Abstract
Expression of the mTORC1 repressor, Regulated in DNA Damage and Development 1 (REDD1), is elevated in skeletal muscle during various catabolic conditions including fasting, hindlimb immobilization, and sepsis. Conversely, REDD1 expression is suppressed by anabolic stimuli such as resistance exercise or nutrient consumption following a fast. Though it is known that nutrient consumption reduces REDD1 expression, it is largely unknown how nutrients and hormones individually contribute to the reduction in REDD1 expression. Therefore, the purpose of the present study was to determine how nutrients and hormones individually regulate REDD1 expression. HeLa cells were deprived of leucine or serum for 10 h, after which either leucine or serum was reintroduced to cell culture medium for 60 min. Re-supplementation of either leucine or serum resulted in a reduction in REDD1 protein levels by 34.8±5.8% and 54.1±3.4%, respectively, compared to the deprived conditions. Re-supplementation of leucine or serum to deprived cells also led to a reduction in REDD1 mRNA content by 49.1±2.7% and 65.0±1.4%, respectively, compared to the deprived conditions. Interestingly, rates of REDD1 protein degradation were unaffected by either leucine or serum re-supplementation, as assessed in cells treated with cycloheximide to block protein synthesis. Likewise, addition of leucine- or serum to cells treated with Actinomycin D to inhibit gene transcription failed to alter the rate of REDD1 mRNA degradation. The data indicate that the leucine or serum-induced suppression of REDD1 expression occurs independent of changes in the rate of degradation of either the REDD1 protein or mRNA. Thus, the leucine- or serum-induced suppression likely occurs through alternative mechanism(s) such as reduced REDD1 gene transcription and/or mRNA translation.
Keywords: REDD1, mTOR, Transcriptional/translational regulation, Amino acids
Highlights
-
•
Deprivation of leucine or serum induces REDD1 mRNA and protein expression.
-
•
Re-supplementation of leucine or serum reduces REDD1 mRNA and protein expression.
-
•
Nutrient deplete or replete conditions do not affect the degradation rate of REDD1.
-
•
REDD1 expression is controlled through altered rates of transcription.
1. Introduction
Single- and multi-cellular organisms have evolved unique nutrient sensing mechanisms that allow them to quickly respond to the ever-changing nutrient status in their extracellular environment. This bestows upon them the ability to respond to conditions in which anabolic stimuli trigger intracellular signaling cascades that promote cellular growth and proliferation. Conversely, catabolic conditions trigger specific intracellular signals that are growth restrictive and promote energy conservation [1]. Integration of a variety of cellular stimuli including hormone and nutrient availability to regulate various cellular processes such as mRNA translation and ribosome biogenesis is mediated through phosphorylation of downstream substrates, e.g. p70S6K1 on Thr389 [2], by a serine/threonine protein kinase referred to as the mechanistic target of rapamycin in complex 1 (mTORC1). Two independent pathways consisting of 1) anabolic hormone signaling (i.e. insulin and IGF-1) that activates the GTPase, ras-homolog enriched in brain (Rheb) [3], and 2) nutrient sensing mechanisms like the Rag GTPases [4], work in coordination for complete activation of mTORC1 signaling.
Activation of downstream anabolic pathways by mTORC1 is energetically demanding [5] and thus during catabolic or stressful conditions cells have evolved mechanisms to govern its activation state. Regulated in Development and DNA Damage (REDD1) functions as an upstream repressor of mTORC1 signaling and is upregulated under various catabolic conditions including long-term nutrient deprivation [6], [7], [8], [9]. Alternatively, REDD1 expression is rapidly suppressed upon refeeding a previously fasted animal in association with activation of mTORC1 signaling [10]. A common mechanism of REDD1 expression under the aforementioned conditions is through induction of the endoplasmic reticulum (ER) stress response, which allows for the translation of select transcripts, including activating transcription factor 4 (ATF4), leading to REDD1 mRNA and protein expression [11]. Our group has previously demonstrated that ATF4 is necessary and sufficient for the ER stress induced induction of REDD1 mRNA and protein expression [12].
A recent report demonstrates that REDD1 also plays a role in limiting the nutrient-induced stimulation of protein synthesis [10]. Accordingly, REDD1 is a critical regulator of the cellular response to nutrient status. Understanding the factors that regulate intracellular processes related to fasting and re-feeding will be critical in expanding our knowledge of the intimate relationship between the cellular response to fasting and re-feeding and may provide insight into contributors of metabolic disease development and progression. While most studies have focused on conditions of REDD1 induction and its mechanism of action, very few have investigated the factors contributing to the resolution of REDD1 mRNA or protein expression. Therefore, the goal of the present study is to further understand the regulation of REDD1 mRNA and protein expression in response to in vitro deprivation and re-supplementation of either leucine or serum, two potent activators of mTORC1.
2. Materials and methods
2.1. Cell culture
HeLa cells (ATCC, Manassas, VA, Cat. # CCL-2) were maintained in culture at 37 °C in a humidified incubator with 5% CO2 in high glucose Dulbecco's modified Eagle's Medium (DMEM, Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO) and 1% penicillin-streptomycin (Invitrogen). On the day of the experiment, cells were placed in serum-free medium (DMEM) or leucine-free medium (Atlanta Biologicals, Lawrenceville, GA) for 10 h. After 10 h of starvation, 10% serum or 0.76 mM leucine (Sigma, St. Louis, MO) was re-introduced into the culture medium for the indicated times. For experiments using Actinomycin D (Sigma) or cycloheximide (A.G. Scientific, San Diego, CA), cells were prepared as described above with the exception that either Actinomycin D (2 μg/mL) or cycloheximide (100 μg/mL) was added to the medium for 15 min or 2 min, respectively, prior to the re-introduction of serum or leucine.
2.2. RNA isolation and quantitative real-time PCR
Cells were treated as described above and total RNA was extracted with TRIzol reagent according to the manufacturer's protocol (Invitrogen). RNA (1 μg) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and subjected to quantitative real-time PCR using QuantiTect SYBR Green master mix (Qiagen, Germantown, MD). Human specific primers used for amplification of REDD1 and GAPDH mRNA were previously described [12]. Mean cycle threshold (CT) values for REDD1 and GAPDH were determined for control and experimental samples. Changes in REDD1 mRNA expression levels were normalized to GAPDH mRNA expression using the calculations as previously described [13].
2.3. Western blot analysis
Cells were treated as described above, washed once in phosphate-buffered saline, lysed and harvested in Laemmli sample buffer, and boiled for 5 min. An equal volume of each sample was fractionated by SDS-PAGE using Bio-Rad precast Criterion gels (Bio-Rad, Hercules, CA). Fractionated samples were transferred onto PVDF membranes and blocked for 1 h with 5% nonfat dry milk (NFDM) in Tris-buffered saline with 0.1% Tween20 (TBST). Membranes were incubated overnight at 4 °C with one of the following primary antibodies: anti-REDD1 (ProteinTech Group Inc., Chicago, IL, Cat. # 10638), p70S6K1 T389 (Cell Signaling, Danvers, MA, Cat. # 9205), alpha-Tubulin (Santa Cruz, Dallas, TX, Cat. # sc-32293). Membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibody (Bethyl, Montgomery, TX) in 5% NFDM in TBST at room temperature for one hour then washed thrice in TBST. Blots were developed with ECL Plus reagents (Bio-Rad, Hercules, CA) using a FluorChem M Multifluor System (ProteinSimple, San Jose, CA). Western blot densitometry was quantitated using Image J Software (National Institutes of Health).
2.4. Statistics
Data are expressed as the mean±standard error of the mean (SEM). Statistical analyses are described within the figure legends. Statistical significance was set at p<0.05.
3. Results
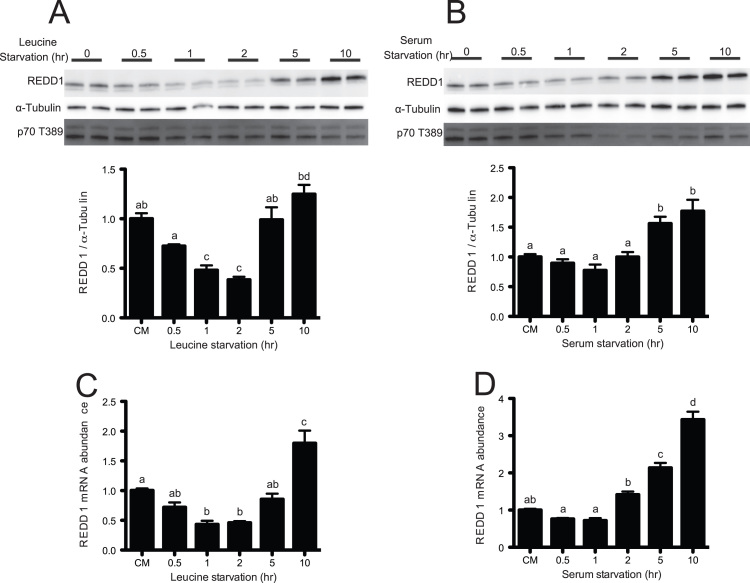
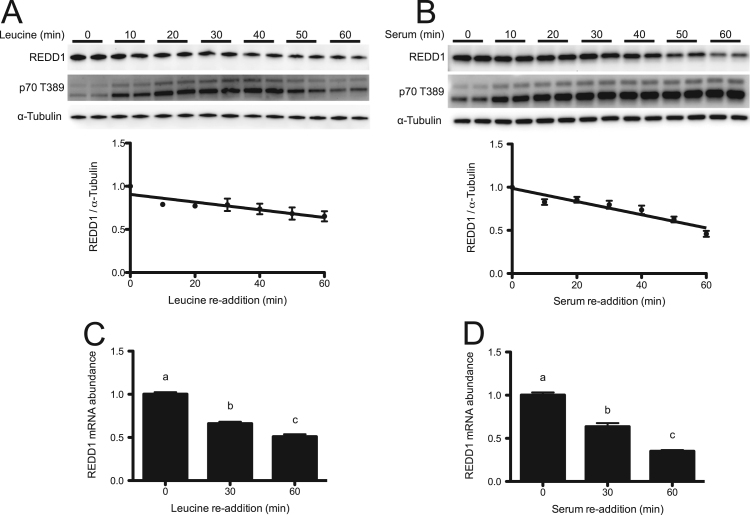
As an in vitro experimental model of an overnight fast, HeLa cells were deprived of leucine or serum for up to 10 h prior to quantitation of REDD1 expression by Western blot analysis. REDD1 expression in leucine-deprived cells exhibited a bi-phasic response, with an initial drop in expression during the first 2 h of deprivation followed by a subsequent, prolonged increase in expression (Fig. 1A). The response to serum deprivation exhibited a similar biphasic response (Fig. 1B). The time course of leucine- or serum-deprivation induced changes in REDD1 mRNA expression temporally mirrored those for the protein (Fig. 1C and D, respectively). Consistent with previous reports [10], [14], elevated REDD1 protein expression was associated with a concomitant reduction in signaling through mTORC1 as assessed by reduced phosphorylation of p70S6K1 on T389 (Fig. 1A, B). Upon re-supplementation of either leucine or serum to a final concentration of 0.76 mM or 10%, respectively, for one hour, REDD1 protein expression was reduced by 34.8±5.8% and 54.1±3.4% respectively, compared to the deprived conditions (Fig. 2A, B). As expected, the reduction in REDD1 protein expression was associated with an increase in phosphorylation of p70S6K1 on T389 (Fig. 2A, B). Interestingly, re-supplementation of either leucine or serum also led to a reduction in REDD1 mRNA expression by 49.1±2.7% and 65.0±1.4%, respectively, compared to the deprived conditions (Fig. 2C, D).
Fig. 1.
Prolonged leucine and serum deprivation increase REDD1 protein and mRNA expression. HeLa cells were deprived of leucine (A, C) or serum (B, D) for varying times up to 10 h. (A, B) Phosphorylation of p70S6K1 on T389 and REDD1 protein was assessed by Western blot analysis. REDD1 mRNA abundance (C, D) was assessed by SYBR Green qRT-PCR and normalized to GAPDH. Results represent the mean±SEM of 3 independent experiments with 2 replicates performed in each experiment. Experimental groups with means that are statistically different from one another are indicated with different letters above the bars (One-Way ANOVA and Tukey's post hoc test for multiple comparisons, p<0.05).
Fig. 2.
Leucine and serum re-supplementation increase mTORC1 signaling and reduce REDD1 protein and mRNA expression. HeLa cells were deprived of leucine (A, C) or serum (B, D) for 10 h after which leucine or serum were added to the respective culture media to a final concentration of 0.76 mM or 10%, respectively. (A, B) Phosphorylation of p70S6K1 on T389 and REDD1 protein was assessed by Western blot analysis. REDD1 mRNA abundance (C, D) was assessed by SYBR Green qRT-PCR and normalized to GAPDH. Results represent the mean±SEM of 3 independent experiments with 2 replicates performed in each experiment. In panels A and B linear regression was performed for visualization of the decline in REDD1 expression. In panels C and D, experimental groups with means that are statistically different from one another are indicated with different letters above the bars (One-Way ANOVA and Tukey's post hoc test for multiple comparisons, p<0.05).
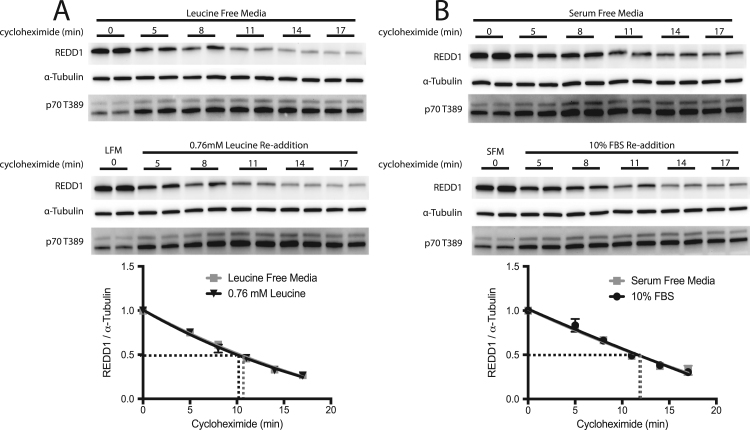
We next sought to evaluate the rate of REDD1 protein turnover in the presence or absence of leucine or serum. Previous reports have demonstrated that the half-life of the endogenous REDD1 protein is approximately five minutes [15] while ectopically expressed REDD1 protein has a half-life of approximately 30 min [16]. With such a relatively short half-life it is possible that REDD1 protein expression is controlled through mechanisms that regulate its stability. To assess whether REDD1 protein stability is altered in the presence or absence of leucine or serum, cycloheximide was added to the culture medium to block de novo protein synthesis. Consistent with previous results, the half-life of REDD1 protein was approximately 10 min and there were no significant differences in its rate of turnover between cells maintained in the presence or absence of leucine or serum (Fig. 3A and B). These data demonstrate that REDD1 protein degradation is not affected by the re-supplementation of leucine or serum to cells in culture.
Fig. 3.
Leucine and serum do not alter the rate of REDD1 protein degradation. HeLa cells were deprived of leucine (A) or serum (B) for 10 h. The cell culture medium was then supplemented with cycloheximide to a final concentration of 100 μg/mL, and two minutes later one-half of the cells in leucine-free medium (LFM) were supplemented with leucine and one-half of the cells in serum-free medium (SFM) were supplemented with serum to final concentrations of 0.76 mM or 10%, respectively. Samples were collected immediately before cycloheximide addition and at three-minute intervals after the re-addition of leucine or serum. REDD1 protein was assessed by Western blot analysis. Non-linear regression using one-phase decay and least squares fit of each condition; results represent the mean±SEM of 3 independent experiments with 2 replicates performed in each experiment. R2 values with and without leucine are 0.9442 and 0.9316, respectively. R2 values with and without serum are 0.8772 and 0.9116, respectively.
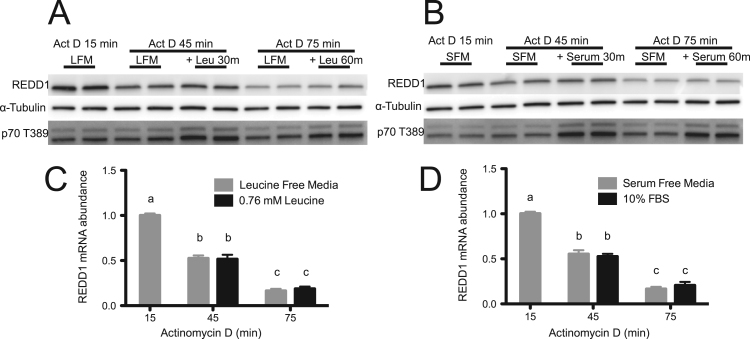
It is also possible that REDD1 mRNA expression is regulated post-transcriptionally. To investigate if the leucine- or serum-induced reductions in REDD1 mRNA expression were due to differences in mRNA degradation, the transcriptional inhibitor, actinomycin D, was added to HeLa cells in the presence or absence of leucine or serum. Similar to results obtained regarding protein stability, re-supplementation of culture medium with 0.76 M leucine led to no significant difference in the rate of REDD1 mRNA degradation when compared to leucine deprived cells (Fig. 4C). Likewise, no significant difference was observed when 10% serum was re-introduced to the culture medium compared to serum deprived cells (Fig. 4D). These data are consistent with previous reports wherein REDD1 mRNA half-life is approximately 30–60 min, depending on culture conditions [17]. Interestingly, under the aforementioned conditions, REDD1 protein expression mirrored REDD1 mRNA expression regardless of the presence or absence of leucine or serum (Fig. 4 A, B), demonstrating that neither REDD1 mRNA nor protein degradation rate is affected by the re-supplementation of leucine or serum to cells in culture.
Fig. 4.
Leucine and serum do not alter the rate of REDD1 mRNA degradation. HeLa cells were deprived of leucine (A, C) or serum (B, D) for 10 h. The cell culture medium was then supplemented with Actinomycin D (Act D) to a final concentration of 2 μg/mL, and 15 min later one-half of the cells in leucine-free medium (LFM) were supplemented with leucine and one-half of the cells in serum-free medium (SFM) were supplemented with serum to final concentrations of 0.76 mM or 10%, respectively. Samples were collected immediately before and at 30 min intervals after the re-addition of leucine or serum. Phosphorylation of p70S6K1 on T389 and REDD1 protein was assessed by Western blot analysis (A, B). REDD1 mRNA abundance was assessed by SYBR Green qRT-PCR and normalized to GAPDH (C, D). Results represent the mean±SEM of 3 independent experiments with 2 replicates performed in each experiment. Experimental groups with means that are statistically different from one another are indicated with different letters above the bar (Two-Way ANOVA and Tukey's post hoc test for multiple comparisons, p<0.05).
4. Discussion
Quickly sensing changes in the extracellular milieu and responding accordingly by altering gene transcription and mRNA translation is critical to the ability of cells to maintain homeostasis during various stressful conditions. Under conditions of nutritional stress, nutrient sensing mechanisms exist to divert resources away from energy consuming to energy conserving processes, a hallmark of the integrated stress response (ISR) [18].
While very little has been done with regard to understanding the post-translational regulation of REDD1, a recent study showed that GSK3β phosphorylates REDD1 at T23/25 and that the half-life of a REDD1 variant in which T23/25 are mutated to Ala is significantly longer compared to the wild type protein [16]. These findings suggest that phosphorylation of REDD1 by GSK3β results in its destabilization. However, re-addition of growth factors to serum starved cells in culture activates the phosphatidylinositol 3-kinase (PI3K)/AKT signaling cascade resulting in an inhibitory phosphorylation of GSK3β [19]. Thus, readdition of serum to serum-deprived cells would repress GSK3β activity, resulting in stabilization of REDD1. However, we show that there are no differences in rates of REDD1 protein degradation in the presence or absence of serum, suggesting that the effect of GSK3β did not play a role in the stabilization of REDD1 under conditions of serum deprivation and re-supplementation in the present study.
The data regarding post-transcriptional regulation of REDD1 is also very limited, but several groups have shown that microRNA's (miR) play a role in the regulation of REDD1 expression. For example, ectopically expressed miR-495 can directly suppress REDD1 expression by binding to the 3'-untranslated region of REDD1 mRNA. This led to an approximate 40% reduction in both REDD1 mRNA and protein expression that contributes to the resistance of breast cancer stem cells to hypoxia [20]. Similar results were obtained in a model of gamma-irradiated hematopoetic and osteoblastic cells, where gamma-irradiation induced upregulation of miR-30c and its overexpression led to a reduction in REDD1 expression. A potential binding site for miR-30c was also identified in the 3'-UTR of the REDD1 gene [21]. These data demonstrate differential expression of miRNA's under distinct physiological and pathological conditions and suggest that novel mechanisms of REDD1 suppression exist to promote cell survival and will likely warrant further investigation. Though we cannot speculate on the post-transcriptional repression of REDD1 by miRNAs under the conditions of leucine and serum deprivation used in this study, their role should not be discounted.
The results of the present study corroborate previous findings that REDD1 induction is a consequence of nutritional stress through serum deprivation [6] and extend those data to show leucine deprivation alone can induce REDD1 protein expression. They also show that re-addition of either leucine or serum to the culture medium restores REDD1 mRNA and protein expression to near baseline levels within one hour. Given that REDD1 mRNA levels decline at a similar rate in the presence or absence of the transcriptional inhibitor, actinomycin D (Fig. 2C, D vs Fig. 4C, D), and the extraordinarily short half-life of REDD1 protein (Fig. 3), these data suggest that REDD1 is largely regulated by rates of transcription. This idea is also supported by a similar decline in REDD1 protein expression during a one-hour actinomycin D treatment independent of the presence or absence or leucine or serum (Fig. 4A, B). Frost and colleagues reported a similar finding where the insulin-like growth factor 1-induced increase in REDD1 mRNA and protein required new transcription and translation of the REDD1 gene and mRNA [22]. In a recent publication from our laboratory, REDD1+/+ mice refed for 15 min after an overnight fast showed no noticeable decreases in REDD1 protein expression, whereas 60 min of refeeding was sufficient to reduce REDD1 protein expression [10]. It is tempting to speculate that even though REDD1 gene transcript may have been repressed in mice refed for 15 min, insufficient time had elapsed to allow for a detectable change in the amount of the protein. In contrast, by 60 min of refeeding, mRNA levels had declined allowing changes in expression of the protein to be detected. Further support for the idea that REDD1 is primarily regulated through rates of transcription is obtained from studies in skeletal muscle of hindlimb-immobilized rats that were fasted overnight and later administered an oral dose of either saline or leucine as a nutrient stimulus. Accordingly, REDD1 mRNA was elevated in soleus muscle as soon as one day following immobilization and continued to increase at day two and three post-immobilization. Interestingly, 15 min after receiving an oral gavage of leucine REDD1 mRNA levels were significantly lower compared to their respective controls, regardless of the rat's immobilization status [8].
Experimental approaches to investigate rates of REDD1 gene transcription and mRNA translation were unsuccessful. We hypothesize that the relatively low abundance and short half life of REDD1 mRNA and protein combined with REDD1's potent inhibitory effects on protein synthesis makes it problematic to determine in vitro conditions that are amicable to study its rate of transcription or translation. However, the tight association between leucine- and serum-induced changes in REDD1 mRNA and protein expression strongly suggest that regulation of gene expression plays a key role in controlling REDD1 protein expression. Thus, it seems reasonable to conclude that during times of nutrient deplete conditions either through leucine- or serum-deprivation, induction of the ISR leads to a concomitant induction of REDD1 mRNA and protein expression and repletion of nutrients by re-addition of leucine or serum to the culture medium resolves the stress allowing for a reduction in REDD1 transcription. Combined, these data suggest that the leucine- or serum-induced suppression of REDD1 expression occurs independent of rates of degradation of the REDD1 protein or mRNA and suggest a mechanism in which REDD1 expression is primarily controlled through rates of transcription.
Acknowledgements
The work in this study was supported by the NIH [DK-13499, DK-15658, and EY023612] and the American Diabetes Association Pathway to Stop Diabetes [1–14-INI-04].
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.10.003.
Appendix A. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter A.K., Anthony T.G., Anthony J.C., Jefferson L.S., Kimball S.R. The mTOR signaling pathway mediates control of ribosomal protein mRNA translation in rat liver. Int. J. Biochem. Cell Biol. 2004;36:2169–2179. doi: 10.1016/j.biocel.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Inoki K., Guan K.L. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 2004;24:7965–7975. doi: 10.1128/MCB.24.18.7965-7975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis M.D., McGhee N.K., Jefferson L.S., Kimball S.R. Regulated in DNA damage and development 1 (REDD1) promotes cell survival during serum deprivation by sustaining repression of signaling through the mechanistic target of rapamycin in complex 1 (mTORC1) Cell. Signal. 2013;25:2709–2716. doi: 10.1016/j.cellsig.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeYoung M.P., Horak P., Sofer A., Sgroi D., Ellisen L.W. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelleher A.R., Kimball S.R., Dennis M.D., Schilder R.J., Jefferson L.S. The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am. J. Physiol. Endocrinol. Metab. 2013;304:E229–E236. doi: 10.1152/ajpendo.00409.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGhee N.K., Jefferson L.S., Kimball S.R. Elevated corticosterone associated with food deprivation upregulates expression in rat skeletal muscle of the mTORC1 repressor, REDD1. J. Nutr. 2009;139:828–834. doi: 10.3945/jn.108.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon B.S., Williamson D.L., Lang C.H., Jefferson L.S., Kimball S.R. Nutrient-induced stimulation of protein synthesis in mouse skeletal muscle is limited by the mTORC1 repressor REDD1. J. Nutr. 2015;145:708–713. doi: 10.3945/jn.114.207621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin H.O., Seo S.K., Woo S.H., Kim E.S., Lee H.C., Yoo D.H., Choe T.B., Hong S.I., Kim J.I., Park I.C. SP600125 negatively regulates the mammalian target of rapamycin via ATF4-induced Redd1 expression. FEBS Lett. 2009;583:123–127. doi: 10.1016/j.febslet.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 12.Whitney M.L., Jefferson L.S., Kimball S.R. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem. Biophys. Res Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Dennis M.D., Coleman C.S., Berg A., Jefferson L.S., Kimball S.R. REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2005103. (ra68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimball S.R., Do A.N., Kutzler L., Cavener D.R., Jefferson L.S. Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. J. Biol. Chem. 2008;283:3465–3475. doi: 10.1074/jbc.M706643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katiyar S., Liu E., Knutzen C.A., Lang E.S., Lombardo C.R., Sankar S., Toth J.I., Petroski M.D., Ronai Z., Chiang G.G. REDD1, an inhibitor of mTOR signalling, is regulated by the CUL4A-DDB1 ubiquitin ligase. EMBO Rep. 2009;10:866–872. doi: 10.1038/embor.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otulakowski G., Duan W., Sarangapani A., Gandhi S., O’Brodovich H. Glucocorticoid-mediated repression of REDD1 mRNA expression in rat fetal distal lung epithelial cells. Pedia. Res. 2009;65:514–519. doi: 10.1203/PDR.0b013e3181998db6. [DOI] [PubMed] [Google Scholar]
- 18.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D.F., Bell J.C., Hettmann T., Leiden J.M., Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty A., Koldobskiy M.A., Bello N.T., Maxwell M., Potter J.J., Juluri K.R., Maag D., Kim S., Huang A.S., Dailey M.J., Saleh M., Snowman A.M., Moran T.H., Mezey E., Snyder S.H. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang-Verslues W.W., Chang P.H., Wei P.C., Yang C.Y., Huang C.K., Kuo W.H., Shew J.Y., Chang K.J., Lee E.Y., Lee W.H. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 21.Li X.H., Ha C.T., Fu D., Xiao M. Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS One. 2012;7:e48700. doi: 10.1371/journal.pone.0048700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost R.A., Huber D., Pruznak A., Lang C.H. Regulation of REDD1 by insulin-like growth factor-I in skeletal muscle and myotubes. J. Cell. Biochem. 2009;108:1192–1202. doi: 10.1002/jcb.22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material