Abstract
TBX5 is a member of the T-box transcription factor family and is primarily known for its role in cardiac and forelimb development. Human patients with dominant mutations in TBX5 are characterized by Holt–Oram syndrome, and show defects of the cardiac septa, cardiac conduction system, and the anterior forelimb. The range of cardiac defects associated with TBX5 mutations in humans suggests multiple roles for the transcription factor in cardiac development and function. Animal models demonstrate similar defects and have provided a useful platform for investigating the roles of TBX5 during embryonic development. During early cardiac development, TBX5 appears to act primarily as a transcriptional activator of genes associated with cardiomyocyte maturation and upstream of morphological signals for septation. During later cardiac development, TBX5 is required for patterning of the cardiac conduction system and maintenance of mature cardiomyocyte function. A comprehensive understanding of the integral roles of TBX5 throughout cardiac development and adult life will be critical for understanding human cardiac morphology and function.
1. TBX5, A MEMBER OF THE T-BOX FAMILY OF TRANSCRIPTION FACTORS
Ever since the association of TBX5 mutations with Holt–Oram syndrome (Basson et al., 1997; Li et al., 1997), the TBX5 gene has been a source of study, particularly with respect to cardiac and limb development. This review covers progress since those initial reports, published two decades ago.
The T-box family of transcription factors share a common T-box DNA-binding domain (Bollag et al., 1994) and are named after the founding member, T, which encodes the transcription factor Brachyury (Herrmann, Labeit, Poustka, King, & Lehrach, 1990; Pflugfelder, Roth, & Poeck, 1992). The T-box domain is approximately 170–200 amino acids in length (Agulnik, Bollag, & Silver, 1995; Agulnik et al., 1996; Bollag et al., 1994; Papaioannou, 2014), binds DNA directly (Pflugfelder et al., 1992), and is required for transcriptional activity (Kispert, Koschorz, & Herrmann, 1995). All members of the T-box gene family appear to bind the DNA consensus motif AGGTGHBA (Conlon, Fairclough, Price, Casey, & Smith, 2001; He, Kong, Ma, & Pu, 2011; Jolma et al., 2013; Kispert et al., 1995; Mathelier et al., 2016; Waldron et al., 2016; Wilson & Conlon, 2002). While some members, such as T and EOMES, are only able to bind palindromic sequences as dimers (Conlon et al., 2001; Ghosh et al., 2001; Kispert et al., 1995; Muller & Herrmann, 1997), others, including TBX2, TBX3, and TBX5, can bind individual motifs as monomers (Bruneau et al., 2001; Carreira, Dexter, Yavuzer, Easty, & Goding, 1998; Ghosh et al., 2001; He, Wen, Campbell, Wu, & Rao, 1999).
In the human genome, there are 17 coding genes that fall within five subfamilies of the T-box family (Agulnik et al., 1996; Papaioannou & Silver, 1998), most of which show sequence similarity between vertebrates and invertebrates (Agulnik et al., 1995, 1996; Papaioannou & Silver, 1998; Pflugfelder et al., 1992; Ruvinsky, Silver, & Gibson-Brown, 2000; Wilson & Conlon, 2002). The human genes TBX2, TBX3, TBX4, and TBX5 belong to one subfamily, and are homologous to the Drosophila gene omb (Agulnik et al., 1996). These four genes likely originated through tandem duplication of the ancestral gene through unequal crossover to form the ancestral TBX2/3 and TBX4/5 (Agulnik et al., 1996; Ruvinsky et al., 2000), followed by cluster duplication prior to the divergence of bony fish and tetrapods approximately 400 million years ago (Agulnik et al., 1996; Ruvinsky & Silver, 1997). This second duplication event generated one cluster containing TBX2 and TBX4 and a second containing TBX3 and TBX5, which are located on human chromosomes 17 and 12, respectively (Agulnik et al., 1996; Ruvinsky & Silver, 1997).
The T-box family gene TBX5 encodes a 518-amino acid protein with a 180-amino acid T-box domain located between amino acid residues 56 and 236 (Basson et al., 1997; Li et al., 1997). TBX5 contains two nuclear localization sequences (NLS): NLS1 located within the T-box domain (amino acids 78–90), and NLS2 located outside the T-box domain on the C-terminal end (325–340) (Collavoli et al., 2003; Zaragoza et al., 2004). While each NLS is sufficient to drive nuclear localization, they appear to work cooperatively (Collavoli et al., 2003). As a transcription factor, TBX5 also contains a transactivation domain located from amino acids 339–379 with functional requirement of amino acids 349–351 (Zaragoza et al., 2004). The sequence of amino acids 152–160 has been proposed to act as a nuclear export signal through the CRM1 export pathway by which TBX5 subcellular localization can be regulated through binding with the PDLIM7 protein (Camarata et al., 2006, 2010; Kulisz & Simon, 2008); however, this theory remains controversial as the crystal structure of TBX5 suggests this domain would be located on the inside of the protein and inaccessible without major protein rearrangements (Stirnimann, Ptchelkine, Grimm, & Muller, 2010). Aside from these domains, TBX5 also contains several other protein–protein interaction domains, discussed in detail later.
In addition to the best-described isoform, sometimes referred to as TBX5a (Georges, Nemer, Morin, Lefebvre, & Nemer, 2008), there have been four additional isoforms described in the literature (Georges et al., 2008; Yamak et al., 2015). These isoforms are derived from alternative splicing within the TBX5 locus and result in proteins of varying lengths, including either an N-terminal or a C-terminal truncated form as well as forms with varying C-terminal modifications (Georges et al., 2008; Yamak et al., 2015). Interestingly, all described isoforms retain the T-box domain (Yamak et al., 2015). The alternative isoforms show differential expression and activity, including antagonism of TBX5a, and further investigation into these isoforms will be important for understanding the roles of TBX5 in human development and health (Yamak et al., 2015).
2. TBX5 EXPRESSION
The broad spatiotemporal expression domains of TBX5 during development appear to be generally conserved throughout vertebrate evolution and consist of the heart, forelimb, and retina (Bruneau et al., 1999; Chapman et al., 1996; Gibson-Brown et al., 1996; Gibson-Brown, Agulnik, Silver, & Papaioannou, 1998; Horb & Thomsen, 1999; Showell, Christine, Mandel, & Conlon, 2006; Takabatake, Takabatake, & Takeshima, 2000); however, some tissue-specific expression differences occur between species. First, we will examine the expression domains of TBX5 in the common tetrapod models as well as humans, with the greatest emphasis on subcardiac domains, and then we will discuss the regulation of TBX5 gene expression based on evidence from mouse and human studies.
2.1 TBX5 Expression Domains in the Embryonic and Adult Heart
The cardiac expression patterns of TBX5 in human, mouse, chick, and frog are very similar. In human hearts, TBX5 is expressed in the epicardium, myocardium, and endocardium of embryonic and adult hearts (Hatcher, Goldstein, Mah, Delia, & Basson, 2000). Human TBX5 is expressed in the free walls and septa of all four chambers during development; however, atrial expression is much greater than ventricular, as seen in mouse and chicken (Hatcher et al., 2000). TBX5 is expressed in the embryonic atrioventricular (AV) node, and is excluded from the AV valves (Hatcher et al., 2000). TBX5 is expressed throughout the epicardium, but not in the endocardium of the left ventricle (Hatcher et al., 2000). Much like in animal models, TBX5 expression is absent from the developing outflow tract of the heart (Hatcher et al., 2000). In human adults, TBX5 expression is highest in the atrial appendages, followed by the lungs, left ventricle, and esophagus (GTEx Consortium, 2013; Mele et al., 2015).
In mice, Tbx5 becomes abundantly expressed around E8.0 throughout the cardiac crescent (Bruneau et al., 1999), and this expression becomes restricted to the posterior portion of the forming heart tube, corresponding to the sinus venosa and future atria, between E8.25 and E8.5 (Bruneau et al., 1999; Chapman et al., 1996). At E9.0, Tbx5 expression expands throughout the future left ventricle (Bruneau et al., 1999). Additionally, atrial expression of Tbx5 is stronger than in the left ventricle, and in the ventricular free wall it is higher than in the trabeculae (Bruneau et al., 1999). Tbx5 is also expressed in the right ventricular trabeculae, but not free wall (Bruneau et al., 1999). Expression of Tbx5 in the left ventricle and atria is maintained throughout embryonic development (Chapman et al., 1996). During maturation of the mouse heart, like humans, Tbx5 is expressed in and colocalizes with markers of the cardiac conduction system, including the AV bundle and bundle branch (Moskowitz et al., 2004). Genetic inducible fate mapping demonstrated that left ventricular Tbx5 expression arises from the first heart field, specified prior to morphogenesis of the heart, whereas atrial and atrial septum Tbx5 expression arises from Mef2cAHF+ second heart field domain, suggesting potential independent roles of Tbx5 in the first and second heart fields during development (Devine, Wythe, George, Koshiba-Takeuchi, & Bruneau, 2014).
In cardiac development of the chick, Tbx5 expression is first detected throughout the entire bilateral cardiac primordia (Bruneau et al., 1999). This expression is maintained following fusion of the heart tube along the entire rostrocaudal length (Gibson-Brown et al., 1998), but adopts an anterior-to-posterior gradient shortly after (Bruneau et al., 1999). Although there appears to be a gradient to the expression, Tbx5 is expressed throughout the whole heart during cardiac looping (Gibson-Brown et al., 1998). After looping is complete, Tbx5 expression remains in the entire heart except the outflow tract, and this is the only major difference between mouse and chick heart expression. However, as cardiac development and maturation proceed, expression of Tbx5 is restricted from the right ventricle, similar to the expression pattern in the mouse embryo (Bruneau et al., 1999).
In Xenopus, the earliest expression domains of tbx5 are in two lateral stripes, corresponding to the cardiac primordia, on either side of the embryo and continue to be expressed in the migrating precardiac mesoderm (Horb & Thomsen, 1999; Showell et al., 2006). Similar to chick cardiogenesis, after fusion at the midline and formation of the early heart tube, tbx5 is expressed throughout most of the cardiac tissue including the sinus venosus/inflow tract of the heart (Horb & Thomsen, 1999; Showell et al., 2006). As development continues, tbx5 expression is lost from the most anterior structure, the bulbus cordis, and is strongly detected in the posterior regions of the heart, while it is also maintained in the ventricle (Horb & Thomsen, 1999; Showell et al., 2006). tbx5 is expressed robustly in both the endocardium and myocardium and is detected in the epicardium (Horb & Thomsen, 1999).
2.2 Extracardiac TBX5 Expression
In addition to the cardiac expression domains, TBX5 is expressed in many noncardiac tissues. Perhaps the best-studied expression domain outside of the heart is that of the developing forelimb. Tbx5 is expressed in the lateral plate mesoderm giving rise to the forelimb starting at E8.8 of mouse embryonic development (Gibson-Brown et al., 1996) and is robustly expressed in the forelimb bud at E9.5 (Chapman et al., 1996; Gibson-Brown et al., 1996). Expression throughout the developing limb is maintained until E11.5, when it then becomes restricted to the proximal portion of the forelimb (Gibson-Brown et al., 1996). Tbx5 is also expressed in the periochondrium of the forelimb at E13.5 (Gibson-Brown et al., 1996).
Outside of the heart and forelimb, TBX5 expression has been reported in several notable domains during development. The earliest reported expression domain for Tbx5 during mouse development is in the allantois at E7.5 where it is transiently coexpressed with Tbx4 (Chapman et al., 1996). Expression of Tbx5 in the allantois has been suggested to be a mammalian-specific trait, as transcription of Tbx5 is never observed in the allantois of chick embryos (Gibson-Brown et al., 1998). Tbx5 is also expressed in the optic vesicle and the neural retina of the developing eye in mouse, chick, and Xenopus where it is coexpressed with the other members of the omb family of T-box genes (Chapman et al., 1996; Gibson-Brown et al., 1998; Horb & Thomsen, 1999; Showell et al., 2006). Additionally, Tbx5 is expressed in the mesenchyme of the mandibular arch, the trachea, and the lung, as well as the body wall of the thorax (Chapman et al., 1996; Gibson-Brown et al., 1998). In both mouse and chicken, expression of Tbx5 has been reported in the genital papilla (Chapman et al., 1996; Gibson-Brown et al., 1998). Specific to avian development, Tbx5 expression is observed in the notochord during midembryonic development (Gibson-Brown et al., 1998).
2.3 Transcriptional Regulation of TBX5
The mechanisms governing spatiotemporal regulation of TBX5 have begun to be addressed by defining cis-regulatory elements driving TBX5 expression in the embryo and adult. Preliminary investigations have identified several elements that drive distinct spatial domains during development; however, this area is ripe for future investigations. Tiling experiments have identified three enhancers associated with in vivo expression of Tbx5 in the mammalian heart (Minguillon et al., 2012; Smemo et al., 2012). The first enhancer, corresponding to hg19 chr12:114,463,712–114,464,080, drives expression of a reporter construct in both ventricular and atrial myocardium of E11.5 mouse hearts (Smemo et al., 2012). The second enhancer, hg19 chr12:114,701,207–114,704,691, drives expression in the posterior portion of the heart, including the ventricles, interventricular septum, and AV canal (Smemo et al., 2012). Additionally, this second enhancer contains a low-frequency SNP that abrogates the enhancer’s ability to drive expression (Smemo et al., 2012). While this SNP was predicted to disrupt a TAL1 binding site, Tal1 is not expressed in the myocardium, suggesting other members of the basic helix–loop–helix E-box-binding transcription factors may be driving expression of this enhancer (Smemo et al., 2012). The third Tbx5 enhancer, hg19 chr12:114,853,271–114,858,238, is sufficient to drive expression in the ventricles, interventricular septum, and AV canal as well as the atria (Smemo et al., 2012).
In addition to the identified cardiac enhancers, there have been two additional enhancers identified that regulate limb expression. The first is located within intron 2 of Tbx5 and drives expression within the lateral plate mesoderm of the forelimb, but not the heart (Minguillon et al., 2012). This forelimb enhancer is regulated in part through Hox4/5 genes, expressed in the region of the lateral plate mesoderm that gives rise to the forelimb, and has been proposed as the mechanism by which forelimb Tbx5 expression is positionally defined along the anterior–posterior body axis (Minguillon et al., 2012). The second forelimb enhancer identified is known as CNS12 and is located approximately 120 kbp downstream of the Tbx5 coding region (Adachi, Robinson, Goolsbee, & Shubin, 2016). The CNS12 enhancer drives expression in the lateral plate mesoderm and is sufficient to drive Tbx5 expression for forelimb formation (Adachi et al., 2016).
Taken together, Tbx5 expression in the developing heart and forelimb appears to be driven by distinct cis-regulatory elements, although the factors and transcriptional complexes that control the expression of these enhancers have yet to be uncovered.
Recent insight into the regulation of Tbx5 expression has come from investigations of the local three-dimensional architecture of the Tbx5 locus and its neighboring genes: Rbm19, Tbx3, and Med13l (Jin et al., 2013; van Weerd et al., 2014). Circular chromosome conformation capture with sequencing (4C-seq) data from the viewpoints of the Tbx3 and Tbx5 promoters, and the CTCF binding site between the two loci, suggests that the loci are in contact and that putative cis-regulatory elements for each gene are located almost exclusively within their own loci with CTCF sites acting as a regulatory barrier (Fig. 1) (van Weerd et al., 2014). Additionally, 4C-seq from the viewpoint of Rbm19, the nearest gene 3′ of Tbx5, suggests partially overlapping regulatory elements (van Weerd et al., 2014). Together, this suggests that most of the cis-regulatory information for Tbx5 expression is located in the 375 kbp region between the CTCF sites demarcating the boundaries of the Tbx3/Tbx5 and the Tbx5/Rbm19 loci (Fig. 1) (van Weerd et al., 2014).
Fig. 1.
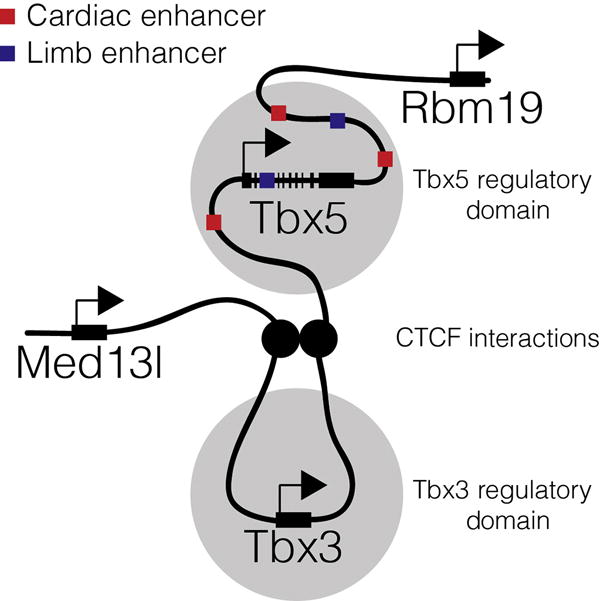
Representation of Tbx5 regulatory domain. Chromatin capture techniques suggest interactions between the promoter of Tbx5 and cis-regulatory elements are limited to a 375-kbp region demarcated by functional CTCF sites located between Rbm19 and Tbx5 and Tbx5 and Tbx3 (van Weerd et al., 2014).
In addition to transcriptional regulation, Tbx5 has also been shown to be regulated through microRNA-dependent mechanisms (Wang et al., 2014). In a screen of candidate human microRNAs, MiR-10a and MiR-10b were shown to bind to the 3′-UTR of the Tbx5 and inhibit its translation (Wang et al., 2014). More recently, it has been shown that regulation through these two microRNAs may play a role in adult conduction defects and pathological remodeling in disease-state hearts (Torrado et al., 2015).
3. TBX5 HAPLOINSUFFICIENCY: HOLT–ORAM SYNDROME
Holt–Oram syndrome is an autosomal dominant disorder caused primarily by dominant mutations in TBX5. Holt–Oram syndrome is a clinical diagnosis that includes completely penetrant, variably expressed upper-limb malformations including preaxial radial ray anomalies and congenital heart defects, typically septal and/or conduction defects (Holt & Oram, 1960). Although sometimes difficult to detect, upper-limb malformations are fully penetrant, while structural cardiac defects occur in 76% of patients with Holt–Oram syndrome (Basson et al., 1994, 1999; Holt & Oram, 1960; Newbury-Ecob, Leanage, Raeburn, & Young, 1996). Holt–Oram syndrome affects 1 in 100,000–135,000 live births in European populations (Barisic et al., 2014; Elek, Vitez, & Czeizel, 1991), although defects can occur in any population (Al-Qattan & Abou Al-Shaar, 2015; Ekure, Okoromah, Briggs, & Ajenifuja, 2004; Kimura, Kikuchi, Ichinoi, & Kure, 2015; Najjar, Mardini, Tabbaa, & Nyhan, 1988). Holt–Oram syndrome exhibits classic Mendelian inheritance for a dominant trait (Basson et al., 1997, 1994; Holt & Oram, 1960; McDermott et al., 2005), and the risk of nonaffected parents with an affected proband giving rise to a second child with a de novo pathogenic mutation is the same as the average population (McDermott, Fong, & Basson, 1993). Manifestations of upper-limb defects can include single or combinatorial abnormalities of the hand and digits, bones of the lower arm, humerus, or shoulder girdle (Basson et al., 1994; Newbury-Ecob et al., 1996). Defects of the hand and digits must include defects of the thumb for the Holt–Oram diagnosis, while defects of the lower arm are associated primarily with the radius (Basson et al., 1994; Holt & Oram, 1960; McDermott et al., 2005; Newbury-Ecob et al., 1996). Structural abnormalities of the heart can include secundum-type atrial septal defects, primum-type atrial septal defects, and/or ventricular septal defects (Basson et al., 1994; Holt & Oram, 1960; Newbury-Ecob et al., 1996). Conduction system defects manifest as long PR interval, AV block, bundle branch block, bradycardia, sick sinus syndrome, and atrial fibrillation (Basson et al., 1994; Holt & Oram, 1960; Newbury-Ecob et al., 1996), and these conduction defects can occur in the absence of overt structural defects (Basson et al., 1994; Newbury-Ecob et al., 1996). Holt– Oram syndrome is not associated with defects of the lower limb, postaxial upper limb, gastrointestinal system, genitourinary, or nervous system, which if present suggest an alternative diagnosis (Basson et al., 1994; Debeer, Race, Gewillig, Devriendt, & Frijns, 2007; Holt & Oram, 1960; McDermott et al., 2005; Newbury-Ecob et al., 1996).
Current evidence supports a model in which Holt–Oram syndrome is caused by TBX5 haploinsufficiency (Basson et al., 1997; Li et al., 1997) with genetic abnormalities associated with TBX5 coding or splice regulatory sequences underlying approximately 70% of patients meeting strict clinical diagnoses (Debeer et al., 2007; McDermott et al., 2005). In the latest collection of the Human Gene Mutation Database, there have been 103 reported mutations in coding, splicing, or regulatory sequences of TBX5, which result in Holt–Oram syndrome or other cardiac defects (Stenson et al., 2014), recently reviewed in Yamak et al. (2015). Additionally, there have been 44 pathologic point mutations reported in the coding region of TBX5 (Stenson et al., 2014). Similar to the eight reported gross deletions (Stenson et al., 2014), some of these are nonsense mutations resulting in highly truncated proteins that are thought to act as null alleles (Basson et al., 1997; Fan et al., 2003; Gruenauer-Kloevekorn & Froster, 2003; McDermott et al., 2005). Missense mutations have been reported throughout much of the T-box domain, typically resulting in transcriptional decrements (Basson et al., 1999; Boogerd et al., 2010; McDermott et al., 2005; Postma et al., 2008). There are also several reported mutations that result in missplicing or alternative splicing (Basson et al., 1999; Borozdin et al., 2006; Cross et al., 2000; Heinritz et al., 2005; McDermott et al., 2005; Vianna, Miura, Pereira, & Jatene, 2011). Interestingly, duplications of TBX5 are pathogenic, resulting in atypical Holt–Oram syndrome (Kimura et al., 2015; Patel, Silcock, McMullan, Brueton, & Cox, 2012). Additionally, a patient with a homozygous, single-base-pair mutation within a cis-regulatory element controlling TBX5 displayed decreased TBX5 expression and non-syndromic congenital heart disease, raising the possibility that the etiology of some of the remaining 30% of Holt–Oram patients may result from cis-regulatory element mutations (Smemo et al., 2012).
While all cases of Holt–Oram syndrome result in both cardiac and forelimb defects, several cases have been reported in which defects in either the heart (Basson et al., 1997; Brassington et al., 2003; Li et al., 1997; Yang et al., 2000) or the limb (Brassington et al., 2003; Li et al., 1997; Yang et al., 2000) appear more severe in one tissue than the other, suggesting a potential tissue-specific role for distinct domains of the protein (Isphording, Leylek, Yeung, Mischel, & Simon, 2004). While the underlying mechanisms of these biased defects have not been identified, some proposed models include differences in binding partners and binding motif recognition (Basson et al., 1999; Camarata et al., 2006; Garg et al., 2003; Isphording et al., 2004; Krause et al., 2004).
4. ANIMAL MODEL OF HOLT–ORAM SYNDROME
Investigations into the role of TBX5 in cardiac development have been undertaken in most major animal model systems: mouse, chick, frog, and zebrafish. Each system provides a unique set of tools for investigating the role of TBX5 in the developmental etiology of Holt–Oram syndrome. The most well-characterized model of Holt–Oram syndrome is the mouse heterozygous for a Tbx5 knockout allele. The Tbx5tm1Jse mouse allele contains loxP sites surrounding exon 3, which encodes a portion of the T-box DNA-binding domain, and upon Cre-mediated recombination, will generate truncated Tbx5 transcripts (Bruneau et al., 2001). Germline deletion of exon 3 generates the Tbx5tm1.1Jse mouse (Bruneau et al., 2001). Heterozygous Tbx5tm1.1Jse mice exhibit the characteristic haploinsufficient phenotype of Holt–Oram syndrome, including anterior defects of the forelimb, septal defects of the heart, and defects of cardiac conduction (Bruneau et al., 2001; Moskowitz et al., 2004). The mouse animal model has provided a robust system in which to study Holt–Oram syndrome in vivo and will continue to provide a platform by which to study the disease and the role of Tbx5 in cardiac and limb development.
5. TBX5 IN CARDIAC MORPHOLOGIC DEVELOPMENT
The morphologic cardiac defects associated with Holt–Oram syndrome are most commonly malformations of the septa dividing the left and right sides of the heart (Basson et al., 1994; Holt & Oram, 1960; McDermott et al., 2005; Newbury-Ecob et al., 1996). From our current understanding, the ontogeny of the septa dividing the ventricular and atrial chambers is quite different (Anderson, Webb, Brown, Lamers, & Moorman, 2003), and yet defects in both arise from haploinsufficiency of Tbx5 (Basson et al., 1997; Bruneau et al., 1999, 2001; Hoffmann et al., 2014; Koshiba-Takeuchi et al., 2009; Takeuchi et al., 2003; Xie et al., 2012).
5.1 Ventricular Septum
The morphology of the ventricular septum depends heavily on the localization of Tbx5 expression during development. Both the left and the right ventricles contribute equally toward the formation of the interventricular septum, suggesting that a balance of left and right contributions may underlie development of the septum (Franco et al., 2006). During development of the ventricular chambers, Tbx5 is unilaterally expressed on the left side, including the left side of the ventricular septum (Bruneau et al., 1999, 2001; Takeuchi et al., 2003). Overexpression of Tbx5 bilaterally results in malformation of the ventricular chambers and absence of the ventricular septum (Koshiba-Takeuchi et al., 2009; Liberatore, Searcy-Schrick, & Yutzey, 2000; Takeuchi et al., 2003). However, it remains unclear whether Tbx5-dependent transcriptional regulation alone controls ventricular septum formation (Franco et al., 2006; Koshiba-Takeuchi et al., 2009; Takeuchi et al., 2003). Two additional T-box family genes, Tbx18 and Tbx20, are unilaterally expressed in the left and right ventricles, respectively (Franco et al., 2006; Takeuchi et al., 2003). The boundary between Tbx5-positive, Tbx20-negative, and Tbx5-negative, Tbx20-positive myocardium appears to demarcate the location of ventricular septation and shifts in the expression levels can result in ventricular septum abnormalities; however, the exact mechanism by which the interface between Tbx5 and the other T-box family genes instructs formation of the ventricular septum remains unclear (Koshiba-Takeuchi et al., 2009; Takeuchi et al., 2003).
5.2 Atrial Septum
Haploinsufficiency of TBX5 in Holt–Oram patients results in atrial septal defects in approximately half of cases (Bruneau et al., 1999), and Tbx5 haploinsufficient mice exhibit atrial septal defects approximately 40% of the time (Bruneau et al., 2001). The atrial septum is derived from the second heart field contributing to the inflow tract of the heart (Goddeeris et al., 2008; Hoffmann, Peterson, Friedland-Little, Anderson, & Moskowitz, 2009; Mommersteeg et al., 2006; Snarr, Wirrig, Phelps, Trusk, & Wessels, 2007; Wessels et al., 2000). Sonic hedgehog (Shh), secreted from the pulmonary endoderm, signals through GLI-dependent transcription factors and is essential for atrial septation (Goddeeris et al., 2008; Hoffmann et al., 2009, 2014; Xie et al., 2012). Gli1 genetically interacts with Tbx5 in the second heart field to coactivate downstream targets, including Osr1 and Foxf1 (Goddeeris et al., 2008; Hoffmann et al., 2009, 2014; Xie et al., 2012). Furthermore, deletion of one or both copies of Tbx5 from Shh-receiving cells results in primum-type atrial septal defects, which can be rescued through constitutive activation of hedgehog signaling (Xie et al., 2012). This supports a model in which Tbx5 acts upstream of active Shh signaling in the second heart field and that both Tbx5 and activating GLI factors coregulate transcription at the top of a hierarchy of atrial septation genes (Hoffmann et al., 2014; Xie et al., 2012).
6. TBX5 IN CARDIAC CONDUCTION SYSTEM DEVELOPMENT
The cardiac conduction system is a highly specialized network of cardiomyocytes within the heart that generate and transmit electrical impulses throughout the heart to coordinate contraction. A majority of Holt–Oram syndrome patients present with conduction system abnormalities (Basson et al., 1994; Holt & Oram, 1960; Newbury-Ecob et al., 1996). Evidence suggests that Tbx5 plays three key roles in the cardiac conduction system: specification of the conduction system during development, regulation of the conduction system transcriptome, and maintenance of conduction system identity in the adult (Arnolds et al., 2012; Moskowitz et al., 2007, 2004). Tbx5 and Nkx2-5 genetically interact to specify the ventricular cardiac conduction system in a ventricular subdomain with the highest expression of both factors (Moskowitz et al., 2004; Thomas et al., 2001). Furthermore, Tbx5 and Nkx2-5 are required to coregulate the transcriptional repressor Id2, which is required for proper formation and function of the conduction system (Moskowitz et al., 2007). Although the Tbx5-dependent transcriptome in the conduction system has not been well characterized to date, Tbx5 is required for the regulation of critical conduction system ion channels, Gja5 and Scn5a, and removal of Tbx5 from the adult ventricular conduction system results in loss of these ion channels and altered ventricular conduction system function (Arnolds et al., 2012; Bruneau et al., 2001; Moskowitz et al., 2004; van den Boogaard et al., 2014).
7. HOMOZYGOUS TBX5 MUTATIONS REVEAL NOVEL ROLES OF TBX5
While much attention has been focused on understanding Tbx5 haploinsufficiency as it relates to human disease, homozygous deletion of Tbx5 in animal models reveals novel requirements for Tbx5 not uncovered by haploinsufficiency.
7.1 Complete Loss of Mammalian Tbx5
As Tbx5 haploinsufficiency is associated with the phenotype of Holt–Oram syndrome in mice and Tbx5 null embryos die in utero around E10.5, most studies examining the role of Tbx5 in cardiac morphogenesis and transcription have focused on the Tbx5 null heterozygote. In contrast, the null state provides the opportunity to understand critical roles of Tbx5 not observed in heterozygotes (Agarwal et al., 2003; Bruneau et al., 2001; Hoffmann et al., 2014; Luna-Zurita et al., 2016; Mori et al., 2006; Moskowitz et al., 2004; Rallis et al., 2003; Xie et al., 2012). In the mouse, germline deletion of both copies of Tbx5, Tbx5tm1.1Jse/tm1.1Jse, results in embryonic lethality by E10.5 (Bruneau et al., 2001). These animals exhibit a grossly abnormal, linear heart tube (Bruneau et al., 2001) and complete absence of the forelimb buds (Agarwal et al., 2003; Bruneau et al., 2001). The complete absence of the forelimb buds indicates a role for Tbx5 in limb bud initiation, shown to be downstream of fibroblast growth factor signaling (Agarwal et al., 2003; Hasson, Del Buono, & Logan, 2007; Rallis et al., 2003). Similar to the germline homozygous null embryos, homozygous hypomorphs for Tbx5 show embryonic lethality prior to E11.5, with hypoplastic left ventricles and sinoatrial structures (Mori et al., 2006). Distinctly, unlike the Tbx5 homozygous null phenotype, homozygous hypomorphs still undergo heart looping and rudimentary formation of the left and right atrial chambers (Mori et al., 2006). These observations indicate important roles for Tbx5 at sequential stages of cardiac development, although the distinctions between these roles have yet to be elucidated.
7.2 Zebrafish tbx5a/tbx5b
The heartstrings mutation is the first published mutation of tbx5a in zebrafish and was found as part of a screen for recessive lethal mutations affecting cardiac function (Garrity, Childs, & Fishman, 2002). The tbx5a/heartstrings mutants or morpholino knockdown of tbx5a recapitulate some aspects of Holt–Oram syndrome including forelimb defects and conduction defects (Ahn, Kourakis, Rohde, Silver, & Ho, 2002; Garrity et al., 2002). The hearts of these animals appear to develop normally during early cardiac development, only later displaying defects starting with the failure of heart looping and subsequent deterioration of chamber myocardium and heart failure (Ahn et al., 2002; Garrity et al., 2002).
More recently, a second copy of tbx5 (tbx5b) was found in the genome of zebrafish (Albalat, Baquero, & Minguillon, 2010). While tbx5a is expressed in the eye, heart, and forelimb during early development, tbx5b is only expressed robustly in the eye and heart (Albalat et al., 2010), suggesting possible redundant functions for the two tbx5 genes during early heart and eye development. Using morpholinos, tbx5a, tbx5b, or tbx5a/tbx5b double knockdowns all result in the heartstrings phenotype, i.e., normal heart tube formation, bradycardia, and progressive deterioration/heart failure (Garrity et al., 2002; Parrie, Renfrew, Wal, Mueller, & Garrity, 2013). Different downstream targets have been identified for tbx5a and tbx5b (Parrie et al., 2013). While tbx5b knockdown does not result in patterning defects of chamber formation or sinus venosus seen in either the tbx5a mutants or knockdown experiments, tbx5b knockdown results in abnormal expansion of two morphogenesis markers, hand2 and vcana, similar to tbx5a (Garrity et al., 2002; Parrie et al., 2013). However, known direct targets of mammalian TBX5 or zebrafish tbx5a, such as bmp4, nppa, tbx2b, and hey2 (Bruneau et al., 2001; Camarata et al., 2010; Chi et al., 2008; Plageman & Yutzey, 2004; Puskaric et al., 2010), were not disrupted with tbx5b knockdown, and neither tbx5a overexpression nor tbx5b overexpression rescues the reciprocal knockdown, suggesting the role of tbx5b is different than that of tbx5a (Parrie et al., 2013). The apparent differences in tbx5a and tbx5b function suggest that following gene duplication there may have been evolutionarily beneficial subfunctionalization of the two copies.
8. THE TBX5 GENE REGULATORY NETWORK
As a T-box transcription factor, the primary role of TBX5 is thought to be the regulation of target gene transcription. Historically, TBX5 has thought to act a positive regulator of transcriptional activity; however, recent evidence suggests that TBX5 may have a role in both transcriptional activation and repression (Fig. 2). In this section, we will explore what is known about both roles.
Fig. 2.
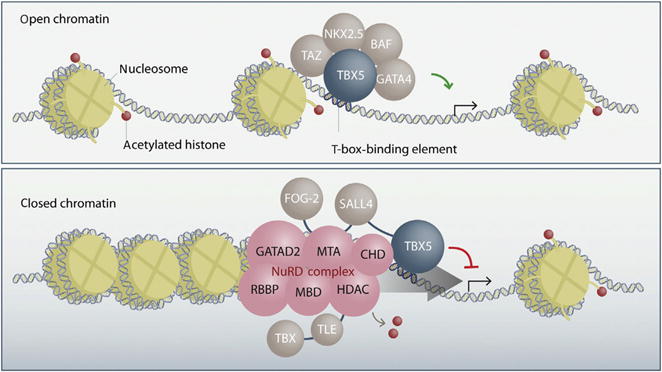
Transcriptional regulation by TBX5. TBX5, through its interactions with other cardiac transcription factors, such as GATA4 and NKX2-5, and the BAF chromatin-remodeling complex drive active transcription of target cardiac genes in regions of open chromatin (top panel). TBX5, through its interactions with the NuRD complex and other transcriptional repressors, such as SALL4, remodel chromatin to a closed state, which represses gene expression of noncardiac genes (bottom panel). Reprinted from Boogerd, C. J., & Evans, S. M. (2016). TBX5 and NuRD divide the heart. Developmental Cell, 36(3), 242–244, with permission from Elsevier.
8.1 Positive Transcriptional Activation by TBX5 and Cofactors: Cardiomyocyte Specific Factors, Chromatin Modification, and Maturation
TBX5 has long been known to act as a positive regulator of transcription in heart development and cardiomyocyte maturation (Bruneau et al., 2001; Goetz, Brown, & Conlon, 2006; Hiroi et al., 2001; Moskowitz et al., 2004). Some of the earliest identified direct targets of TBX5 were NPPA (encoding ANF) and GJA5 (encoding cx40), both of which are expressed in differentiating cardiomyocytes during development and are markers of cardiac chamber differentiation (Bruneau et al., 2001; Christoffels et al., 2000; Delorme et al., 1997; Hiroi et al., 2001). Gja5 and Nppa are both highly sensitive markers of TBX5 activity with a nearly complete loss of Gja5 in Tbx5tm1.1Jse/+ embryos and a graded response of Nppa across a Tbx5 allelic series (Bruneau et al., 2001; Mori et al., 2006).
The first identified interaction partner of TBX5 was the tinman transcription factor NKX2-5 (Hiroi et al., 2001). Identified by a classic yeast two-hybrid screen, TBX5 and NKX2-5 interact through the highly conserved C-terminus of NKX2-5, relying on four key amino acids in an α-helix, P139, D140, R150, and Q151 (Bruneau et al., 2001; Hiroi et al., 2001; Luna-Zurita et al., 2016). The interaction between TBX5 and NKX2-5 allows the proteins to synergistically activate targets such as Nppa through tandem transcription factor binding motifs (Bruneau et al., 2001; Hiroi et al., 2001). This interaction of TBX5 and NKX2-5 at tandem binding motifs induces bending in the DNA for transcriptional activity, a molecular mechanism by which synergistic activities of TBX5 and NKX2-5 interactions are determined at specific cis-regulatory elements (Luna-Zurita et al., 2016). Furthermore, the interaction between TBX5 and NKX2-5 is required to maintain fidelity in transcription factor binding throughout the genome, and the absence of either factor allows for inappropriate localization and activation of noncardiac genes by the other (Luna-Zurita et al., 2016). To this point, several sites within TBX5 are important for the synergistic activation of Nppa, which can be abrogated by HOS mutation in the α-helix mentioned earlier as well as G80R and R237W (Boogerd et al., 2010; Garg et al., 2003; Hiroi et al., 2001).
Besides NKX2-5, TBX5 also shows direct interaction with other major cardiac transcription factors, including GATA4 (Garg et al., 2003; Maitra et al., 2009), GATA6 (Maitra et al., 2009), TBX20 (Brown et al., 2005), MEF2C (Ghosh et al., 2009), and Myocardin (Wang, Cao, Wang, & Wang, 2011). The interaction between TBX5 and GATA4 was diminished by GATA4 mutations causing nonsyndromic congenital heart defects (Garg et al., 2003), as well as by TBX5 Holt–Oram mutations causing heart defects but not those causing only limb defects (Boogerd et al., 2010; Garg et al., 2003; Luna-Zurita et al., 2016). Both GATA4-TBX5 and MEF2C-TBX5 interactions are required for synergistic activation of α-cardiac myosin heavy chain encoded by MYH6 (Ghosh et al., 2009; Maitra et al., 2009), though GATA6-TBX5 protein interactions are not, suggesting that TBX5 interaction partners generate tissue- and context-specific gene expression (Maitra et al., 2009).
TBX5 as a transcription factor appears to act as part of a multifactor transcriptional complex for the activation and maintenance of cardiac lineage genes; however, the ability of transcription factors to regulate gene targets requires the ability of the factors to bind open chromatin. Addition of Gata4, Mef2c, and Tbx5 to fibroblasts is sufficient to drive reprogramming toward a cardiomyocyte fate (Ieda et al., 2010; Qian et al., 2012). This ability to reprogram cells suggests that this core set of transcription factors may drive chromatin accessibility. In support of this supposition, it has been shown that TBX5 interacts with Baf60c and Brg1, encoded by Smarcd3 and Smarca4, respectively, members of the SWI/SNF family of proteins involved in chromatin remodeling to drive mesodermal cells to cardiomyocyte fate in vitro and in vivo (Lickert et al., 2004; Takeuchi & Bruneau, 2009; Takeuchi et al., 2011). Furthermore, in Tbx5 haploinsufficient mice, there is a loss of chromatin remodeling complexes at the promoters of Tbx5-dependent cardiac genes (Takeuchi et al., 2011). For another T-box family member, T-bet, encoded by Tbx21, it has been previously shown that T-bet, the Brg1 chromatin-remodeling complex, and H3K27 demethylases physically interact (Miller, Mohn, & Weinmann, 2010), suggesting potential shared mechanisms with Tbx5 in chromatin transitions seen in cardiac development (Wamstad et al., 2012).
8.2 TBX5-Mediated Repression: Inhibition of Noncardiomyocyte Fate Through Chromatin Remodeling
In addition to its positive role driving cardiac gene regulatory networks, recent genomic studies indicate that TBX5 acts as a direct transcriptional repressor during cardiac development where it is required for inhibition of inappropriate gene expression (Lewandowski et al., 2014; Waldron et al., 2016). While transcriptional repression by T-box factors in cardiac development and function has been well documented in the cases of TBX2 (Carreira et al., 1998; Christoffels et al., 2004) and TBX3 (He et al., 1999; Hoogaars et al., 2007, 2004; Lingbeek, Jacobs, & van Lohuizen, 2002), and it has been shown that TBX20 has roles in both transcriptional repression and activation (Kaltenbrun et al., 2013; Sakabe et al., 2012; Stennard et al., 2005), only recently has a repressive role for TBX5 been elucidated. Waldron et al. (2016) demonstrated that Tbx5 inhibits noncardiac gene regulatory programs, including neuronal networks during early cardiac development. Through biochemical and genetic interaction studies, it was shown that TBX5 protein interacts with the nucleosome remodeling and deacetylase (NuRD) complex during embryonic development (Waldron et al., 2016). Similar to TBX20, the TBX5–NuRD interaction complex acts as an inhibitory mechanism by which TBX5 is able to repress noncardiogenic gene expression in the heart (Kaltenbrun et al., 2013; Waldron et al., 2016). TBX5 physically interacts with the NuRD complex through an evolutionarily conserved α-helix domain located from amino acids 255–264 and disruption of this domain can result in Holt–Oram syndrome (e.g., S261C; Brassington et al., 2003; Waldron et al., 2016). Using cardiomyocytes derived from murine embryonic stem cell differentiation, Luna-Zurita et al. (2016) demonstrated that TBX5 imparts specificity in cardiac transcription factor complexes by preventing off-target binding of other cardiac transcription factors. These findings suggest that the dual roles of T-box factors (i.e., TBX5 and TBX20 repression of noncardiomyocyte fate) may be a more common theme in cardiac development than previously thought.
8.3 Direct Targets of TBX5 Regulation
As a member of the T-box family of transcription factors, TBX5 regulates transcription through direct interaction with DNA. In recent years, multiple groups have turned to chromatin immunoprecipitation with sequencing (ChIP-sequencing) to identify direct targets of TBX5 binding and regulation (He et al., 2011; Luna-Zurita et al., 2016). The first ChIP-seq dataset was generated in the HL-1 atrial cardiomyocyte cell line using an overexpression construct of biotinylated TBX5, which identified over 56 k binding sites within the genome (He et al., 2011). The second set of data was generated using ChIP-exo technology in the context of mouse ES cell differentiation in both cardiac progenitors and cardiomyocytes, resulting in approximately 5 and 9 k bindings sites, respectively (Luna-Zurita et al., 2016). While these datasets share many of the same locations, each also identifies many unique sites, suggesting that binding site information will need to be generated in each Tbx5 expression context in order to understand the direct Tbx5 transcriptome. For example, whereas approximately 60% and 40% sites identified in the cardiac progenitor and cardiomyocyte mouse ES cell datasets are shared, only 4% of sites identified in the HL-1 dataset are shared with the mouse ES cell datasets. It is currently unclear to what degree the HL-1 dataset overestimates and the mouse embryonic stem cell datasets underestimate the total number of relevant binding sites, or whether both datasets overestimate functional binding events, HL-1 to a greater extent. Inclusion of additional markers, such as open chromatin, histone marks, and known TBX5-binding partners may allow the broad utilization of current datasets for identification of truly functional TBX5-binding sites.
To date, functionally confirmed direct targets of TBX5 are almost exclusively in genes implicated in cardiac proliferation, maturation, and function, including Nppa, Gja5, and Scn5a (Arnolds et al., 2012; Bruneau et al., 2001; Goetz et al., 2006; Hatcher et al., 2001; Hiroi et al., 2001; Mori et al., 2006; Moskowitz et al., 2007, 2004; Puskaric et al., 2010; Xie et al., 2012). Interestingly, the direct targets mediating the morphogenesis requirement for TBX5 remain unknown. While some candidate target genes may act prior to morphological changes (Hoffmann et al., 2014; Xie et al., 2012), no direct mediators of morphology have been uncovered. TBX5 may therefore indirectly regulate these processes. With advances in genome-wide technology, understanding the basis by which TBX5 regulates morphological change will be key to understanding the role of TBX5 in cardiac development.
9. CONCLUDING REMARKS
The requirement of TBX5 for normal human cardiac structure was identified over 20 years ago (Basson et al., 1997; Li et al., 1997); however, surprisingly, the mechanistic role of TBX5 in cardiac development remains unclear. Little is known about the essential downstream targets of TBX5-mediated transcription in the context of cardiac development. Similarly, the complex temporal and spatial gene expression of TBX5 has been mapped throughout development and into adult life; however, the cis-regulatory architecture governing this expression is just beginning to be described. From a biochemical perspective, significant strides have been made in recent years to understand how TBX5 activates gene expression. However, the mechanisms by which TBX5 and its cofactors are targeted to specific loci, the temporal recruitment of TBX5 and its cofactors, the interplay between TBX5 and it cotranscriptional partners, and the mechanisms distinguishing active and repressive TBX5 activity are just recently coming into focus, and provide opportunities for exciting mechanistic studies. These areas of investigation will contribute to a broader understanding of the mechanisms underlying the requirement for TBX5 in cardiac morphogenesis and more generally the transcriptional control of metazoan development.
References
- Adachi N, Robinson M, Goolsbee A, Shubin NH. Regulatory evolution of Tbx5 and the origin of paired appendages. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(36):10115–10120. doi: 10.1073/pnas.1609997113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, et al. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130(3):623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Agulnik SI, Bollag RJ, Silver LM. Conservation of the T-box gene family from Mus musculus to Caenorhabditis elegans. Genomics. 1995;25(1):214–219. doi: 10.1016/0888-7543(95)80128-9. [DOI] [PubMed] [Google Scholar]
- Agulnik SI, Garvey N, Hancock S, Ruvinsky I, Chapman DL, Agulnik I, et al. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144(1):249–254. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417(6890):754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- Albalat R, Baquero M, Minguillon C. Identification and characterisation of the developmental expression pattern of tbx5b, a novel tbx5 gene in zebrafish. Gene Expression Patterns. 2010;10(1):24–30. doi: 10.1016/j.gep.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Al-Qattan MM, Abou Al-Shaar H. A novel missense mutation in the TBX5 gene in a Saudi infant with Holt-Oram syndrome. Saudi Medical Journal. 2015;36(8):980–982. doi: 10.15537/smj.2015.8.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RH, Webb S, Brown NA, Lamers W, Moorman A. Development of the heart: (2) Septation of the atriums and ventricles. Heart. 2003;89(8):949–958. doi: 10.1136/heart.89.8.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. Journal of Clinical Investigation. 2012;122(7):2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic I, Boban L, Greenlees R, Garne E, Wellesley D, Calzolari E, et al. Holt Oram syndrome: A registry-based study in Europe. Orphanet Journal of Rare Diseases. 2014;9:156. doi: 10.1186/s13023-014-0156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nature Genetics. 1997;15(1):30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Basson CT, Cowley GS, Solomon SD, Weissman B, Poznanski AK, Traill TA, et al. The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome) New England Journal of Medicine. 1994;330(13):885–891. doi: 10.1056/NEJM199403313301302. [DOI] [PubMed] [Google Scholar]
- Basson CT, Huang T, Lin RC, Bachinsky DR, Weremowicz S, Vaglio A, et al. Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(6):2919–2924. doi: 10.1073/pnas.96.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag RJ, Siegfried Z, Cebra-Thomas JA, Garvey N, Davison EM, Silver LM. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nature Genetics. 1994;7(3):383–389. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- Boogerd CJ, Dooijes D, Ilgun A, Mathijssen IB, Hordijk R, van de Laar IM, et al. Functional analysis of novel TBX5 T-box mutations associated with Holt-Oram syndrome. Cardiovascular Research. 2010;88(1):130–139. doi: 10.1093/cvr/cvq178. [DOI] [PubMed] [Google Scholar]
- Borozdin W, Bravo Ferrer Acosta AM, Bamshad MJ, Botzenhart EM, Froster UG, Lemke J, et al. Expanding the spectrum of TBX5 mutations in Holt-Oram syndrome: Detection of two intragenic deletions by quantitative real time PCR, and report of eight novel point mutations. Human Mutation. 2006;27(9):975–976. doi: 10.1002/humu.9449. [DOI] [PubMed] [Google Scholar]
- Brassington AM, Sung SS, Toydemir RM, Le T, Roeder AD, Rutherford AE, et al. Expressivity of Holt-Oram syndrome is not predicted by TBX5 genotype. American Journal of Human Genetics. 2003;73(1):74–85. doi: 10.1086/376436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BM, Smith JC, et al. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132(3):553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, et al. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Developmental Biology. 1999;211(1):100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Camarata T, Bimber B, Kulisz A, Chew TL, Yeung J, Simon HG. LMP4 regulates Tbx5 protein subcellular localization and activity. The Journal of Cell Biology. 2006;174(3):339–348. doi: 10.1083/jcb.200511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarata T, Krcmery J, Snyder D, Park S, Topczewski J, Simon HG. Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish heart atrioventricular boundary and valve formation. Developmental Biology. 2010;337(2):233–245. doi: 10.1016/j.ydbio.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Dexter TJ, Yavuzer U, Easty DJ, Goding CR. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Molecular and Cellular Biology. 1998;18(9):5099–5108. doi: 10.1128/mcb.18.9.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Developmental Dynamics. 1996;206(4):379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes & Development. 2008;22(6):734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, et al. Chamber formation and morphogenesis in the developing mammalian heart. Developmental Biology. 2000;223(2):266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Developmental Dynamics. 2004;229(4):763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- Collavoli A, Hatcher CJ, He J, Okin D, Deo R, Basson CT. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. Journal of Molecular and Cellular Cardiology. 2003;35(10):1191–1195. doi: 10.1016/s0022-2828(03)00231-1. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC. Determinants of T box protein specificity. Development. 2001;128(19):3749–3758. doi: 10.1242/dev.128.19.3749. [DOI] [PubMed] [Google Scholar]
- Cross SJ, Ching YH, Li QY, Armstrong-Buisseret L, Spranger S, Lyonnet S, et al. The mutation spectrum in Holt-Oram syndrome. Journal of Medical Genetics. 2000;37(10):785–787. doi: 10.1136/jmg.37.10.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeer P, Race V, Gewillig M, Devriendt K, Frijns JP. Novel TBX5 mutations in patients with Holt-Oram syndrome. Clinical Orthopaedics and Related Research. 2007;462:20–26. doi: 10.1097/BLO.0b013e3181123ffe. [DOI] [PubMed] [Google Scholar]
- Delorme B, Dahl E, Jarry-Guichard T, Briand JP, Willecke K, Gros D, et al. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circulation Research. 1997;81(3):423–437. doi: 10.1161/01.res.81.3.423. [DOI] [PubMed] [Google Scholar]
- Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife. 2014;3:1–23. doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekure EN, Okoromah CN, Briggs E, Ajenifuja OA. Holt-Oram syndrome with hypoplastic left heart syndrome in an African child. The Nigerian Postgraduate Medical Journal. 2004;11(3):190–192. [PubMed] [Google Scholar]
- Elek C, Vitez M, Czeizel E. Holt-Oram syndrome. Orvosi Hetilap. 1991;132(2):73–74. 77–78. [PubMed] [Google Scholar]
- Fan C, Duhagon MA, Oberti C, Chen S, Hiroi Y, Komuro I, et al. Novel TBX5 mutations and molecular mechanism for Holt-Oram syndrome. Journal of Medical Genetics. 2003;40(3):e29. doi: 10.1136/jmg.40.3.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco D, Meilhac SM, Christoffels VM, Kispert A, Buckingham M, Kelly RG. Left and right ventricular contributions to the formation of the interventricular septum in the mouse heart. Developmental Biology. 2006;294(2):366–375. doi: 10.1016/j.ydbio.2006.02.045. [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129(19):4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- Georges R, Nemer G, Morin M, Lefebvre C, Nemer M. Distinct expression and function of alternatively spliced Tbx5 isoforms in cell growth and differentiation. Molecular and Cellular Biology. 2008;28(12):4052–4067. doi: 10.1128/MCB.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Human Molecular Genetics. 2001;10(18):1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, et al. Physical interaction between TBX5 and MEF2C is required for early heart development. Molecular and Cellular Biology. 2009;29(8):2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Silver LM, et al. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mechanisms of Development. 1996;56(1–2):93–101. doi: 10.1016/0925-4773(96)00514-x. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Papaioannou VE. Expression of T-box genes Tbx2-Tbx5 during chick organogenesis. Mechanisms of Development. 1998;74(1–2):165–169. doi: 10.1016/s0925-4773(98)00056-2. [DOI] [PubMed] [Google Scholar]
- Goddeeris MM, Rho S, Petiet A, Davenport CL, Johnson GA, Meyers EN, et al. Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development. 2008;135(10):1887–1895. doi: 10.1242/dev.016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133(13):2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenauer-Kloevekorn C, Froster UG. Holt-Oram syndrome: A new mutation in the TBX5 gene in two unrelated families. Annales de G en etique. 2003;46(1):19–23. doi: 10.1016/s0003-3995(03)00006-6. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nature Genetics. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P, Del Buono J, Logan MP. Tbx5 is dispensable for forelimb outgrowth. Development. 2007;134(1):85–92. doi: 10.1242/dev.02622. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Goldstein MM, Mah CS, Delia CS, Basson CT. Identification and localization of TBX5 transcription factor during human cardiac morphogenesis. Developmental Dynamics. 2000;219(1):90–95. doi: 10.1002/1097-0177(200009)219:1<90::AID-DVDY1033>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Kim MS, Mah CS, Goldstein MM, Wong B, Mikawa T, et al. TBX5 transcription factor regulates cell proliferation during cardiogenesis. Developmental Biology. 2001;230(2):177–188. doi: 10.1006/dbio.2000.0134. [DOI] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Wen L, Campbell CE, Wu JY, Rao Y. Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10212–10217. doi: 10.1073/pnas.96.18.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinritz W, Moschik A, Kujat A, Spranger S, Heilbronner H, Demuth S, et al. Identification of new mutations in the TBX5 gene in patients with Holt-Oram syndrome. Heart. 2005;91(3):383–384. doi: 10.1136/hrt.2004.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343(6259):617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nature Genetics. 2001;28(3):276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP. Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development. 2009;136(10):1761–1770. doi: 10.1242/dev.034157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, et al. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genetics. 2014;10(10):e1004604. doi: 10.1371/journal.pgen.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M, Oram S. Familial heart disease with skeletal malformations. British Heart Journal. 1960;22:236–242. doi: 10.1136/hrt.22.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes & Development. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovascular Research. 2004;62(3):489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126(8):1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isphording D, Leylek AM, Yeung J, Mischel A, Simon HG. T-box genes and congenital heart/limb malformations. Clinical Genetics. 2004;66(4):253–264. doi: 10.1111/j.1399-0004.2004.00314.x. [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152(1–2):327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, et al. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. Journal of Proteome Research. 2013;12(12):5395–5409. doi: 10.1021/pr400818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kikuchi A, Ichinoi N, Kure S. Novel TBX5 duplication in a Japanese family with Holt-Oram syndrome. Pediatric Cardiology. 2015;36(1):244–247. doi: 10.1007/s00246-014-1028-x. [DOI] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO Journal. 1995;14(19):4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Mori AD, Kaynak BL, Cebra-Thomas J, Sukonnik T, Georges RO, et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461(7260):95–98. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, et al. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Developmental Biology. 2004;273(1):106–120. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Kulisz A, Simon HG. An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Molecular and Cellular Biology. 2008;28(5):1553–1564. doi: 10.1128/MCB.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski SL, Janardhan HP, Smee KM, Bachman M, Sun Z, Lazar MA, et al. Histone deacetylase 3 modulates Tbx5 activity to regulate early cardiogenesis. Human Molecular Genetics. 2014;23(14):3801–3809. doi: 10.1093/hmg/ddu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nature Genetics. 1997;15(1):21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Developmental Biology. 2000;223(1):169–180. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lingbeek ME, Jacobs JJ, van Lohuizen M. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. Journal of Biological Chemistry. 2002;277(29):26120–26127. doi: 10.1074/jbc.M200403200. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, et al. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell. 2016;164(5):999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, et al. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Developmental Biology. 2009;326(2):368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, et al. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Research. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DA, Bressan MC, He J, Lee JS, Aftimos S, Brueckner M, et al. TBX5 genetic testing validates strict clinical criteria for Holt-Oram syndrome. Pediatric Research. 2005;58(5):981–986. doi: 10.1203/01.PDR.0000182593.95441.64. [DOI] [PubMed] [Google Scholar]
- McDermott DA, Fong JC, Basson CT. Holt-Oram syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, et al., editors. GeneReviews(R) Seattle, WA: University of Seattle; 1993. [Google Scholar]
- Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Molecular Cell. 2010;40(4):594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguillon C, Nishimoto S, Wood S, Vendrell E, Gibson-Brown JJ, Logan MP. Hox genes regulate the onset of Tbx5 expression in the forelimb. Development. 2012;139(17):3180–3188. doi: 10.1242/dev.084814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommersteeg MT, Soufan AT, de Lange FJ, van den Hoff MJ, Anderson RH, Christoffels VM, et al. Two distinct pools of mesenchyme contribute to the development of the atrial septum. Circulation Research. 2006;99(4):351–353. doi: 10.1161/01.RES.0000238360.33284.a0. [DOI] [PubMed] [Google Scholar]
- Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Developmental Biology. 2006;297(2):566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, et al. A molecular pathway including Id2, Tbx5, and Nkx2–5 required for cardiac conduction system development. Cell. 2007;129(7):1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, et al. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131(16):4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- Muller CW, Herrmann BG. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389(6653):884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- Najjar H, Mardini M, Tabbaa R, Nyhan WL. Variability of the Holt-Oram syndrome in Saudi individuals. American Journal of Medical Genetics. 1988;29(4):851–855. doi: 10.1002/ajmg.1320290415. [DOI] [PubMed] [Google Scholar]
- Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID. Holt-Oram syndrome: A clinical genetic study. Journal of Medical Genetics. 1996;33(4):300–307. doi: 10.1136/jmg.33.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou VE. The T-box gene family: Emerging roles in development, stem cells and cancer. Development. 2014;141(20):3819–3833. doi: 10.1242/dev.104471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou VE, Silver LM. The T-box gene family. Bioessays. 1998;20(1):9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Parrie LE, Renfrew EM, Wal AV, Mueller RL, Garrity DM. Zebrafish tbx5 paralogs demonstrate independent essential requirements in cardiac and pectoral fin development. Developmental Dynamics. 2013;242(5):485–502. doi: 10.1002/dvdy.23953. [DOI] [PubMed] [Google Scholar]
- Patel C, Silcock L, McMullan D, Brueton L, Cox H. TBX5 intragenic duplication: A family with an atypical Holt-Oram syndrome phenotype. European Journal of Human Genetics. 2012;20(8):863–869. doi: 10.1038/ejhg.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugfelder GO, Roth H, Poeck B. A homology domain shared between Drosophila optomotor-blind and mouse Brachyury is involved in DNA binding. Biochemical and Biophysical Research Communications. 1992;186(2):918–925. doi: 10.1016/0006-291x(92)90833-7. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. Journal of Biological Chemistry. 2004;279(18):19026–19034. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, et al. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circulation Research. 2008;102(11):1433–1442. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, et al. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Human Molecular Genetics. 2010;19(23):4625–4633. doi: 10.1093/hmg/ddq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, et al. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130(12):2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Silver LM. Newly identified paralogous groups on mouse chromosomes 5 and 11 reveal the age of a T-box cluster duplication. Genomics. 1997;40(2):262–266. doi: 10.1006/geno.1996.4591. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Silver LM, Gibson-Brown JJ. Phylogenetic analysis of T-Box genes demonstrates the importance of amphioxus for understanding evolution of the vertebrate genome. Genetics. 2000;156(3):1249–1257. doi: 10.1093/genetics/156.3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, et al. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Human Molecular Genetics. 2012;21(10):2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C, Christine KS, Mandel EM, Conlon FL. Developmental expression patterns of Tbx1, Tbx2, Tbx5, and Tbx20 in Xenopus tropicalis. Developmental Dynamics. 2006;235(6):1623–1630. doi: 10.1002/dvdy.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Human Molecular Genetics. 2012;21(14):3255–3263. doi: 10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snarr BS, Wirrig EE, Phelps AL, Trusk TC, Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Developmental Dynamics. 2007;236(5):1287–1294. doi: 10.1002/dvdy.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132(10):2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Human Genetics. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann CU, Ptchelkine D, Grimm C, Muller CW. Structural basis of TBX5-DNA recognition: The T-box domain in its DNA-bound and -unbound form. Journal of Molecular Biology. 2010;400(1):71–81. doi: 10.1016/j.jmb.2010.04.052. [DOI] [PubMed] [Google Scholar]
- Takabatake Y, Takabatake T, Takeshima K. Conserved and divergent expression of T-box genes Tbx2-Tbx5 in Xenopus. Mechanisms of Development. 2000;91(1–2):433–437. doi: 10.1016/s0925-4773(99)00329-9. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459(7247):708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nature Communications. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, et al. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130(24):5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- Thomas PS, Kasahara H, Edmonson AM, Izumo S, Yacoub MH, Barton PJ, et al. Elevated expression of Nkx-2.5 in developing myocardial conduction cells. Anatomical Record. 2001;263(3):307–313. doi: 10.1002/ar.1106. [DOI] [PubMed] [Google Scholar]
- Torrado M, Franco D, Lozano-Velasco E, Hernandez-Torres F, Calvino R, Aldama G, et al. A microRNA-transcription factor blueprint for early atrial arrhythmogenic remodeling. BioMed Research International. 2015;2015:263151. doi: 10.1155/2015/263151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJ, Klous P, et al. A common genetic variant within SCN10A modulates cardiac SCN5A expression. Journal of Clinical Investigation. 2014;124(4):1844–1852. doi: 10.1172/JCI73140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weerd JH, Badi I, van den Boogaard M, Stefanovic S, van de Werken HJ, Gomez-Velazquez M, et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circulation Research. 2014;115(4):432–441. doi: 10.1161/CIRCRESAHA.115.303591. [DOI] [PubMed] [Google Scholar]
- Vianna CB, Miura N, Pereira AC, Jatene MB. Holt-Oram syndrome: Novel TBX5 mutation and associated anomalous right coronary artery. Cardiology in the Young. 2011;21(3):351–353. doi: 10.1017/S1047951111000072. [DOI] [PubMed] [Google Scholar]
- Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, et al. The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Developmental Cell. 2016;36(3):262–275. doi: 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cao D, Wang Q, Wang DZ. Synergistic activation of cardiac genes by myocardin and Tbx5. PloS One. 2011;6(8):e24242. doi: 10.1371/journal.pone.0024242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yang XY, Zhao JY, Yu LW, Zhang P, Duan WY, et al. miR-10a and miR-10b target the 3′-untranslated region of TBX5 to repress its expression. Pediatric Cardiology. 2014;35(6):1072–1079. doi: 10.1007/s00246-014-0901-y. [DOI] [PubMed] [Google Scholar]
- Wessels A, Anderson RH, Markwald RR, Webb S, Brown NA, Viragh S, et al. Atrial development in the human heart: An immunohistochemical study with emphasis on the role of mesenchymal tissues. Anatomical Record. 2000;259(3):288–300. doi: 10.1002/1097-0185(20000701)259:3<288::AID-AR60>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Wilson V, Conlon FL. The T-box family. Genome Biology. 2002;3(6):1–7. doi: 10.1186/gb-2002-3-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Developmental Cell. 2012;23(2):280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamak A, Georges RO, Sheikh-Hassani M, Morin M, Komati H, Nemer M. Novel exons in the tbx5 gene locus generate protein isoforms with distinct expression domains and function. Journal of Biological Chemistry. 2015;290(11):6844–6856. doi: 10.1074/jbc.M114.634451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hu D, Xia J, Yang Y, Ying B, Hu J, et al. Three novel TBX5 mutations in Chinese patients with Holt-Oram syndrome. American Journal of Medical Genetics. 2000;92(4):237–240. doi: 10.1002/(sici)1096-8628(20000605)92:4<237::aid-ajmg2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Zaragoza MV, Lewis LE, Sun G, Wang E, Li L, Said-Salman I, et al. Identification of the TBX5 transactivating domain and the nuclear localization signal. Gene. 2004;330:9–18. doi: 10.1016/j.gene.2004.01.017. [DOI] [PubMed] [Google Scholar]


