Abstract
Human papillomavirus (HPV) L1 gene methylation deeply involved in the progression and heterogeneity of cervical cell epithelial lesions. The DNA ploidy also represented the early lesions of cervical cell, and it was associated with different HPV infection status in different ethnic women. So, the research was to explore whether it was possible that HPV L1 gene methylation and HPV infection status as the risk factors to lead to the differences of cervical epithelial cells’ lesions in different ethnics women.
The flow-through hybridization and gene chip for HPV genotypes test, general characteristics, and cervical exfoliated cell samples were collected from 94 Uygur and 79 Han women with HPV-16 infection. The cases were divided into the single HPV-16 (sHPV-16) infection group and multiple HPV-16 (mHPV-16) infection group in each ethnic women. The DNA ploidy was analyzed by flow cytometry, and the methylation-sensitive high resolution melting (MS-HRM) was used to test the HPV-16 L1 gene methylation, the results of methylation was segmented into mild methylation, moderate methylation, and severe methylation groups. Multifactor logistic analysis explored the relation between DNA heteroploid and HPV-16 infection status, HPV-16 L1 gene methylation in different ethnic women.
The higher proportion of mHPV-16 infection in Uygur than Han women (61.7% vs 38.0%). L1 gene methylation had statistic difference between single and mHPV-16 infection under the same ethnic women. The proportion of DNA heteroploid had statistic difference between different HPV-16 infection status or different L1 gene methylation grades in Han or Uygur women. Both L1 gene methylation and HPV infection status were the risk factors of DNA heteroploid. Compared to the sHPV-16 infection, the odds ratio (OR) of mHPV-16 infection were 4.409 (CI: 1.398–13.910) and 3.279 (CI: 1.069–10.060) in Han and Uygur women. Compared the mild L1 gene methylation, the OR of moderate L1 gene methylation were 3.313 (CI: 1.002–10.952) and 5.075 (CI: 1.385–18.603) in Han and Uygur women, the OR of severe L1 gene methylation were 20.592 (CI: 3.691–114.880) and 63.634 (CI: 10.400–389.368) in Han and Uygur women.
The study first reported that HPV L1 gene methylation and HPV infection status were the risk factors to the DNA heteroploid of cervical cell in different ethnics women, HPV L1 gene methylation and infection status should be recommended to the existing system of cervical lesion screening in order to provide better serves for the HPV infected women, especially for the ethnic women with high proportion of severe L1 gene methylation and multiple infection status.
Keywords: cervical epithelial cell, DNA ploidy, human papillomavirus, L1 gene, methylation
1. Introduction
Cervical cancer occurs was in the womb malignant tumors of the vagina and cervix tube. In the developing countries, cervical cancer has the highest incidence in gynecological tumors.[1] It was the 8th high incidence cancer for women in the People's Republic of China, the general trend is higher incidence in rural than urban area, and the prevalence shows younger trend.[2] Xinjiang region had the highest incidence of cervical cancer in China, the incidences of cervical cancer were different in 2 major ethnic of Xinjiang, including Han and Uygur ethnic.[3]
Human papillomavirus (HPV) infection especially high-risk type HPV infection was a major cause of cervical lesions. There were numerous studies of HPV about cervical lesions, which focused on the relationship between cervical lesions and HPV-related gene and protein, such as L1 protein, L2 protein, E6, and E7 gene.[4–7] The L1 protein as the main capsid protein of HPV played an important role to recognize the host cell and keep persistent infection, which was a good index to evaluate the infection state in host cell.[8] Previous studies showed that the quantity of L1 protein was declining with aggravate of cervical cell lesion, L1 gene was the coding gene of L1 protein, its methylation was the major reason of L1 protein decreasing, which showed positive correlation to the degree of cervical lesions.[9,10] So, L1 gene methylation deeply involved in the progression and heterogeneity of cervical lesions, which was the potential clinical molecular target of cervical lesions to early diagnose and monitor the prognosis.[11]
DNA ploidy of cervical epithelial cells was contributed to monitor the lesion of HPV infected cervical cells and the prognosis of treatment.[12–14] Our previous studies had proved that single and multiple HPV infection status could influenced on the DNA ploidy of cervical exfoliated cells in Xinjiang women.[15] Meanwhile, we also found that, when the proportion of DNA heteroploid had no difference between Uygur and Han women in Xinjiang when they were in the same HPV infection status, but DI and S-phase cells’ peak percentage (SPF) as quantitative index of DNA ploidy had differences,[16] which was contradictory. We speculated that the persistent/transient infection and single/multiple infection primary lead to the contradictory. Because L1 gene methylation reflected the persistent or transient infection of HPV infection in the host cell, so it was speculated that, the L1 gene methylation and single/multiple infection should explain the contradictory.
In conclusion, the research was to explore whether it was possible that HPV L1 gene methylation and HPV infection status as the risk factors to lead to the differences of cervical epithelial cell lesions in different ethnics women.
2. Methods
2.1. Patients
The sample cases sourced from Xinjiang Uygur and Han women, who initially visited the gynecology department of the Tumor Hospital Affiliated to Xinjiang Medical University from July 2015 to October 2016. The chosen women must not accept any HPV-related treatment and HPV vaccine. A total of 173 HPV-16 genotype infected cases were collected, including 94 Uygur women and 79 Han women. At the same time, their general case characteristics were also collected. The ethics committee of the tumor hospital affiliated to Xinjiang Medical University approved the study and the consent procedure. The samples of flow cytometry DNA ploidy analysis, HPV genotype test, and HPV-16 L1 gene methylation were cervical exfoliated cells, which were collected as required by clinicians. The insufficient or polluted samples were ruled out. The research related to human had been complied with all the relevant national regulations, institutional policies, and in accordance with the tenets of the Helsinki Declaration, and had been approved by The Tumor Hospital Affiliated to Xinjiang Medical University institutional review board.
2.2. Reagents and instruments
The HPV genotype test used the 21 HPV GenoArray Diagnostic Kit from ChaoZhou Hybribio Biological Chemical Co. Ltd. (People's Republic of China). The method of HPV genotype test was flow-through hybridization and gene chip, the equipments for the test such as Thermal Cycler and HybriMax devices (Flow-through Hybridization HybriMax).
The DNA ploidy analysis kit was from the Beckman Coulter; the flow cytometer was Beckman CytomicsTM FC500. The DNA cell cycle analysis software was also from Beckman Coulter.
The L1 gene methylation level was tested by the methylation-sensitive high resolution melting (MS-HRM). The major instrument was Roche LightCycler type 480 sensitivity analyzer. The completely methylated and unmethylated HPV-16 L1 gene standards of MS-HRM were synthesized (Genscript, Nanjing, China), the specific primers also were synthesized (Genscript, Nanjing, China). The EpiTect Bisulfite Kit and EpiTect HRM PCR Kit were bought from Germany QIAGEN company.
2.3. Experimental procedure
2.3.1. Flow cytometry DNA ploidy analysis
-
(1)
Collected the exfoliative cytology specimens, which were in cell preservation solution. Then through the 300 mesh nylon mesh filter, 1500 r/s centrifugal for 10 minutes, discarded the liquid supernatant, and then repeated this process by adding PBS fluid to the sediment, finally, suspended the exfoliated cells with 1 mL phosphate buffered saline (PBS) solution.
-
(2)
Added 200 μL DNA-Prep LPR reagent into the above solution that blended immediately, and placed it for 5 seconds; then added 2 mL of the DNA-Prep Stain reagent into it. Incubated for 20 minutes in dark place. Last, tested the specimen by Flow Cytometry DNA Ploidy Analysis System of FC500 flow cytometer.
-
(3)
Applied the DNA ploidy analysis software (DNA cell cycle analysis software) to analyze the results and obtained the DNA index (DI) and SPF of each specimen.
2.3.2. Flow-through hybridization and gene chip for HPV genotype test
HPV genotype test was carried out by the steps of HPV GenoArray Diagnostic Kit, which could detect 21 HPV genotypes, including 6, 11, 16, 18, 31, 33, 35, 39, 42, 43,44, 45, 51, 52, 53, 56, 58, 59, 66, and 68 types, and CP8304 types.
-
(1)
Extracted the HPV viral DNA by DNA extraction kit.
-
(2)
Took 1 μL of extracted DNA solution and then did PCR amplification according to the instructions in the reaction system by PCR amplification.
-
(3)
Made diversion hybridization amplification for amplified DNA samples.
-
(4)
Made hybridization results analysis of hybrid membrane after coloration; corresponding color parts’ classification is the result.
2.3.3. MS-HRM analysis of the HPV-16 L1 gene
The sequence HPV-16 L1 localized from nucleotide (nt) 5576 to (nt) 5636 (NCBI accession no. NC_001526.2), which contains 4 CpG sites (nt 5602, nt 5608, nt5611, and nt 5617) were tested.
-
1.
Mixed the completely methylated and unmethylated HPV-16 L1 gene standards in 0%, 10%, 25%, 50%, 75% and 100% methylated to unmethylated template ratios, which served as the methylation standards for MS-HRM.
-
2.
Extracted the HPV viral DNA by DNA extraction kit.
-
3.
The methylation standards and all extracted HPV viral DNA were bisulfite modificated, the detailed steps referred to the instruction book of EpiTect Bisulfite Kit.
-
4.
The specific PCR primers used were that, forward primer: 5′ GCGCATTATTGTTGATGTAGGTGATTTTTATTTATATTTTAG3′, reverse prime: 5′ GCCGCACTAAACAACCAAAAAAACATCTAAAAAAAAATA 3′. The detailed steps of MS-HRM PCR referred to the handbook of EpiTect HRM PCR.
-
5.
The HRM data were analyzed using the Genescanning Software (Roche).
2.4. Statistical analysis
The result was showed by mean ± standard, if the data were normally distributed; the statistical analysis was processed by SPSS 18.0 software. Comparison of count data models was by chi-square test. Multivariate logistic regression analysis was used to evaluate the risk factors. α = 0.05 is the inspection level, and P < 0.05 was received as having statistical differences.
3. Results
3.1. The general characteristics of 173 cases
The 4 characteristics factors between 2 ethnics were collected and compared such as age, marriage status, childbearing history, and abortion history, which was related to the HPV infection. The detail result is given in Table 1.
Table 1.
The general characteristics of 173 cases.

3.2. The HPV infection situation of 173 cases
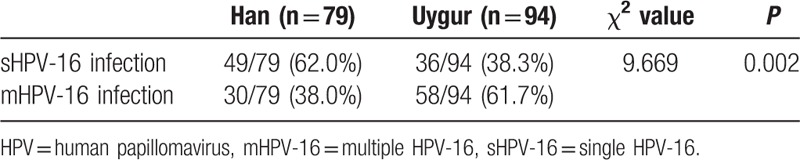
The HPV-16 infected women were divided into 2 groups in each ethnic women, including single HPV-16 (sHPV-16) infection (only HPV-16 infection) and multiple HPV-16 (mHPV-16) infection (existing HPV-16 infection and other HPV genotype infection at the same time). Then, the differences of infection status between 2 ethnics were compared. The results are shown in Table 2.
Table 2.
Comparison of the HPV infection status in Han and Uygur women.
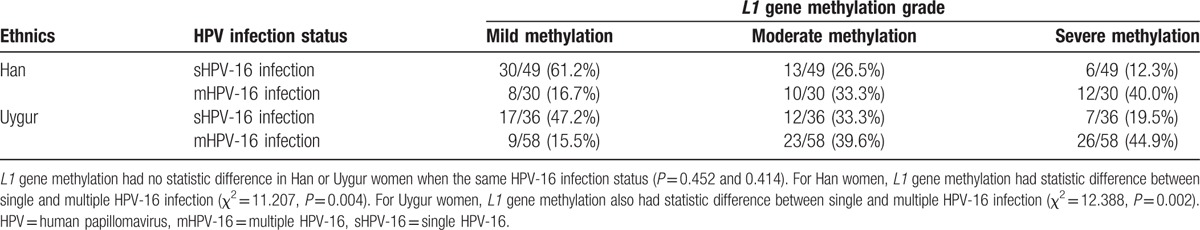
3.3. Comparison of HPV-16 L1 gene methylation in different HPV infection status between Han and Uygur women
The result of L1 gene methylation was divided into 3 grades, including mild methylation group (L1 gene methylation less than 25%), moderate methylation group (L1 gene methylation between 25% and 50%), and severe methylation group (L1 gene methylation more than 50%). The differences between 2 ethnics and 2 HPV-16 infection status were compared. The results are shown in Table 3.
Table 3.
Comparison of L1 methylation status between 2 ethnics and 2 HPV-16 infection status.
3.4. Comparison of DNA ploidy in different HPV-16 infection status between Han and Uygur women
The result of DNA ploidy was shown as DI and SPF, DI = 1.10 was the threshold of the DNA ploidy results; if a sample's DI was more than 1.10, it was seen as positive of DNA ploidy analysis, which meant heteroploid. If not, the sample was seen as negative of DNA ploidy analysis. The differences between 2 ethnics and 2 HPV-16 infection status were also compared. The results are shown in Table 4.
Table 4.
Comparison of DNA ploidy between 2 ethnics and 2 HPV-16 infection status.

3.5. Comparison of DI, SPF in different L1 gene methylation grades
Because DI and SPF had statistic difference between Han and Uygur women in the same HPV-16 infection status, so respectively compared the DI and SPF in different L1 gene methylation grades to prove that L1 gene methylation effected the DNA ploidy of host cells. The results are shown in Table 5.
Table 5.
Comparison of DI, SPF in different L1 gene methylation grades.

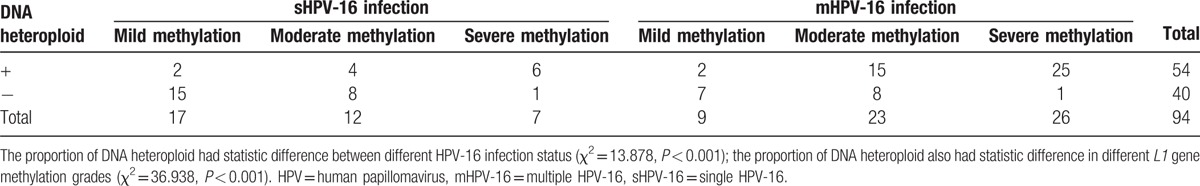
3.6. Comparison of HPV-16 L1 gene methylation and HPV-16 infection status in different DNA ploidy status
Because DI or SPF had statistic difference in different L1 gene methylation grades; therefore, respectively further compared the HPV-16 L1 gene methylation and HPV-16 infection status in different DNA ploidy status, the results show in Tables 6 and 7.
Table 6.
Comparison of HPV-16 L1 gene methylation and HPV-16 infection status in different DNA ploidy status for Han women.

Table 7.
Comparison of HPV-16 L1 gene methylation and HPV-16 infection status in different DNA ploidy status for Uygur women.
3.7. Multifactor analysis between HPV-16 infection status, HPV-16 L1 gene methylation, and heteroploid of DNA ploidy
Because both L1 gene methylation and HPV-16 infection status had statistic differences in different DNA ploidy status for Uygur women or Han women, so respectively discussed the relationship between HPV-16 infection status, HPV-16 L1 gene methylation, and heteroploid of DNA ploidy by logistic regression analysis in Han or Uygur women. The corresponding logistic regression expression was:
For Han women:
 |
For Uygur women:
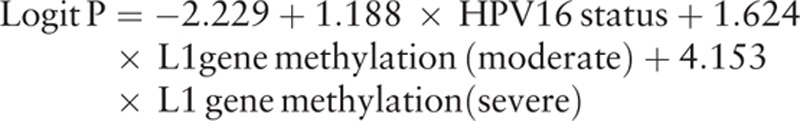 |
Tables 8 and 9 show the results of multivariate logistic regression analysis. Therefore, HPV-16 infection status, HPV-16 L1 gene methylation were the risk factors which were significantly associated with increased risk of DNA heteroploid. The odds ratio (OR) is displayed in Figs. 1–3.
Table 8.
Results of multivariate logistic regression analysis for Han women.
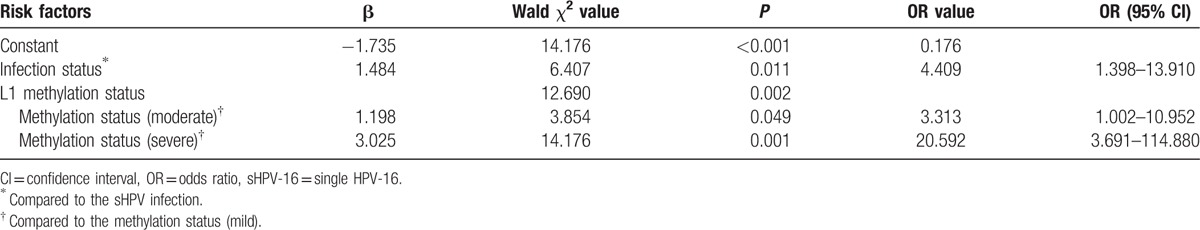
Table 9.
Results of multivariate logistic regression analysis for Uygur women.
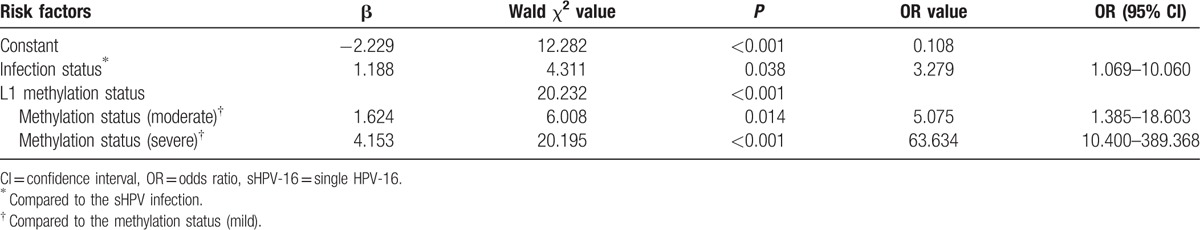
Figure 1.

The odds ratio of mHPV-16 infection status compared to sHPV-16 infection status for DNA heteroploid in Han and Uygur women. mHPV-16 = multiple HPV-16, sHPV-16 = single HPV-16.
Figure 3.

The odds ratio of severe L1 gene methylation compared to mild L1 gene methylation for DNA heteroploid in Han and Uygur women.
Figure 2.

The odds ratio of moderate L1 gene methylation compared to mild L1 gene methylation for DNA heteroploid in Han and Uygur women.
4. Discussion
The study researched 173 HPV-16 infected women in Xinjiang region by completely randomized design, including 94 Uygur and 79 Han women. It was found that there were differences in the risk factors of age and childbearing history by analyzing the general characteristics. The age of Uygur infected HPV-16 women was younger than Han women, meanwhile, the Uygur women showed more childbearing time than Han women, all of these fit on the common characteristics of Uygur and Han women in Xinjiang.[17]
The infection status of HPV-16 genotype included sHPV-16 and mHPV-16 infection. In this study, the proportion of mHPV-16 infection in Uygur women was much higher than Han women (61.7% vs 38.0%). So, the mHPV-16 infection was more common in Uygur, because high-risk multiple HPV infection was easy likely to cause the lesion of cervical epithelial cells in the previous report.[15] So, the cervical lesion because of HPV infection also was often occurred. The present study has found that DNA methylation was common in the process of cervical cancer as the molecular biology marks,[18] both host gene methylation and HPV gene methylation played the important role in the process of cervical lesion.[19–21] HPV L1 gene was the coding gene of major capsid protein, it was found that a high level of L1 gene methylation should mean the integration status of HPV genome and host cell genome, the low level of L1 gene methylation proved that HPV genome was in free status from the host cell genome.[8] Bryant et al[22] reported that a consistent trend was existed between HPV L1 gene methylation and cell morphology changes, it could be used as a molecular target of cervical lesions’ diagnosis and treatment. So, L1 gene methylation level not only represented the status of HPV infection in cervical exfoliated cells, but also had a good correction with the cervical cell lesion. HPV-16 L1 gene methylation was tested by MS-HRM in the research, MS-HRM was a promising technology to detect the gene methylation, which could make semiquantitative detection of methylation situation. It also be reported that the MS-HRM was a feasible method to detect HPV-16 L1 gene methylation.[23,24] According to the results of MS-HRM, the degree of the L1 gene methylation was divided into mild, moderate, and severe grades, the methylation status had no significant difference between 2 ethics in the same HPV-16 infection, but it existed significant difference in different HPV-16 status of the same ethnic women. So, L1 gene methylation level was severity as the seriousness of infection status, which was in accordance with the present reports.
Flow cytometry DNA ploidy analysis could reflect the DNA replication of specimens by DI value and SPF value, meanwhile, could indicate the cell lesion by heteroploid situation, when DI and SPF reached the critical value, means the appearance of cell lesion for an abnormal proliferation cells.[25] DI and SPF value had differences between Han and Uygur women neither in sHPV-16 infection nor mHPV-16 infection, but DNA heteroploid had no difference. The phenomenon proved that, compared to Han women, DNA replication of HPV-16 host cells were active in Uygur women, but most cases only stayed in hyperplasia replication active phase, also did not achieve the level of heteroploid. HPV-16 infection as a type of high-risk HPV infection, which could promote the host cell to immortalize, also enhances the activity of cell metabolism in order to make the DNA duplicate activity. When L1 gene methylation degree deepening, which represented the HPV DNA was integrated into the genome of host cell, meant the persistent infection, which was easy to result the DNA heteroploid of host cell. So, combined the results from Table 4 to Table 5, it could be thought that infection status and L1 gene methylation lead to the differences of DNA ploidy between Han and Uygur women.
From Table 6 and Table 7, it proved that infection status and L1 gene methylation could effect the DNA heteroploid, so logistic multifactor regression analysis was respectively done for Han and Uygur women. DNA heteroploid as dependent variable, with infection status and L1 gene methylation grades as potential influence factors, it was explored the risk level of infection status and L1 gene methylation to generate the DNA heteroploid. Infection status and L1 gene methylation were the risk factors to cause DNA heteroploid. When L1 gene methylation unchanged, it was 4.409 times to appear DNA heteroploid in multiple infection than single infection in Han women, in Uygur women, the OR was 3.279, the results accord with previous studies.[15,26] Compared to the Uygur women, HPV-16 infection status seemed to be more influence on Han women (OR: 3.297 vs 4.409). And L1 gene methylation created larger influence on Uygur women than Han women, in the same HPV infection, with a mild degree of L1 gene methylation as reference, Uygur women increase 5.075 and 63.634 times to appear DNA heteroploid when L1 gene methylation changed to moderate and severe grade, but the 2 OR values were 3.313 and 20.592 in Han women, which was smaller than Uygur women.
In a word, the research explored the influence of HPV-16 infection status and HPV-16 L1 gene methylation on DNA ploidy of cervical cell, discovered that both HPV-16 infection status and L1 gene methylation should be the risk factors of DNA ploidy. Especially L1 gene methylation had the greatest influence on the DNA heteroploid of cervical cell in Uygur women, it should be recommended to introduce the L1 gene methylation as a potential index to the existing system of cervical lesion screening and treatment standard of HPV infection in order to provide better serves for the masses of HPV-16 infected women.
Footnotes
Abbreviations: DI = DNA index, HPV = human papillomavirus, mHPV-16 = multiple HPV-16, MS-HRM = methylation-sensitive high resolution melting, OR = odds ratio, sHPV-16 = single HPV-16, SPF = S-phase cells’ peak percentage.
Funding/support: This research was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region, People's Republic of China (No. 2015211C133).
The authors have no conflicts of interest to disclose.
References
- [1].Word Health Organization: World Cancer Report 2014. WHO Report. Geneva, WHO, 2014, Chapter 5.12. [Google Scholar]
- [2].Chen WQ, Zheng RS, Zhang SW, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [3].Niyazi M, Sui S, Zhu K, et al. Correlation between methylation of human papillomavirus-16 L1 gene and cervical carcinoma in Uyghur women. Gynecol Obstet Invest 2017;82:22–9. [DOI] [PubMed] [Google Scholar]
- [4].Zine El Abidine A, Tomaić V, Bel Haj Rhouma R, et al. A naturally occurring variant of HPV-16 E7 exerts increased transforming activity through acquisition of an additional phospho-acceptor site. Virology 2016;500:218–25. [DOI] [PubMed] [Google Scholar]
- [5].Zhen S, Lu JJ, Wang LJ, et al. In vitro and in vivo synergistic therapeutic effect of cisplatin with human papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line. Transl Oncol 2016;9:498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fontecha N, Basaras M, Hernáez S, et al. Assessment of human papillomavirus E6/E7 oncogene expression as cervical disease biomarker. BMC Cancer 2016;16:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Anderson EL, Banister CE, Kassler S, et al. Human papillomavirus type 16 L2 DNA methylation in exfoliated cervical cells from college-age women. J Low Genit Tract Dis 2016;20:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oka N, Kajita M, Nishimura R, et al. L1 gene methylation in high-risk human papillomaviruses for the prognosis of cervical intraepithelial neoplasia. Int J Gynecol Cancer 2013;23:235–43. [DOI] [PubMed] [Google Scholar]
- [9].Wang J, Tian Q, Zhang S, et al. Clinical significance of HPV L1 capsid protein detection in cervical exfoliated cells in high-risk HPV positive women. Zhonghua Fu Chan Ke Za Zhi 2015;50:253–7. [PubMed] [Google Scholar]
- [10].Xu X, Yang J, Lin N, et al. Semi-quantitative detection of HPV L1 capsid protein in exfoliative cytological examination facilitates the differential diagnosis of cervical lesions. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2014;30:1194–7. [PubMed] [Google Scholar]
- [11].Turan T, Kalantari M, Calleja-Macias IE, et al. Methylation of the human papilloma virus-18 L1 gene: a biomarker of neoplastic progression? Virology 2006;49:175–83. [DOI] [PubMed] [Google Scholar]
- [12].Bollmann R, Méhes G, Torka R, et al. Determination of features indicating progression in atypical squamous cells with undetermined significance: human papillomavirus typing and DNA ploidy analysis from liquid-based cytologic samples. Cancer 2003;99:113–7. [DOI] [PubMed] [Google Scholar]
- [13].Bollmann R, Méhes G, Torka R, et al. Human papillomavirus typing and DNA ploidy determination of squamous intraepithelial lesions in liquid-based cytologic samples. Cancer 2003;99:57–62. [DOI] [PubMed] [Google Scholar]
- [14].Garner D. Clinical application of DNA ploidy to cervical cancer screening: a review. World J Clin Oncol 2014;5:931–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Feng YC, Yang J, Liu CM, et al. DNA ploidy of cervical epithelial cells should be a cure criterion of high-risk HPV infection in Xinjiang Uygur women. Onco Targets Ther 2015;8:827–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Feng YC, Zhang D, Huang YC. The effects of human papilloma infection status on the DNA ploidy of cervical epithelial cells in Uygur and Han women. Chongqing Medicine 2016;45:5–8. [Google Scholar]
- [17].Wang L, Wang P, Ren Y, et al. Prevalence of high-risk human papillomavirus (HR-HPV) genotypes and multiple infections in cervical abnormalities from Northern Xinjiang, China. PLoS One 2016;11:e0160698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang HJ. Aberrant DNA methylation in cervical carcinogenesis. Chin J Cancer 2013;32:42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lorincz AT, Brentnall AR, Vasiljević N, et al. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int J Cancer 2013;133:637–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bryant D, Hibbitts S, Almonte M, et al. Human papillomavirus type 16 L1/L2 DNA methylation shows weak association with cervical disease grade in young women. J Clin Virol 2015;66:66–71. [DOI] [PubMed] [Google Scholar]
- [21].Sun Y, Li S, Shen K, et al. DAPK1, MGMT and RARB promoter methylation as biomarkers for high-grade cervical lesions. Int J Clin Exp Pathol 2015;8:14939–45. [PMC free article] [PubMed] [Google Scholar]
- [22].Bryant D, Tristram A, Liloglou T, et al. Quantitative measurement of human papilloma virus type 16 L1/L2 DNA methylation correlates with cervical disease grade. J Clin Virol 2014;59:24–9. [DOI] [PubMed] [Google Scholar]
- [23].Qiu C, Zhi Y, Shen Y, et al. High-resolution melting analysis of HPV-16L1 gene methylation: a promising method for prognosing cervical cancer. Clin Biochem 2015;48:855–9. [DOI] [PubMed] [Google Scholar]
- [24].Qiu C, Zhi Y, Shen Y, et al. Performance of the HPV-16 L1 methylation assay and HPV E6/E7 mRNA test for the detection of squamous intraepithelial lesions in cervical cytological samples. J Virol Methods 2015;224:35–41. [DOI] [PubMed] [Google Scholar]
- [25].Pinto AE, Pires A, Silva G, et al. Ploidy and S-phase fraction as predictive markers of response to radiotherapy in cervical cancer. Pathol Res Pract 2011;207:623–7. [DOI] [PubMed] [Google Scholar]
- [26].De Brot L, Pellegrini B, Moretti ST, et al. Infections with multiple high-risk HPV types are associated with high-grade and persistent low-grade intraepithelial lesions of the cervix. Cancer 2016;doi: 10.1002/cncy.21789. [DOI] [PubMed] [Google Scholar]


