Abstract
Due to its high biomass yield, low environmental impact, and widespread adaptability to poor soils and harsh conditions, switchgrass (Panicum virgatum L.), a warm-region perennial herbaceous plant, has attracted much attention in recent years. However, little is known about microRNAs (miRNAs) and their functions in this bioenergy grass. Here, we identified and characterized a miRNA gene, Pvi-MIR319a, encoding microRNA319a in switchgrass. Transgenic rice lines generated by overexpressing the Pvi-MIR319a precursor gene exhibited broader leaves and delayed flowering compared with the control. Gene expression analysis indicated at least four putative target genes were downregulated. Additionally, we cloned a putative target gene (PvPCF5) of Pvi-MIR319a from switchgrass. PvPCF5, a TCP transcription factor, is a nuclear-localized protein with transactivation activity and control the development of leaf. Our results suggest that Pvi-MIR319a and its target genes may be used as potential genetic regulators for future switchgrass genetic improvement.
Keywords: switchgrass (Panicum virgatum), leaf development, microRNA319 (miR319), bioenergy, TCP transcription factor
Introduction
Switchgrass (Panicum virgatum L.), which is highly adaptable and particularly resistant to stress, can survive in many climatic conditions, including in a variety of soil conditions (Vogel et al., 2010). It is used in several bioenergy conversion processes, such as ethanol production, biogas, and thermal energy applications (McLaughlin and Adams Kszos, 2005). Generally, switchgrass can be grouped into two main ecotype categories: lowland, with tall height and high output, and upland, with short stature and low biomass output (Gunter et al., 1996). Switchgrass has been increasingly regarded as a model energy crop with huge potential in the United States and worldwide (McLaughlin and Adams Kszos, 2005; David and Ragauskas, 2010; Monti, 2012). Genetic engineering can improve plant traits and enhance stress tolerance (Hisano et al., 2011; Li and Qu, 2011). Application of transgenic biotechnologies and genomic analyses will contribute to the development of bioenergy using switchgrass (Sticklen, 2006; Bouton, 2007).
In plants, microRNAs (miRNAs), one of the prominent posttranscriptional regulators, are a class of ∼19–24 nucleotide (nt) endogenous non-coding small RNAs (Reinhart et al., 2002; Chen, 2008; Fabian et al., 2010; Liu and Chen, 2016). These small RNAs form an imperfect stem-loop structure in single-stranded primary miRNAs through recognition by a Dicer-like protein, DCL1, and mature miRNAs are recruited into an RNA-induced silencing complex (RISC) to control target gene expression by either direct mRNA cleavage or translational repression (Bartel, 2004). Recent evidence indicates that miRNAs are important genetic regulators and play critical roles in plant development, nutrient homeostasis, and various biotic and abiotic stress responses, including salinity, drought, and extreme temperatures, via interaction with their targets (Chen, 2008; Chuck et al., 2011; Zhou and Luo, 2013; Zhou et al., 2013; He et al., 2014).
Studies on switchgrass include examinations of the establishment and optimization of a transformation system (Li and Qu, 2011), gene expression analysis, and identification of miRNAs (Chuck et al., 2011; Fu et al., 2012; Li et al., 2014). For example, by utilizing bioinformatics and experimental approaches, Matts et al. (2010) identified ∼20 conserved miRNA families, predicted 37 miRNA target genes, and confirmed 4 target mRNAs using a modified 5′ RACE (Rapid Amplification of cDNA Ends) strategy. They also found that miR395 and miR399 have distinct expression patterns compared with other species, and this may be related to the wide adaptability of switchgrass growth. Furthermore, using a well-defined computational approach, Xie et al. (2010) identified 121 predicted miRNAs from 44 miRNA families and 839 potential targets in switchgrass. Transgenic switchgrass overexpressing miR156 exhibited different morphological phenotypes compared with wild-type, and the degree of alteration depended on miR156 levels, where relatively low miR156 levels could increase biomass yield (Chuck et al., 2011; Fu et al., 2012).
In plants, microRNA319 (miR319) is one of the most conserved and ancient microRNA families, which was found in diverse plant species from moss to flowering plants (Palatnik et al., 2003, 2007; Pandey and Baldwin, 2007; Cuperus et al., 2011). The miR319s have been demonstrated to target TCP (TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTORS) genes, which encode plant-specific transcription factors (Nag et al., 2009; Martín-Trillo and Cubas, 2010). The TCP transcription factors have a conserved TCP domain with a basic helix-loop-helix structure and this family is known to play essential roles in diverse biological processes, including various aspects of plant growth and development, responses to biotic and abiotic stresses (Palatnik et al., 2003; Ori et al., 2007; Nag et al., 2009; Danisman, 2016).
The recent studies showed that overexpression of miR319 led to abnormal development and tolerance to stress in several transgenic plants (Yang et al., 2013; Zhou et al., 2013; Wang et al., 2014; He et al., 2014; Zhou and Luo, 2014; Li et al., 2016). High levels of Osa-miR319 in transgenic rice led to wider leaf blades and significantly enhanced cold tolerance in acclimated transgenic plants (Yang et al., 2013). Furthermore, other researchers suggested that osa-miR319b may play a role in the response to cold stress via control of OsPCF6 and OsTCP21 (Wang et al., 2014). In addition, Zhou et al. (2013) found that transgenic creeping bentgrass (Agrostis stolonifera) overexpressing Osa-miR319a displayed morphological changes in leaves and stems with a parallel increased tolerance to salinity and drought stress (Zhou et al., 2010, 2013; Yang et al., 2013). However, few studies have examined the roles played by several important microRNAs, such as miR319, in switchgrass. The genetic engineering and application of miRNAs provides an opportunity to make effective improvements in switchgrass (Zhou et al., 2013; Li et al., 2014).
In this study, we cloned and characterized Pvi-MIR319a, which encodes a microR319 in switchgrass. Pvi-MIR319a-overexpressing transgenic rice lines were used to investigate the role of miR319 in plant morphological development. We also isolated and identified a putative target gene (PvPCF5) of Pvi-MIR319a in switchgrass; this is a nuclear localization protein with transcriptional activation activity that is negatively regulated by Pvi-MIR319a. These data suggest that Pvi-MIR319a and its targets may possess important biological functions in switchgrass.
Materials and Methods
Plant Materials and Growth Conditions
Switchgrass (P. virgatum L.) cultivar Alamo was used in this study. The seeds were provided by Dr. Wanjun Zhang (Department of Grassland Science, China Agricultural University, Beijing, China). The switchgrass were planted in the greenhouse and the experimental station of the Institution of Genetics and Developmental Biology, Chinese Academy of Sciences (IGDB, CAS), China.
Rice (Oryza sativa L.) subspecies japonica cultivar Zhonghua 11 (ZH-11) was used as transgenic materials in this study. The transgenic and wild type control rice plants were grown in the greenhouse at 28°C for 14 h (day) and 10 h (night) and the field of experimental station.
Cloning of the Pvi-MIR319a Gene, Construction of Plant Expression Vectors, and Rice Transformation
The Osa-MIR319a sequence (ID: MI0001098) was downloaded from the miRNA database1 (Kozomara and Griffiths-Jones, 2013). By searching with this sequence in the NCBI, we obtained the putative Pvi-MIR319a EST sequence from switchgrass.
To overexpress the switchgrass Pvi-MIR319a gene in rice, Pvi-MIR319a cDNA containing a stem-loop structure was cloned via RT-PCR amplification using the gene-specific primers 5′-TCTAGATTGAGTTTATGGCTTCTCTGGAAGA-3′ and 5′-GTCGACTGAGGTGTTCTATTGGTAGCCCAA -3′; the cloned DNA fragment was then inserted into the Xba I and Sal I sites of the binary vector pZH01, producing p35S-Pvi-MIR319a/p35S-hyg. The construct, containing the CaMV 35S promoter, was transferred into Agrobacterium tumefaciens strain LBA4404 by electroporation. Rice was transformed using the method of Hiei et al. (1994).
Phylogenetic Analysis of TCPs
TCP (TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR) protein sequence data were downloaded from the Arabidopsis Information Resource2 and the MSU Rice Genome Annotation Project Database and Resource3, respectively. Multiple alignments of TCP protein family members from rice and arabidopsis were constructed using Clustal X (Wagner et al., 2008). A phylogenetic tree was constructed using the MEGA5 program (Tamura et al., 2011).
RNA Isolation, cDNA Preparation, and Quantitative RT-PCR
Total RNA was extracted from switchgrass using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Genomic DNA was removed using DNase I (Takara, Japan). For real-time PCR, synthesis of the first-strand cDNA was terminated by applying M-MLV reverse transcriptase (Promega, Madison, WI, USA). Genomic DNA was removed using DNase I (Takara, Japan). Quantitative RT-PCR was performed on a Bio-Rad Chromo 4 Real-Time PCR System (Bio-Rad, Hercules, CA, USA) using a TransStart SYBR Green qPCR SuperMix Kit (TransGen, Beijing, China). A switchgrass ubiquitin gene was used as a control (Wuddineh et al., 2015) (Switchgrass Unitranscript ID: AP13CTG25905). Expression levels of examined genes were quantified by a relative quantitation method, each sample was subjected to at least three biological replicates in this experiment. The specific primers used in this study were listed in Supplementary Table S1.
Pvi-MIR319a and PvPCF5 Transient Expression
To observe whether PvPCF5 is a Pvi-MIR319a target gene, Nicotiana benthamiana leaves were used in this experiment (Llave and Carrington, 2002). The PvPCF5 coding regions were inserted into an pEZR(K)-LC vector (LC) containing the CaMV35S promoter and green fluorescence protein (GFP) to generate 35S::PvPCF5-GFP (LC-PCF5), while the Pvi-MIR319a was inserted into the LC vector that lack GFP. We separated these into four groups: (a) LC-vector; (b) LC-PCF5; (c) LC-PCF5 and LC-319; and (d) LC-319. These four groups of plasmids were injected into 28-day-old tobacco leaves and observed 3 days later. The specific primers used in this study were listed in Supplementary Table S1.
Subcellular Localization in Switchgrass
Young etiolated seedlings leaves were cut and digested with 20 ml of enzyme solution to prepare the protoplasts. The fusion gene (LC-PCF5) and the negative control pEZR(K)-LC vector were used for PEG (polyethylene glycol)-mediated transformation of switchgrass protoplasts (Liu et al., 2015). After 18 h of incubation, GFP fusion protein fluorescence signals were visualized using a fluorescence confocal microscope (Zeiss Axio Imager.Z2)4.
Transactivation Assay in Yeast Cells
The complete ORF of PvPCF5, N-terminal putative activation domain (1–299 amino acids), and C-terminal area (300–439 amino acids) were amplified and digested with EcoR I and BamH I and fused to the pGBKT7-GAL-4 vector. An empty pBD vector (pGBKT7) was used as a negative control. These constructs and the empty vector were each introduced into yeast strain Y2HGold containing the AUR1-C and MEL1 reporter genes. Yeast cell transformation was carried out using the instructions in the YeastmakerTM Yeast Transformation System 2 User Manual5. The yeast transformants were selected on SD/-Trp and SD/-Trp/A/X-α-gel plates for 2–4 days at 30°C to identify transactivation activity (Yeast Protocols Handbook; Clontech, USA).
Results and Discussion
Identification and Characterization of Pvi-MIR319a in Switchgrass
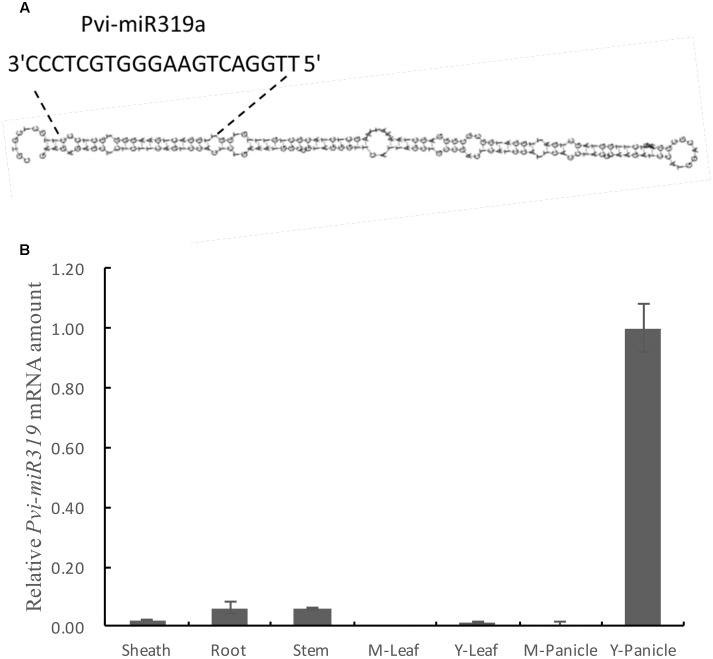
Considering the close phylogenetic relationship between switchgrass and rice, we hypothesized that the sequence and biological functions of miR319s have conserved in the two species. We could use the known information of the rice miR319 to identify and clone its homologs in switchgrass. Based on the miRNA database6, two members (Osa-miR319a and Osa-miR319b) of the rice miR319 family have common mature miRNA sequences (Yang et al., 2013). Using the stem-loop sequences of the rice miR319 precursor genes as a reference, we found an EST (ID: FL985594) with a length of 748 bp from switchgrass (P. virgatum) by BLAST search7 (Supplementary Figure S1). The miR319 sequences from rice and switchgrass showed that the two mature regions were in complete agreement (Supplementary Figure S2). Sequence similarity analysis showed that EST has substantial similarity with the osa-miR319a precursor gene (Supplementary Figure S3). This suggests that the EST of switchgrass contains the miR319a precursor sequence. And the stem-loop structure of this miR319a was found (Figure 1A and Supplementary Figure S4), So, we named it as Pvi-MIR319a (P. virgatum MIR319a).
FIGURE 1.
Predicted stem-loop structure and expression profile analysis of pvi-miR319a in switchgrass. (A) A putative structure diagram of Pvi-miR319a stem-loop area. The enlarged site is complete sequence of mature Pvi-miR319a. (B) Expression levels of Pvi-miR319a in different switchgrass tissues were checked by qRT-PCR analysis. Root from 2-week-old seedlings was harvested to be used as experimental materials, while other tissues were collected from switchgrass at heading stage. Rice ACTIN 1 gene was involved in this experiment as a internal reference.
Pvi-MIR319a Expression Analysis
In plants, a long primary miRNA is first transcripted under the guidance of the miRNA gene. It is then processed into pre-miRNA (precursor miRNA) and, finally, into mature miRNA. The mature miRNA recognizes and regulates its target through complementarity to direct RISC-mediated cleavage (Liu and Chen, 2016).
To determine whether Pvi-MIR319a plays roles in different plant tissues and developmental stages, we conducted quantitative real-time PCR of Pvi-MIR319a transcripts in roots, stems, leaf sheaths, leaves, and panicles. Pvi-MIR319a transcript showed a higher expression level in young panicles, indicating that this gene may plays an important role in inflorescence development (Figure 1B). This result is consistent with the findings reported by Nag et al. (2009) with regard to the role of miR319a in flower development (Nag et al., 2009).
The expression level of Pvi-MIR319a was low in roots, stems, and mature panicles. This is similar to the case in rice, where the expression level of miR319 was high only in young panicles, low in other tissues, and lowest in mature leaves and mature panicles (Yang et al., 2013).
Generation of Transgenic Rice Plants Overexpressing Pvi-MIR319a
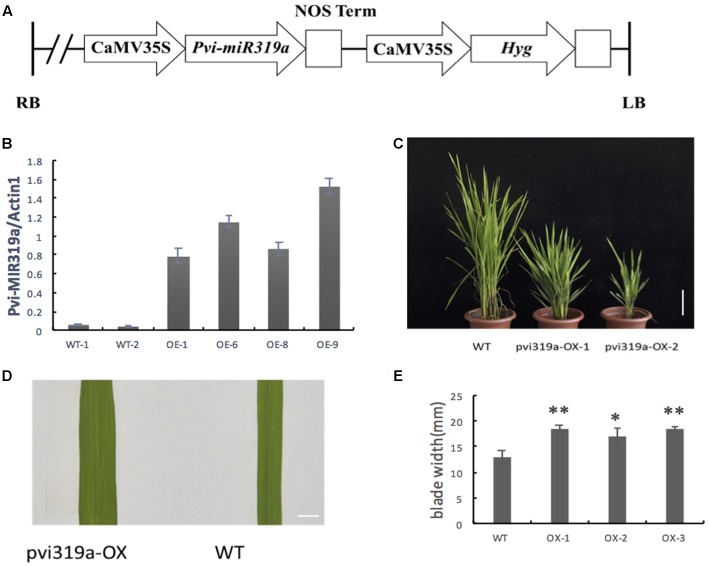
To investigate the function of Pvi-MIR319a, Pvi-MIR319a gene under the control of 35S promoter, were introduced into rice (Figure 2A). Transgenic rice plants overexpressing Pvi-MIR319a were obtained to identify whether overexpressing plants had different phenotypes and growth conditions compared with wild-type plants. Positive transgenic plants were identified using RT-PCR method with genomic DNA extracted from transgenic leaves as a template.
FIGURE 2.
Identification and phenotypes analysis of the Pvi-MIR319a-overexpressing transgenic plants. (A) A schematic of the T-DNA region of the binary construct for rice transformation. (B) Pvi-MIR319a expression in transgenic lines and wild type rice was detected by qRT-PCR analysis. Transcripts of Pvi-MIR319a showed a significantly higher accumulation in transgenic rice plants than that in WT controls. (C) Two-month-old transgenic plants showed lower plant height and slower growth compared with the wild-type rice. Bar = 10 cm. (D) Comparison blade width between the transgenic plants over-expressed Pvi-MIR319a and WT controls. Bar = 10 mm. (E) Quantitative measurement and remarkable analysis of average blade width of Pvi-MIR319a overexpressing lines and WT control. ∗represent a significant difference between transgenic and control plants at P < 0.05 by Student’s t-test. ∗∗represents a remarkable difference between the overexpressing plants and WT control at P < 0.01 applying t-test method.
A total of 17 independent positive transgenic lines were tested and evaluated. To determine the expression level of Pvi-MIR319a, four transgenic lines were analyzed using qRT-PCR. Two wild-type plants were used as controls. The abundance of Pvi-MIR319a precursors in transgenic plants was higher than that in control plants (Figure 2B), suggesting the Pvi-MIR319a fusion plasmids, driven by the 35S promoter, were successfully expressed in rice. It is possible that the ectopic expression of Pvi-MIR319a cloned from switchgrass could affect the normal growth and development of transgenic rice plants, which would help in examining the biological function of the switchgrass miRNA using reverse genetic analysis.
Overexpression of Switchgrass Pvi-MIR319a Results in Pleiotropic Phenotype Changes in Transgenic Rice Plants
As a highly conserved ancient miRNA, miR319 plays important roles in plant development (Jones-Rhoades et al., 2006; Yang et al., 2013; Zhou et al., 2013; Zhou and Luo, 2014). High levels of miR319 or downregulation of its target genes leads to a broader leaf phenotype in arabidopsis (Palatnik et al., 2003), and similar results were found in snapdragons, tomatoes, rice, and other plants (Ori et al., 2007; Yang et al., 2013). Although the role of miR319 in leaf morphogenesis is highly conserved, there are several differences in leaf phenotype between dicotyledonous and monocotyledonous plants. Crinkled leaves appear in transgenic dicotyledonous plants ectopically expressing miR319 but not in transgenic monocotyledonous plants. Based on the highly conserved roles of miR319, we hypothesize that Pvi-MIR319a may play a similar role in leaf morphogenesis and lead to similar leaf phenotypes in monocotyledonous plants (Yang et al., 2013; Zhou et al., 2013). Overexpression of Pvi-MIR319a in rice increased the number of leaf veins and led to broader leaves with a larger leaf area (Figures 2C–E). Very similar leaf phenotypes were observed in transgenic rice/bentgrass overexpressing osa-miR319 (Yang et al., 2013; Zhou et al., 2013; Zhou and Luo, 2014). These results suggest that the function of miR319 in leaf morphogenesis is high conversed in grass family (Poaceae).
Analysis of transgenic rice plants (Figure 2) constitutively expressing Pvi-MIR319a showed that transgenic plants exhibit very distinct phenotypes during the life cycle, including broader and slightly curly leaf blades, shorter plants, and delayed development. These different phenotypes are first observed when transgenic plants initially enter the vegetative growth phase. Measurements were taken from the widest point on the blades at the same position. Statistical analysis showed that leaf blades were significantly wider compared with wild-type (WT) plants and controls. Another important feature is growth retardation, in which overexpressing (OE) lines are shorter than the wild-type. The heading date of transgenic rice plants was also delayed. Pvi-MIR319a overexpression transgenic rice plants delayed flowering by about 14–20 days compared with those of wild type lines. We noticed that the similar report was appear in Arabidopsis thaliana, overexpression miR319 (jaw-D mutants) lead the late flowering in Arabidopsis thaliana (Schommer et al., 2008). Those results suggest that the function of miR319 in flowering may conservative, and it deserves study deeper in the future.
Switchgrass Pvi-MIR319a Regulates Expression of Putative Target Genes in Transgenic Rice Plants
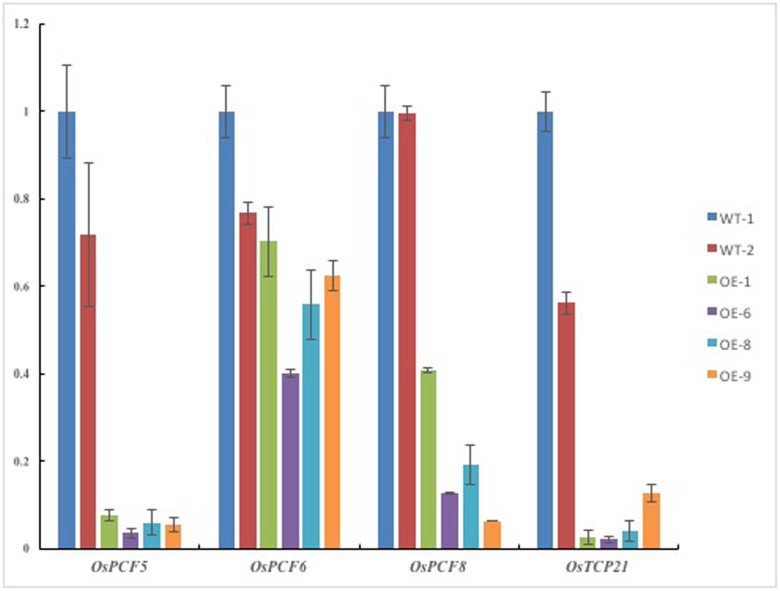
We performed a series of qRT-PCR tests to analyze the transcript levels of five TCP genesin Pvi-MIR319a-overexpressing rice. Expression analysis showed that the levels of the four genes (OsPCF5, OsPCF6, OsPCF8, and OsTCP21) were downregulated compared with those of WT, in which OsPCF5, OsPCF8, and OsTCP21 were significantly reduced (Figure 3). The obvious decrease in the expression of the predicted targets was similar to that observed in osa-miR319a-overexpressing rice (Yang et al., 2013). We hypothesize that mRNAs from the predicted target genes may be directly degraded by Pvi-MIR319a due to the high degree of homology between Pvi-MIR319a and Osa-miR319a. Based on the abnormal leaf phenotype of transgenic plants and the early functional studies on TCP transcription factors, we think that the continuous expression of Pvi-MIR319a leads to a series of pleiotropic phenotypic changes in transgenic rice plants due to the negative regulation of its possible target genes. In this study, higher levels of target transcripts enabled leaves to maintain normal growth, whereas target mRNAs were cleaved by overexpressing Pvi-MIR319a, thereby leading to broad leaves and abnormal developmental patterns. It has been reported that miR319-targeted TCPs are involved in leaf development in eudicot species, such as arabidopsis and tomato (Nag et al., 2009; Martín-Trillo and Cubas, 2010). However, the biological functions of both PvPCF5 and OsPCF5 genes are largely unknown. It is worthy to do further study. Taken together, the results imply that Pvi-MIR319a and its targets play critical roles in plant development.
FIGURE 3.
Expression levels of putative targets were analysed in Pvi-MIR319a-overexpressing rice plants and wild types by real-time quantitative PCR. Compared with WT controls, expression of three predicted target genes were decreased in different degree in transgenic lines. Rice ACTIN 1 (LOC_Os10g36650) is used as a reference gene.
PvPCF5 is a miR319a Target Gene in Switchgrass
The gene sequence of PvPCF5 was cloned and identified. Sequence analysis showed that PvPCF5 encodes a protein with a TCP DNA-binding domain at the N-terminus, and the complete domain was obtained by 5′ RACE assay. We proposed that PvPCF5, with a length of 444 amino acids and a molecular mass of 46.4 kilodaltons, is a transcription factor belonging to the TCP family (Supplementary Figure S5).
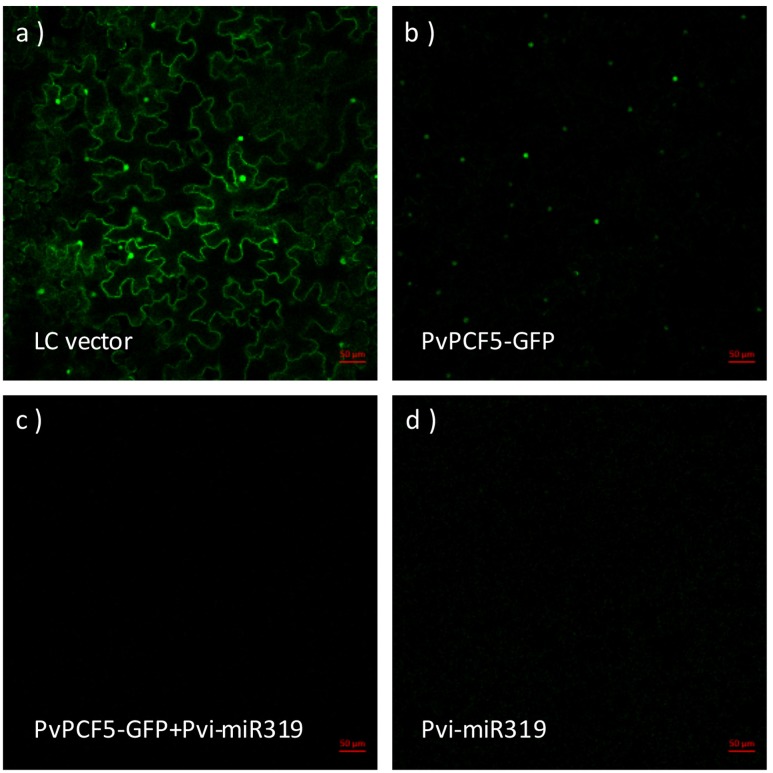
To determine that PvPCF5 is an important target of Pvi-MIR319a, a rapid Agrobacterium-mediated transient expression assay was performed in N. benthamiana. Fusion plasmids (35S::PvPCF5-GFP and 35S::Pvi-MIR319a) were constructed and introduced into Agrobacterium strain EH105 with a control vector. Transient transformation was carried out by co-injection into tobacco leaves. After 48 h of recovery growth, a GFP signal was observed by the UV visualization of green fluorescence under a confocal microscope. The results showed that only tobacco leaves injected with single 35S::PvPCF5-GFP constructs or empty vectors generated GFP fluorescence, whereas co-expression with 35S::Pvi-MIR319a led to a notably weaker GFP signal (Figure 4). This shows that the PvPCF5 levels were markedly decreased when co-expressed with 35S::Pvi-MIR319a. These results provide direct evidence that PvPCF5 is a target of Pvi-MIR319a and that Pvi-MIR319a can regulate the expression of PvPCF5, leading to a significantly lower expression level by direct cleavage (Supplementary Figure S6). Additionally, Figure 3 shows that the GFP signal was localized around the cell nucleus in tobacco leaves, indicating that PvPCF5 is a nuclear-localized protein.
FIGURE 4.
PvPCF5 is a miR319a target supported by transient expression assay in Nicotiana benthamiana. N. benthamiana leaves were injected with Agrobacterium, respectively, carrying the empty vector (LC) (a); 35S::PvPCF-GFP (b); 35S::PvPCF-GFP plus 35S::Pvi-MIR319a (c); and 35S::Pvi-MIR319a (d). GFP green fluorescent signals were observed by using a confocal microscope. Bar = 50 μm.
Expression Profile of PvPCF5
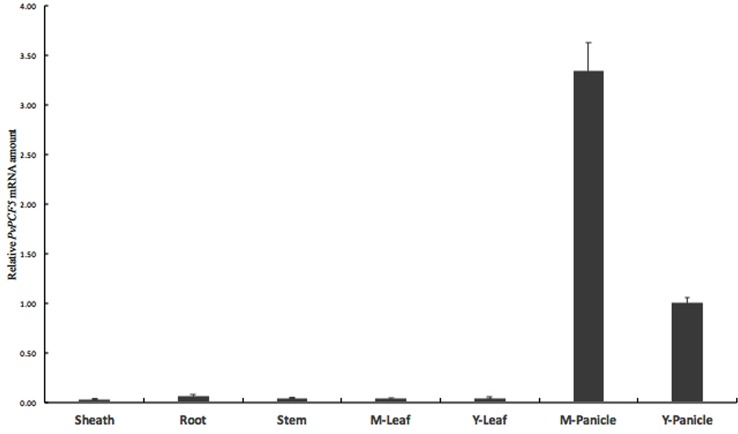
PvPCF5 was identified as one of the target gene of Pvi-MIR319a in switchgrass. The abundance of this target gene in various tissues was examined by quantitative real-time PCR (Figure 5). PvPCF5 is highly expressed in mature panicles, but it is weakly expressed in young panicles, leaf sheaths, stems, and roots, with the lowest expression levels in leaves. The expression level of PvPCF5 had a relatively negative correlation with the expression of Pvi-MIR319a, at least in young and mature panicles.
FIGURE 5.
Expression pattern of PvPCF5 gene in various organs by qRT-PCR assay. Switchgrass organs include sheath, root, stem, mature leaf, mature panicle, and young panicle were harvested from plants at heading stage. Young leaves were collected before tillering stage. Young panicles were harvested at less than 5 cm.
PvPCF5 Encodes a TCP Transcription Factor
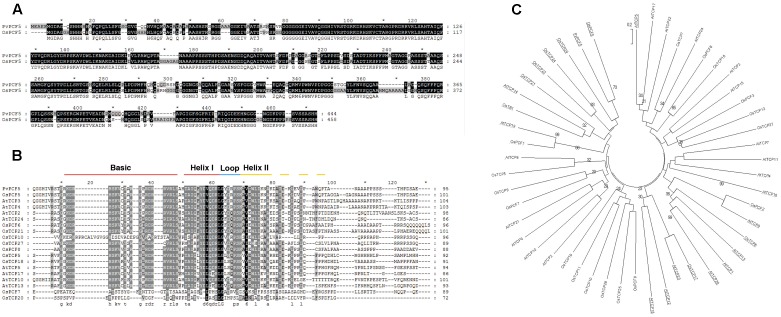
Based on the similarity of the amino acid sequences of the TCP domain region, TCP subfamily proteins are classified into three subgroups: PCF, CIN, and CYC/TB1 (Martín-Trillo and Cubas, 2010). Almost all TCP family members with miR319-binding sites belong to the CIN subgroup, including OsPCF5 (Yang et al., 2013). Previous studies also showed that most CIN subfamily members are involved in leaf development in various plant species. Sequence alignment analysis of PvPCF5 protein from switchgrass and OsPCF5 cloned from rice was performed to determine their homology (Figure 6). Results revealed that PvPCF5 had high homology to OsPCF5. To investigate the evolutionary relationships between PvPCF5 and other TCP subfamily TFs from rice and arabidopsis, a series of related TCP transcription factors, including 26 from arabidopsis and 25 from rice, were collected for phylogenetic analysis. The phylogenetic tree (Figure 6C) indicated that PvPCF5, targeted by Pvi-MIR319a, was clustered in the CIN subfamily, and AtTCP4 was most closely related to PvPCF5. The OsPCF5 transcription factor is important in rice leaf development, and there is high conservation between PvPCF5 and OsPCF5 indicating that PvPCF5 may be primarily involved in leaf morphological development (Yang et al., 2013). Multiple alignments among the 51 TCP proteins confirmed the presence of the TCP domain in PvPCF5 near the N-terminus. However, there were major differences in other areas of the TCP proteins, suggesting that these differences may be due to protein functional specificity.
FIGURE 6.
PvPCF5 is a TCP transcript factor and has the highest homology with rice OsPCF5. (A) Amino acid sequence alignment of switchgrass PvPCF5 and rice OsPCF5. (B) Parts of PvPCF5 protein sequence was comparatively analyzed with other TCPs selected from CIN subfamily. PvPCF5 and other TCP members are conserved in TCP DNA-binging domain. bHLH structure position was marked in this figure. (C) The phylogenetic relationship between PvPCF5 and other TCP transcription factor subfamily members from rice and arabidopsis. The phylogenetic tree was constructed by using MEGA 5.0 software. The number in the diagram represent the bootstrap value from 500 replicates.
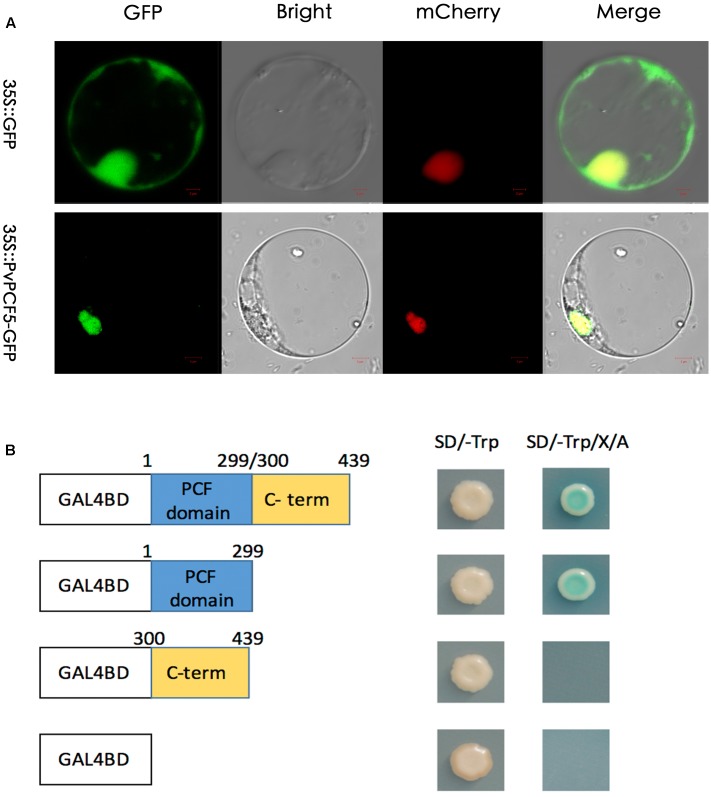
To investigate the subcellular localization of the PvPCF5 protein, a 35S::PvPCF5-GFP fusion and a GFP control were obtained and transformed into prepared switchgrass protoplast cells. The PvPCF5-GFP gene was driven by the CaMA35S expression promoter (Figure 7A). Based on the observed fluorescence signal, 35S:: PvPCF5-GFP was targeted to the cell nucleus, whereas the fluorescence signal of the negative control was observed over the entire protoplast. These results show that PvPCF5 is a nuclear-localized protein, which is a strong indicator of a transcription factor.
FIGURE 7.
Subcellular localization and transcriptional activation analysis of PvPCF5. (A) Subcellular localization of PvPCF5 was performed in switchgrass protoplasts. Confocal images showing the PvPCF5-GFP fusion protein driven by the 35S promoter was specifically expressed in the protoplasts. 35S::GFP was used as a control. Bar = 10 μm. (B) Transactivation activity of the PvPCF5 protein in yeast. The full-length, N-terminal region, C-terminal region of the PvPCF5 were fused into DNA sequences containing a GAL4 DNA-binding domain with the negative control plastid (pGBKT7). These four kinds of constructs were expressed in the yeast strain Y2HGold. The transformants were streaked on the SD/-trp and SD/-trp/A/X-α-gel plates, plates were cultured for 2–4 days at 30°C to check a possible transcriptional activation function of PvPCF5.
We use a yeast system to determined whether PvPCF5 has transactivation activity (Figure 7B). The pGBKT7-PvPCF5, pGBKT7-N (aa 1–299), or pGBKT7-C (aa 300–439) fusion constructs or an empty BD vector (pGBKT7) were transformed into the Y2HGold yeast strain. As shown in Figure 7B, all yeast cells showed good growth on the SD/-Trp plates. However, only cells containing the pGBKT7-PvPCF5 and pGBKT7-N (aa 1–299) constructs could form colonies on SD/-Trp/A/X-α-gel plates. In the presence of abscisic acid and X-α-gel, yeast colonies turned blue, indicating that other reporter genes, such as AUR1-C and MEL1, were activated. These phenomena illustrate that the PvPCF5 has transcription activation activity.
Conclusion
We cloned Pvi-MIR319a from switchgrass and employed reverse genetics to investigate its function. Overexpression of Pvi-MIR319a in transgenic rice increased leaf width, reduced plant height, and delayed heading time. It suggests that Pvi-MIR319a and its target genes may be used as potential genetic regulators for future switchgrass genetic improvement. As a model energy plant, the high effective Agrobacterium-mediated transformation systems of switchgrass has been established (Liu et al., 2015). Next, we plan to express the Pvi-MIR319a and PvPCF5 into switchgrass itself, with its own promoters, constitutive promoters or tissue-specific expression promoters, respectively. The gene editing approaches forPvi-MIR319a and PvPCF5 by CRISPR/CAS9 would also be considered. And then we will screen the elite agronomic traits from the transgenic lines to improve the quality of switchgrass.
Author Contributions
Designed the experiments: GS, LH, XL, and DL. Preformed the experiments: QX, YZ, XL, DY, BF, and JT. Analyzed the data: YZ, XL, LZ, DL, and JT. Wrote the paper: YZ, QX, DL, and XL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Wanjun Zhang (Department of Grassland Science, China Agricultural University) for providing the seeds of switchgrass and helpful discussion. We also thank Dr. Man Zhou (Department of Genetics and Biochemistry, Clemson University, USA) for her critical reading on the manuscript.
Funding. This work was supported by National Natural Science Foundation of China (No. 31672477, 81403090), the National High Technology Research and Development Program of China (863 Program) (2013AA102607), the Major Project of New Varieties of Genetically Modified Organism of China (2016ZX08009-001), and the State Key Laboratory of Plant Genomics, China (2015B0129-03).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00396/full#supplementary-material
FIGURE S1 | The information of Pvi-MIR319 sequence.
FIGURE S2 | Alignment of mature miR319s.
FIGURE S3 | Alignment of the stem-loop sequences.
FIGURE S4 | Phylogenetic tree analysis of mature miR319a.
FIGURE S5 | The structure and sequences of PvPCF5.
FIGURE S6 | The target site between Pvi-MIR319a and PvPCF5.
TABLE S1 | The information of primers.
References
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis mechanism, and function. Cell 116 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bouton J. H. (2007). Molecular breeding of switchgrass for use as a biofuel crop. Curr. Opin. Genet. Dev. 17 553–558. 10.1016/j.gde.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Chen X. (2008). MicroRNA metabolism in plants. Curr. Top. Microbiol. Immunol. 320 117–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G. S., Tobias C., Sun L., Kraemer F., Li C., Dibble D., et al. (2011). Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108 17550–17555. 10.1073/pnas.1113971108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J. T., Fahlgren N., Carrington J. C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23 431–442. 10.1105/tpc.110.082784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7:1930 10.3389/fpls.2016.01930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David K., Ragauskas A. J. (2010). Switchgrass as an energy crop for biofuel production: a review of its ligno-cellulosic chemical properties. Energy Environ. Sci. 3 1182–1190. 10.1039/B926617H [DOI] [Google Scholar]
- Fabian M. R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Fu C., Sunkar R., Zhou C., Shen H., Zhang J. Y., Matts J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10 443–452. 10.1111/j.1467-7652.2011.00677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter L. E., Tuskan G. A., Wullschleger S. D. (1996). Diversity among populations of switchgrass based on RAPD markers. Crop Sci. 36 1017–1022. 10.2135/cropsci1996.0011183X003600040034x [DOI] [Google Scholar]
- He H., He L., Gu M. (2014). Role of microRNAs in aluminum stress in plants. Plant Cell Rep. 33 831–836. 10.1007/s00299-014-1565-z [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hisano H., Nandakumar R., Wang Z. Y. (2011). “Genetic modification of lignin biosynthesis for improved biofuel production,” in Biofuels: Global Impact on Renewable Energy, Production Agriculture, and Technological Advancements, eds Tomes D., Lakshmanan P., Songstad D. (New York, NY: Springer; ), 223–235. [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Plant Biol. 57 19–53. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2013). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42 D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhou M., Li Z., Luo H. (2014). “MicroRNAs and their potential applications in switchgrass improvements,” in Compendium of Bioenergy Plants: Switchgrass, eds Luo H., Wu Y. (Boca Raton, FL: CRC Press, Tailor & Francis Group; ), 214–227. [Google Scholar]
- Li L., Xue M., Yi H. (2016). Uncovering microRNA-mediated response to SO2 stress in Arabidopsis thaliana by deep sequencing. J. Hazard. Mater. 316 178–185. 10.1016/j.jhazmat.2016.05.014 [DOI] [PubMed] [Google Scholar]
- Li R., Qu R. (2011). High throughput Agrobacterium-mediated switchgrass transformation. Biomass Bioenergy 35 1046–1054. 10.1016/j.biombioe.2010.11.025 [DOI] [Google Scholar]
- Liu L., Chen X. (2016). RNA quality control as a key to suppressing RNA silencing of endogenous genes in plants. Mol. Plant 9 826–836. 10.1016/j.molp.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. R., Cen H. F., Yan J. P., Zhang Y. W., Zhang W. J. (2015). Inside out: high-efficiency plant regeneration and Agrobacterium-mediated transformation of upland and lowland switchgrass cultivars. Plant Cell Rep. 34 1099–1108. 10.1007/s00299-015-1769-x [DOI] [PubMed] [Google Scholar]
- Llave C., Carrington J. C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Matts J., Jagadeeswaran G., Roe B. A., Sunkar R. (2010). Identification of microRNAs and their targets in switchgrass, a model biofuel plant species. J. Plant Physiol. 167 896–904. 10.1016/j.jplph.2010.02.001 [DOI] [PubMed] [Google Scholar]
- McLaughlin S. B., Adams Kszos L. (2005). Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 28 515–535. 10.1016/j.biombioe.2004.05.006 [DOI] [Google Scholar]
- Monti A. (2012). Switchgrass: A Valuable Biomass Crop for Energy. New York, NY: Springer. [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 22534–22539. 10.1073/pnas.0908718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39 787–791. [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., et al. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13 115–125. 10.1016/j.devcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Pandey S. P., Baldwin I. T. (2007). RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant J. 50 40–53. 10.1111/j.1365-313X.2007.03030.x [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. 10.1101/gad.1004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticklen M. (2006). Plant genetic engineering to improve biomass characteristics for biofuels. Curr. Opin. Biotechnol. 17 315–319. 10.1016/j.copbio.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K. P., Sarath G., Saathoff A. J., Mitchell R. B. (2010). “Chapter 17 switchgrass,” in Energy Crops, eds Halford N. G., Karp A. (London: The Royal Society of Chemistry; ), 341–380. [Google Scholar]
- Wagner H., Morgenstern B., Dress A. (2008). Stability of multiple alignments and phylogenetic trees: an analysis of ABC-transporter proteins family. Algorithms Mol. Biol. 3:15 10.1186/1748-7188-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. T., Sun X. L., Hoshino Y., Yu Y., Jia B., Sun Z. W., et al. (2014). MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 9:e91357 10.1371/journal.pone.0091357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuddineh W. A., Mazarei M., Zhang J., Poovaiah C. R., Mann D. G., Ziebell A., et al. (2015). Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol. J. 13 636–647. 10.1111/pbi.12287 [DOI] [PubMed] [Google Scholar]
- Xie F., Frazier T. P., Zhang B. (2010). Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta 232 417–434. 10.1007/s00425-010-1182-1 [DOI] [PubMed] [Google Scholar]
- Yang C., Li D., Mao D., Liu X., Ji C., Li X., et al. (2013). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36 2207–2218. 10.1111/pce.12130 [DOI] [PubMed] [Google Scholar]
- Zhou L., Liu Y., Liu Z., Kong D., Duan M., Luo L. (2010). Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 61 4157–4168. 10.1093/jxb/erq237 [DOI] [PubMed] [Google Scholar]
- Zhou M., Li D., Li Z., Hu Q., Yang C., Zhu L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161 1375–1391. 10.1104/pp.112.208702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Luo H. (2013). MicroRNA-mediated gene regulation: potential applications for plant genetic engineering. Plant Mol. Biol. 83 59–75. [DOI] [PubMed] [Google Scholar]
- Zhou M., Luo H. (2014). Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant Signal. Behav. 9 1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | The information of Pvi-MIR319 sequence.
FIGURE S2 | Alignment of mature miR319s.
FIGURE S3 | Alignment of the stem-loop sequences.
FIGURE S4 | Phylogenetic tree analysis of mature miR319a.
FIGURE S5 | The structure and sequences of PvPCF5.
FIGURE S6 | The target site between Pvi-MIR319a and PvPCF5.
TABLE S1 | The information of primers.