Abstract
Soluble starch synthase (SS) is one of the key enzymes involved in amylopectin biosynthesis in plants. However, no information is currently available about this gene family in the important fruit crop banana. Herein, we characterized the function of MaSSIII-1 in amylopectin metabolism of banana fruit and described the putative role of the other MaSS family members. Firstly, starch granules, starch and amylopectin content were found to increase during banana fruit development, but decline during storage. The SS activity started to increase later than amylopectin and starch content. Secondly, four putative SS genes were cloned and characterized from banana fruit. Among them, MaSSIII-1 showed the highest expression in banana pulp during fruit development at transcriptional levels. Further Western blot analysis suggested that the protein was gradually increased during banana fruit development, but drastically reduced during storage. This expression pattern was highly consistent with changes in starch granules, amylopectin content, and SS activity at the late phase of banana fruit development. Lastly, overexpression of MaSSIII-1 in tomato plants distinctly changed the morphology of starch granules and significantly increased the total starch accumulation, amylopectin content, and SS activity at mature-green stage in comparison to wild-type. The findings demonstrated that MaSSIII-1 is a key gene expressed in banana fruit and responsible for the active amylopectin biosynthesis, this is the first report in a fresh fruit species. Such a finding may enable the development of molecular markers for banana breeding and genetic improvement of nutritional value and functional properties of banana fruit.
Keywords: banana (Musa acuminata L.), soluble starch synthase, amylopectin metabolism, expression analysis, SSIII-1 function
Introduction
Starch is the most widespread carbohydrate storage molecule in plants and plays a vital role in human nutrition, food industry and chemical manufacturing. The physicochemical properties of starch are strongly affected by its two key components, amylose and amylopectin (Zeeman et al., 2010; Bischof et al., 2013). In higher plants, there are at least six classes of enzymes that are involved in amylose and amylopectin biosynthesis: ADP-Glc pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), soluble starch synthase (SS), starch branching enzyme (SBE), starch debranching enzyme (DBE), and starch phosphorylase (SP) (Fujita et al., 2008; Szydlowski et al., 2009; Jeon et al., 2010; Subasinghe et al., 2014).
Starch synthase uses ADP-glucose for chain elongation via α-1,4-glycosidic linkages, and directly catalyzes the amylopectin biosynthesis (Szydlowski et al., 2009, 2011). A plant typically carries at least four SS subfamilies, termed as SSI, II, III, and IV, respectively (Dian et al., 2005). Comparison of the deduced amino acid sequences showed that these four subfamilies are highly similar in a span of approximately 450 amino acid residues in the C-terminus that comprises the catalytic and starch-binding domains (Schwarte et al., 2013). The sizes of SSI, II, and III proteins were estimated to be approximately 67, 81, and 112 kDa, respectively, in Arabidopsis leaves (Delvallé et al., 2005; Zhang et al., 2008; Gámez-Arjona et al., 2014). SS gene expression was recently profiled in a numbers of plants including rice (Crofts et al., 2015), maize (Liu et al., 2015; Huang et al., 2016), potato (Edwards et al., 1999), Arabidopsis (Schwarte et al., 2013), wheat and taro (Li et al., 2000; Lin and Jeang, 2005). However, different SSs may predominate in storage organs of different species e.g., SSI in rice endosperm, SSII in wheat (Li et al., 2000; Crofts et al., 2015). In common wheat, SSIII expression was detected from very early to the middle stages of endosperm development (Li et al., 2000). Rice SSIII-2 and SSIV-1 showed the highest expression level at the middle and late development stages of rice endosperm, respectively (Dian et al., 2005).
Amylopectin chain length distribution and amylopectin/ amylase ratio affected starch functionality (Dian et al., 2005; Zhang et al., 2005). Biochemical and gene overexpression experiments revealed that SSI, II, and III are involved in the elongation of short (dp 8–12) (Fujita et al., 2008), medium (dp 13–25), and long (dp > 30) starch chains (Szydlowski et al., 2011; Zhu et al., 2014), respectively. SSIV is closely related to SSIII; both have similar structures (Leterrier et al., 2008). Further reverse genetic studies showed that impaired SSI expression was reported to result in structurally altered amylopectin in Arabidopsis leaves (Delvallé et al., 2005) and rice endosperm (Takemoto-Kuno et al., 2006; Fujita et al., 2008). Inhibition of SSII gene expression gave rise to lower pasting temperature as the result of alterations in amylopectin structure (Edwards et al., 1999). Loss of SSIII expression reduced the proportion of amylopectin with very long chains and affected the amylopectin/amylase ratio in maize (Zhu et al., 2014), Arabidopsis (Zhang et al., 2005), and rice (Dian et al., 2005). A defective mutant of SSIV displayed a severely compromised growth phenotype with fewer but larger starch granules within the plastid in Arabidopsis (Roldán et al., 2007). However, Zhang et al. (2008) reported that some SSs, such as SSII and SSIII, may overlap in amylopectin biosynthesis in Arabidopsis. In addition, GBSSI from other SS isoforms also contributed to amylopectin synthesis in rice and Chlamydomonas reinhardtii (Ral et al., 2006; Hanashiro et al., 2008). These evidences suggested the important roles of the SS genes in regulating plant amylopectin metabolism.
Banana (Musa spp.) is not only one of the most highly consumed fruits in the world but also widely used as a staple food in the tropical and subtropical regions (D’Hont et al., 2012). The typical starch content of a green dessert banana fruit accounts for 20∼25% of fresh weight or 60∼75% of dry weight (DW), and the starch granules are relatively large (8∼48 μm in diameter) compared to those of cereals (Hubbard et al., 1990; de Barros Mesquita et al., 2016). Hence banana is also a potential excellent model plant for studying fresh fruit starch metabolism. Current research on banana starch has mainly focused on granule structure (Low et al., 2015) and antioxidant capacities (Sarawong et al., 2014), as well as physicochemical properties of starch (Zhang and Hamaker, 2012; Utrilla-Coello et al., 2014). Several key genes involved in starch biosynthesis or conversion of starch to sucrose have been isolated and characterized in banana, including MaGBSSI (Miao et al., 2014), DBE (Bierhals et al., 2004), SUCROSE PHOSPHATE SYNTHASE (SPS), SUCROSE SYNTHASE (SuSy), and INVERTASE (Hubbard et al., 1990). However, to our knowledge, the function of SS genes has not been characterized in banana.
Amylopectin is the major component of immature banana fruit (Torre-Gutiérrez et al., 2008), which has a number of remarkable features, including higher retrogradation and lower pasting temperature (Sarawong et al., 2014), relative to those in rice (Syahariza et al., 2013), maize (Huang et al., 2016), and potato (Wikman et al., 2014). Due to the important role of SS in amylopectin metabolism, there is a need to investigate the function of SS in banana. In this study, we revealed that MaSSIII-1 is a key gene expressed in banana fruit and responsible for the active amylopectin biosynthesis by expression, enzyme activity, and functional analyses. This result could be used in the genetic manipulation of banana fruit for genetic improvement of its nutritional values as well as value-added industrial applications, such as high starch foods, higher retrogradation and lower pasting temperature raw processing materials.
Materials and Methods
Plant Materials and Treatments
Banana (M. acuminata L. AAA group, cv. ‘Dwarf Cavendish’; ITC 0002) seedlings were obtained from the banana tissue culture center (Chinese Academy of Tropical Agricultural Sciences, Danzhou, China) and were grown at 28°C with 70% humidity, 200 μmol⋅m-2⋅s-1 light intensity, and long day condition (16 h light/8 h dark cycle). When banana seedlings produced five leaves, they were planted in the Institute of Tropical Bioscience and Biotechnology banana plantation (Chengmai, Hainan, 20N, 110E) until harvest. Roots, stems, leaves, bracts, flowers, peels, and pulps at 60 days after emergence from the pseudostem (DAF) were collected separately using tweezers, frozen immediately in liquid nitrogen, and stored at -80°C until expression analysis in different tissues. For each biological replicate, two banana hands having a similar developmental stage from two plants were selected and six fingers from the hands were obtained. Banana pulps at 0, 10, 20, 30, 40, 50, and 60 DAF were collected, immediately frozen in liquid nitrogen, and stored at -80°C until starch/amylopectin quantification, mRNA and protein expression analysis.
Banana hands (60 DAF) were separated into individual fingers representing the same developmental stage. For natural ripening treatment, banana samples were kept at 22°C and allowed to ripen in open air. In accordance with the previously published banana ripening stages (Miao et al., 2014), fruits stored for 0, 5, 10, 15, 20, 25, and 30 days period of post-harvest (DPH) were frozen in liquid nitrogen and stored at -80°C until starch/amylopectin quantification, mRNA and protein expression analysis.
Scanning Electron Microscopy (SEM) Observation
Pulp samples were fixed in stubs using double-faced tape and coated with a 10 nm thick platinum layer using a Bal-tec MED 020 Coating system (Kettleshulme, UK) before analysis with a FEI Quanta 600 FEG Scanning Electron Microscope (FEI Company, Hillsboro, OR, USA). SEM observations were performed using the secondary electron mode operating at 15 kV.
Determination of Total Starch Content, Amylopectin Content, and SS Activity
Banana pulp was immersed in 0.5% (w/v) sodium bisulfite solution for 10 min to prevent browning, and then dried at 40°C for 20∼24 h. The dried pulp was milled to powder and suspended in 5 mL 80% (w/v) Ca(NO3)2, placed in a boiling water bath for 10 min, and centrifuged for 4 min at low speed (3,800 g). The supernatant was then transferred to a 20 mL volumetric flask. The total starch, amylose, and amylopectin contents of the extract were determined following Yang et al. (1992) and Miao et al. (2014). The amylopectin content was calculated by subtracting the amylose content from the total starch content. Enzymatic analysis of SS activity was carried out following Nakamura et al. (1989). A unit represents increasing 0.01 OD value per min at 340 nm.
Cloning and Sequence Analysis of Genes Encoding SS in Banana Fruit
RNA purification and cDNA synthesis were performed as previously described by Li et al. (2015). To obtain full-length cDNAs, sequences of four SS family members, includingSSI (GSMUA_Achr3G03290_00), SSII (GSMUA_Achr6G23190_001), SSIII-1 (GSMUA_Achr11G18570_001), and SSIII-2 (GSMUA_Achr5G00700_001), were retrieved from the banana DH-Pang (AA group) genome sequence database1. Four primer pairs (Supplementary Table S1) were designed based on these sequences. The SS coding sequences were submitted to BLAST analysis to recover their corresponding genomic DNA sequences. Exon lengths were calculated by alignment of genomic DNA sequences with cDNA sequences, and introns were determined according to the “GC-AG” rule (Miao et al., 2014). The deduced amino acid sequences were aligned using Clustal W, and a phylogenetic tree was constructed based on the neighbor-joining (NJ) method with a Kimura 2-parameter model using MEGA5.0 software (Arizona State University, Tempe, AZ, USA). The numerical value for each interior branch is the percent bootstrap value calculated from 1,000 replicates.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Specific primer pairs were designed using Primer 5.0 software. Primers that had high specificity and efficiency on the basis of melting curve analysis were used to conduct quantification analysis (Supplementary Table S1). Moreover, PCR products were sequenced to confirm the specificity of primer pairs. Amplification efficiencies of primer pairs ranged from 0.9 to 1.1. The levels of MaSS and SISSIII-1 expression were quantified by qRT-PCR using an iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) with the SYBR ExScript RT-PCR Kit (TaKaRa, Dalian, China). ACTIN or GAPDH that were verified to be constitutive in expression and hence suitable to be used as internal controls were used as reference genes to normalize transcriptional levels of each MaSS gene and SISSIII-1 (Supplementary Table S1). Relative expression levels of four MaSS genes were analyzed in three technical replicates and calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Each sample contains three biological replicates.
Detection of MaSSIII-1 Protein Levels by Western Blot Analysis
Pulp samples (0.5 g) were homogenized with a set of mortar and pestle on ice in an equal volume of solution, which consisted of 50 mM HEPES-NaOH (pH 7.4), 2 mM MgCl2, 50 mM β-mercaptoethanol, and 12.5% (v/v) glycerol (Nishi et al., 2001). A 20 μL aliquot of the supernatant was incubated with 36 μL of solution I consisting of 50 mM HEPES-NaOH (pH 7.4), 1.6 mM adenosine diphosphate glucose (ADPG), 0.7 mg amylopectin, and 15 mM DL-dithiothreitol (DTT) for 20 min at 30°C. Following the reaction termination at 100°C for 30 s, 20 μL solution II was added, which consisted of 50 mM HEPES-NaOH, 4 mM phosphoenolpyruvate (PEP), 200 mM KCl, 10 mM MgCl2, and 1.2 U pyruvate kinase, and incubation on ice for 5 min. The reaction was incubated at 30°C for 20 min before it was terminated at 100°C for 30 s. A 60 μL aliquot of the supernatant was mixed with 43 μL of solution III (50 mM HEPES-NaOH, 10 mM glucose, 20 mM MgCl2, 2 mM nicotinamide adenine dinucleotide phosphate, 1.4 U hexokinase, 0.35 U glucose-6-phosphate dehydrogenase) and incubated at 30°C for 10 min. The supernatant (SS enzyme solution) was used to Western blot analysis.
For Western blot analysis, 30 μL of the SS enzyme solution from banana pulps at different developmental stages extracted by the above methods was loaded on each lane and separated on a 12% polyacrylamide gel. Proteins were then transferred onto HybondTM-N+ membranes (Amersham Biosciences, Buckinghamshire, UK). Membranes were probed with rabbit anti-MaSSIII-1 polyclonal antibody (Abmart, Shanghai, China) and banana actin antibody (control) in 1:1,000 dilution in PBS-Tween 20 plus 3% BSA, respectively, followed by alkaline phosphatase-conjugated anti-rabbit IgG secondary antibody (Sigma, St. Louis, MO, USA) in 1:1,000 dilution. Positive signals on the membranes were detected with a 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (BCIP/NBT) solution (Amresco, Solon, OH, USA).
Plant Transformation, Generation and Southern Blot Analysis of MaSSIII-1 Transgenic Plants
The entire MaSSIII-1 coding region was inserted into the pCAMBIA-1302 vector under the transcriptional control of CaMV 35S promoter following a double digestion with Nco I and Spe I. The pCAMBIA-1302-MaSSIII-1 was transferred into A. tumefaciens strain LBA4404. Transgenic tomato (Solanum lycopersicum L.) plants were generated using the Agrobacterium-mediated transformation as previously described (Arshad et al., 2014). Kanamycin-resistant transgenic tomato lines were selected and the transgene integration was determined by Southern blot analysis. Genomic DNA (10 μg per sample) from transgenic tomato leaves was isolated using a CTAB method (Li et al., 2015) and digested with EcoR I overnight at 37°C, separated on a 0.8% (w/v) agarose gel and transferred onto Hybond-N+ nylon membranes (Hybond N+, Amersham, UK) (Li et al., 2015). Probes were prepared from the PCR product amplified using the primers (5′-gagagagaagatggtggaatctat-3′ and 5′-aggagcactagaccagtcatgac-3′) and labeled with DIG-dUTP according to the manufacturer’s instructions (Roche Applied Science, Mannheim, Germany). Fruits from two single-copy transgenic plants L4 and L11 were used for expression analysis and functional investigation of MaSSIII-1 in comparison to wild-type.
Statistical Analysis
Three biological replicates were performed for each sample, unless specified otherwise. Statistical analyses were performed using Microsoft Excel and SPSS (Chicago, IL, USA). Analysis of variance was used to compare the statistical difference based on Student’s t-tests, at significant levels of p < 0.05 (∗), and p < 0.01 (∗∗).
Results
Change in Starch Granules, Total Starch Content, Amylopectin Content, and SS Enzyme Activity at Different Developmental and Storage Stages of Banana Fruit
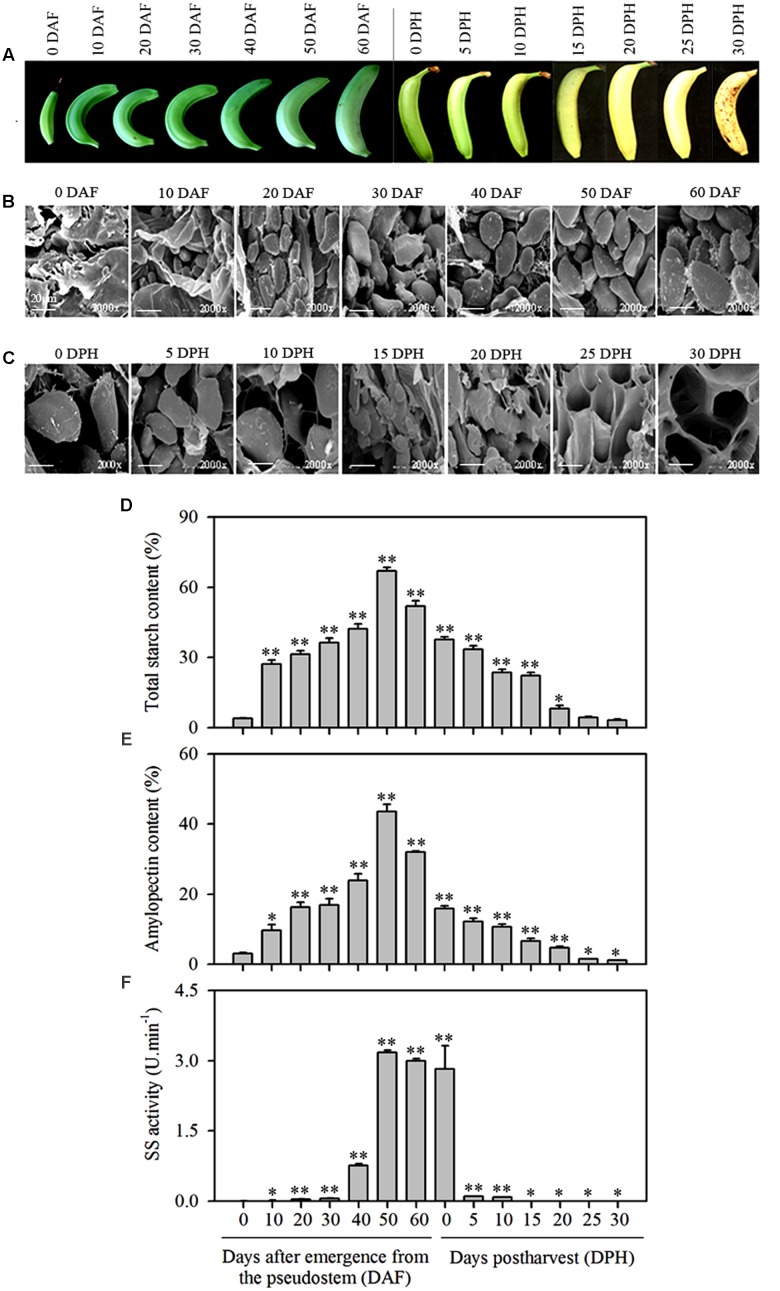
During banana fruit development the fruit size gradually increases and it becomes curled in a crescent shape as fruit develops (Figure 1A). As observed using SEM, starch granules in banana pulp were barely detectable at the initial sampling time, i.e., 0 DAF, but the oval-shaped starch granules become clearly visible and were significantly increased in size as the fruit developed from 10 (6.0∼8.2 μm) to 60 DAF (28.0∼35.7 μm). Morphological comparison of starch granules during this period of time indicated that the shape and number of starch granules remained consistent, but the size was significantly increased and maximized at 60 DAF (Figure 1B).
FIGURE 1.
Changes in starch granules, total starch content, amylopectin content, and SS activity in banana pulp at different stages of development and during storage. (A) Banana fruit at different development stages (0, 10, 20, 30, 40, 50, and 60 DAF) and following storage for varying amounts of time (0, 5, 10, 15, 20, 25, and 30 DPH), (B) SEM of starch granules in pulp during banana fruit development, (C) SEM of starch granules in pulp during banana fruit storage, (D) Total starch content in banana pulp at different stages of development and during storage, (E) Amylopectin content in banana pulp at different stages of development and during storage, (F) SS activity in banana pulp at different stages of development and during storage. DAF: days after emergence from the pseudostem; DPH: days of post-harvest. The vertical bars represent the mean ± SD of three replicates. Asterisks indicate significant difference from 0 DAF and 0 DPH vs. the following days (∗p < 0.05; ∗∗p < 0.01). Scale bar = 20 μm.
Starch granules in mature banana pulp were also observed using SEM at the 30 DPH during storage at 22°C, at 5 days’ intervals. Compared to the first time point (0 DPH), the quantity of starch granules per unit volume decreased rapidly over time. After 15 DPH of storage, starch granules were hardly detectable (Figure 1C).
The total starch content and amylopectin content in pulp increased as fruit developed and peaked at 50 DAF (these two polymers reached to 67 and 44% DW, respectively) before decline at the 60 DAF time point and continued to decrease gradually during storage (Figures 1D,E), implying that after maturity starch and amylopectin may have experienced a rapid degradation process. However, the SS enzyme activity increased later than starch and amylopectin accumulation at maturity and had an abrupt decline at the very beginning of storage (Figure 1F).
Nucleotide Sequence Characteristics, Chromosomal Localization, and Phylogenetic Analysis of Banana MaSS Genes
Full-length cDNAs encoding MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 were 2,076 bp, 1,851 bp, 2,397 bp, and 3,267 bp, respectively. All these sequences were deposited at GenBank and their accession numbers were listed in Figure 2. BLAST analysis against the banana DH-Pang (AA group) genome sequence database2 revealed that MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 are located on chromosome 3, 6, 11, and 5, respectively (Supplementary Figure S1). The sequence analysis also revealed markedly different primary structures as MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 contain 25, 6, 5, and 9 exons, respectively. The stop codon usage is also different as TAA was used by MaSSI and MaSSIII-1, but TGA was used by MaSSII and MaSSIII-2 (Supplementary Figure S1).
FIGURE 2.
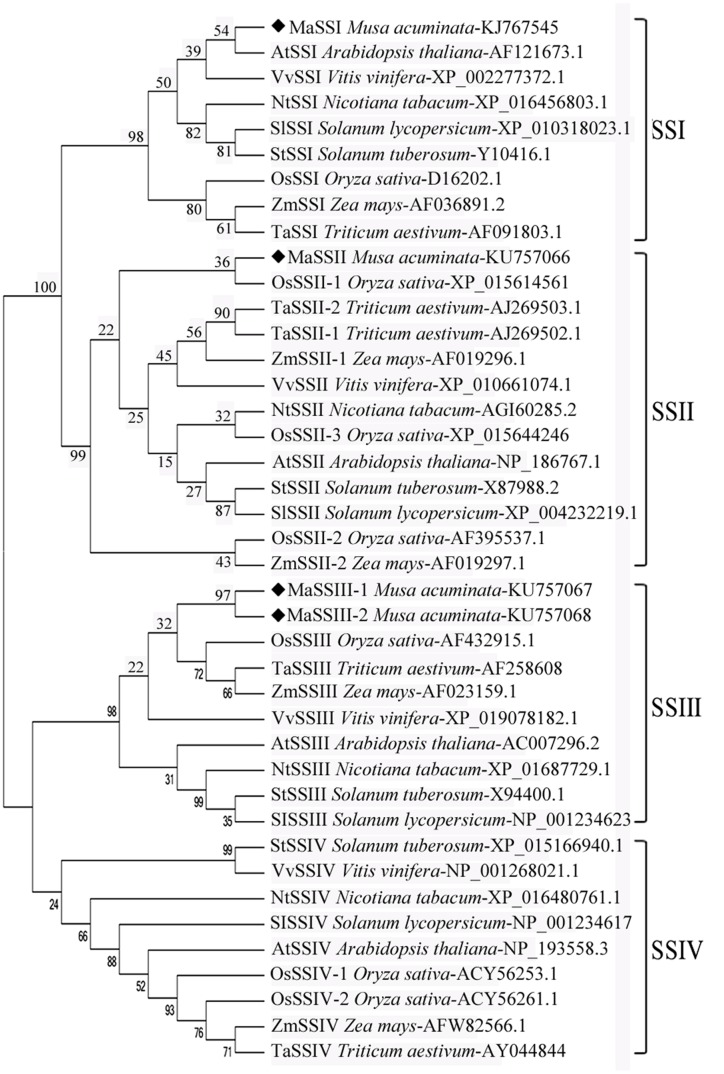
Phylogeny of MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 amino acid sequences in relation to other known SS sequences. Bootstrapping was performed with 1000 replicates. The numbers indicated for each clade represent bootstrap support values given as percentages. Scale bar represents 0.2 substitutions per amino acid position.
The deduced amino acid sequences of the MaSSI and MaSSII shared three conserved regions, referred to as Domain I, II, and III (Supplementary Table S2), as previously identified in SS enzymes from amaranth (Park et al., 2012). The MaSSIII-1 and MaSSIII-2 amino acid sequences contained four distinct regions: a transit peptide region, a variable repeat region, a SSIII specific region, and a C-terminal catalytic domain (Supplementary Table S3), as previously identified in SSIII enzymes from wheat (Li et al., 2000). According to pI/MW software analysis3, the predicted molecular weights of the MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 proteins were 76.69, 69.85, 90.69, and 119.26 kDa, respectively, and their theoretical pIs were 5.24, 5.36, 6.44, and 8.21, respectively.
To elucidate phylogenetic relationship of banana MaSS genes, the four deduced polypeptide sequences were aligned with orthologous SS sequences and a NJ tree was constructed (Figure 2). MaSSIII-1 and MaSSIII-2 are aligned next to each other, forming a side branch in association with other SSIII sequences from monocot species. Similarly, MaSSI and MaSSII are clustered together with their respective orthologs from other species. This may also indicate that the divergence of the SSI, SSII and SSIII occurred prior to the speciation of these plant species, but the separation of SSIII-1 and SSIII-2 might be of a more recent history likely through a gene duplication and subsequent divergence.
Spatially and Temporally Differential Expression of Four MaSS Genes
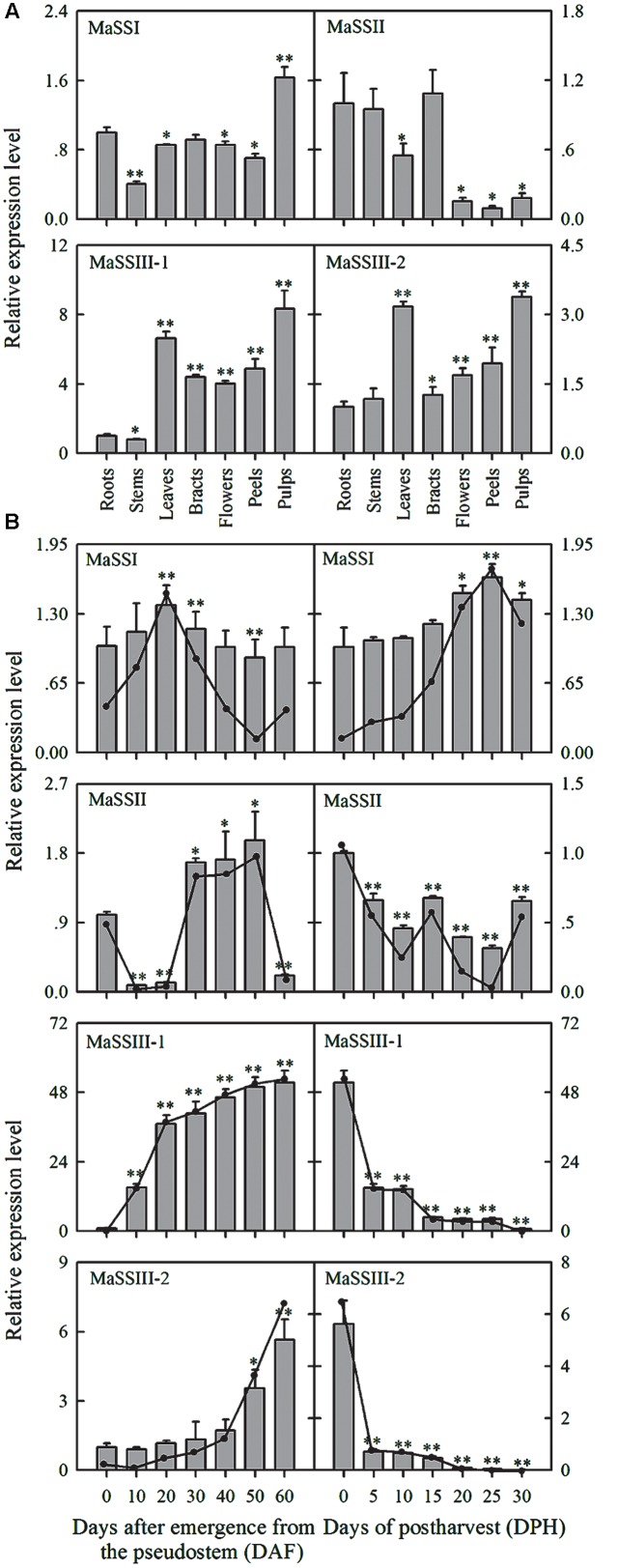
The spatial expression patterns of the four MaSS genes were analyzed by qRT-PCR in seven different tissues, including roots, stems, leaves, bracts, flowers, peels, and pulp (60 DAF) (Figure 3A). MaSSI was expressed at low levels in all tissues examined with the highest expression in pulps. MaSSII was also found to express low in all the vegetative tissues, including roots, stems, leaves, and bracts, and particularly low in flowers and fruit tissues. The expression patterns of MaSSIII-1 and MaSSIII-2 were very similar, with highest expression in pulps and leaves (starch content in leaves was approximately 22.67 ± 1.32%), but that MaSSIII-1 had the highest levels of expression.
FIGURE 3.
Expression of MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 genes in different banana tissues (A) and in fruit at different stages of development and after various storage times (B). The y-axis represents the relative fold-difference in mRNA level, which was calculated using the 2-ΔΔCt formula with MaActin and MaGAPDH as internal controls. The vertical bars represent the mean ± SD of three replicates. Asterisks indicate significant difference from 0 DAF and 0 DPH vs. the following days (∗p < 0.05; ∗∗p < 0.01).
The temporal expression of MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 during banana fruit development was also determined by qRT-PCR (Figure 3B). Despite of being highly variable, the expression levels of MaSSI and MaSSII genes were found to be generally low throughout the entire banana pulp development. Both MaSSIII-1 and MaSSIII-2 showed a sharp increase of expression during the fruit development, the increase in expression occurred earlier in MaSSIII-1 than in MaSSIII-2, with the highest expression detected at the last sampling stage when banana had reached its maturity. However, the expression of MaSSIII-1 was about 10∼20-fold higher than that of MaSSIII-2 during the fruit development.
When the naturally ripen banana fruits were subjected to varying period of storage, the MaSS genes were also found to be differentially expressed (Figure 3B). MaSSII showed consistently low levels of expression throughout the storage period, but the MaSSI expression was slightly increased during storage. The seemingly negative association between MaSSI expression and starch degradation needs to be further investigated. Although there can be a lack of congruency between transcript and activity, it is possible that there can be some starch biosynthesis occurring even during net degradation (Luengwilai and Beckles, 2009). In contrast, a drastic reduction of expression was detected at 5 days after storage, as observed in both MaSSIII-1 and MaSSIII-2.
Western Blot Analyses of MaSSIII-1 Protein
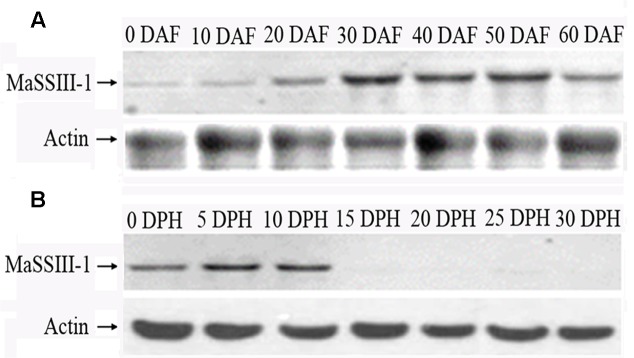
Western blot analysis with rabbit anti-MaSSIII-1 polyclonal antibody as a probe indicated that the size of MaSSIII-1 is approximately 90.0 kDa, which is consistent with the molecular weight (90.69 kDa) as predicted by the PeptideMass program. The expression of MaSSIII-1 protein was gradually increased during banana fruit development, but drastically reduced from 0 to 30 DPH of storage (Figures 4A,B). The Western blot results was consistent with changes in starch granules (Figures 1B,C), amylopectin content (Figure 1E) and SS activity (Figure 1F) during banana development and ripening, suggesting the expression of MaSSIII-1 protein might play an important role in regulating amylopectin metabolism in banana fruit during development and ripening.
FIGURE 4.
Western blot analyses of the MaSSIII-1 protein. (A) Western blot analyses of MaSSIII-1 proteins in banana pulp at different stages of development, (B) Western blot analyses of MaSSIII-1 proteins in banana pulp at different stages of storage. DAF: days after emergence from the pseudostem; DPH: days of post-harvest.
Overexpression of MaSSIII-1 in Tomato Changes the Morphology of Starch Granules and Increases the Amylopectin Content and SS Activity
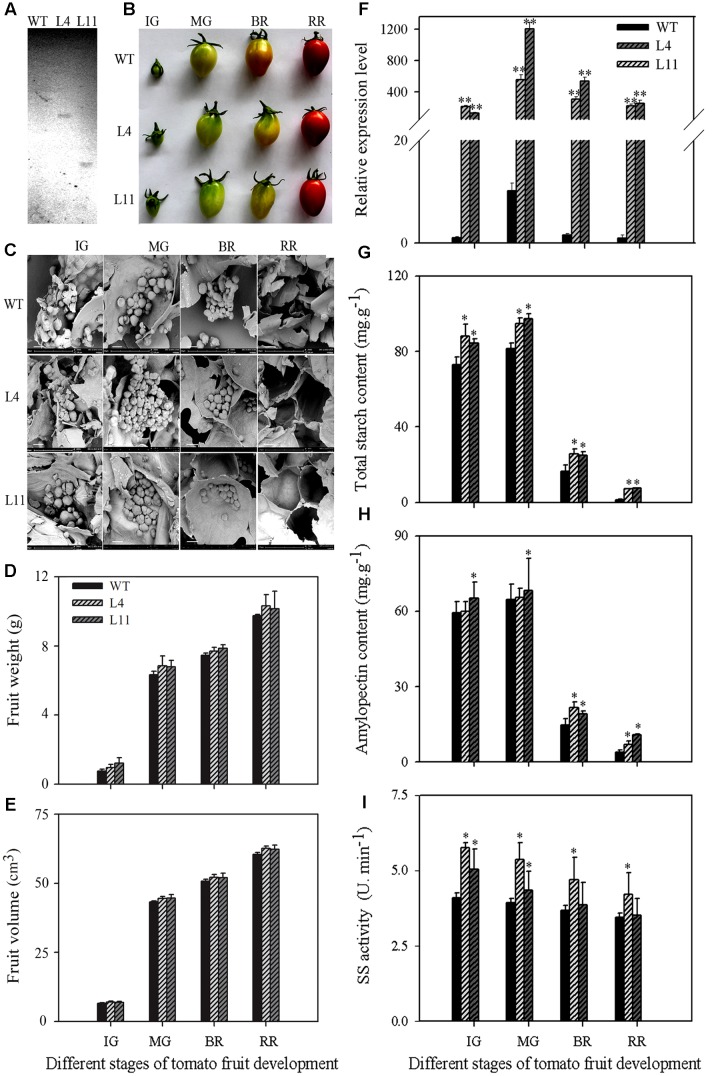
To further examine the function of MaSSIII-1 during fruit development and ripening, MaSSIII-1 was introduced into a pCAMBIA-1302 vector under the transcriptional control of the CaMV 35S promoter. Two single-copy transgenic plants (named L4 and L11) were identified by Southern blot analysis (Figure 5A). The fruit shape, starch granule morphology, gene expression, total starch content, amylopectin content, and SS activity were investigated in the wild-type (WT) and the MaSSIII-1 overexpressing transgenic plants.
FIGURE 5.
Southern blot analysis (A) and change of fruit shape (B), starch granule (C), fruit weight (D), fruit volume (E), gene expression (F), total starch content (G), amylopectin content (H), and SS enzyme activity (I) at different developmental stages of MaSSIII-1 transgenic plants in tomato. WT: wild-type; L4, L11: MaSSIII-1 transgenic plants; IG: immature green; MG: mature-green; BR: orange-breaker; RR: red ripening stage. Asterisks indicate significant difference between the WT and MaSSIII-1 transgenic plants (∗p < 0.05; ∗∗p < 0.01). Three biological experiments were performed, which produced similar results. Scale bar = 15 μm.
Compared to WT, the fruit shape of MaSSIII-1 transgenic plants did not change (Figure 5B), but the morphology of starch granules was significantly altered from immature green (IG) to orange-breaker (BR) in transgenic tomato plants, severe cracks in the surface of starch granules were observed (Figure 5C). For fruit weight and volume, there are no significant differences between MaSSIII-1 transgenic plants and WT (Figures 5D,E). Overexpression of MaSSIII-1 in tomato plants significantly increased the gene expression (Figure 5F), SISSIII-1 (NP_001234623; MaSSIII-1 orthologs) expression in tomato fruit was lower than that of MaSSIII-1 transgenic plants (Figure 5F and Supplementary Figure S2). At IG and mature-green (MG) stages, total starch content, amylopectin content, and SS activity showed significant differences between transgenic plants and WT (Figures 5G–I). Especially at the MG stage, the gene expression, total starch content, amylopectin content, and SS activity significantly increased 55∼120-fold, 13.47∼15.83 mg⋅g-1, 6.36∼9.17 mg⋅g-1, and 0.54∼0.80 U⋅min-1 in the transgenic fruits compared to WT, respectively.
Discussion
Despite of the extensive studies on cereal starch, little information is available regarding the dynamics of starch accumulation (net result of starch synthesis and degradation), SS activity, and amylopectin metabolism during fruit development and storage in fresh starchy fruits, such as banana. In tomato, starch is transiently accumulated during fruit development and degraded within the lifecycle of that organ, while starch synthesis and degradation are occurring simultaneously (Luengwilai and Beckles, 2009). However, change characteristics of SS activity are unclear. In this study, a temporal modulation in starch content was found concomitant to banana fruit development and storage. The result was consistent with the report of tomato (Luengwilai and Beckles, 2009). SS activity increased as the starch granule expanded in size and amylopectin content increased during fruit development, but decreased significantly along with the degradation of amylopectin and starch granule during storage. Moreover, we found that SS activity increased later than starch and amylopectin accumulation, the same was true for transcript and protein analyses, implying SS act at later stages of fruit development while other enzymes may act in the early phases.
The SS isoforms comprise at least four families (namely SSI, SSII, SSIII, and SSIV) in plants and play important roles in amylopectin biosynthesis (Park et al., 2012). GBSS as a separate from the other SS isoforms also influences biosynthesis of extra-long unit chains of amylopectin in rice (Hanashiro et al., 2008). Six GBSS genes have been cloned and identified in banana (Miao et al., 2014). In this report, we characterized the function of MaSSIII-1 in amylopectin metabolism of banana fruit and described the putative role of the other MaSS family members. Four MaSSs could be categorized into three classes, including MaSSI, MaSSII, and MaSSIIIs (MaSSIII-1 and MaSSIII-2). This scenario seems to apply in some lower green algae species where the gene distribution of SSIV is patchy (Deschamps et al., 2008). Moreover, in Arabidopsis the role of SSIV in starch granule seeding can be replaced, in part, by the phylogenetically related SSIII (Szydlowski et al., 2011). Sequence analysis indicated that the MaSSI, II, III-1, and III-2 genes showed characteristics of a typical plant SS gene (Li et al., 2000; Park et al., 2012). The MaSSI and MaSSII amino acid sequences shared three conserved regions encoding Domain I, II, and III, respectively (Park et al., 2012). Domain I is a putative N-terminal transit peptide, Domains II and III are the C-terminal catalytic domain (Park et al., 2012). The amino acid sequences of MaSSIII-1 and MaSSIII-2 contained four distinct regions: a transit peptide region, a variable repeat region, a SSIII specific region, and a C-terminal catalytic domain. The SSIII domain organization was reported in wheat (Li et al., 2000).
Starch synthase paralogs show varying spatial-temporal expression (Senoura et al., 2004; Dian et al., 2005). MaSSI and MaSSII were found to express in low levels in vegetative organs, flower and banana fruits, similar to their homologs in kidney bean (Senoura et al., 2004) and rice (Jiang et al., 2004). In comparison, MaSSIII-1 and MaSSIII-2 were abundantly expressed in the late phase of developing fruit pulp, with the expression level of MaSSIII-1 being significantly higher (Figure 3A). Such an observation is consistent with previous studies in rice, wheat, and Arabidopsis (Li et al., 2000; Dian et al., 2005; Busi et al., 2008; Valdez et al., 2008). Taken together, it is proposed that the four MaSS genes cloned from banana fruit may play divergent roles, with MaSSI and MaSSII being house-keeping, but MaSSIII-1 and MaSSIII-2 expression at transcription level clearly occurs at later stages of fruit development, they’re not involved in amylopectin synthesis during early phase of fruit development.
SSIII protein as a catalytic factor plays an important role in amylopectin metabolism (Zhu et al., 2014; Huang et al., 2016). SSIII contributes the major activity (80% of the total) in potato tubers (Abel et al., 1996). In Arabidopsis, loss of both SSII and SSIII caused slower plant growth and dramatically reduced starch content (Zhang et al., 2008). Defective SSIII mutations resulted in change of starch structure into large clusters with more singly branched building blocks in amylopectin in maize (Zhu et al., 2014). Inhibition of SSIII expression resulted in a drastic drop in the overall rate of starch accumulation and the proportion of amylopectin with very long chains in potato (Abel et al., 1996; Edwards et al., 1999). SSIII activity was found to affect starch phosphorylation in potato tubers (Carpenter et al., 2015). In fresh fruits, whether starch metabolism was affected by SS, as far as we know there has not been reported. In this study, MaSSIII-1 transcript in both transgenic lines was 100-fold higher than WT. SS activity, total starch content, and amylopectin content were enhanced significantly by overexpressing MaSSIII-1 gene in tomato transgenic plants at MG stage (Figure 5), suggesting these parameter increases may be attributed to MaSSIII because of increased transcript and polypeptide. Similar results were reported in wheat (Li et al., 2000) and rice (Dian et al., 2005), but this is the first report in a fresh fruit species. In addition, we found that overexpression of MaSSIII-1 in tomato fruit showed severe cracks in the surface of starch granules and changed the starch granules morphology. This study suggests that MaSSIII-1 is a key gene in the amylopectin biosynthesis and therefore it could be used as a useful tool for marker assisted molecular breeding in banana.
Conclusion
Starch synthase activity increased along with the amylopectin accumulation at later stages of banana fruit development, but declined during storage. Four SS genes encoding MaSSI, MaSSII, MaSSIII-1, and MaSSIII-2 were cloned and characterized. Expression pattern of only MaSSIII-1 was highly consistent with dynamic changes in starch granules, amylopectin content, and SS activity. MaSSIII-1 transgenic lines distinctly changed the morphology of starch granules. Overexpression of MaSSIII-1 in tomato plants significantly increased the amylopectin accumulation and SS activity in comparison to WT. This is the first report about the MaSSIII-1 gene involved in amylopectin metabolism in a fresh fruit species. These findings establish a solid foundation to further regulate the amylopectin metabolism in banana fruits or other fresh fruits using the MaSSIII-1 or its homologous genes.
Author Contributions
ZJ and BX conceived and designed the experiments. HM, PS, QL, CJ, JL, and WH performed the experiments and carried out the analysis. HM, PS, and QL designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
ZJ and BX conceived and designed the experiments. HM, PS, QL, CJ, JL, and WH performed the experiments and carried out the analysis. HM, PS, and QL designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401843 and 31071788), the Modern Agro-industry Technology Research System (CARS-32), the 973 Projects (2014CB160314), and the Natural Science Foundation of Hainan Province (314100).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00454/full#supplementary-material
FIGURE S1 | Sequence motifs and chromosomal localization of four MaSS genes in banana. (A) Structural organization of banana MaSS genes, solid boxes indicate exons, and bold lines represent introns, (B) Chromosomal localization of banana MaSS genes.
FIGURE S2 | Expression of SISSIII-1 at different stages of development in tomato fruit. IG: immature green; MG: mature-green; BR: orange-breaker; RR: red ripening stage. The y-axis represents the relative fold-difference in mRNA level, which was calculated using the 2-ΔΔCt formula. The vertical bars represent the mean ± SD of three replicates. Asterisks indicate significant difference from IG stage vs. the following stages (∗p < 0.05; ∗∗p < 0.01).
TABLE S1 | Primers uxsed in this study.
TABLE S2 | Comparison of the deduced amino acid sequences of banana fruit MaSSI and MaSSII. The three consensus regions (Domain I, II, and III) for both starch synthases are highlighted in bold and box.
TABLE S3 | Comparison of the deduced amino acid sequences of banana fruit MaSSIII-1 and MaSSIII-2. The four consensus regions (Transit peptide region, Variable repeat region, SSIII specific region, and Catalytic domain) for both starch synthases are highlighted in bold and box.
References
- Abel G. J., Springer F., Willmitzer L., Kossmann J. (1996). Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J. 10 981–991. 10.1046/j.1365-313X.1996.10060981.x [DOI] [PubMed] [Google Scholar]
- Arshad W., Haq I. U., Waheed M. T., Mysore K. S., Mirza B. (2014). Agrobacterium-mediated transformation of tomato with rolB gene result in enhancement of fruit quality and foliar resistance against fungal pathogens. PLoS ONE 9:e96979 10.1371/journal.pone.0096979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhals J. D., Lajolo F. M., Cordenunsi B. R., Oliveira do Nascimento J. R. (2004). Activity, cloning, and expression of an isoamylase-type starch-debranching enzyme from banana fruit. J. Agric. Food Chem. 52 7412–7418. 10.1021/jf049300g [DOI] [PubMed] [Google Scholar]
- Bischof S., Umhang M., Eicke S., Streb S., Qi W., Zeeman S. C. (2013). Cecropia peltata accumulates starch or soluble glycogen by differentially regulating starch biosynthesis genes. Plant Cell 25 1400–1415. 10.1105/tpc.113.109793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi M. V., Palopoli N., Valdez H. A., Formasari M. S., Wayllace N. Z., Gomez-Casati D. F., et al. (2008). Functional and structural characterization of the catalytic domain of the starch synthase III from Arabidopsis thaliana. Proteins 70 31–40. 10.1002/prot.21469 [DOI] [PubMed] [Google Scholar]
- Carpenter M. A., Joyce N. I., Genet R. A., Cooper R. D., Murray S. R., Noble A. D., et al. (2015). Starch phosphorylation in potato tubers is influenced by allelic variation in the genes encoding glucan water dikinase, starch branching enzymes I and II, and starch synthase III. Front. Plant Sci. 6:143 10.3389/fpls.2015.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts N., Abe N., Oitome N. F., Matsushima R., Hayashi M., Tetlow I. J., et al. (2015). Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 66 4469–4482. 10.1093/jxb/erv212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros Mesquita C., Leonel M., Franco C. M., Leonel S., Garcia E. L., Dos Santos T. P. (2016). Characterization of banana starches obtained from cultivars growth in Brazil. Int. J. Biol. Macromol. 89 632–639. 10.1016/j.ijbiomac.2016.05.040 [DOI] [PubMed] [Google Scholar]
- Delvallé D., Dumez S., Wattebled F., Roldán I., Planchot V., Berbezy P., et al. (2005). Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 43 398–412. 10.1111/j.1365-313X.2005.02462.x [DOI] [PubMed] [Google Scholar]
- Deschamps P., Moreau H., Worden A. Z., Dauvillée D., Ball S. G. (2008). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts. Genetics 178 2373–2387. 10.1534/genetics.108.087205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A., Denoeud F., Aury J. M., Baurens F. C., Carreel F., Garsmeur O., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488 213–217. 10.1038/nature11241 [DOI] [PubMed] [Google Scholar]
- Dian W., Jiang H. W., Wu P. (2005). Evolution and expression analysis of starch synthase III and IV in rice. J. Exp. Bot. 56 623–632. 10.1093/jxb/eri065 [DOI] [PubMed] [Google Scholar]
- Edwards A., Fulton D. C., Hylton C. M., Jobling S. A., Gidley M., Rössner U., et al. (1999). A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 17 251–261. 10.1046/j.1365-313X.1999.00371.x [DOI] [Google Scholar]
- Fujita N., Goto S., Yoshida M., Suzuki E., Nakamura Y. (2008). The function of rice starch synthase I expressed in E. coli. J. Appl. Glycosci. 55 167–172. 10.1093/jxb/eri065 [DOI] [Google Scholar]
- Gámez-Arjona F. M., Raynaud S., Ragel P., Mérida A. (2014). Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 80 305–316. 10.1111/tpj.12633 [DOI] [PubMed] [Google Scholar]
- Hanashiro I., Itoh K., Kuratomi Y., Yamazaki M., Igarashi T., Matsugasako J., et al. (2008). Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiol. 49 925–933. 10.1093/pcp/pcn066 [DOI] [PubMed] [Google Scholar]
- Huang B., Keeling P. L., Hennen-Blerwagen T. A., Myers A. M. (2016). Comparative in vitro analyses of recombinant maize starch synthases SSI, SSIIa, and SSIII reveal direct regulatory interactions and thermosensitivity. Arch. Biochem. Biophys. 16 30051–30060. 10.1016/j.abb.2016.02.032 [DOI] [PubMed] [Google Scholar]
- Hubbard N. L., Pharr D. M., Huber S. C. (1990). Role of sucrose phosphate synthase in sucrose biosynthesis in ripening bananas and its relationship to the respiratory climacteric. Plant Physiol. 94 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. S., Ryoo N., Hahn T. R., Walia H., Nakamura Y. (2010). Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 48 383–392. 10.1016/j.plaphy.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Jiang H., Dian W., Liu F., Wu P. (2004). Molecular cloning and expression analysis of three genes encoding starch synthase II in rice. Planta 218 1062–1070. 10.1007/s00425-003-1189-y [DOI] [PubMed] [Google Scholar]
- Leterrier M., Holappa L. D., Broglie K. E., Beckles D. M. (2008). Cloning, characterization and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol. 8:98 10.1186/1471-2229-8-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Miao H. X., Ma Y. W., Wang L., Hu G. B., Ye Z. X., et al. (2015). CrWSKP1, an SKP1-like gene, is involved in the self-incompatibility reaction of “Wuzishatangju” (Citrus reticulate Blanco). Int. J. Mol. Sci. 16 21695–21710. 10.3390/ijms160921695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Mouille G., Kosar-Hashemi B., Rahman S., Clarke B., Gale K. R., et al. (2000). The structure and expression of the wheat starch synthase III gene. Motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol. 123 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. G., Jeang C. L. (2005). Cloning, expression, and characterization of soluble starch synthase I cDNA from taro (Colocasoa esculenta Var. esculenta). J. Agric. Food Chem. 53 7985–7990. 10.1021/jf0504566 [DOI] [PubMed] [Google Scholar]
- Liu H., Yu G., Wei B., Wang Y., Zhang J., Hu Y., et al. (2015). Identification and phylogenetic analysis of a novel starch synthase in maize. Front. Plant Sci. 6:1013 10.3389/fpls.2015.01013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Low D. Y., Williams B. A., D’Arcy B. R., Flanagan B. M., Gidley M. J. (2015). In vitro fermentation of chewed mango and banana: particle size, starch and vascular fibre effects. Food Funct. 6 2464–2474. 10.1039/c5fo00363f [DOI] [PubMed] [Google Scholar]
- Luengwilai K., Beckles D. M. (2009). Starch granules in tomato fruit show a complex pattern of degradation. J. Agric. Food Chem. 57 8480–8487. 10.1021/jf901593m [DOI] [PubMed] [Google Scholar]
- Miao H. X., Sun P. G., Liu W. X., Xu B. Y., Jin Z. Q. (2014). Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit. PLoS ONE 9:e88077 10.1371/journal.pone.0088077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Yuki K., Park S. Y., Ohya T. (1989). Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol. 30 833–839. 10.1104/pp.108.125633 [DOI] [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N., Satoh H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127 459–472. [PMC free article] [PubMed] [Google Scholar]
- Park Y. J., Nishikawa T., Tomooka N., Nemoto K. (2012). Molecular cloning and expression analysis of a gene encoding soluble starch synthase I from grain amaranth. Mol. Breed. 30 1065–1076. [Google Scholar]
- Ral J. P., Colleoni C., Wattebled F., Dauvillée D., Nempont C., Deschamps P., et al. (2006). Circadian clock regulation of starch metabolism establishes GBSSI as a major contributor to amylopectin synthesis in Chlamydomonas reinhardtii. Plant Physiol. 142 305–317. 10.1104/pp.106.081885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán I., Wattebled F., Lucas M. M., Delvallé D., Planchot V., Jiménez S., et al. (2007). The phenotype of soluble starch synthase IV defective mutant of Arabidopsis thaliana suggests a novel function of elongation enzyme in the control of starch granule formation. Plant J. 49 492–504. 10.1111/j.1365-313X.2006.02968.x [DOI] [PubMed] [Google Scholar]
- Sarawong C., Schoenlechner R., Sekiguchi K., Berghofer E., Ng P. K. (2014). Effect of extrusion cooking on the physicochemical properties, resistant starch, phenolic content and antioxidant capacities of green banana flour. Food Chem. 143 33–39. 10.1016/j.foodchem.2013.07.081 [DOI] [PubMed] [Google Scholar]
- Schwarte S., Brust H., Steup M., Tiedemann R. (2013). Intraspecific sequence variation and differential expression in starch synthase genes of Arabidopsis thaliana. BMC Res. Notes 6:84 10.1186/1756-0500-6-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoura T., Isono N., Yoshikawa M., Asao A., Hamada S., Watanabe K., et al. (2004). Characterization of starch synthase I and II expressed in early developing seeds of kidney bean (Phaseolus vulgaris L.). Biosci. Biotechnol. Biochem. 68 1949–1960. [DOI] [PubMed] [Google Scholar]
- Subasinghe R. M., Liu F., Polack U. C., Lee E. A., Emes M. J., Tetlow I. J. (2014). Multimeric states of starch phosphorylase determine protein-protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 83 168–179. 10.1016/j.plaphy.2014.07.016 [DOI] [PubMed] [Google Scholar]
- Syahariza Z. A., Sar S., Hasjim J., Tizzotti M. J., Gilbert R. G. (2013). The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem. 136 742–749. 10.1016/j.foodchem.2012.08.053 [DOI] [PubMed] [Google Scholar]
- Szydlowski N., Ragel P., Hennen-Bierwagen T. A., Planchot V., Myers A. M., Mérida A., et al. (2011). Integrated functions among multiple starch synthases determine both amylopectin chain length and branch linkage location in Arabidopsis leaf starch. J. Exp. Bot. 62 4547–4559. 10.1093/jxb/err172 [DOI] [PubMed] [Google Scholar]
- Szydlowski N., Ragel P., Raynaud S., Lucas M. M., Roldán I., Montero M., et al. (2009). Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 21 2443–2457. 10.1105/tpc.109.066522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto-Kuno Y., Suzuki K., Nakamura S., Satoh H., Ohtsubo K. (2006). Soluble starch synthase I effects differences in amylopectin structure between indica and japonica rice varieties. J. Agric. Food Chem. 54 9234–9240. 10.1021/jf061200i [DOI] [PubMed] [Google Scholar]
- Torre-Gutiérrez L., Chel-Guerrero L. A., Betancur-Ancona D. (2008). Functional properties of square banana (Musa balbisiana) starch. Food Chem. 106 1138–1144. 10.1016/j.foodchem.2007.07044 [DOI] [Google Scholar]
- Utrilla-Coello R. G., Rodríguez-Huezo M. E., Carrillo-Nava H., Hernández-Jaimes C., Vernon-Carter E. J., Alvarez-Ramirez J. (2014). In vitro digestibility, physicochemical, thermal and rheological properties of banana starches. Carbohydr. Polym. 101 154–162. 10.1016/j.carbpol.2013.09.019 [DOI] [PubMed] [Google Scholar]
- Valdez H. A., Busi M. V., Wayllace N. Z., Parisi G., Uqalde R. A., Gomez-Casati D. F. (2008). Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry 47 3026–3032. 10.1021/bi702418h [DOI] [PubMed] [Google Scholar]
- Wikman J., Blennow A., Buléon A., Putaux J., Pérez S., Seetharaman K., et al. (2014). Influence of amylopectin structure and degree of phosphorylation on the molecular composition of potato starch lintners. Biopolymers 101 257–271. 10.1002/bip.22344 [DOI] [PubMed] [Google Scholar]
- Yang J. H., Bi C. G., Song W. C. (1992). A half-grain method for analyzing the amylose content in rice grains and its application. Acta Agron. Sin. 18 366–372. [Google Scholar]
- Zeeman S. C., Kossmann J., Smith A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61 209–234. 10.1146/annurev-arplant-042809-112301 [DOI] [PubMed] [Google Scholar]
- Zhang P., Hamaker B. R. (2012). Banana starch structure and digestibility. Carbohydr. Polym. 87 1552–1558. 10.1016/j.carbpol.2011.09.053 [DOI] [Google Scholar]
- Zhang X., Szydlowski N., Delvallé D., D’Hulst C., James M. G., Myers A. M. (2008). Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 8:96 10.1186/1471-2229-8-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. L., Myers A. M., James M. G. (2005). Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138 663–674. 10.1104/pp.105.060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Bertoft E., Seetharaman K. (2014). Distribution of branches in whole starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 62 4577–4583. 10.1021/jf500697g [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Sequence motifs and chromosomal localization of four MaSS genes in banana. (A) Structural organization of banana MaSS genes, solid boxes indicate exons, and bold lines represent introns, (B) Chromosomal localization of banana MaSS genes.
FIGURE S2 | Expression of SISSIII-1 at different stages of development in tomato fruit. IG: immature green; MG: mature-green; BR: orange-breaker; RR: red ripening stage. The y-axis represents the relative fold-difference in mRNA level, which was calculated using the 2-ΔΔCt formula. The vertical bars represent the mean ± SD of three replicates. Asterisks indicate significant difference from IG stage vs. the following stages (∗p < 0.05; ∗∗p < 0.01).
TABLE S1 | Primers uxsed in this study.
TABLE S2 | Comparison of the deduced amino acid sequences of banana fruit MaSSI and MaSSII. The three consensus regions (Domain I, II, and III) for both starch synthases are highlighted in bold and box.
TABLE S3 | Comparison of the deduced amino acid sequences of banana fruit MaSSIII-1 and MaSSIII-2. The four consensus regions (Transit peptide region, Variable repeat region, SSIII specific region, and Catalytic domain) for both starch synthases are highlighted in bold and box.