Abstract
Different olfactory cortical regions are thought to harbor distinct sensory representations, enabling each area to play a unique role in odor perception and behavior. In the piriform cortex (PCx), spatially-dispersed sensory inputs evoke activity in distributed ensembles of neurons that act as substrates for odor learning. In contrast, the posterolateral cortical amygdala (plCoA) receives hardwired inputs that link specific odor cues to innate olfactory behaviors. Here we show that, despite stark differences in the patterning of plCoA and PCx inputs, odor-evoked neural ensembles in both areas are equally capable of discriminating odors, and exhibit similar odor tuning, reliability and correlation structure. These results demonstrate that brain regions mediating odor-driven innate behaviors can, like brain areas involved in odor learning, represent odor objects using distributive population codes; these findings suggest both alternative mechanisms for the generation of innate odor-driven behaviors and additional roles for the plCoA in odor perception.
Introduction
Sensory systems contain multiple internal representations of the external world. A hierarchy of interconnected areas constructs these representations by performing sequential transformations that extract progressively higher-order stimulus features. However, this ordered flow can branch into parallel streams that format sensory representations in distinct ways optimized for different computational and behavioral functions.
For example, in the olfactory system, information is transformed in serial and parallel as it flows from the nose to the brain. In the main olfactory system of rodents, odors are detected by odor receptors expressed by olfactory sensory neurons (OSNs). Each mature OSN expresses a single receptor gene, and elaborates an axon that innervates a single glomerulus (of thousands) within the main olfactory bulb (OB); the specific glomerulus innervated by a given OSN is determined by the receptor expressed by that OSN (Axel, 1995). The convergence of OSN axons into glomeruli both organizes odor information into receptor-specific channels and sorts those channels in space based, at least in part, upon the behavioral significance of the odor: innately-relevant odors (e.g., the fox odor trimethylthiazoline (TMT), the spoiled food odor 2-methylbutyrate, and aversive and appetitive amines) activate glomeruli within dorsal and dorsolateral bulbar subdomains, while more “neutral” odors are encoded by glomeruli distributed across the bulb surface (Dewan et al., 2013; Kobayakawa et al., 2007; Mori and Sakano, 2011).
This singular (albeit tessellated) stream of olfactory information is then projected forward by neurons called mitral and tufted (MT) cells. MT cells innervate multiple higher brain targets in parallel, each of which is thought to play a unique function in odor perception and behavior (Haberly, 2001). Perhaps the best studied of these is the piriform cortex (PCx), which both neural recordings and functional experiments have implicated in various forms of odor learning. For example, disruption of PCx function interferes with retrieval of odor memories, whereas optogenetic actuation of ensembles of PCx neurons can substitute for odor cues in an odor-context pairing task (Choi et al., 2011; Sacco and Sacchetti, 2010). In contrast, the posterolateral cortical amygdala (plCoA), which also receives inputs from MT cell axons, is thought to mediate innate odor-driven behaviors, as plCoA neurons are necessary and sufficient for innate appetitive and avoidance responses to specific monomolecular odorants (Root et al., 2014).
How are sensory representations within PCx and plCoA organized to support their distinct behavioral functions? The PCx receives MT cell axons that are splayed across the surface of the PCx in a distributive pattern. This spatial mixing of inputs from all glomeruli, together with broad associational connectivity intrinsic to the PCx, has two main consequences: individual PCx pyramidal neurons can represent information about discontinuous subsets of odor space (with respect to chemical structure, behavioral meaning and glomerular identity), and both monomolecular odorants and odor mixtures activate distributed and overlapping ensembles of PCx neurons whose activity signifies odor identity (Ghosh et al., 2011; Illig and Haberly, 2003; Miyamichi et al., 2011; Sosulski et al., 2011; Stettler and Axel, 2009). The PCx therefore represents odor objects through a population code, in which the coordinated activity of groups of neurons affords downstream brain regions more information about the sensory environment than possible if these neurons were considered individually.
Although neurons in plCoA have been shown to respond to odorants, their tuning properties have not yet been defined (Bergan et al., 2014; Govic and Paolini, 2015; Root et al., 2014; Staples et al., 2008). The plCoA receives input from both the olfactory bulb and the PCx, and harbors intrinsic associational connections that distribute information locally (Price, 1973; Schwabe et al., 2004). However, in contrast to the PCx, the axonal inputs from the olfactory bulb to the plCoA target patch-like subdomains whose location is glomerulus-specific and stereotyped from animal to animal, suggesting that connections between MT cells in the bulb and recipient cells in the plCoA are hardwired during development (Sosulski et al., 2011). The plCoA is also preferentially innervated by glomeruli in the dorsal olfactory bulb, raising the possibility that sensory responses to innately-relevant cues may be over-represented in the plCoA (Miyamichi et al., 2011). Furthermore, immediate early gene staining has suggested that aversive odors (including TMT, isopentyl amine and 4-methylthiazole) evoke activity in a spatially-distinct subset of plCoA neurons that do not respond to appetitive odors (such as 2-phenylethanol (2-PE)) (Root et al., 2014).
These observations suggested a working model in which the plCoA mediates innate odor-driven behaviors through hardwired and spatially-segregated labeled lines that couple information from the periphery to circuits that drive innate behaviors. In principle, neurons participating in labeled lines in the plCoA could exhibit specific sensory responses to innately-relevant odors (e.g., TMT, 2-PE), to behaviorally-relevant chemical classes (e.g., thiazoles, amines), or to odor valences (e.g., appetitive or aversive), as each of these response types could be meaningfully used to generate an appropriate behavioral response.
However, because no large-scale recordings of the plCoA have been previously performed, it is unclear how odor representations are functionally organized in the plCoA to support the generation of innate behaviors. To address this question, here we systematically record neural responses in plCoA to a variety of ethologically-relevant and neutral monomolecular odors and odor mixtures. We find that odor-driven responses in plCoA strikingly resemble those apparent in PCx, suggesting that odor objects are represented in the plCoA via a population code. Like their counterparts in PCx, the responses of individual plCoA neurons convey little reliable information about odor identity, chemical class or odor valence. Rather, the plCoA represents sensory information through the activity of neural ensembles that can signify odor identity and odor concentration. These findings argue that odor-driven innate behaviors can be supported by a distributive mode of information coding in the olfactory system.
Results
To characterize responses in plCoA neurons, awake head-fixed mice were exposed to different odors as extracellular records were obtained using four-shank silicon probes targeted to layers II and III (Figures S1A–C). As a control, similar recordings were obtained in independent animals from neurons in the anterior PCx. Naïve mice without any prior odor training or reward association were exposed to four distinct odor panels, which included appetitive, aversive and neutral monomolecular odorants as well as natural odor mixtures (see STAR Methods for odor panel definitions); three panels included the aversive odor TMT and the appetitive odor 2-PE, whose behavioral effects depend upon the activity of specific neurons in plCoA (Root et al., 2014). Mice were free to run on a treadmill during the experiment, and exhibited both reliable odor-specific locomotor responses during recordings and typical approach and avoidance behaviors to innately-relevant odors after the recording session (Figures S1D, S1F). Spike sorting identified a total of 1080 single units from the plCoA of 39 mice and 868 single units from the PCx of 31 mice, yielding a total of 23,492 cell-odor pairs.
Raster plots in Figure 1A illustrate typical patterns of activity observed in neurons in the plCoA and PCx before, during and after the presentation of distinct monomolecular odorants. Baseline levels of neural activity were different in these two brain regions, with the plCoA exhibiting significantly lower firing rates than the PCx (Figure 1B). While a substantial fraction of cells in PCx exhibited spontaneous firing rates of 5 Hz or more, few such highly spontaneously-active cells were observed in plCoA (Figure 1B).
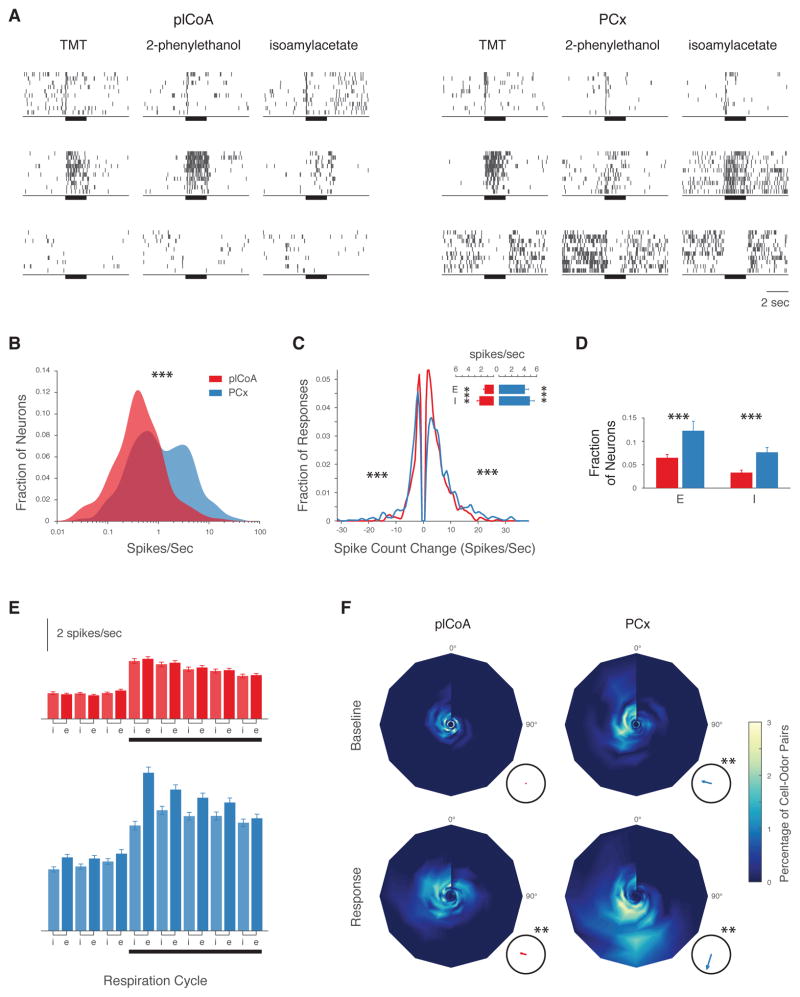
Figure 1. The Posterolateral Cortical Amygdala Exhibits Dynamic and Diverse Odor Responses Like Piriform Cortex.
A. Example raster plots of odor responses (y axis = 10 trials) in plCoA and PCx. Odors were presented for 2 seconds (bar). B. Spontaneous firing rate distributions in plCoA (535 neurons, red) and PCx (339 neurons, blue; median plCoA: 0.52 spikes/sec, median PCx: 1.32 spikes/sec, p<0.001, Wilcoxon rank sum test). C. Odor response magnitude histogram (firing rate change during the first second of odor presentation) in plCoA (red) and PCx (blue); median response amplitude plCoA: 3.9 spikes/sec, median PC: 5.7 spikes/sec; p < 0.001, Wilcoxon rank sum test. Inset: Average baseline firing rates (± SEM) for neurons exhibiting significant excitatory (E) and inhibitory (I) responses in plCoA (red) and PCx (blue). D. Average response fraction of neurons in plCoA (red) or PCx (blue) exhibiting excitatory (E) or inhibitory (I) odor responses (excitatory responses: 6% in plCoA vs 11% in PCx; inhibitory responses: 3% in plCoA vs 8% in PCx; p<0.001, χ2 test). E. Mean (±SEM) baseline and odor-evoked firing in plCoA (red) and PCx (blue) during the inspiratory (i) or expiratory (e) phases of the sniff cycle. F. Phase-intensity plots illustrating the distribution of the phase and firing rate of neuronal activity relative to the onset of the last respiration cycle before odor onset (baseline) and of the first cycle after onset (response). The plot angle indicates peak phase; radius indicates peak firing rate (spikes/sec, maximum is 15 Hz); color map indicates the proportion of cell-odor pairs exhibiting any given phase and rate. Insets: the mean phase of the population (arrow angle) and the concentration of the data around the mean (arrow length, where perfect concentration = 1). The mean phase in both brain areas is not uniformly distributed across the respiration cycle during odor responses (p<0.01, Raleigh’s uniformity test).
Despite this difference in spontaneous firing rates, the overall character of neural responses to odors was remarkably similar in both brain regions. Odor presentation modulated the activity of plCoA and PCx neurons, with most responses being excitatory (Figures 1C, 1D). Inspection of odor-evoked responses revealed a diversity of temporal response patterns, varying from conventional odor-locked excitation to “off”-responses in which neurons responded to the cessation of odor delivery (Figure 1A). Sorting of neurons into clusters based upon the dynamics of their odor responses revealed that the distribution of response dynamics was similar in the plCoA and PCx, as was the average response onset (data not shown); furthermore, in both areas odor-driven responses were modulated by respiration (Figures 1E, 1F) (Litaudon et al., 2003; Miura et al., 2012). Spectral analysis of the local field potential in the plCoA revealed odor-elicited beta activity, which is characteristic of sensory responses in PCx and other olfactory cortical areas (Figures S2A–C) (Neville and Haberly, 2003; Poo and Isaacson, 2009). However, the density and amplitude of odor responses were lower in plCoA than in PCx, and the peak of odor responses in plCoA were phase delayed with respect to PCx (Figures 1D, 1G, S3A, S3B). These differences, together with the lower observed spontaneous activity in plCoA, suggest that plCoA neurons may have lower membrane excitability than PCx neurons, or may be systematically subjected to lower levels of excitation and/or greater levels of inhibition.
Response Reliability is Similar in plCoA and PCx
Given that projections from a single glomerulus to the plCoA are not dispersed but spatially clustered — and that plCoA neurons may therefore be capable of averaging signals from multiple MT cells representing the same glomerulus — it is possible that individual plCoA neurons respond to odors more reliably than those in PCx; such increased reliability, in turn, could enable small numbers of plCoA neurons to accurately convey the presence of an innately-relevant cue on each trial. We therefore assessed the reliability of binary responses in both plCoA and PCx on a trial-by-trial basis, using a threshold to identify neurons that responded to a given presentation of odor. Individual neurons in PCx responded on a subset of trials to individual odorants; on average, single neurons did not reliably report the presence of a given odorant on every trial (Figure 2A). The observed distribution of response reliabilities in plCoA was indistinguishable from that in PCx, both in general and when neutral, appetitive and aversive odorants were independently compared (Figures 2A, 2B). In addition, on those trials in which a given neuron responded, the magnitude of the response did not vary systematically with odor valence (which here we take as synonymous with appetitive or aversive) in either PCx (as has been previously shown) or plCoA (Figures 2C, 2D) (Stettler and Axel, 2009).
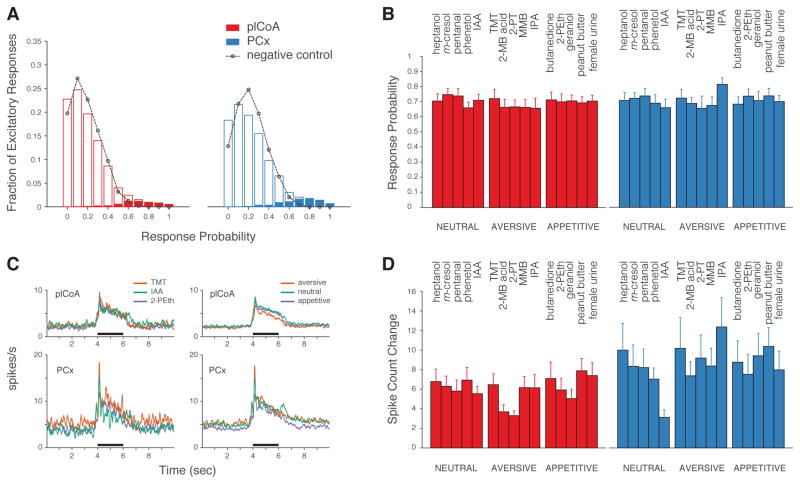
Figure 2. Similar Reliability of Odor-Evoked Responses in plCoA and PCx Regardless of Odor Identity or Valence.
A. Histogram depicting the probability of an excitatory response out of ten trials (see STAR Methods). Open bars (red = plCoA, blue = PCx) depict the fraction of cell-odor pairs with a given excitatory response probability; filled bars represent only those cell-odor pairs whose responses are considered significant by auROC analysis. Black dotted lines: distribution of false-positive responses in absence of odor presentation; note that this rate reflects the level of spontaneous activity in each brain area. B. Response probabilities for cell-odor pairs with a significant excitatory response for five neutral, aversive and appetitive odors; no significant differences were observed (three factors: valence, odor identity and area, three-way ANOVA). C. (Left) Grand averages of peri-stimulus time histograms of excitatory responses to isoamyl acetate/neutral (green), TMT/aversive (red) and 2-phenylethanol/appetitive (blue). (Right) Same as (Left) with all odors considered, grouped by valence: neutral (green), aversive (red) and appetitive (blue). Odor period (black) is demarked. D. Spike count change during presentation of five neutral, aversive and appetitive odorants for those neurons that had an excitatory response. Firing rate was not significantly modulated by the innate valence of odors (three factors: valence, odor identity and area, p<0.05 only for difference between areas, three-way ANOVA).
Consistent with these findings, the average Fano Factor (spike count variance divided by the mean) of plCoA and PCx neurons both before and during odor exposure was significantly above one (the value expected for a Poissonian process), indicating substantial across-trial variability of spike counts when assessed using this threshold-free metric; this variability fell during the odor presentation, as has been previously observed (Churchland et al., 2010; Miura et al., 2012), and did not differ based upon odor valence (Figures S4A, S4B, data not shown). Importantly, throughout the baseline, odor response and recovery periods the Fano Factor was statistically indistinguishable between the plCoA and PCx. Although response habituation can contribute to observed across-trial variability — and the number of odor-evoked spikes in both plCoA and PCx decreased across trials (Figure S4C) — odor-responsive neurons were only slightly less likely to respond at the end of the experiment than at the beginning, suggesting that the across-trial response variability was not dominated by habituation effects (Figure S4D).
Taken together, these data demonstrate that the across-trial variability that characterizes responses in awake PCx is present and qualitatively similar in the plCoA, and that the degree of this response variability does not depend upon the innate behavioral meaning of the odor. These observations further demonstrate that (under naïve conditions) odor identity is not generally represented in plCoA via reliable firing of neurons that faithfully represent the presence of a given cue on every trial; our results suggest that odor identity may instead be represented during single trials using a population code, as pooling information from multiple individual neurons could be used to reliably identify odors on each trial.
Individual Neurons in plCoA and PCx Exhibit Similar Odor Tuning Properties
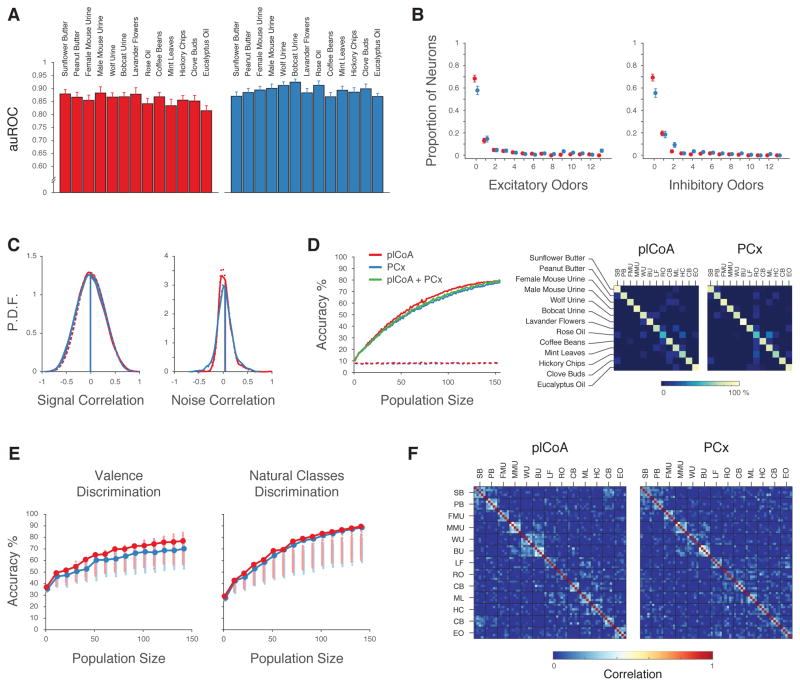
Neurons that participate in population codes can be broadly tuned to odor cues, narrowly tuned, or a mixture of both, while neurons that are part of labeled lines respond to a limited subset of odor space. We therefore characterized and compared the odor tuning properties of individual neurons in plCoA and PCx. To identify neurons whose responses distinguish the presence of a specific odor from background when all trials are considered, we performed an Area-Under-the-Receiver-Operator-Curve (auROC) analysis. Consistent with the results observed by thresholding single-trial responses, the average auROCs of odor-responsive plCoA and PCx neurons (which can be taken as a surrogate for “responsiveness” as it incorporates both overall reliability and response magnitude) were similar regardless of odor identity or valence (Figures 3A, 3B).
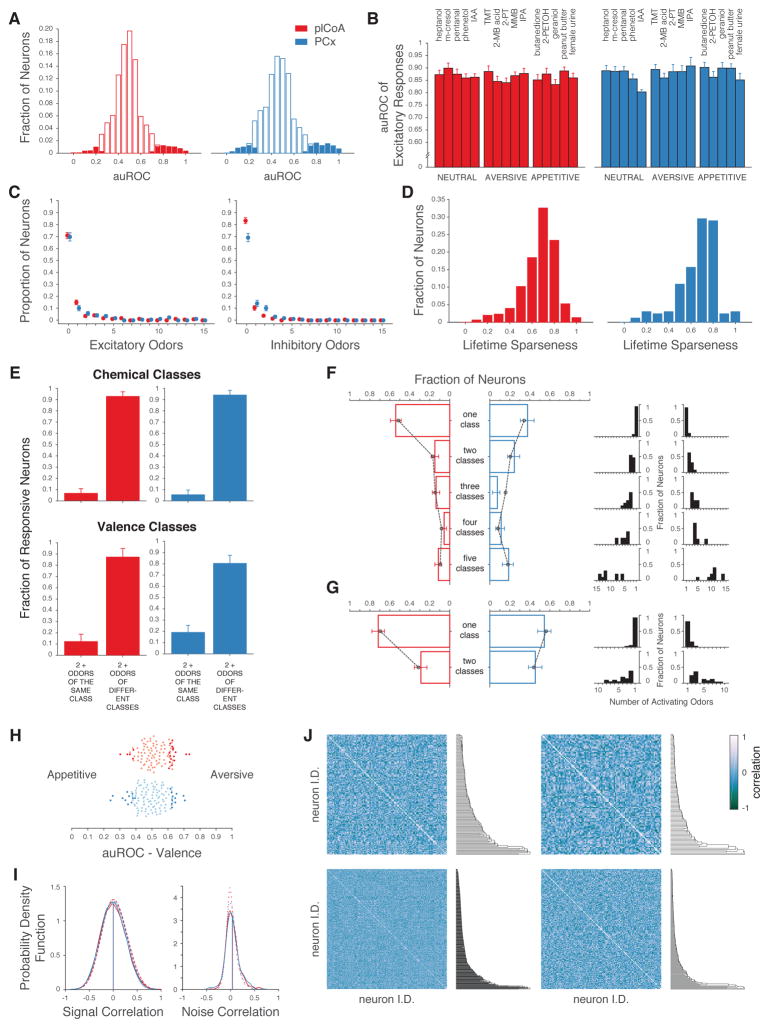
Figure 3. plCoA and PCx Neurons Respond to Limited Subsets of Odor Space.
A. Histograms of the discriminability of olfactory responses in plCoA (red) and PCx (blue) as assessed by response auROC (in which an auROC of 1 indicates a perfectly discriminable excitatory response, and 0 indicates a perfectly discriminable inhibitory response, see STAR Methods). Filled bars: significant responses. Odor response discriminability in plCoA and PCx were similar (p>0.05, permutation test). B. auROCs of significant responses (mean ± SEM) to five neutral, aversive and appetitive odors in plCoA (red) and PCx (blue). Response discriminability was not modulated by the innate valence of odors (three factors: valence, odor identity and area, three-way ANOVA). C, left. Number of monomolecular odorants that significantly activate a given neuron (29 percent of plCoA neurons and 32 percent of PCx neurons activated by at least one odor; 14 percent of plCoA neurons and 11 percent of PCx neurons activated by only one odor out of 15). C, right. Same as C, left. but for odor suppression (16 percent of plCoA neurons and 29 percent of PCx neurons inhibited by at least one odor; 9 percent of plCoA neurons and 13 percent of PCx neurons inhibited by only one odor). The excitatory and inhibitory tuning breadth of neurons was not statistically different between areas (Kolmogorov-Smirnov test). D. Lifetime sparseness (1 = more odor selective) distributions of plCoA (red) and PCx (blue) neurons. E. Fraction of neurons activated by two or more odors of the same chemical or valence class, compared to the fraction activated by two or more odors of different classes in plCoA (red) and PCx (blue). F, left. Fraction of neurons that respond to the indicated number of chemical classes. Black dotted line: null distribution obtained by reshuffling odor labels across classes. F, right. Distributions of the number of odors to which each neuron responded in associated panel on the left. Note that most of the neurons that respond to one class of odorant respond to a single odor, suggesting they may not be “class” specific as they do not generalize across odors within a class. G. Similar to F. but with respect to odor valence (appetitive or aversive). H. Odor valence discriminability of plCoA (red) and PCx (blue) neurons; dark colored dots represent discriminability greater than expected by chance (permutation test). No differences in the significant auROCs between the plCoA and PCx were observed (permutation test). I. Probability density function of signal (left) and noise (right) correlations between neurons that responded to at least one odor in plCoA (red) and PCx (blue); signal and noise correlations observed after shuffling odor labels indicated with the dashed lines. Signal correlations were computed between all pairs of neurons from different experiments using the same odor panel of 15 monomolecular odorants. Noise correlations were computed only between neurons recorded in the same experiment. Observed distributions were not significantly different (t-test, mean signal correlation plCoA = 0.005 ± 0.003, PCx = 0.00025 ± 0.004, noise correlation plCoA = 0.02 ± 0.003, PCx = 0.04 ± 0.007). J. Correlation matrices (Pearson’s r) of plCoA (left panels) and PCx (right panels) of the tuning curves for individual neurons. Neurons are ordered via hierarchical clustering (metric: Pearson’s r, see STAR Methods) as shown by the dendrogram to the right. Bottom panels are for all neurons; top panels include only odor-responsive neurons.
Neurons identified as responsive via the auROC analysis in plCoA and PCx shared similar tuning breadths — most individual neurons in both areas were excited (or inhibited) by a limited number of specific odors, although neurons could be identified that responded to many odorants (Figures 3C, 3D). This distribution of observed tuning breadths was not different between the plCoA and PCx, and was not dissimilar from the tuning breadth distribution observed previously in the PCx of awake mice confronted with novel odors (Zhan and Luo, 2010). Neither plCoA nor PCx neurons exhibited enriched responses to specific chemical classes (e.g., alcohols, aldehydes, amines, phenols, and thiazoles) or odor valences. For example, neurons in both the plCoA and PCx were much less likely to respond to multiple odors of the same class than of different classes (either with respect to chemical class or valence, Figure 3E), and the distribution of number of chemical classes (or valence classes) to which a given neuron responded was neither statistically distinguishable between the plCoA and PCx nor was it greater than that expected by chance (Figures 3F, 3G). Finally, only a small (and similar) number of neurons in plCoA and PCx were capable of specifically discriminating odor valence (as assessed by auROC analysis, Figure 3H). Taken together, these results demonstrate that individual plCoA and PCx neurons generally respond to limited subsets of odor space, and that there are not proportionally more neurons that respond to specific odor categories in the plCoA than there are in the PCx.
This analysis of categorical responses requires a priori specification of the categories; however, it is possible that neurons in the plCoA or PCx respond to specific groups of odors (potentially in a manner relevant to innate behavior) based upon properties other than chemical class or odor valence. To address this, we asked whether neurons exhibited similar tuning profiles (“signal correlations”), or similar amounts of trial-to-trial variability about the mean of the response (“noise correlations”), which can reflect shared inputs (Averbeck et al., 2006; Cohen and Kohn, 2011; Pouget et al., 2000). As had been shown previously, neural responses in PCx to multiple odors exhibited both low signal and noise correlations (Figure 3I) (Miura et al., 2012). Odor-responsive neurons within the plCoA also exhibited near-zero signal and noise correlations whose distribution was indistinguishable from that expected by chance (Figures 3I, 3J). These findings demonstrate that the responses of individual plCoA neurons are highly decorrelated, and therefore are not enriched for responses to subsets of odor space; this decorrelation further suggests that plCoA neurons (like those in PCx) may be well suited to participate in population codes for odor identity.
Population Coding for Odor Identity in both plCoA and PCx
To directly assess whether population codes enable plCoA and PCx to efficiently represent sensory information, we asked how well odor identity, chemical class or valence could be decoded from ensembles of neurons in each area. Decoders were instantiated using a support vector machine with a linear kernel, which acts as a classifier whose performance is a surrogate for the ability of neural populations to encode information about stimulus features like odor identity; we favored this linear classification approach because of its technical simplicity and biological plausibility. Linear classifiers were trained using single-trial responses of pseudo-populations of neurons pooled from several individual animals, and then tested using held-out data (with the training and test data randomly selected in 9:1 proportions, and reported accuracies reflecting the average performance of 500 such classifiers for every data point, see STAR Methods for classification details).
We first asked if decoders could correctly identify individual odors (from a set of 15 monomolecular odors) based upon the responses of randomly-chosen neurons from plCoA and PCx, and if so how classification performance changed as the decoder gained access to progressively more neurons in each area. While classifiers trained with small numbers of neurons (either from plCoA or PCx) were ineffective at identifying odors, increasing the number of neurons in the population improved the performance of both plCoA and PCx classifiers (Figure 4A). No systematic confusions between odors were observed in the classifier predictions (Figure 4B). The rate at which classifier performance improved as individual neurons were added to the PCx and plCoA classifiers was nearly identical; this observation suggests that neurons from plCoA and PCx might be interchangeable from the perspective of a decoder, and therefore encode information about odor identity in a similar manner. Consistent with this possibility, a classifier built using randomly selected neurons from both PCx and plCoA exhibited similar performance characteristics to classifiers built using neurons from plCoA or PCx separately (Figure 4A). These findings did not depend the specific implementation of the classifier, as classifiers using a non-linear kernel or least-squares multiple regression gave qualitatively similar results (Figures S5A, S5B).
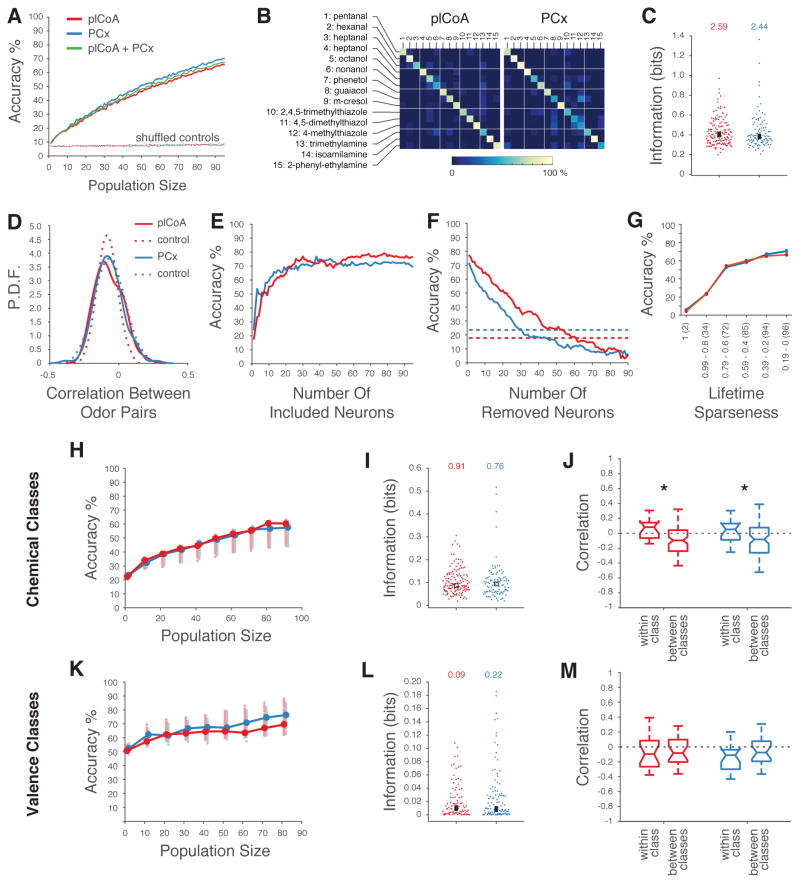
Figure 4. Decoding of Odor Identity from plCoA and PCx Population Activity.
A. Mean accuracy of linear decoders trained to discriminate 15 different monomolecular odors (see STAR Methods). Dashed curves indicate performance after shuffling odor labels for all trials. B. Confusion matrices based upon the linear classifier in A reveal no systematic confusions between odors. C. Distribution of mutual information about odor identity in individual neurons for 15 monomolecular odors in plCoA and PCx (see STAR Methods, no significant differences observed using a Wilcoxon rank sum test). The number indicated on top is the information content of the population; square box indicates the mean. D. Distribution of the pairwise correlation coefficients (Pearson’s r) between ensemble neural representations (using the average response for each neuron) for 15 odors in plCoA (red) and PCx (blue). Control distributions (dashed lines) were obtained by reshuffling odor labels 500 times for each neuron. E. Classification performances obtained after sorting neurons based on their informativeness about odor identity (highest to lowest, as in C.). F. Classification performances in which populations of 90 plCoA and PCx neurons were systematically depleted of neurons in order of their informativeness; dashed lines indicate the performance of the most informative single neurons in plCoA (red) and PCx (blue). Note the sharp initial drop in accuracy in PCx was caused by removing the two highly informative neurons depicted in figure 1C, followed by the equalization of the slopes of the decrementing curves. G. Classification performances obtained after sorting neurons based on their lifetime sparseness (highest to lowest). Note that the number of neurons added at each step was equal for both plCoA and PCx (total number of at each lifetime sparseness indicated within parentheses). H. Linear discriminator accuracies (as in A) of the chemical class of an odor, plotted as a function of the size of plCoA (red) and PCx (blue) populations. 15 odors were grouped in five classes based on their main chemical moiety (e.g., alcohols, aldehydes, amines, phenols, and thiazoles). Circles indicate means. Shaded circles indicate the mean accuracies obtained after randomly grouping the 15 odors in 5 arbitrary classes; this represents chance performance in this experiment. The performances for populations of 70, 80 and 90 plCoA neurons are just above the 97.5th percentile of the controls. I. Distribution of chemical class information in individual neurons across 15 odors in plCoA and PCx, with total population information indicated at the top, and with the box indicating the mean (no significant differences, Wilcoxon rank sum test). J. Pairwise correlation between population vectors representing two odors belonging to the same chemical class or to two different chemical classes. Medians are reported and whiskers represent the interquartile range (p<0.05, t-test). K.– M. Like H. – J. but with respect to odor valence.
Consistent with the nearly identical performance of decoders trained using plCoA or PCx ensembles, the mean amount of information encoded by each plCoA and PCx neuron was similar (Figure 4C). Correlations between individual neurons (which on average were near-zero, see Figures 3I, 3J) did not affect the ability of classifiers to distinguish odors, as classifier performance was nearly identical before and after elimination of signal and noise correlations (Figure S5C). Furthermore, pairwise comparisons of odor representations in plCoA and PCx revealed similar (and near chance) levels of ensemble correlation, a feature that likely supports classifier performance (Figures 4D). Importantly, the observed correlation distributions were similar in plCoA and PCx, consistent with odor-evoked neural ensembles in each of these areas having an equivalent capacity to distinguish odors.
Because randomly selecting the neurons used for classification might understate the relative influence of particularly informative neurons, we also built a series of classifiers in which the most informative individual neurons (as defined by their information content in Figure 4C) were provided to the classifier first. This analysis demonstrates that small numbers of neurons in both areas can drive classification performance (hence odor representations are compact), but that even the most informative neurons contribute fractionally to the ability of the classifier to disambiguate odor identity; importantly, nearly all of the most informative neurons are broadly tuned (Figure 4E, data not shown). Conversely, removing neurons from a classifier in the order of their informativeness only modestly affected classifier performance, consistent with the capacity of plCoA and PCx ensembles to discriminate odors even when composed of relatively less informative neurons (Figure 4F).
Classifiers in which all discriminative information is provided by labeled-line neurons should not benefit from incorporating additional broadly-tuned neurons into the population; thus, observing improvement in classifier performance when broadly-tuned neurons are included in the population would support the argument that important information is not passed entirely through labeled lines composed of specifically-tuned neurons. We therefore built classifiers in which neurons were added in order of tuning breadth, with those neurons that exhibited the highest lifetime sparseness (i.e., are the most odor selective) fed to the classifier first. The performance of plCoA and PCx-based classifiers was nearly identical for all numbers of included neurons (Figure 4G). Critically, incorporating more broadly-tuned neurons monotonically increased odor prediction accuracy at a similar rate in both plCoA and PCx-based classifiers; this result directly demonstrates that both plCoA and PCx harbor population codes for odor identity.
Finally, we asked how the performance of classifiers built with plCoA and PCx neurons compared to the performance of classifiers built with neurons that respond selectively to a single odor, as would be expected for labeled lines for odor identity. We therefore synthetically generated a population of labeled-line neurons from our recording data by replacing the responses of each neuron to all but its preferred odor with baseline levels of activity. As shown in Figure S5D, classifier performance using data derived from plCoA and PCx neurons is higher than that observed for those same neurons after converting them into synthetic labeled lines; this enhanced performance likely reflects the ability of broadly-tuned neurons to more parsimoniously convey information about odor identity that labeled lines that are tuned to a single odorant. Taken together, these data strongly suggest that PCx and plCoA share similar modes of population-based odor identity coding.
Population Responses to both Purified Odors and Natural Mixtures are Similar in both plCoA and PCx
Specific chemical features of odorants are detected by odor receptors, which induce patterns of odor-evoked activity in olfactory bulb glomeruli. However, to date there is little evidence that ensembles of neurons in the mouse PCx are organized according to either lower-order (i.e., chemical features) or higher-order (i.e., odor valence or innate behavioral meaning) olfactory categories; rather, PCx neural populations are thought to encode odor identity in a manner that maximally separates representations of encountered odors (Leinwand and Chalasani, 2011; Wilson and Sullivan, 2011). If ensembles of plCoA neurons instead distinguish odor categories (either chemical or behavioral), decoding circuits downstream of the plCoA could use this information to support the generation of innate patterns of action.
We therefore generated classifiers trained to distinguish the chemical class to which an odor belonged based upon its main functional group; as for classifiers for odor identity, the performance of classifiers trained to discriminate odor classes monotonically increased as additional neurons were added to the population in both plCoA and PCx. However, the observed decoding accuracy may reflect the ability of a classifier to discriminate any group of odors, regardless of their chemical category. We therefore generated a null distribution of decoding accuracy by training classifiers to distinguish any subset of three odors, which is equal to the number of odors that belong to each chemical class within our experiment. Classifiers were unable to distinguish chemical classes using smaller groups of neurons, although decoders built from larger sets of neurons just exceeded the statistical threshold for classification (Figure 4H, compare filled circles to associated shaded circles). Consistent with these observations, the information content of individual neurons about odor class in plCoA and PCx were similar (Figure 4I); furthermore, the correlation distance between odor representations was near zero, although a modest and statistically significant difference within and between chemical classes was observed in both plCoA and PCx (Figures 4J, S5E).
We also used responses to an odor panel composed entirely of appetitive or aversive odorants to train classifiers to discriminate odor valence. In neither plCoA nor PCx, were populations of neurons effective at distinguishing appetitive versus aversive odorants at a rate above chance (as defined using a null distribution) (Figure 4K). Indeed, individual neurons in both plCoA and PCx had similar levels of information about odor valence, and the correlation distances separating odor representations were similar (and near zero) both within and between valence classes (Figures 4L–M, S5F). These results demonstrate that neural populations in plCoA and PCx are not better able to discriminate odor valence than arbitrary, similarly-sized groups of odors of mixed valence, suggesting that information about odor valence is not privileged at the level of population codes in either brain area.
The characterization of odor representations described above was performed using odor panels whose constituents were largely composed of purified monomolecular odors. However, nearly all odors encountered by mice in the wild are complex mixtures derived from natural sources. We therefore repeated both the single-neuron and ensemble-level analyses using natural odor mixtures, including predator and conspecific urines, food-derived odors, and complex odors from natural sources (like mint, coffee and lavender; see STAR Methods for valence and class assignments). As shown in Figure 5, all the key features of natural odor encoding — including response reliability, tuning curves, signal and noise correlation, and odor identity and odor valence classifier performance — were indistinguishable in plCoA and PCx neurons. As was true for chemical classes in the monomolecular odor experiment, a linear classifier trained to distinguish the “class” of natural odor (e.g., predator odors, conspecific odors, food odors, or other) was unable to outperform a control classifier except at the largest population sizes (Figure 5E, right); correlation matrices for the ensemble responses reveal that representations for most odor mixtures are decorrelated, although modest cross-correlations were observed between predator urines and between nut butters in the plCoA and PCx (Figure 5F). These data demonstrate that odor encoding of natural mixtures, like that for monomolecular odors, is similar in both plCoA and PCx.
Figure 5. Similar Encoding for Natural Mixtures in plCoA and PCx.
A. auROCs of responses to 13 natural odor mixtures in plCoA (red) and PCx (blue). Responses were not distinguishable between these areas (two-way ANOVA). B. Number of natural odor mixtures that significantly activate (left) or inhibit (right) a given neuron. The excitatory and inhibitory tuning breadth of neurons were not statistically different between areas (Kolmogorov-Smirnov test). C. Probability density function of signal (left) and noise (right) correlations between individual neurons in plCoA (red) and PCx (blue) in response to natural mixtures. Correlation distribution observed after shuffling odor labels is indicated with the dashed lines. D, left. Accuracy of linear decoders trained to discriminate 13 different natural odor mixtures, with classification accuracy after odor label shuffling indicated in dashed lines. D, right. Confusion matrices of the classifier shown on the left. E. Classifier accuracy at discriminating either valence (left) or ethological class (right) of natural mixtures and controls, computed as in 4H and 4K (see STAR Methods for assignment of individual mixtures to valences or ethological classes). Shaded circles indicate the mean accuracies obtained after randomly grouping the odors in arbitrary classes; this represents chance performance in this experiment. The classification of the ethological class of an odor for population sizes of 20 – 140 neurons is just above the 97.5th percentile of the control distribution. F. Correlation matrices of ensemble odor representations for natural odor mixtures in plCoA and PCx in response to natural odor mixtures; five pseudo-trials (average of two consecutive trials) of each odor (whose identity is indicated by a letter code, and which are in the same order as the odors in panel A) are independently depicted here to reveal cross-trial variability as well as across-odor correlations.
Multiplexed Representations of both Identity and Concentration in plCoA and PCx
Sensory representations in PCx are subject to concentration normalization, a computation in which increases in odor concentration recruit sublinear increases in the total number of activated neurons (Stettler and Axel, 2009). In addition, axonal imaging suggests that changes in odor concentration can have complex non-linear and non-monotonic effects on the firing patterns of individual PCx neurons (Otazu et al., 2015). While labeled lines in principle can signal both odor identity and concentration simultaneously (if, for example, firing rate is related to the concentration of an optimal stimulus), it is not immediately obvious how non-monotonic relationships between concentration and firing rate (like those that have been observed in PCx axons) are consistent with the function of labeled lines for odor identity (Cleland, 2010).
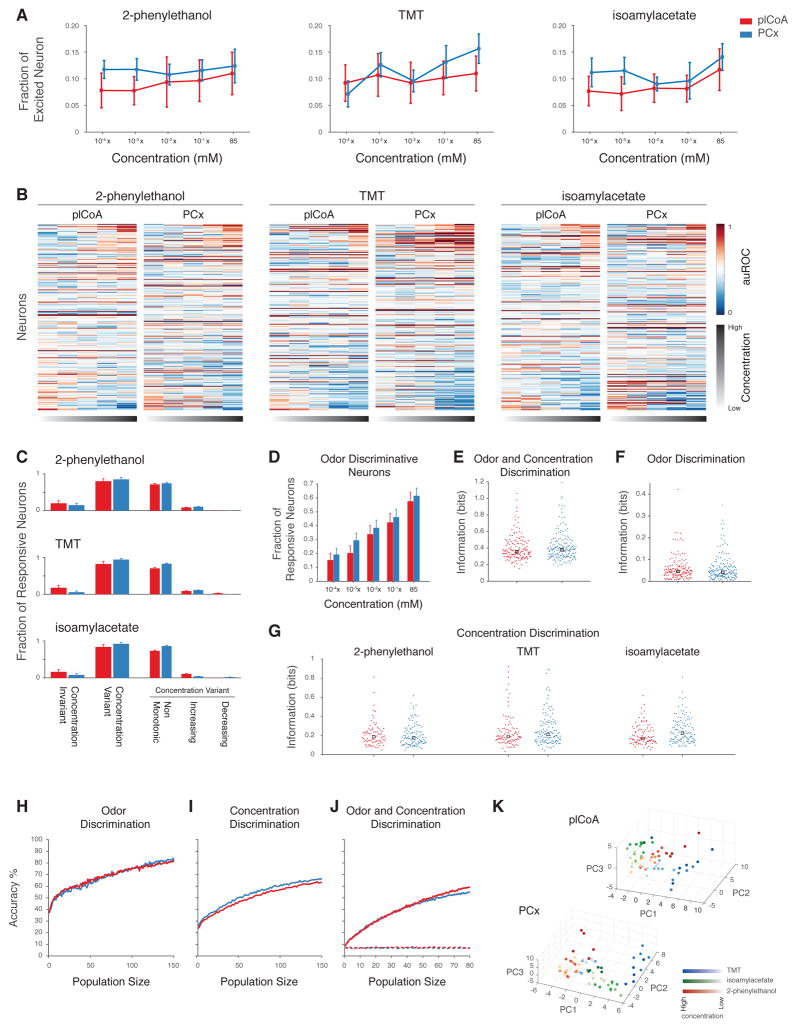
To explore how sensory representations evolve in plCoA and PCx as stimulus concentrations are varied, we exposed mice to three separate odors delivered at concentrations spanning five orders of magnitude. One of the chosen odors (2-phenylethanol, 2-PE) has been previously shown to be appetitive, one has been shown to be appetitive at low concentrations and aversive at higher concentrations (TMT), and one (isoamylacetate) is a neutral control (Root et al., 2014; Saraiva et al., 2016). In general, the total number of responding neurons was stable in response to logarithmic changes in odor concentration in both plCoA and PCx, with no systematic differences observed between brain areas (Figure 6A). Most neural responses in both plCoA and PCx changed as concentrations rose (Figures 6B), but these changes generally unfolded in a non-monotonic manner regardless of the odor tested, suggesting that information about odor identity and concentration may be in part decoupled at the single neuron level in both brain areas (Figure 6C).
Figure 6. Individual plCoA and PCx Neurons Respond Non-Linearly and Non-Monotonically to Increasing Odor Concentrations.
A. Proportion of neurons activated by increasing concentrations of three odors (mean and SEM indicated, no significant changes as assessed by χ2 test). B. Single neuron responses to different concentrations of three odors in plCoA and PCx; the auROC of each cell-odor pair response is depicted (color bar). C. Fraction of plCoA (red) and PCx (blue) neurons that significantly responding to distinct concentrations of the same odor. Of those neurons that are concentration variant, the fraction of plCoA and PCx neurons whose responses change monotonically or non-monotonically to increasing concentrations of an odor is also depicted. See STAR Methods for definitions of concentration variance and monotonicity. D. Fraction of responsive neurons distinguishing odor identity at each concentration in plCoA (red) and PCx (blue). E.–F. Distribution of mutual information about odor (F), concentration (G) and odor and concentration (E) in individual neurons across 3 odors and 5 concentrations in plCoA and PCx (see STAR Methods, no significant differences, Wilcoxon rank sum test). H.–J. Accuracy of linear classifiers using the indicated numbers of neurons at discriminating odor identity (H), odor concentration (I) or both (J) in plCoA (red) or PCx (blue). Dotted line indicates chance performance exhibited after odor label shuffling. K. Principal component plot of ensemble responses (limited to the first three principal components) to the three indicated odors across five concentrations (5 pseudotrials/odor, see STAR Methods).
Consistent with this apparent complexity, individual neurons in both areas can similarly discriminate information about odor identity, odor concentration or both (Figures 6E–G). Interestingly, as odor concentrations increase the number of neurons that can discriminate between different odors rises (Figure 6D) even though the size of the odor-evoked ensembles remains roughly the same (Figure 6A). This discriminative capacity is apparent at the population level as well, as linear classification reveals that neural ensembles from the plCoA and PCx can discriminate odor concentration, can discriminate odor identity, and can discriminate both odor identity and concentration simultaneously (Figures 6H–J). Plotting odor representations in principal components space suggests that the overlap between odor representations decreases as the concentrations of individual odors increases, potentially providing a basis for simultaneous decoding of identity and concentration (Figure 6K).
These data demonstrate that ensembles in both plCoA and PCx respond similarly to changes in odor concentration (regardless of the behavioral meaning of the odor), and that these regions harbor population-level odor representations that can convey information about both odor identity and concentration. Furthermore, most individual neurons in both brain areas exhibit non-linearities and non-monotonicities in their responses to stimuli at different concentrations. While these complex response properties would not be expected from circuits in which information is faithfully conveyed using labeled lines, they can be observed in circuits in which population codes represent multiplexed sensory information (Fusi et al., 2016).
Spatial Isotropy for Odor Responses in plCoA and PCx
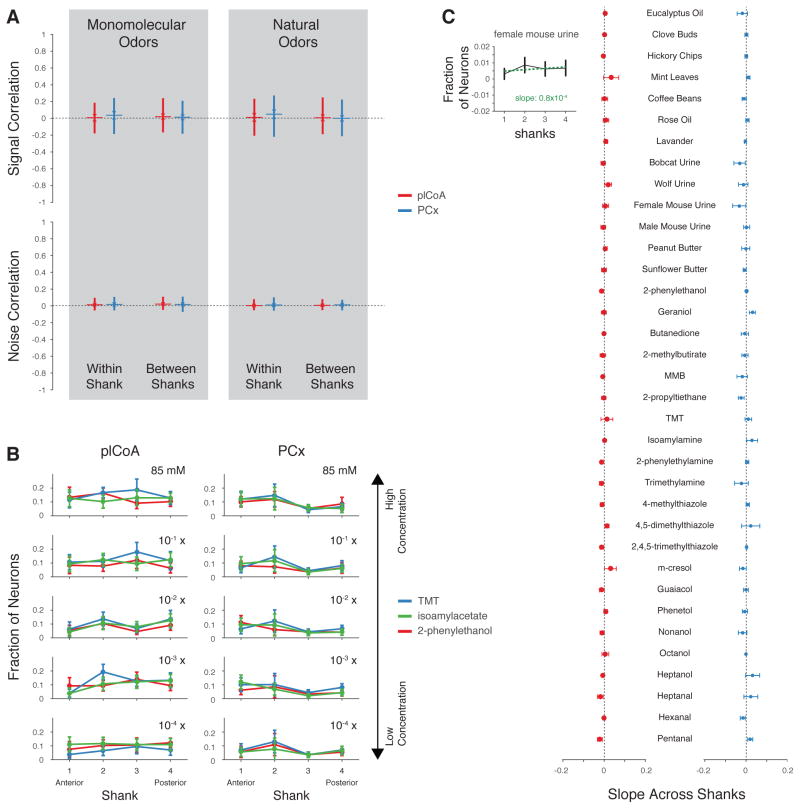
Both anatomical and functional data have suggested that neurons in plCoA, unlike those in PCx, are organized in space based upon the behavioral meaning of the odor to which an animal is exposed (Miyamichi et al., 2011; Root et al., 2014; Sosulski et al., 2011). Neurons in which aversive odors induce immediate early gene expression, for example, appear at least partially segregated from those activated by appetitive odors along the anteroposterior axis of the plCoA; such anisotropies could provide the basis for a coding scheme in which different labeled lines are distinguished based upon their anatomic position in the plCoA (Root et al., 2014). We therefore assessed the spatial distribution of responses across the four shanks of our silicon probe, which were oriented anteroposteriorly and cover 0.6 mm, in both plCoA (which is 1.2 mm in the anteroposterior axis) and PCx.
In both areas, similar levels of pairwise correlations were observed between neurons on the same shank and on different shanks (Figure 7A). Importantly, overt spatial biases in the tuning of neurons to odorants along the anteroposterior axis of plCoA or PCx were not observed, as neurons responding to the appetitive odorant 2-PE and the aversive odorant TMT were isotropically distributed amongst all the shanks in single animals across multiple odor concentrations (Figure 7B). Consistent with this finding, the average slope of the line formed by plotting neural responses to a given odor across the four shanks (which would be expected to be zero if no spatial response biases are present, see Figure 7C, inset) was near-zero for all odors (Figure 7C). These results suggest that neurons do not appear generally clustered in space in plCoA or PCx based on the valence of the odors to which they respond.
Figure 7. Signal Correlations Are Low at Multiple Spatial Scales.
A. Pairwise signal correlation (similarity in odor tuning between pairs of neurons, Pearson’s r) and noise correlations between neurons recorded from the same and different shank of the silicon probe (shanks separated by 200, 400 and 600 μm, plCoA = red, PCx = blue) for the indicated odor sets. B. Observed fraction of responsive neurons on each shank preferring one of the three odors across 5 concentrations. The across shanks slopes (see inset in C) are not statistically different from zero for each odor at all concentrations (permutation test). C. Average slope of the line fit to the distribution of responsive neurons preferring the indicated odor across shanks (see inset) for natural odor mixtures and monomolecular odorants (p>0.05, ANOVA).
Discussion
A major goal of sensory neuroscience is to understand how internal representations of the external world enable perception and behavior. Within the olfactory bulb, the first waystation for olfactory information in the brain, the receptive fields of individual odor receptors are organized into segregated glomerular information channels (Axel, 1995). This discretized odor representation is then reformatted (via spatially dispersed inputs) into a radically different form within the PCx, where the activity of spatially interwoven but highly decorrelated ensembles of neurons represent odor identity (Illig and Haberly, 2003; Miura et al., 2012; Stettler and Axel, 2009). This representational mode is thought to enhance the discriminability of odor objects in the environment (regardless of their specific chemical constituents), and to facilitate various forms of odor learning (Choi et al., 2011; Sacco and Sacchetti, 2010). The plCoA, in contrast, has been functionally implicated in the generation of innate olfactory-driven behaviors and receives hardwired inputs from the bulb, suggesting that it harbors labeled lines directly coupled to specific behavioral output centers (Root et al., 2014; Sosulski et al., 2011).
Here we show that sensory responses in plCoA are organized in a manner that strikingly resembles that apparent in PCx. Individual neurons in both PCx and plCoA exhibit similar odor response reliability and dynamics; odor tuning properties; signal and noise correlations; and information content about odor identity, chemical class and odor valence. Furthermore, activated neural ensembles in both plCoA and PCx are almost identically decorrelated, and have a similar capacity to encode information about odor identity and odor concentration. Finally, at least within the level of resolution of our silicon probes (about half the total width of the plCoA in the anteroposterior axis), there is no apparent spatial order to the tuning properties of neurons within either plCoA or PCx. These findings suggest that the plCoA, like the PCx, uses a distributed population code to create discriminable and holistic representations for odor objects in the environment. This representational strategy emphasizes the distinctions between different odor objects, rather than any similarities those odor objects may have with respect to higher-order chemical, perceptual or behavioral features like chemical class or odor valence.
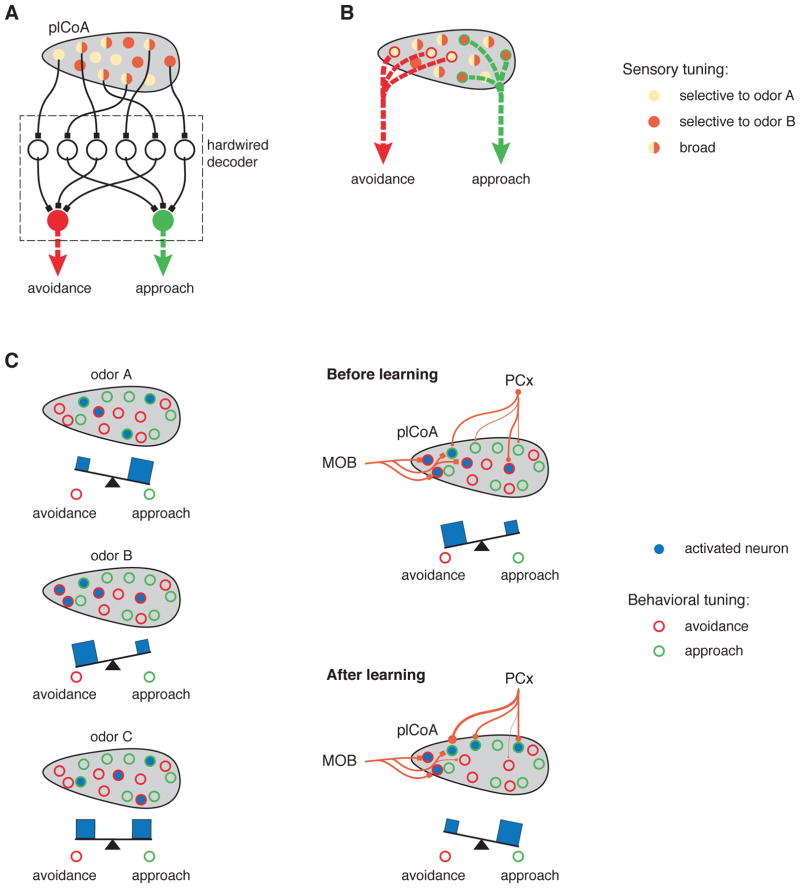
How, then, might a population code in the plCoA support the generation of innate odor-driven behaviors? One extreme model posits that the neurons that decode representations in plCoA do so using precisely balanced, genetically-programmed synaptic weights (i.e., there is a hardwired population code, Figure 8A). In this model, highly refined and innately-specified patterns of connectivity between populations of plCoA neurons and decoding neurons substitute for both the narrow tuning properties and the anatomic segregation that traditionally characterize labeled lines, enabling the plCoA to connect information about specific odors to defined innate behaviors. Because each neuron within plCoA contains relatively low information content about odor identity, in this model decoding neurons would require access to many plCoA neurons in parallel; furthermore, for this hardwiring to mediate stimulus-behavior relationships that are invariant from animal to animal, the neurons that participate in the code for any given odor or odor category would have to (at some point) be genetically marked to allow for the generation of appropriate connectivity.
Figure 8. Models for Decoding Innately-Relevant Odor Information in Cortical Amygdala.
Three models for the generation of innate behaviors by the plCoA in response to odors. Neurons have the tuning properties indicated by the legend on the right. The assigned behavioral meaning of neurons is indicated with external circles (appetitive = green, aversive = red). Model A and model B represent two extremes: in A, odor identity is decoded through precise and developmentally-specified hardwiring; in B, odor identity is decoded using rare labeled lines that are embedded within a broader population code for odor identity. In model C (left), the behavioral consequence of plCoA activation depends upon the balance between neurons mediating attraction and avoidace. After odor learning (C, right), however, the relative strength of the PCx to pCoA afferents are altered, causing changes in the tuning properties of the plCoA neurons. For example, now odor B elicits activity in more approach neurons than avoidance neurons, thereby changing the effective valence of odor B. This model provides an explanation for both how hardwiring from the bulb can elicit an innate behavior from what appears to be a population code, and how the plCoA could take advantage of its access to the PCx and downstream decoders to act as a switchboard, re-routing information about odors to appropriate behavioral centers in an adaptive fashion.
At the other extreme, rare odor- or category- specific neurons in the plCoA may mediate innate odor-driven behaviors by hardwired connections to decoders; in other words, these specifically-tuned neurons may represent classic labeled lines embedded within a neural structure that appears to generally represent odor identity using a population code (Figure 8B). These neurons may have privileged access to specific decoding circuits in known (and behaviorally-relevant) plCoA targets like the medial amygdala and the olfactory tubercle (although the PCx also innervates both those targets directly) (Agustín-Pavón et al., 2014; Keshavarzi et al., 2015; Li and Liberles, 2015; Novejarque et al., 2011; Ubeda-Bañon et al., 2007). In our recordings, we were unable to identify a statistical excess of such neurons; nevertheless, our experiments cannot definitively rule out the possibility that labeled lines, built from such neurons, exist in plCoA. It is important to note that in this embedded labeled-line model, the function of the bulk of the neurons in plCoA, which respond either to general odors, or to combinations of general and innately-relevant odors, is unexplained.
Although in principle the plCoA could mediate many distinct types of odor-driven innate behaviors (such as feeding or aggression), the only innate behaviors definitively assigned to plCoA thus far are attraction and aversion, which themselves can be highly diverse in form. Recent data demonstrates that nearly all odors elicit simple approach or avoidance to some degree in mice, with a few odors (like TMT, female urine and 2-PE) driving stronger (but still relatively modest) biases (Kermen et al., 2016; Root et al., 2014; Saraiva et al., 2016; Wiltschko et al., 2015). These observations, taken with the results reported here, suggest a third model, one in which the plCoA can both innately and flexibly assign odors to some degree of behavioral attraction or avoidance (Figure 8C).
In this model, plCoA neurons each fractionally contribute to attraction or avoidance behavior (perhaps through differential access to downstream effector circuits). Under naïve conditions, the combined effect of distributed afferents from PCx and subtly-biased hardwired connectivity from the olfactory bulb would allow odors to recruit decorrelated plCoA ensembles that could nevertheless specify the degree to which a given odor was appetitive or aversive. However, during odor learning, specific synapses between PCx and plCoA neurons (depending on whether those neurons were “appetitive” or “aversive”) could be altered, allowing the strength of attraction or avoidance associated innately with any given odor object to be adaptively reassigned. Indeed, learning-based reassignment of valence has been observed for multiple innately-relevant odors including TMT (Sarrafchi et al., 2013). In this manner, the plCoA could act as a kind of neural switchboard that routes information from PCx to striatal or amygdalar behavioral centers; under baseline conditions this routing occurs based upon biases built into the system by hardwired afferents from the bulb (which represent a given odor object), but these biases can be overridden by learning-driven plasticity in PCx afferents that represent that same odor object. Population codes may therefore be present and function within the plCoA to facilitate odor discrimination, allowing the plCoA to adaptively reassign specific odors new behavioral meanings as a consequence of experience. Because PCx axons also innervate plCoA targets (like the medial amygdala and olfactory tubercle), in an alternative formulation of this model the relevant site of learning-based modulation may reside downstream of the plCoA.
It is important to note that the models articulated above presume that plCoA and PCx neurons respond similarly to odors. Although our analysis supports this viewpoint, there are observable (albeit subtle) differences between the plCoA and PCx — particularly in terms of the relative amounts of observed excitation and inhibition — that may play an important role in odor perception under circumstances not explored herein. For example, the relatively low levels of odor-driven activation observed in the plCoA could in principle lead to sparser representations for certain odors (perhaps apparent under conditions in which many more odors are tested; note the trend in Figure 3F); this could in turn generate a relative excess of odor-selective neurons, a feature that may be useful for generating odor-specific behaviors.
Our conclusions regarding the structure and nature of odor representations in plCoA are also tempered by several caveats that reflect technical limitations to our experiments. Although our mice were freely behaving on a circular treadmill during the neural recordings and behaviorally responded to the presentation of odors, they were still subject to conditions of restraint during the recordings themselves. To our knowledge, meaningful innate behavioral responses to odors in head-fixed mice on a treadmill have not been previously observed, perhaps because such responses require closed-loop modulation of odor dynamics during approach and avoidance-type behaviors. It is therefore possible that behavioral restraint — or other state-dependent differences that might impinge upon the olfactory mantle — rewrites primary odor representations within the plCoA as to obscure the tuning properties of plCoA neurons apparent during behavioral engagement. Even if this is the case, however, collectively our observations argue against the possibility that labeled lines represent the default organizational mode of the plCoA.
A second caveat pertains to the geometry and placement of our probes: it is possible that our probes did not sample sufficient anatomical space to capture subregions in which olfactory information was categorically organized within the plCoA. The anterior region of the plCoA has been proposed to be enriched for neurons that respond to aversive odors (Root et al., 2014); although in many experiments our targeting coordinates appeared to partially cover this area (Figure S1), it is possible the subregion containing “aversive” neurons was missed, or that unavoidable variability in shank placement prevented us from identifying enriched populations of neurons due to spatial averaging across experiments.
Finally, our experiments were performed under conditions that did not allow us to definitively assign cellular identities to the recorded neurons. This limitation leaves open the possibility that different cell types in both the plCoA and PCx may non-uniformly represent sensory features relevant to innate behaviors. This limitation is potentially relevant given the hints we observe that at least some information regarding the chemical or ethological class of specific odors is present within populations of neurons in both plCoA and PCx.
Our findings that odor identity is represented as a population code in plCoA is reminiscent of findings by Ben-Shaul and colleagues, who recently characterized neural responses to natural odor mixtures within the accessory olfactory bulb (AOB), a peripheral neural center that regulates a variety of odor-driven innate behaviors (Kahan and Ben-Shaul, 2016). In those experiments, reliable decoding of chemosensory information about the strain or reproductive state of a conspecific required pooling of information from populations of AOB neurons. Thus, the use of population codes may be a general strategy for encoding information relevant to both innate and learned odor-driven behaviors at multiple levels of the mammalian olfactory system.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Sandeep Robert Datta (www.dattalab.org).
Experimental Model and Subject Details
Mice
Adult male C57BL/6 mice were used for experiments (age: 5–7 weeks). Mice were co-housed with their littermates (2–5/cage) and maintained on a 12 hrs/12 hrs light/dark cycle (dark hours: 10:00 – 22:00). Electrophysiological and behavioral experiments were performed during the dark cycle. Prior to experiments, food and water were available ad libitum. For the natural mixture experiments, mice were food deprived for 24 hrs before the recording session to normalize their hunger state.
All experimental manipulations were performed in accordance with protocols approved by the Harvard Standing Committee on Animal Care following guidelines described in the National Institute of Health Guide for the Care and Use of Laboratory Animals.
Method Details
Surgical Procedures
Five to six days before recording, mice were fitted with custom-made aluminum head plates. Mice were first anaesthetized using 0.1mg/kg/2% isoflurane. The skin was then infiltrated with bupivacaine and the head-plate was fixed in place with acrylic cement (Lang). A well was created over the head-plate with acrylic cement, and a craniotomy was performed over the plCoA (a.p.: −1.65 from bregma, m.l.: 2.8 from medial suture) or the anterior PCx (a.p.: 0.8 from bregma, m.l: 3.10 from medial suture) leaving the dura mater intact. Kwik-Cast (WPI) was then used to cover the skull. Animals were injected with 0.1 mg/kg buprenorphine to ensure pain management post-surgically for three consecutive days.
Set-up Familiarization
Two days before the recording, mice were familiarized to head-fixation over a spherical treadmill and to the mask used for odor delivery and respiration monitoring for two 30–60 minutes sessions per day. Purified and humidified air was delivered during the familiarization procedure to habituate the mouse to airflow during the experiment. Before and after each familiarization session, each mouse was gently handled for 2 x 10 minutes.
In Vivo Awake Extracellular Recordings
Mice were injected with dexamethasone (0.2 mg/kg IP) and the head-plate was clamped to a metal post over the treadmill. NeuroNexus 32-channel silicon probes (either A4x2-tet-7mm-150-200-121 or A32-BuzsakiL) were then implanted. Both electrode arrays have a four-shank configuration (distance between adjacent shanks: 200 μm; max vertical distance spanned by the 8 electrodes in each shank: 150 μm; see Figure S1A). The array was lowered at a speed of 2 μm/sec until a slight bending was observed via stereoscope, due to contact with the inner surface of the skull. The array was then slowly retracted by 50–250 μm. Recordings started after 45–60 min to ensure stabilization of the probe. A drop of Kwik-Cast (WPI) was then applied to minimize brain movement. The well over the skull was filled with Ringer’s solution and a ground wire was immersed in the solution. A recording session typically lasted 80 min. Acquisition was done at 20 KHz with an RHD2000 Series Intan Amplifier. In a subset of experiments (24 out of 70) an additional recording session was performed to minimize the number of mice used. To this end, the array was either retracted or lowered by 200 μm depending on the depth of the prior experiment. This travel distance guaranteed that the electrodes remained within the ventral and dorsal boundaries of the plCoA and PCx (~500 μm) while ensuring that a new set of neurons was sampled. The new recording session was started after an interval of 45–60 min to allow the array to stabilize.
Post-mortem Verification of Recording Sites
Recording sites in plCoA and PCx were verified by post-mortem inspection of the recording probe track (Figures S1B, S1C). Recording sites were chosen within the anterior PCx, as this area is thought to contain neural populations that represent odor identity (as opposed to the posterior PCx which may contain stronger representations of task-related features). After recordings were concluded, the probe was slowly retracted, painted with DiI dissolved in ethanol and then reinserted to the last recording site. The probe remained in the brain for at least 20 min before being retracted. The mouse was then sacrificed and the brain incubated in 4% PFA for 24 hours. 100 μm sections were cut with a vibratome, counter-stained with DAPI to reveal cortical layering and inspected under a fluorescence microscope at 10x magnification.
Odor Presentation
A custom Arduino-controlled 16-valve olfactometer that can separately deliver up to 15 odorants was used to present odors. The 16th valve was used to deliver a blank stimulus (no odor) between odor presentations. Custom Arduino software controlled valve opening and closing, thereby enabling switching between odor vials and the blank vial. This software also controlled the output of two mass flow controllers (MFC). The first MFC delivered a constant carrier flow at 1.8 L/min of purified and humidified air into a final common channel; the second MFC supplied a constant flow at 0.2 L/min of clean air that was injected into an odor vial (see below) and then merged with the carrier flow in a plastic mask placed in front of the mouse’s nose. A vacuum line continuously drew air/odor out of the mask’s airspace. A larger exhaust fan drew air from the Faraday cage that enclosed the rig to further prevent cross-contamination. Monomolecular odors were diluted in di-propylene glycol (DPG) according to individual vapor pressures to give a nominal concentration of 100 ppm. This vapor-phase concentration was further diluted 1:10 by the carrier airflow inside the nose mask. For the concentration series experiments, odorants were initially diluted to a nominal concentration of 85 mM and then serially diluted 1:10, 1:100, 1:1000 and 1:10000. Natural mixtures, whose relative concentrations were indeterminate, were not diluted in the vial. For all odors, a common, final 1:10 dilution in air (due to carrier flow) occurred in the nose mask. The blank vial contained only DPG.
Odor Panels
Four different panels of odorants were used in independent experiments. One panel included 15 monomolecular compounds: pentanal, hexanal, heptanal, heptanol, octanol, nonanol, phenetol, guaiacol, m-cresol, 2,4,5-trimethylthiazole, 4,5-dimethylthiazole, 4-methylthiazole, trimethylamine, isoamilamine and 2-phenyl-ethylamine. The second panel consisted of 8 monomolecular odorants: 2,3,5-trimethyl-3-thiazoline (TMT), 2-methylbutyric acid (2-MB), 2-propyltiethane (2-PT), 3-mercapto-3-methylbutan-1-ol (MMB), isopentylamine (IPA), 2,3-butanedione, geraniol and 2-phenylethanol; and two natural odors: peanut oil and estrus female urine (collected 1–3 days before an experiment and stored at 4°C in the dark). The third panel included 2-phenylethanol (2-PEth), isoamylacetate (IAA) and TMT at five different dilutions (described above). The fourth panel included 13 natural odors (referred to herein as natural odor mixtures): non-homogenized sunflower seed butter, non-homogenized peanut butter, urine from a male mouse (who was a resident in a different cage from the mouse being recorded), urine from estrus female mice, wolf urine, bobcat urine, lavender flowers, rose oil, coffee beans, fresh mint leaves, hickory chips, clove buds and eucalyptus oil.
Odorants were defined as “appetitive,” “aversive” or “neutral” based upon innate place preference assays previously published in the literature; all of the odors defined as appetitive or aversive used herein drive both changes in the position of mice with respect to the odor source and egocentric changes in behavior (like freezing) in our laboratory as well (data not shown and (Wiltschko et al., 2015)). Of the odorants used in these experiments, TMT, 2-MB, 2-PT, MMB, and IPA have been shown to drive innate aversive or avoidance responses (Kobayakawa et al., 2007; Root et al., 2014). 2,3-butanedione, geraniol, 2-PE, peanut oil and estrus female urine have been previously shown to elicit approach and appetitive behaviors (Mandairon et al., 2009; Root et al., 2014). For the natural odor panel, two distinct odor classifications were adopted. One classification was based on the presumptive valence of the individual odors, whereas the other classification was based on a natural, albeit arbitrary, grouping of odors based on their possible ethological meaning. Within the valence categorization, sunflower butter, peanut butter, female urine and rose oil were assigned to the appetitive class; wolf urine, bobcat urine and mint leaves were assigned to the avoidance class as peppermint oil has been shown to elicit avoidance (Saraiva et al., 2016); the remaining odors were grouped in “neutral” class because their valence is unknown. Within the natural ethological categorization, sunflower and peanut butters were categorized as food odors, male and female mouse urines were classified as conspecific odors, wolf and predator urines were classified as predator odors and the remaining odors were classified as “neutral” odors.
Odor Delivery
Odors presentations lasted for two seconds and were interleaved by 28 seconds of blank (DPG) delivery. The order of presentation of odors was pseudo-randomized for each experiment, and thus odors were not presented in a predictable order or twice in a row. Each odor was presented 10 times in each experiment. Respiration was monitored using a flow sensor AWM3100V Honeywell) connected to the nose mask used to deliver odors.
In a subset of experiments (n = 18), we measured the locomotion speed of the mouse through a 1024 P/R Quadrature rotary encoder attached to the treadmill shaft.
Open Field Behavioral Characterization
Innate olfactory avoidance of a predator odor (wolf urine), innate attraction to estrus female urine and the relative preference between these two odors were measured in 20 male mice after the electrophysiological recordings were completed; during these specific experiments mice had been exposed to natural odors including wolf urine and estrus urine, thereby allowing this experiment to test whether odor exposures during recording caused behavioral habituation.
After the electrophysiological experiment, mice were moved to a recovery cage where they could rest for 20 minutes before being placed in a white acrylic box (20 cm wide x 35 cm long x 35 cm high) for the behavioral test. Each wall on the short sides of the box contained a port (diameter: 2 cm) with a U-shaped IR break beam opto-sensor that was connected to an Arduino microcontroller. A small tube for odor delivery was placed behind each opto-sensor, with a valve connected to an Arduino microcontroller controlling airflow. Beam interruption by nose pokes triggered the opening of the valve and the delivery of an odor, which was contained in a small vial. The duration of each beam interruption was recorded through a Matlab custom script. Mice were allowed to familiarize with the box for 10 minutes, during which both ports delivered only DPG during each nose poke. All but two mice showed an idiosyncratic preference for one port as assessed by the total amount of time spent exploring the port. After 10 minutes of familiarization, the behavioral test was automatically started. For avoidance testing, wolf urine was delivered during the exploration of the port that was preferred during the familiarization phase. DPG was delivered during exploration of the other port. For approach testing, female estrus urine was delivered during exploration of the port that was less preferred and DPG was presented upon investigation of the other port. This approach was taken to maximally challenge the mouse to generate a given appetitive or aversive behavior. The test was automatically terminated after 10 minutes. A preference index for the familiarization phase and one for the test phase were calculated as
where P1 = investigation time of port 1 and P2= investigation time of port 2 (note that ports 1 and 2 are the same for both phases). For 5 mice we adopted a variation of this assay to test the preference between wolf urine and estrus female mouse urine odors that were delivered during the same experiment. Wolf urine was delivered during investigation of the port that was more explored during the familiarization phase whereas female urine was delivered during investigation of the less explored port. A preference index was calculated as
Data Analysis
LFP signals
LFP signals were extracted by band-pass filtering the raw traces of one channel of the most anterior shank in each experiment (0.1–300 Hz). LFP power in the beta band was estimated by taking the average power in the 10–30 Hz frequency range in 3 second window in absence of odors and in a 3 second window starting at the onset of the first inhalation after the onset of the odor delivery. The odor evoked fractional change of beta power was computed as
Spike Sorting and Criteria for Single Unit Inclusion
Spikes were sorted using a semi-supervised method. The open-source software Klusta was used to detect and cluster spikes into putative single units (Rossant et al., 2016). Only spikes with amplitude larger than 4 times the standard deviation of the background noise were detected and sorted. Clusters were manually curated to correct for any errors made by the automated algorithms. The quality of clusters was evaluated post-hoc by visual inspection of waveform shape, temporal stability, violations of the refractory period (2 ms), cross-correlation among units. Only units with a L-ratio > 0.5 (isolation distance >30, average S.N.R.: 5.2) were included in the analyses. Furthermore, units that fired less than one action potential in more than five trials for all odors in the 1000 ms baseline window or response window were excluded. Repeating the same analyses performed after including units with L-ratio < 1 gave similar results.
Criteria for Olfactory Response Identification
For each odor presentation (trial), spike times were aligned to the onset of the first inhalation (t0) after the opening of the odor valve. The number of spikes in 1000 ms windows before and after t0 was counted for each trial. These spike counts were used to compute the area-under-the-Receiver-Operating-Curve (auROC) of the responses. Spike counts during a 1 second baseline window (from −2 second to −1 second before the onset of the response window) and a 1 second response window in each trial were permuted 1000 times to generate a null distribution of auROC values. The actual auROC value was considered significant if it was either below the 2.5th percentile or above the 97.5th percentile of the null distribution (p<0.05). An excitatory olfactory response in each trial was considered significant if the maximum spike count in a 50 ms bin (bin counts were smoothed through a 5 ms sliding window) exceeded 5 standard deviations of the baseline firing rate in the same trial.
A Gaussian Mixture Model (GMM, gmfit in Matlab) was used to cluster response types. This procedure was only used for summarizing the data and was not intended to rigorously define response- or cell-types.
PSTH and Assessment of Phase Locking of Single Unit Activity to the Respiration Cycle
Smoothed peri-stimulus time histograms (PSTH) were obtained by convolving spike rasters with a Gaussian kernel with 50 ms standard deviation. Onset latencies of olfactory responses were determined as the first time point after the spike density exceeded the mean of the baseline spike density calculated over a 1000 ms window. To analyze whether neural activity was respiration modulated, single trial rasters were partitioned into bins delimited by the onset and offset of each consecutive inhalation and exhalation. The spike count in each bin was normalized by the inhalation (or exhalation) duration to obtain a firing rate. Cycle-matching bins were averaged across trials to obtain an inhalation-exhalation based PSTH for each olfactory response. To examine the distribution of the phases of the maximum firing rates during baseline and during the first second of odor presentation, the phase of each spike relative to the current respiratory cycle was determined. The lengths of each inhalation and exhalation cycle were linearly mapped on an 180° window and spike times were then transformed into the respective angle.
Fano Factor
The Fano Factor of olfactory responses is computed as the variance of the spike count change (relative to baseline) normalized by the mean spike count change over all trials.
Lifetime Sparseness
Lifetime sparseness (Willmore and Tolhurst, 2001) was computed as
where rj was the spike count change of a neuron in response to odorant j and N was the total number of odorants. Only for this analysis, negative spike count changes were zeroed.
Information about Odor/Class Identity for Single Units
Mutual information (MI) about odor/class identity for single units and pseudo-population of neurons was obtained from the respective confusion matrix previously obtained through linear SVM decoding as follows
where x is the actual class of the observation presented to the classifier, y is the predicted class for that observation, p(x) and p(y) are marginal probability and p(x,y) is the joint probability obtained from the confusion matrix. Details about the linear SVM decoding procedure are provided in the paragraph “Classifier Analysis”.
Odor Valence Discriminability for Single Units
The discriminability of the valence of an odor from the spike count responses of a neuron was calculated using auROC analysis. The auROC was obtained from two spike count vectors x and y. x and y are 50-dimensional vectors of spike counts in a one second response window (5 odors/valence class, 10 trials). To assess the significance of each index, the elements of x and y were reciprocally shuffled 200 times, and a distribution of index values was generated to assign a percentile to the actual vector value. Indexes below the 2.5th or above the 97.5th percentiles of the null distribution were considered significant.
Signal and Noise Correlation
The signal correlation between two neurons indicates the similarity of their tuning profiles. The pairwise signal correlation was computed as the Pearson’s correlation coefficient between the two vectors of mean spike count responses (change from baseline) to odors. The noise correlation is defined as the correlation of the trial-to-trial variability around the mean response to an odor. To compute the pairwise noise correlation between two neurons, the mean spike count response (change from baseline) to a given odor was subtracted from the spike count response (change from baseline) to the same odor on each trial. The vectors of these residuals for each odor were concatenated to obtain a single vector for each neuron. This vector was z-scored. The pairwise noise correlation was computed as the Pearson’s correlation coefficient of the two vectors of residuals of two neurons.
Neurons were grouped by agglomerative hierarchical clustering (method: single, metric: correlation) to generate a dendrogram of tuning profiles (see Figure 3J). For each neuron, the spike count responses to each odor (relative to baseline) were aligned in an odor-tuning vector. The distance between the tuning profiles of pairs of odors was computed as 1 – Pearson’s correlation coefficient of the two odor tuning vectors. Pairs of neurons were hierarchically linked together based on the shortest correlation distance.
Analysis of Concentration Series Experiment
For the concentration series experiment, a cell-odor pair with at least one excitatory response was deemed as invariant if the neuron exhibited significant excitatory responses to a given odor at all concentrations and these responses were not significantly different among them as assessed by an ANOVA test (p>0.05). To assess the monotonicity of the responses of a cell-odor pair, a line was fit to the average responses of the responsive neuron to all concentrations of a given odor sorted by the concentration magnitude. Note that this specific method was used to assess both concentration invariance and response monotonicity in work exploring odor responses in PCx axons in the olfactory bulb (Otazu et al., 2015). To test whether the slope of the fit line was significantly different from 0, the order of the average responses to distinct concentrations of given odor was permuted 500 times to obtain a control distribution of 500 slopes. If the actual slope was within the 2.5th and 97.5th percentiles of the control distribution, the neuron-odor pair responses were deemed as non-monotonic.
For each concentration, a unit that responded to at least one odor was defined as odor discriminant if the responses to different odors were different as assessed by an ANOVA test (p<0.05).
Classifier Analysis
To estimate the information carried by plCoA and PCx ensembles we took a decoding-based approach. Either a support vector machine or a least-square multiple regression model was used to identify a hyperplane that segregated neural activity data based upon odor identity, chemical class or odor valence, and we asked how well this hyperplane segregated these categories given increasing number of neurons as data points. Specifically, all units (L-ratio < 1) that responded to at least one odor in each odor panel were pooled in plCoA and PCx to build two pseudo-populations of neurons. Then responses (z-scored spike-counts) of a population of up to N randomly selected neurons (the maximum common number of neurons recorded across the four different odor panel experiments performed) were considered, given t presentations of j odors as a matrix X with N rows (neurons/features) and t x j columns (trials/observations x odors/classes). Each column of this matrix is thus a vector of N spike counts, one for each cell in response to a given odor in each trial.
Each decoding session started with a split of the matrix in two sets of vectors: the training set included 0.9 x t randomly chosen trials for each class and the test set comprised the 0.1 x t held out trials for each class (i.e., a standard 9:1 training:testing split). Note that here, “class” depends upon the specific experiment being analyzed, and can refer to odor identity, chemical class, ethological class or odor valence. In case of unequal number of trials for different classes, the number of trials across all classes was equalized by randomly selecting a number of trials equal to that available for the least represented class. Regardless of the number of different classes in an experiment (e.g., 15 monomolecular odors), all classifications were performed using binary classifiers in which the ability of the classifier to distinguish two specific classes was assessed. Distinct classifiers were generated for all possible binary combinations of classes within a given experiment. The models obtained from each trained binary classifier were then probed using the test data set. All classifiers classified each trial, and the class label assigned to a given trial was that which the individual binary classifiers choose most often. In the case of ties, the trial was randomly assigned to a class.
This procedure, which is instantiated as part of the standard SVM library (http://www.csie.ntu.edu.tw/~cjlin/libsvm/), allows us to use a binary classification algorithm (such as an SVM) to compare multiple classes. To show that this method is robust to the details of the binary comparison, we also implemented an alternative classification strategy previously used for neural classification, in which the binary classification is not performed between two odors but rather between one odor and all other odors considered as a separate class (Rust and DiCarlo, 2010). This alternative procedure yielded nearly identical results to the one-vs-one binary classifier (data not shown), which we therefore chose to use because of its simplicity of implementation.
In any given experiment, the train-test procedure was iterated 500 times (with training and test data randomly chosen on each iteration) to cross-validate classifier performance, and at the end of this procedure the outcomes of each individual iteration (of the 500) were averaged to generate a measure of classification accuracy across all restarts; this is the overall measure that is reported in the main text.
If the decoding procedure was performed on a subpopulation of neurons, a randomly selected subset of neurons was used for each cross-validation cycle. The hyperplanes for each classifier were determined using the LIBSVM library (http://www.csie.ntu.edu.tw/~cjlin/libsvm/) with a linear kernel, the C-SVC algorithm, and cost c. Cost c is the only free parameter for a linear kernel, and it was found by a grid search on an initial dataset including 50 randomly chosen neurons from plCoA and 50 randomly chosen neurons from PCx in order to maximize the accuracy of the decoder’s classification. Alternatively, a least square multiple regression model (Shen et al., 2013) or a non-linear, Gaussian Radial Basis Support Vector Machine was used.
To assess the influence of correlated activity, which has been shown to affect linear read-out population performance, in some analyses noise correlations and signal correlations were removed by shuffling trial order for each neuron in a pseudo-population and the odor label for each neuron; note that most of these pairwise correlations are naturally absent because the data are pooled from multiple animals, but since each animal contributes multiple neurons to the dataset, removing noise correlations from the dataset eliminates any residual correlation structure due to neurons recorded from the same mouse.
To test the hypothesis that the chemical class of an odor (15 odors, 3 odors/class) can be decoded from the responses of a population of neurons, a null distribution was built by pooling the decoding accuracy of decoders trained to classify 5 random combinations of 3 odors. To this aim, the actual sequence of 15 odors sorted by chemical class was permuted 150 times and consecutive triplets of odors in the new sequence were assigned to a specific class. A new decoder was trained for each permutation of the odor labels.
Similarly, the hypothesis that valence (appetitive/aversive) of an odor (10 odors, 5 odors/class) can be decoded from the responses of a population of neurons was tested by generating a null distribution including the decoding accuracy for all possible bi-partitions of the 10 odor labels.
To test the statistical significance of the decoding accuracies of the valence and the ethological class of natural mixtures of odorants, null distributions were generated by adopting the same method used for the chemical class experiment.
Partitioning the 13 natural mixtures of odorants in 3 valence classes (neutral, aversive and appetitive) or 4 ethological classes (nut butters, conspecific urines, predator urines and other) results in an unbalanced number of observations per class. To correct for such unbalance, the number of trials in each class was forced to equal the number of trials in the smallest class for each training and testing iteration.
Correlation Analysis
Pairwise similarity between the population representations of two odors was assessed as the Pearson’s correlation coefficient of two population vectors of z-scored mean spike count responses (change from the baseline) to the two odors. For each unit that responded to at least one odor, the mean spike count responses to each odor were z-scored across odors. For correlation matrices shown in Figure 5F and Supplementary Figures 5E–F, pairs of single trial population vectors were averaged.
PCA analysis
PCA analysis was used to visualize odor ensemble representations in the reduced coding space of plCoA and PCx in Figure 6K. N-dimensional vector representation of each stimulus for each trial were built from the responses of all units that were activated by at least one stimulus in each area. Pairs of these population vectors were then averaged to obtain pseudo-trial odor representations. The responses of each neuron were z-scored across all stimuli. Finally, Matlab function pca was applied to the matrix composed of all population vectors in each area to obtain a reduced representation of the odor representation (first 3 principal components) in that area.
Quantification and Statistical Analysis
All statistical analyses were performed with Matlab. Sample sizes were not estimated in advance. Data groups were tested for normality using the Kolmogorov-Smirnov test and then compared using the appropriate test (t-test, N-ways ANOVA, Wilkoxon rank-sum test, permutation test, χ2 test). All tests were two sided. Bootstrap estimates of percentiles were calculated by re-sampling with replacement from the original data. Statistical parameters including the measurements of arithmetic mean standard error of the mean (mean ± S.E.M.), median, 2.5th–97.5th percentiles, statistical tests and significance are reported in the Figures and the Figure Legends. In figures, asterisks denote statistical significance as calculated by the specified statistical tests (*, p < 0.05; **, p < 0.01; ***, p < 0.001). For statistical analysis on correlation coefficients a Fisher transformation of the coefficients was performed. Further details for quantification and analysis of behavior and electrophysiology are provided in the corresponding sections.
Data and Software Availability
Data are available at https://dattalab.github.io/Population-coding-in-an-Innately-Relevant-Olfactory-Area/. Custom Arduino code for olfactometer control, Matlab scripts and functions for exploring the data set and wrapper code for classification analysis and concentration series analysis are available at https://dattalab.github.io/Population-coding-in-an-Innately-Relevant-Olfactory-Area/
Additional Resources
N/A
Supplementary Material
Acknowledgments
We thank Stefano Panzeri, Sean X. Luo, Venkatesh Murthy and members of the Datta Lab (particularly Jeffrey Markowitz and Maria Lissitsyna Bloom) for helpful comments on the manuscript, Paola Patella for assistance with figures, and Alexandra Nowlan and Neha Bhagat for laboratory assistance. We thank Ofer Mazor and Pavel Gorelik from the Research Instrumentation Core Facility for engineering support. Core facility support is provided by NIH grant P30 HD18655. GI is supported by the Human Frontiers Science Program. SRD is supported by fellowships from the Burroughs Wellcome Fund, the Vallee Foundation, the Khodadad Program, by grant RO11DC011558 from the National Institutes of Health, and by the Global Brain Initiative from the Simons Foundation.
Footnotes
Author Contributions
Investigation and Software: GI; Conceptualization, Formal Analysis and Writing: GI and SRD.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agustín-Pavón C, Martínez-García F, Lanuza E. Focal lesions within the ventral striato-pallidum abolish attraction for male chemosignals in female mice. Behavioural brain research. 2014;259:292–296. doi: 10.1016/j.bbr.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Averbeck BB, Latham PE, Pouget A. Neural correlations, population coding and computation. Nature Reviews Neuroscience. 2006;7:358–366. doi: 10.1038/nrn1888. [DOI] [PubMed] [Google Scholar]
- Axel R. The molecular logic of smell. Sci Am. 1995;273:154–159. doi: 10.1038/scientificamerican1095-154. [DOI] [PubMed] [Google Scholar]
- Bergan JF, Ben-Shaul Y, Dulac C. Sex-specific processing of social cues in the medial amygdala. eLife. 2014;3:e02743–e02743. doi: 10.7554/eLife.02743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Stettler DD, Kallman BR, Bhaskar ST, Fleischmann A, Axel R. Driving Opposing Behaviors with Ensembles of Piriform Neurons. Cell. 2011;146:1004–1015. doi: 10.1016/j.cell.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland MM, Yu BM, Cunningham JP, Sugrue LP, Cohen MR, Corrado GS, Newsome WT, Clark AM, Hosseini P, Scott BB, et al. Stimulus onset quenches neural variability: a widespread cortical phenomenon. Nature Publishing Group. 2010;13:369–378. doi: 10.1038/nn.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland TA. Early transformations in odor representation. Trends in neurosciences. 2010;33:130–139. doi: 10.1016/j.tins.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MR, Kohn A. Measuring and interpreting neuronal correlations. Nature Neuroscience. 2011;14:811–819. doi: 10.1038/nn.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013:1–5. doi: 10.1038/nature12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi S, Miller EK, Rigotti M. Why neurons mix: high dimensionality for higher cognition. Current opinion in neurobiology. 2016;37:66–74. doi: 10.1016/j.conb.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Larson SD, Hefzi H, Marnoy Z, Cutforth T, Dokka K, Baldwin KK. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 2011;472:217–220. doi: 10.1038/nature09945. [DOI] [PubMed] [Google Scholar]
- Govic A, Paolini AG. In vivo electrophysiological recordings in amygdala subnuclei reveal selective and distinct responses to a behaviorally identified predator odor. Journal of Neurophysiology. 2015;113:1423–1436. doi: 10.1152/jn.00373.2014. [DOI] [PubMed] [Google Scholar]
- Haberly LB. Parallel-distributed processing in olfactory cortex: new insights from morphological and physiological analysis of neuronal circuitry. Chemical Senses. 2001;26:551–576. doi: 10.1093/chemse/26.5.551. [DOI] [PubMed] [Google Scholar]
- Illig KR, Haberly LB. Odor-evoked activity is spatially distributed in piriform cortex. J Comp Neurol. 2003;457:361–373. doi: 10.1002/cne.10557. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan A, Ben-Shaul Y. Extracting Behaviorally Relevant Traits from Natural Stimuli: Benefits of Combinatorial Representations at the Accessory Olfactory Bulb. PLoS computational biology. 2016;12:e1004798–1004725. doi: 10.1371/journal.pcbi.1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermen F, Midroit M, Kuczewski N, Forest J, Thévenet M, Sacquet J, Benetollo C, Richard M, Didier A, Mandairon N. Topographical representation of odor hedonics in the olfactory bulb. Nature Neuroscience. 2016;19:876–878. doi: 10.1038/nn.4317. [DOI] [PubMed] [Google Scholar]
- Keshavarzi S, Power JM, Albers EHH, Sullivan RKS, Sah P. Dendritic Organization of Olfactory Inputs to Medial Amygdala Neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35:13020–13028. doi: 10.1523/JNEUROSCI.0627-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Leinwand SG, Chalasani SH. Olfactory networks: from sensation to perception. Current Opinion in Genetics \& Development. 2011;21:806–811. doi: 10.1016/j.gde.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Li Q, Liberles SD. Aversion and attraction through olfaction. Current biology: CB. 2015;25:R120–129. doi: 10.1016/j.cub.2014.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litaudon P, Amat C, Bertrand B, Vigouroux M, Buonviso N. Piriform cortex functional heterogeneity revealed by cellular responses to odours. The European journal of neuroscience. 2003;17:2457–2461. doi: 10.1046/j.1460-9568.2003.02654.x. [DOI] [PubMed] [Google Scholar]
- Mandairon N, Sultan S, Rey N, Kermen F, Moreno M, Busto G, Farget V, Messaoudi B, Thevenet M, Didier A. A computer-assisted odorized hole-board for testing olfactory perception in mice. J Neurosci Methods. 2009;180:296–303. doi: 10.1016/j.jneumeth.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Min S, Ai M, Shin SA, Suh G. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S. 2013;110:1321–1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Mainen ZF, Uchida N. Odor Representations in Olfactory Cortex: Distributed Rate Coding and Decorrelated Population Activity. Neuron. 2012;74:1087–1098. doi: 10.1016/j.neuron.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sakano H. How is the Olfactory Map Formed and Interpreted in the Mammalian Brain? Annual review of neuroscience. 2011;34:467–499. doi: 10.1146/annurev-neuro-112210-112917. [DOI] [PubMed] [Google Scholar]
- Neville KR, Haberly LB. Beta and gamma oscillations in the olfactory system of the urethane-anesthetized rat. Journal of Neurophysiology. 2003;90:3921–3930. doi: 10.1152/jn.00475.2003. [DOI] [PubMed] [Google Scholar]
- Novejarque A, Gutiérrez-Castellanos N, Lanuza E, Martínez-García F. Amygdaloid projections to the ventral striatum in mice: direct and indirect chemosensory inputs to the brain reward system. Frontiers in neuroanatomy. 2011;5:54. doi: 10.3389/fnana.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otazu GH, Chae H, Davis MB, Albeanu DF. Cortical Feedback Decorrelates Olfactory Bulb Output in Awake Mice. Neuron. 2015;86:1461–1477. doi: 10.1016/j.neuron.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri S, Macke JH, Gross J, Kayser C. Neural population coding: combining insights from microscopic and mass signals. Trends in cognitive sciences. 2015;19:162–172. doi: 10.1016/j.tics.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo C, Isaacson JS. Odor representations in olfactory cortex: “sparse” coding, global inhibition, and oscillations. Neuron. 2009;62:850–861. doi: 10.1016/j.neuron.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget A, Dayan P, Zemel R. Information processing with population codes. Nature Reviews Neuroscience. 2000;1:125–132. doi: 10.1038/35039062. [DOI] [PubMed] [Google Scholar]
- Price JL. An autoradiographic study of complementary laminar patterns of termination of afferent fibers to the olfactory cortex. J Comp Neurol. 1973;150:87–108. doi: 10.1002/cne.901500105. [DOI] [PubMed] [Google Scholar]
- Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515:269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant C, Kadir SN, Goodman DFM, Schulman J, Hunter MLD, Saleem AB, Grosmark A, Belluscio M, Denfield GH, Ecker AS, et al. Spike sorting for large, dense electrode arrays. Nature Neuroscience. 2016;19:634–641. doi: 10.1038/nn.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust NC, DiCarlo JJ. Selectivity and Tolerance (“Invariance”) Both Increase as Visual Information Propagates from Cortical Area V4 to IT. The Journal of Neuroscience. 2010;30:12978–12995. doi: 10.1523/JNEUROSCI.0179-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco T, Sacchetti B. Role of secondary sensory cortices in emotional memory storage and retrieval in rats. Science (New York, NY) 2010;329:649–656. doi: 10.1126/science.1183165. [DOI] [PubMed] [Google Scholar]
- Saraiva LR, Kondoh K, Ye X, Yoon KH, Hernandez M, Buck LB. Combinatorial effects of odorants on mouse behavior. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E3300–E3306. doi: 10.1073/pnas.1605973113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrafchi A, Odhammer AME, Hernandez Salazar LT, Laska M. Olfactory Sensitivity for Six Predator Odorants in CD-1 Mice, Human Subjects, and Spider Monkeys. PLoS ONE. 2013;8:e80621. doi: 10.1371/journal.pone.0080621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe K, Ebert U, Löscher W. The central piriform cortex: anatomical connections and anticonvulsant effect of gaba elevation in the kindling model. Neuroscience. 2004;126:727–741. doi: 10.1016/j.neuroscience.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Shen K, Tootoonian S, Laurent G. Encoding of mixtures in a simple olfactory system. Neuron. 2013;80:1246–1262. doi: 10.1016/j.neuron.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosulski DL, Lissitsyna Bloom M, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472:213–216. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples LG, McGregor IS, Apfelbach R, Hunt GE. Cat odor, but not trimethylthiazoline (fox odor), activates accessory olfactory and defense-related brain regions in rats. NSC. 2008;151:937–947. doi: 10.1016/j.neuroscience.2007.11.039. [DOI] [PubMed] [Google Scholar]
- Stettler DD, Axel R. Representations of odor in the piriform cortex. Neuron. 2009;63:854–864. doi: 10.1016/j.neuron.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Ubeda-Bañon I, Novejarque A, Mohedano-Moriano A, Pro-Sistiaga P, de la Rosa-Prieto C, Insausti R, Martínez-García F, Lanuza E, Martinez-Marcos A. Projections from the posterolateral olfactory amygdala to the ventral striatum: neural basis for reinforcing properties of chemical stimuli. BMC neuroscience. 2007;8:103. doi: 10.1186/1471-2202-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore B, Tolhurst DJ. Characterizing the sparseness of neural codes. Network (Bristol, England) 2001;12:255–270. [PubMed] [Google Scholar]
- Wilson DA, Sullivan RM. Cortical Processing of Odor Objects. Neuron. 2011;72:506–519. doi: 10.1016/j.neuron.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, Datta SR. Neuron. Elsevier Inc; 2015. Mapping Sub-Second Structure in Mouse Behavior; pp. 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan C, Luo M. Diverse patterns of odor representation by neurons in the anterior piriform cortex of awake mice. Journal of Neuroscience. 2010;30:16662–16672. doi: 10.1523/JNEUROSCI.4400-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.