Abstract
Protein hydrolysates are an emerging class of crop management products utilized for improving nutrient assimilation and mitigating crop stress. They generally consist of a mixture of peptides and free amino acids derived from the hydrolysis of plant or animal sources. The present work was aimed at studying the effects and the action mechanisms of a protein hydrolysate derived from animal residues on maize root growth and physiology in comparison with the effects induced by either free amino acids or inorganic N supply. The application of the protein hydrolysate caused a remarkable enhancement of root growth. In particular, in the protein hydrolysate-treated plants the length and surface area of lateral roots were about 7 and 1.5 times higher than in plants treated with inorganic N or free amino acids, respectively. The root growth promoting effect of the protein hydrolysate was associated with an increased root accumulation of K, Zn, Cu, and Mn when compared with inorganic N and amino acids treatments. A microarray analysis allowed to dissect the transcriptional changes induced by the different treatments demonstrating treatment-specific effects principally on cell wall organization, transport processes, stress responses and hormone metabolism.
Keywords: biostimulant, ionomic analysis, hormone metabolism, maize, microarray analysis, protein hydrolysates, root, transport
Introduction
The availability of mineral nutrients in the soil represents one of the most important limiting factors for crop productivity which is therefore highly dependent on the vast use of fertilizers (Tilman et al., 2002). However, excessive application of fertilizers as well as agrochemicals is causing severe environmental problems resulting in massive ecological degradation throughout the world (Tilman et al., 2002) Hence, the improvement of crop nutrient use efficiency that could reduce the use of fertilizers without any penalty on productivity, is worldwide an important goal (Baligar et al., 2001).
In this scenario, biostimulants are an emerging class of crop management products aiming at the mitigation of crop stress and improvement of nutrient assimilation (Halpern et al., 2015). The European Biostimulant Industry Council (EBIC) define these products as “materials which contain substance(s) and/or microorganisms whose function when applied to plants or the rhizosphere is to stimulate natural processes to benefit nutrient uptake, nutrient use efficiency, tolerance to abiotic stress, and/or crop quality, independently of its nutrient content” (http://www.biostimulants.eu). The emerging biostimulants market is estimated to grow of 10.4% from 2016 to 2021, reaching a value of 2.91 billion USD and an area of application of 24.9 million hectares by 2021 (http://www.marketsandmarkets.com/search.asp?Search=biostimulants).
A variety of biostimulant compounds are available in the market (reviewed in Calvo et al., 2014). They are classified as microbial inoculants, humic substances, fulvic acids, protein hydrolysates and amino acids, and seaweed extracts. These formulations are usually composed by different molecules and therefore their effect can be the result of many components that may work synergistically. The positive effects of biostimulants on plants include: yield increase (Ertani et al., 2009), increase of abiotic stress tolerance (Zhang et al., 2003; El Hadrami et al., 2010) and nutrient assimilation (Varanini and Pinton, 1995; Canellas et al., 2002), enhancement of fruit quality (Masny et al., 2004; Karakurt et al., 2009) and soil microbial activity (Chen et al., 2002). Their positive influence on plant growth is not due to a direct fertilization effect because they are active at very low concentration (Calvo et al., 2014). They indeed exhibit auxin-like and gibberellin-like activities and thus they are thought to function as signaling molecules (Ertani et al., 2009, 2013).
The protein hydrolysates have been proven to stimulate root growth and leaf biomass of several crops. du Jardin (2015) reviewed various effects resulting from the application of these compounds to crops. Direct effects on plants include modulation of N uptake and assimilation by regulation of enzymes involved in N metabolism and by acting on the signaling pathway of N acquisition in roots (Ertani et al., 2009, 2013). They can also regulate enzymes of the TCA cycle, contributing to the interplay of C and N metabolisms (Schiavon et al., 2008). Protein hydrolysates can improve plant antioxidant defense against free radicals thus mitigating environmental stress (du Jardin, 2015). They are also known to increase microbial biomass and activity, soil respiration and soil fertility (du Jardin, 2015). The application of protein hydrolysates can modify the morphology of the roots, facilitating nutrient uptake as a consequence of the increased absorptive surface area (Ertani et al., 2013). Moreover, the chelating and complexing activities of specific amino acids and peptides in the substrates are supposed to enhance nutrients availability and acquisition by roots (Colla et al., 2014).
In perspective of a circular economy, the use of protein hydrolysates can contribute to environment protection. Indeed, protein hydrolysates are generally produced from industrial and agricultural organic waste, turning them into high value-added products and, at the same time, reducing the costs derived from their disposal.
Although the effects of protein hydrolysates on crop performance have been documented, the scientific basis of their action has partially been elucidated mainly due to the complex nature of these products. The present work aims at shedding light on the effects and the action mechanisms of a commercial protein hydrolysates. The product used in this work is obtained by chemical hydrolysis of animal by-products and consists of a mixture of small peptides and a low percentage of free amino acids. We assessed the effects of the protein hydrolysates on maize root growth in comparison with the effects produced by an equal amount of N supplied either as a free amino acids mixture mimicking the biostimulant composition, or as N inorganic compound (NH4H2PO4). It is noteworthy that free amino acids are also included among the biostimulant compounds (Calvo et al., 2014). In order to elucidate the mechanisms underlying the effects observed in the root apparatus, root micro- and macro-nutrient accumulation was evaluated. Furthermore, we performed a transcriptome analysis that allowed identifying differential gene expression patterns in maize roots in response to the different forms of N supply highlighting global changes in gene transcription across multiple metabolic processes.
Materials and methods
Plant materials and growth conditions
Maize seeds (P0423 Hybrid, Pioneer Italia S.p.A.) were soaked in water for 24 h and germinated in the dark on wet filter paper for 72 h. The seedlings were then transferred to plastic pots containing 2 L of a 0.05 mM CaSO4 solution and grown for 24 h under a 16/8 h light/dark regime at 22–26°C, 40–50% relative humidity, 125 μE m−2s−1 light intensity. Each pot contained 12 seedlings that were grown in a diluted nutrient solution (Pinton et al., 1999) containing 100 mM MgSO4, 5 μM KCl, 200 μM K2SO4, 175 μM KH2PO4, 400 μM CaSO4, 25 μM NH4H2PO4, 2.5 μM H3BO3, 0.2 μM MnSO4, 0.2 μM ZnSO4, 0.05 μM CuSO4, 0.05 μM NaMoO4, 2 μM Fe-EDTA and supplemented with either protein hydrolysates (SICIT2000 S.p.A.) or inorganic nitrogen (NH4H2PO4) or a mixture of free amino acids mimicking the amino acids content of the protein hydrolysates (see also Results and Discussion section for treatment description). In all the treatments, the total N amount was kept constant at 5.65 or 11.3 mgL−1. After 3 days, roots of 24 seedlings for each treatment (protein hydrolysate, inorganic N and free amino acids) at 5.65 or 11.3 mgL−1 total N dose were collected for further analysis. The experiment was run three times obtaining three independent biological replicates.
Phenotypic analysis of maize seedlings
Primary, seminal and lateral root average length was evaluated using ImageJ software. For the measurement of lateral roots length, the 10 longest roots per plant were considered. Primary, seminal and lateral root total length and surface area were measured with the aid of WinRHIZO™ scanner and automated software (Arsenault et al., 1995).
Macro- and micro-nutrients quantification
The nitrogen concentration of the root samples was determined using the EA-IRMS Delta V (Thermo Fisher Scientific). The calibration curve for %N determination in dried tissues was performed using atropine (%N = 4.84).
Other macro- and micro-nutrients were quantified by ICP-MS analysis. Dried root samples (about 5 mg) were weighted and digested in a TFM microsampling insert using 250 μl of 69% ultrapure HNO3. Three inserts were put into 100-ml oven vessel containing 10 ml of water (milliQ, 18.2 M cm) and 1 ml of 30% H2O2. In addition, 5 mg of the following reference material were digested: NIST 1515 (apple leaves). Sample digestion was performed using a microwave oven (Milestone StartD® microwave). A 20-min ramping period was used to reach a digestion temperature of 180°C, which thereupon was maintained for 20 min. At the end, sample were diluted with water (milliQ, 18.2 M cm) to a final concentration of 3% HNO3. Multi-elemental analysis was carried out using the Agilent 7500cx ICP-MS (Agilent). The instrument was tuned using tuning solution (Agilent tuning solution 1 ppb) in a standard mode checking the sensitivity of masses 7Li, 89Y, and 205Tl and the oxide and double charged ion levels (<2%). Each macro- and micronutrient were quantified using a multi-element standard solution.
RNA extraction and microarray analyses
Total RNA was isolated from plants treated with protein hydrolysates, inorganic N and free aminoacid mixture at the highest N concentration (11.3 mgL−1) using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich) and quantified by spectrophotometry using NanoDrop™ 1000 (Thermo Scientific). RNA quality was evaluated using Agilent 2100 Bioanalyzer (Agilent). For each sample, the reactions of cRNA synthesis and labeling were carried out using 200 ng of total RNA and the Low Input Quick Amp Labeling Kit, One-Color (Agilent) and Cyanine 3 (Cy3)-CTP fluorescent dye according to the Agilent technical manual (http://www.agilent.com). Cy3-labeled cRNA (1.65 μg) of each sample was hybridized on a custom 4x44K Agilent array according to manufacturer's manual for 17 h at 65°C and scanned on Agilent G2565CA Microarray Scanner System (Agilent). Array hybridizations and washing were performed according to manufacturer's manual (One-Color Microarray-Based Gene Expression Analysis—Low Input Quick Amp Labeling—Protocol). Each “subarray” allow analyzing the expression of 39,372 maize transcripts predicted from the B73 reference genome (ftp://ftp.gramene.org/pub/gramene/maizesequence.org/release-5b/). Probe design was performed using Agilent eArray (http://www.genomics.agilent.com). Complete description of chip is available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entry (GPL22578). Feature intensities were extracted using Agilent's Feature Extraction Software 10.5.1.1 (Agilent). The hybridization data all samples were normalized using the value of the 75th percentile. Differentially expressed transcripts between Bio vs. N, Aa vs. N, and Bio vs. Aa were identified through Student's t-test using MeV software (http://mev.tm4.org/#/welcome) setting with the following parameters: p-value based on permutation with critical p-value of 0.01 and adjusted Bonferroni correction. Differentially expressed transcripts were filtered on the basis of fold changes value (|FC|≥2). All microarray expression data are available at the GEO (http://www.ncbi.nlm.nih.gov/geo) under the series entry (GSE89535).
Quantitative RT-PCR analysis
For the quantitative RT-PCR (qRT-PCR) we used the same RNA samples extracted as described above. Three cDNA samples derived from 3 independent RNA samples were analyzed. DNase treatment and reverse transcription were performed as described in Molesini et al. (2014). cDNA amplification and PCR cycling conditions and product dissociation curve were also performed as indicated in Molesini et al. (2014). Data from qRT-PCR experiments were analyzed according to the 2−ΔΔCt method. The list of primers adopted for qRT-PCR is reported in Supplementary Table 1. UBCE gene, coding for ubiquitin-conjugating enzyme, was used as reference gene (Manoli et al., 2012).
Results and discussions
Protein hydrolysates and free amino acids display different stimulatory effects on root growth
To investigate the effects of protein hydrolysates, we grew maize seedlings for 72 h after the emergence of the primary root in a N-free nutrient solution supplemented with the protein hydrolysate (Bio), inorganic nitrogen (NH4H2PO4;N) or a mixture of amino acids (Aa) mimicking the composition in amino acids of the protein hydrolysate. The protein hydrolysate is a liquid formulate derived from the hydrolysis of cow connective tissue, a by-product of tanning industry. It contains 30% (w/w) organic matters (C), 11.3% (w/w) total nitrogen (N), 10% (w/w) organic N, of which 62.5% (w/w) total amino acids and 10% (w/w) free amino acids (the detailed amino acids composition is summarized in Supplementary Table 2). The molecular weights of the peptides present in the protein hydrolysates range from 1,500 to 2,000 Da. To evaluate the dose response and the relative effects of Bio treatment on roots and shoots, we supplied the seedling with increasing doses of the biostimulant from 0.001 to 0.1 mlL−1. The growth of the shoots was not affected by the treatments, whereas the protein hydrolysate at 0.05 and 0.1 mlL−1 promoted the growth of the roots (Supplementary Figure 1). To analyze the effects of Bio treatments in terms of their contribution to N supply in the growth medium, we compared the root growth of maize seedlings treated with Bio at 0.05 and 0.1 mlL−1 and seedlings treated with equivalent amounts of total N (5.65 and 11.3 mgL−1, respectively) supplied either as inorganic N (NH4H2PO4) or free Aa. The Aa treatment consisted of a mixture of free amino acids identical in composition and concentration to the amino acids present in the protein hydrolysates described in Supplementary Table 2. Both Bio treatments induced root growth (Figures 1A–C), this effect was particularly evident for the lateral roots whose average length was approximately 2 and 3 times higher than that of seedlings supplied with 5.65 and 11.3 mgL−1 inorganic N, respectively. Also the Aa treatments showed the capacity to promote root growth compared to N. The effect was detectable in the primary and seminal roots at the higher Aa concentration, whereas the average length of lateral roots was increased also with the lower concentration (Figures 1A–C). Interestingly, the protein hydrolysates, containing only 10% of free amino acids, had always a stronger effect on root growth than a treatment consisting of free amino acids only.
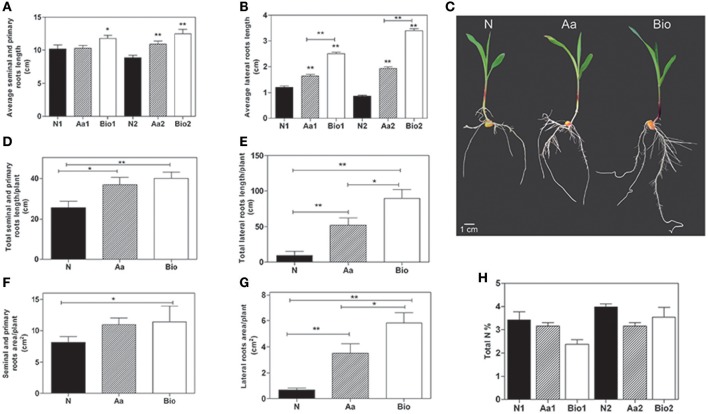
Figure 1.
Phenotypic analysis of maize roots after 3 days of treatment with inorganic nitrogen (N), amino acids (Aa), or protein hydrolysates (Bio). Average seminal and primary root length (A) and average lateral root length (B) of seedlings treated with protein hydrolysates (0.05 and 0.1 mlL−1) and seedlings treated with equivalent amounts of total N (5.65 and 11.3 mg L−1, respectively) supplied either as inorganic nitrogen (N) or as a mixture of free amino acids mimicking the composition in amino acids of the protein hydrolysate product. Root length was evaluated using ImageJ software (http://imagej.net). For lateral root length determination, the 10 longest roots were chosen manually. (C) Representative maize seedlings from N2, Aa2, and Bio2 treatments. Total length of seminal and primary roots (D), total length of lateral roots (E), total surface area of primary and seminal roots (F), and total surface of lateral roots (G) of seedlings treated with a concentration of protein hydrolysates, free amino acids and inorganic nitrogen equal to11.3 mgL−1 of total N measured with WinRHIZO™ software. (H) Total N content in roots of seedlings treated with protein hydrolysates, free amino acids and inorganic N at two doses. In (A,B,H) N1, Aa1, and Bio1 refer to the lowest N dose (5.65 mg L−1) and N2, Aa2, and Bio2 refer to the highest one (11.3 mg L−1). The average values are reported. Bars represent the standard error (SEM) [n = 24, except for data in (H), where n = 3], if not otherwise specified, Student's t-test was applied vs. N-treated plants, *P < 0.05; **P < 0.01.
A rapid and efficient growth of root apparatus can be advantageous during the first phases of seedling emergence after sowing, increasing the seedling capacity to absorb water and mineral elements (Lynch, 1995). Therefore, we calculated the total root length and area per plant of seedlings subjected to the different treatments using the WinRHIZO™ apparatus. The total length of both primary and seminal and lateral roots per plant was significantly higher in Bio (0.1 mlL−1) and Aa-treated seedlings than in those supplied with N (Figures 1D–G). The most striking effect was observed as expected, on total lateral root length. Considering the total surface area of the primary and seminal roots per plant, the highest value was measured in the Bio-treated seedlings, whereas the Aa treatment was ineffective (Figure 1F). The total surface area of the lateral roots increased by ~3 and 5-fold in Aa and Bio-treated seedlings, respectively as compared with seedlings treated with N (Figure 1G). Collectively, these results demonstrate the marked promoting effect of low doses of Bio on lateral root development; this effect is superior to that produced by free amino acids and inorganic N treatments equalized for N content.
To assess whether the effects of protein hydrolysate and free amino acids were due to increased N accumulation, we determined the total N concentration in roots of seedlings treated with Bio, N and Aa and N at two doses, corresponding to the addition of 5.65 and 11.3 mgL−1 N, respectively. The total N concentration in Bio-, Aa-, and N-treated roots did not differ (Figure 1H).
Protein hydrolysates increase the uptake of specific nutrients
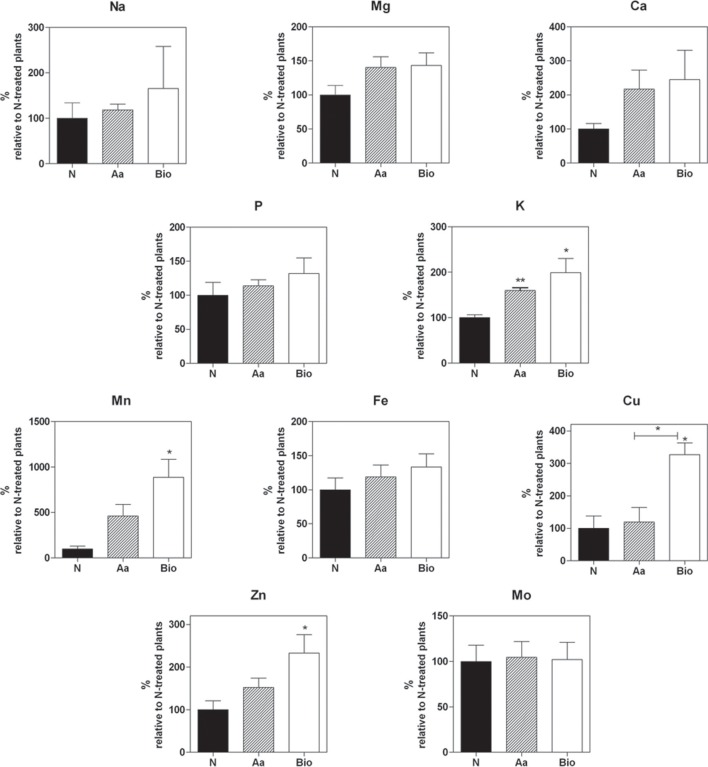
To study the effects of Bio and Aa on root nutrient accumulation, we quantified the macro- and micro-nutrient concentrations in the roots of seedlings treated with Bio, N, and Aa; in all the treatments, the total N supply was equal to 11.3 mgL−1 (Figure 2). The root concentration of Ca, Mg, Na, and P did not show statistically significant variations irrespectively from the treatment applied (Figure 2). Among the macronutrients, only K concentration was significantly increased in Bio- and Aa-treated seedlings compared with N-treated ones (Figure 2). Regarding the micronutrients, the concentration of Fe and Mo was not modified by the different treatments, whereas Cu, Mn, and Zn concentrations increased Bio-treated roots, but not in Aa-treated roots. The strongest effect of the Bio treatment was observed for Mn whose concentration was more than 8-fold higher in Bio-treated than in N-treated roots (Figure 2). The increased level of K in roots treated with organic N might be related to its function in maintaining ion balance and stabilizing cellular pH. The improved accumulation of Cu, Mn, and Zn in protein hydrolysate-treated roots might be the result of a specific action on metal trasporters (see Section Transport Processes) or the consequence of the peptide metal binding capacity that might facilitate nutrient availability.
Figure 2.
Concentrations of macro- and micro-nutrients in the roots of seedling treated with protein hydrolysate, free amino acids and inorganic N. Mg, K, Mn, Fe, Cu, Zn, Na, Ca, Mo, P root concentration was measured by means of high throughput inductively coupled plasma-mass spectroscopy (ICP-MS). In all the treatments, the total N supply was equal to 11.3 mgL−1. The nutrient concentrations are expressed as percentage of concentrations measured in seedlings treated with inorganic nitrogen. The average values are reported. Bars represent the standard error (SEM) (n = 3). If not otherwise specified, Student's t-test was applied vs. N-treated plants, *P < 0.05; **P < 0.01.
Global changes in the root transcriptome in response to protein hydrolysates and free amino acids
The transcriptional changes in maize roots subjected to 3-day treatment with Bio, N, or Aa were analyzed by means of genome-wide microarray hybridization analysis. For this aim, we used an Agilent chip that allowed to analyse the expression of 39,372 among the predicted maize transcripts (Schnable et al., 2009; Release 5b; http://www.maizesequence.org/index.html).
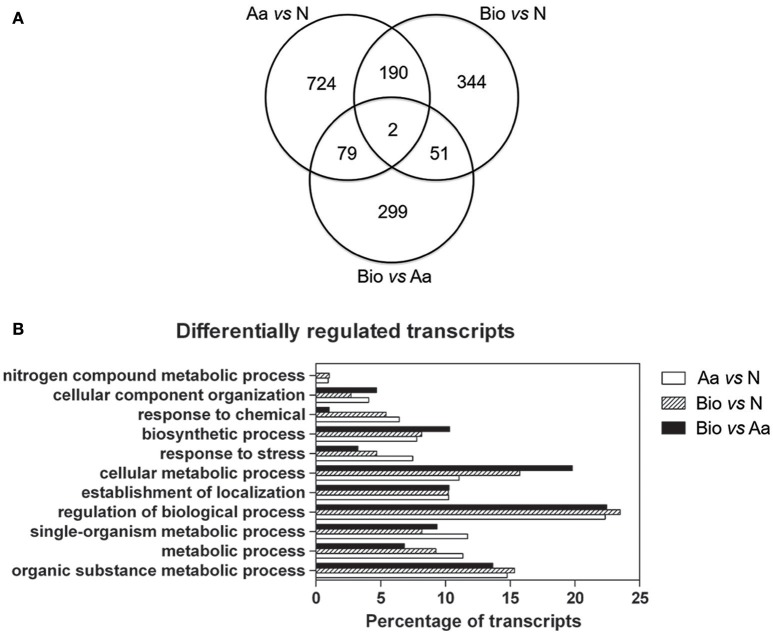
Differentially expressed transcripts between roots supplied with different N forms were identified through a t-test (adjusted p-value ≤ 0.01 and |FC| ≥ 2). The analysis revealed that 995 transcripts were differentially expressed between Aa- and N-treated (Supplementary Table 3), 587 between Bio- and N-treated roots (Supplementary Table 4) and 431 between Bio- and Aa-treated roots (Supplementary Table 5) (Figure 3A), indicating high dissimilarity between the three transcriptional profiles. Moreover, 79 transcripts were differentially expressed both in the comparisons Aa vs. N and Bio vs. Aa, 51 were in common between Bio vs. Aa and Bio vs. N and 190 between Bio vs. N and Aa vs. N (Figure 3A) (Supplementary Tables S3–S5). Only two transcripts were differentially expressed in all the three comparisons (Supplementary Tables S3–S5). The transcriptional profile of 5 differentially expressed transcripts (GRMZM2G347457_T01, GRMZM2G096958_T01, GRMZM2G429955_T01, GRMZM2G030036_T01, and GRMZM2G024996_T01, coding respectively for peptide transporter, nicotianamine aminotransferase1, chlorophyll a-b binding protein 2, nicotianamine synthase 2, glycine-rich cell wall structural protein genes) was validated through qRT-PCR (Supplementary Figure 2). The annotation of all the up- and down-regulated transcripts was hand-curated, assigning them a “Gene Ontology” (GO) biological process term on the basis of a BlastP analysis. The transcripts were then grouped in main functional categories. In all the comparisons about 50% of the transcripts encode proteins with an unknown function and were assigned to the “biological process” class (Figure 3B, Supplementary Tables S3–S5). The other most abundant categories are “regulation of biological process,” “organic substance metabolic process” and “cellular metabolic process.” Interestingly, the transcripts belonging to “response to stress” were highly represented in Aa vs. N comparison while they were less abundant in Bio vs. Aa and Bio vs. N. Noticeably, the “nitrogen compound metabolic process” category was poorly represented, whereas the transcripts related to cellular transport (“establishment of localization”) were quite abundant.
Figure 3.
Distribution of differentially regulated genes in the three comparisons and main functional categories of differentially expressed transcripts. (A) Venn diagrams showing the shared and the specific differentially regulated transcripts in the different treatments (|FC| ≥ 2; adjusted p-value ≤ 0.01). (B) Distribution of differentially regulated transcripts in the three comparisons Bio vs. Aa, Bio vs. N and Aa vs. N grouped into main functional categories. For each functional category, the transcript percentage is calculated on the total of the differentially expressed transcripts minus those belonging to the “biological process” category.
Transcription factors
The “regulation of biological process” category included several transcripts encoding transcription factors (TFs). We identified 61 TF transcripts in the Bio vs. N, 35 in Bio vs. Aa and 89 in the Aa vs. N comparisons (Supplementary Tables S3–S5).
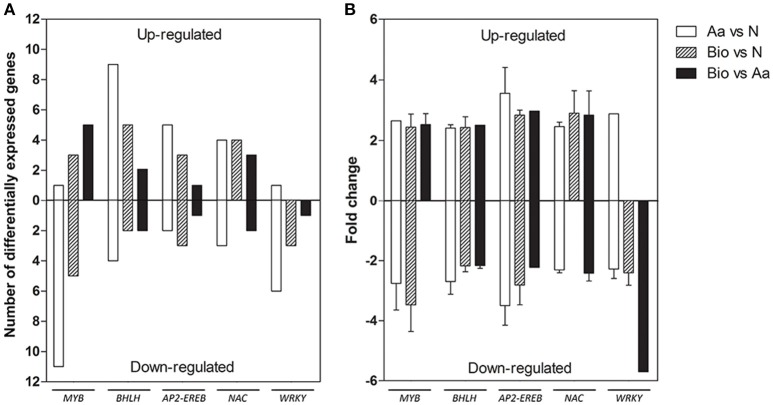
Concerning the distribution of these transcripts in TF gene families, AP2-EREB, bHLH, MYB, WRKY, NAC were the most represented families (Figure 4). Recently, AP2-EREBP, bHLH, MYB, and WRKY TF families have been shown to participate in the response to nutrient stress playing a major role in controlling regulatory network related to root development and N-deficiency response (Zhao et al., 2005; Rushton et al., 2012; Takehisa et al., 2013; He et al., 2016). Tai et al. (2016) also reported that members of some of these TF families were differentially expressed in primary, crown and seminal roots of maize, suggesting functional specialization of the different root types.
Figure 4.
Differentially expressed genes belonging to the most represented transcription factors families detected in the three comparisons, divided into up- and down-regulated. (A) Number of differentially expressed genes. (B) Expression fold change. The data reported are means of FC values ± SD calculated for each transcription family except for those transcription factor families comprising a single transcript.
AP2-EREB transcripts were both up- and down-regulated in the different comparisons, whereas those encoding WRKYs were mostly down-regulated in all the three comparisons. A member of AP2-EREB family, preferentially expressed in coleoptile nodes during maize root development, plays a role in the formation of crown roots (Muthreich et al., 2013). PLETHORA2 in Arabidopsis (baby boom1, the homolog in maize) encodes another AP2-EREBP TF required for the formation of root stem cells (Aida et al., 2004). AP2-EREBP and WRKY family were proven to be involved in the differential root response to N limitation of two Chinese maize inbred lines (Chen et al., 2015). WRKY transcripts were also induced in rice root after N, P, and K deficiency (Takehisa et al., 2013).
MYB proteins are important regulator of different physiological processes in plants, including development, metabolism and responses to environmental stresses (Li et al., 2015). In our work, a large number of MYB transcripts were modulated in response to the different N sources, resulting all up-regulated in the comparisons Bio vs. Aa, whereas in the comparison Aa vs. N, 11 members of this family were down-regulated. Some members of MYB family in Arabidopsis and rice regulate lateral root development and modulate auxin-mediated progression of lateral root development (Dai et al., 2012; Gibbs et al., 2014). MYB TFs can also respond to N deficiency; a member of this family in cucumber showed a rapid induction after N deprivation and functions in ethylene and auxin signaling (Zhao et al., 2015). Interestingly, these results well fit with the enhanced lateral growth in roots of Bio-treated seedlings (Figure 1C).
Gene family encoding bHLH TFs was largely up-regulated in all the three comparisons. This TF family is implicated in the control of various biological pathways, including the plant response to nutrient deprivation (Yang et al., 2016). A rice bHLH family member (OsPTF1) mediates tolerance to P deprivation in rice (Yi et al., 2005). Another member of this family plays an important role in adaptation to the P- and N-starvation in Triticum aestivum regulating genes involved in the uptake of P and N (Yang et al., 2016).
Several NAC transcripts were also differentially modulated in all three treatments. A variety of NAC members isolated from different species participate in both biotic and abiotic stress signaling pathways enhancing drought and salt stress tolerance (Liu et al., 2014; Su et al., 2015). Lu et al. (2015) analyzed the function of 7 maize NAC demonstrating their role in ABA-dependent abiotic stress responses. Interestingly, a NAC member of Populus tremula x Populus alba, is implicated in roots response to N deficiency, probably regulating root growth under low N conditions (Wei et al., 2013). Similarly, NAC29 in Arabidopsis showed an elevated expression after N deprivation, and it was also responsive to chronic low N (Peng et al., 2007).
An interesting TF gene family is that of the LATERAL ORGAN BOUNDARIES domain (LBD) proteins. Intriguingly, every transcript belonging to this family was down-regulated in all the pairwise comparisons (AC149818.2_FGT009 and GRMZM2G092542_T01 in Bio vs. N; GRMZM2G092483_T01 in Bio vs. Aa; GRMZM2G092542_T01, AC234149.1_FGT002, AC149818.2_FGT009 and GRMZM2G073044_T01 in Aa vs. N) (Supplementary Tables S3–S5). These proteins display high functional diversity including regulation of lateral root formation and N metabolism (Xu et al., 2016). In Arabidopsis, LBD16, LBD18, and LBD29 regulate lateral root organogenesis acting on auxin signaling pathway (Feng et al., 2012). The maize LBD protein RTCS (down-regulated in Aa vs. N and Bio vs. N) the closest homologs of AtLBD16/29, is involved in seminal and shoot-borne root initiation (Taramino et al., 2007). Moreover, lateral root emergence requires LDB-dependent activation of EXPANSIN (Kim and Lee, 2013). Members of the LBD family were shown to negatively regulate genes involved in response to N limitation in Arabidopsis (Rubin et al., 2009).
Finally, two members of TEOSINTE BRANCHED1/CYCOIDEA/PROLIFERATING CELL FACTOR (TCP) family were up-regulated in the comparisons Bio vs. N (GRMZM2G180568_T01) and Bio vs. Aa (GRMZM2G060319_T01). A member of this family (TCP20) expressed in root tips and vascular tissue of Arabidopsis, was shown to modulate lateral root growth in response to N supply and to regulate the nitrate transporter NRT1.1 expression (Guan et al., 2014).
Overall, this analysis may suggest that the different N sources produce different effects on root growth and metabolism by distinct modulation of TFs involved in the control of root development and N availability.
Cell wall components
In the present work, we found numerous differentially expressed genes belonging to “cellular component organization” category in all the pairwise comparisons (18 in Aa vs. N, 6 in Bio vs. N and 7 in Bio vs. Aa) (Table 1). Almost all the differentially expressed transcripts of this category encode extensins, expansins, pectinesterases, Casparian strip membrane proteins, xyloglucanendotransglucosylases/hydrolases, and glycine-rich cell wall structural proteins. In particular, the transcript GRMZM2G024996_T01, coding for a glycine-rich cell wall structural protein (GRP), showed the highest level of down-regulation in Bio vs. N (-19) and Aa vs. N (-63). This transcript showed high homology with a Petunia hybrida gene encoding GRP1 that is expressed during cell expansive growth and repressed during lignification (Condit, 1993). Genes involved in cell wall remodeling can regulate root growth and lateral root formation. Transcripts coding for expansins were modulated in all the three comparisons. It is well documented that these enzymes loosen the network of wall polysaccharides, allowing turgor-driven cell enlargement (Cosgrove, 2000). In addition, two transcripts for root Casparian strip proteins were up-regulated specifically in Aa vs. N. These proteins apparently regulate the transition of the lateral root primordia from flat to rounded morphology during root development (Lucas et al., 2013). Four and one transcripts coding for xyloglucanendotransglucosylases/hydrolases were differentially expressed in Aa vs. N and Bio vs. Aa, respectively. These enzymes play an important role in the remodeling of the xyloglucan/cellulose framework in the wall during cell growth and differentiation (Hara et al., 2014). Similarly, pectinesterase enzymes (modulated in Aa vs. N and Bio vs. N) are involved in the process of cell wall extension.
Table 1.
Differentially expressed transcripts involved in cell wall organization.
| Genome ID | UniProt ID | Description | Aa vs. N (FC) | Bio vs. N (FC) | Bio vs. Aa (FC) |
|---|---|---|---|---|---|
| GRMZM2G070913_T01 | K7VZR1 | Pectinesterase | 3.77 | 2.81 | |
| GRMZM2G112619_T01 | K7U9U0 | Xyloglucan endotransglucosylase/hydrolase | 3.28 | ||
| GRMZM2G083888_T01 | C5Y9U6 | Casparian strip membrane protein 4 | 2.93 | ||
| GRMZM5G886185_T01 | A0A096UH62 | Xyloglucan endotransglucosylase/hydrolase | 2.80 | ||
| GRMZM5G858456_T01 | B6SRP0 | Fucosyltransferase 7 | 2.80 | ||
| GRMZM2G455564_T01 | K7TZQ3 | Pectinesterase | 2.41 | ||
| GRMZM2G127184_T01 | B6SRP0 | Fucosyltransferase 7 | 2.39 | ||
| GRMZM2G073079_T01 | Q9ZT66 | Endo-1,3;1,4-beta-D-glucanase | 2.24 | ||
| GRMZM2G110832_T01 | B6T959 | Casparian strip membrane protein 1 | 2.23 | ||
| GRMZM2G144898_T01 | Q1ZYQ8 | Expansin-B10 | 2.00 | ||
| GRMZM2G114322_T01 | Q8H274 | Expansin-like A3 | −2.02 | −2.41 | |
| GRMZM2G435380_T01 | M8BPN6 | Polygalacturonase | −2.06 | ||
| GRMZM2G168651_T01 | P14918 | Extensin | −2.11 | ||
| GRMZM5G870571_T01 | M7ZVY5 | Galactoside 2-alpha-L-fucosyltransferase | −2.16 | ||
| GRMZM2G392125_T01 | A0A096TJQ7 | Xyloglucan endotransglucosylase/hydrolase | −2.22 | ||
| GRMZM2G167637_T01 | A0A096SUN3 | Pectinesterase | −2.37 | ||
| GRMZM2G113761_T01 | A0A096RUI5 | Xyloglucan endotransglucosylase/hydrolase | −2.74 | ||
| GRMZM2G024996_T01 | P27483 | Glycine-rich cell wall structural protein | −63.07 | −19.32 | |
| GRMZM2G127072_T01 | B6UAK6 | Expansin-like 3 | 2.06 | ||
| GRMZM2G043943_T01 | A0A096QHT7 | Pectinesterase | −2.01 | ||
| GRMZM2G109842_T01 | P35082 | Profilin-2 | −3.45 | ||
| GRMZM2G164785_T01 | P0C1Y5 | Expansin-B11 | 5.68 | ||
| GRMZM2G152189_T01 | B6STF8 | Vegetative cell wall protein gp1 | 2.71 | ||
| GRMZM2G153666_T01 | B6UB39 | Polygalacturonase | 2.54 | ||
| GRMZM2G413006_T01 | A0A096TNC3 | Xyloglucan endotransglucosylase/hydrolase | −2.19 | ||
| GRMZM2G118759_T01 | B6TEE0 | Glycine-rich cell wall structural protein 2 | −2.24 | ||
| GRMZM2G469701_T01 | M7ZJB5 | Expansin-A22 | −3.29 | ||
| GRMZM5G870571_T01 | K7V5L2 | Uncharacterized protein | −3.58 |
Genome ID, Maize transcript ID (ZmB73_5b_FGS_cdna.fasta.gz); FC, fold change value.
Taken together these results suggest that transcriptional changes in genes encoding cell wall modifying enzymes induced by Bio and Aa, may results in cell wall remodeling that in turn affects root growth and architecture.
Stress-related transcripts
A relatively high number (38) of stress-related transcripts were differentially regulated in Aa vs. N comparison, whereas they were 14 and 6 in Bio vs. N and Bio vs. Aa, respectively (Table 2). About 50% of the stress related transcripts in Aa vs. N were represented by those coding for peroxidases (12 up-regulated, 5 down-regulated). At first glance, it is clear how the Aa treatment compared with N caused a higher stress response in the roots than Bio did. A possible explanation is that Aa caused an increase in ROS activity followed by an increase in expression of peroxidase genes for preventing H2O2 damage. Up-regulation of stress-related genes such as those for peroxidases and superoxide dismutases was observed in the root transcriptome of rice grown under P, K, and N deficiency (Takehisa et al., 2013). Moreover, ROS, such as H2O2 and are known secondary messages in several pathways associated with responses to biotic and abiotic stresses in plants (Apel and Hirt, 2004). Aside from their anti-oxidative activity, peroxidases are the most abundant enzymes in the cell wall where they display a multifunctional activity, also related to growth regulation (Vuletic et al., 2014). Peroxidases exist in multiple forms exhibiting different cellular localization and playing numerous biological functions. Therefore, to identify the specific involvement of peroxidases in the response of roots to Aa and Bio would require further investigation. Among the other differentially expressed genes in this category, the majority are transcripts involved in biotic stress response (Table 2).
Table 2.
Differentially expressed stress-related transcripts.
| Genome ID | UniProt ID | Description | Aa vs. N (FC) | Bio vs. N (FC) | Bio vs. Aa (FC) |
|---|---|---|---|---|---|
| GRMZM2G138450_T01 | A0A096SAM6 | Peroxidase | 19.08 | ||
| GRMZM2G410175_T01 | D7NLB3 | Peroxidase | 15.97 | ||
| GRMZM2G150731_T01 | B4FYH1 | Peroxidase | 6.40 | ||
| GRMZM2G407740_T01 | C0P3T3 | Peroxidase | 5.32 | ||
| GRMZM2G133475_T01 | A5H454 | Peroxidase 66 | 4.86 | ||
| GRMZM2G027217_T01 | A0A096Q770 | Peroxidase | 4.76 | ||
| GRMZM2G010640_T01 | A0A096PWP5 | Peroxidase | 4.27 | ||
| GRMZM2G404676_T01 | A0A096TLS6 | Peroxidase | 4.19 | 2.11 | |
| GRMZM2G023840_T01 | K7VCN5 | Peroxidase | 3.46 | ||
| GRMZM2G009792_T01 | A0A096PVZ5 | Uncharacterized protein | 3.41 | ||
| GRMZM2G037156_T01 | K7UT08 | Peroxidase | 3.29 | ||
| GRMZM2G089982_T01 | B6T3V1 | Peroxidase | 2.17 | ||
| GRMZM2G405459_T01 | A0A096TLY3 | Peroxidase | 2.16 | ||
| GRMZM2G033665_T01 | C5XUZ2 | MLO-like protein | −2.06 | −2.54 | |
| GRMZM2G017116_T01 | B6TR53 | Defense-related protein | −2.07 | −2.75 | |
| GRMZM2G103342_T01 | A0A096RME9 | Peroxidase | −2.18 | ||
| GRMZM2G108847_T01 | A3FMA3 | Putative serine type endopeptidase inhibitor | −2.22 | −2.01 | |
| GRMZM2G112538_T01 | Q29SB6 | Pathogenesis-related protein 10 | −2.33 | ||
| GRMZM2G112488_T01 | D4HR93 | Pathogenesis-related protein 10 | −2.45 | ||
| GRMZM2G112524_T01 | B6TR52 | Pathogenesis-related protein 1 | −2.52 | −2.11 | |
| GRMZM2G430500_T01 | A0A096TRQ6 | Uncharacterized protein | −2.60 | ||
| GRMZM2G374971_T01 | P33679 | Zeamatin | −2.61 | −3.07 | |
| AC197758.3_FGT004 | K7U6B6 | Peroxidase | −2.67 | ||
| GRMZM2G108207_T01 | B4FH68 | Peroxidase | −2.67 | ||
| GRMZM2G466563_T01 | Q6YYA1 | Putative calmodulin-binding protein | −2.72 | ||
| GRMZM2G117971_T01 | A0A059Q1C7 | Pathogenesis-related protein | −2.72 | ||
| GRMZM2G422240_T01 | B4G197 | 16.9 kDa class I heat shock protein 1 | −2.73 | ||
| GRMZM2G061766_T01 | K7V347 | Uncharacterized protein | −2.88 | ||
| GRMZM2G171078_T01 | A0A096SWX6 | Peroxidase | −2.89 | ||
| GRMZM5G899188_T01 | Q08275 | 17.0 kDa class II heat shock protein | −3.36 | ||
| AC214360.3_FGT001 | Q6Z5J6 | Ent-pimara-8(14),15-diene synthase | −3.44 | ||
| GRMZM2G176085_T01 | B4FYD8 | Peroxidase | −3.46 | ||
| GRMZM2G158232_T01 | B4G197 | 16.9 kDa class I heat shock protein 1 | −3.49 | −2.22 | |
| GRMZM2G335242_T01 | B6TQD6 | 17.4 kDa class I heat shock protein | −3.50 | ||
| GRMZM2G028306_T01 | Q1EG72 | (S)-beta-macrocarpene synthase | −3.78 | −4.81 | |
| GRMZM2G073510_T01 | B6UFK1 | Mating-type switching protein swi10 | −3.79 | ||
| GRMZM2G419675_T01 | B6TRF5 | Win1 | −4.10 | −5.31 | |
| GRMZM2G117989_T01 | B6TRF5 | Win1 | −4.64 | ||
| GRMZM2G078013_T01 | W0NT67 | NBS-LRR disease resistance protein | 2.45 | ||
| GRMZM2G037146_T01 | Q8W0Q8 | Small heat shock-like protein | 2.14 | ||
| GRMZM2G168447_T01 | M8B2N3 | Pathogenesis-related protein 1A | −2.10 | ||
| GRMZM2G080183_T01 | B4FVT4 | Peroxidase | −3.14 | ||
| GRMZM2G117942_T01 | A0A059Q1C7 | Pathogenesis-related protein | −3.25 | ||
| GRMZM2G312597_T01 | N1QRC0 | Subtilisin inhibitor 1 | 2.55 | ||
| GRMZM2G085934_T01 | B6U175 | 17.5 kDa class II heat shock protein | 2.23 | ||
| GRMZM2G063438_T01 | AAMT3 | Anthranilate O-methyltransferase 3 | 2.12 | ||
| GRMZM2G008740_T01 | D9J101 | Benzoate O-methyltransferase | −2.20 | ||
| GRMZM2G080689_T01 | K7VH54 | Peroxidase | −2.39 | ||
| GRMZM2G145552_T01 | M7YYQ7 | Cucumber peeling cupredoxin | −3.45 |
Genome ID, Maize transcript ID (ZmB73_5b_FGS_cdna.fasta.gz); FC, fold change value.
Transport processes
Differentially expressed transcripts grouped in “establishment of localization” are involved in several transport processes (Table 3). We focused on genes playing a role in transport processes of amino acids, peptides, and . Considering amino acid transport, only in the Aa vs. N comparison, we found the up-regulation of a transcript encoding a putative amino acid permease (AAP; GRMZM2G180547_T01) that could be involved into transport across membranes. This result suggests that amino acid supply as N-source positively affected components involved in their uptake and/or translocation (Tegeder, 2012; Tegeder and Ward, 2012). Members of Amino Acid-Polyamine-Organocation (APC), Drug/Metabolite Transporter (DMT), ATP Binding Cassette (ABC), and Major Facilitator (MFS) super families play a role in amino acid export from the cytosol to apoplastic space or into intracellular compartments such as the vacuole (Okumoto and Pilot, 2011). Aa and Bio treatments modulated the expression of transcripts encoding protein belonging to DMT, ABC, and MFS families that can mediate these transport processes (Supplementary Tables S3, S4). In addition, members of the ABC and PTR families can be involved also in peptide transport (Koh et al., 2002; Waterworth and Bray, 2006). Other peptide transporters belong to the oligopeptide transporter (OPT) family (Koh et al., 2002; Waterworth and Bray, 2006). Concerning the PTR transporters, we observed a prevailing negative modulation of transcripts caused by Aa and Bio (GRMZM2G026523_T01, GRMZM2G122712_T01, GRMZM2G347457_T01, GRMZM2G057611_T01, GRMZM2G015767_T01). Focusing on OPT family, only the comparison Bio vs. N underlined the up-regulation of a transcripts encoding a putative transporter (GRMZM2G152555_T01) suggesting a role of this gene in transport of peptides in Bio-treated maize roots.
Table 3.
Differentially expressed transcripts involved in transport processes.
| Genome ID | UniProt ID | Description | Aa vs. N (FC) | Bio vs. N (FC) | Bio vs. Aa (FC) |
|---|---|---|---|---|---|
| GRMZM2G010251_T01 | B4FSV9 | High affinity nitrate transporter | 9.37 | ||
| GRMZM2G156599_T01 | Q9AY27 | Iron-phytosiderophore transporter yellow stripe 1 | 8.60 | 2.25 | |
| GRMZM2G000614_T01 | Q7FMW4 | ABC transporter G family member 38 | 3.94 | ||
| GRMZM2G135291_T01 | G3XDL3 | Putative iron-phytosiderophore transporter | 3.82 | ||
| GRMZM2G180547_T01 | Q53LH2 | Amino acid carrier, putative, expressed | 3.53 | ||
| GRMZM2G099382_T01 | B6T0F4 | Tonoplast dicarboxylate transporter | 3.30 | ||
| GRMZM2G072071_T01 | B6U4J2 | ATP-binding cassette sub-family B member 10 | 2.76 | ||
| GRMZM2G148060_T01 | K7VD92 | Putative ferroportin-domain family protein | 2.70 | ||
| GRMZM2G118507_T01 | K7VD86 | Uncharacterized protein | 2.65 | ||
| GRMZM2G024196_T01 | Q7XKF4 | Probable metal-nicotianamine transporter YSL13 | 2.53 | 3.89 | |
| GRMZM2G064437_T01 | B6TDG1 | Proton myo-inositol cotransporter | 2.53 | ||
| GRMZM2G059465_T01 | K7TWC7 | Calcium-transporting ATPase | 2.49 | ||
| GRMZM2G129843_T01 | B6U7Q9 | Lipid binding protein | 2.41 | ||
| GRMZM2G362848_T01 | V9SBV7 | Nucleobase cation symporter 1 | 2.32 | ||
| GRMZM2G029951_T01 | A0A096Q8Z7 | Uncharacterized protein | 2.29 | ||
| GRMZM2G123884_T01 | Q7XVB3 | Probable sodium/metabolite cotransporter BASS1, chloroplastic | 2.21 | ||
| GRMZM2G056908_T01 | Q9ATL8 | Aquaporin TIP2-2 | 2.19 | ||
| GRMZM2G053991_T01 | Q5W7C1 | UPF0014 membrane protein STAR2 | 2.04 | ||
| GRMZM2G142924_T01 | A0A096SDC7 | Uncharacterized protein | −2.00 | ||
| GRMZM2G024808_T01 | B6U7W3 | Nitrate and chloride transporter | −2.02 | ||
| GRMZM2G072955_T01 | M8CTF4 | Chloride channel protein | −2.06 | −2.02 | |
| GRMZM2G153920_T01 | B4FQN6 | Sorbitol transporter | −2.07 | ||
| GRMZM2G137421_T01 | B6TSV4 | Peptide transporter PTR2 | −2.09 | −2.12 | |
| GRMZM2G457523_T01 | Q2QLJ1 | Sodium/hydrogen exchanger family protein, expressed | −2.10 | ||
| GRMZM5G872392_T01 | B6T9U6 | Bidirectional sugar transporter SWEET | −2.15 | ||
| AC186166.3_FGT008 | A0A096PGB1 | Uncharacterized protein | −2.16 | ||
| GRMZM2G112456_T01 | C0HIN0 | Oligopeptide transporter 4 | −2.17 | ||
| GRMZM2G047431_T01 | Q69EY5 | GDP dissociation inhibitor protein | −2.17 | ||
| GRMZM2G080178_T01 | B6UC24 | Sulfate transporter 1.2 | −2.21 | ||
| GRMZM2G122712_T01 | Q6AU97 | Putative proton-dependent oligopeptide transporter (POT) | −2.26 | −2.96 | |
| GRMZM2G009344_T01 | B4FET6 | ATPUP3 | −2.28 | ||
| GRMZM2G026523_T01 | B4FQ14 | Peptide transporter PTR2 | −2.30 | ||
| GRMZM2G167758_T01 | Q9FMC7 | Nuclear transport factor 2 (NTF2) family protein | −2.32 | ||
| AC185254.4_FGT002 | Q6ZIV9 | Putative ABC transporter | −2.39 | ||
| GRMZM2G311401_T01 | A0A096T5V7 | Uncharacterized protein | −2.48 | ||
| GRMZM2G027891_T01 | Q0JB23 | Os04g0561000 protein | −2.54 | ||
| GRMZM2G091478_T01 | K7TVU7 | Uncharacterized protein | −2.68 | ||
| GRMZM2G036631_T01 | K7V7R6 | Uncharacterized protein | −2.69 | ||
| AC235544.1_FGT004 | Q2QP91 | Dor1-like family protein, expressed | −2.75 | ||
| GRMZM2G040871_T01 | B6SKF6 | Hexose transporter | −2.77 | ||
| GRMZM2G139639_T01 | B6TJ37 | Inorganic phosphate transporter 1-5 | −2.78 | 2.72 | |
| GRMZM2G319781_T01 | M8CDC8 | Phosphatidylinositol transfer protein 2 | −2.85 | ||
| GRMZM2G348726_T01 | B4FJ28 | Signal recognition particle 9 kDa protein | −2.93 | −2.73 | |
| GRMZM2G137108_T01 | Q9ATN2 | Aquaporin NIP2-2 | −3.02 | ||
| GRMZM2G154845_T01 | A0A096SLG9 | Protein DETOXIFICATION | −3.19 | −2.31 | |
| GRMZM2G168439_T01 | B4F9E1 | Aquaporin TIP1-2 | −3.49 | −3.14 | |
| GRMZM2G168365_T01 | B6T9U6 | Bidirectional sugar transporter SWEET | −3.57 | ||
| GRMZM2G010779_T01 | B6U903 | Vacuolar cation/proton exchanger 2 | −3.60 | −2.44 | |
| GRMZM2G351347_T01 | C4J1B8 | Calcium-activated outward-rectifying potassium channel 1 | −3.67 | ||
| GRMZM2G305446_T01 | Q9ATL7 | Aquaporin TIP3-1 | −3.70 | ||
| GRMZM2G144581_T01 | A0A096SEB4 | Bidirectional sugar transporter SWEET4 | −4.01 | ||
| GRMZM2G060742_T01 | B6SSI8 | Citrate transporter family protein | −4.44 | −2.55 | |
| GRMZM2G101958_T01 | B6SGP7 | Non-specific lipid-transfer protein | 7.01 | ||
| GRMZM2G037229_T01 | A0A096QDL2 | Probable magnesium transporter | 5.44 | ||
| GRMZM2G107239_T01 | Q0IQZ4 | Os11g0695900 protein | 4.87 | ||
| GRMZM2G173669_T01 | B4FTL9 | Sugar transporter SWEET | 3.38 | ||
| GRMZM2G476069_T01 | M7ZN84 | Nitrate transporter 1.4 | 3.01 | ||
| GRMZM2G098088_T01 | K7UTZ0 | Hexose transporter | 2.75 | ||
| GRMZM2G384661_T01 | K7WBE5 | Uncharacterized protein | 2.71 | ||
| GRMZM2G425683_T01 | B6TCP1 | Carbohydrate transporter/sugar porter/transporter | 2.37 | ||
| GRMZM2G152555_T01 | W9R1V3 | Oligopeptide transporter 1 | 2.28 | ||
| GRMZM2G092780_T01 | C0P4H8 | Phosphate transporter | 2.22 | ||
| GRMZM2G130454_T01 | B6UDW9 | Lipid transfer protein | 2.07 | ||
| GRMZM2G005293_T01 | B6U0S4 | Patellin-5 | −2.06 | ||
| GRMZM2G150468_T01 | Q0J9C7 | Os04g0660900 protein | −2.09 | ||
| GRMZM2G093276_T01 | E3WCP2 | Zinc transporter | −2.32 | ||
| GRMZM2G047762_T01 | A0A096QKI7 | Uncharacterized protein | −2.32 | ||
| GRMZM2G009045_T01 | Q67UA2 | Putative phosphate transport protein, mitochondrial | −2.46 | ||
| GRMZM2G477872_T01 | AB45G | ABC transporter G family member 45 | −2.70 | ||
| GRMZM2G075150_T01 | Q7XEN0 | Exocyst complex component Sec15 | −2.71 | ||
| GRMZM2G335218_T01 | K7V706 | Ammonium transporter | −2.80 | −4.26 | |
| GRMZM2G055545_T01 | A0A096QQK0 | Uncharacterized protein | −2.88 | ||
| GRMZM2G347457_T01 | B6TSV4 | Peptide transporter PTR2 | −3.47 | ||
| GRMZM2G070500_T01 | B6TIX8 | Nodulin-like protein | 3.62 | ||
| GRMZM2G061495_T01 | Q7XMZ2 | OSJNBa0027G07.3 protein | 3.20 | ||
| GRMZM5G865543_T01 | B6SUB5 | Electron carrier/electron transporter/iron ion binding protein | 3.16 | ||
| GRMZM2G476069_T01 | M8C905 | Nitrate/chlorate transporter | 2.89 | ||
| GRMZM2G057611_T01 | Q67VA9 | Putative oligopeptide transporter | 2.85 | ||
| GRMZM2G423884_T01 | K7V9U9 | Protein detoxification | 2.79 | ||
| GRMZM2G519761_T01 | K7UHM7 | Uncharacterized protein | 2.64 | ||
| GRMZM2G055834_T01 | Q852B2 | Os03g0823500 protein | 2.64 | ||
| GRMZM2G020859_T01 | B6SV43 | Potassium channel AKT2/3 | 2.36 | ||
| GRMZM2G153961_T01 | M7YVY5 | ABC transporter B family member 11 | 2.34 | ||
| GRMZM5G806774_T01 | K3XV44 | Glutamate receptor | 2.23 | ||
| GRMZM2G032899_T01 | K7V706 | Ammonium transporter | 2.20 | ||
| GRMZM2G011636_T01 | A0A096PXD7 | 25.3 kDa vesicle transport protein | −2.12 | ||
| GRMZM5G850455_T01 | B6T2C9 | Lipid transfer protein | −2.15 | ||
| GRMZM2G015767_T01 | B6U0T7 | Peptide transporter PTR2 | −2.21 | ||
| AC234152.1_FGT007 | K7UVH5 | Potassium channel2 | −2.28 | ||
| AC203966.5_FGT006 | M8CDC1 | 25.3 kDa vesicle transport protein | −5.56 |
Genome ID, Maize transcript ID (ZmB73_5b_FGS_cdna.fasta.gz); FC, fold change value.
Aa and Bio treatments caused also different modulations of transcripts involved in the uptake of N inorganic forms ( and ). Aa up-regulated (Aa vs. N) the ZmNRT2.2 (GRMZM2G010251_T01; Plett et al., 2010), a well-known gene involved in the inducible high affinity transport systems (iHATS) in maize roots (Garnett et al., 2013; Zamboni et al., 2014; Pii et al., 2016). Both Aa and Bio treatments down-regulated the ZmNRT1.2 transcript encoding a low affinity transporter (GRMZM2G137421_T01; Garnett et al., 2013), while the expression of another low affinity transporter, ZmNRT1.4B (GRMZM2G476069_T01) which is very low expressed during plant development (Garnett et al., 2013), seems to be Bio-specific (up-regulated both in Bio vs. Aa and Bio vs. N). Another Bio-specific transcript encodes a putative transporter (AMT2; GRMZM2G335218_T01, down-regulated both in Bio vs. Aa and Bio vs. N).
Our analysis showed that transcripts involved in the uptake systems of other mineral nutrients were selectively affected by Aa and Bio. Organic N sources stimulated the expression of transcripts encoding putative yellow stripe-like (YSL) transporters (GRMZM2G156599_T01, GRMZM2G024196_T01, and GRMZM2G135291_T01) involved into the uptake of iron-phytosiderophore and distribution of metals within the whole plant (Curie et al., 2009). ZmYS1 (GRMZM2G156599_T01) encodes a protein that acts as a proton-coupled symporter of metals chelated to phytosiderophore and to nicotianamine (Schaaf et al., 2004) playing a key role in Strategy II of Fe acquisition utilized by the graminaceous species such as maize (Kobayashi and Nishizawa, 2012). A noticeable difference in the responses to organic N forms relies in the behavior of genes involved in nicotianamine and phytosiderophore biosynthesis. In particular, only in the Aa vs. N comparison we could observe a positive modulation of transcripts involved into nicotianamine synthesis (nicotianamine synthase, NAS; GRMZM2G030036_T01, AC233955.1_FGT003, GRMZM2G124785_T01, GRMZM2G034956_T01, GRMZM2G385200_T01, GRMZM2G312481_T01) and into phytosiderophore biosynthesis (deoxymugineic acid synthase, DMAS; GRMZM2G060952_T01) (Kobayashi and Nishizawa, 2012). However, in the comparison Bio vs. N only one gene related to phytosiderophore biosynthesis (nicotianamine aminotransferase, NAAT; GRMZM2G096958_T01) was found up-regulated. Taken together, even with defined differences these results indicate a strong impact of Aa and Bio with respect to Fe-stress responses of maize roots. Furthermore, besides Fe, nicotianamine forms transport coordination complexes with divalent transition metal cations, i.e., Mn2+, Zn2+, Cu2+ (Benes et al., 1983) whose concentrations were increased in Bio-treated roots.
The category of “establishment of localization” grouped also transcripts involved in S, P, and K transport processes. We observed a down-regulation of a transcript for a sulfate transporter (SULTR; GRMZM2G080178_T01; Hawkesford, 2002) when we compared the transcriptional profile of Aa- with that of N-treated roots. Concerning P, transcripts encoding putative Pi transporters (PHT; Raghothama, 2000) were down-regulated by Bio (GRMZM2G009045_T01) and Aa treatment (GRMZM2G139639_T01). Only in the comparison Bio vs. Aa two transcripts encoding for K channel AKT (Hirsch et al., 1998) were up- (GRMZM2G020859_T01) and down-regulated (AC234152.1_FGT007), respectively. Due to the presence of several members belonging to transporter families that have specific roles in nutrient uptake and translocation, and considering that transporter proteins are subjected to multiple forms of regulation (e.g., post-translational modifications), the transcriptional data do not allow a full explanation of the observed changes in tissue nutrient concentrations displayed by the different treatments.
Interestingly, a glutamate receptor (GRMZM5G806774_T01) functioning as non-selective cation channel is induced in Bio vs. Aa. This receptor, regulated by a broad range of amino acids, is involved in different physiological processes such as C/N sensing, resistance against fungal infection, root growth and response to wounding (De Bortoli et al., 2016).
Hormonal metabolism and signaling
A number of genes related to hormonal metabolism and signaling displayed expression changes in Bio- and Aa-treated seedlings vs. seedlings supplied with N (Table 4). A set of 9 genes whose expression profiles distinguished the Bio and Aa treatment vs. N represented the signature of organic N. Three of these genes coding for gibberellin 3-beta-dioxygenase 1 (GRMZM2G046669_T01), gibberellin 2-oxidase (GRMZM2G022679_T01) and gibberellin-regulated protein 2 (GRMZM2G164090_T01), respectively were related to gibberellin action. Gibberellin 3-beta-dioxygenase which converts inactive gibberellins (GAs) in their active form was up-regulated, whereas gibberellin 2-oxidase, implicated in GAs deactivation, was down-regulated in both Bio vs. N and Aa vs. N, suggesting that organic N forms induced the increase of active GAs in the roots. The other 5 genes that characterized the organic N supply were all related to auxin signaling or transport and were down-regulated in both Aa vs. N and Bio vs. N.
Table 4.
Differentially expressed transcripts involved in hormonal metabolism.
| Genome ID | UniProt ID | Description | Aa vs. N (FC) | Bio vs. N (FC) | Bio vs. Aa (FC) |
|---|---|---|---|---|---|
| GRMZM2G046669_T01 | M8AKK4 | Gibberellin 3-beta-dioxygenase 1 | 4.77 | 3.07 | |
| GRMZM2G035156_T01 | Q0DUR2 | Transcription factor ILI6 | 4.68 | ||
| GRMZM2G011463_T01 | B4FC68 | SAUR37-auxin-responsive SAUR family member | 3.83 | ||
| GRMZM2G462883_T01 | N1R055 | Putative gibberellin receptor GID1L3 | 2.63 | ||
| GRMZM2G093173_T01 | K7U964 | WAT-1-related protein | 2.32 | 2.12 | |
| GRMZM2G034917_T01 | N1R055 | Putative gibberellin receptor GID1L3 | 2.10 | ||
| GRMZM2G301932_T01 | B6TXN5 | Gibberellin receptor GID1L2 | 2.08 | ||
| GRMZM2G012546_T01 | M8BFB1 | Putative gibberellin receptor GID1L3 | 2.07 | ||
| GRMZM2G422419_T01 | A0A096TQ86 | Uncharacterized protein; response to auxin | 2.0 | ||
| GRMZM2G364328_T01 | B6TBZ6 | WAT1-related protein | −5.57 | ||
| GRMZM2G050997_T01 | Q709Q5 | Cytokinin oxidase 2 | −4.48 | ||
| GRMZM2G164090_T01 | B6TLZ8 | Gibberellin-regulated protein 2 | −4.33 | −3.34 | |
| GRMZM2G150688_T01 | B6TWS8 | Gibberellin-regulated protein 1 | −4.0 | ||
| GRMZM2G030790_T01 | B6TLX4 | Jasmonate-induced protein | −3.18 | ||
| GRMZM2G702564_T01 | K7V9I8 | Cytokinin oxidase 3 | −3.04 | 2.02 | |
| GRMZM2G173732_T01 | A2Z6Z0 | Protein BIG GRAIN 1-like | −2.98 | −3.01 | |
| GRMZM2G065230_T01 | B7ZXT3 | WAT-1 related protein | −2.75 | −2.7 | |
| GRMZM2G420812_T01 | B6T2P5 | SAUR31-auxin-responsive SAUR family member | −2.74 | −2.49 | |
| GRMZM2G117878_T01 | B6STN8 | Cytokinin-N-glucosyltransferase 1 | −2.51 | ||
| GRMZM2G013448_T01 | C0PEP2 | 1-aminocyclopropane-1-carboxylate oxidase 1 | −2.50 | ||
| GRMZM2G025742_T01 | I3RWV5 | Auxin efflux carrier component | −2.49 | ||
| GRMZM2G062019_T01 | B6TWT9 | Gibberellin receptor GID1L2 | −2.49 | ||
| GRMZM2G368591_T01 | K7V647 | WAT 1- related protein | −2.44 | −3.26 | |
| GRMZM2G107900_T01 | A0A096RQH9 | Uncharacterized protein; response to auxin | −2.39 | ||
| GRMZM2G171822_T01 | Q2QM77 | Protein kinase PINOID | −2.15 | ||
| GRMZM2G022679_T01 | Q8S0S6 | Gibberellin 2-oxidase | −2.10 | −2.34 | |
| GRMZM2G068701_T01 | Q0D4Z6 | Probable indole-3-acetic acid-amido synthetase GH3.8 | −2.06 | ||
| GRMZM2G141473_T01 | O23888 | Indole-3-acetaldehyde oxidase | −2.0 | ||
| AC233864.1_FGT009 | Q7XTN9 | OSJNBa0093O08.8 protein; response to auxin | −11.81 | −6.49 | |
| GRMZM2G471931_T01 | K7TM25 | Cytokinin riboside 5′-monophosphate phosphoribohydrolase | 2.38 | ||
| GRMZM2G136567_T01 | K7VFP2 | WAT-1 related protein | 3.43 | ||
| GRMZM2G330012_T01 | A0A0B4J3E8 | Uncharacterized protein; response to auxin | 3.64 | ||
| GRMZM2G001977_T01 | B6UC04 | Gibberellin receptor GID1L2 | −2.05 | ||
| GRMZM2G053338_T01 | B6U4E2 | Indole-3-acetic acid-amido synthetase GH3.8 | −2.11 | ||
| GRMZM2G050321_T01 | B6SZU3 | Jasmonate O-methyltransferase | −2.69 | ||
| GRMZM2G024131_T01 | Q41819 | Indole-3-acetate beta-glucosyltransferase | −4.34 | ||
| GRMZM2G155680_T01 | A0A096SLZ9 | Cytokinin riboside 5′-monophosphate phosphoribohydrolase | 2.04 | ||
| GRMZM2G384762_T01 | A0A096S9W7 | Auxin-responsive protein | 16.7 |
Genome ID, Maize transcript ID (ZmB73_5b_FGS_cdna.fasta.gz); FC, fold change value.
A characteristic feature of the Bio treatment vs. Aa and N was the induction of genes (GRMZM2G471931_T01 and GRMZM2G155680_T01) coding for the enzyme cytokinin (CK) riboside 5′- monophosphate phosphoribohydrolase that converts CK ribosides in free CK. The up-regulation of these genes would result in an increased release of CKs from conjugates. CKs play an important role in root response to N supply coordinating root growth and N availability in the soil (Kiba et al., 2011; Kiba and Krapp, 2016). CKs also interact with auxin in determining root growth and architecture (Mi et al., 2008; Pacifici et al., 2015; Schaller et al., 2015). A second characteristic feature of the response of root to Bio was the down-regulation of GRMZM2G053338_T01 and GRMZM2G024131_T01 genes coding for a indole-3-acetic acid-amido synthetase and indole-3-acetate beta-glucosyltransferase, respectively. As these enzymes mediate the formation of IAA conjugate, their down-regulation indicates that Bio-treated roots retains a higher level of active indol-3acetic acid (IAA). The GRMZM2G050321_T01 gene coding for a jasmonate O-methyltransferase was also down-regulated in Bio vs. N. Jasmonate O-methyltransferase catalyzes the formation of volatile methyl jasmonate that plays different roles in plant development and stress-response. In particular, high level of methyl jasmonate inhibits root growth. For instance, maize jasmonic acid-deficient opr7opr8 double-mutant showed much longer lateral roots compared with wild type (Yan et al., 2014), phenotype that resembles the root morphology of Bio-treated plants (Figure 1C).
The analyses of the differentially expressed genes of the Aa vs. N comparison revealed the strong involvement of the gibberellin signaling pathway in the free amino acids action. Four genes (GRMZM2G012546_T01, GRMZM2G462883_T01, GRMZM2G034917_T01, GRMZM2G301932_T01) coding for putative GA receptors were up-regulated and one (GRMZM2G062019_T01) down-regulated in Aa vs. N comparison. Another interesting feature was the down-regulation of two genes involved in auxin transport (GRMZM2G025742_T01 and GRMZM2G171822_T01), the first coding for a component of an auxin efflux carrier and the second coding for a PINOID kinase that regulates the membrane localization of the auxin efflux transporters PIN (Christensen et al., 2000). One of the effects of the Aa treatment was also the restraint of IAA and CK catabolism as the results of the down regulation of a gene coding for indole-3-acetaldehyde oxidase (GRMZM2G141473_T01) and two genes (GRMZM2G702564_T01 and GRMZM2G050997_T01) coding for a CK oxidase 3 and a cytokinin oxidase 2, respectively.
The growth and architecture of the root apparatus is the result of the action of several phytohormones as well as their interplay. Therefore, it is not surprising that the activity of Bio and Aa on the root apparatus leaded to modifications in the metabolism and signaling pathways of different phytohormones. However, although some transcriptional changes were common to both treatments (i.e., the changes in the expression of genes coding for enzymes involved in GA metabolism), differences in the involvement of hormones in Bio and Aa root growth promoting effects have emerged. For instance, CK release from conjugated and inhibition of IAA conjugation together with a lower synthesis of methyl JA appeared the main effects of Bio, whereas the activity of Aa seems to result principally from altered GA synthesis and signaling and restrain of CK and IAA degradation.
A few transcripts coding for CLE peptide hormones were down-regulated in the analyzed comparisons (GRMZM2G468688_T01 in Bio vs. N and Aa vs. N, GRMZM2G165836_T01 in Bio vs. A and GRMZM2G466532_T01 in A vs. N) and one was up-regulated (GRMZM2G114127_T01 in Aa vs. N). On the other hand, GRMZM2G438840_T01, coding for CLE receptor kinase CLAVATA1 (CLV1) was up-regulated in Bio vs. N and Aa vs. N.
CLE signaling peptides and CLV1 play a notable role in sensing N (Miyawaki et al., 2013). It has been shown that lateral roots stop growing under severe deficiency of N, while the expression of CLE peptides is induced (Araya et al., 2014). This regulation serves as a mechanism to prevent the expansion of the lateral root system into N-poor environments (Gruber et al., 2013). The clv1 mutant exhibits progressive growth of lateral roots under N-deficient conditions (Wang et al., 2016).
Interestingly, the transcripts GRMZM2G055607_T01, coding for a sulfotransferase which catalyzes post-translational tyrosine sulfation of secreted peptides, was over-expressed in the Bio vs. Aa comparison. In Arabidopsis, the loss-of-function mutant for tyrosyl-protein sulfotransferase, shows a short-root phenotype (Shinohara et al., 2016). Thus, this finding may represent another clue to unravel the action mechanisms of root growth stimulation exerted by Bio.
Conclusions
The present study dissected the biostimulatory activity of short peptides and free amino acids on maize seedlings. We demonstrated that protein hydrolysates containing peptides and a very low fraction of free amino acids were more efficient in stimulating root growth and micronutrient accumulation than free amino acid mixture with the same amino acid composition, suggesting a specific role for small peptides in controlling root growth. The genome-wide transcriptional analysis of maize root responses to Bio and Aa as compared with inorganic N, allowed to shed light on the similarities and differences in the mechanisms of action of the two biostimulants. The Aa produced a stronger modification of transcriptional networks than Bio, 995 and 587 differentially expressed genes were detected in Aa vs. N and Bio vs. N, respectively. Both treatments displayed effects on genes related to oligopeptide and induced modifications of genes involved in transport, demonstrating that N organic forms can interfere with inorganic N uptake, although the root total N was unchanged. On the other hand, a specific action of Bio seemed to be related to the regulation of the glutamate receptor which is involved in root growth and C/N signaling. Modification in genes implicated in metal ion transport were also detected in both treatments, although with some distinctive features. Even if plants were grown in the presence of Fe-EDTA and the content of Fe in the roots was unmodified, Aa and Bio positively affected components involved in Strategy II responses to Fe deficiency. In particular, Aa treatment caused the up-regulation of several transcripts involved in the synthesis of metal chelators (nicotianamine and mugineic acids) and in their transport. On the contrary, only three genes (one related to phytosiderophore synthesis and two to metal-phytosiderophore uptake and translocation) were up-regulated in Bio-treated roots. We might hypothesize that peptides could chelate metals facilitating their uptake and making in turn the biosynthesis of phytosiderophores less crucial. This might explain the higher contents of Cu, Mn, and Zn detected in maize roots treated with Bio. The stimulation of root growth was associated as expected, with perturbations in hormone balance. Both biostimulants modulated genes involved in GAs metabolism thus likely leading to increased GAs levels, and in auxin signaling and transport. In addition, Bio specifically modulated CKs release from conjugates and jasmonate metabolism. Future investigations aiming at studying the effects of protein hydrolysate and free amino acid applications on the content and distribuition of phytohormones in the plant, would be useful to deepen these findings. Noticeably, Aa treatment modified the expression of a high number of genes involved in the response to oxidative stress, whereas Bio caused only a modest modulation of stress-related genes. This might suggest that the lower growth promoting capacity of Aa respect to Bio might also be linked to the different metabolic engagement in stress responses.
Author contributions
ZV and TP designed the experimental set up. CS and AZ performed the experiments. CS, AZ, ZV, and TP carried out the analysis of the data and draft the manuscript. TP coordinated the project. All the authors approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been supported by a Joint Project grant of the University of Verona and SICIT 2000 S.p.A. We thank SICIT 2000 S.p.A. for the effective collaboration and scientific support. We are grateful to Dr. Youry Pii for his help in phosphate analysis.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00433/full#supplementary-material
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y. N., et al. (2014). CLE-CLV1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 2029–2034. 10.1073/pnas.1319953111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenault J.-L., Pouleur S., Messier C., Guay R. (1995). WinRHIZO™, a root-measuring system with a unique overlap correction method. Horticult. Sci. 30:906. [Google Scholar]
- Baligar V. C., Fageria N. K., He Z. L. (2001). Nutrient use efficiency in plants. Commun. Soil Sci. Plant Anal. 32, 7–8. 10.1081/CSS-100104098 [DOI] [Google Scholar]
- Benes I., Schreiber K., Ripperger H., Kircheiss A. (1983). Metal complex formation by nicotianamine, a possible phytosiderophore. Experientia 39, 261–262. 10.1007/BF01955293 [DOI] [Google Scholar]
- Calvo P., Nelson L., Kloepper J. (2014). Agricultural uses of plant biostimulants. Plant Soil 383, 3−41. 10.1007/s11104-014-2131-8 [DOI] [Google Scholar]
- Canellas L. P., Olivares F. L., Okorokova-Facanha A. L., Facanha A. R. (2002). Humic acids isolated from earthworm compost enhance root elongation, lateral root emergence, and plasma membrane H+-ATPase activity in maize roots. Plant Physiol. 130, 1951–1957. 10.1104/pp.007088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Liu Z., Wang B., Wang X., Lai J., Tian F. (2015). Transcriptome sequencing reveals the roles of transcription factors in modulating genotype by nitrogen interaction in maize. Plant Cell Rep. 34, 1761–1771. 10.1007/s00299-015-1822-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. K., Subler S., Edwards C. A. (2002). Effects of agricultural biostimulants on soil microbial activity and nitrogen dynamics. Appl. Soil Ecol. 19, 249–260. 10.1016/S0929-1393(02)00002-1 [DOI] [Google Scholar]
- Christensen S. K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478. 10.1016/S0092-8674(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Colla G., Rouphael Y., Canaguier R., Svecova E., Cardarelli M. (2014). Biostimulant action of a plant-derived protein hydrolysate produced through enzymatic hydrolysis. Front. Plant Sci. 5:448. 10.3389/fpls.2014.00448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit C. M. (1993). Developmental expression and localization of petunia glycine-rich protein 1. Plant Cell 5, 277–288. 10.1105/tpc.5.3.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321–326. 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M., et al. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103, 1–11. 10.1093/aob/mcn207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Wang Y., Yang A., Zhang W. H. (2012). OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol. 159, 169–183. 10.1104/pp.112.194217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bortoli S., Teardo E., Szabò I., Morosinotto T., Alboresi A. (2016). Evolutionary insight into the ionotropic glutamate receptor superfamily of photosynthetic organisms. Biophys. Chem. 218, 14–26. 10.1016/j.bpc.2016.07.004 [DOI] [PubMed] [Google Scholar]
- du Jardin P. (2015). Plant biostimulants: definition, concept, main categories and regulation. Sci. Hortic. 196, 3–14. 10.1016/j.scienta.2015.09.021 [DOI] [Google Scholar]
- El Hadrami A., Adam L. R., El Hadrami I., Daayf F. (2010). Chitosan in plant protection. Mar. Drugs 8, 968–987. 10.3390/md8040968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertani A., Cavani L., Pizzeghello D., Brandellero E., Altissimo A., Ciavatta C., et al. (2009). Biostimulant activities of two proteins hydrolysates on the growth and nitrogen metabolism in maize seedlings. J. Plant Nutr. Soil Sci. 172, 237–244. 10.1002/jpln.200800174 [DOI] [Google Scholar]
- Ertani A., Pizzeghello D., Altissimo A., Nardi S. (2013). Use of meat hydrolyzate derived from tanning residues as plant biostimulant for hydroponically grown maize. J. Plant Nutr. Soil Sci. 176, 287–295. 10.1002/jpln.201200020 [DOI] [Google Scholar]
- Feng Z., Sun X., Wang G., Liu H., Zhu J. (2012). LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann. Bot. 110, 1–10. 10.1093/aob/mcs019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett T., Conn V., Plett D., Conn S., Zanghellini J., Mackenzie N., et al. (2013). The response of the maize nitrate transport system to nitrogen demand and supply across the lifecycle. New Phytol. 198, 82–94. 10.1111/nph.12166 [DOI] [PubMed] [Google Scholar]
- Gibbs D. J., Voβ U., Harding S. A., Fannon J., Moody L. A., Yamada E., et al. (2014). AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol. 203, 1194–1207. 10.1111/nph.12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber B. D., Giehl R. F. H., Friedel S., von Wiren N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. 10.1104/pp.113.218453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Wang R., Nacry P., Breton G., Kay S. A., Pruneda-Paz J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 111, 15267–15272. 10.1073/pnas.1411375111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern M., Bar-Tal A., Ofek M., Minz D., Muller T., Yermiyahu U. (2015). The use of biostimulants for enhancing nutrient uptake. Adv. Agron. 130, 141–174. 10.1016/bs.agron.2014.10.001 [DOI] [Google Scholar]
- Hara Y., Yokoyama R., Osakabe K., Toki S., Nishitani K. (2014). Function of xyloglucan endotransglucosylase/hydrolases in rice. Ann. Bot. 114, 1309–1318. 10.1093/aob/mct292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkesford M. J. (2002). Transporter gene families in plants: the sulphate transporter gene family—redundancy or specialization? Physiol. Plant. 117, 115–163. 10.1034/j.1399-3054.2003.00034.x [DOI] [Google Scholar]
- He X., Ma H., Zhao X., Nie S., Li Y., Zhang Z., et al. (2016). Comparative RNA-seq. analysis reveals that regulatory network of maize root development controls the expression of genes in response to N stress. PLoS ONE 11:e0151697. 10.1371/journal.pone.0151697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R. E., Lewis B. D., Spalding E. P., Sussman M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. 10.1126/science.280.5365.918 [DOI] [PubMed] [Google Scholar]
- Karakurt Y., Unlu H., Padem H. (2009). The influence of foliar and soil fertilization of humic acid on yield and quality of pepper. Acta Agricult. Scand. 59, 233–237. 10.1080/09064710802022952 [DOI] [Google Scholar]
- Kiba T., Krapp A. (2016). Plant nitrogen acquisition under low availability: regulation of uptake and root architecture. Plant Cell Physiol. 57, 707–714. 10.1093/pcp/pcw052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Kudo T., Kojima M., Sakakibara H. (2011). Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 4, 1399–1409. 10.1093/jxb/erq410 [DOI] [PubMed] [Google Scholar]
- Kim J., Lee H. W. (2013). Direct activation of EXPANSIN14 by LBD18 in the gene regulatory network of lateral root formation in Arabidopsis. Plant Signal. Behav. 8:e22979 10.4161/psb.22979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. 10.1146/annurev-arplant-042811-105522 [DOI] [PubMed] [Google Scholar]
- Koh S., Wiles A. M., Sharp J. S., Naider F. R., Becker J. M., Stacey G. (2002). An oligopeptide transporter gene family in Arabidopsis. Plant Physiol. 128, 21–29. 10.1104/pp.010332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ng C. K.-Y., Fan L. M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. 10.1016/j.envexpbot.2014.06.014 [DOI] [Google Scholar]
- Liu T., Song X., Duan W., Huang Z., Liu G., Li Y., et al. (2014). Genome-wide analysis and expression patterns of NAC transcription factor family under different developmental stages and abiotic stresses in Chinese cabbage. Plant Mol. Biol. Rep. 32, 1041–1056. 10.1007/s11105-014-0712-6 [DOI] [Google Scholar]
- Lu M., Sun Q. P., Zhang D. F., Wang T. Y., Pan J. B. (2015). Identification of 7 stress-related NAC transcription factor members in maize (Zea mays L.) and characterization of the expression pattern of these genes. Biochem. Biophys. Res. Commun. 462, 144–150. 10.1016/j.bbrc.2015.04.113 [DOI] [PubMed] [Google Scholar]
- Lucas M., Kenobi K., von Wangenheim D., Voβ U., Swarup K., De Smet I., et al. (2013). Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc. Natl. Acad. Sci. U.S.A. 110, 5229–5234. 10.1073/pnas.1210807110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995). Root architecture and plant productivity. Plant Physiol. 109, 7–13. 10.1104/pp.109.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli A., Sturaro A., Trevisan S., Quaggiotti S., Nonis A. (2012). Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 169, 807–815. 10.1016/j.jplph.2012.01.019 [DOI] [PubMed] [Google Scholar]
- Masny A., Basak A., Zurawicz E. (2004). Effects of foliar applications of Kelpak SL and Goëmar BM 86® preparations on yield and fruit quality in two strawberry cultivars. J. Fruit Ornament. Plant Res. 12, 23–27. [Google Scholar]
- Mi G., Chen F., Zhang F. (2008). Multiple signaling pathways control nitrogen-mediated root elongation in maize. Plant Signal. Behav. 3, 1030–1032. 10.4161/psb.6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Tabata R., Sawa S. (2013). Evolutionarily conserved CLE peptide signaling in plant development, symbiosis and parasitism. Curr. Opin. Plant Biol. 16, 598–606. 10.1016/j.pbi.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Molesini B., Cecconi D., Pii Y., Pandolfini T. (2014). Local and systemic proteomic changes in Medicago truncatula at an early phase of Sinorhizobium meliloti infection. J. Proteome Res. 13, 408–421. 10.1021/pr4009942 [DOI] [PubMed] [Google Scholar]
- Muthreich N., Majer C., Beatty M., Paschold A., Schützenmeister A., Fu Y., et al. (2013). Comparative transcriptome profiling of maize coleoptilar nodes during shoot-borne root initiation. Plant Physiol. 163, 419–430. 10.1104/pp.113.221481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S., Pilot G. (2011). Amino acid export in plants: a missing link in nitrogen cycling. Mol. Plant. 4, 453–463. 10.1093/mp/ssr003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici E., Polverari L., Sabatini S. (2015). Plant hormone cross-talk: the pivot of root growth. J. Exp. Bot. 66, 1113–1121. 10.1093/jxb/eru534 [DOI] [PubMed] [Google Scholar]
- Peng M. S., Bi Y. M., Zhu T., Rothstein S. J. (2007). Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 65, 775–797. 10.1007/s11103-007-9241-0 [DOI] [PubMed] [Google Scholar]
- Pii Y., Alessandrini M., Dall'Osto L., Guardini K., Prinsi B., Espen L., et al. (2016). Time-resolved investigation of molecular components involved in the induction of high affinity transport system in maize roots. Front. Plant Sci. 7:1657. 10.3389/fpls.2016.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton R., Cesco S., Iacolettig G., Astolfi S., Varanini Z. (1999). Modulation of uptake by water-extractable humic substances: involvement of root plasma membrane H+-ATPase. Plant Soil 215:155 10.1023/A:1004752531903 [DOI] [Google Scholar]
- Plett D., Toubia J., Garnett T., Tester M., Kaiser B. N., Baumann U. (2010). Dichotomy in the NRT gene families of dicots and grass species. PLoS ONE 5:e15289. 10.1371/journal.pone.0015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K. (2000). Phosphorus acquisition: plant in the driver's seat! Trends Plant Sci. 5, 412–413. 10.1016/S1360-1385(00)01746-5 [DOI] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.-R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584. 10.1105/tpc.109.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D. L., Tripathi P., Rabara R. C., Lin J., Ringler P., Boken A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. 10.1111/j.1467-7652.2011.00634.x [DOI] [PubMed] [Google Scholar]
- Schaaf G., Ludewig U., Erenoglu B. E., Mori S., Kitahara T., von Wirén N. (2004). ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J. Biol. Chem. 279, 9091–9096. 10.1074/jbc.M311799200 [DOI] [PubMed] [Google Scholar]
- Schaller G. E., Bishopp A., Kieber J. J. (2015). The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63. 10.1105/tpc.114.133595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavon M., Ertani A., Nardi S. (2008). Effects of an alfalfa protein hydrolysate on the gene expression and activity of enzymes of TCA cycle and N metabolism in Zea mays L. J. Agric. Food Chem. 56, 11800–11808. 10.1021/jf802362g [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Shinohara H., Mori A., Yasue N., Sumida K., Matsubayashi Y. (2016). Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 3897–3902. 10.1073/pnas.1522639113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H. Y., Zhang S. Z., Yin Y. L., Zhu D. Z., Han L. Y. (2015). Genome-wide analysis of NAMATAF1, 2-CUC2 transcription factor family in Solanum lycopersicum. J. Plant Biochem. Biotechnol. 24, 176–183. 10.1007/s13562-014-0255-9 [DOI] [Google Scholar]
- Tai H., Lu X., Opitz N., Marcon C., Paschold A., Lithio A., et al. (2016). Transcriptomic and anatomical complexity of primary, seminal, and crown roots highlight root type-specific functional diversity in maize (Zea mays L.). J. Exp. Bot. 67, 1123–1135. 10.1093/jxb/erv513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa H., Sato Y., Antonio B. A., Nagamura Y. (2013). Global transcriptome profile of rice root in response to essential macronutrient deficiency. Plant Signal Behav. 8:e24409. 10.4161/psb.24409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G., Sauer M., Stauffer J., Multani D., Niu X., Sakai H., et al. (2007). The rtcs gene in maize (Zea mays L.) encodes a lob domain protein that is required for postembryonic shoot-borne and embryonic seminal root initiation. Plant J. 50, 649–659. 10.1111/j.1365-313X.2007.03075.x [DOI] [PubMed] [Google Scholar]
- Tegeder M. (2012). Transporters for amino acids in plant cells: some functions and many unknowns. Curr. Opin. Plant Biol. 15, 315–321. 10.1016/j.pbi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Tegeder M., Ward J. M. (2012). Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 3:21. 10.3389/fpls.2012.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D., Cassman K. G., Matson P. A., Naylor R., Polasky S. (2002). Agricultural sustainability and intensive production practices. Nature 418, 671–677. 10.1038/nature01014 [DOI] [PubMed] [Google Scholar]
- Varanini Z., Pinton R. (1995). Humic substances and plant nutrition, in Progress in Botany, Vol. 56, ed Luttge U.(Berlin: Springer; ), 97–117. [Google Scholar]
- Vuletic M., Hadsi-Taskovic Sukalovic V., Markovic K., Kravic N., Vucinic Z., Maksimovic V. (2014). Differential response of antioxidative systems of maize roots cell walls to osmotic and heavy metal stress. Plant Biol. 16, 88–96. 10.1111/plb.12017 [DOI] [PubMed] [Google Scholar]
- Wang G., Zhangand G., Wu M. (2016). CLE peptide signaling and crosstalk with phytohormones and environmental stimuli. Front. Plant Sci. 6:1211. 10.3389/fpls.2015.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth W. M., Bray C. M. (2006). Enigma variations for peptides and their transporters in higher plants. Ann. Bot. 98, 1–8. 10.1093/aob/mcl099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Yordanov Y. S., Georgieva T., Li X., Busov V. (2013). Nitrogen deprivation promotes Populus root growth through global transcriptome reprogramming and activation of hierarchical genetic networks. New Phytol. 200, 483–497. 10.1111/nph.12375 [DOI] [PubMed] [Google Scholar]
- Xu C., Luo F., Hochholdinger F. (2016). LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167. 10.1016/j.tplants.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Yan Y., Huang P. C., Borrego E., Kolomiets M. (2014). New perspectives into jasmonate roles in maize. Plant Signal. Behav. 9:e970442. 10.4161/15592316.2014.970442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Hao L., Yao S., Zhao Y., Lu W., Xiao K. (2016). TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to Pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiol. Biochem. 104, 99–113. 10.1016/j.plaphy.2016.03.023 [DOI] [PubMed] [Google Scholar]
- Yi K., Wu Z., Zhou J., Du L., Guo L., Wu Y., et al. (2005). OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 138, 2087–2096. 10.1104/pp.105.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A., Astolfi S., Zuchi S., Pii Y., Guardini K., Tononi P., et al. (2014). Nitrate induction triggers different transcriptional changes in a high and a low nitrogen use efficiency maize inbred line. J. Integr. Plant Biol. 56, 1080–1094. 10.1111/jipb.12214 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ervin E. H., Schmidt R. E. (2003). Effects of liquid application of a seaweed extract and a humic acid on creeping bentgrass (Agrostis palustris Huds. A.). J. Am. Soc. Horticult. Sci. 128, 492–496. [Google Scholar]
- Zhao C., Craig J. C., Petzold H. E., Dickerman A. W., Beers E. P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803–818. 10.1104/pp.105.060202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Yang X., Yu H., Jiang W., Sun N., Liu X., et al. (2015). RNA-Seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol. 56, 455–467. 10.1093/pcp/pcu172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.