressed sensing into CSI. The real-time under sampling CSI sequence was implemented on the MRI scanner and tested in vivo in rats. CSI of the rat kidney was chosen not only because kidney shows better 13C metabolite contrast for CS evaluation, but also because HP 13C has been applied in various kidney diseases [4, 5]. The results demonstrate that when the entire CSI data set is treated as a single object, compressed sensing can be satisfactorily applied to spectroscopic CSI.
METHODS
Random Under-Sampling Pulse Sequence for 13C CSI
To implement compressed sensing for CSI in our MRI scanner, the starting point was a conventional 2D CSI sequence. The phase encoding was reordered to start from the center of k-space and spiral out to the edge in order to preserve the highest HP 13C signal at the center of k-space. As most of the omitted phase encodings are located in the outer k-space region, the artifact caused by reduction will be minimal thereby making the CS reconstruction more efficient. Random under-sampling patterns were generated in the 2D phase encoding directions. After a 2D random matrix was generated, the threshold for different reduction ratios was set by a 2D Gaussian plus a flat plane. Therefore the center of k-space is weighted higher than the edge [9]. The data acquisition on the frequency or spectroscopic direction was kept intact in order to take advantage of conventional CSI of having a large spectral bandwidth and accurate spectral interpolation by zero filling. During the CSI scan, the sequence executes only phase encodings marked ON according to the pattern. Because of this real time reduction in total scan time a portion of the HP 13C signal remains even after a 2D CSI. Therefore, multiple sets of CSI can be performed through “multi-phase” options implanted in the sequence for dynamic studies. Proton anatomy reference images were acquired using a GRE sequence with a field of view (FOV) of 8 cm, 256×256 matrix, 3mm slice thickness, and 6 averages. The 13C 2D-CSI spectra were acquired with the implemented CSI pulse sequence using the same FOV as the proton reference image. 16 × 16 phase encoding was used for this experiment.
Data Processing and Analysis
The hyperpolarized 13C CSI raw data were processed offline. The un-acquired phase encoding readouts were given values of zero as initial values for the compressed sensing optimization processing. All variables and operators in the optimization process are in 3D format for full 3D compressed sensing reconstruction. This insures that the entire “3D image” is compressible and treated as unity. Figure 1 shows the proton reference image of the phantom, a reduction matrix (R=2.5) and a symbolic 3D Wavelet object. After the compressed sensing reconstruction, 16×16 spectra were analyzed with the singular value decomposition (SVD) based decomposition method [10–11]. The intensity of each metabolite from each CSI voxel was then quantified. A 16×16 low resolution image for every metabolite was then interpolated using the Fourier method to match the corresponding 1H anatomy image and overlapped onto the 1H anatomy image.
Figure 1.

left: proton image of the phantom containing 13C-enriched lactate, alanine, formic acid and bicarbonate in 4 individual cylinders; middle: sparse phase encoding matrix with R=2.5; right: symbolic 3D Wavelet objects used in 3D compressed sensing process.
Phantom and Feasibility Study
For the feasibility and validation study, a phantom containing 4 metabolites, [1-13C]lactate, [1-13C]alanine, [1-13C]formate and [1-13C]bicarbonate, was used as shown in Figure 1a. Each metabolite was contained in a cylinder 40mm in length and 10mm to 15mm in diameter. Each metabolite chamber has different diameter for intuitive identification on the 1H and 13C images. 16×16 spatial phase encoding with spectrum length of 256 points and spectral bandwidth of 2 KHz were fully acquired. Sparseness of the k-space data was applied in two phase encoding directions to obtain a total reduction rate of 10X, 5x, 3x, 2.5x and 2x corresponding to 10%, 20%, 33%, 40% and 50% of fully sampled data respectively by zeroing the designated phase encoding data after the scan. The fully acquired data were reconstructed normally for reference purposes. The quality of the compressed sensing reconstruction for each reduction rate was quantified by normalized root of mean square error (nRMSE):
| (1) |
where yi and ŷi are the CS reconstructed and the reference CSI element i respectively; n is the total element.
Animal Preparation
Sprague Dawley rats (200–300g) fed a normal rat chow diet were imaged 7–10 days after arrival. During the imaging experiment, each rat was placed in a 75 mm birdcage coil at the isocenter of the MR scanner. A catheter was placed into the tail vein for the intravenous administration of hyperpolarized [1-13C]pyruvate. Anesthesia was continued with a constant delivery of isoflurane (2%–3%) through a long tube to a cone placed over the rat’s nose and mouth while the rat was in the scanner. The respiration and temperature of the rat were continuously monitored. The body temperature was maintained at 37°C throughout the imaging procedures by maintaining a flow of heated air through the coil cylinder.
Polarization Procedure
A mixture of 22.8 μl of neat [1-13C]pyruvic acid (ρ=1.25g/ml, 89.06g/mol) containing 15 mM OX63 trityl radical was hyperpolarized in a homemade DNP polarizer in a field of 4.6 T at approximately 1.2 K by irradiation with 129GHz microwaves. After approximately 60 minutes of microwave irradiation, the hyperpolarized pyruvic acid was rapidly dissolved with a phosphate buffered saline heated to 37°C at 10 bars to yield a ~ 4mL solution at pH ~7.4. This yielded a final pyruvate concentration of 80mM (0.0228ml*1.25g/ml/89.06g/mol/4ml=80mM) with ~15% polarization. Approximately 2.5 mL of this solution was then injected into the tail vein of a rat within 10 seconds after removal from the polarizer. The injection lasted ~10 seconds.
1H and 13C MR Imaging
Experiments were performed in a 3T GE MR750W scanner (GE Healthcare, Milwaukee, Wisconsin) equipped with the multinuclear spectroscopy hardware upgrade. The RF coil used in these experiments was a GE Healthcare 1H/13C dual-tuned birdcage coil. A high-resolution axial anatomical 1H image was obtained following the localization scan. Two-dimensional (2D) 13C imaging was conducted with a 16 × 16 Chemical-Shift Imaging (CSI) with centric k-space encoding, 10° flip angle RF excitation, 15 mm axial slice thickness to cover both kidneys, 2.5 kHz spectral bandwidth (BW) and 256 spectral points. An 8cm FOV gave an in-plane resolution of 5×5 mm. Data acquisition started 10s after injection of hyperpolarized [1-13C]-pyruvate is completed. With TR=125ms, a fully sampled CSI scan takes 32 seconds. Hyperpolarized [1-13C]pyruvate was injected into the rat via tail vein. Five sampling rates, 100%, 50%, 33%, 25% and 10% which correspond to a reduction factor of R=1, 2, 3, 4 and 10 on five individual rats were scanned, one rat for each sampling rate. The acquisition time for each scan was 32s, 16s, 11s, 8s and 3.3s respectively. With the real-time saving in time and excitations, multiphase CSI were performed for R=3 & 4 for the dynamic study. The same phase encoding under-sampling pattern was applied for each series of multiphase CSI. The fully sampled rat serves the purpose of extra quality validation as in-vivo images along with the phantom images. The under sampled data sets with different reduction ratio served to demonstrate the feasibility of the 3D CS on real-time sparsely acquired data, even without reference for each individually reduced CSI data.
RESULTS
Phantom experiments
The reconstructed metabolite images ([1-13C]lactate, [1-13C]alanine, 2 peaks of [1-13C]formic acid and [1-13C]bicarbonate) with the 3D compressed sensing are shown in Figure 2. The compressed sensing reconstructed CSI were essentially identical to the reference image for reduction factors R less than or equal to 3. Minor artifacts appeared with reduction factors R = 5 but theses reflect significant time and excitation savings for a CSI acquisition. In the R = 10 case (10% sampling rate), the errors became larger and artifacts appeared. However the phantom metabolite features were still identifiable although noise was noticeable in the reconstructed images. The normalized root mean square error (nRMSE), as a function of sampling rate from the validation study with the phantom is shown in Figure 3. Low sampling rates, R = 5 or R = 10 (corresponding to 10 or 20% acquisition of total available data), are associated with increased error.
Figure 2.
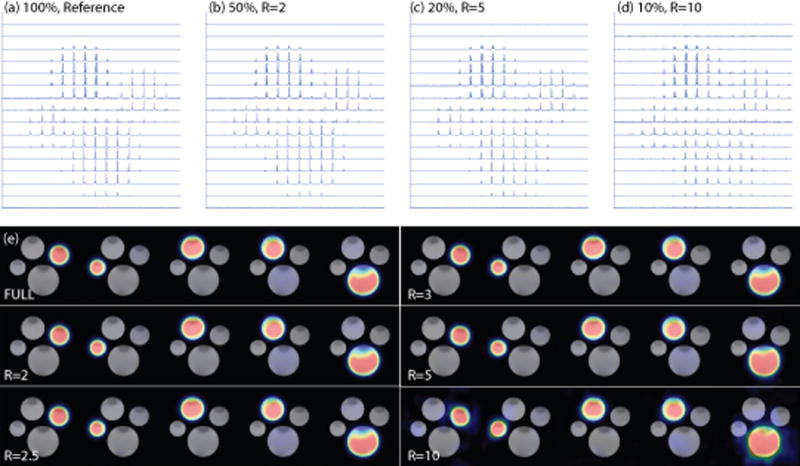
Compressed sensing reconstructed CSI and metabolite images of the phantom containing four different 13C-enriched substrates. a) to d) show reconstructed CSI images in spectroscopic form with 100%, 50%, 20% and 10% sampling rates. Noise artifacts became noticeable for R=5 and became more severe in R=10 case. e) shows the reconstructed metabolite images, from left to right: lactate, alanine, formate-1, formate-2 and bicarbonate (refer to Figure 1). Although noisy, the metabolite subjects are well distinguished in the R=10 images.
Figure 3.
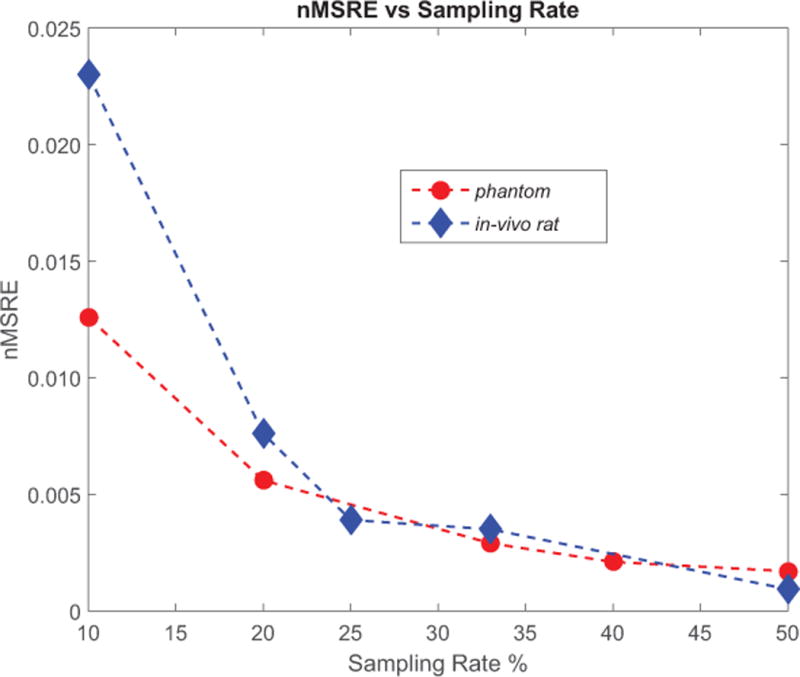
Normalized root-of-mean-square error (nRMSE) of compressed sensing reconstructed CSI as function of artificially under sampled data; red dots represent that of the 4-metabolite phantom and the blue diamond the in-vivo rat scanning. The low nRMSE values for reduction rate up to 5 (20% sampling rate) reflected the good quality of compressed sensing reconstructed phantom images in Figure 2 and the rat #1 kidney images in Figure 4.
In-vivo animal experiments
After the injection of HP [1-13C]pyruvate, axial images of the abdomen were obtained at the level of the kidneys. The 2D CSI raw data from four individual rats with real time under-sampling were directly reconstructed using the 3D compressed sensing method. The raw data from the fully sampled rat was treated in two ways. First, it was directly reconstructed as the fully-sampled data set. Each metabolite signal was decomposed from the 16×16 spectra and zero filled images were displayed along with under-sampled rat metabolite images from the compressed sensing results. Second, a set of under-sampled data were produced by artificially zeroing out some phase encoding acquisitions. The pattern of the zeroed phase encoding was the same as real-time under sampling pattern. These artificially under-sampled data sets were reconstructed using the compressed sensing procedure as the real time under-sampled in-vivo data. This procedure also serves as extra validation for the real-time under-sampled results. The calculated nRMSE values were similar to those from the phantom validation experiment, except for the 10% case as shown in Figure 3. Figure 4 shows the 13C images of rat kidneys acquired with centric CSI with reduction factors R=1, 2, 3, 4 and 10 as well as the artificially under-sampled compressed sensing reconstruction results from the fully-sampled in-vivo data. Like the compressed sensing reconstructed phantom, the rat images showed that minor noise only started to appear at R=4. The noise only became noticeable in R=5 case and severe at R=10 with only 10% of the data sampled. Finally, the under-sampled CSI sequence was applied in a dynamic study of pyruvate/lactate exchange in a live rat after injection of HP-[1-13C]pyruvate (Figure 5). For R=3, the scan time is reduced from 32 seconds for a full 16 × 16 phase encoding CSI to 11 seconds using the under-sampled CSI sequence. Figure 5 shows the [1-13C]lactate image of a rat kidney and the lactate-pyruvate ratio dynamics over a period of 60s. In general the results from the phantom validation and the under-sampled in vivo data showed that the 3D CS method provides a real-time savings of time and spin polarization without significant loss of biological information.
Figure 4.
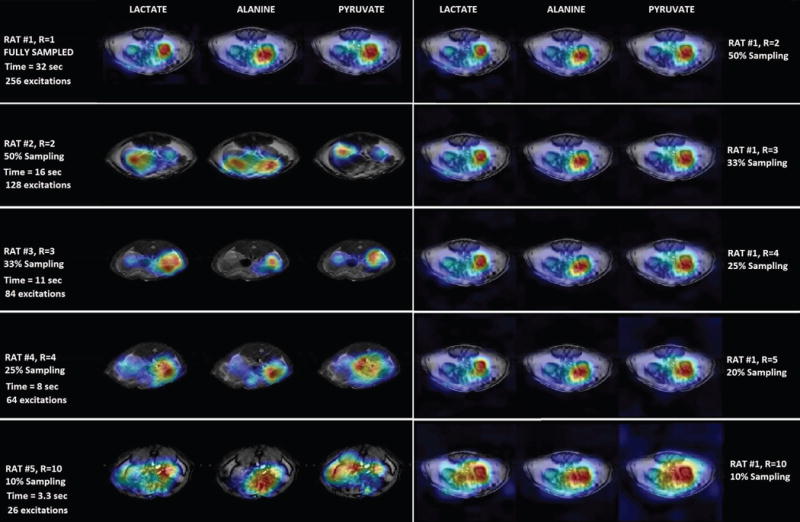
Compressed sensing reconstructed in-vivo hyperpolarized 13C CSI images of a slice that contains rat kidneys. The five rows of the left column reflect images from real-time reduction rates of R=1, 2, 3, 4 and 10 from five individual rats; These under sampled rats with different reduction ratio demonstrated the feasibility of the 3D CS reconstruction on real-time sparsely acquired data, even without reference for each individually reduced CSI data; the five rows of the right column show the images reconstructed from artificially under-sampled data from the fully sampled rat #1 with reduction rate R=2, 3, 4, 5 and 10 which reflected the CS quality along with the CS reconstructed phantom data.
Figure 5.
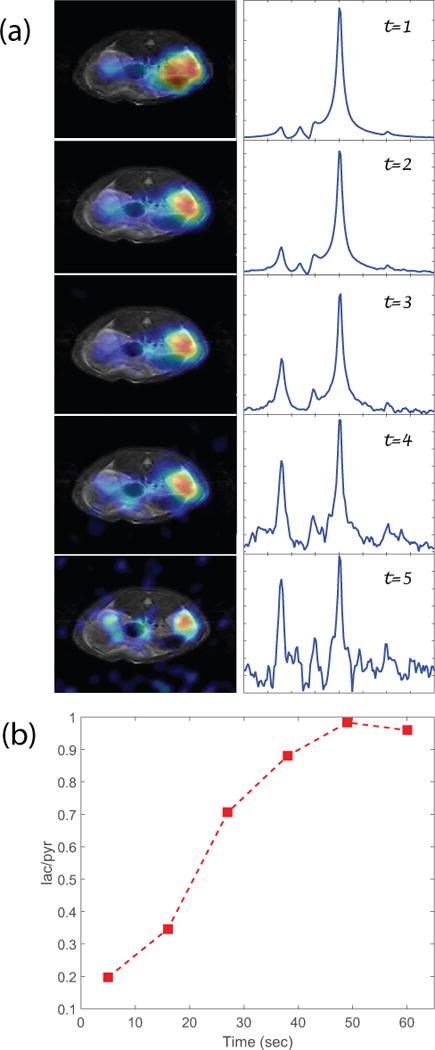
(a) Multi-phase CSI scans of R=3, left: lactate images; right: spectrum; shows the metabolic dynamics. With R=3, each 16×16 phase encoding CSI scan time of 11s. The spectra show evolution of the lactate-pyruvate ratio during the first minute; (b) Lactate to pyruvate ratio vs time from the multi-phase CSI. The intensity is the integration from the kidney area.
DISCUSSION
The results demonstrated here show that true 3D compressed sensing is feasible for chemical-shift imaging acceleration of hyperpolarized 13C metabolic imaging. The results show that this true 3D compressed sensing method can be applied successfully with reduction factor up to 4 or 5 (25% or 20% of sampling). Although better algorithms can be developed, even greater reduction rates will be possible by using multi-channel coils combined with parallel imaging, so that dynamic data can be collected over longer periods of time without total loss of the polarized signal. Combining parallel imaging with compressed sensing to further accelerate the imaging speed has been reported previously [15 & 16] and we believe that the application of this method to CSI has significant advantages for in vivo hyperpolarized 13C dynamic metabolic imaging.
The use of compressed sensing for chemical-shift imaging has been reported earlier using flyback 3D-MRSI [13–14]. The flyback CSI sequence works the same way as the conventional CSI to phase encode one (or two) direction with multiple excitations but adds another phase encoding in another direction while acquiring the FID signal. This method can accelerate CSI by factor of one dimension which reduces the scan time for CSI. However, due to the added phase encoding during the acquisition which combines the z-phase encoding and frequency into one, the spectral bandwidth is limited to about 581 Hz on a 3T clinic scanner and the raw spectral length is limited to 59 points [13–14]. This becomes limiting for those applications requiring collection of data over a wider range of chemical-shift values. In these cases, conventional CSI is still the best alternative. Hu et al [13] also demonstrated that sparseness of the CSI using 2D Wavelet in the frequency and one physical dimension. This sparseness may not be satisfied along the edge of the object and in weak signal areas. When the 2D CS method is applied to 3D or 4D CSI, the compressed sensing optimization and reconstruction process must be applied n or n × n times independently. Obviously, this disconnects the intrinsic links of the CSI. The work presented in this paper demonstrates that 3D compressed sensing with the subject and all operators (Wavelet, TV, etc.) in 3D (physical x-y and frequency dimensions) and treats the entire 2D CSI subject as an unit is a suitable CSI acceleration method for hyperpolarized 13C metabolic imaging of complex metabolic systems. The robustness of this pilot study using true 3D applied to 2D CSI also opens a possibility for single acquisition of 3D CSI. That is a major goal of this pilot study. In a typical 3D CSI, for example 16×16×16, 4096 acquisitions will be required. However, this large number of excitations is not possible for hyperpolarized nuclei. In the acceptable 2D CSI case with a reduction factor of 5, we can estimate a feasible reduction rate of 11 for 3D CSI by compressed sensing alone. By combining a better algorithm and parallel imaging, a reduction rate of 16 would reduce the number of required excitations to 256 which is equivalent to the number of excitations used for typical hyperpolarized 2D CSI acquisitions.
Acknowledgments
This work was supported by NIH grants P41 EB015908 and R37-HL034557. The authors acknowledge the hyperpolarization operation and animal handling team Dr. L. Fidelino, Dr. X.D. Wen and T. Hever.
References
- 1.Golman K, Zandt R, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic Imaging by Hyperpolarized 13C Magnetic Resonance Imaging for In vivo Tumor Diagnosis. Cancer Res. 2006;66(22):10855–60. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 2.Nelson SJ, et al. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1-13C]Pyruvate, Science. Translational Medicine. 2013 Aug 14;5:198RA108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golman K, Petersson JS, Magnusson P, Johansson E, Åkeson P, Chai CM, Hansson G, Månsson S. Cardiac Metabolism Measured Noninvasively by Hyperpolarized 13C MRI. Magnetic Resonance in Medicine. 2008;59:1005–1013. doi: 10.1002/mrm.21460. [DOI] [PubMed] [Google Scholar]
- 4.Laustsen C, Lycke S, Palm F, Østergaard JA, Bibby BM, Norregaard R, Flyvbjerg A, Pedersen M, Ardenkjaer-Larsen JH. High altitude may alter oxygen availability and renal metabolism in diabetics as measured by hyperpolarized [1–13C]pyruvate magnetic resonance imaging. Kidney International. 2014;86:67–74. doi: 10.1038/ki.2013.504. [DOI] [PubMed] [Google Scholar]
- 5.Clatworthy MR, Kettunen MI, Hu DE, Mathews RJ, Witney TH, Kennedy BWC, Bohndiek SE, Gallagher SA, Jarvis LB, Smith KGC, Brindle KM. Magnetic resonance imaging with hyperpolarized [1,4–13C2]fumarate allows detection of early renal acute tubular necrosis. PNAS. 2012;109(33):13379. doi: 10.1073/pnas.1205539109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candès EJ, Romberg J, Tao T. Robust uncertainty principles: Exact signal reconstruction from highly incomplete frequency information. Inform Theory IEEE Trans. 2006;52(2):489–509. [Google Scholar]
- 7.Lustig M, Donoho D, Pauly JM. Sparse mri: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–95. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 8.Wang JX. Optimizing Spars 3D-MRI using Cubic Compressed Sensing Reconstruction Method. Proceeding of International Society for Magnetic Resonance in Medicine. 2012:4231. [Google Scholar]
- 9.Kim SJ, Koh K, Lustig M, Boyd S, Gorinevsky D. An Interior-Point Method for Large-Scale 11-Regularized Least Squares. IEEE Journal of Selected Topics in Signal Processing. 2007;1(4) [Google Scholar]
- 10.Barkhuysan H, De Beer R, Van Ormondt D. Improved Algorithm for Noniterative Time-Domain Model Fitting to Exponentially Damped Magnetic Resonance Signals. Journal of Magnetic Resonance. 1987;73:553–557. [Google Scholar]
- 11.Pijnappela WWF, Van Den Boogaart De, Beer R, Van Ormondt D. SVD-Based Quantification of Magnetic Resonance Signals. Journal Of Magnetic Resonance. 1992;97:122–134. [Google Scholar]
- 12.Levin YS, Mayer D, Yen YF, Hurd RE, Spielman DM. Optimization of Fast Spiral Chemical Shift Imaging Using Least Squares Reconstruction: Application for Hyperpolarized 13C Metabolic Imaging. Magnetic Resonance in Medicine. 2007;58:245–252. doi: 10.1002/mrm.21327. [DOI] [PubMed] [Google Scholar]
- 13.Hu S, Lustig M, Chen AP, Crane J, Kerr AB, Kelley DAC, Hurd R, Kurhanewicz J, Nelson SJ, Pauly JM, Vigneron DB. Compressed sensing for resolution enhancement of hyperpolarized 13C flyback 3D-MRSI. Journal of Magnetic Resonance. 2008;192:258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu S, Lustig M, Balakrishnan A, Larson PEZ, Bok R, Kurhanewicz J, Nelson SJ, Goga A, Pauly JM, Vigneron DB. 3D Compressed Sensing for Highly Accelerated Hyperpolarized 13C MRSI With In Vivo Applications to Transgenic Mouse Models of Cancer. Magnetic Resonance in Medicine. 2010;63:312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang D, Liu B, Wang J, Ying L. Accelerating SENSE using compressed sensing. Magnetic Resonance in Medicine. 2009;62(6):1574–84. doi: 10.1002/mrm.22161. [DOI] [PubMed] [Google Scholar]
- 16.Lai P, Lustig M, Brau AC, Vasanawala, Beatty PJ, Alley M. Efficient L1SPIRiT Reconstruction (ESPIRiT) for Highly Accelerated 3D Volumetric MRI with Parallel Imaging and Compressed Sensing. Proc Intl Soc Mag Reson Med. 2010;18 [Google Scholar]


