Abstract
Objective
We sought to investigate the effects of HIV infection on the vaginal microbiota, and associations with treatment and demographic factors. We thus compared vaginal microbiome samples from HIV+ and HIV− women collected at two Chicago area hospitals.
Design
We studied vaginal microbiome samples from 178 women analyzed longitudinally (n=324 samples), and collected extensive data on clinical status and demographic factors.
Methods
We used 16S rRNA gene sequencing to characterize the bacterial lineages present, then UniFrac, Shannon Diversity and other measures to compare community structure to sample metadata.
Results
Differences in microbiota measures were modest in the comparison of HIV+ and HIV− samples, in contrast to several previous studies, consistent with effective antiretroviral therapy. Proportions of healthy Lactobacillus species were not higher in HIV-negative subjects overall, but were significantly higher when analyzed within each hospital in isolation. Rates of bacterial vaginosis (BV) were higher among African American women and HIV+ women. BV was associated with higher frequency of HIV+. Unexpectedly, African-American women were more likely to switch BV status between sampling times; switching was not associated with HIV+ status.
Conclusions
The influence of HIV infection on the vaginal microbiome was modest for this cohort of well-suppressed urban American women, consistent with effective anti-retroviral therapy. HIV+ was found to be associated with BV. Although BV has previously been associated with HIV transmission, most of the women studied here became HIV+ many years before our test for BV, thus implicating additional mechanisms linking HIV infection and BV.
Keywords: microbiome, bacterial vaginosis, lentivirus, cervicovaginal mucus, cervical mucus, transmission
Introduction
As the human microbiome comes to be better understood, interest has shifted to understanding differences between human populations and associations with disease. Multiple factors affect microbiome assembly, including stochastic acquisition of bacterial strains, diet, age, pet ownership and antibiotic use [1–6]. These complexities often leave dissecting the effects of individual factors challenging, yet these complex interpersonal differences represent the background on which real-world treatment of disease takes place. Many of modulators of the microbiota are expected to vary with demographic and socioeconomic status. Here we investigate longitudinal vaginal microbiota samples from HIV+ and HIV− women in the city of Chicago, where sampling took place at two hospitals, which are separated by less than 5 miles but serve quite different socioeconomic groups.
HIV infection rates differ with economic status, and HIV infection is known to affect the human microbiota [7–14]. Advanced disease and AIDS are associated with immunodeficiency, which can result in opportunistic infections and alterations in the microbiome [8, 15–19]. Some of the expanded microbes are readily detected in the healthy human microbiome and are only pathogenic in immunodeficient states. The microbiota has been implicated in HIV transmission and disease progression [20–23]. Immune responses to the normal microbiota may shape the possible immune responses to new infections with HIV, and inflammation associated with microbial action may promote transmission by inducing proliferation of cells that support HIV infection. After infection, leakage of microbial antigens across the damaged gut may promote inflammation and associated morbidity [24, 25]. These observations and others have motivated detailed studies of the interaction of HIV and the human microbiome.
Associations between composition of the vaginal microbiota and HIV status have varied among studies. A 2008 meta-analysis showed that bacterial vaginosis (BV) was associated with a 1.6 times increased risk of HIV acquisition [26]. A study of 174 Rwandan sex workers showed a strong association between the structure in the vaginal microbiome and HIV+ status [27]. In a study of at-risk women in Kenya, BV and HSV2 infection were the two strongest risk factors measured for HIV acquisition over a 20-year period [28]. BV is known to be more common in African American women, motivating studies of associations with HIV infection [29]. In contrast, a study of the vaginal microbiome in 64 women in Chicago showed no differences associated with HIV infection [30]. Another study, comparing vaginal microbiota in women from Rwanda and the US together with HIV status, found modest differences for both geographic site and infection status [31]. Thus reported associations between HIV+ status and structure of the vaginal microbiota have varied with cohort and geographical location.
Here we study the vaginal microbiome in women sampled at two hospitals in Chicago – Stroger Hospital of Cook County (CC) and Northwestern Memorial Hospital (NMH) which serve quite different socioeconomic groups, one relatively affluent (NMH) and the other less advantaged (CC) (Figure 1A). The data could thus be explored to interrogate the relationship between demographic status, HIV status and the vaginal microbiota.
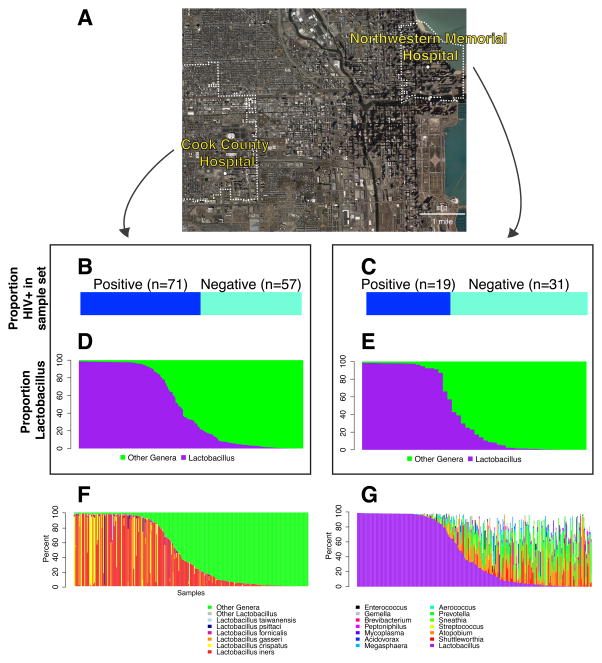
Figure 1.
Overview of the two hospitals studied and the metagenomic sample set. A) Map showing the locations and zip code boundaries of the two hospitals. B) Proportion of HIV+ in the sample from Cook County Hospital. C) Proportion of HIV+ in the sample from Northwestern Memorial Hospital. D) Proportion of Lactobacillus in 16S rRNA gene sequence data from Cook County Hospital. E) Proportion of Lactobacillus in 16S rRNA gene sequence data from Northwestern Memorial Hospital. F) Proportions of Lactobacillus lineages in sequence data from both Northwestern Memorial Hospital and Cook County Hospital, with colors (key at bottom) indicating the lineages detected. G) Proportions of non-Lactobacillus in sequence data from Northwestern Memorial Hospital and Cook County Hospital, with colors (key at bottom) indicating the lineages detected.
Results
Cohorts studied
Women from Stroger Hospital of Cook County (CC) or Northwestern Memorial Hospital (NMH) were studied, using an extensive sample set collected previously to analyze the biophysics of HIV particle interactions with the vaginal epithelium [32]. Demographic data is summarized in Supplemental Table 1. Women at CC were on average 40 years of age (range 20 to 50), while women at NMH were on average 31 years of age (range 22 to 51). A total of 76% (98/119) of women at CC were self-reported African-American compared to 54% (27/50) at NMH; microbiome correlates with race are investigated below. The average CC HIV+ woman studied was diagnosed in 2002; the most common treatment was Atripla. The average year of diagnosis for NMH HIV+ women was 2003, and the most common treatment was the combination of Norvir, Reyataz, and Truvada.
The CC cohort was derived from a less advantaged socioeconomic background than the NMH cohort. To compare income levels, IRS individual income tax information from 2013 ((https://www.irs.gov/uac/soi-tax-stats-individual-income-tax-statistics-2013-zip-code-data-soi) was used to calculate income for all taxpayers in the zip code of each hospital. The median income of subjects at CC hospital was inferred to be between $1 to $25,000, while that of those from NMH was between $75,000 to $100,000.
The HIV status of the women in these two cohorts is shown in Figure 1A–C. 71/128 (55%) women included in this study from CC were HIV positive, while 19/50 (38%) of the NMH women were HIV positive.
Several clinical measures differed between the two hospital cohorts (Table 1). The vaginal pH was higher in the CC cohort than in the NMH cohort (p<0.001, Table 1). The average Nugent score, an indication of BV, was also higher in the CC cohort than in the NMH cohort (p<0.001, Supplemental Table 1). Similar to previous observations [33], the average Nugent score was higher in older women (p<0.001).
Table 1.
Association of subject metadata with hospital of origin and HIV infection status. Median values are listed; the parentheses contain the range showing the minimum and maximum values. P-values were calculated using the Wilcoxon signed-rank test and bolded if less than 0.05.
| Hospital CC (128 women, 231 samples) | Hospital NMH (50 women, 93 samples) | P-value | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| HIV Negative (n=57) | HIV Positive (n=71) | HIV Negative (n=31) | HIV Positive (n=19) | Hospital | HIV Status | |
|
|
||||||
| Age | 39 (20–50) | 40 (26–48) | 28 (23–48) | 35 (22–51) | <0.0001* | <0.0001* |
| pH | 4.47 (3.78–8.4) | 4.685 (3.83–8.32) | 3.97 (3.37–7.72) | 4.84 (3.56 – 6.01) | <0.0001* | <0.0001* |
| Progesterone | 0.67 (0.01–15.73) | 0.705 (0.02–21.63) | 0.75 (0.01–16.67) | 0.655 (0.09–16.41) | 0.787 | 0.744 |
| Nugent Score | 5 (0–10) | 7 (0–10) | 1 (0–10) | 8 (0–10) | <0.0001* | <0.0001* |
| Estradiol | 70.5 (13–567) | 96 (21–549) | 71 (21–1132) | 85.5 (13–494) | 0.131 | 0.002* |
| CD4 Count | Not measured | 515 (21–1292) | Not measured | 429.5 (2–1300) | 0.561 | - |
| Shannon Diversity | 2.376 (0.467–5.678) | 3.137 (0.416–5.388) | 2.330 (0.334–4.947) | 1.003 (0.372–4.405) | 0.003* | 0.186 |
| Lactobacillus | 0.449 (0–0.985) | 0.279 (0–0.987) | 0.087 (0–0.988) | 0.917 (0.006–0.986) | 0.786 | 0.693 |
| Lactobacillus iners | 0.159 (0–0.979) | 0.077 (0–0.980) | 0.019 (0–0.985) | 0.260 (0–0.982) | 0.096 | 0.255 |
| Lactobacillus crispatus | 0 (0–0.982) | 0 (0–0.979) | 0 (0–0.977) | 0 (0–0.975) | 0.009* | 0.0874 |
| Lactobacillus gasseri | 0 (0–0.708) | 0 (0–0.977) | 0 (0–0.939) | 0 (0-.976) | 0.001* | 0.505 |
| Profinflammatory Genera | 0.081 (0–0.928) | 0.140 (0–0.667) | 0.069 (0–0.945) | 0.002 (0–0.701) | 0.077 | 0.464 |
Several clinical parameters differed between HIV+ and HIV− women within each cohort (Table 1). The CD4 counts did not differ between hospitals (mean of 516 versus 561 cells/mm3) and were well above the AIDS-defining threshold of 200 cells/mm3. Viral loads were <40 copies per ml. The Nugent score was significantly higher for HIV+ women (p<0.001, Table 1), indicating a link between BV and HIV status as reported previously [26]. Vaginal pH was also higher (p<0.001)—high pH is associated with BV. For unknown reasons, estradiol concentration, measured in pg/mL from the blood at the time of mucus collection, also differed between HIV+ and HIV− (p=0.002), with higher values associated with HIV infection in both hospitals.
DNA purification and sequencing
The vaginal microbiome was characterized using 16S rRNA gene tag sequencing. DNA was purified from cervical or cervicovaginal mucus, then samples amplified with 16S rRNA gene primers targeting the V1V2 region [34–37]. This primer set has been used in previous studies of the vaginal microbiome [30], and is generally effective at recovering 16S sequences from vaginal organisms. Sequence data was acquired using the Illumina MiSeq platform (paired end sequencing, average of 50,127 sequences per sample).
Samples were analyzed using the QIIME pipeline [38]. Sequences were clustered into operational taxonomic units (OTUs) using UCLUST at 97% identity. An average of 483 OTUs were detected per sample. There were no systematic differences in the numbers of OTUs per sample between the hospitals or associated with HIV infection status (both p>0.1).
Bacterial lineages detected in the sample sets
The bacterial lineages present in each sample were then investigated (Figure 1D–G). The OTUs were assigned to the Greengenes bacterial taxonomy using UCLUST. To assign Lactobacillus sequences to the species level, sequences were compared to the 16S Lactobacillus type strains downloaded from Bacterio.net. As was seen in previous studies [3, 9, 11, 39, 40], many samples showed high proportions of bacteria from the genus Lactobacillus (Figure 1D–E). A total of 99.3% of Lactobacillus sequences were classified to the species level (using BLAST with e-value threshold of 10−5 and percent identity > 97%). Species present included L. iners, crispatus, gasseri, fornicalis, taiwanensis, psittaci, jensenii, helveticus, vaginalis, ultunensis, and rhamnosus (Figure 1F). The proportions of Lactobacillus varied widely, with some samples composed nearly entirely by Lactobacillus while others were almost free of Lactobacillus. The proportions of the different Lactobacillus genera in general did not differ between the hospitals studied or HIV infection status (p=0.786 and p=0.693 respectively), though the relatively rare Lactobacillus crispatus and Lactobacillus gasseri were more common in samples from CC hospital (p=0.009 and p=0.001 respectively, Table 1). The significance of this difference is unknown and could be either due to happenstance of colonization in differences in geographical locations or factors such as race, diet, health status, or hygiene practices.
Genera detected in addition to Lactobacillus, in order of total abundance, included Shuttleworthia, Atopobium, Streptococcus, and lineages suggested to be proinflammatory [41] including Sneathia, Prevotellla, Aerococcus, and Gemella (Figure 1G).
Relationship of vaginal microbial patterns and sample metadata
We next explored relationships of the vaginal microbiota and clinical data available for the subjects studied. Distances were calculated based on 16S rRNA gene sequence proportions between all pairs of samples, and the resulting matrix then used for cluster analysis to allow visualization of microbial communities relative to metadata (Figure 2). Distances were calculated using UniFrac, which involves placing pairs of communities on a common phylogenetic tree and calculating the unique and shared branch length [42, 43]. UniFrac distances were calculated based on the relative abundance of each taxa (Figure 2, weighted analysis), or based on presence-absence information only (Supplemental Figure 1, unweighted analysis).
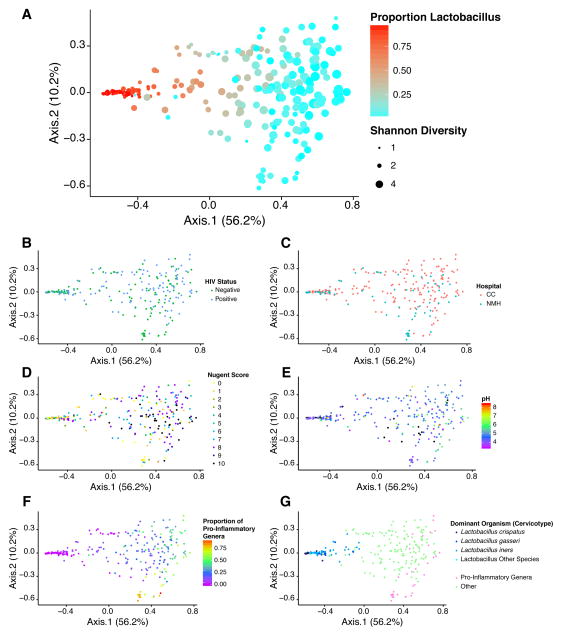
Figure 2.
Analysis of the data set using Weighted Unifrac. A) Ordination based on Principal Coordinate Analysis showing the distribution of samples, where color indicates the proportion of Lactobacillus in the sample, and the size of the point indicates Shannon diversity (indicated by black dots in legend to the right). The same ordination is colored based on B) HIV status, C) hospital of origin, D) Nugent score, E) vaginal pH, F) the proportion of pro-inflammatory genera, and G) cervicotype based on the dominant Lactobacillus species or pro-inflammatory genera.
Communities were compared using Principal Coordinate Analysis for data visualization and PERMANOVA for statistical testing. Axes 1 and 2 explained 56.2% and 10.2% of the variation, respectively in the weighted analysis, and axis 1 correlated with the proportion of Lactobacillus (Figure 2A). No significant difference in community structure was observed associated with HIV infection status (Figure 2B) or Hospital (Figure 2C). Higher Nugent scores (Figure 2D) and higher pH values (Figure 2E) trended with lower proportions of Lactobacillus (Figure 2A).
Several genera have been suggested to be pro-inflammatory, including Fusobacterium, Aerococcus, Sneathia, Gemella, Mobiluncus, and Prevotella. Pooling the proportions of these genera showed that they partitioned into a specific part of the low-Lactobacillus group and separated along axis 2 (Figure 2F). Gardnerella, another proinflammatory lineage previously identified in studies of the vaginal microbiome, is not resolved at the Genus level using the V1V2 16S gene primer set used here.
Several previous studies have suggested vaginal bacterial communities may partition into “cervicotypes” based on community membership—that is, vaginal communities may show reproducible stable states. In Figure 2G, communities are colored according to cervicotypes proposed previously (greater than 50% Lactobacillus or pro-inflammatory organisms). Samples studied here also separate based on their content of these organisms, though cervicotypes tended to grade into one-another and were not sharply distinguished.
In summary, vaginal communities separated by proportions of Lactobacillus and diversity (Figures 2A). Diversity and pH were associated with Nugent score (Pearson’s correlation r=0.45 and r=0.44, p<0.001 and p<0.001 respectively, Supplemental Figure 2), paralleling several studies that have shown that BV is associated with low Lactobacillus, high diversity, and high pH [9, 11, 18, 39, 40]. The high diversity state was associated with increased representation of proinflammatory lineages (Figure 2G), including Mobiluncus.
The Shannon diversity and proportion of certain Lactobacillus species differed between the two hospitals, likely due to the differences in rates of BV (Table 1). No statistically significant difference was found in the overall proportion of Lactobacillus between hospitals or HIV status (p=0.786 and p=0.693 respectively), but within each hospital, higher proportions of Lactobacillus were observed in HIV negative individuals (NMH HIV+ versus HIV− p=0.047, CC HIV+ versus HIV− p=0.052) (Supplemental Figure 3).
We next investigated microbial community structure in the women most affected by HIV infection. For women with the highest viral loads (>10,00 copies per mm3), there was a trend toward higher diversity (p=0.188). For women with CD4 counts below 200 cells per mm3, there was no significant difference in Shannon Diversity (comparison of samples with < 200 versus > 200 yielded p=0.8256). Thus the most HIV-affected women were not detectably different, though our power to detect differences was low due to the small numbers.
Global analysis of patient metadata and bacterial measurements
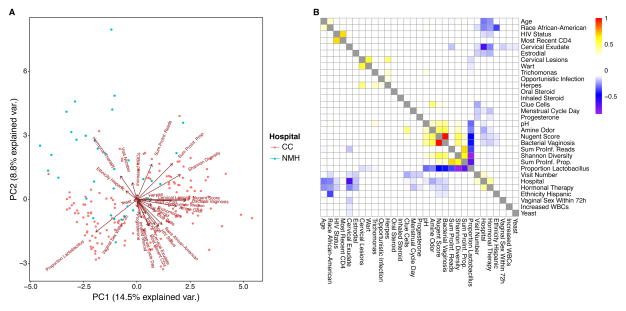
Given the large amount of patient metadata collected, we were able to carry out a global investigation of associations between patient information and bacterial measurements, including proportion of Lactobacillus, sum of pro-inflammatory genera, and Shannon diversity (Figure 3). We converted variables such HIV status, presence of an opportunistic infection, and oral steroids, into binary variables. Race was split into African-American or not and ethnicity into Hispanic or not. Excluding the hospital information, we built principle components from the multi-parameter data (Figure 3A), and compared correlations among data types (Figure 3B).
Figure 3.
Associations between patient information and bacterial community measures. A) PCA-biplot showing the relationship between patient information and bacterial community measures. The biplot shows the relationship between patient demographic, hospital records, sampling visit number, and microbial community assessments. The samples are colored by hospital (key at right). The direction and length of each arrow indicates its importance in separating the samples. First and second principal components PC1 and PC2 explained 14.5 and 8.8% of total variation. B) Heat map of p-values for comparisons among variables. Significant positive correlations are shown yellow and red; while negative correlations are shown in blue and purple. Non-significant correlations are colored in white. Three statistical tests were used. For continuous variables versus continuous variables, the Pearson correlation was used; for continuous variables versus categorical variables, the Kruskal-Wallis test was used; for categorical variables versus categorical variables, Fisher’s exact test was used.
We observed the expected relationship of Nugent score, BV, sum of pro-inflammatory genera and Shannon diversity, which were positively correlated with each other and negatively correlated with proportion of Lactobacillus (Figure 3A and B). BV was positively associated with age (BV – median age: 35, BV + median age=40, p=9.04×10−5). In both cohorts Shannon diversity was higher for the HIV+ women, but the range was quite wide, so the p-value only trended toward significance (p=0.186).
Associations of BV, HIV, and race
The data showed an association between BV, HIV, and race. Self identified African American women were diagnosed with BV more frequently (Fisher’s exact test p=0.00112; odds ratio=3.48), consistent with previous literature [29]. African American women were also more likely to be HIV+ (Fisher’s exact test p=0.00014; odds ratio=3.77).
We then asked whether there wasFa positive association between BV and HIV+. For the cohort, analyzed over all time points, the association was highly significant (Fisher’s exact test p=5.62×10−6). When the test was conducted using only the first time point, so that each woman contributed only one measurement, results were still significant (Fisher’s exact test p=0.046). Following previous literature on BV and health disparities [29], we investigated associations of HIV, BV and race. We found that when all data points were analyzed as a pool, HIV and BV were positively associated within both self-identified African Americans (Fisher’s exact test p=0.0087) and other racial groups (Fisher’s exact test p=0.009) independently. When samples were compared using only the first time point, which reduced power, significance was lost for African Americans, but still detectable for other races (Fisher’s exact test p=0.020). Thus HIV+ is associated with BV+; we return to possible mechanisms explaining this correlation in the Discussion.
Switching BV status as a measure of community stability
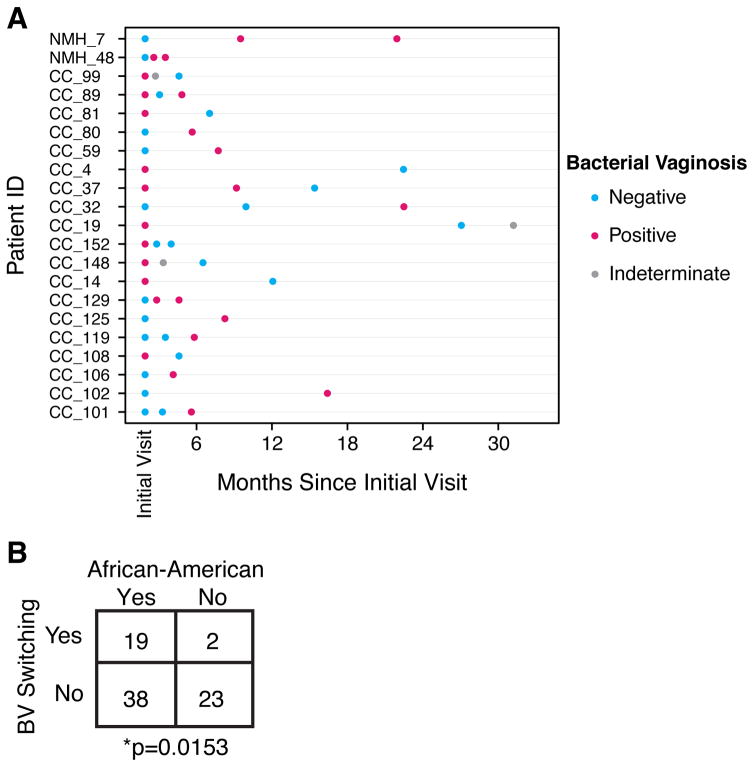
Longitudinal data for each woman provided an opportunity to investigate individual changes in BV status. Eighty-two women provided at least 2 samples with a known BV status for several samples, excluding instances when the examining physician categorized the BV state as “indeterminate”. The timing between samples was comparable in women from both hospitals.
To investigate longitudinal dynamics, we thus compared the frequency of switching between BV and non-BV states. Of the 82 women, 21 switched BV status in the study. 19 of the 21 women that switched BV status were from CC while only 2 were from NMH (Figure 4, Fisher’s Exact Test p=0.007). Both NMH women switched from being BV negative to positive; 11 of the 19 (58%) CC women switched from BV negative to positive.
Figure 4.
Association between BV switching and self-identified race. A) Each patient who switched BV status is shown. The dots are arranged based on time since initial visit. B) Contingency table showing the frequency of BV status switching, comparing African American women to other women in the study.
The propensity to switch states was next queried relative to sample metadata. Self-identified African American women were found to switch BV status more frequently (Figure 4B, Fisher’s Exact Test p=0.015). This was found to account for the greater frequency of switching in the comparison between hospitals, because the CC cohort included a higher percentage of African American women. Within African American women, HIV status did not correlate with switching frequency (Fisher’s Exact Test p=0.6).
Comparison of cervical mucus and cervicovaginal mucus
This study is the first to compare cervical mucus and cervicovaginal mucus within the same women (n=146). These two sample types differ in site and procedure of collection (described in methods and [32]). We first investigated the bacterial composition of the cervical mucus samples (Supplemental Figure 4A). Both cervical mucus and cervicovaginal mucus were dominated by Lactobacillus and showed similar microbial diversity (Supplemental Figure 4B). No systematic differences were detected between sample types. When we attempted to assess differences between CM and CVM based on bacterial abundance using Random Forest, we were unable to predict sample type better than guessing.
We investigated the extent of differences by comparing the within-subject differences between sample types to between subject differences within a sample type. Samples were compared using unweighted (Supplemental Figure 4C) and weighted Unifrac (Supplemental Figure 4D). We found that samples of either of the two types were more similar if they came from the same subject than samples of the same type from different subjects (p<0.001 for weighted and p<0.001 for unweighted). Furthermore, samples within the same subject taken at the same time point, but from different sites, were the most similar. Thus no significant differences between sample types were detectable by 16S rRNA gene sequencing.
Discussion
Here we report a study of the association of HIV infection and the vaginal microbiota in women sampled at two Chicago hospitals. The cohorts sampled, though from the same city, differed in median income, health, racial and demographic factors. Women treated at both hospitals achieved good virologic control (<40 viral RNA copies per ml of blood). Thus the HIV+ subjects were relatively healthy, likely accounting for the relatively modest differences seen in the vaginal microbiota associated with HIV status. Analysis of the vaginal microbiome showed notable associations between high diversity states, high pH, and BV as inferred from Nugent scores, paralleling previous studies [9, 11, 18, 39]. Previous studies have differed on associations between HIV status and BV[26–28, 30, 31]—here we found that HIV+ status was correlated with BV status.
Our most unexpected finding came from the longitudinal analysis, in which switching of BV status turned out to be associated with self-identified African-American race, though this was not detectably linked to HIV status. This finding accounted for more frequent switching by the women sampled from the CC hospital. For some women, the vaginal microbiota has been reported to show longitudinal variation, in some cases associated with the menstrual cycle but also on other time scales as well [40, 44–46]. The factors driving longitudinal dynamics have not been fully clarified. The fluctuations in BV status were not associated with HIV infection, though the numbers of switchers identified was low and so limited power to detect association. The basis of longitudinal instability in vaginal microbiome structure among African-American women is unknown and was not obviously associated with any of our metadata. Candidate explanations include genetic differences or cultural differences such as hygenic practices.
This study has several limitations. We did not collect detailed data on diet, behavior, and hygiene practices which might have influenced microbial community structure. Although our sample size is relatively large, the heterogeneity of the subjects limited power. Finally, the fact that patients were well cared for limited the number of subjects with low CD4 counts and high viral loads, which would have been useful to compare to other studies that reported larger differences in the vaginal microbiota associated with HIV infection.
BV has been associated with de novo HIV infection in previous work[29]—in contrast, most of the women studied here had been HIV positive for many years, but still a positive association between HIV and BV was detectable. Various models may explain this observation. It is possible that women can develop a persistent long term state where BV is frequent, and that this state is associated with more frequent HIV infection. Alternatively, HIV infection may promote BV, despite effective antiretroviral therapy. Still another possibility is that high frequencies of BV are markers for socio-economic, genetic, or other factors that are positively associated with HIV infection as well without being directly causal. At present it seems simplest to invoke the first model, since inflammation associated with BV may result in recruitment of HIV infection target cells to the mucosa, and so provides a simple explanation for the data [29]. If correct, this emphasizes that interventions designed to reprogram the persistent frequent BV state may help prevent HIV transmission.
Methods
Human Subjects
CVM and CM samples were obtained from donors after written informed consent, under a protocol approved by the Institutional Review Board at Northwestern University, as per a previously described protocol [32]. Premenopausal women were enrolled throughout the menstrual cycle and returned for up to 3 visits, with a minimum of one month between sampling times. CVM was collected using an Instead SoftCup (EvoFem, San Diego, CA), which was inserted into the vagina for a minimum of 30 minutes by each donor, removed, and placed into a 50ml centrifuge tube. CM samples were aspirated with a mucus collection device, Mucat (Sepal Reproductive Devices, Boston, MA), directly from the cervical os. All CM and CVM samples were centrifuged at 780 × g for 10 min, and the clear top layer was collected using a positive displacement pipette. The pH of all mucus samples was measured using a small-volume pH electrode (Metrohm, Riverview, FL). All samples were stored at −80°C until DNA was extracted in batches. The Nugent score was determined by Gram stain of CVM smears and microscopic assessment of the abundance of large, small, or curved Gram-positive rods.
Sequence analysis
Isolated DNA was quantified using the Picogreen system and 50 ng of DNA was amplified in each PCR reaction. Primers were barcoded to label each sample as described previously [47, 48]. PCR reactions were carried out in triplicate using Accuprime (Invitrogen, Carlsbad, CA, USA). Each reaction contained 50 nanograms of DNA and 10 pM of each primer. Primers annealing to the V1V2 region of the 16S bacterial gene were used for amplification. The PCR protocol for 16S amplicons was described previously [34]. Amplified 16S rDNA fragments were purified using a 1:1 volume of Agencourt AmPure XP beads (Beckman-Colter, Brea, CA, USA). The purified products were sequenced on an Illumina MiSeq. All DNA sequence data has been deposited at the NCBI SRA under accession number SRP062720.
Data analysis
Sequence data was processed using QIIME [38], augmented by the R package QIIMER (http://cran.r-project.org/web/packages/qiimer). The 16S rRNA sequences reads were clustered into 97% OTUs using UClust [49], yielding 157726 OTUs. All pairwise distances between samples were calculated using Weighted and Unweighted UniFrac [42]. Results were not corrected for multiple comparisons. Data was displayed using Principal Coordinate Analysis and colored based on different metadata variables. All statistical calculations and plots were carried out using R.
Supplementary Material
Supplemental Figure 1. Analysis of the data set using unweighted Unifrac. A) Ordination based on Principal Coordinate Analysis showing the distribution of samples, where color indicates the proportion of Lactobacillus in the sample, and the size of the point indicates Shannon diversity (black dots in legend to the right). The same ordination is colored based on B) HIV status, C) hospital of origin, D) Nugent score, E) pH, F) the proportion of pro-inflammatory genera, and G) cervicotype based on the dominant Lactobacillus species or pro-inflammatory genera.
Supplemental Figure 2. Associations of BV and sample metadata. A) Association of Nugent score and Shannon diversity. The 16S data from each microbiome sample was quantified using the Shannon diversity index, and the levels plotted by Nugent score. B) Association of Nugent score and vaginal pH.
Supplemental Figure 3. Proportion of Lactobacillus across hospital and HIV infection status. Histograms show the distribution of the proportions of Lactobacillus, stratified by patients’ HIV infection status and hospital, Stroger Hospital of Cook County (CC) or Northwestern Memorial Hospital (NMH).
Supplemental Figure 4. Microbiome structure in cervical mucus (CM) samples. A) Heat map showing the types of bacteria present. B) Comparison of the Shannon diversity in CM versus CVM samples. C) Comparison of unweighted UniFrac distances within and between sample types. D) Comparison of weighted UniFrac distances within and between sample types.
Supplemental Table 1. Demographics of the women studied. Where appropriate, median values are listed with parentheses showing the minimum and maximum values. Counts, of patients or samples, are listed for the remaining categories.
Acknowledgments
We are grateful to members of the Hope and Bushman laboratories for help and suggestions. We thank Laurie Zimmerman for her assistance with the figures. This work was supported by R01 AI 052845 to F.D.B., the Penn Center for AIDS Research (P30 AI 045008), the PennCHOP Microbiome Program, P01 AI082971 to A.L.F and T.J.H., R33 AI094584 to T.J.H., and OPP1031734 from the Bill and Melinda Gates foundation (BMFG) to T.J.H. Hope. IRB approval numbers were STU00041328 and STU00025456. C.C. and S. S.-M. were supported by T32 AI007632.
References
- 1.Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microbial ecology in health and disease. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu GD, Compher C, Chen EZ, Smith SA, Shah RD, Bittinger K, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 2016;65(1):63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352(6285):544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams B, Landay A, Presti RM. Microbiome alterations in HIV infection a review. Cellular microbiology. 2016 doi: 10.1111/cmi.12588. [DOI] [PubMed] [Google Scholar]
- 8.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell host & microbe. 2016;19(3):311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB, et al. Deep sequencing of the vaginal microbiota of women with HIV. PloS one. 2010;5(8):e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal immunology. 2014;7(4):983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10(2):e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1(1):26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gori A, Tincati C, Rizzardini G, Torti C, Quirino T, Haarman M, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46(2):757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, Poles MA, Fisch GS, Ma Y, Nossa C, Phelan JA, et al. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30(1):19–29. doi: 10.1097/QAD.0000000000000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michelet C, Arvieux C, Francois C, Besnier JM, Rogez JP, Breux JP, et al. Opportunistic infections occurring during highly active antiretroviral treatment. AIDS. 1998;12(14):1815–1822. doi: 10.1097/00002030-199814000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Crowe SM, Carlin JB, Stewart KI, Lucas CR, Hoy JF. Predictive value of CD4 lymphocyte numbers for the development of opportunistic infections and malignancies in HIV-infected persons. J Acquir Immune Defic Syndr. 1991;4(8):770–776. [PubMed] [Google Scholar]
- 17.Moore RD, Chaisson RE. Natural history of opportunistic disease in an HIV-infected urban clinical cohort. Ann Intern Med. 1996;124(7):633–642. doi: 10.7326/0003-4819-124-7-199604010-00003. [DOI] [PubMed] [Google Scholar]
- 18.Masur H, Ognibene FP, Yarchoan R, Shelhamer JH, Baird BF, Travis W, et al. CD4 counts as predictors of opportunistic pneumonias in human immunodeficiency virus (HIV) infection. Ann Intern Med. 1989;111(3):223–231. doi: 10.7326/0003-4819-111-3-223. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan JE, Hanson D, Dworkin MS, Frederick T, Bertolli J, Lindegren ML, et al. Epidemiology of human immunodeficiency virus-associated opportunistic infections in the United States in the era of highly active antiretroviral therapy. Clin Infect Dis. 2000;30(Suppl 1):S5–14. doi: 10.1086/313843. [DOI] [PubMed] [Google Scholar]
- 20.Nunn KL, Wang YY, Harit D, Humphrys MS, Ma B, Cone R, et al. Enhanced Trapping of HIV-1 by Human Cervicovaginal Mucus Is Associated with Lactobacillus crispatus-Dominant Microbiota. mBio. 2015;6(5):e01084–01015. doi: 10.1128/mBio.01084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5(193):193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buve A, Jespers V, Crucitti T, Fichorova RN. The vaginal microbiota and susceptibility to HIV. AIDS. 2014;28(16):2333–2344. doi: 10.1097/qad.0000000000000432. [DOI] [PubMed] [Google Scholar]
- 23.Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev. 2013;37(5):762–792. doi: 10.1111/1574-6976.12029. [DOI] [PubMed] [Google Scholar]
- 24.Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Current opinion in HIV and AIDS. 2008;3(3):356–361. doi: 10.1097/COH.0b013e3282f9ae9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 26.Salas JT, Chang TL. Microbiome in human immunodeficiency virus infection. Clinics in laboratory medicine. 2014;34(4):733–745. doi: 10.1016/j.cll.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgdorff H, Tsivtsivadze E, Verhelst R, Marzorati M, Jurriaans S, Ndayisaba GF, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. The ISME journal. 2014;8(9):1781–1793. doi: 10.1038/ismej.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masese L, Baeten JM, Richardson BA, Bukusi E, John-Stewart G, Graham SM, et al. Changes in the contribution of genital tract infections to HIV acquisition among Kenyan high-risk women from 1993 to 2012. AIDS. 2015;29(9):1077–1085. doi: 10.1097/QAD.0000000000000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcendor DJ. Evaluation of Health Disparity in Bacterial Vaginosis and the Implications for HIV-1 Acquisition in African American Women. Am J Reprod Immunol. 2016;76(2):99–107. doi: 10.1111/aji.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta SD, Donovan B, Weber KM, Cohen M, Ravel J, Gajer P, et al. The vaginal microbiota over an 8- to 10-year period in a cohort of HIV-infected and HIV-uninfected women. PloS one. 2015;10(2):e0116894. doi: 10.1371/journal.pone.0116894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benning L, Golub ET, Anastos K, French AL, Cohen M, Gilbert D, et al. Comparison of lower genital tract microbiota in HIV-infected and uninfected women from Rwanda and the US. PloS one. 2014;9(5):e96844. doi: 10.1371/journal.pone.0096844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukair SA, Allen SA, Cianci GC, Stieh DJ, Anderson MR, Baig SM, et al. Human cervicovaginal mucus contains an activity that hinders HIV-1 movement. Mucosal immunology. 2013;6(2):427–434. doi: 10.1038/mi.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann JN, You HM, Hedberg EC, Jordan JA, McClintock MK. Prevalence of bacterial vaginosis and Candida among postmenopausal women in the United States. J Gerontol B Psychol Sci Soc Sci. 2014;69(Suppl 2):S205–214. doi: 10.1093/geronb/gbu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS pathogens. 2008;4(2):e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic acids research. 2007;35(13):e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36(18):e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith SB, Ravel J. The vaginal microbiota, host defence and reproductive physiology. J Physiol. 2016 doi: 10.1113/JP271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42(5):965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaban B, Links MG, Jayaprakash TP, Wagner EC, Bourque DK, Lohn Z, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014;2:23. doi: 10.1186/2049-2618-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112(35):11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert JA, John S, Sobel JD, Akins RA. Longitudinal analysis of vaginal microbiome dynamics in women with recurrent bacterial vaginosis: recognition of the conversion process. PloS one. 2013;8(12):e82599. doi: 10.1371/journal.pone.0082599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4(2):e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35(13):e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Analysis of the data set using unweighted Unifrac. A) Ordination based on Principal Coordinate Analysis showing the distribution of samples, where color indicates the proportion of Lactobacillus in the sample, and the size of the point indicates Shannon diversity (black dots in legend to the right). The same ordination is colored based on B) HIV status, C) hospital of origin, D) Nugent score, E) pH, F) the proportion of pro-inflammatory genera, and G) cervicotype based on the dominant Lactobacillus species or pro-inflammatory genera.
Supplemental Figure 2. Associations of BV and sample metadata. A) Association of Nugent score and Shannon diversity. The 16S data from each microbiome sample was quantified using the Shannon diversity index, and the levels plotted by Nugent score. B) Association of Nugent score and vaginal pH.
Supplemental Figure 3. Proportion of Lactobacillus across hospital and HIV infection status. Histograms show the distribution of the proportions of Lactobacillus, stratified by patients’ HIV infection status and hospital, Stroger Hospital of Cook County (CC) or Northwestern Memorial Hospital (NMH).
Supplemental Figure 4. Microbiome structure in cervical mucus (CM) samples. A) Heat map showing the types of bacteria present. B) Comparison of the Shannon diversity in CM versus CVM samples. C) Comparison of unweighted UniFrac distances within and between sample types. D) Comparison of weighted UniFrac distances within and between sample types.
Supplemental Table 1. Demographics of the women studied. Where appropriate, median values are listed with parentheses showing the minimum and maximum values. Counts, of patients or samples, are listed for the remaining categories.