Abstract
The regulation of stem cell proliferation in plants is controlled by intercellular signaling pathways driven by the diffusible CLAVATA3 (CLV3p) peptide. CLV3p perception is thought to be mediated by an overlapping array of receptors in the stem cell niche including the transmembrane receptor kinase CLV1, Receptor-Like Protein Kinase 2 (RPK2), and a dimer of the receptor-like protein CLV2 and the CORYNE (CRN) pseudokinase. Mutations in these receptors have qualitatively similar effects on stem cell function but it is unclear if this represents common or divergent signaling outputs. Previous work in heterologous systems has suggested that CLV1, RPK2 and CLV2/CRN could form higher order complexes but it is also unclear what relevance these putative complexes have to in vivo receptor functions. Here I use the in vivo regulation of a specific transcriptional target of CLV1 signaling in Arabidopsis to demonstrate that, despite the phenotypic similarities between the different receptor mutants, CLV1 controls distinct signaling outputs in living stem cell niches independent of other receptors. This regulation is separable from stem cell proliferation driven by WUSCHEL, a proposed common transcriptional target of CLV3p signaling. In addition, in the absence of CLV1, CLV1-related receptor kinases are ectopically expressed but also buffer stem cell proliferation through the auto-repression of their own expression. Collectively these data reveal a unique in vivo role for CLV1 separable from other stem cell receptors and provides a framework for dissecting the signaling outputs in stem cell regulation.
Author summary
The proliferation of plant stem cells in above ground tissues is controlled by a suite of receptors in response to the CLAVATA3 peptide ligand. Receptor signaling in response to CLAVATA3 prevents over-proliferation of stem cells. It is unclear what the functional relationship is between the proposed CLAVATA3 receptors or if they impact common signaling outputs. Here I demonstrate that CLAVATA1 signals independently of the other receptors kinases to control distinct transcriptional outputs independent of stem cell proliferation. Stem cell proliferation is buffered by a two-step mechanism which transcriptionally regulates receptor levels in the stem cell niche. This mechanism helps explain the strict control of stem cell proliferation and could provide new avenues for improving plant growth.
Introduction
Co-operative receptor kinase function is a common feature in both animal and plant signaling systems. Receptor kinase mutants are frequently genetically additive in plants but the molecular mechanisms underlying this effect are often different. For instance, double mutants between the EF-TU RECEPTOR and FLAGELLIN SENSITIVE2 receptor kinases display enhanced susceptibility to bacterial infection above each single mutant [1], reflecting differences in pathogen derived ligands, followed by quantitative activation of common downstream outputs. On the other hand, additive genetic interactions among mutants in SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE family co-receptor kinases in response to specific ligands reflect quantitative redundancy as co-receptors [2]. Dissecting the molecular basis of redundancy in gene families in plants is often also complicated by unequal contribution from distinct genes and often requires in vivo analysis of signaling outputs or component interactions [3]. Balanced stem cell production in shoot (SAM) and floral meristems (FMs) is mediated by cell-to-cell signaling pathways initiated by the CLAVATA3 (CLV3) peptide ligand, a founding member of the CLE family of peptides [4]. Mutations in CLV3 lead to excess accumulation of stem cells in both SAM and FMs [5]. CLV3p is thought to be perceived by a series of overlapping receptor kinases which signal to dampen stem cell production. To date, much of the analysis of these receptor-ligand mutants has been at the gross morphological level. Mutations in the different proposed receptors vary considerably in their strength and genetic interactions. It is unclear if the morphological similarities, strength differences, or genetic interactions are due to co-operative cross talk, convergence on a common signaling output, or divergent pathways.
CLV3 is secreted from stem cells in the growing tip of the meristem, proteolytically processed and modified to a 13 amino acid diffusible glycopeptide (CLV3p) which diffuses broadly throughout the SAM [6–10]. Genetically, CLV3p perception is mediated by the transmembrane receptor kinase CLV1 [11–13], but also by a heterodimer of the receptor like protein CLV2 and the transmembrane pseudokinase CORYNE (CRN, [14–17]). Additionally, the receptor kinase RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2) may also function in CLV3p perception [18]. All four mutants are resistant to exogenous CLV3p treatment to different degrees suggesting that they act as receptors for CLV3p in vivo. Previous data using overexpression in differentiated tobacco leaf cells has suggested that CLV1 could form higher order complexes with CLV2/CRN leading to the hypothesis that they may signal co-operatively in the SAM [19–22]. However conflicting results have been obtained by different groups and it is not clear if such complexes form in SAM tissues, or when receptors are expressed at endogenous levels in the appropriate cell types. While CLV3p has been reproducibly demonstrated to bind the CLV1 ectodomain [12, 23], differing results have been obtained for the ability of CLV2 to bind CLV3p [20, 23]. In addition, these studies have not tested for potential co-receptor binding of CLV3p. As such it is unclear how, or if, these receptors control stem cell proliferation co-operatively in vivo.
Strong alleles of clv1, such as clv1-4 and clv1-8, contain missense mutations in the LRR domain of CLV1 [11]. The residues effected in these mutants are highly conserved among related LRR-RKs and structural and biochemical studies with PXY/TDIF receptor ligand pair have shown that these residues direct ligand binding [24–28]. In contrast, clv1 null mutants are significantly weaker [29]. The weak stem cell defects in clv1 null mutant plants can partially be explained by the compensatory up-regulation of CLV1-related receptor kinases BARELY ANY MERISTEM1(BAM1), BAM2 and BAM3 [28]. In wild type plants, BAM receptor expression in the center of the SAM, where CLV1 is highly expressed, is undetectable. Consistent with this observation, triple mutants in bam1 bam2 bam3 have no defects in stem cell regulation on their own. In clv1 mutant SAMs BAM receptors are ectopically expressed in the center of the SAM and partially compensate for clv1. However it is unclear why null clv1 alleles are only weakly compensated by ectopic BAM expression. It is also unclear why strong clv1 alleles are phenotypically more severe. Previous work has suggested that strong clv1 mutant receptors may interfere with CRN/CLV2 signaling [16]. Alternatively, strong clv1 mutant receptors have been suggested to interfere with BAM signaling [30]. It is not clear how this relates to the feedback regulation of BAM expression by CLV1.
CLV1, and the other putative CLV3p receptors, are proposed to negatively regulate WUSCHEL (WUS) expression in the center of the SAM [13, 31]. WUS is a homeodomain transcription factor and de-repression of WUS in clv3 mutants is thought to drive excess stem cell proliferation [31, 32]. Despite this, the expression of WUS is robust and co-incident with CLV1 in wild type plants in the center of the SAM and WUS levels do not change dramatically at the cellular levels in response to loss of CLV1 signaling [11, 28, 32, 33]. Unlike WUS, CLV3p-CLV1 signaling fully represses BAM expression in the center of the SAM in wild type plants [28]. Plants expressing up to 300 fold higher levels of CLV3 have a wild type appearance, suggesting that repression of WUS is most effective outside of the physiological range of CLV3p concentration [34]. Interestingly, expression of CLV1 from the WUS promoter is necessary and sufficient to fully complement both clv1 null mutants and clv1 bam1 bam2 bam3 quadruple null mutants back to wild type levels of stem cell regulation [28]. As such, CLV1 operates exclusively in WUS expressing cells of the SAM, despite WUS being a target for transcriptional repression. It is not clear where in the SAM other proposed CLV3p receptors function or if WUS-mediated cell proliferation is linked to BAM transcriptional regulation by CLV1 in vivo.
Here I use quantitative genetics, and the highly specific transcriptional repression of BAM3 by CLV1 to demonstrate that CLV1 signals independent of CRN, CLV2 and RPK2 in response to CLV3p in vivo. In clv1 null mutants, ectopic BAM receptors compensate for CLV1 but also act in an additional feedback loop to dampen their own expression in the SAM and buffer stem cell proliferation. Strong alleles of clv1 specifically interfere with this process and have no impact on CLV2/CRN function. Despite their proposed ability to repress WUS, CRN/CLV2 function exclusively in WUS expressing cells of the SAM like CLV1. Consistent with this, WUS-induced stem cell proliferation is genetically separable from BAM3 regulation by CLV1. My data demonstrate that despite the qualitative phenotypic similarities, CLV1 signaling outputs diverge from other receptors, and from WUS, and support a model in which CLV1 is functionally independent of other proposed receptors in vivo.
Results
Generation of a crn null mutant
In order to determine the functional relationship between the proposed CLV3p receptors I used previously published null alleles in CLV1, CLV2, BAM1, BAM2 and BAM3 in the Col-0 background [28, 35]. To date there are no null EMS generated alleles of CRN in any ecotype and no T-DNA insertions in the CRN coding sequence. I therefore used Cas9 to target CRN and create a null mutant. I created a gRNA targeting the signal sequence encoding region of the CRN gene and used the pCUT series of Cas9 vectors to create indels in the CRN gene in the Col-0 ecotype [36]. One of these, hereby referred to as crn-10, introduced a single thymine base between bases 20 and 21 in the CRN CDS. The mutation introduces a frameshift in the protein after amino acid 6 in the 33 amino acid signal sequence, leading to three in-frame stop codons four amino acids downstream. No other in frame methionine residues are present in, or before, the predicted CRN transmembrane sequence. Thus, the crn-10 allele retains 6 amino acids out of the original 402 in CRN and creates an early stop codon in the CRN signal sequence and deletes all downstream domains of CRN. crn-10 was segregated away from the Cas9 transgene for all subsequent work. crn-10 plants are qualitatively similar to other published crn alleles [16], with no new phenotypes noted. Wild type FMs give rise to stem cell populations that support stereotypical numbers of floral organs, culminating in the production of two central fused carpels. In clv mutant FMs, the enhanced rate of stem cell production results in increases in floral organ numbers, providing a quantitative measure of stem cell defects [29]. crn-10 plants displayed increased carpel number with a strength comparable to existing clv2 null alleles in Col-0 (Fig 1A). As such crn-10 behaves similarly to recessive non-null EMS alleles of crn such as crn-1 in the La-er ecotype [16]. crn-10 was fully complemented by a CRN-2xmCherry fusion protein expressed from the endogenous CRN promoter (44/44 lines, S1A Fig) as expected [17]. Previous work aimed at testing the hypothesis that CRN encoded a pseudokinase demonstrated that expression of a CRNK146E mutant protein, which further mutates the conserved active site lysine in CRN, fully complements crn-1 when expressed from the native CRN promoter [22]. At the time crn-1 was the only allele available and encodes a protein with a missense mutation in the CRN transmembrane domain (G70E) [16]. It is formally possible that if CRN homodimerized, the crn-1 and CRNK146E proteins could cross complement. I therefore transformed crn-10 with a pCRN::CRNK146E-2xmCherry transgene. Again I observed full complementation supporting the previous designation of CRN as a pseudokinase (26/26 lines, S1A Fig). Recent work has suggested that phosphorylation of CRN at serine 156 is important for function and that S156A substitutions fail to complement crn-1 when expressed from the 35S promoter [37]. In contrast, expression of either a S156A or a S156D CRN variant fully complemented the crn-10 null mutant when expressed from the CRN native promoter (26/27 and 24/24 line displaying full complementation for the S156A and S156D CRN variants respectively, S1A Fig). The reason for the different complementation results are unknown but could reflect either the use of different crn mutant plants or different transgene promoters in the complementation experiments.
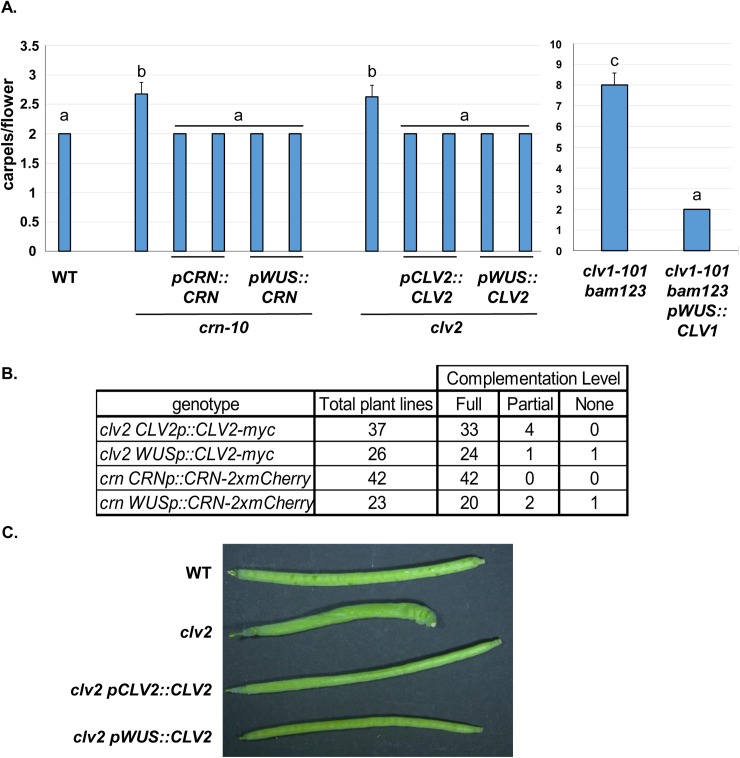
Fig 1. CLV3p receptors function in WUS-expressing cells is sufficient to control stem cell proliferation.
(A) CRN, CLV2 and CLV1 control stem cell production in WUS expressing cells. Previous work demonstrated that CLV1 function in WUS expressing cells is necessary and sufficient for stem cell regulation in vegetative and floral meristems in clv1-101 null mutants and clv1-101 bam1 bam2 bam3 quadruple mutant plant (clv1-101 bam123, [28]). Like CLV1, crn-10 and clv2 null mutants were complemented fully by expression of CRN-2xmCherry or CLV2-Myc fusion proteins from either their native promoter or the WUS promoter. Y-axis, mean number of carpels per flower for different genotypes, with two independent transgenic lines per genotype displayed. Error bars represent the calculated 95% C.I. N = 40 flowers counted per plant. Letters above bars indicate significance groups as determined by Tukey HSD test/ANOVA. (B) Complementation rates for clv2 and crn transgenic lines. Full complementation was defined as no flower containing more than 2 carpels as in wild type (WT). (C) Example of full complementation in carpels from clv2 transgenic lines. Similar results obtained with crn-10 complementation.
CLV1, CRN and CLV2 control stem cell regulation exclusively in WUS expressing cells of the SAM
I previously demonstrated that CLV1 function in WUS expressing cells is necessary and sufficient for stem cell regulation [28]. Both CLV2 and CRN are expressed broadly in inflorescence tissue (see S1B Fig, [15, 16]) but it is not clear if they function in WUS expressing cells of the SAM like CLV1. I therefore tested the ability of CLV2-myc and CRN-2xmCherry fusion proteins to complement their respective null mutants when expressed from either their native promoters or the WUS promoter. Like CRN, expression of CLV2-myc from the native CLV2 promoter in clv2 null mutant plants (rpl10-1, [35]) fully complemented stem cell defects in the majority of lines (Fig 1). Both CRN and CLV2 expression from the WUS promoter also fully complemented their respective null mutant plants in the majority of T1 lines (Fig 1). Fully complemented lines contained no flowers with more than two carpels, a level of complementation equivalent to complementation of the clv1 bam1 bam2 bam3 quadruple provided by the pWUS::CLV1-2xGFP transgene as previously reported [28]. Collectively these data indicate that like CLV1, CRN and CLV2 function exclusively in WUS expressing cells of the SAM and FM.
CRN, CLV2, and RPK2 are dispensable for CLV1-mediated repression of BAM receptors
My data demonstrate that CRN, CLV2 and CLV1 all function exclusively in WUS-expressing cells in the center of the meristem, however, this observation does not address if they act together at the biochemical level to converge on similar signaling outputs. In wild type plants CLV1 represses the expression of the related BAM receptors in the center of the SAM in response to CLV3p [28]. I therefore asked if CRN and CLV2 participated in CLV1-mediated repression of BAM3 in the SAM center. For simplicity, BAM3 expression was analyzed since BAM1, BAM2 and BAM3 are all targets of CLV1 in the SAM center, but BAM1 and BAM2 display expression in the SAM epidermis and floral primorida which is clv1-independant [28]. I introgressed the previously characterized pBAM3::Ypet-N7 transgenic line [28] from Col-0 into the null alleles of clv2 and crn, and isolated homozygous transgenic lines in each mutant background. For rpk2, a CRISPR null (rpk2-cr) was generated directly in the homozygous pBAM3::Ypet-N7 wild type transgenic line, and segregated away from the Cas9 transgene for analysis (see Materials and Methods). The pBAM3::Ypet-N7 reporter generates a tandem Ypet fusion protein that is targeted to the nucleus. I then compared the expression of the BAM3 reporter in the L3-L5 cells of the SAM center (see S2 Fig for image calibration). CLV1 is expressed strongly in L3-L5 cells in wild type, clv3, and clv1-8 meristems and expression in these cells is sufficient to account for all stem cell regulation by CLV1 [25]. Consistent with previous imaging, BAM3 reporter expression was undetectable in the center of the SAM in wild type plants, but was robustly detectable in clv3 mutants and strong alleles of clv1 (Fig 2A). Similar results were found in FMs (Fig 2A), consistent with CLV3p-CLV1 repression of BAM3 expression in both meristems [28]. In contrast, BAM3 was not expressed in the center of SAMs or FMs in either clv2 or crn mutants (Fig 2A, see S2 Fig for imaging calibration). BAM3 is expressed in phloem lineage cells independent of CLV3-CLV1 signaling [28, 38]. In all plants examined, BAM3 reporter expression was observed in the phloem linage cells of the vasculature outside of the SAM, consistent with previous work [28], demonstrating that lack of signal in the SAM was not due to reporter silencing in any one line (for example see S4 Fig). Similarly, no ectopic expression of BAM3 was observed in the L3-L5 cells from SAMs or FMs of rpk2 null mutant plants Fig 2A, see Materials and Methods for construction of rpk2 null mutant) [18]. These data demonstrate that CLV1 signals to repress BAM3 in response to CLV3p independent of CLV2, CRN, and RPK2.
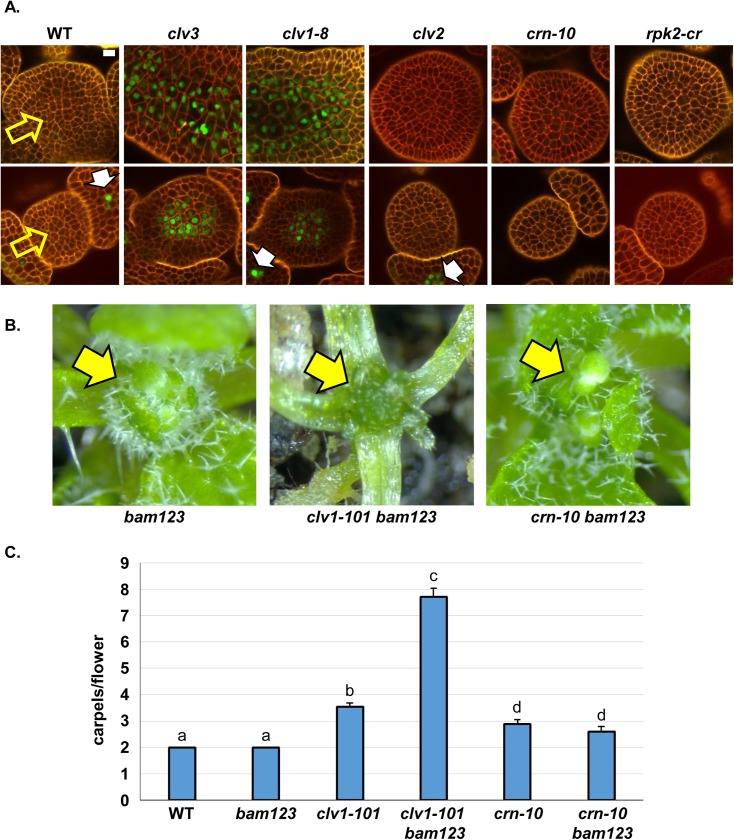
Fig 2. CLV1 signals independent of other proposed CLV3p receptors to control BAM3 expression.
(A) CRN and CLV2 are dispensable for CLV3-CLV1 mediated BAM3 repression. BAM3::Ypet-N7 signal is off in the center of wild type (WT) SAMs (top row, yellow arrow) or FMs (bottom row, yellow arrow), but is de-repressed specifically in clv3 or clv1-8 plants. Note the phloem derived BAM3::Ypet-N7 signal in developing sepals (white arrows) in some imaging planes. Phloem nuclei in other images are not in image plane but still express Ypet-N7. Tissues were stained with propidium iodide (red) to visualize cell walls. SAMs and FMs were imaged by CSLM and each image slice represents the L5 cell layer as determined by counting cell layers from the surface of the SAM or FM. pBAM3::Ypet-N7 fluorescence signal was false colored green to provide better contrast with the red channel. For each genotype, 10 SAMs or FMs were imaged and experiments were repeated three times. White bars, 10 μM. All subpanels are at the same image magnification. See methods for details on rpk2-cr null allele. (B) CRN is not required for CLV1 mediated repression of BAM receptors. clv1-101 bam1 bam2 bam3 (clv1-101 bam123) quadruple mutant plants display uncontrolled vegetative SAM proliferation (yellow arrow), not seen in bam123 triple mutant plants, crn-10 bam123 quadruple mutant plants (yellow arrows). (C) CRN is not required for CLV1 mediated repression of BAM receptors in floral meristems. Y-axis, mean carpels number per flower for different genotypes. Ectopic BAM expression masks full loss of clv1 in clv1-101 null mutants, and clv1-101 null mutants are strongly enhanced by simultaneous mutation of bam1 bam2 bam3 (bam123), In contrast, crn-10 null mutations are strictly additive with bam123 triple mutants and resemble crn-10 alone. N = 60, experiment repeated twice. Error bars represent the calculated 95% C.I. Letters above bars indicate significance groups as determined by Tukey HSD test/ANOVA.
I sought to test this observation genetically by creating higher order receptor mutants. Owing to their repression by CLV1 in the center of the SAM, BAM receptors do not normally participate in stem cell regulation leading to an invariant two carpels per flower in bam1 bam2 bam3 triple mutants as in wild type Col-0 plants. However, when ectopically expressed in the center of FMs and SAMs in clv1-101 null mutants, BAM receptors partially compensate for the lack of CLV1 and correspondingly clv1-101 bam1 bam2 bam3 mutants greatly enhance carpel numbers of clv1-101 null mutants and display massive SAM over-proliferation during vegetative growth (Fig 2B and 2C, [28]). crn null mutants are phenotypically weaker than clv1-101 null mutants (Fig 2C). However, unlike clv1-101, crn null mutants were strictly additive with bam1 bam2 bam3 triple mutants in FMs and crn-10 bam1 bam2 bam3 plants were identical to crn alone (Fig 2C). In addition, crn-10 bam1 bam2 bam3 displayed a bam1 bam2 bam3 vegetative SAM phenotype, and lacked the unregulated SAM over-proliferation seen in clv1-101 bam1 bam2 bam3 plants (Fig 2B) [39]. This observation is consistent with CRN being dispensable for CLV1 mediated regulation of BAM expression and signaling in vivo.
Ectopic BAM receptors auto-dampen their own expression in the SAM in clv1 null mutants
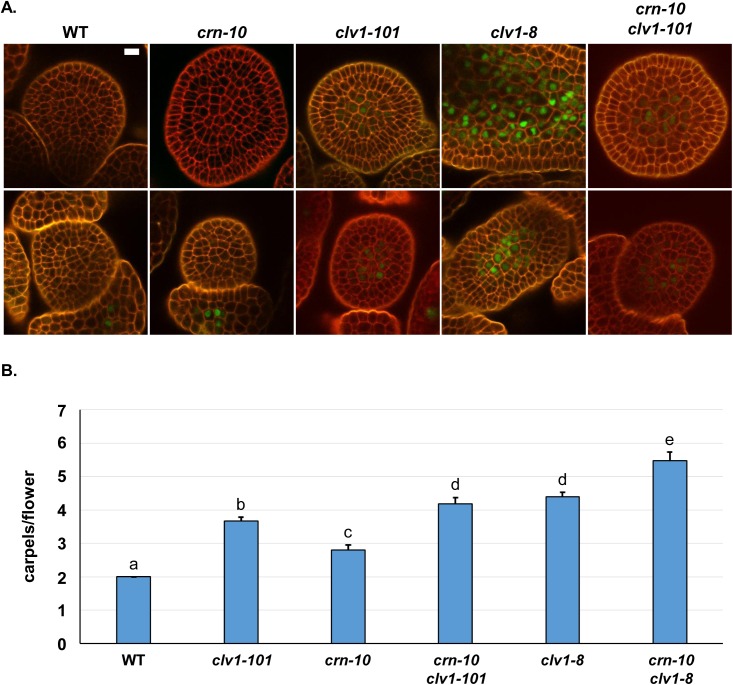
I previously assessed BAM3 reporter expression in strong alleles of clv1 [28], using the clv1-8 allele in Col-0 which contains a D295N mutation implicated in CLV3p binding [11]. Strong alleles of clv1 are weakly dominant negative and the molecular basis of this remains unclear [29]. Previous work has suggested strong clv1 mutant receptors could be interfering with CRN [16], or perhaps BAM1 and BAM2 [30]. To test these possibilities I first examined BAM3 expression in the clv1-101 null allele in Col-0. BAM3 reporter expression was considerably lower in L3-L5 cells of clv1-101 null SAMs compared to both clv3 null and clv1-8 strong alleles (Fig 3 and Fig 2A, S3 Fig). In some clv1-101 null plants, BAM3 expression was nearly undetectable in the SAM center. Expression of BAM3 in FMs was more reliably detected in clv1-101 null plants, but never approached the levels seen in clv3 null and clv1-8 strong alleles (Fig 3, Fig 2A). Unlike the low levels of BAM3 expression in clv1-101 null plants, BAM3 reporter expression in clv2, crn or rpk2 was not observed (Fig 2A, S3 Fig). Therefore, while BAM3 expression is de-repressed in the center of the SAM in both null and strong clv1 alleles, the level of de-repression of BAM3 is higher in the strong clv1-8 allele. This result implies that in clv1-101 null mutants, unknown receptor(s) signaling is still effective at repressing BAM3 expression. This reduced repression is not due to co-operative receptor function with CRN, as crn clv1-101 double null mutants contained levels of the BAM3 reporter equivalent to the clv1-101 null alone (Fig 3A).
Fig 3. BAM3 repression is weak in clv1-101 null mutants.
(A) pBAM3::Ypet-N7 signal (green) is not detectable in the center of WT or crn-10 SAMs (top row) or FMs (bottom row) but is strongly de-repressed in strong clv1-8 mutant plants which encode an interfering clv1 protein. In contrast, BAM3 expression is weakly de-repressed in clv1-101 null mutant plants. This weak de-repression is not due to redundancy with CRN, as crn-10 clv1-101 plants display similar low BAM3 levels as seen in clv1-101 alone. Imagining as in Fig 2. White bars, 10 μM. All subpanels are at the same image magnification. Experiment repeated twice. (B) Additive genetic interactions between crn-10 null mutants and clv1 null and strong alleles. Y-axis, mean number of carpels per flower for different genotypes. N = 100, experiment repeated twice. Error bars represent the calculated 95% C.I. Letters above bars indicate significance groups as determined by Tukey HSD test/ANOVA.
Since BAM receptors are ectopically expressed in clv1-101 null SAMs and partially compensate for clv1 they are capable at some level of signaling like CLV1 [28]. I therefore reasoned that ectopic BAM receptors might be dampening their own expression in the center of the SAM in clv1-101 null mutants. To test this I generated bam1 bam2 bam3 and clv1-101 bam1 bam2 bam3 quadruple mutant pBAM3::Ypet-N7 transgenic lines. Consistent with the previous observation that bam1 bam2 bam3 are dispensable for CLV1 function [28], BAM3 was fully repressed in the center of either SAMs or FMs in bam1 bam2 bam3 mutant plants. In contrast, the weak expression of the BAM3 reporter in the center of SAMs and FMs in clv1-101 null plants was greatly enhanced in clv1-101 bam1 bam2 bam3 quadruple mutants (Fig 4A). This result demonstrates that ectopically expressed BAM receptors in the center of clv1-101 null mutant SAMs signal to dampen their own expression. The level of BAM3 reporter de-repression in clv1-101 bam1 bam2 bam3 plants was comparable to, and occasionally stronger, than in clv1-8 alleles [28]. Previous work has suggested that clv1 missense receptors could interfere with BAM function [30] or perhaps CRN [16]. To test these hypotheses, I generated crn-10 clv1-8 double mutants and clv1-8 bam1 bam2 bam3 mutants. Consistent with the BAM3 imaging, crn-10 clv1-8 mutants were additive with respect to carpel number compared to single mutant plants (Fig 3B, S5 Fig), indicating that clv1-8 receptors do not interfere with CLV2/CRN signaling genetically. If clv1-8 receptors were interfering with the function of unknown receptors, and not BAM receptors, then clv1-8 bam1 bam2 bam3 should be as strong, or stronger, than quadruple null mutants. However, I found that clv1-8 bam1 bam2 bam3 were comparable to clv1-101 bam1 bam2 bam3 quadruple null mutants (Fig 4B). These data support the hypothesis that clv1-8, and presumably other strong missense clv1 receptors, function exclusively by interfering with the signaling of ectopic BAM receptors in the SAM. This result is consistent with the BAM3 reporter imaging demonstrating that CRN is dispensable for CLV1 signaling, and also implies that like CLV1, BAM receptors signal in vivo in a CLV2/CRN independent manner.
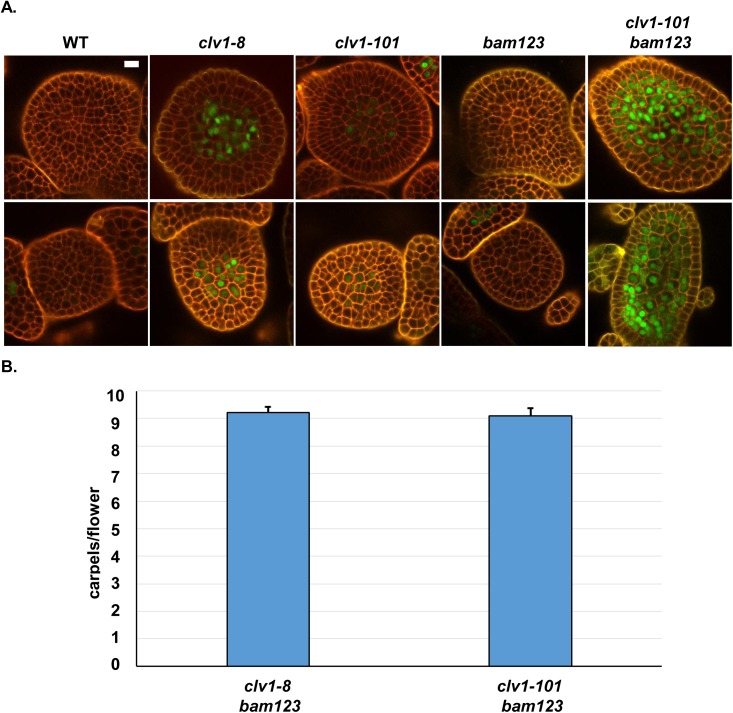
Fig 4. Ectopic BAM receptors feedback to dampen their own expression in the absence of CLV1.
(A) Ectopic BAM receptors dampen their own expression in clv1-101 null mutants. In WT or bam1 bam2 bam3 (bam123) triple mutants SAMs (top row), or FMs (bottom row), BAM3 (green) is effectively repressed by CLV3-CLV1 signaling in the meristem centers. In clv1-101 null mutant plants, BAM3 is partially repressed compared to strong clv1-8 mutant allele plants. Removal of all BAM function from clv1-101 null mutant plants (clv1-101 b123) results in de-repression of BAM3 to levels occasionally higher than clv1-8 plants, indicating that BAM receptor signaling in clv1-101 null mutants partially represses BAM target genes in the absence of CLV1. Imaging as in Fig 2. White bars, 10 μM. All subpanels are at the same image magnification. Experiment repeated 3 times. (B) clv1 strong alleles act via interfering with ectopic BAM receptors. Y-axis, mean number of carpels per flower for different genotypes. N = 80 flowers per genotype, experiment repeated three times. Error bars, represent calculated 95% C.I. Difference not significant at 95% as determined by T-test.
Functional independence of WUS-induced stem cell proliferation and BAM3 repression
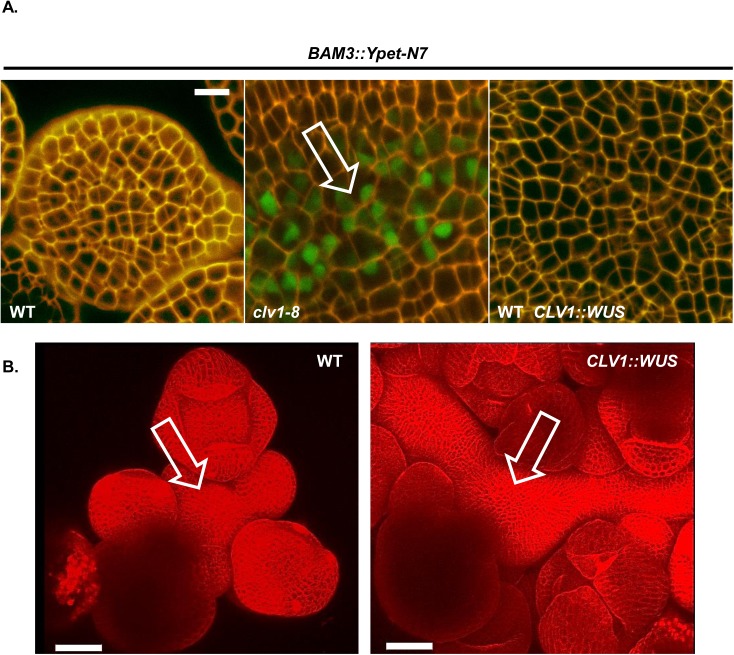
CLV1 signaling strongly represses BAM expression, but has a considerably weaker effect on WUS in physiological ranges of CLV3p [11, 28, 33, 34]. The current model in the field postulates that upregulation of WUS drives the excess cell proliferation in clv class meristems. I sought to test if WUS up-regulation was causally connected to BAM de-repression in the SAM of clv1 mutants. To do this I repeated the experiments in Schoof et al [31] and used the CLV1 promoter to express WUS in the SAM of Col-0 pBAM3::Ypet-N7 plants. In those experiments, ectopic expression of WUS using a two-component inducible system drove SAM enlargement and resulted in flowers with extra carpels, phenocopying clv1 mutants. Of the 36 T1 CLV1p::WUS lines generated in the Col pBAM3::Ypet-N7 background, only 11 plants displayed increases in SAM size and stem fasciation. Despite having enlarged SAMs reminiscent of clv1 SAMs (Fig 5B), no de-repression of BAM3 was observed in the SAM or FMs of any of the enlarged meristem plants examined (N = 10, Fig 5A). This indicates that WUS-induced over-proliferation of the stem cell niche is genetically separable from BAM3 transcriptional regulation in the CLV1 pathway.
Fig 5. CLV1/CLV3 regulation of BAM expression is separable from WUS- mediated stem cell proliferation.
(A) WUS-induced SAM proliferation is separable from BAM3 repression by CLV1. Wild type (WT) pBAM3::Ypet-N7 plants were imaged and compared to clv1-8 pBAM3::Ypet-N7 and WT pBAM3::Ypet-N7 plants transformed with pCLV1::WUS transgene. No ectopic BAM3 signal was seen in pCLV1::WUS lines (N = 10), despite the massive over-proliferation of SAM tissue in some lines (B). White bars, 10 μM. All subpanels are at the same image magnification. B) 3D surface reconstruction of image stack obtained in red channel (propidium iodide, cell wall stain). White bars, 50 μM. All subpanels are at the same image magnification.
Discussion
Despite being cloned nearly 20 years ago, we have little understanding about of how CLV1 signals in planta or its relationship to other proposed CLV3p receptors. Here I demonstrate, using BAM3 repression as a readout, that CLV1 signals to control at least some transcriptional outputs independent of CLV2/CRN and RPK2 but fully dependent on CLV3 (Fig 6). CLV2/CRN have no effect on BAM3 reporter expression by themselves, or in combination with CLV1, and genetically do not participate in BAM feedback compensation. As such, despite the different receptor mutants having similar qualitative stem cell defects and resistance to CLV3p, CLV1, RPK2 and CLV2/CRN are functionally separate and converge on distinct signaling outputs in vivo. This result implies that CLV2/CRN are dispensable for CLV3p mediated perception by CLV1, and that putative CLV2/CRN/CLV1 complexes seen in tobacco overexpression studies are dispensable for CLV1 signaling in vivo. This is consistent with previous data showing that CLV1 traffics from the plasma membrane to the lytic vacuole in response to CLV3p in a CLV2-independent manner [10], and consistent with additive genetic interactions with clv2, crn and clv1 [16]. As such, every readout for CLV1 function would suggest that CLV2/CRN are dispensable for CLV3p perception and signaling in vivo. Previously it was suggested that strong clv1 receptors could interfere with CLV2/CRN function in vivo [16], however I found that there is a significant enhancement of the strong clv1-8 allele in clv1-8 crn-10 double mutants. Genetic analysis and BAM3 repression analysis suggests that the strength of the clv1-8 mutant receptor can be accounted for solely by interfering with ectopic BAM receptors in SAM, supporting previous studies [30].
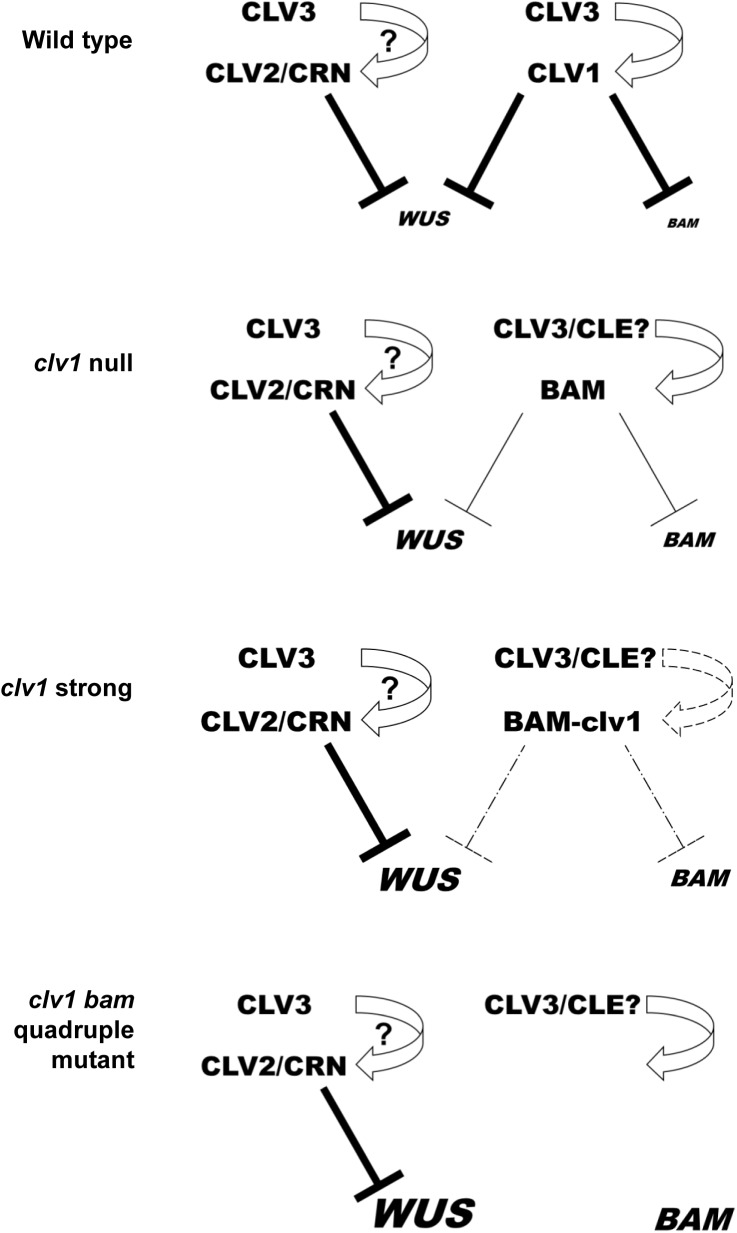
Fig 6. Model for CLV3-receptor signaling circuits in planta.
Model for CLV3p mediated regulation of WUS and BAM target genes in planta. CRN/CLV2 dimers do not participate in BAM regulation which is governed by CLV3-CLV1 signaling. In clv1 null mutants, ectopic BAM receptors partially replace CLV1 but also dampen their own expression. In clv1 strong alleles, mutant clv1 receptor interfere with BAM receptor signaling. In clv1 bam1 bam2 bam3 quadruple receptor mutant plants repression of BAM is lifted. The strength of repression is represented by the repression line thickness and target gene expression levels are depicted by font size. Question marks reflect the potential for direct binding CLV3p to CLV2/CRN dimers [23,19] and the existence of postulated CLV3p redundant CLE peptides based on observation that clv1 bam1 bam2 bam3 quadruple receptors mutants are stronger than clv3 null mutants. [28].
Despite their independence from CLV1, clv2 and crn mutants are resistant to ectopic CLE peptide-mediated SAM or RAM termination in several species of plants, and have not been identified in genetic screens for other peptide responses to date. This suggests that there is either a tight relationship between CLV2/CRN and CLV3/CLE ligand function, or that CLV2/CRN impact a developmental process which superficially resembles CLV3p function. Interestingly, while CLV2 and CRN mutants display resistance to CLV3/CLE peptide induced root termination [16, 40], they do not display conspicuous root growth or patterning defects in the absence of peptide ligand [16]. I previously demonstrated that mutants that do not function in the CLV3 pathway, but have a higher rates of SAM cell proliferation and an expanded SAM, display resistance to CLE peptide mediated SAM termination [28]. This suggests that CLE peptide resistance per se is insufficient to determine gene function in CLV3p signaling or perception. Based on this, it is formally possible that CLV2/CRN do not function in CLE mediated perception in vivo, as has been suggested by peptide binding assays [23]. The function of CLV2/CRN in CLE mediated signaling, if any, remains enigmatic, but current data demonstrates that CLV1 ligand perception and binding, stability, endomembrane trafficking, and signaling are all independent of CLV2/CRN in planta.
clv1 null mutants are phenotypically weak, despite having ectopic BAM expression in the SAM center. However, like CLV1, BAM receptors also repress BAM expression in the center of the SAM (Fig 6). As such, clv1 null mutant SAMs contain low levels of BAM expression, potentially explaining why ectopic BAM expression is not sufficient to fully compensate for clv1. In the organizing center, CLV1 is proposed to also repress WUS expression. Despite this, CLV1 and WUS expression is largely co-incident and expression of either CLV1 or CRN/CLV2 in WUS-expressing cells is sufficient to account for all functions in stem cell regulation. WUS induced SAM over-proliferation can be uncoupled from CLV1-mediated BAM repression. As such, clv1 and clv3 SAMs are functionally different from WUS-induced clv-like SAMs at some level. WUS itself has been proposed to repress CLV1 [33], but the significance of this is unclear due to the co-incident expression patterns of both genes, the lack of de-repression of BAM3 in enlarged SAMs ectopically expressing WUS, and the fact that uncoupling CLV1 expression from the native CLV1 promoter in SAM cells has no phenotypic consequence other than to complement clv1 null mutants [10, 28–30, 39]. In addition, CLV1 mediated repression of WUS is quantitatively different from that for BAM receptors. In the center of wild type SAMs BAM gene expression is nearly undetectable and becomes robustly detectable in clv1 or clv3 mutants. In contrast, WUS is robustly detected in wild type SAMs [28, 31]. Overexpression of CLV3 represses WUS, however plants expressing up to 300 fold more CLV3 are wild type in appearance [34]. Thus, at endogenous levels CLV3p strongly suppresses BAM expression in a CLV1-dependent manner, but has less of an effect on WUS, which requires considerably higher and potentially non-physiological levels of CLV3p for full repression. Understanding the regulation of BAM expression by CLV1 could lead to new insights into this signaling pathway.
Materials and methods
Plant growth, genetics and selection
Plant growth and transgenic plant selection was performed as described in [28]. All clv1, bam1, bam2, bam3 and clv2 alleles are in the isogenic Col-0 background and have been characterized previously. Genotyping of plants from crosses were performed using appropriate primers selecting for mutant alleles or T-DNA insertions [28, 35]. Carpel counts were performed as described with the exception of comparisons using crn-10 or clv2. In these lines early termination of the SAM and the cessation of flower production was observed after 5 flowers on average as noted before for clv2 mutations in the Col-0 background [35]. Therefore, I counted flowers 6 and beyond for all comparisons using clv2 or crn-10 alleles for all genotypes in those experiments. The transient termination phenotype of crn-10 was not altered by mutations in clv1, clv3 or in bam1 bam2 bam3 triple null mutants. In bam1 bam2 bam3 clv1 plants stems elongation is highly distorted as is SAM production as described in [28], making inferences about floral primordia order difficult. As such, all flowers were counted for comparisons between quadruple mutants.
Confocal imaging
Confocal imaging was performed using an inverted Zeiss 710 confocal. Briefly, an inverter adaptor (LSM Tech, Etters, PA, USA) was used to allow upright imaging of shoot meristems when attached to the Zeiss 710. Details on the configuration of the inverter are available on request and will be published elsewhere. Meristem staging, dissection and mounting were performed as described in [28], with each presented photo being a mean of eight scan cycles. For each experiment a minimum of 6 meristems were imaged and all imaging experiments were repeated with different plant populations two to four times. Imaging settings for the Ypet channel were kept constant across all experiments, but the gain on the red channel for propidium iodide was altered to account for staining differences as necessary. Image settings were calibrated to capture the dynamic linear range in most plants. At these image settings YFP signal in clv1 bam1 bam2 bam3 quadruple mutant SAMs was occasionally saturating in some nuclei (S2 Fig) but no signal was detectable in wild type, clv2, crn or bam1 bam2 bam3 SAM at these settings (S2 Fig, Figs 2–5).
Generation of binary vectors
The CRN promoter binary vectors used were described in [17]. For the CLV2 promoter binary, 1250 bp and 588 bp of the 5' and 3' promoter and UTR regions were amplified from Col-0 and fused together using recombinant PCR to introduce a unique BamH1 site and cloned into pBJ shuttle vector. A Gateway cloning cassette was inserted into the BamH1 site and the entire promoter cassette was transferred into the pMOA33 binary vector as a Not1 fragment [41]. Col-0 CLV2 CDS was amplified using primers that allowed cloning into the pENTRD (Invitrogen) vector and fused with an in frame MYC epitope to the C-terminus. This was then recombined into either pMOA33 CLV2p or pMOA33 WUSp. The pMOA33 WUSp vector was described as in [28]. For the generation of pMOA33 CLV1p::WUS, a pENTRD WUS CDS clone was recombined into the pMOA33 CLV1p vector used previously [10, 28].
Creation of the crn-10 and rpk2-cr null allele mutants
The pCUT vector system was used to generate the crn-10 allele and rpk2-cr allele used here [36]. Briefly, this vector series co-expresses nuclear targeted Cas9 from the UBQUITIN10 promoter and gRNAs from the U6 promoter. A 20 base pair gRNA target site (bold), including upstream G and downstream PAM (underlined) was selected gaagcaaagaagaagaagaaatgg near the initiator methionine codon in the CRN genomic signal sequence encoding region. This target site was used to generate a gRNA that was cloned into pCUT3 as described in [36]. Kanamycin resistant plants were selected in the T1. In a couple of T1 plants, branches were observed with flowers which all contained elevated levels of carpels relative to wildtype. Seeds were collected specifically from one of these branches and sequencing in the next generation revealed that these branches arose as somatic bi-allelic sectors containing either an A or T insertion (bold) at the same location downstream of the initiator ATG (uppercase) (ATGaagcaaagaagaagaagTaaatgg) leading to equivalent truncated CRN proteins with only 7 amino acids remaining in the signal sequence of CRN (MKQRRRRKWMstop). Plants with the homozygous T insertion event were selected as this provides resistance to Hph1 digestion using the dCAPs primers CRN-10 F gtagaagcagcaatgaagcaaagaagaaggtg and CRN-10 R gttgaagttgtggataagtg [42], and segregated away from the pCUT3 transgene. Complementation analysis was performed using vectors described in [17]. For rpk2-cr mutant creation, a tandem array of two different U6 promoter gRNA cassettes targeting the RPK2 CDS were gene synthesized by Invitrogen and cloned into pCUT3 as described in [36]. The RPK2 gRNA target sites chosen were aagattactgctcctggtttgg and tcatggctcttaacattagtgg, with the PAM sequence in bold. This vector was transformed directly into the Col-0 pBAM3::Ypet-N7 line. In the T2 generation from a select T1 line, multiple plants displaying an rpk2 phenotype were identified based on male sterility and extra carpels [18, 43]. Imaging was performed on 10 plants with an rpk2 phenotype. One line lacking the pCUT3 vector was identified by PCR and backcrossed to Col-0 to maintain as a heterozygous owing to the male sterility of rpk2 mutants [43]. This line was then imaged again in the next generation to confirm BAM3 expression patterns. This line, termed rpk2-cr, contains a +A insertion between nucleotides 229 and 230 and an additional +T insertion between nucleotides 279 and 290 in the RPK2 CDS. This results in the production of an RPK2 protein that contains a stop codon directly after amino acid 76 (serine76) downstream of the signal sequence resulting in the deletion of all LRR repeats and downstream transmembrane, juxtamembrane and kinase domains [43].
Supporting information
(A) crn-10 is fully complemented by CRN, kinase enzymatically dead CRN (CRNK146E) and serine 156 substitution mutants (CRNS156A and CRNS156D). Full complementation was designated as every flower containing two carpels as in wild plants. (B) CRN is expressed in the SAM center and center of developing FMs. crn-10 mutant plant complemented with pCRN::CRN-2xGFP transgene. Images of the meristem center, determined by appearance of chloroplasts in L3 [10]. CRN-2xGFP signal appears at presumptive PM and signal was not seen in tonoplast or perinuclear ER. CRN-2xGFP fully complemented crn-10 in all lines examined (n = 20) like 2x mCherry, however, signal in CRN-2xmCherry plants was below detection limit in the SAM most lines likely owing to lower intrinsic brightness of mCherry.
(TIF)
At the calibrated imaging settings (3.5% 514 nm laser power, pinhole 121 μM, scans averaged) in this study saturating levels of nuclear Ypet signal are seen in some clv1 bam1 bam2 bam3 nuclei (yellow arrow) but signal is undetectable in wild type plants at same setting.
(TIF)
Detail from Fig 4. Note that images were brightened and enlarged equally in PowerPoint relative to Fig 4 in order to magnify detail and illustrate the intensity differences between pBAM3::Ypet-N7 signal (green nuclei) in clv1-8 and clv1-101.
(TIF)
Imaging of bam1 bam2 bam3 pBAM3::Ypet-N7 (green) plants as an example of genotypes in which BAM3 signal is below detection in the SAM center (left, white bars = 10 μM), but still expressed in developing vasculature/phloem below the SAM proper (right, green arrows, white bars = 20 μM).
(TIF)
Distribution of flowers with specific carpel numbers. clv1-8, blue bars; clv1-8 crn-10 double, orange bars. Data from Fig 3B. N = 100, experiment repeated twice. Y-axis, total number of flowers. X-axis, carpel number per flower.
(TIF)
Acknowledgments
I wish to thank Tony Perdue for assistance with the Inverter configuring in the UNC Confocal Imaging Facility, James Garzoni and members of the UNC Greenhouse Facility for providing greenhouse support, Susan Whitfield for help with illustration software, and Brenda Peterson and Jamie Winshell for help maintaining common lab stocks, and members of the Nimchuk lab for their comments on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported grants to ZLN from the National Science Foundation (IOS-1455607 and IOS-1546837; https://www.nsf.gov/bio/ios/about.jsp) and the National Institutes of Health (R35GM119614-01; https://www.nigms.nih.gov/research/mechanisms/MIRA/pages/default.aspx). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7(4):e1002046 PubMed Central PMCID: PMCPMC3085482. 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma X, Xu G, He P, Shan L. SERKing Coreceptors for Receptors. Trends Plant Sci. 2016. [DOI] [PubMed] [Google Scholar]
- 3.Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends Plant Sci. 2006;11(10):492–8. 10.1016/j.tplants.2006.08.005 [DOI] [PubMed] [Google Scholar]
- 4.Soyars CL, James SR, Nimchuk ZL. Ready, aim, shoot: stem cell regulation of the shoot apical meristem. Current opinion in plant biology. 2016;29:163–8. 10.1016/j.pbi.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 5.Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121(7):2057–67. [Google Scholar]
- 6.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283(5409):1911–4. [DOI] [PubMed] [Google Scholar]
- 7.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. The Plant cell. 2002;14(5):969–77. 10.1105/tpc.002196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313(5788):845–8. 10.1126/science.1128439 [DOI] [PubMed] [Google Scholar]
- 9.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5(8):578–80. 10.1038/nchembio.182 [DOI] [PubMed] [Google Scholar]
- 10.Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011;21(5):345–52. PubMed Central PMCID: PMC3072602. 10.1016/j.cub.2011.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89(4):575–85. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319(5861):294 10.1126/science.1150083 [DOI] [PubMed] [Google Scholar]
- 13.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289(5479):617–9. [DOI] [PubMed] [Google Scholar]
- 14.Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant physiology. 2010;152(1):166–76. 10.1104/pp.109.149930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. The Plant cell. 1999;11(10):1925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant cell. 2008;20(4):934–46. 10.1105/tpc.107.057547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. The Plant cell. 2011;23(3):851–4. PubMed Central PMCID: PMC3082267. 10.1105/tpc.110.075622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137(22):3911–20. 10.1242/dev.048199 [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Clark SE. Membrane distributions of two ligand-binding receptor complexes in the CLAVATA pathway. Plant Signal Behav. 2010;5(11):1442–5. PubMed Central PMCID: PMCPMC3115250. 10.1111/j.1365-313X.2010.04295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. The Plant journal for cell and molecular biology. 2010;63(6):889–900. PubMed Central PMCID: PMC2974754. 10.1111/j.1365-313X.2010.04295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somssich M, Ma Q, Weidtkamp-Peters S, Stahl Y, Felekyan S, Bleckmann A, et al. Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci Signal. 2015;8(388):ra76 10.1126/scisignal.aab0598 [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Wan Y, Lin J. Multiple receptor complexes assembled for transmitting CLV3 signaling in Arabidopsis. Plant Signal Behav. 2010;5(3):300–2. PubMed Central PMCID: PMCPMC2881284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinohara H, Matsubayashi Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. The Plant journal for cell and molecular biology. 2015;82(2):328–36. 10.1111/tpj.12817 [DOI] [PubMed] [Google Scholar]
- 24.Morita J, Kato K, Nakane T, Kondo Y, Fukuda H, Nishimasu H, et al. Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide. Nature communications. 2016;7:12383 PubMed Central PMCID: PMCPMC4979064. 10.1038/ncomms12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Lin X, Han Z, Qu LJ, Chai J. Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 2016;26(5):543–55. PubMed Central PMCID: PMCPMC4856767. 10.1038/cr.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, et al. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 2015;517(7534):377–80. PubMed Central PMCID: PMC4297503. 10.1038/nature13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. The Plant journal for cell and molecular biology. 2012;70(5):845–54. 10.1111/j.1365-313X.2012.04934.x [DOI] [PubMed] [Google Scholar]
- 28.Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development. 2015;142(6):1043–9. PubMed Central PMCID: PMC4360179. 10.1242/dev.119677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dievart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. The Plant cell. 2003;15(5):1198–211. 10.1105/tpc.010504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeYoung BJ, Clark SE. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics. 2008;180(2):895–904. 10.1534/genetics.108.091108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–44. [DOI] [PubMed] [Google Scholar]
- 32.Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–15. [DOI] [PubMed] [Google Scholar]
- 33.Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, et al. Transcriptional control of a plant stem cell niche. Developmental cell. 2010;18(5):849–61. 10.1016/j.devcel.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 34.Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. The Plant cell. 2006;18(5):1188–98. 10.1105/tpc.105.040444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant physiology. 2008;147(2):503–17. PubMed Central PMCID: PMCPMC2409048. 10.1104/pp.108.119487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson BA, Haak DC, Nishimura MT, Teixeira PJ, James SR, Dangl JL, et al. Genome-Wide Assessment of Efficiency and Specificity in CRISPR/Cas9 Mediated Multiple Site Targeting in Arabidopsis. PloS one. 2016;11(9):e0162169 PubMed Central PMCID: PMCPMC5021288. 10.1371/journal.pone.0162169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somssich M, Bleckmann A, Simon R. Shared and distinct functions of the pseudokinase CORYNE (CRN) in shoot and root stem cell maintenance of Arabidopsis. Journal of experimental botany. 2016;67(16):4901–15. PubMed Central PMCID: PMCPMC4983110. 10.1093/jxb/erw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Depuydt S, Rodriguez-Villalon A, Santuari L, Wyser-Rmili C, Ragni L, Hardtke CS. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(17):7074–9. PubMed Central PMCID: PMC3637694. 10.1073/pnas.1222314110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. The Plant journal for cell and molecular biology. 2006;45(1):1–16. 10.1111/j.1365-313X.2005.02592.x [DOI] [PubMed] [Google Scholar]
- 40.Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. The Plant cell. 2005;17(9):2542–53. 10.1105/tpc.105.034009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrell PJ, Conner AJ. Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. Biotechniques. 2006;41(6):708–10. [DOI] [PubMed] [Google Scholar]
- 42.Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. The Plant journal for cell and molecular biology. 1998;14(3):387–92. [DOI] [PubMed] [Google Scholar]
- 43.Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, et al. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. The Plant journal for cell and molecular biology. 2007;50(5):751–66. 10.1111/j.1365-313X.2007.03083.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) crn-10 is fully complemented by CRN, kinase enzymatically dead CRN (CRNK146E) and serine 156 substitution mutants (CRNS156A and CRNS156D). Full complementation was designated as every flower containing two carpels as in wild plants. (B) CRN is expressed in the SAM center and center of developing FMs. crn-10 mutant plant complemented with pCRN::CRN-2xGFP transgene. Images of the meristem center, determined by appearance of chloroplasts in L3 [10]. CRN-2xGFP signal appears at presumptive PM and signal was not seen in tonoplast or perinuclear ER. CRN-2xGFP fully complemented crn-10 in all lines examined (n = 20) like 2x mCherry, however, signal in CRN-2xmCherry plants was below detection limit in the SAM most lines likely owing to lower intrinsic brightness of mCherry.
(TIF)
At the calibrated imaging settings (3.5% 514 nm laser power, pinhole 121 μM, scans averaged) in this study saturating levels of nuclear Ypet signal are seen in some clv1 bam1 bam2 bam3 nuclei (yellow arrow) but signal is undetectable in wild type plants at same setting.
(TIF)
Detail from Fig 4. Note that images were brightened and enlarged equally in PowerPoint relative to Fig 4 in order to magnify detail and illustrate the intensity differences between pBAM3::Ypet-N7 signal (green nuclei) in clv1-8 and clv1-101.
(TIF)
Imaging of bam1 bam2 bam3 pBAM3::Ypet-N7 (green) plants as an example of genotypes in which BAM3 signal is below detection in the SAM center (left, white bars = 10 μM), but still expressed in developing vasculature/phloem below the SAM proper (right, green arrows, white bars = 20 μM).
(TIF)
Distribution of flowers with specific carpel numbers. clv1-8, blue bars; clv1-8 crn-10 double, orange bars. Data from Fig 3B. N = 100, experiment repeated twice. Y-axis, total number of flowers. X-axis, carpel number per flower.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.