Abstract
Bacteria with an actinomycetes-like morphology have recently been discovered, and the class Ktedonobacteria was created for these bacteria in the phylum Chloroflexi. They may prove to be a valuable resource with the potential to produce unprecedented secondary metabolites. However, our understanding of their diversity, richness, habitat, and ecological significance is very limited. We herein developed a 16S rRNA gene-targeted, Ktedonobacteria-specific primer and analyzed ktedonobacterial amplicons. We investigated abundance, diversity, and community structure in forest and garden soils, sand, bark, geothermal sediment, and compost. Forest soils had the highest diversity among the samples tested (1181–2934 operational taxonomic units [OTUs]; Chao 1 estimate, 2503–5613; Shannon index, 4.21–6.42). A phylogenetic analysis of representative OTUs revealed at least eight groups within unclassified Ktedonobacterales, expanding the known diversity of this order. Ktedonobacterial communities markedly varied among our samples. The common mesic environments (soil, sand, and bark) were dominated by diverse phylotypes within the eight groups. In contrast, compost and geothermal sediment samples were dominated by known ktedonobacterial families (Thermosporotrichaceae and Thermogemmatisporaceae, respectively). The relative abundance of Ktedonobacteria in the communities, based on universal primers, was ≤0.8%, but was 12.9% in the geothermal sediment. These results suggest that unknown diverse Ktedonobacteria inhabit common environments including forests, gardens, and sand at low abundances, as well as extreme environments such as geothermal areas.
Keywords: Ktedonobacteria, Chloroflexi, Ktedonobacteria-specific primer
Many studies have examined the relationship between the complex morphological differentiation of actinomycetes and their capacity to produce secondary metabolites (26). The class Ktedonobacteria, the members of which have an actinomycetes-like morphology, was recently proposed (5) and placed in the phylum Chloroflexi (41), which is known to be a deep-branching lineage of the domain Bacteria. This class currently contains only six named species in two orders: in Ktedonobacterales, Ktedonobacter racemifer SOSP1-21T (isolated from soil under black locust in Italy; [5, 6]), which is the first pure culture in the class, Thermosporothrix hazakensis SK20-1T (isolated from ripe compost in Japan; [41]), and T. narukonensis F4T (isolated from fallen leaves deposited on geothermal soil; [43]); and in Thermogemmatisporales, Thermogemmatispora onikobensis ONI-1T and T. foliorum ONI-5T (isolated from fallen leaves deposited on geothermal soil; [42]), and T. carboxidivorans PM5T (isolated from a geothermally heated biofilm; [19]). Other cultivated isolates for which valid names have not yet been proposed include ten isolates (strain SOSP series, from the same source as K. racemifer SOSP1-21T; [5]) in Ktedonobacterales; and four isolates (strains P-352, P-359, T104, and T81 from geothermal soils; [32]) and two isolates (strains BPP55 and PM6 from the same source as T. carboxidivorans PM5T; [19]) in Thermogemmatisporales. Furthermore, we identified five isolates (strains FA, FB, F7, MYC, and I1 from the same source as T. narukonensis F4T; [43]) within the genus Thermosporothrix.
The known Ktedonobacteria have a complex morphology and differentiation. All are Gram-positive and aerobic, and form branched mycelia with spores, similar to those of mycelia-forming actinomycetes. Moreover, all type strains sporulate via the formation of multiple exospores per cell by budding, which is unique among bacteria (40). Strains SOSP1-9, from the strain SOSP series, may form sporangia (5). Genome sizes are 13.7 Mbp in K. racemifer SOSP1-21T (6), 7.3 Mbp in T. hazakensis SK20-1T (27), and 5.6 Mbp in T. carboxidivorans PM5T (Integrated Microbial Genomes & Microbiomes database: http://img.jgi.doe.gov/). The genome of K. racemifer SOSP1-21T encodes 11,453 putative proteins, which is the largest number reported for a prokaryotic genome to date (6). The genomes of K. racemifer SOSP1-21T and T. hazakensis SK20-1T also contain 15 and 21 secondary metabolite-related gene clusters, respectively, as assessed by the antiSMASH online tool (https://antismash.secondarymetabolites.org/) (unpublished).
These unique morphological and genomic characteristics led us to speculate that this class may constitute a valuable new microbial resource for the discovery of novel compounds. We previously identified new secondary metabolites from T. hazakensis SK20-1T (27, 28). However, in order to use a taxon as a source for screening novel compounds, it is important to know the diversity and habitat of the organisms in question.
Previous studies based on clone libraries or next-generation sequencing of the 16S rRNA gene of bacteria showed that environmental clone sequences assigned to Ktedonobacteria are prominent in extreme environments such as volcanic, Antarctic, and cave ecosystems (2, 12, 14, 17, 25, 29, 34, 38). A few studies have also investigated the detection of these clone sequences at low abundances in non-extreme environments such as sandy soils (1) and soils of the Flooding Pampa of Argentina (24). These studies did not focus on Ktedonobacteria in various habitats. Therefore, the abundance, diversity, richness, habitat preference, and community structure of the class are poorly understood.
In order to obtain detailed information on minor communities in samples from a range of environments, it is more effective to use taxon-specific primers than universal primers applicable to all bacteria when analyzing the pool of PCR amplicons. Specific primers may also be used for the rapid and sensitive screening of a particular taxonomic affiliation from among cultured organisms and microbial DNA. However, although many group-specific primers for 16S rRNA genes already exist, a Ktedonobacteria-specific (KS) primer has not yet been developed.
In order to obtain a tool for the rapid screening of sites at which Ktedonobacteria occur and to investigate their diversity and community structure in situ, we developed a KS primer for the 16S rRNA gene. We then surveyed the diversity and abundance of the bacteria in soils, sand, bark, geothermal sediment, and ripe compost using the Illumina MiSeq sequencing of amplicons with the specific primer.
Materials and Methods
Site description and sample collection
We collected eight samples from six sites: three forest soils, one garden soil, one sand, and one bark as common mesic terrestrial environments; and a geothermal sediment and compost as less common environments from which several ktedonobacterial strains have already been isolated.
Soil and bark samples
Forest soil samples 1A and 1B were collected from depths of 0–10 cm (1A, upper layer, 16°C) and 15–30 cm (1B, lower layer, 15°C) at a site under a cherry tree (Cerasus jamasakura) in the town of Murata, Miyagi Prefecture, northeastern Japan (38°07′36″N, 140°42′21″E) in May 2015. We removed the aboveground litter before sampling. Approximately 5 g of bark was collected from the cherry tree.
Forest soil sample 2 was collected from a depth of 0–10 cm (22°C) under large bamboo plants (Bambusoideae) in the town of Toyohashi, Aichi Prefecture, central Japan (34°47′49″N, 137°29′07″E) in May 2015.
The garden soil sample was collected from a depth of 0–10 cm (26°C) under a loquat tree (Eriobotrya japonica) in a campus garden at Tohoku University, Sendai City, Miyagi Prefecture (38°16′34″N, 140°52′22″E) in May 2015.
Sand sample
Sand was collected from a depth of 0–10 cm (24°C) on a riverbank in the town of Murata, Miyagi Prefecture (38°07′56″N, 140°42′55″E) in May 2015.
Geothermal sediment sample
Sediment was collected at the same site from which the T. onikobensis and T. foliorum isolates were previously collected (42) in the Jigokudani geothermal area, near Onikobe, in the Naruko hot springs region of Miyagi Prefecture (38°48′18″N, 140°40′24″E). We sampled fallen leaves deposited on steaming geothermal soil at a depth of 10–15 cm within the litter layer (72°C) in November 2015.
Compost sample
The sample was collected from ripe compost produced by a field-scale manure composter (Hazaka Plant, Kennan Eisei Kogyo, Murata Town, Japan; [39]) in May 2015. A species in the genus Thermosporothrix was previously isolated from compost produced by the same system (41).
All samples were collected in plastic press-seal bags with autoclaved scoops, transported to the laboratory, and stored at –80°C until DNA extraction and soil analyses. The characteristics of all samples are summarized in Table 1.
Table 1.
Sample details.
| Sample name | Depth (cm) | Latitude, longitude | Temperature (°C) | Moisture content (%) | pH | Total carbon (%) | Total nitrogen (%) |
|---|---|---|---|---|---|---|---|
| Forest soil 1 (A) | 0–15 | 38°07′36″N, 140°42′21″E | 16 | 55.0 | 5.4 | 11.79 | 0.83 |
| (B) | 15–30 | 15 | 19.6 | 6.0 | 0.32 | 0.08 | |
| Forest soil 2 | 0–10 | 34°47′49″N, 137°29′07″E | 22 | 23.3 | 5.8 | 7.56 | 0.59 |
| Garden soil | 10–20 | 38°16′34″N 140°52′22″E |
22 | 9.3 | 5.9 | 1.89 | 0.31 |
| Sand | 0–10 | 38°07′56″N, 140°42′55″E | 24 | 2.7 | 5.7 | 0.05 | 0.03 |
| Bark | — | 38°07′36″N, 140°42′21″E | — | 21.3 | 5.4 | 48.92 | 0.84 |
| Geothermal sediment | 10–15 | 38°48′18.28N 140°40′24.23E |
72 | 82.4 | 6.8 | 32.0 | 1.6 |
| Compost | — | — | 34 | 51.2 | 7.9 | 21.3 | 2.4 |
Soil physicochemical analysis
Samples were dried at 105°C in an oven for 5 h in order to assess moisture content. The pH of a suspension of 10 g of wet soil in 100 mL of deionized water was evaluated with a pH electrode. Air-dried samples were sieved through a 2-mm mesh, and total C and N contents were measured with a CN analyzer (JM3000N/CN, J Science Lab, Kyoto, Japan). The results obtained are presented in Table 1.
Design of a 16S rRNA gene-targeted KS primer
We designed a KS primer to amplify the 16S rRNA gene. We randomly selected 121 16S rRNA sequences (isolates and uncultured sequences, >1,200 bp and good quality) from each class in the phylum Chloroflexi, including Ktedonobacteria, from the Ribosomal Database Project II (RDP-II) (22). Multiple alignments of the sequences were performed using CLUSTALW (v. 1.83) (35), and consensus sequences were elucidated in BioEdit sequence alignment software (15). A KS forward primer was designed by comparing regions of nucleotide mismatches at the 3′ end of the oligonucleotide alignments of the consensus sequences by eye with those from other classes in the phylum (Fig. S1). We selected regions that had no mismatches at the 3′ end in the Ktedonobacteria class, but had mismatches in other classes. The primer was assessed in silico using the Probe Match tool in RDP-II (Differences Allowed: 0)
We named it KTED161F (E. coli position: 140–161). Since we were unable to find an appropriate KS reverse primer in the alignments, we used GNSB941R (E. coli position: 941–957), which was designed specifically for Chloroflexi (13).
We confirmed the specificity of the primer using all type strains within the class Ktedonobacteria and Ktedonobacteria isolates (strains FA, FB, F7, MYC, and I1). We performed the same tests using Escherichia coli NBRC3972, Bacillus subtilis NBRC3134, and Micrococcus luteus NBRC13867 as negative controls. In the results obtained, we confirmed that PCR bands were detected in the ktedonobacterial strains, but were absent in the negative controls.
DNA extraction
DNA was isolated from 0.15 g of each sample using a Power Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA) following the manufacturer’s protocols, and eluted in 50 μL of nuclease-free water. The quality of DNA was examined on agarose gels, and samples were quantified using the QuantiFluor dsDNA System (Promega, Fitchburg, WI, USA). DNA samples were stored at −20°C until further analyses.
Amplification of the 16S rRNA gene by PCR for Illumina MiSeq sequencing
KS amplicons were generated by nested PCR and additional PCR was used for the barcoding of amplicons. The first round used the primers KTED161F and GNSB941R (Table 2). The predicted products (ca. 800 bp) were too long for sequencing on an Illumina MiSeq desktop sequencer and the detection sensitivity of one round of PCR was low; therefore, we conducted a second round (nested PCR), which also increased the likelihood of detecting products missed in the first round. The second round used KTED161F and UNIV538R; the latter targeted the 16S rRNA genes of the entire domain Bacteria (23) and was linked via an Illumina sequencing primer-binding site (Table 2). The third round used primers containing Illumina adapter sequences, an index sequence, and a sequencing primer-binding site without target gene-specific sequences. The index sequence was a unique sequence designed to differentiate sequencing reads from different samples (Table S1).
Table 2.
Primer sequences used in this study.
| Primer | Sequences (5′-3′) | Target taxon | References |
|---|---|---|---|
| KTED161F: | ATACCGGBGMGAAAKYGYCGAC | Ktedonobacteria (class) | This study |
| GNSB941R: | AAACCACACGCTCCGCT | Chloroflexi (phylum) | (14) |
| UNIV27F | AGAGTTTGATCMTGGCTCAG | Bacteria (domain) | (16) |
| UNIV538R | GTATTACCGCGGCTGCTGG | Bacteria (domain) | (25) |
| MiSeq-adapter (5′–3′)1) | Index2) | Sequence site (5′-3′)3) | Target site | ||
|---|---|---|---|---|---|
| Primer set for class Ktedonobacteria amplicons | |||||
| 1st PCR | F | — | — | — | KTED161F |
| R | — | — | — | GNSB941R | |
| 2nd PCR | F | — | — | ACACTCTTTCCCTACACGACGC | KTED161F |
| R | — | — | GTGACTGGAGTTCAGACGTGTG | UNIV538R | |
| 3rd PCR | F | AATGATACGGCGACCACCGCGCTCTACAC | Table S1 | ACACTCTTTCCCTACACGACG | — |
| R | CAAGCAGAAGACGGCATACGAGAT | Table S1 | GTGACTGGAGTTCAGACGTGTG | — | |
| Primer set for domain Bacteria amplicons (universal) | |||||
| 1st PCR | F | — | — | ACACTCTTTCCCTACACGACGC | UNIV27F |
| R | — | — | GTGACTGGAGTTCAGACGTGTG | UNIV538R | |
| 2nd PCR | F | AATGATACGGCGACCACCGCGCTCTACAC | Table S1 | ACACTCTTTCCCTACACGACG | — |
| R | CAAGCAGAAGACGGCATACGAGAT | Table S1 | GTGACTGGAGTTCAGACGTGTG | — | |
Flow cell-binding site for Illumina MiSeq sequencing.
Unique sequence designed to differentiate sequencing reads from different samples. The sequence of each index is shown in Table S1.
Sequence primer-binding site for Illumina MiSeq sequencing.
PCR was performed in 50 μL containing 1 μL of DNA (adjusted to 1 ng μL−1 for the first round, 1 μL of the first PCR product for the second round, and 1 μL of the second PCR product for the third round), 5 μL of 10×PCR buffer, 4 μL of dNTP mixture (2.5 mM each dNTP), 1 μL of each primer (0.2 μM), 38.5 μL of deionized distilled water, and 0.5 μL of Ex-Taq DNA polymerase (5 U μL−1) (Takara, Otsu, Japan). The first- and second-round PCR programs included an initial denaturation step at 94°C for 2 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 2 min. The third-round PCR program included an initial 94°C for 2 min; 10 cycles at 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s; and a final 72°C for 5 min.
In order to evaluate the sensitivity of the new primer, we performed a control experiment using two rounds of PCR to amplify the 16S rRNA gene sequence (V1–V3) of the domain Bacteria. The first round used the universal primers UNIV27F and UNIV538R, linked via an Illumina sequencing primer-binding site (Table 2). The second round used primers containing Illumina adapter sequences, an index sequence, and a sequencing primer-binding site. The conditions for the first and second rounds were the same as those for the first and third rounds described above. All PCR products were separated by electrophoresis on 1.0% agarose gels, stained with a GelRed fluorescent DNA stain (Biotium, Hayward, CA, USA), and purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA). The products were quantified with a QuantiFluor dsDNA System (Promega, Fitchburg, WI, USA), then mixed (50 ng each) and sent to FASMAC (Kanagawa, Japan) for Illumina MiSeq sequencing.
Sequence analysis and evaluation of the new primer
We compared the KS amplicon sequences of the same samples between the new and universal primers. The raw reads obtained from the sequencer were sorted by the index sequence (one mismatch was allowed), trimmed for the primer sequences with the FASTX-Toolkit v. 0.0.13.2 tools (Gordon A., and G.J. Hannon. 2010. FASTX-toolkit. FASTQ/A short-reads preprocessing tools. [unpublished] http://hannonlab.cshl.edu/fastx_toolkit), filtered according to quality scores (threshold: 20) in Sickle v. 1.33 software (https://github.com/najoshi/sickle), and merged with forward and backward sequences by FLASH v. 1.2.10 software (21) at default conditions using the Quantitative Insights Into Microbial Ecology (QIIME) v. 1.8.0 software package (4). The universal amplicons were used as single reads. Processed high-quality reads were assigned to taxa by the RDP classifier (36) on the basis of RDP data, with a minimum confidence level of 80% using the RDP online tool (http://pyro.cme.msu.edu/index.jsp). Reads that were taxonomically unclassified at the domain level were removed. We evaluated the specificity of the new primer and the abundance of KS amplicons in the samples using the processed high-quality reads (Table 3).
Table 3.
Specificities of primer sets and abundances of reads assigned to the class Ktedonobacteria by the RDP classifier.
| Forest soil 1 (A) | Forest soil 1 (B) | Forest soil 2 | Garden soil | Sand | Bark | Geothermal sediment | Compost | |
|---|---|---|---|---|---|---|---|---|
| Ktedonobacteria class-specific amplicons (primers KTED161F-GNSB941, KTED161F-UNIV538R) | ||||||||
| Domain Bacteria | 127731 | 104688 | 250365 | 127779 | 90988 | 150662 | 81296 | 160067 |
| Class Ktedonobacteria | 125540 | 104333 | 250228 | 127465 | 90983 | 150611 | 81293 | 159997 |
| % of reads in the class Ktedonobacteria | 98.285 | 99.661 | 99.945 | 99.754 | 99.995 | 99.966 | 99.996 | 99.956 |
| Universal amplicons (primers UNIV27F-UNIV538R) | ||||||||
| Domain Bacteria | 77615 | 88875 | 79132 | 83650 | 61188 | 80793 | 77039 | 200801 |
| Class Ktedonobacteria | 4 | 741 | 15 | 12 | 47 | 57 | 9912 | 19 |
| % of reads in the class Ktedonobacteria | 0.005 | 0.80 | 0.02 | 0.014 | 0.08 | 0.07 | 12.9 | 0.01 |
Phylogenetic and statistical analysis of KS amplicons
The high-quality reads that were assigned to Ktedonobacteria with high confidence (more than 70%) were used to analyze the diversity and phylogeny of the taxon. They were clustered into operational taxonomic unit (OTU) sequences that shared ≥97% similarity. We obtained representative sequences from each OTU using the functions pick_otus and pick_rep in QIIME and the UCLUST clustering algorithm (10) with default conditions. Ktedonobacterial singletons were not removed. The phylogenetic tree was constructed using OTUs that accounted for >0.5% of the relative abundance of Ktedonobacteria reads in each sample, together with the 16S rRNA genes of other Ktedonobacteria strains and clone sequences obtained from GenBank and RDP-II (8). Multiple alignments of the sequences were performed using CLUSTALW v. 1.83 (35), and the alignment was manually improved or unaligned regions trimmed with BioEdit software (15). The phylogenetic tree was constructed using the neighbor-joining method (30) in MEGA 5 software (33) with bootstrap values based on 100 replications (11). Evolutionary distances were computed using the Kimura two-parameter method (18). The taxonomic position of each OTU was assessed from the phylogenetic tree and compared with the ktedonobacterial composition of the samples. The compositions of the samples according to the clusters on the phylogenetic tree are compared in Fig. 1.
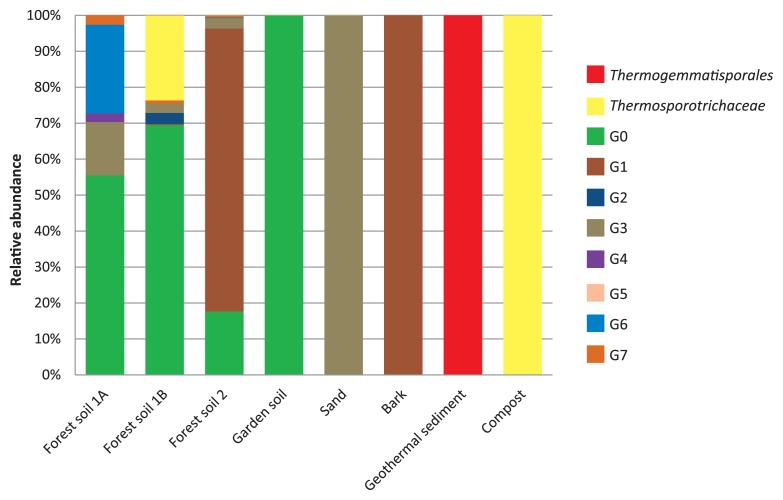
Fig. 1.
Comparison of ktedonobacterial community structures among soils, sand, bark, geothermal sediment, and compost. This figure shows OTUs that occupy >0.5% in at least one of the samples sequenced.
In the statistical analysis, we standardized the number of sequences per sample to 80,000 by random subsampling of the OTUs in order to avoid bias. Rarefaction curves, Shannon indices (31), and Chao 1 indices (7) were calculated using QIIME scripts (Table 4).
Table 4.
Ktedonobacterial diversity and richness values in samples.
| OTU richness | OTU diversity | |||
|---|---|---|---|---|
|
|
|
|||
| No. of OTUs | Chao 1 | Simpson | Shannon | |
| Forest soil 1 (A) | 1181 | 2503 | 0.926 | 5.00 |
| Forest soil (B) | 2934 | 5613 | 0.955 | 6.42 |
| Forest soil 2 | 1762 | 3947 | 0.754 | 4.21 |
| Garden soil | 343 | 742 | 0.698 | 2.52 |
| Sand | 77 | 506 | 0.003 | 0.03 |
| Bark | 462 | 1259 | 0.550 | 1.56 |
| Geothermal sediment | 402 | 746 | 0.634 | 2.30 |
| Compost | 217 | 806 | 0.109 | 0.49 |
The OTUs used in the statistical analysis were standardized to 80, 000 sequences.
Nucleotide sequence accession numbers
The sequencing data obtained in this study were deposited in the DDBJ Sequence Read Archive (DRA) under accession numbers DRR068692 to DRR068699 (universal amplicons) and DRR067880 to DRR067887 (ktedonobacterial amplicons).
Results
Physicochemical characteristics of samples
Differences among the eight samples were evident (Table 1). The temperature of the geothermal sediment sample (72°C) was markedly higher than those of the other samples (15–34°C). pH ranged from slightly alkaline in compost (pH 7.9) to slightly acidic in the other samples (pH 5.4–6.8). The total carbon contents of the upper-layer soils (forest soils 1A and 2) were high (7.56%–11.79%), whereas those of the lower-layer soils (forest soil 1B and garden soil) were low (0.32%–1.89%). The total nitrogen contents of the upper-layer soils (forest soils 1A and 2), the geothermal sediment, and compost were high (0.59%–2.4%), whereas those of the lower-layer soils (forest soil 1B) and sand were low (0.03%–0.08%).
In silico analysis of KS primers
We selected a primer sequence (5′-ATACCGGBGMGAAAKYGYCGAC-3′) that showed dissimilarities to the 3′ end of the non-target sequences using the consensus sequence of the representative sequences in each class within the phylum Chloroflexi (Fig. S1). According to the Probe Match tool in RDP-II, this sequence completely matched 275 out of 1,459,456 sequences from the domain Bacteria, of which 273 (99%) were assigned to Ktedonobacteria. It matched 61% (273/448) of all known ktedonobacterial sequences in the database. However, since the important first two positions from the 3′ end of the primer matched 91% (403/448) of these sequences (Fig. S1), we decided to use this region as the KS primer, and named it KTED161F (Table 2).
The Chloroflexi-specific primer GNSB941R (5′-AAACCACACGCTCCGCT-3′) (25) completely matched 19,461 out of 1,459,456 domain Bacteria sequences, of which 19,244 (99%) were assigned to Chloroflexi. It also matched 94% (422/448) of all known ktedonobacterial sequences in the database.
We also evaluated the reverse primer UNIV538R (3′-GTATTACCGCGGCTGCTGG-5′) for second PCR (nested PCR), paired with KTED161F. UNIV538R completely matched 1,271,351 out of 1,459,456 domain Bacteria sequences and 89% (400/448) of all known ktedonobacterial sequences in the database.
Evaluation of primer sets used to amplify 16S rRNA genes from environmental DNA samples
After the first KS amplification using the primer set KTED161F and GNSB941R, the PCR bands of the DNA samples obtained from the soils, sand, bark, and compost were either not visible or very weak, whereas those of the geothermal sediment sample were clearly visible. After second PCR using the primer set KTED161F and UNIV538R, the PCR bands of all samples were clearly visible. The PCR bands of all samples using the universal primer set (UNIV27F–538R) from the same DNAs were also clearly visible. After filtering, quality control, assembly of paired-end reads for ktedonobacterial amplicons, and removal of primers, chimeras, and low-confidence leads, 1,093,576 high-quality 16S rRNA sequence reads remained for ktedonobacterial PCR (81,296 to 250,365 per sample) in 1,430,264 raw reads and 749,093 for universal PCR (61,188 to 200,801 per sample) in 868,828 raw reads.
Most of the KS PCR amplicons (98.285%–99.996% of each total read) from each sample were assigned to Ktedonobacteria (Table 3). Very few (0.005%–0.80%) of the universal PCR amplicons were assigned to Ktedonobacteria (Table 3), except for 12.9% from the geothermal sediment. Thus, the primer set KTED161F-GNSB941R exhibited high specificity for the class Ktedonobacteria, which formed a small proportion of the samples tested in this study, except for the geothermal sediment. This low presence explains why the bands from first PCR were either not visible or very weak, despite the use of the KS primer, for all samples, except the geothermal sediment.
Statistical and phylogenetic analysis of KS amplicons
We estimated the diversity of ktedonobacterial communities in samples from the KS amplicons. The number of OTUs markedly varied among the standardized sequences, ranging from 77 to 2,934 (Table 4). This analysis also indicated that the ktedonobacterial richness of these forest soils was higher than those of the other samples. The highest diversity was found in lower-layer forest soil (1B: Chao 1=5613, Shannon index=6.42, Simpson index=0.955), while the lowest was found in sand (Chao 1=506, Shannon index=0.03, Simpson index=0.003; Table 4).
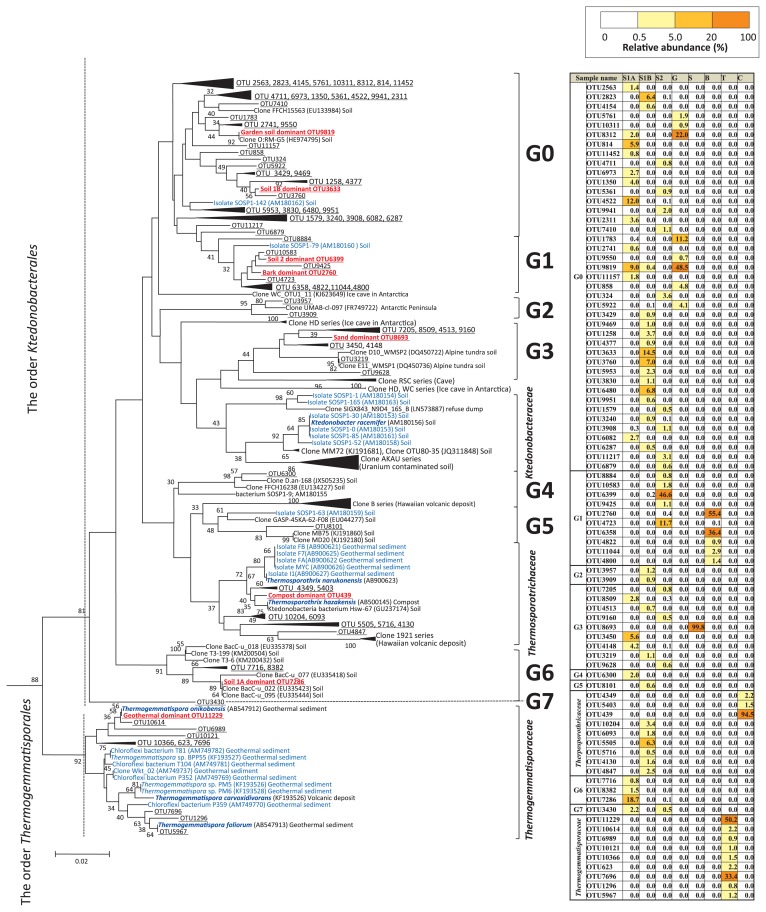
We constructed a phylogenetic tree (Fig. 2) by assigning the OTUs that accounted for >0.5% of the relative abundance in each sample to the three known families and eight unclassified Ktedonobacterales groups (G0–7) on the tree. Groups 1–7 were supported at >30% of bootstrap values and Group 0 at <30%. We used this phylogenetic tree to compare the ktedonobacterial community composition of the eight samples.
Fig. 2.
(A) Maximum likelihood phylogenetic analysis of representative 16S rRNA OTUs of ktedonobacterial amplicons from environmental DNA obtained from soils, sand, bark, geothermal sediment, and compost. The OTUs detected in this study are shown as underlines. The dominant OTU in each sample is shown in red. Cultured isolates of Ktedonobacteria are shown in blue (type strains are shown in bold blue). Numbers at nodes are bootstrap percentages based on 100 replicated data sets; only values >30% are shown. Scale bar, 2% sequence dissimilarity. Caldilinea aerophila (AB067647), Litorilinea aerophila (JQ733906), Anaerolinea thermolimosa (AB109437), Anaerolinea thermophila (AB046413), Leptolinea tardivitalis (AB109438), Longilinea arvoryzae (AB243673), Bellilinea caldifistulae (AB243672), Levilinea saccharolytica (AB109439), Roseiflexus castenholzii (CP000804), Oscillochloris trichoides (AF093427), Chloroflexus aurantiacus (D38365), Chloroflexus aggregans (CP001337), Herpetosiphon geysericola (AF039293), Herpetosiphon aurantiacus (CP000875), Thermomicrobium roseum (M34115), and Sphaerobacter thermophilus (AJ420142). Aquifex pyrophilus (M83548) and Hydrogenobacter thermophilus (Z30214) were used as an outgroup. (B) The relative abundance of each OTU within each community in the tree. The relative values of OTU abundances are indicated by fill intensities at the bottom right corner. Abbreviated sample names: S1A; Forest soil 1A, S1B; Forest soil 1B, S2; Forest soil 2, G; Garden soil, S; Sand, B; bark, T; Geothermal soil, C; Compost.
In the geothermal sediment sample, representative sequences from the family Thermogemmatisporaceae in the order Thermogemmatisporales dominated (93.5%; Fig. 2 and 3). Very few representative sequences from the other samples were assigned to this cluster.
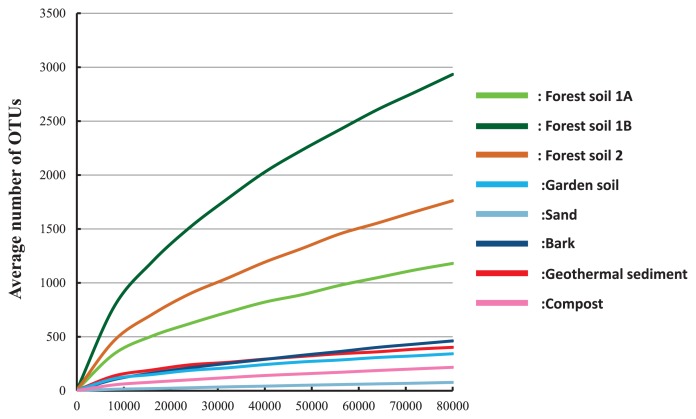
Fig. 3.
Rarefaction curves for reads normalized to 80,000 for each sample using 0.03 distance OTUs (n=10).
In the compost sample, sequences from the family Thermosporotrichaceae in the order Ktedonobacterales were dominant (98.3%). In addition, OTU 439 was particularly dominant in this sample (94.5%), and, thus, diversity was low (Shannon=0.49; Table 4).
In the remaining samples (forest soils, garden soil, sand, and bark), most of the representative sequences were assigned to unknown Ktedonobacterales groups (52.4%–99.9%). The ktedonobacterial community in forest soil 1A was dominated by three unknown groups: group 0 (47.3%), group 6 (21.0%, including the dominant OTU 7286), and group 3 (12.6%; Fig. 1 and 2). In forest soil 1B, the community was dominated by group 0 (47.7%, including the dominant OTU 3633) and Thermosporotrichaceae (16.2%; Fig. 1 and 2). In forest soil 2, the community was dominated by group 1 (62.4%, including the dominant OTU 6399) and group 0 (14.0%). In garden soil, the community was dominated by group 0 (94.1%, including the dominant OTU 9819). The sand sample was dominated by group 3 (99.8%, including the dominant OTU 8693). In bark, the community was dominated by group 1 (97.1%, including the dominant OTU 2760; Fig. 1 and 2).
Discussion
The class Ktedonobacteria was proposed ten years ago (5), and was more recently added to genetic databases. Therefore, few studies have discussed Ktedonobacteria. Prior to their proposal, environmental clones that have since been assigned to this class were described as unclassified bacteria or, in some cases, as unclassified Chloroflexi. For example, in 2006, ktedonobacterial clones that were detected in uranium-contaminated soil were reported as novel Chloroflexi clones with low homology to previously sequenced Chloroflexi (3), which were positioned within Ktedonobacteraceae in this study (Fig. 2). Likewise, environmental clones from an alpine tundra soil in the US Rocky Mountains (9) and from Hawaiian volcanic deposits (Clone 1921 series) (14) that were reported as new subdivisions of Chloroflexi were positioned within groups 3 and Thermosporotrichaceae, respectively, of Ktedonobacteria in this study (Fig. 2).
Since 2010, taxa within Ktedonobacteria are becoming more common in studies on microbial ecosystems in extreme environments. Many of these studies have described Ktedonobacteria as being among the most abundant types of bacteria in such environments. For example, they predominate in the cinder deposits of the Kilauea volcano (38), in lava caves (25), in high-elevation mineral soils (20), in soil from CO2 gas vents in the Calatrava volcanic field in Spain (29), in orthoquartzite caves in Venezuela (2), in the dark oligotrophic volcanic ice caves of Mt. Erebus in Antarctica (34), in soil from northern Victoria Land in Antarctica (17), and in geothermal soils on Pantelleria Island in Italy (12). Furthermore, most of the environments in which Ktedonobacteria have been found to predominate are acidic (2, 12, 17, 20, 29, 34); a survey of microbial ecosystems in the soil of Antarctica showed that they markedly decreased with increases in pH in the range of pH 5 to 7 (17).
The primary analytical method recently used to investigate the microbiota of various environments has changed from the 16S rRNA gene clone library method to the sequencing of millions of fragments using next-generation sequencing technologies from Roche and Illumina. These new methods allow researchers to investigate low-abundance taxonomic groups, and have facilitated the detection of ktedonobacterial phylotypes as a minor taxonomic group in terrestrial environments such as mesic soils (1, 24, 37, 44). These studies have led us to suspect that Ktedonobacteria only predominate in extreme acidic oligotrophic environments, such as volcanic fields and caves, and that they only occur at low abundances in more common terrestrial environments. The results of the present study have partially confirmed this: we found Ktedonobacteria in some common, mesic terrestrial environments at a low relative abundance (0.005–0.80%; Table 3), whereas they dominated in a hot geothermal sediment (12.9%; Table 3). However, the geothermal sediment was near-neutral (pH 6.8) and rich in organic matter (total C, 32.0%; total N, 1.6%); therefore, Ktedonobacteria may be dominant not only in acidic oligotrophic environments. In addition, this is the first time that Ktedonobacteria have been detected in bark.
This study is the first attempt to estimate their diversity and richness and to compare the community structure of their phylotypes in any environment using a specific primer. Our analysis showed that the diversity and richness indices of Ktedonobacteria in forest soils are very high (Table 4). In forest soils, the failure of the OTU rarefaction curves to plateau indicates that these communities were incompletely sampled (Fig. 3). Therefore, a large proportion of ktedonobacterial diversity remains to be sampled from these forest soils. Although these indices were lower in the other samples, each community was dominated by a different group (garden soil, G0; sand, G3; bark, G1; geothermal sediment, Thermogemmatisporales; compost, Thermosporotrichaceae; Fig. 1). These results suggest a diversity of phylotypes of Ktedonobacteria highly adapted to local environments, and so the overall diversity of the class may be extremely high. These results support our hypothesis that Ktedonobacteria are diverse in most environments.
The class Ktedonobacteria currently consists of two orders and three families. Almost all of the representative OTUs from soils, sand, and bark were not assigned to the known families. In our phylogenetic tree, OTUs were divided into eight groups, which lacked validly proposed cultured isolates (Fig. 1 and 2). Therefore, difficulties are associated with predicting their physiology, morphology, and ecological functions. However, strains SOSP1-142, 1-79, 1-9, and 1-63, isolated from soil in Italy (5), which have not yet been validly proposed as taxa, were positioned in groups 0, 1, 4, and 5, respectively, with low bootstrap values. Their known characteristics (5) are only that they were isolated at 30°C, form mycelia with spores, similar to typical actinomycetes, hydrolyze starch, casein, and gelatin, and are catalase-positive, Gram-positive, and resistant to some antibiotics. These characteristics may be common to the groups to which they belong. Groups 0 and 1 were the dominant groups in forest soils, garden soil, and bark (Fig. 1 and 2), implying that diverse undiscovered phylotypes with an actinomycetes-like morphology and not belonging to the class Actinobacteria may inhabit common terrestrial environments. All isolates previously assigned to Ktedonobacteria have an actinomycetes-like morphology (5, 19, 32, 40–43) even though they are distributed across two orders, three families, and groups 0, 1, 4, and 5 in the phylogenetic tree (Fig. 2). Therefore, it is possible that morphological features similar to those of actinomycetes are highly conserved across all phylotypes in Ktedonobacteria.
Groups 2, 3, and 5 include clones detected from extreme environments. Group 2 includes the unpublished clone UMAB-cl-097 (FR749722) from soil collected on the Antarctic Peninsula. This group formed a cluster with the OTUs from forest soil 1B at a high bootstrap value (95%), suggesting that the phylotypes of group 2 are not restricted to Antarctica.
Group 3 includes the clones D10_WMSP2 (DQ450722) and E11_WMSP1 (DQ450736), which were predominant in cold, acidic soil (pH 4.3–5.3, near 0°C) in the alpine tundra in the Rocky Mountains (9). These bacteria may play an active biogeochemical role in cold, anoxic soils (9). However, group 3 was dominated by OTUs from the sand sample (24°C), with markedly different characteristics from the cold soil collected in the alpine tundra. It also included 12.6% of the representative OTUs from forest soil 1A. Therefore, the phylotypes in this group occur in a wide range of environments, including extreme habitats and common environments such as forest soils and sand.
Group 5 includes the clone cluster (Clone B series) of the Hawaiian volcanic deposit (Fig. 2), predominantly comprising clones from unvegetated cinder deposits in the Hawaii Volcanoes National Park (38). Sequences most closely related to that of a gene for the large subunit of the carbon monoxide dehydrogenase (coxL) from K. racemifer dominated there (38), and some strains belonging to Thermosporotrichaceae and Thermogemmatisporaceae oxidize CO (19). However, it is not yet clear whether the phylotypes in group 5 have this ability because no strains belonging to group 5 have yet been isolated. In addition, 0.6% of the representative OTUs from forest soil 1B were affiliated with group 5. However, since this is a small proportion, and the other members of group 5 originated in a very different environment, it is difficult to make predictions about the characteristics of this group.
Groups 6 and 7 each formed a clade that was clearly distinguished from the other groups. Group 6 was predominant in forest soil 1A, at a relative abundance of 21.0%. It also included the clones BacC-u_018 (EU335378) and BacC-u_077 (EU335418), which had been found in soil with unsaturated C horizons in Tennessee, USA. Therefore, the phylotypes within group 6 may be widely distributed in soil ecosystems. Group 7 contained 2.2% of the representative OTUs in forest soil 1A.
In contrast, almost all the representative OTUs from the geothermal sediment and compost were assigned to known families in Ktedonobacteria. Compost was dominated by Thermosporotrichaceae (98.3%; Fig. 1). Species of Thermosporothrix form branched aerial mycelia with spores, and are Gram-positive, thermophilic, aerobic, and heterotrophic. Thermosporotrichaceae were isolated from compost produced by the same composting system (41); therefore, we expected this family to be predominant in the compost sample. However, Ktedonobacteria were rare in that sample (Table 3). T. narukonensis was isolated from fallen leaves deposited on geothermal soil and proposed as a novel species in the genus (43). These isolates commonly have an actinomycetes-like morphology and the ability to hydrolyze polysaccharides such as CM-cellulose, microcrystalline cellulose, and xylans (41, 43). Therefore, we consider this genus to contribute to the decomposition of polysaccharides in fallen leaves and wood fragments in compost and geothermal ecosystems. This family was also the predominant taxon in the OTUs of forest soil 1B (16.2%), indicating that it may include mesophilic species.
The geothermal sediment was the only sample in which Ktedonobacteria were predominant, at 12.9% relative abundance in the universal amplicons (Table 3). The KS amplicons of this sample were dominated by Thermogemmatisporaceae (93.5%) in Thermogemmatisporales. This order was detected almost solely in the geothermal sediment, although Ktedonobacterales are widely distributed in various environments. Furthermore, the sequences assigned to Thermogemmatisporales in the RDP database comprised only the three type strains and seven isolates described above and the environmental clone Wkt_02 (AM749737), all of which were isolated from the geothermal sediment. Therefore, the phylotypes in this order may be strongly adapted to environments characteristic of geothermal sediments. These isolates are commonly thermophilic bacteria with an actinomycetes-like morphology, which are able to grow at 40–74°C (19, 42). T. onikobensis, T. foliorum, and T. carboxidivorans hydrolyze cellulose and xylans (19, 42). Thermobifida and Acidothermus are well-known thermophilic cellulolytic actinomycetes, but cannot grow at >65°C. Therefore, phylotypes in the order Thermogemmatisporales may play a role in decomposing polysaccharides in very high temperature environments.
In summary, the results of the present study indicate that diverse phylotypes of the class Ktedonobacteria inhabit a range of environments, including mesic and extreme habitats, nutrient-rich and oligotrophic conditions, high and low pH, and high and low temperatures. This diversity suggests that these phylotypes produce unique metabolites, particularly secondary metabolites, in order to survive in this range of environments because secondary metabolism is commonly associated with complex differentiation similar to that of the actinomycetes (2). It also strongly supports our hypothesis that Ktedonobacteria may contain valuable new resources for the discovery of novel compounds.
Few strains in the class Ktedonobacteria been isolated and cultured to date. It is easy to cultivate strains in the genera Thermosporothrix and Thermogemmatispora because their growth is rapid: they form aerial mycelia in less than a week on common media such as ISP1 and ISP3 (41–43). In contrast, the growth of K. racemifer SOSP1-21T is slow (5), but does not require special culture conditions. Therefore, few strains may have been isolated due to their very low relative abundance in common mesic environments.
This study provides an insight into ktedonobacterial abundance, diversity, richness, and habitats in several terrestrial environments. However, further studies are needed in order to fully reveal the diversity and abundance of this class. It is important to develop selective methods for culturing Ktedonobacteria, which will contribute to the investigation of this class as a valuable new microbial resource and also as a substitute for actinomycetes, and will reveal the roles they play in their ecosystems.
Supplemental data
Acknowledgements
This work was supported by MEXT/JSPS KAKENHI grant number 16K20903. We thank Hiroshi Katsumata (Bioengineering Lab) for his help with MiSeq sequencing and the data analysis.
References
- 1.Acosta M.V., Dowd S.E., Bell C.W., Lascano R., Booker J.D., Zobeck T.M., Upchurch D.R. Microbial community composition as affected by dryland cropping systems and tillage in a semiarid sandy soil. Diversity. 2010;2:910–931. [Google Scholar]
- 2.Barton H.A., Giarrizzo J.G., Suarez P., Robertson C.E., Broering M.J., Banks E.D., Vaishampayan P.A., Venkateswaran K. Microbial diversity in a Venezuelan orthoquartzite cave is dominated by the Chloroflexi (Class Ktedonobacterales) and Thaumarchaeota Group I.1c. Front Microbiol. 2014;5:1–14. doi: 10.3389/fmicb.2014.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodie E.L., DeSantis T.Z., Joyner D.C., et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caporaso J.G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavaletti L., Monciardini P., Bamonte R., Schumann P., Rohde M., Sosio M., Donadio S. New lineage of filamentous, spore-forming, Gram-positive bacteria from soil. J Appl Environ Microbiol. 2006;72:4360–4369. doi: 10.1128/AEM.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y.J., Land M., Hauser L., et al. Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1–21T) Stand Genomic Sci. 2011;5:97–111. doi: 10.4056/sigs.2114901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao A., Bunge J. Estimating the number of species in a stochastic abundance model. Biometrics. 2002;58:531–539. doi: 10.1111/j.0006-341x.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- 8.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:633–642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello E.K., Schmidt S.K. Microbial diversity in alpine tundra wet meadow soil: Novel Chloroflexi from a cold, water-saturated environment. Environ Microbiol. 2006;8:1471–1486. doi: 10.1111/j.1462-2920.2006.01041.x. [DOI] [PubMed] [Google Scholar]
- 10.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 12.Gagliano A.L., Tagliavia M., D’Alessandro W., Franzetti A., Parello F., Quatrini P. So close, so different: geothermal flux shapes divergent soil microbial communities at neighbouring sites. Geobiology. 2015;14:150–162. doi: 10.1111/gbi.12167. [DOI] [PubMed] [Google Scholar]
- 13.Gich F., Garcia-Gil J., Overmann J. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch Microbiol. 2001;177:1–10. doi: 10.1007/s00203-001-0354-6. [DOI] [PubMed] [Google Scholar]
- 14.Gomez A.V., King G.M., Nüsslein K. Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol Ecol. 2007;60:60–73. doi: 10.1111/j.1574-6941.2006.00253.x. [DOI] [PubMed] [Google Scholar]
- 15.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 16.Jiang H., Dong H., Zhang G., Yu B., Chapman L.R., Fields M.W. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol. 2006;72:3832–3845. doi: 10.1128/AEM.02869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M., Cho A., Lim H.S., Hong S.G., Kim J.H., Lee J., Choi T., Ahn T.S., Kim O.S. Highly heterogeneous soil bacterial communities around Terra Nova Bay of Northern Victoria Land, Antarctica. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.King C.E., King G.M. Description of Thermogemmatispora carboxidivorans sp. nov., a carbon-monoxide oxidizing member of the class Ktedonobacteria isolated from a geothermally heated biofilm, and analysis of carbon monoxide oxidation by members of the class Ktedonobacteria. Int J Syst Evol Microbiol. 2014;64:1244–1251. doi: 10.1099/ijs.0.059675-0. [DOI] [PubMed] [Google Scholar]
- 20.Lynch R.C., King A.J., Farías M.E., Sowell P., Vitry C., Schmidt S.K. The potential for microbial life in the highest-elevation (>6000 m. a.s.l.) mineral soils of the Atacama region. J Geophys Res Biogeosci. 2012;117:1–10. [Google Scholar]
- 21.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak B.L., Cole J.R., Lilburn T.G., Parker C.T., Saxman P.R., Farris R.J., Garrity G.M., Olsen G.J., Schmidt T.M., Tiedje J.M. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyzer G., de Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieva A.S., Bailleres M.A., Corriale M.J., Llames M.E., Menéndez A.B., Ruiz O.A. Herbicide-mediated promotion of Lotus tenuis (Waldst. & Kit. ex Wild.) did not influence soil bacterial communities, in soils of the Flooding Pampa, Argentina. Appl Soil Ecol. 2016;98:83–91. [Google Scholar]
- 25.Northup D.E., Melim L.A., Spilde M.N., Hathaway J.J.M., Garcia M.G., Moya M., Stone F.D., Boston P.J., Dapkevicius M.L.N.E., Riquelme C. Lava cave microbial communities within mats and secondary mineral deposits: implications for life detection on other planets. Astrobiology. 2011;11:601–618. doi: 10.1089/ast.2010.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi Y., Yamazaki H., Kato J., Tomono A., Horinouchi S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. J Biosci Bioeng. 2005;69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 27.Park J.S., Kagaya N., Hashimoto J., Izumikawa M., Yabe S., Shin-ya K., Nishiyama M., Kuzuyama T. Identification and biosynthesis of new acyloins from the thermophilic bacterium Thermosporothrix hazakensis SK20-1T. Chembiochem. 2014;15:527–532. doi: 10.1002/cbic.201300690. [DOI] [PubMed] [Google Scholar]
- 28.Park J.S., Yabe S., Shin-ya K., Nishiyama M., Kuzuyama T. New 2-(1′H-indole-3′-carbonyl)-thiazoles derived from the thermophilic bacterium Thermosporothrix hazakensis SK20-1T. J Antibiot (Tokyo) 2014;68:60–62. doi: 10.1038/ja.2014.93. [DOI] [PubMed] [Google Scholar]
- 29.Sáenz de Miera L.E., Arroyo P., de Luis Calabuig E., Falagán J., Ansola G. High-throughput sequencing of 16S RNA genes of soil bacterial communities from a naturally occurring CO2 gas vent. Int J Greenhouse Gas Control. 2014;29:176–184. [Google Scholar]
- 30.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Shannon C.E., Weaver W. The mathematical theory of information. AT&T Tech J. 1949;27:359–423. [Google Scholar]
- 32.Stott M.B., Crowe M.A., Mountain B.W., Smirnova A.V., Hou S., Alam M., Dunfield P.F. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ Microbiol. 2008;10:2030–2041. doi: 10.1111/j.1462-2920.2008.01621.x. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tebo B.M., Davis R.E., Anitori R.P., Connell L.B., Schiffman P., Staudigel H. Microbial communities in dark oligotrophic volcanic ice cave ecosystems of Mt. Erebus, Antarctica. Front Microbiol. 2015;6:1–14. doi: 10.3389/fmicb.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Wang X., Zhang W., et al. Invariant community structure of soil bacteria in subtropical coniferous and broadleaved forests. Sci Rep. 2016;6:19071. doi: 10.1038/srep19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber C.F., King G.M. Distribution and diversity of carbon monoxide-oxidizing bacteria and bulk bacterial communities across a succession gradient on a Hawaiian volcanic deposit. Environ Microbiol. 2010;12:1855–1867. doi: 10.1111/j.1462-2920.2010.02190.x. [DOI] [PubMed] [Google Scholar]
- 39.Yabe S., Kato A., Hazaka M., Yokota A. Thermaerobacter composti sp. nov., a novel extremely thermophilic bacterium isolated from compost. J Gen Appl Microbiol. 2009;55:323–328. doi: 10.2323/jgam.55.323. [DOI] [PubMed] [Google Scholar]
- 40.Yabe S., Aiba Y., Sakai Y., Hazaka M., Yokota A. A life cycle of branched aerial mycelium- and multiple budding spore-forming bacterium Thermosporothrix hazakensis belonging to the phylum Chloroflexi. J Gen Appl Microbiol. 2010;56:137–141. doi: 10.2323/jgam.56.137. [DOI] [PubMed] [Google Scholar]
- 41.Yabe S., Aiba Y., Sakai Y., Hazaka M., Yokota A. Thermosporothrix hazakensis gen. nov., sp. nov., isolated from compost, description of Thermosporotrichaceae fam. nov. within the class Ktedonobacteria Cavaletti et al. 2007 and emended description of the class Ktedonobacteria. Int J Syst Evol Microbiol. 2010;60:1794–1801. doi: 10.1099/ijs.0.018069-0. [DOI] [PubMed] [Google Scholar]
- 42.Yabe S., Aiba Y., Sakai Y., Hazaka M., Yokota A. Thermogemmatispora onikobensis gen. nov., sp. nov. and Thermogemmatispora foliorum sp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. nov. and Thermogemmatisporales ord. nov. within the class Ktedonobacteria. Int J Syst Evol Microbiol. 2011;61:903–910. doi: 10.1099/ijs.0.024877-0. [DOI] [PubMed] [Google Scholar]
- 43.Yabe S., Sakai Y., Yokota A. Thermosporothrix narukonensis sp. nov., belonging to the class Ktedonobacteria, isolated from fallen leaves on geothermal soil, and emended description of the genus Thermosporothrix. Int J Syst Evol Microbiol. 2016;66:2152–2157. doi: 10.1099/ijsem.0.001004. [DOI] [PubMed] [Google Scholar]
- 44.Zeng Q., Dong Y., An S. Bacterial community responses to soils along a latitudinal and vegetation gradient on the Loess Plateau, China. PLoS One. 2016;11:e0152894. doi: 10.1371/journal.pone.0152894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.