Abstract
The PIN-FORMED (PIN) protein, the most important polar auxin transporter, plays a critical role in the distribution of auxin and controls multiple biological processes. However, characterizations and functions of this gene family have not been identified in cotton. Here, we identified the PIN family in Gossypium hirsutum, Gossypium arboreum, and Gossypium raimondii. This gene family was divided into seven subgroups. A chromosomal distribution analysis showed that GhPIN genes were evenly distributed in eight chromosomes and that the whole genome and dispersed duplications were the main duplication events for GhPIN expansion. qRT-PCR analysis showed a tissue-specific expression pattern for GhPIN. Likely due to the cis-element variations in their promoters, transcripts of PIN6 and PIN8 genes from the At (tetraploid genome orginated from G. arboreum) subgenome and PIN1a from the Dt (tetraploid genome orginated from G. raimondii) subgenome in G. hirsutum was significantly increased compared to the transcripts in the diploids. The differential regulation of these PIN genes after the polyploidization may be conducive to fiber initiation and elongation. Exogenously applied auxin polar transport inhibitor significantly suppressed fiber growth, which is consistent with the essential function of these PIN genes for regulating cotton fiber development. Furthermore, the overexpression of GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At in Arabidopsis promoted the density and length of trichomes in leaves.
Keywords: cotton, PIN-formed, auxin efflux carriers, fiber development, expression patterns
Introduction
The plant phytohormone, auxin, plays a crucial role in various developmental processes, such as root development, apical dominance, embryo formation, vascular differentiation, tropism, and plant response to internal and external stimuli (Dubrovsky et al., 2008; Mashiguchi et al., 2011; Rakusova et al., 2015; Wang H.Z. et al., 2015; Xi et al., 2016). Auxin is primarily synthesized in the apical meristems via several different pathways and then transported to other tissues via the plant vascular system (Zhao, 2010; Remy et al., 2013; Chen et al., 2014). The asymmetric distribution of auxin, which is largely dependent on an active process called directional polar auxin transport (PAT) mediated by influx and efflux carriers, has been confirmed to play a vital role in controlling plant growth and development (Friml, 2003; Wang et al., 2008; Band et al., 2014; D’Alessandro et al., 2015; Yang et al., 2015). The influx and efflux carriers of the auxin transporter that are distributed in the plasma membrane are grouped into three major categories: auxin resistant 1/like aux1 (AUX1/LAX) influx carriers, P-glycoprotein (MDR/PGP/ABCB) efflux/conditional transporters, and plant specific PIN-FORMED (PIN) efflux carriers. The PINs are known as the most important auxin carriers that control the auxin PAT process (Péret et al., 2012; Adamowski and Friml, 2015; Yue et al., 2015; Row et al., 2016). Recently, PIN-LIKES (PILS) family, which are localized to the endoplasmic reticulum and involved in the regulation of intracellular auxin homeostasis, was characterized as a novel putative auxin carriers (Barbez et al., 2012; Feraru et al., 2012).
In Arabidopsis, the PIN gene family, including eight members, is divided into two types: the PIN proteins with long hydrophilic loop (long PIN proteins) and with short hydrophilic loop (short PIN proteins). Both PIN types have been reported to participate in various developmental processes (Petrásek et al., 2006; Habets and Offringa, 2014; Wang H.Z. et al., 2015; Omelyanchuk et al., 2016). The long PIN proteins (AtPIN1–4 and AtPIN7) are situated in the plasma membrane and participate in root development, apical shoot establishment, and various tropic responses (Friml et al., 2002; Robert et al., 2013). AtPIN5 and AtPIN8, are localized in the endoplasmic reticulum and are involved in the homeostasis of intracellular auxin (Mravec et al., 2009; Ding et al., 2012). Interestingly, AtPIN6 is found to localize in both the plasma membrane and endoplasmic reticulum (Simon et al., 2016). Moreover, the PIN genes have been reported to be involved in the hormone signaling and abiotic stress responses, such as drought, salt and dehydration (Willige et al., 2011; Guo et al., 2015; Li F. et al., 2015).
The PIN gene family has been extensively studied using phylogenetic analyses and expression profiles in monocots and eudicots. In rice, 12 PIN genes have been identified, including four PIN1 and one PIN2, which belong to the long PINs, and three PIN5 and one PIN8, which belong to the short PINs and three monocot-specific PINs (PIN9, PIN10a, PIN10b) (Wang et al., 2009; Lu et al., 2015). Until now, 10 PINs in potato, 23 PINs in soybean, and 12 PINs in maize have been identified, and their expressions exhibit a tissue-specific pattern (Yue et al., 2015). ZmPIN1d is predominantly expressed in the shoot apical meristem and inflorescence meristem during the flowering stage, whereas the ZmPIN9 transcript was primarily detected in the root endodermis and pericycle (Forestan et al., 2012). GmPIN9, a legume-specific PIN gene, may contribute to auxin redistribution in the soybean root under various abiotic stresses (Wang Y.Q. et al., 2015). StPINs play a major role in the swelling stolon during tuber development (Roumeliotis et al., 2013). In Sorghum bicolor (a monocot), 11 PIN genes have been characterized, including at least three PIN1 and three PIN5 genes, suggesting that not only monocot-specific PINs but also long PINs and short PINs exist together in monocots (Shen et al., 2010).
Cotton is one of the most important industrial crops in the world and its fibers are the main resource for the textile industry, which contributes more than 10 billion dollars annually to the global economy. Despite the extreme importance of PINs, the origins, characters and functions of these genes in cotton are still largely unknown. Taking advantage of the whole genome sequence of three cotton species that were recently completed (Paterson et al., 2012; Wang K. et al., 2012; Li et al., 2014; Li K. et al., 2015; Zhang T.Z. et al., 2015), we analyzed the PIN gene family comprehensively, including the phylogenic relationship, chromosomal distribution and gene duplication of the PIN genes. Furthermore, we suggest that GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At may have played an important role in the fiber growth during the evolution and domestication of cotton. This is because higher transcriptional levels have only been observed in longer fiber cells in allotetraploid cotton Gossypium hirsutum, rather than in its diploid ancestors, Gossypium arboreum and Gossypium raimondii, which might result from the differential regulation of GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At. The overexpression of GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At in Arabidopsis enhanced the density and lengths of trichomes in leaves. Our study provides a basis to systematically elucidate the evolution and function of PINs in cotton and to effectively characterize the precise biological roles of PINs and their utilization in future cotton breeding programs.
Materials and Methods
Plant Materials and Growth Condition
Gossypium hirsutum (Xuzhou 142), G. arboreum (Shixiya 1), and G. raimondii (CMD10) that were used in this study were grown in a climate-controlled greenhouse with a 16 h light and 8 h dark cycle at 30°C, as previously reported (Shi et al., 2006). For tissue-specific expression patterns, the roots, stems, leaves, and flowers that were collected from the same growth stage of G. hirsutum and G. arboreum were immediately frozen and stored in liquid nitrogen after harvest.
Sequence Retrieval, Multiple Sequence Alignment, and Phylogenetic Analysis
The cotton genome sequences were downloaded from the CottonGen website1. The Arabidopsis genome sequence was acquired from TAIR 102. Other plant genome sequences that were used in this study were retrieved from Phytozome3. We used the HMMER software with default parameters and the conserved PIN transmembrane domain (PF03547.13) to search for corresponding protein sequences. The PIN sequences were further identified based on a homology search using the BLAST program. Multiple sequence alignments of all identified PINs in this study were performed using ClustalX (Thompson et al., 1997). A phylogenetic tree of deduced amino acid sequences was constructed using the neighbor-joining algorithm with default parameters and 1000 bootstrap replicates in MEGA 5.04. The transmembrane topologies of the PINs were predicted using the TMHMM web server with corresponding parameters5.
Gene Structure and Chromosoman Mapping
For the exon–intron structural analysis of the PIN genes, the coding sequences were compared with their genomic DNA sequences and the structure diagrams were generated using the GSDS server6. The conserved domains were predicted using the Simple Modular Architecture Research Tool (SMART7) and Pfam8 (Hu et al., 2015). The chromosome positions of all PINs in G. hirsutum were retrieved from gene annotation files that were downloaded from the CottonGen website. The map of the PIN gene distribution along the chromosomes was drawn from the top to the bottom using the CIRCOS program (Zou et al., 2013). We used MCScanX software for the detection and evolutionary analysis of PIN gene synteny and collinearity (Wang Y. et al., 2012).
RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis
The plant materials that were harvested as described above were frozen in liquid nitrogen and then ground to a fine powder with a mortar and pestle using a modified method (Jin et al., 2013; Liu et al., 2015). The total RNA was extracted using the PureLinkTM RNA mini kit (Invitrogen, Lot no.1687455) according to the manufacturer’s instructions, and the cDNA was reverse-transcribed from 5 μg of the total RNA as previously reported (Ji et al., 2003). In the qRT-PCR experiments, each gene was run in three biological and three technical replicates with the following reaction parameters: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 56°C for 30 s. A melting curve was generated from 65 to 95°C. SigmaStat software was used for one-way statistical variance analysis. The UBQ7 cotton gene was employed as the internal control for all related results.
In vitro Ovule Culture
The in vitro ovule culture was performed as previously reported (Xiao et al., 2016). N-1-naphthylphtha-lamic acid (NPA, Sigma) was added to the culture medium with different concentrations as in the previous experimental design. Fiber lengths from cultured ovules were manually measured under a bright field microscope.
Vector Construction and Plant Transformation
The full length (from the start codon to the stop codon) of each of the GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At genes was amplified from the cDNA using the following gene-specific primers, respectively: 5′ primer: ACGGGGGACTCTAGAGGAT CCATGATCACTTTAACAGATTTTTACC 3′ primer: TCATA TACCCAATAAAATGTAGTACAGCTCTGTCGACTGTGGTA CCTTA 5′ primer: ACGGGGGACTCTAGAGGATCCATGATA ACAGGGGGTGATTTTTACA 3′ primer: GTATTACATACTTT TAGGCATATGAAGCTCTGTCGACTGTGGTACCTTA 5′ primer: ACGGGGGACTCTAGAGGATCCATGATTTCCCTGG CAGATGTTTATC 3′ primer: TCACAATGCTAAAAGAAGATA GTAGAGCTCTGTCGACTGTGGTACCTTA.
The pQG110 vector was digested using BamHI and KpnI. The product of each gene was then cloned into the digested pQG110 vector driven by the constitutive cauliflower mosaic virus 35S promoter via recombinase vazyme according to the manual, and the construct was further transformed into wild-type Arabidopsis plants. Transgenic plants were generated via the floral dip method and were selected on solid half-strength MS medium plates containing 50 mg/mL of the appropriate antibiotics (Zhang Y.Z. et al., 2015). The trichomes in the transgenic plant leaves were examined under a dissecting microscope (Leica MZ APO).
Southern Blots Assay
Southern blots were performed as described previously (Jin et al., 2013). 15 μg genomic DNA was digested thoroughly using BamHI or EcoRI. After purification, the digested DNA was loaded onto electrophoretic gels for 4 h, then blotted prior to hybridization and detected using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) with GhPIN gene sequences as probe sequences.
Results
Genome-Wide Identification of PIN Proteins in Three Different Cotton Species
Two diploid cottons, which were designated as AA genome and DD genome, diverged from a common ancestor approximately 2 to 13 million years ago (MYA) and developed genomes that differ approximately twofold in size. Approximately 1 to 2 MYA, these two genomes reunited in a common nucleus through allopolyploidization (Supplementary Figure S1), which led to the progenitor of the modern polyploidy cotton, G. hirsutum. This is the world’s primary commercial cotton species. Thus, allotetraploid cotton G. hirsutum contains two ancestral genomes, named the At and Dt subgenome. To identify all of the PIN proteins in G. hirsutum (AADD genome) and its two diploid ancestors, G. arboreum (AA genome) and G. raimondii (DD genome), we used the Arabidopsis PIN protein sequences to query the three reference genomes to screen out candidate PIN-like proteins in cotton. The collected PIN-like candidates were subjected to a further selection process based on their conserved domain using SMART. The PINs without the auxin efflux carrier domain were discarded. After this strict two-steps selection process, 17 deduced PINs were identified in G. hirsutum, along with 12 in G. arboreum and 10 in G. raimondii. Among the 17 PIN proteins identified in the G. hirsutum genome, seven originated from the At subgenome, nine from the Dt subgenome, and one was located on the scaffold (Supplementary Table S1). The lengths of the identified GhPIN proteins ranged from 127 (GhPIN8a_At) to 678 (GhPIN2_Dt) amino acids (aa) with an average length of 511 aa. The GaPIN proteins ranged from 124 (GaPIN3b) to 681 (GaPIN3) aa with an average length of 495 aa, and the GrPIN proteins ranged from 355 (GrPIN5) to 648 (GrPIN3) aa with an average length of 539 aa. We also analyzed orthologous PIN gene pairs in G. hirsutum and their corresponding diploid ancestors. Five genes from the A genome and two from the D genome were lost in G. hirsutum, whereas the orthologous genes of GhPIN8a_At and GhPIN8a_Dt in G. hirsutum were lost in its diploids, G. arboreum and G. raimondii, respectively.
Analysis of Gene Loss and Duplication of PIN Genes in Cotton
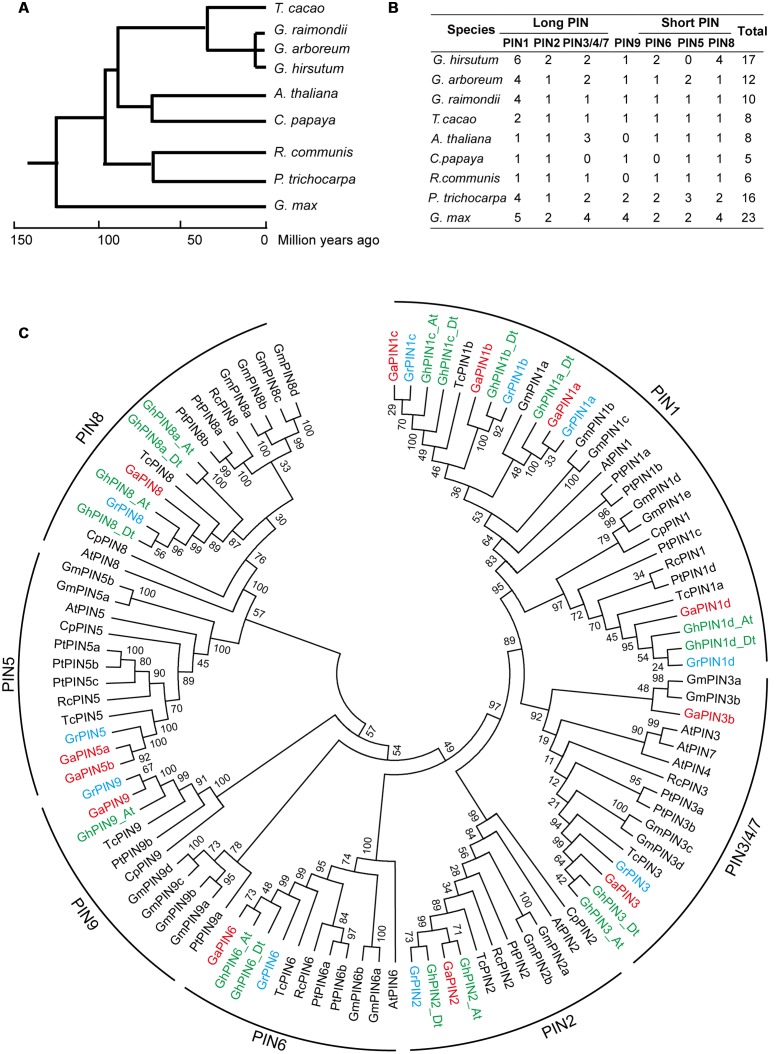
To investigate the evolutionary relationships of PINs among different plant species, nine representative genome-sequenced dicotyledons that exhibited close evolutionary relationships to the three cotton species were used to construct a phylogenetic tree (Figure 1A). The evolutionary analysis showed that the PIN family genes in cotton are divided into long PINs (PIN1, PIN2, PIN3/4/7), short PINs (PIN5, PIN6, PIN8) and PIN9 (Figure 1B). According to the phylogenetic tree, the PIN family proteins can be divided into seven subgroups (Figure 1C). The PIN1 subgroup was most prominent in cotton. In contrast to most of the plant species analyzed, the PIN1 subgroup was extensively expanded in cotton, Populus trichocarpa and Glycine max. The PIN8 subfamily, contributing to auxin homeosatstic and facilitating the male gametophyte development in Arabidopsis (Ding et al., 2012), was extensively expanded in G. hirsutum, which may correspond to its better agronomic. PIN5 subgroup gene, which act antagonistically with PIN8, were missing in allotetraploid cotton, suggesting the loss of the gene occurred during the evolution of the G. hirsutum after polyploidization.
FIGURE 1.
Phylogenetic analysis of PIN auxin transporters of nine representative plant species. (A) Phylogenetic analysis of nine representative species. (B) Number of PIN genes in the nine plant species. (C) Phylogenetic relationships of the PINs. The phylogenetic tree was constructed in MEGA 5.0 software using the neighbor-joining (NJ) method. The seven subfamilies of the PINs are represented by different colors. Numbers on the branches are bootstrap proportions of 1000 replicates. Ga, Gossypium arboretum; Gr, Gossypium raimondii; Gh, Gossypium hirsutum; Pt, Populus trichocarpa; Cp, Carica papaya; At, Arabidopsis thaliana; Rc, Ricinus Communis; Tc, Theobroma cacao; Gm, Glycine max.
Compared with Arabidopsis, eight other plant species contained an additional PIN9 subgroup, which suggests a loss of this PIN in Arabidopsis during the evolutional course. Both PIN4 and PIN7 subgroups were only found in Arabidopsis, but none were found in the other species studied, indicating that these genes may have been acquired in Arabidopsis after it diverged from the common ancestor of the nine plant species used for analysis in the Figure 1A.
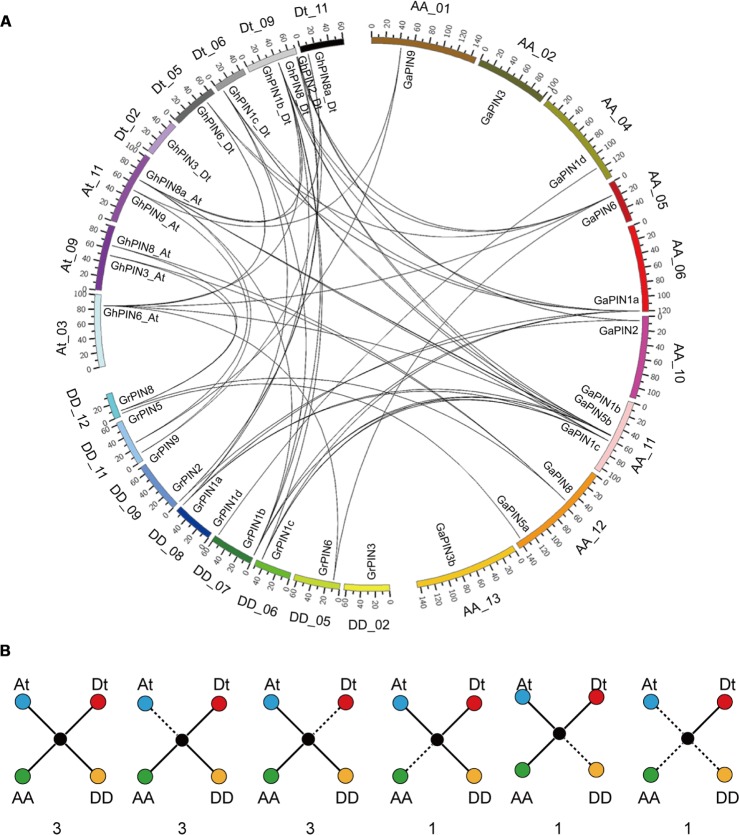
With the A-genome G. arboreum, D-genome G. raimondii, and the allotetraploid genome sequenced, we further explored the evolutionary history of the cotton PIN genes involved in gene duplication and loss. Our results showed that 12 GhPINs were evenly distributed on eight chromosomes, three from the At subgenome, and five from the Dt subgenome (Figure 2A). According to a whole genome analysis of gene duplications, we found that the whole genome duplication (WGD) and dispersed duplication, rather than the proximal duplication and tandem duplication, were the main driving forces of the GhPIN gene expansion (Supplementary Table S2). Specifically, nine GhPINs were produced via WGD, and three genes were produced via dispersed duplication among the analyzed genes.
FIGURE 2.
Chromosomal distribution and evolutionary analysis of the PIN genes. (A) Chromosomal location and analysis of PIN gene duplication. The CIRCOS genome visualization tool was used. Each chromosome number is provided next to the corresponding chromosome. The black lines represent duplicated gene pairs. (B) Scenarios and statistics of gene conservation of the three different genomes. The solid lines indicate observed genes, and the dotted lines indicate lost genes. The number of gene pairs found in the three different genomes that fit the specific model is provided below each graphic.
Then, we analyzed the PIN gene conservation and loss based on the best match and the syntenic gene finder in MCScan. Three PINs were ultra-conserved homologous gene pairs in G. hirsutum. Of the orthologous gene pairs, six were lost from either the At or the Dt subgenome of G. hirsutum and two were lost from either G. arboreum or G. raimondii (Figure 2A). This indicated that the GhPIN genes experienced a higher frequency of genic sequence losses and only retained either the At or Dt homologous genes during the formation of G. hirsutum. Compared with one gene lost in the A genome and one lost in the D genome (Figure 2B), the rate of gene losses was higher in the allotetraploid cotton than in both diploid species. Additionally, one gene was only present in the Dt subgenome (Figure 2B).
Gene Structure and Transmembrane Topology among PIN Proteins
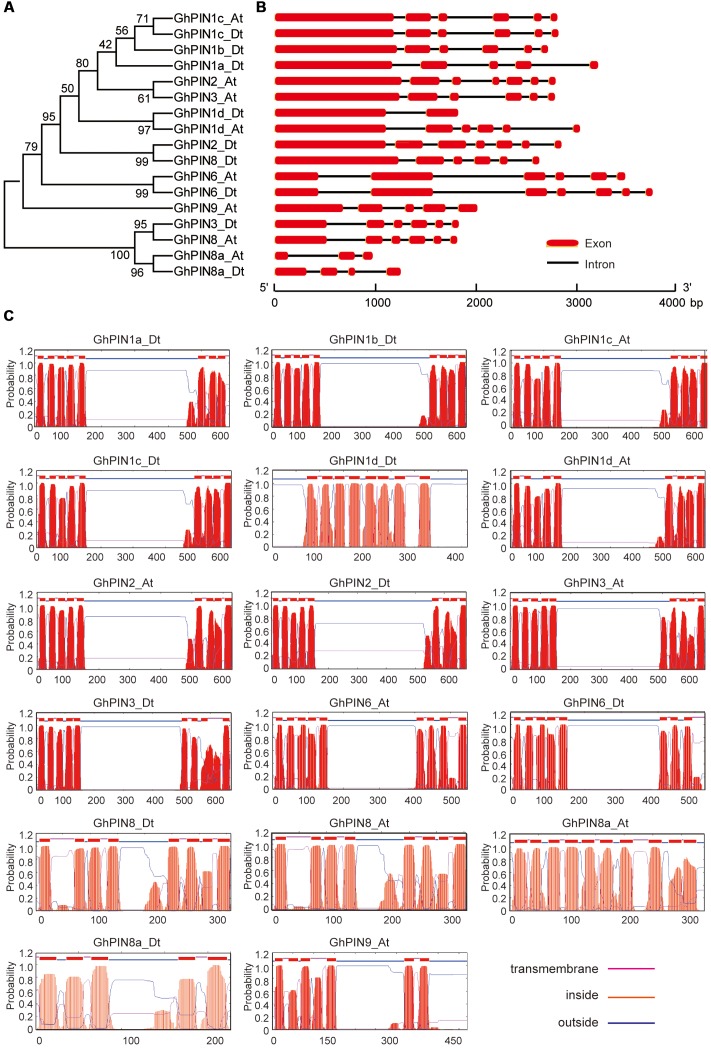
To better understand the similarity and diversity of the gene structures and transmembrane topology of GhPINs, we constructed a phylogenetic tree with the deduced amino acids of GhPINs (Figure 3A) and investigated the exon/intron structures of GhPIN genes. In general, GhPINs possessed at least one intron and the structural patterns were highly conserved between long PINs and short PINs (Figure 3B). Although GhPIN6_At and GhPIN6_Dt genes belong to short PINs, both had five introns, including the two largest introns, in their open reading frame. The predicted transmembrane topology showed that GhPIN proteins, except for GhPIN1d_Dt, had the typical structure with two highly conserved hydrophobicity segments at the N- and C- terminus and a central hydrophilic loop within the two termini (Figure 3C). All GhPIN proteins possessed 8–10 transmembrane segments, except GhPIN8a_Dt, which presented six transmembrane helices. Notably, GhPIN8a_At possessed identical transmembrane segments as the other GhPINs, but these segments were evenly distributed across the entire protein, which resulted in the loss of the central hydrophilic loop.
FIGURE 3.
Phylogenetic relationships, gene structures, and transmembrane topology prediction for the GhPINs. (A) Phylogeny of 17 PIN proteins in Gossypium hirsutum. The phylogenetic tree (left panel) was constructed in MEGA 5.0 using the neighbor-joining (NJ) method with 1000 bootstrap replicates. (B) The gene structures (right panel) were drawn with the Gene Structure Display Server 2.042. The exons and introns are marked with red boxes and black lines, respectively. The scale bar is shown at the bottom. (C) Transmembrane topology prediction of the GhPIN proteins. The transmembrane helices of the GhPIN proteins were predicted using the TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The red peaks indicate the predicted transmembrane helices.
Tissue-Specific Expression Profiles of Cotton PIN Genes
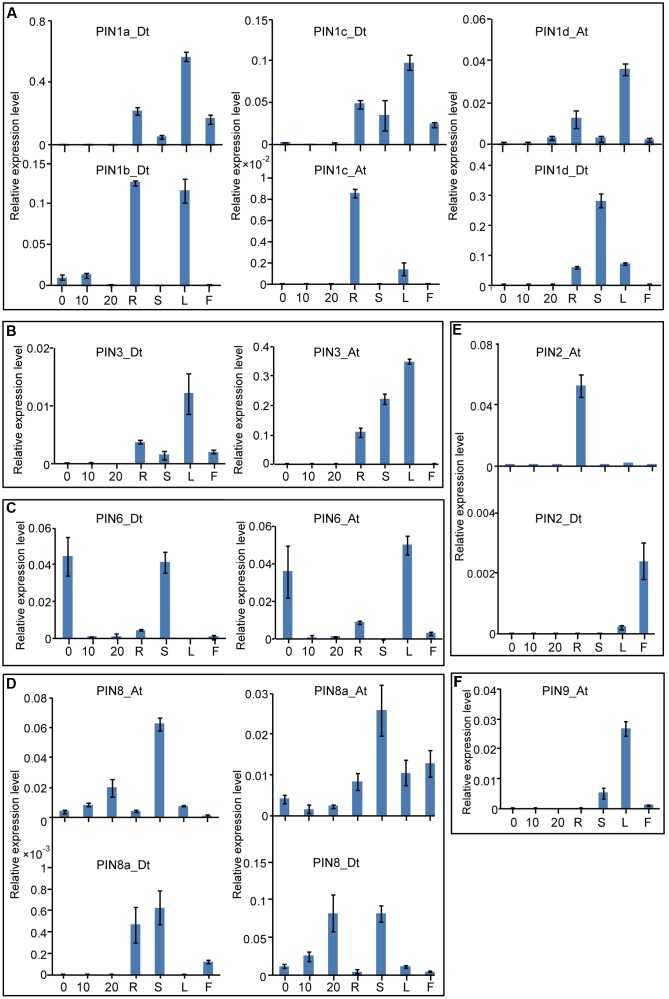
Systematic analyses of gene expression patterns can help us understand the possible physiological functions of the cotton genes. First, we investigated the expression patterns of the GhPIN genes in different tissues, including the root, stem, leaf, flower, and fibers at various developmental stages. As shown in Figure 4, GhPIN transcripts were widely detected and exhibit a tissue-specific expression pattern in cotton. Genes from the PIN1, PIN2, PIN3, and PIN9 subgroups were predominantly expressed in vegetative tissues, but they were barely detected in the fibers, indicating that these genes may play specific roles in regulating the development of vegetative tissues. The transcripts from the PIN6 and PIN8 subgroup genes were abundant in diversified tissues. PIN1c_At from the PIN1 subgroup showed a root-specific expression, whereas the other four genes from the PIN1 subgroup were notably more highly expressed in the leaf (Figure 4A). Two PIN3 subgroup genes, PIN3_Dt and PIN3_At, were expressed in a leaf-preferential manner (Figure 4B). PIN6_Dt and PIN6_At, two genes from the PIN6 subgroup, were highly expressed in the 0 day post-anthesis (DPA) ovule (Figure 4C). Four PIN8 subgroup genes were significantly expressed in the stem (Figure 4D). The transcripts of PIN2_At genes were significantly higher in the root, whereas PIN2_Dt was specifically expressed in the flower (Figure 4E). PIN9_At was specifically expressed in the leaf.
FIGURE 4.
Tissue-specific expression profiles of the GhPIN genes. (A) Three GhPIN genes were expressed significantly higher in the fibers. (B) Three GhPIN genes were expressed significantly higher in the stem. (C) Six GhPIN genes were expressed significantly higher in the leaf. (D) Four GhPIN genes were expressed significantly higher in the root. (E) PIN2a-At was expressed significantly higher in the flower. The qRT-PCR analyses were performed in three biological replicates, and the error bars represent their mean values ± SE. The expression levels are given relative to cotton UBQ7. (F) PIN9-At was expressed significantly higher in the leaf. R, root; S, stem; L, leaf; F, flower. The primers for the qRT-PCR analyses are listed in Supplementary Table S3.
Analysis of the Link between PIN and Fiber Elongation
Fiber length is one of the key agronomic traits that determine cotton yield and quality. First, we analyzed the fiber length in G. hirsutum and its diploid ancestors and found pronounced differences in G. hirsutum, G. arboreum, and G. raimondii (Figure 5A). The fiber length of G. hirsutum was more than 3 cm, whereas G. arboreum and G. raimondii showed a substantial reduction in fiber length (Figure 5B). To reveal the potential clues regarding whether the differential expression of PIN genes, which were generated during the origin and evolution of cotton, influenced the fiber development, we performed qRT-PCR to analyze the expression level of PIN genes during the different growth periods of the fibers from G. hirsutum and G. arboreum. Among the seven PIN genes analyzed, the PIN6 and PIN8 genes that originated from the At subgenome of G. hirsutum, rather than their homologs from the A genome of G. arboreum, were preferentially expressed in the fiber initiation and elongation stages (Figure 5C and Supplementary Figure S2). A comparative analysis of promoter sequences provides insights into the possible regulation mechanisms for the differential expression of PIN6 and PIN8 between G. hirsutum and G. arboreum. Compared with the GaPIN6 promoter, a large DNA fragment deletion was observed in the GhPIN6_At promoter. Additionally, a point mutation was identified in the GaPIN6 promoter, which resulted in the loss of an auxin response element (Figure 5D and Supplementary Figure S3). Although the GhPIN8_At promoter displayed a 98.1% similarity with the GaPIN8 promoter, we observed the loss of a pyrimidine box in the GaPIN8 promoter (Figure 5D and Supplementary Figure S4), which was reported to play an important role in regulating the expression of Ces3–4 gene from the At subgenome (Li F. et al., 2015).
FIGURE 5.
The emergence of differential regulation of PIN expression in the fiber indicates its important role in fiber elongation during the evolution of cotton. (A) Comparisons of mature fibers of G. hirsutum, G. arboreum, and G. raimondii. Bar = 1.5 cm. (B) Final statistics of mature fibers of G. hirsutum, G. arboreum, and G. raimondii. Each data is the mean of three independent experiments with a total of 10 measured samples. The error bars indicate the mean values ± SE. (C) Quantitative RT-PCR analysis of the GaPIN6 and GaPIN8 genes. ∗P < 0.05. (D) Dot plots showing consensus in the promoter regions of GaPIN6 (left) and GaPIN8 (right) and their orthologs from the At subgenome. T, putative TATA box; AuxRE, putative auxin response element; P box, putative pyrimidine box. (E) Quantitative RT-PCR analysis of the PIN genes using the Dt (indicated by blue lines) or D-originated (red lines) copies at the 10 DPA as the template. Three biological replicates were analyzed via qRT-PCR in (C,E), and the error bars represent the mean values ± SE. The expression levels are relative to cotton UBQ7. DPA, day after anthesis. ∗∗∗P < 0.001. (F) Promoter sequence analysis of GrPIN1a and its orthologs from the Dt subgenome. The blue box marks the putative HD-ZIP binding site, and the red color represents the putative ZF-HD binding site.
We further compared the transcripts of the other 10 GhPIN genes with their corresponding orthologs in G. raimondii. In the qRT-PCR analysis, nine PIN genes showed no substantial differences in their transcriptional levels between the two cotton species, and only the expression level of PIN1a was approximately fivefold higher than its ortholog in G. raimondii (Figure 5E). Further promoter scrutiny revealed that several short sequences were missing in the GrPIN1a promoter (indels of 8–16 bp) compared with the GhPIN1a_Dt promoter, which resulted in the loss of a putative HD-ZIP binding site and a putative ZF-HD binding site (Figure 5F). Our results imply that the cis-element variations generated in these gene promoters during evolution may be coupled with the high-level expressions of the GhPIN6_At, GhPIN8_At, and GhPIN1a_Dt genes in allotetraploid cotton.
Increased expressions of PIN genes in longer fiber cells suggest that the auxin transport may promote fiber elongation. Therefore, we needed to confirm whether PINs play important roles in cotton fiber development. We utilized an auxin polar transport inhibitor to block the functions of the PINs and measured the fiber lengths with an ovule culture system in vitro. When NPA, an inhibitor of auxin polar transport, was added to the standard ovule culture media for 13 days, we observed a significant reduction in fiber elongation (Supplementary Figure S5A). Furthermore, the fiber lengths were reduced in a dose-dependent manner and the addition of 80 μM NPA to the culture media almost entirely blocked fiber growth (Supplementary Figure S5B). However, the application of different concentrations of NPA did not affect the final size of the cultured ovules (Supplementary Figure S5C). qRT-PCR analysis showed that the transcription levels of most PIN genes decreased after the application of NPA (Supplementary Figure S5D). These results suggest that the PIN proteins are essential for cotton fiber cell elongation.
The Overexpression of the GhPIN1a_Dt, GhPIN6_At and GhPIN8_At Genes Increase the Density and Lengths of Trichomes in Leaves
To further investigate the functions of the GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At genes identified through their differential expression patterns of the three cotton species, we generated three transgenic lines with GhPIN1a_Dt, GhPIN6_At or GhPIN8_At overexpressed in Arabidopsis that was driven by the constitutive 35S promoter (Figure 6A), and the expression level of GhPIN1a_Dt, GhPIN6_At and GhPIN8_At was 123, 48, and 87% of the Arabidopsis UBQ5 gene, respectively (Supplementary Figure S6). Meanwhile, the three transgenic lines used for phenotypic analysis were with single copy verified by southern blotting experiment (Figure 6B). In contrast to the transgenic lines carrying the empty vector, the three transgenic plants that carried the 35S::GhPIN1a_Dt, 35S::GhPIN6_At or 35S::GhPIN8_At constructs showed a considerable increase of number and length of trichomes, an organ similar to cotton fiber, in the leaves (Figures 6C,D). Furthermore, the calculated density of trichomes per leaf in the 35S::GhPIN1a_Dt, 35S::GhPIN6_At or 35S::GhPIN8_At transgenic lines increased to 250, 220, or 210% of the level in the leaf of transgenic line carrying the empty vector, respectively (Figure 6E). Compared with the transgenic line carrying the empty vector, the 35S::GhPIN1a_Dt, 35S::GhPIN6_At or 35S::GhPIN8_At displayed longer trichomes in the leaves (Figure 6F). These results suggest that the GhPIN1a_Dt, GhPIN6_At and GhPIN8_At genes possess biological activity for cell elongation in vivo, which strongly suggests that the high expression of the three PINs in G. hirsutum contributes to cotton fiber development.
FIGURE 6.
Density and lengths of trichomes in leaves from GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At transgenic plants. (A) PCR verification of GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At transgenic plants. (B) Southern blot analysis of GhPIN1a_Dt, GhPIN6_At and GhPIN8_At transgenic plants. (C) Trichome phenotypes of transgenic plants. The scale bars in the upper and lower panels are 0.5 mm and 100 μm, respectively. (D) Trichome lengths of the transgenic and control plants. The scale bars is 50 μm. (E) Comparison of the densities of trichomes in the leaves from the transgenic and control plants. Each experiment was performed in three biological replicates, and the error bars represent the mean ± SE. ∗∗∗P < 0.001. (F) Comparison of the lengths of trichomes in the leaves from the transgenic and control plants. Each experiment was performed in three biological replicates, and the error bars represent the mean ± SE. ∗∗∗P < 0.001. Vector, the construct without PIN gene as the control.
Discussion
For the first time, we simultaneously identified the PIN genes in three representative types of cotton, allotetraploid cotton G. hirsutum and its diploid ancestors, G. arboreum and G. raimondii, which provides significant insight into the evolution and functions of in PINs in cotton. This study showed that the PIN1 and PIN8 subgroup genes in cotton were significantly expanded (Figure 1B), indicating the functional innovation of these PIN subgroup genes in cotton. Conversely, the PIN5 subgroup genes were only lost in G. hirsutum but were found in G. arboreum and G. raimondii and in other plant species, showing a close evolutionary relationship to Gosspium. This result implies that the PIN5 subgroup genes were present in the common ancestor of these plant species and even in the most recently common ancestors of cotton (ancestral Gossypium) before the divergence of the diploid cottons (Figure 1C). After the polyploidization of the two ancestral diploid cotton types, the PIN5 subgroup gene was gradually lost during the evolution or domestication history of allotetraploid cotton, indicating that the PIN5 gene might be not essential for G. hirsutum development. Although two whole genomic duplication events occurred in cotton, many PIN subgroups contain a similar amount of genes compared with other plant species that have a close evolutional relationship.
The chromosomal distribution of the PIN genes showed that 12 of the 17 genes were evenly located on eight chromosomes (Figure 2A). Among the 12 analyzed genes, nine PINs were produced by WGD and three by dispersed duplication (Supplementary Table S3). This implies that the two types of duplication events are the main contributors of the PIN gene expansion in cotton. More PIN genes were lost in G. hirsutum instead of in the two diploid ancestors, suggesting that a higher rate of gene losses occurred in G. hirsutum. This confirms the previous notion that the allotetraploid genome experienced a higher frequency of genic sequence losses than the diploid genomes during polyploidization (Li F. et al., 2015; Zhang T.Z. et al., 2015; He et al., 2016). Therefore, PINs may play a different role in allotetraploid cotton than in the diploid ancestors.
The transcription profiling of cotton fibers showed that the transcriptomes of cotton fibers are extraordinarily complex, involving 1000s of genes that vary in expression level between G. hirsutum and its diploid ancestors, G. arboreum and G. raimondii (Hovav et al., 2008). Previous work revealed that the ethylene biosynthetic gene, 1-aminocyclopropane-1-carboxylic acid oxidase (ACO), and the cell wall biosynthetic gene, cellulose synthase (Ces), play a vital role in cotton fiber elongation (Li F. et al., 2015). A comparative analysis showed different expression patterns of these genes in the three representative cotton species, resulting in cis-element variations in their promoter, including the indel and nucleotide transitions, which generate new transcriptional factor binding sites. The analysis of PINs in cotton showed that, apart from the genes concerned with ethylene and cell wall synthesis, the elevated expression patterns of PIN genes also contribute to the cotton fiber elongation in G. hirsutum (Figure 5), especially, PIN1a, PIN6, and PIN8. These are driven by the promoter with new cis-elements that were acquired during evolution and domestication. These results suggest that the differential expression pattern of these genes that are related to the fiber elongation, which were derived from the force of the evolution or domestication, may be a common phenomenon and facilitate the emergence of fibers in cotton.
No fiber was found in G. raimondii, and the fiber length in G. arboreum was only 1.3–1.5 cm. However, in G. hirsutum, the two genomes reunited in a common nucleus via allopolyploidization, resulting in its fiber length of more than 3 cm. Our results showed that in the three functional PINs identified, the G. hirsutum PIN1a originated from the Dt subgenome, whereas PIN6 and PIN8 originated from the At subgenome. This suggests that allopolyploidization facilitates the acquisition of both advantages of the two diploid cotton types, which contributes to fiber initiation and elongation in G. hirsutum.
Auxin is known to play essential roles in enhancing cotton fiber cell initiation (Zhang et al., 2011), which also rely on the auxin transporters. On that basis, we further showed that PIN genes may also have a positive effect on the fiber elongation in addition to initiation. This suggests PINs play bifunctional roles in fiber development. Our work provides evolutionary and functional insights into the roles of PINs in cotton fiber development from both evolutionary and functional aspects and thus may help to improve both cotton fiber yields and the quality of future research and development efforts for improving cotton.
Author Contributions
YZ and GX conceived all of the experiments and analyzed the data. PH, ZY, and GH performed the experiments, drafted the manuscript, and prepared the figures. GX and JY wrote and reviewed the manuscript. LW and HX analyzed the data. PZ and CP contributed to the manuscript preparation. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (Grant No. GK201603066, Grant No. GK201403007) and the State Key Laboratory of Cotton Biology Open Fund (Grant No. CB2016A02).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00461/full#supplementary-material
References
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L. R., Wells D. M., Fozard J. A., Ghetiu T., French A. P., Pound M. P., et al. (2014). Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26 862–875. 10.1105/tpc.113.119495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E., Kubes M., Rolcik J., Beziat C., Pencik A., Wang B., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485 119–122. 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Chen X., Grandont L., Li H., Hauschild R., Paque S., Abuzeineh A., et al. (2014). Inhibitor of cell expansion by rapid ABP1-mediated auxin effect on microtubles. Nature 516 90–93. 10.1038/nature13889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro S., Golin S., Hardtke C. S., Lo-Schiavo F., Zottini M. (2015). The co-chaperone p23 controls root development through the modulation of auxin distribution in the Arabidopsis root meristem. J. Exp. Bot. 66 5113–5122. 10.1093/jxb/erv330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Wang B., Moreno I., Duplakova N., Simon S., Carraro N., et al. (2012). ER localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3:941 10.1038/ncomms1941 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J. G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M. G., Friml J., Shishkova S., et al. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. U.S.A. 105 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Vosolsobě S., Feraru M. I., Petrášek J., Kleine-Vehn J. (2012). Evolution and structural diversification of PILS putative auxin carriers in plants. Front. Plant Sci. 3:227 10.3389/fpls.2012.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C., Farinati S., Varotto S. (2012). The maize PIN gene family of auxin transporters. Front. Plant Sci. 3:16 10.3389/fpls.2012.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. (2003). Auxin transport-shaping the plant. Curr. Opin. Plant Biol. 6 7–12. [DOI] [PubMed] [Google Scholar]
- Friml J., Benkova E., Blilou I., Wisniewska J., Hamann T., Ljung K., et al. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Guo X. L., Qin Q. Q., Yan J., Niu Y. L., Huang B. Y., Guan L. P., et al. (2015). TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiol. 167 1058–1075. 10.1104/pp.114.249904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets M. E., Offringa R. (2014). PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytol. 203 362–377. 10.1111/nph.12831 [DOI] [PubMed] [Google Scholar]
- He Q. L., Jones D. C., Li W., Xie F. L., Ma J., Sun R. R., et al. (2016). Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 6:22980 10.1038/srep22980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R., Udall J. A., Chaudhary B., Rapp R., Flagel L., Wendel J. F., et al. (2008). Partitioned expression of duplicated gene during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. U.S.A. 105 6191–6195. 10.1073/pnas.0711569105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J. P., Guo A. Y., Zhang H., Luo J. C., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S. J., Lu Y. C., Feng J. X., Wei G., Li J., Shi Y. H., et al. (2003). Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31 2534–2543. 10.1093/nar/gki702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Li Q., Xiao G. H., Zhu Y. X. (2013). Using genome-reference expressed sequence tag assembly to analyze the origin and expression patterns of Gossypium hirsutum transcripts. J. Integr. Plant Biol. 55 576–585. 10.1111/jipb.12066 [DOI] [PubMed] [Google Scholar]
- Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33 524–530. 10.1038/nbt.3208 [DOI] [PubMed] [Google Scholar]
- Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46 567–572. 10.1038/ng.2987 [DOI] [PubMed] [Google Scholar]
- Li K., Kamiya T., Fujiwara T. (2015). Differential roles of PIN1 and PIN2 in root meristem maintenance under low-B conditions in Arabidopsis thaliana. Plant Cell Physiol. 56 1205–1214. 10.1093/pcp/pcv047 [DOI] [PubMed] [Google Scholar]
- Liu G. J., Xiao G. H., Liu N. J., Liu D., Chen P. S., Qin Y. M., et al. (2015). Targeted lipidomics studies reveal that linolenic acid promotes cotton fiber elongation by activating phosphatidylinositol and phosphatidylinositol monophosphate biosynthesis. Mol. Plant 8 911–921. 10.1016/j.molp.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Lu G. W., Coneva V., Casaretto J., Ying S., Mahmood K., Liu F., et al. (2015). OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83 913–925. 10.1111/tpj.12939 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J., Skupa P., Bailly A., Hoyerova K., Krecek P., Bielach A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459 1136–1140. 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Omelyanchuk N. A., Kovrizhnykh V. V., Oshchepkova E. A., Pasternak T., Palme K., Mironoca V. V. (2016). A detailed expression map of the PIN1 auxin transporter in Arabidopsis thaliana root. BMC Plant Biol. 16:5 10.1186/s12870-015-0685-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., Jin D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers. Nature 492 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- Péret B., Swarup K., Ferguson A., Seth M., Yang Y. D., Dhondt S., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24 2874–2885. 10.1105/tpc.112.097766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Mravec J., Bouchard R., Blakeslee J. J., Abas M., Seifertova D., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918. 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Rakusova H., Fendrych M., Friml J. (2015). Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Curr. Opin. Plant Biol. 23 116–123. 10.1016/j.pbi.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Remy E., Cabrito T. R., Baster P., Batista R. A., Teixeira M. C., Friml J., et al. (2013). A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25 901–926. 10.1105/tpc.113.110353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H. S., Grones P., Stepanova A. N., Robles L. M., Lokerse A. S., Alonso J. M., et al. (2013). Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 23 2506–2512. 10.1242/dev.115832 [DOI] [PubMed] [Google Scholar]
- Roumeliotis E., Kloosterman B., Ortwijn M., Visser R., Bachem C. (2013). The PIN family of proteins in potato and their putative role in tuberization. Front. Plant Sci. 4:524 10.3389/fpls.2013.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row J. H., Topping J. F., Liu J., Lindsey K. (2016). Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 211 225–239. 10.1111/nph.13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. J., Bai Y. H., Wang S. K., Zhang S. N., Wu Y. R., Chen M., et al. (2010). Expression profile of PIN, AUX/ LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 277 2954–2969. 10.1111/j.1742-4658.2010.07706.x [DOI] [PubMed] [Google Scholar]
- Shi Y. H., Zhu S. W., Mao X. Z., Feng J. X., Qin Y. M., Zhang L., et al. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18 651–664. 10.1105/tpc.105.040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Skupa P., Viaene T., Zwiewka M., Tejos R., Klíma P., et al. (2016). PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 211 65–74. 10.1111/nph.14019 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Z., Yang K. Z., Zou J. J., Zhu L. L., Xie Z. D., Morita M. T., et al. (2015). Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 6:8822 10.1038/ncomms9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. R., Hu H., Wang G. H., Li J., Chen J. Y., Wu P. (2009). Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol. Plant 2 823–831. 10.1093/mp/ssp023 [DOI] [PubMed] [Google Scholar]
- Wang K., Wang Z., Li F., Ye W., Wang J., Song G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44 1098–1103. 10.1038/ng.2371 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W. Z., Song L. F., Zou J. J., Su Z., Wu W. H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148 1201–1211. 10.1104/pp.108.126375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Q., Chai C. L., Valliyodan B., Maupin C., Annen B., Nguyen H. T. (2015). Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genomics 16:951 10.1186/s12864-015-2149-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B. C., Isono E., Richter R., Zourelidou M., Schwechheimer C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23 2184–2195. 10.1105/tpc.111.086355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W. Y., Gong X. M., Yang Q. Y., Yu H., Liou Y. C. (2016). Pin1At regulates PIN1 polar localization and root gravitropism. Nat. Commun. 7:10430 10.1038/ncomms10430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., Wang K., Huang G., Zhu Y. (2016). Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integr. Plant Biol. 58 577–589. 10.1111/jipb.12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li C., Zhao L., Gao S., Lu J., Zhao M., et al. (2015). The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27 1670–1680. 10.1105/tpc.15.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue R. Q., Tie S. G., Sun T., Zhang L., Yang Y. J., Qi J. S., et al. (2015). Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 10:e0118751 10.1371/journal.pone.0118751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zheng X., Song S., Zeng Q., Hou L., Li D., et al. (2011). Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 29 453–458. 10.1038/nbt.1843 [DOI] [PubMed] [Google Scholar]
- Zhang T. Z., Hu Y., Jiang W. K., Fang L., Guan X. Y., Chen J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc.TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 531–537. 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Z., Jiao Y., Liu Z. H., Zhu Y. X. (2015). ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun. 6:6003 10.1038/ncomms7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61 49–64. 10.1146/annurev-arplant-042809-112308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C. S., Lu C. R., Shang H. H., Jing X. R., Cheng H. L., Zhang Y. P., et al. (2013). Genome-wide analysis of the Sus gene family in cotton. J. Integr. Plant Biol. 55 643–653. 10.1111/jipb.12068 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.