Abstract
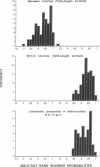
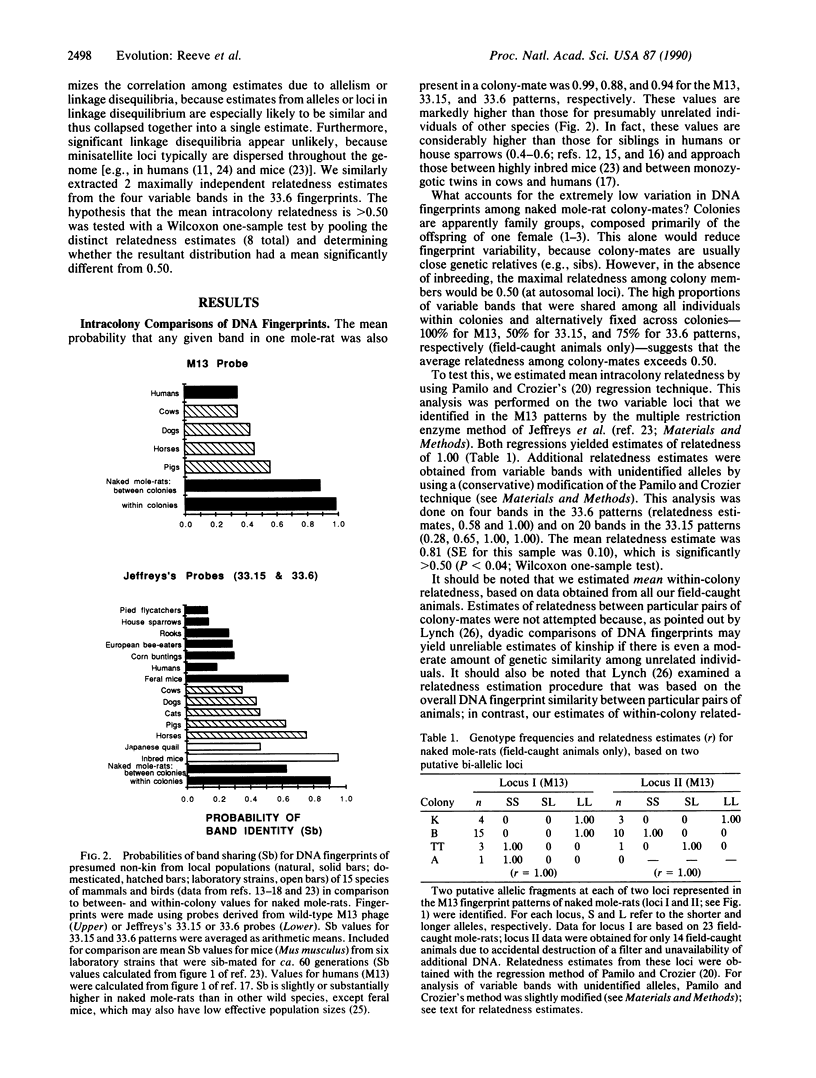
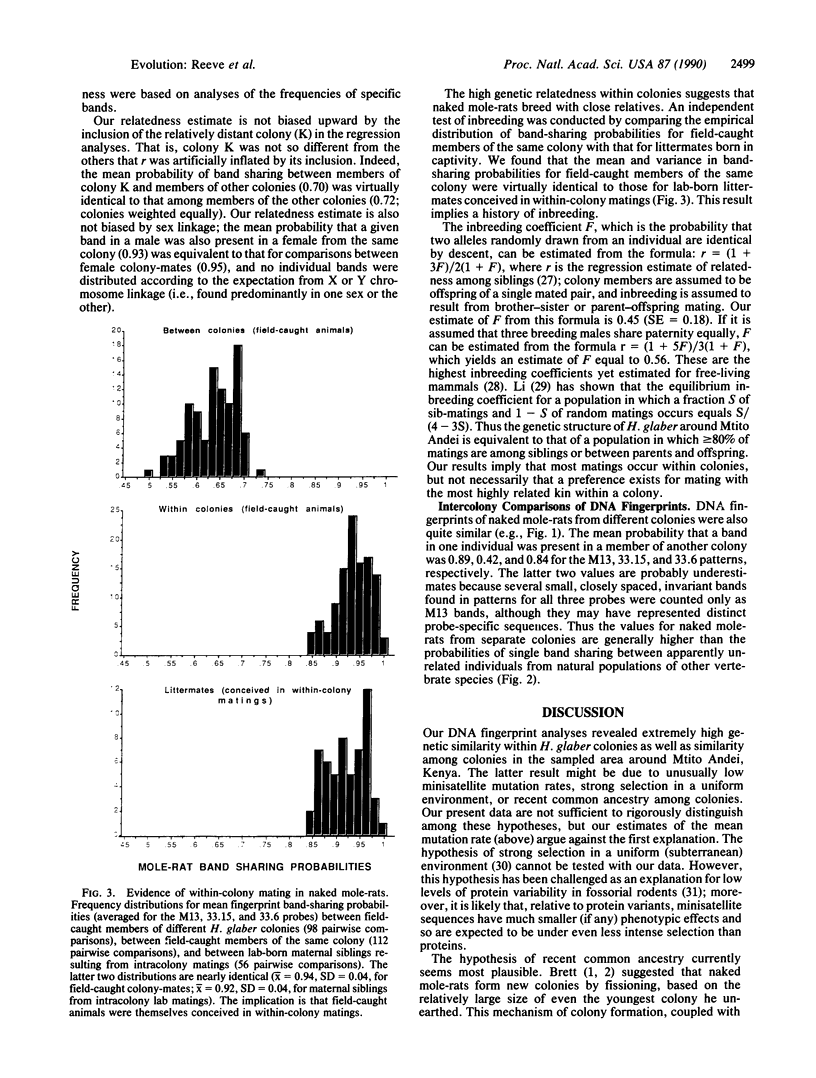
Using the technique of DNA fingerprinting, we investigated the genetic structure within and among four wild-caught colonies (n = 50 individuals) of a eusocial mammal, the naked mole-rat (Heterocephalus glaber; Rodentia: Bathyergidae). We found that DNA fingerprints of colony-mates were strikingly similar and that between colonies they were much more alike than fingerprints of non-kin in other free-living vertebrates. Extreme genetic similarity within colonies is due to close genetic relationship (mean relatedness estimate +/- SE, r = 0.81 +/- 0.10), which apparently results from consanguineous mating. The inbreeding coefficient (F = 0.45 +/- 0.18) is the highest yet recorded among wild mammals. The genetic structure of naked mole-rat colonies lends support to kin selection and ecological constraints models for the evolution of cooperative breeding and eusociality.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartz S. H. Evolution of eusociality in termites. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5764–5768. doi: 10.1073/pnas.76.11.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T., Bruford M. W. DNA fingerprinting in birds. Nature. 1987 May 14;327(6118):149–152. doi: 10.1038/327149a0. [DOI] [PubMed] [Google Scholar]
- Evans J. R., Massler M. The effective clinical teacher. J Dent Educ. 1977 Oct;41(10):613–617. [PubMed] [Google Scholar]
- Georges M., Lequarré A. S., Castelli M., Hanset R., Vassart G. DNA fingerprinting in domestic animals using four different minisatellite probes. Cytogenet Cell Genet. 1988;47(3):127–131. doi: 10.1159/000132529. [DOI] [PubMed] [Google Scholar]
- Hamilton W. D. The genetical evolution of social behaviour. II. J Theor Biol. 1964 Jul;7(1):17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- Jarvis J. U. Eusociality in a mammal: cooperative breeding in naked mole-rat colonies. Science. 1981 May 1;212(4494):571–573. doi: 10.1126/science.7209555. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Brookfield J. F., Semeonoff R. Positive identification of an immigration test-case using human DNA fingerprints. 1985 Oct 31-Nov 6Nature. 317(6040):818–819. doi: 10.1038/317818a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Morton D. B. DNA fingerprints of dogs and cats. Anim Genet. 1987;18(1):1–15. doi: 10.1111/j.1365-2052.1987.tb00739.x. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Kelly R., Taylor B. A., Bulfield G. Mouse DNA 'fingerprints': analysis of chromosome localization and germ-line stability of hypervariable loci in recombinant inbred strains. Nucleic Acids Res. 1987 Apr 10;15(7):2823–2836. doi: 10.1093/nar/15.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Individual-specific 'fingerprints' of human DNA. Nature. 1985 Jul 4;316(6023):76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L., Weatherall D. J., Ponder B. A. DNA "fingerprints" and segregation analysis of multiple markers in human pedigrees. Am J Hum Genet. 1986 Jul;39(1):11–24. [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Estimation of relatedness by DNA fingerprinting. Mol Biol Evol. 1988 Sep;5(5):584–599. doi: 10.1093/oxfordjournals.molbev.a040518. [DOI] [PubMed] [Google Scholar]
- Michod R. E. Evolution of interactions in family-structured populations: mixed mating models. Genetics. 1980 Sep;96(1):275–296. doi: 10.1093/genetics/96.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984 Dec;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassart G., Georges M., Monsieur R., Brocas H., Lequarre A. S., Christophe D. A sequence in M13 phage detects hypervariable minisatellites in human and animal DNA. Science. 1987 Feb 6;235(4789):683–684. doi: 10.1126/science.2880398. [DOI] [PubMed] [Google Scholar]
- Westneat D. F., Noon W. A., Reeve H. K., Aquadro C. F. Improved hybridization conditions for DNA 'fingerprints' probed with M13. Nucleic Acids Res. 1988 May 11;16(9):4161–4161. doi: 10.1093/nar/16.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetton J. H., Carter R. E., Parkin D. T., Walters D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature. 1987 May 14;327(6118):147–149. doi: 10.1038/327147a0. [DOI] [PubMed] [Google Scholar]