Abstract
The redox-regulated transcription factors (TFs) of the bZIP AP1 family, such as yeast Yap1 and fission yeast Pap1, are activated by peroxiredoxin proteins (Prxs) to regulate the antioxidant response. Previously, Aspergillus nidulans mutants lacking the Yap1 ortholog NapA have been characterized as sensitive to H2O2 and menadione. Here we study NapA roles in relation to TFs SrrA and AtfA, also involved in oxidant detoxification, showing that these TFs play different roles in oxidative stress resistance, catalase gene regulation and development, during A. nidulans life cycle. We also uncover novel NapA roles in repression of sexual development, normal conidiation, conidial mRNA accumulation, and carbon utilization. The phenotypic characterization of ΔgpxA, ΔtpxA, and ΔtpxB single, double and triple peroxiredoxin mutants in wild type or ΔnapA backgrounds shows that none of these Prxs is required for NapA function in H2O2 and menadione resistance. However, these Prxs participate in a minor NapA-independent H2O2 resistance pathway and NapA and TpxA appear to regulate conidiation along the same route. Using transcriptomic analysis we show that during conidial development NapA-dependent gene expression pattern is different from canonical oxidative stress patterns. In the course of conidiation, NapA is required for regulation of at least 214 genes, including ethanol utilization genes alcR, alcA and aldA, and large sets of genes encoding proteins involved in transcriptional regulation, drug detoxification, carbohydrate utilization and secondary metabolism, comprising multiple oxidoreductases, membrane transporters and hydrolases. In agreement with this, ΔnapA mutants fail to grow or grow very poorly in ethanol, arabinose or fructose as sole carbon sources. Moreover, we show that NapA nuclear localization is induced not only by oxidative stress but also by growth in ethanol and by carbon starvation. Together with our previous work, these results show that SakA-AtfA, SrrA and NapA oxidative stress-sensing pathways regulate essential aspects of spore physiology (i.e., cell cycle arrest, dormancy, drug production and detoxification, and carbohydrate utilization).
Keywords: ROS, cleistothecia, secondary metabolism, iron scavenging, germination
Introduction
The proposed role of reactive oxygen species (ROS) as essential cell differentiation signals (Hansberg and Aguirre, 1990; Aguirre et al., 2005) led us to study the mechanisms by which eukaryotic cells detoxify ROS, using the model fungus Aspergillus nidulans. To perceive and transmit oxidative stress signals, fungi utilize phosphorelay systems connected to MAP kinases specialized in transducing stress signals or SAPKs. Schizosaccharomyces pombe paradigmatic SAPK Sty1/Spc1 has been characterized as a MAPK involved in cell-cycle control (Shiozaki and Russell, 1995) that is activated by osmotic (Millar et al., 1995; Degols et al., 1996), oxidative (Degols et al., 1996), heat shock (Nguyen and Shiozaki, 1999), nitrogen limitation (Shiozaki and Russell, 1996), and UV light (Degols and Russell, 1997) stress. As indicated in Figure S1 and Table S1, the phosphorelay system linked to Sty1/Spc1 is composed by histidine kinases (HK) Mak1, Mak2, and Mak3 (Buck et al., 2001), the phosphotransfer protein (HPt) Mpr1 and the response regulator (RR) Mcs4. Sty1/Spc1 in turn regulates transcription factor Atf1. Despite the architecture similarity to Saccharomyces cerevisiae Sln1-Ypd1-Ssk1-Hog1 system (de Nadal et al., 2011), S. pombe phosphorelay transmits oxidative, not osmotic stress signals (Nguyen et al., 2000). A second phosphorelay component, the transcription factor Prr1 is also required for oxidative stress responses, independently of Sty1/Spc1 (Quinn et al., 2011). In addition to Atf1 and Prr1, transcription factor Pap1, a homolog of S. cerevisiae Yap1 (Moye-Rowley et al., 1989), is critical for the antioxidant response in this fungus. The oxidation signal is perceived by different peroxiredoxins or Prxs and then transmitted to Pap1 or Yap1, which once oxidized accumulate in the nucleus to regulate the expression of multiple genes involved in the antioxidant response.
All peroxiredoxins belong to a conserved family of peroxidases that reduce peroxide and contain a conserved “peroxidatic” cysteine. Peroxides oxidize this Cys to sulphenic acid, which then reacts with another “resolving” Cys to form a disulfide bond, subsequently reduced by a suitable electron donor to complete a catalytic cycle. Prxs are classified into 2-Cys, atypical 2-Cys and 1-Cys families. 2-Cys are homodimeric and contain peroxidatic and resolving Cys residues in the same subunit. However, the disulfide bond is formed between two different subunits. In atypical 2-Cys an intermolecular disulfide is formed within the same subunit. 1-Cys Prxs form a disulfide with a resolving Cys present in other proteins or small thiol molecules (Rhee, 2016). Until now, typical 2-Cys Prxs have not been found in filamentous fungi. The role of S. cerevisiae peroxiredoxin Gpx3 in Yap1 activation, which also requires Yap1-binding protein Ybp1, was the first description of Prx function in H2O2 sensing (Delaunay et al., 2002). However, under certain conditions peroxiredoxin Tsa1 can also mediate Yap1 activation by H2O2 (Tachibana et al., 2009). In S. pombe, the 2-Cys peroxiredoxin Tpx1 transmits the redox signal to Pap1 (Vivancos et al., 2004, 2005).
A. nidulans contains 15 HKs and the function of most of them is unknown. Genetic evidence indicates that HK NikA transmits osmostress and fungicide signals to (HPt) YpdA and to SrkA RR, which is coupled to the SAPK SakA/HogA (Han and Prade, 2002; Kawasaki et al., 2002), as well as to the SAPK-independent RR SrrA (Hagiwara et al., 2007; Vargas-Perez et al., 2007). Upstream MAPKK PbsB and MAPKKK SskB regulate SakA (Furukawa et al., 2005), which is able to replace Sty1/Spc1 functions in S. pombe, and in A. nidulans is phosphorylated in response to multiple types of stress, including osmotic, oxidative (Kawasaki et al., 2002), nutrient starvation (Lara-Rojas et al., 2011) and hypoxia (Sánchez and Aguirre, unpublished). Stress-activated SakA translocates to the nucleus, where it interacts with transcription factor AtfA, required for induction of multiple genes and both, ΔsakA and ΔatfA mutants are sensitive to oxidative stress (Lara-Rojas et al., 2011). Additionally, SakA and AtfA are required for osmotic-induced gene expression (Hagiwara et al., 2009).
TF SrrA is also needed for oxidative stress resistance (Vargas-Perez et al., 2007) and both, SakA and SrrA play important roles during development. SakA represses sexual development and is activated during asexual development (Kawasaki et al., 2002). ΔsakA intact conidia progressively lose their viability and this is consistent with the fact that phosphorylated SakA accumulates in asexual spores (conidia) in an AtfA-dependent manner, and its dephosphorylation is necessary for germination to take place (Lara-Rojas et al., 2011). Likewise, ΔsrrA mutants show severely decreased asexual sporulation and produce conidia that very rapidly lose their viability (Vargas-Perez et al., 2007). In addition to transcription factors (TFs) AtfA and SrrA, the Yap1/Pap1 functional homolog NapA has been shown to be required for resistance to H2O2 in A. nidulans (Asano et al., 2007). Unrelated protein AN8863, putatively involved in nucleosome assembly, was later also referred to as NapA (Araújo-Bazan et al., 2008). Here we keep using NapA to name the A. nidulans Yap1/Pap1 homolog (AN7513) because it has been used this way in other publications (Lessing et al., 2007; Thön et al., 2010), and because the name “ap” preceded by the first letter of the species name (i.e., nidulans apA) has been widely used in many other filamentous fungi, where the role of Yap1/Pap1 homologs in oxidative stress resistance has been demonstrated (Lessing et al., 2007; Qiao et al., 2008; Temme and Tudzynski, 2009; Tian et al., 2011; Cartwright and Scott, 2013). Notably, in several plant pathogens Yap1/Pap1 homologs are involved not only in regulation of the antioxidant response but also in plant virulence (Molina and Kahmann, 2007; Guo et al., 2011; Huang et al., 2011).
Here we compared the relative contribution of (TFs) AtfA, SrrA and NapA to the antioxidant response and development in A. nidulans and uncovered novel NapA roles in regulation of sexual and asexual development, carbon utilization and gene regulation during asexual sporulation.
Materials and methods
Strains, media, growth conditions, and catalase activity determination
The A. nidulans strains used in this work are listed in Table S2. All strains were grown at 37°C in glucose minimal (MM) nitrate medium (Hill and Käfer, 2001), plus supplements. ΔnapA strains in a veA+ background were obtained from sexual crosses with strain FGSCA4. The presence of wild type veA allele was confirmed by PCR using genomic DNA from selected progeny and the primers veAforward and veAreverse, as reported (Han et al., 2010). Menadione was filter sterilized and like H2O2, added to agar medium at 50°C before solidification. H2O2-containing plates were used the day they were prepared or stored at 4°C for no more than 24 h. Since H2O2 can react with medium components, the actual concentration in plates cannot be estimated. To ensure experimental reproducibility, the same batch of H2O2 containing medium was used when comparing different strains. Spore suspensions containing 1 × 103 or 1 × 104 conidia were used to inoculate plates by dropping the suspension on the center of plates containing different stressors or media. Higher H2O2 resistance is observed at higher spore densities, presumably due to the high catalase A activity levels found in conidia (Navarro et al., 1996; Navarro and Aguirre, 1998). For catalase activity, 30 μg of total protein extracts prepared from conidia or mycelia were separated on native polyacrylamide gels to determine catalase activity as reported (Navarro et al., 1996; Kawasaki et al., 1997). Briefly gels are incubated in 5% methanol with shaking for 5 min and then rinsed with tap water 3 times. After this, the gel is incubated in a 0.03% hydrogen peroxide solution (100 μl of commercial 30% solution in 100 ml of deionized water) for 5 min and rinsed with water. Finally, the gel is incubated in the staining solution until the bands of activity are visible. The staining solution is made by mixing equal volumes of a 2% (w/v) FeCl3 solution and a 2% (w/v) K3Fe(CN)6. The different catalases were mapped before using single, double and triple mutants affected in catA, catB, and catC genes (Kawasaki and Aguirre, 2001).
Deletion of napA, gpxA, tpxA, tpxB and alcA genes, and tagging of NapA
Genomic DNA was used as template to produce the gene-deletion constructs by double joint PCR (Yu et al., 2004). For napA gene (AN7513) replacement construct, the napA ORF was amplified with primers 5′ For-napA and 5′ Rev-napA (see Table S3). The 3′ napA fragment was amplified with primers 3′ For-napA and 3′ Rev-napA. Aspergillus fumigatus pyrG marker was amplified with primers pyrGforward and pyrGreverse, using plasmid PFNO3 as template (Nayak et al., 2006). The three fragments were purified, mixed and used in a fusion PCR with primers 5′nest-napA and 3′nest-napA. The final 4900 bp napA–AfpyrG–napA cassette was purified and used to transform A. nidulans strain 11035 by electroporation (Sanchez and Aguirre, 1996; Sánchez et al., 1998). Five PyrG+ transformants were obtained and analyzed by Southern blot to confirm the elimination of napA. After confirming the proper deletion event (Figure S2), strain TFL9 was chosen and crossed with strain CLK43 to get rid of the kuA deletion, and progeny strain CFL7 was confirmed by PCR and used in further experiments.
To delete the gpxA gene (AN2846), primer pairs 5′For-gpxA/5′Rev-gpxA and 3′For-gpxA/3′Rev-gpxA were used to amplify gpxA 5′ and 3′ regions, respectively. Primers 5′For-nested gpxA and 3′Rev-nested gpxA were used to obtain the final fusion product. A. fumigatus riboB marker was amplified with primers 5Ribo and 6Ribo, using plasmid pAfriboPstE1Skt(ssp1)-37 as template (Nayak et al., 2006). The 5000 bp gpxA-AfriboB-gpxA cassette was purified as before and used to transform A. nidulans strain 11035 by electroporation. Twenty RiboB+ transformants were obtained, and analyzed by PCR to confirm the elimination of gpxA (Figure S3). Six transformants contained the expected event. Strain TAM16 was crossed with strain CLK43 and progeny strain CAM11 was confirmed by PCR and used in further experiments.
A similar strategy was used to delete the tpxA gene (AN10223). Primers pairs 5′For-tpxA/5′Rev-tpxA and 3′For-tpxA/3′Rev-tpxA were used to amplify tpxA 5′ and 3′ regions, respectively. A. fumigatus pyrG marker was amplified with primers pyrGforward and pyrGreverse, using plasmid PFNO3 as template (Nayak et al., 2006). The 3483 bp fusion tpxA-AfpyrG-tpxA PCR product obtained with primers 5′nest tpxA and 3′nest tpxA was used to transform strain 11035 by electroporation. 15 transformants obtained were analyzed by PCR, and 4 transformants were confirmed (Figure S3). Transformant 6 was named TAM17, crossed with strain CLK43 and progeny strain CAM13 confirmed by PCR and chosen for additional experiments.
To delete tpxB gene (AN3973), primer pairs 5′For-tpxB/5′Rev-tpxB and 3′For-tpxB/3′Rev-tpxB were used to amplify tpxB 5′ and 3′ regions, respectively. The A. fumigatus pyrG marker was amplified as before and the 3483 bp fusion PCR product obtained with primers 5′nest tpxB and 3′nest tpxB was used to transform strain 11035 by electroporation. 4 transformants out of 5 obtained were confirmed by PCR (Figure S3). Transformant 4 was named TAM19, crossed with strain CLK43 and progeny strain CAM19 used in further experiments.
The alcA deletion construct containing the riboB gene, as a selective marker, was generated using primers 5′For-alcA and 5′Rev-alcA for 5′ region and 3′For-alcA and 3′Rev-alcA for the 3′ region. A. fumigatus riboB marker was amplified with primers 5Ribo and 6Ribo as before. The 4167 bp band obtained with primers NestForalcA and NestRevaclA was used to transform strain 11035 by electroporation. Six transformants contained the expected event (Figure S4). Transformant TAM20 was crossed with strain CLK43 and progeny strain CAM17 was confirmed by PCR and chosen for further experiments. Sexual crosses generated double, triple and quadruple mutants, as indicated in Table S2.
To generate NapA::GFP C-terminal construct, three PCR products were used. First, 5′ and entire napA ORF were amplified with primers GSP1napA and GSP2napA. Second, a 3′ napA fragment was amplified with primers GSP3napA and GSP4napA. Third, GFP and A. fumigatus pyrG marker were amplified with primers GFP1napA and GFP2napA as before. Purified fragments were used in a fusion PCR with primers GSP1napA and GSP4napA. The 6625 bp napA–GFP–AfpyrG cassette was used to transform A. nidulans strain A1155 by electroporation. Transformant TFL14 strain was confirmed by PCR and used for further experiments (Figure S5).
Microscopy
Fluorescence microscopy images were captured in vivo. Spores from NapA::GFP strain were grown for 18 h on coverslips containing liquid glucose-MM at 37°C with no shaking. After this, samples were shifted to the same medium with or without 2 mM H2O2, incubated for 0–120 min and observed using a NIKON Eclipse E600 microscope to detect DAPI and GFP fluorescence. Images were captured with a cooled camera Neo Andor sCMOS. For DAPI staining, samples of conidia or mycelia were fixed in methanol/acetone for 10 min, washed in water and stained for 5 min with 0.1 mg/ml of 4′,6-diamidino-2-phenylindole (DAPI).
Transcriptomic analysis
Conidia from 6-day old cultures from strains CLK43 (WT) and CFL7 (ΔnapA) were collected and washed 3 times with 10 ml of cold water. Excess liquid was removed by centrifugation and conidia were immediately frozen with liquid nitrogen. Total RNA was extracted by cryogenic grinding using the Tissue Lyser (Qiagen) and purified using the RNAeasy Mini kit (Qiagen), following the manufacturer's protocol. RNA integrity number (RIN) for RNA quality was generated using the Agilent 2100 Bioanalyzer System (Agilent technologies). Two independent samples from each strain (Biological replicates) were processed for cDNA synthesis using Illumina's kit TrueSeqV2 and sequenced at the next-generation sequencing core facility at IBT-UNAM. using Illumina's platform. An average of 10168 reads of 72 bp per sample were obtained, representing nearly 96.3% A. nidulans genome lengths. Biological replicates showed a good level of correlation (r_0.966 for WT and r_0.968 for ΔnapA). Differential gene expression was inferred based on total mapping counts using the EdgeR package. Genes showing a value of log fold change (LFC) ≥2 and a false discovery rate (FDR) ≤ 0.05 were considered as differentially expressed (DE). The differential expression analysis was deposited under the GEO identifier GSE94747, as part of the BioProject PRJNA373914. The sequencing raw reads for all experiments were deposited in the SRA database under the SRP099165 identifier.
Real-time PCR
Total RNA was isolated using the Plant RNA purification kit (Quiagen) according to the manufacturer's instructions and treated with DNase Turbo DNA-free kit (Ambion). The RNA integrity (RIN) and concentration were determinate using the 2200 TapeStation (Agilent Genomics). 1 μg of RNA was used to synthetize cDNA templates for PCR amplification, using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The expression of each gene was measured using 1:10 cDNA sample dilutions. Specific primers used for RT-PCR (Table S3) were devised to produce cDNA amplicons around 130 pb. To estimate genomic DNA contamination, primers were designed to amplify a larger product, including an intron of each gene, from a genomic template. RT-PCR products were observed in 2% agarose gels.
Triplicates from each sample were performed. RT-PCR was done using Platinum SYBR Green qPCR SuperMix-UDG with ROX kit (Invitrogen). RT-PCR was performed in a StepOne Real-Time PCR System (Applied Biosystems). The program used included an initial UDG incubation for 2 min at 50°C, followed by a 2 min denaturation step at 95°C, and 40 amplification cycles at 95° C for 15 s, followed by 30 s at 60°C. For relative quantification, we used the Ct comparative method and the data was analyzed with StepOne Software V2.3 (Applied Biosystems), using histone 2B (H2B) gene (AN3469) as reference.
Results
TFs NapA, SrrA, and AtfA play differential roles in A. nidulans antioxidant response
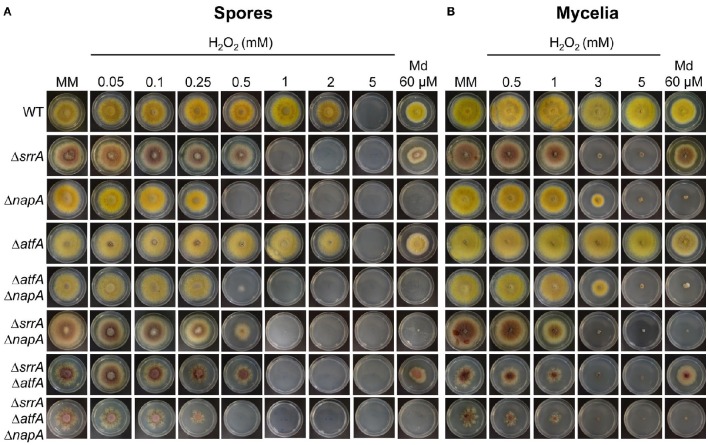
Previous work has shown that (TFs) SrrA (Vargas-Perez et al., 2007), AtfA (Lara-Rojas et al., 2011) and NapA (Asano et al., 2007) are involved in A. nidulans antioxidant response. To analyse the relative contribution of each TF in this process, we compared the sensitivity of conidia and mycelia from ΔnapA, ΔsrrA, and ΔatfA mutants to H2O2 and menadione. Results show that conidia from ΔnapA mutant were the most sensitive to H2O2 followed by ΔsrrA and ΔatfA mutants, while only ΔnapA mutants were sensitive to menadione (Figure 1A). In contrast, the same assay carried out with mycelia showed a slightly higher H2O2 sensitivity for ΔsrrA mutants followed by ΔnapA, while the atfA mutant was as resistant to H2O2 as the WT strain. Again only the ΔnapA mutant was sensitive to menadione (Figure 1B).
Figure 1.
NapA, SrrA and AtfA play differential roles in Aspergillus nidulans antioxidant response. (A) Conidia (1 × 103) from strains CLK43 (WT), COSsrrA3 (ΔsrrA), CFL7 (ΔnapA), TFLΔatfA-04 (ΔatfA), CAM7 (ΔatfA ΔnapA), CAM6 (ΔsrrA ΔnapA), CAM8 (ΔsrrA ΔatfA), and CAM9 (ΔsrrA ΔatfA ΔnapA) were inoculated by dropping spore suspensions on the center of supplemented MM plates containing H2O2 or menadione (Md) at the indicated concentrations, and incubated at 37°C for 4 days. (B) Mycelial plugs cut from the growing edge of 5-day old colonies from strains CLK43, COSsrrA3, CFL7, TFLΔatfA-02, CAM7, CAM6, CAM8, and CAM9 were transferred to plates containing H2O2 or menadione at the indicated concentrations, and incubated at 37°C for 4 days.
To further dissect NapA, SrrA, and AtfA contribution to the antioxidant response, we used ΔnapA, ΔsrrA, and ΔatfA single mutants to perform sexual crosses and obtain double and triple mutants, which were confirmed by PCR analysis (not shown). Results in Figure 1A show that conidia from double and triple mutants did not show additive phenotypes and those carrying the ΔnapA deletion were as sensitive to H2O2 and menadione as the single ΔnapA mutant, except in the case of the ΔsrrA ΔnapA strain whose H2O2 sensitivity was similar to the one displayed by the ΔsrrA mutant (Figure 1A). In the test performed with mycelia, double and triple mutants carrying the ΔsrrA deletion behaved as the single ΔsrrA mutant (Figure 1B). Notably, ΔsrrA mutant growth defects (Vargas-Perez et al., 2007) are enhanced by the presence of the ΔatfA deletion, as colonies from ΔsrrA ΔatfA mutants show a higher decrease in growth and highly irregular colony borders. These growth and conidiation defects were even more drastic in the mutant lacking the 3 TFs (Figures 1A,B).
These results show that TFs NapA, SrrA, and AtfA play different roles at different stages of A. nidulans life cycle. For H2O2 spore resistance NapA plays a more prominent role than SrrA, which in turn is more important than AtfA. For mycelial H2O2 resistance SrrA is somewhat more critical than NapA, while AtfA plays no role in this process, and only NapA is necessary for menadione resistance in both spores and mycelia. During vegetative growth, neither NapA nor AtfA are individually required for normal radial growth. However, ΔsrrA mutant growth defects are enhanced by the deletion of AtfA, indicating that AtfA contributes to normal radial growth. SrrA is almost essential for normal conidiation, NapA is needed for full conidiation (see further) and AtfA is dispensable for the process. Regarding conidial function, SrrA and AtfA are critical for conidial viability, and NapA is necessary for the accumulation of multiple mRNAs in conidia (see further).
NapA, SrrA, and AtfA regulate different catalase genes
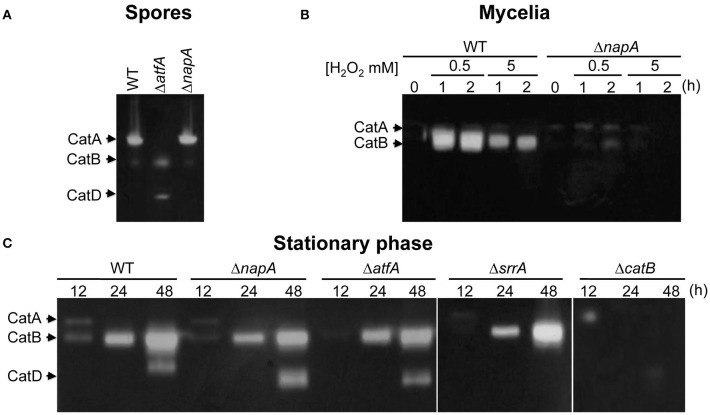
To further understand NapA roles in the antioxidant response, we compared the H2O2 sensitivity of conidia from mutants lacking NapA, the spore-specific catalase CatA (Navarro et al., 1996; Navarro and Aguirre, 1998) or the mycelium inducible catalase CatB (Kawasaki et al., 1997). Notably, ΔnapA conidia were much more sensitive to H2O2 than ΔcatA conidia, while ΔcatB conidia showed only a minor sensitivity at 4 mM H2O2 (Figure S6). As shown in Figure 2A, the presence of CatA activity was completely dependent on AtfA, while being independent of NapA. In contrast, H2O2 induction of mycelial catalase CatB was largely dependent on both NapA (Figure 2B) and SrrA (Vargas-Perez et al., 2007). Since CatB is also highly induced during the stationary phase of growth (Kawasaki and Aguirre, 2001), we asked if NapA, SrrA or AtfA were required for this induction. Unexpectedly, none of these TFs was needed for this process. However, the induction of catalase-peroxidase CatD/CpeA activity (Kawasaki and Aguirre, 2001; Scherer et al., 2002) required of SrrA (Figure 2C). In summary, these results show that NapA and SrrA are both required for CatB induction by H2O2, AtfA is required for CatA expression in conidia and none of them is required for CatB induction during the late stationary phase of growth. This and SrrA regulation of CatD/CpeA activity (Kawasaki and Aguirre, 2001) confirm the differential roles that these TFs perform during A. nidulans antioxidant response.
Figure 2.
Transcriptional factors NapA, SrrA and AtfA play differential roles in catalase regulation. (A) CatA activity levels are not affected in ΔnapA conidia. Conidial protein extracts from strains CLK43 (WT), TFLΔatfA-04 (ΔatfA) and CFL7 (ΔnapA) were analyzed for catalase in-gel activity. (B) CatB activity induction by H2O2 requires NapA. Mycelia from the strains CLK43 (WT) and CFL7 (ΔnapA) were grown for 12 h in liquid MM and then H2O2 was added at the indicated times and concentrations and protein extracts were used for catalase activity determination. (C) NapA, AtfA, and SrrA are not required for CatB induction during the stationary phase of growth. Mycelia from the strains CLK43 (WT), CFL7 (ΔnapA), TFLΔatfA-02 (ΔatfA), COSsrrA3 (ΔsrrA) and TLK12 (ΔcatB) was grown for 12, 24, and 48 h in liquid MM medium. The experiment was repeated at least 3 times; a representative experiment is shown.
NapA localizes in nuclei in response to oxidative stress
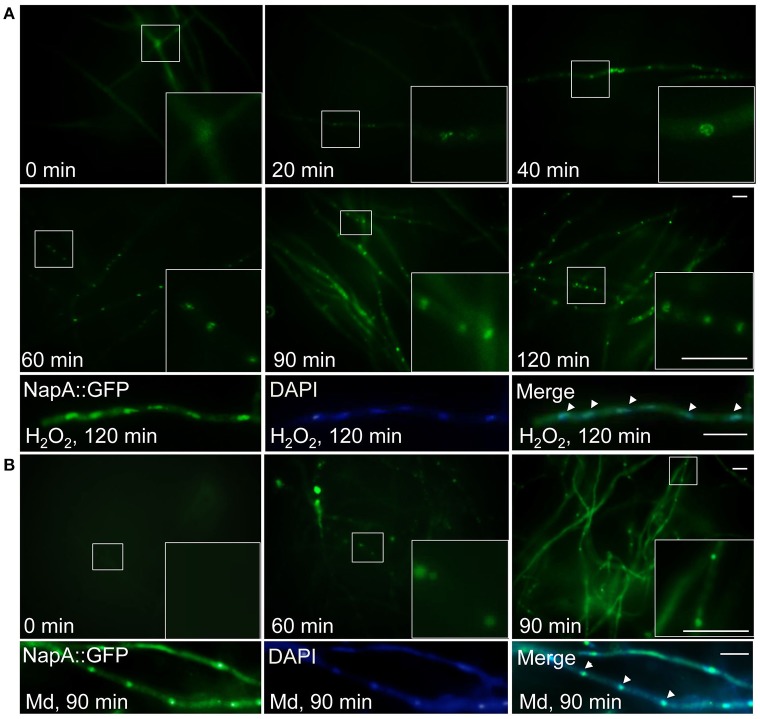
The high sensitivity of ΔnapA conidia to H2O2, not explained by a simple lack of spore catalase activity (Figure 2A), suggests that NapA is required for other conidial functions, including those involved in resistance to menadione. It is well known that S. cerevisiae, S. pombe and other fungal NapA homologs show nuclear accumulation when oxidized in response to H2O2. Peroxiredoxins Gpx3 and Tpx1 perceive the oxidation signal and relay it to Yap1 and Pap1, respectively. To determine NapA localization we introduced a GFP tag at its C-terminus and showed that the H2O2 resistance of the corresponding strain was not affected (Figure S5), indicating the functionality of this NapA::GFP fusion. Then we analyzed NapA::GFP localization using different H2O2 concentrations. Initially, we found a very low basal signal of NapA::GFP expressed from the napA promoter, which increased in nuclei after 30 min of treatment with different H2O2 concentrations (not shown). Under these conditions 2 mM was the minimum concentration needed to induce NapA::GFP nuclear accumulation and therefore we used this concentration for a time-course analysis. As seen in Figure 3A, NapA::GFP starts to show nuclear localization after 20 min of treatment, which gradually increases up to 120 min. When after this time the H2O2 treatment was stop by transferring mycelia to a medium lacking H2O2, it took 6 h to observe that NapA::GFP was no longer observed in nuclei, showing instead a cytoplasm localization. This indicates that under these conditions NapA-mediated adaptation to oxidative stress is a relatively slow process. Since ΔnapA mutants are sensitive to menadione, we also determined if menadione was able to induce NapA::GFP nuclear localization. Indeed, 10 μM menadione induced NapA nuclear accumulation after a 90 min treatment (Figure 3B).
Figure 3.
NapA is induced by oxidative stress and gradually accumulates in nuclei in response to oxidative stress. (A) NapA nuclear localization increases during incubation with H2O2. Conidia from strain CAM20 (NapA::GFP) were grown for 18 h in MM and then exposed to 2 mM H2O2 for the indicated times (0–120 min), observed in vivo and photographed every 10 min using Epifluorescence microscopy. Lower panel shows NapA::GFP and nuclei (DAPI) signal in mycelia treated with H2O2 for 120 min, fixed and photographed. (B) Menadione also induces nuclear localization of NapA. Conidia from strain CAM20 (NapA::GFP) were grown for 18 h in MM and then exposed to 10 μM menadione for the indicated times (0–90 min). Larger square areas in each picture show enlargements of the areas indicated by smaller squares. Bars = 10 μm.
NapA function in the antioxidant response is independent of peroxiredoxins GpxA, TpxA, and TpxB
As indicated before, S. cerevisiae peroxiredoxin Gpx3 and S. pombe Tpx1 relay the oxidation signal to Yap1 and Pap1, respectively. To determine if homologous peroxiredoxins were involved in NapA function, we searched the A. nidulans genome and found genes AN2846, AN10223, and AN3973 (Oh et al., 2010), which according to their closest homolog in S. cerevisiae are named as gpxA (Thön et al., 2010), tpxA and tpxB, respectively. We generated strains in which one of these peroxiredoxin genes was deleted and by sexual crosses generated triple as well as quadruple mutants containing the ΔnapA deletion. In sharp contrast to ΔnapA mutants, mutants lacking either a single or all 3 peroxiredoxins were not sensitive to H2O2 or menadione. However, the simultaneous inactivation of the 3 peroxiredoxins resulted in an enhancement of the sensitivity to H2O2 caused by the inactivation of NapA (Figure 4). This suggests that these 3 proteins might play a partially redundant minor function in H2O2 resistance, different from NapA.
Figure 4.
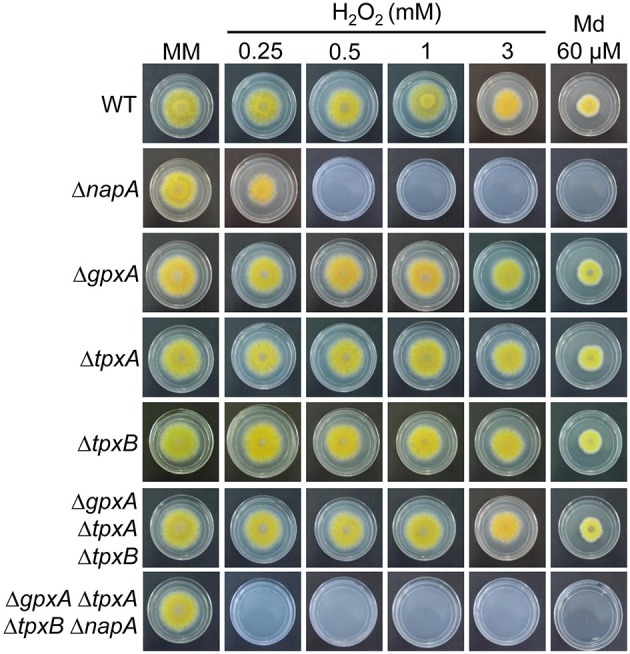
Peroxiredoxins GpxA, TpxA and TpxB are not required for H2O2 or menadione resistance. Conidia (1 × 103) from strains CLK43 (WT), CFL7 (ΔnapA), CAM11 (ΔgpxA), CAM13 (ΔtpxA), CAM19 (ΔtpxB), CAM15 (ΔgpxA ΔtpxA ΔtpxB), and CAM16 (ΔgpxA ΔtpxA ΔtpxB ΔnapA) were inoculated on supplemented MM plates containing H2O2 or menadione (Md) at the indicated concentrations, and incubated at 37°C for 4 days.
NapA is involved in developmental regulation
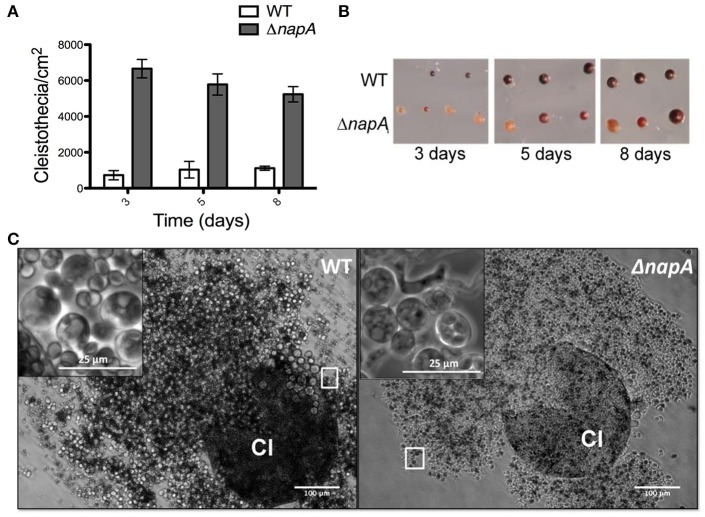
ΔnapA mutants did not show any obvious defects except that they seemed to develop more and paler cleistothecia than the WT strain. To examine this in more detail, we induced sexual development in confluent cultures and determined the number of cleistothecia as reported before (Kawasaki et al., 2002). Indeed, a ΔnapA mutant produced about 3 times more cleistothecia than the WT strain (Figure 5A). Notably, ΔnapA young cleistothecia were white, while older cleistothecia developed some pigmentation (Figure 5B). When observed under the microscope, it became clear that ΔnapA cleistothecia are not pigmented (Figure 5C, right panel) but produce pigmented ascospores, which are viable (not shown). A. nidulans veA gene encodes a member of a fungal protein family required for cleistothecium formation (Kim et al., 2002) and regulation of secondary metabolism (Bayram et al., 2008). Since our laboratory strains contain a veA1 allele that causes higher production of conidia and lower numbers of cleistothecia (Käfer, 1965), we also evaluated ΔnapA sexual development phenotypes in the presence of a wild-type veA allele. As expected, results show higher numbers of cleistothecia in veA+ strain FGSC4 and even higher in ΔnapA veA+ strains. Again, young cleistothecia were not pigmented, while older cleistothecia were non-pigmented but produced pigmented ascospores (not shown). Likewise, the presence of wild type veA gene did not modify ΔnapA oxidative stress sensitivity. These results indicate that NapA represses sexual development and is needed for the expression of genes, possibly a polyketide synthetase gene, associated with the synthesis of cleistothecial melanin. When conidiation levels were determined, we found that a ΔnapA mutant produced half of the conidia of the WT strain (Figure 6), indicating that NapA plays a role in the conidiation process. We also examined the conidiation levels of single, double and triple peroxiredoxin mutants. As shown in Figure 6, the inactivation of gpxA and tpxB had a very minor impact in conidiation. In contrast, the inactivation of tpxA reduced conidiation to levels similar to those observed in ΔgpxA ΔtpxA ΔtpxB and ΔnapA mutants. Moreover, a quadruple ΔgpxA ΔtpxA ΔtpxB ΔnapA mutant showed conidiation numbers slightly higher than those seen in single ΔtpxA and ΔnapA mutants, indicating that TpxA and NapA functions in conidiation are not additive and therefore suggesting that these two proteins work in the same conidiation pathway. Notably, tpxA orthologs are induced by H2O2 and regulated by NapA homologs AfYap1 and Bap1 in A. fumigatus (Lessing et al., 2007) and Botrytis cinerea (Temme and Tudzynski, 2009), respectively. These results suggest that TpxA plays NapA independent and dependent roles in oxidative stress resistance and conidiation, respectively.
Figure 5.
NapA regulates sexual development. (A) The deletion of napA causes premature sexual development. Conidia from strains CLK43 (WT) or CFL7 (ΔnapA) were grown and induced to undergo sexual development as reported (Kawasaki et al., 2002). The total number of cleistothecia per fixed area was counted under a dissection microscope and used to calculate cleistothecia per cm2. Bars represent the standard error of the mean (SEM). (B) Cleistothecia from 3, 5, and 8 days from experiment in (A) were isolated and photographed under a dissection microscope. (C) ΔnapA mutants develop unpigmented cleistothecia containing pigmented ascospores. WT and ΔnapA cleistothecia (Cl) from 5-day old cultures were crushed and photographed under the microscope. Square areas in each picture show enlargements of asci and ascospores.
Figure 6.
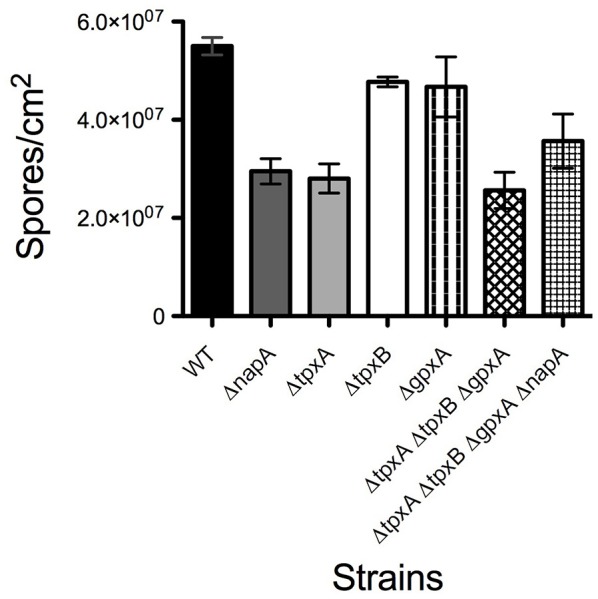
NapA and TpxA are required for normal asexual development. Asexual spores (1 × 103) from strains CLK43 (WT), CFL7 (ΔnapA), CAM13 (ΔtpxA), CAM11 (ΔgpxA), CAM11 (ΔgpxB), CAM15 (ΔgpxA ΔtpxA ΔtpxB), CAM16 (ΔgpxA ΔtpxA ΔtpxB ΔnapA), CAM17 (ΔalcA), and CAM18 (ΔalcA ΔnapA), were inoculated on supplemented MM and incubated at 37°C for 4 days. Total conidia were harvested and counted. Bars indicate standard deviation from three independent experiments.
NapA is required for gene regulation in conidia
To address the role of NapA in the production of fully functional conidia, we decided to carry out a transcriptomic analysis. For this, we isolated total RNA from ΔnapA and WT intact conidia from 6-day old colonies. Two independent samples from each strain (Biological replicates) were processed for cDNA synthesis and DNA sequencing using Illumina's platform. An average of 10168 reads of 72 bp per sample representing nearly 96.3% A. nidulans genome lengths per sample were obtained. The two biological replicates showed a good level of correlation (r_0.966 for WT and r_0.968 for ΔnapA) and principal component analysis showed a clear separation between ΔnapA and WT samples. Results show changes in the expression of 284 genes with a logFC differences higher than 1.5 but lower than 2 (Table S4) and 214 genes with logFC changes higher than 2 (Tables 1, 2). The following analysis will be based mostly on these 214 genes, from which 13 genes were up regulated and 201 were down regulated in ΔnapA conidia, as compared to WT conidia. Using gene ontology (GO) enrichment analysis, Aspergillus genome database AspGD (Cerqueira et al., 2014) and manual annotations, we grouped the genes regulated by NapA as shown in Tables 1, 2.
Table 1.
Up regulated transcripts in ΔnapA conidia.
| Gene | LogFC | Description | Domains |
|---|---|---|---|
| OXIDOREDUCTASES | |||
| AN8449 | −2.1 | Putative role in nitrate assimilation | Oxidoreductase molybdopterin binding domain,Mo-co oxidoreductase dimerisation domain,Cytochrome b5-like Heme/Steroid binding domain,Oxidoreductase FAD-binding domain,Oxidoreductase NAD-binding domain |
| AN8329 | −2.0 | Putative glucose oxidase-related protein | GMC oxidoreductase |
| TRANSPORTER ACTIVITY | |||
| AN7839 | −7.9 | Conserved hypothetical protein | ABC transporter transmembranal, Vacuolar glutathione S-conjugate transporter |
| AN2475 | −2.4 | MFS monosaccharide transporter | SP: MFS transporter, sugar porter (SP) family,Major Facilitator Superfamily, Sugar (and other) |
| AN5323 | −2.1 | MFS sugar transporter | Major Facilitator Superfamily, Major Facilitator Superfamily, Sugar (and other) |
| AN7796 | −2.0 | Conserved hypothetical protein | Major Facilitator Superfamily |
| HYDROLASES | |||
| AN9027 | −2.7 | Conserved hypothetical protein | Hydrolase activity, metallo-beta-lactamase superfamily |
| UNCLASSIFIED | |||
| AN7834 | −10.1 | Conserved hypothetical protein | Heterokaryon incompatibility HET domain |
| AN7836 | −9.4 | Cysteine-rich secreted protein | Not found |
| AN3384 | −2.4 | Conserved hypothetical protein | Acetyltransferase family, possible role in trichothecene biosynthesis |
| AN3557 | −2.0 | Nucleoside metabolic process | Nucleoside phosphorylase |
| HYPOTHETICAL PROTEINS WITH NO IDENTIFIED DOMAIN | |||
| AN2838 | −2.6 | Conserved hypothetical protein | Not found |
| AN0020 | −2.3 | Conserved hypothetical protein | Not found |
Table 2.
Down regulated transcripts in ΔnapA conidia.
| Gene | LogFC | Description | Domains |
|---|---|---|---|
| DNA BINDING | |||
| AN8426 | 3.3 | Conserved hypothetical protein | Fungal Zn(2)-Cys(6) binuclear cluster domain, Fungal specific transcription factor domain |
| AN3769 | 3.0 | C6 transcription factor | Fungal specific transcription factor domain |
| AN7118 | 2.9 | Putative transcription factor | Fungal specific transcription factor domain, Fungal Zn(2)-Cys(6) binuclear cluster domain, Zinc finger, C2H2 type |
| AN8103 | 2.8 | Putative transcription factor | Zn(II)2Cys6 transcription factor |
| AN4720 | 2.5 | conserved hypothetical protein | Zinc finger, C2H2 type |
| AN10897 | 2.4 | C6 zinc finger domain-containing protein | Fungal Zn(2)-Cys(6) binuclear cluster domain |
| AN7061 | 2.3 | Conserved hypothetical protein | Fungal specific transcription factor domain, Fungal Zn(2)-Cys(6) binuclear cluster domain |
| AN11831 | 2.3 | Conserved hypothetical protein | Fungal Zn(2)-Cys(6) binuclear cluster domain, Fungal specific transcription factor domain |
| AN4626 | 2.3 | Putative Zn(II)2Cys6 transcription factor | Has domain(s) with predicted RNA polymerase II transcription factor activity, sequence-specific DNA binding, zinc ion binding activity, role in regulation of transcription, DNA-templated and nucleus localization |
| AN8666 | 2.2 | C6 finger domain-containing protein | Fungal Zn(2)-Cys(6) binuclear cluster domain |
| AN7346 | 2.0 | Conserved hypothetical protein | Fungal Zn(2)-Cys(6) binuclear cluster domain, Fungal specific transcription factor domain |
| AN2667 | 2.0 | C6 transcription factor | Fungal specific transcription factor domain |
| OXIDOREDUCTASES | |||
| AN7418 | 5.5 | FAD monooxygenase | With predicted FAD binding, oxidoreductase activity and role in metabolic process |
| AN7981 | 3.8 | Ferric-chelate reductase | FAD-binding domain |
| AN10388 | 3.6 | Conserved hypothetical protein | Berberine and berberine like, FAD binding domain |
| AN11191 | 3.6 | Polyketide synthase | Beta-ketoacyl synthase, N-terminal domain, Beta-ketoacyl synthase, C-terminal domain, Acyl transferase domain, short chain dehydrogenase, KR domain |
| AN6096 | 3.4 | Conserved hypothetical protein | Aldo/keto reductase family |
| AN8349 | 3.3 | Salicylate hydroxylase | Pyridine nucleotide-disulphide oxidoreductase, FAD binding domain |
| AN0330 | 3.2 | NADH-dependent flavin oxidoreductase | NADH:flavin oxidoreductase / NADH oxidase family |
| AN10099 | 3.2 | Pyridoxamine phosphate oxidase | Pyridoxamine 5′-phosphate oxidase |
| AN2504 | 3.1 | Conserved hypothetical protein | FAD dependent oxidoreductase |
| AN0618 | 3.1 | Oxidoreductase | KR domain, short chain dehydrogenase |
| AN10005 | 3.0 | Short-chain dehydrogenase/reductase SDR | Short chain dehydrogenase |
| AN8979 | 2.8 | AlcA, Alcohol dehydrogenase I | Alcohol dehydrogenase GroES-like domain, Zinc-binding dehydrogenase |
| AN4643 | 2.8 | Conserved hypothetical protein | Cytochrome P450 |
| AN2704 | 2.8 | Conserved hypothetical protein | GMC oxidoreductase |
| AN10296 | 2.8 | FAD dependent oxidoreductase | FAD binding domain, flavo_cyto_c: flavocytochrome c, FAD dependent oxidoreductase, Flavin containing amine oxidoreductase |
| AN6414 | 2.8 | Conserved hypothetical protein | Cytochrome P450 |
| AN2682 | 2.8 | 12-oxophytodienoate reductase | NADH:flavin oxidoreductase / NADH oxidase family |
| AN1548 | 2.7 | Short-chain dehydrogenase | Not found |
| AN10667 | 2.7 | Alcohol dehydrogenase | Alcohol dehydrogenase GroES-like domain, Zinc-binding dehydrogenase |
| AN2723 | 2.7 | Histidinol dehydrogenase | Histidinol dehydrogenase, hisD: histidinol dehydrogenase |
| AN7388 | 2.6 | Catalase-peroxidase | Peroxidase, cat_per_HPI: catalase/peroxidase HPI |
| AN5287 | 2.6 | Acyl-CoA dehydrogenase family member 11 | Acyl-CoA dehydrogenase, middle domain, Acyl-CoA dehydrogenase, C-terminal domain |
| AN5421 | 2.5 | Steroid monooxygenase | Not found |
| AN3400 | 2.5 | Short chain type dehydrogenase | KR domain, short chain dehydrogenase |
| AN3206 | 2.5 | Glucose-methanol-choline oxidoreductase | GMC oxidoreductase |
| AN7415 | 2.5 | Conserved hypothetical protein | FAD binding domain |
| AN1034 | 2.5 | Polyketide synthase | NAD dependent epimerase/dehydratase, short chain dehydrogenase, Methyltransferase domain, Male sterility protein, Acyl transferase domain, Phosphopantetheine attachment site, Beta-ketoacyl synthase, C-terminal domain, Beta-ketoacyl synthase, N-terminal domain |
| AN0027 | 2.5 | Cyclohexanone monooxygenase | Flavin-binding monooxygenase-like, Pyridine nucleotide-disulphide oxidoreductase |
| AN0554 | 2.4 | Aldehyde dehydrogenase ALDH | Aldehyde dehydrogenase family |
| AN5854 | 2.4 | Oxidoreductase | 2-nitropropane dioxygenase, FMN-dependent dehydrogenase, Conserved region in glutamate synthase |
| AN8547 | 2.3 | GMC oxidoreductase | GMC oxidoreductase |
| AN2389 | 2.3 | Ketopantoate reductase | Ketopantoate reductase PanE/ApbA C terminal, apbA_panE: 2-dehydropantoate 2-reductase, Ketopantoate reductase PanE/ApbA |
| AN1825 | 2.3 | Sulfide:quinone oxidoreductase | Pyridine nucleotide-disulphide oxidoreductase |
| AN11096 | 2.3 | Conserved hypothetical protein | Short chain dehydrogenase |
| AN5360 | 2.3 | Conserved hypothetical protein | Cytochrome P450 |
| AN5373 | 2.3 | 3-oxoacyl-(acyl-carrier-protein) reductase | Short chain dehydrogenase, 23BDH: acetoin reductases |
| AN8250 | 2.3 | Conserved hypothetical protein | Cytochrome P450 |
| AN4126 | 2.3 | Aldehyde dehydrogenase | Aldehyde dehydrogenase family |
| AN2396 | 2.3 | Conserved hypothetical protein | NAD dependent epimerase/dehydratase family |
| AN2666 | 2.2 | Sorbitol/xylitol dehydrogenase | Zinc-binding dehydrogenase, bchC: Chlorophyll synthesis pathway, bchC, Alcohol dehydrogenase GroES-like domain |
| AN9315 | 2.2 | Conserved hypothetical protein | Pyridine nucleotide-disulphide oxidoreductase |
| AN2335 | 2.2 | 3-hydroxyisobutyrate dehydrogenase | NAD binding domain of 6-phosphogluconate |
| AN4829 | 2.1 | Alcohol dehydrogenase | Aldo/keto reductase family |
| AN3399 | 2.0 | FAD binding oxidoreductase | FAD binding domain |
| AN5550 | 2.0 | Conserved hypothetical protein | FAD binding domain |
| AN6659 | 2.0 | Short chain dehydrogenase | KR domain, short chain dehydrogenase, NAD dependent epimerase/dehydratase |
| TRANSPORTER ACTIVITY | |||
| AN9000 | 4.4 | MFS transporter | Major Facilitator Superfamily |
| AN10891 | 3.8 | High-affinity glucose transporter | SP: MFS transporter, sugar porter (SP) family, Sugar (and other), Major Facilitator Superfamily |
| AN8981 | 3.6 | AlcS, conserved hypothetical protein | GPR1/FUN34/yaaH family |
| AN6019 | 3.5 | MFS transporter | Major Facilitator Superfamily |
| AN12129 | 3.0 | Conserved hypothetical protein | Major Facilitator Superfamily, Sugar (and other) transporter |
| AN2368 | 3.0 | Membrane transporter | Major Facilitator Superfamily |
| AN9010 | 2.8 | MFS nicotinic acid transporter Tna1 | Major Facilitator Superfamily |
| AN2699 | 2.8 | Conserved hypothetical protein | Major Facilitator Superfamily, Ion channel regulatory protein UNC-93 |
| AN7380 | 2.7 | Conserved hypothetical protein | Major Facilitator Superfamily |
| AN11211 | 2.6 | Uracil permease | Permease for cytosine/purines, uracil, thiamine, allantoin, ncs1: NCS1 nucleoside transporter family |
| AN5275 | 2.6 | Choline transporter | Amino acid permease |
| AN2665 | 2.5 | MFS sugar transporter | Major Facilitator Superfamily, Major Facilitator Superfamily, Sugar (and other) |
| AN2358 | 2.5 | Conserved hypothetical protein | Sugar (and other), Major Facilitator Superfamily |
| AN8995 | 2.5 | Conserved hypothetical protein | Major Facilitator Superfamily |
| AN3207 | 2.5 | Amino acid transporter | Transmembrane amino acid transporter protein |
| AN9165 | 2.4 | Conserved hypothetical protein | Sugar (and other) transporter, Major Facilitator Superfamily |
| AN3352 | 2.4 | Conserved hypothetical protein | ncs1: NCS1 nucleoside transporter family, Permease for cytosine/purines, uracil, thiamine, allantoin |
| AN2201 | 2.4 | Proline permease | Amino acid permease |
| AN8352 | 2.4 | Carbixilic Transporter | Not found |
| AN8941 | 2.4 | Na/K ATPase alpha 1 isoform | Cation transporting ATPase, C-terminus, haloacid dehalogenase-like hydrolase, ATPase_P-type: HAD ATPase, P-type, family IC, E1-E2 ATPase |
| AN2959 | 2.3 | Allantoate transporter | Major Facilitator Superfamily |
| AN6063 | 2.2 | MFS transporter | Major Facilitator Superfamily, 2_A_01_02: Multidrug resistance protein, Sugar (and other) transporter, pump (TRI12) |
| AN12222 | 2.2 | Conserved hypothetical protein | Major Facilitator Superfamily |
| AN8955 | 2.2 | Conserved hypothetical protein | Major Facilitator Superfamily |
| AN3503 | 2.2 | Allantoate permease | Major Facilitator Superfamily |
| AN5187 | 2.1 | Na(+)/H(+) antiporter | Sodium/hydrogen exchanger family |
| AN2814 | 2.1 | MFS lactose permease | Major Facilitator Superfamily, SP: MFS transporter, sugar porter (SP) family |
| AN6960 | 2.1 | Conserved hypothetical protein | WD domain, G-beta repeat, Ankyrin repeat, CorA-like Mg2+ transporter |
| AN6451 | 2.1 | Conserved hypothetical protein | Major Facilitator Superfamily, Sugar (and other) transporter, 2_A_01_02: Multidrug resistance protein, Fungal trichothecene efflux pump (TRI12) |
| AN7067 | 2.0 | Conserved hypothetical protein | Major Facilitator Superfamily, SP: MFS transporter, sugar porter (SP) family, Sugar (and other) |
| AN10305 | 2.0 | Vacuolar membrane ATPase C | V_ATP_synt_C: V-type ATPase, C subunit, ATP synthase subunit C |
| AN9336 | 2.0 | Conserved hypothetical protein | Major Facilitator Superfamily |
| HYDROLASES | |||
| AN2690 | 4.0 | Conserved hypothetical protein | Glycosyl hydrolases family 16 |
| AN10346 | 3.8 | Cutinase | Cutinase, Phospholipase/Carboxylesterase |
| AN3402 | 3.8 | Alpha-amylase | Starch binding domain, Domain of unknown function (DUF1966), Alpha amylase, catalytic domain |
| AN8899 | 3.8 | 1-aminocyclopropane-1-carboxylate deaminase | Pyridoxal-phosphate dependent enzyme, ACC_deam: 1-aminocyclopropane-1- |
| AN5422 | 3.8 | Beta-lactamase | Beta-lactamase |
| AN6097 | 3.7 | Conserved hypothetical protein | 3-carboxy-cis,cis-muconate lactonizing enzyme |
| AN2834 | 3.5 | Conserved hypothetical protein | GDSL-like Lipase/Acylhydrolase |
| AN1792 | 3.5 | GDSL Lipase/Acylhydrolase | GDSL-like Lipase/Acylhydrolase |
| AN2539 | 3.3 | Haloalkanoic acid dehalogenase | HAD_type_II: haloacid dehalogenase, type II, HAD-SF-IA-v2: HAD hydrolase, family IA, variant 2 |
| AN3777 | 3.1 | Endo-beta-1,6-glucanase | Cellulase (glycosyl hydrolase family 5) |
| AN0022 | 3.1 | Alpha-galactosidase/alpha-n-acetylgalactosaminidase | Melibiase |
| AN11051 | 3.0 | Extracellular chitosanase CsnC | Fungal chitosanase |
| AN3613 | 2.8 | Xylanase | Glycosyl hydrolases family 11 |
| AN5060 | 2.7 | Conserved hypothetical protein | Has domain(s) with predicted arylformamidase activity and role in tryptophan catabolic process to kynurenine |
| AN11219 | 2.7 | Glutamyl-tRNA(Gln) amidotransferase subunit A | Amidase |
| AN10333 | 2.7 | Metallopeptidase, putative | Has domain(s) with predicted hydrolase activity and role in metabolic process |
| AN6656 | 2.6 | Endo-polygalacturonase D | Glycosyl hydrolases family 28 |
| AN9518 | 2.5 | Haloalkanoic acid dehalogenase | HAD_type_II: haloacid dehalogenase, type II, HAD-SF-IA-v2: HAD hydrolase, family IA, variant 2 |
| AN7275 | 2.5 | Xylosidase/glycosyl hydrolase | Glycosyl hydrolases family 43 |
| AN0181 | 2.4 | Conserved hypothetical protein | Thioesterase superfamily |
| AN8977 | 2.4 | AlcP, Conserved hypothetical protein | Strictosidine synthase, SMP-30/Gluconolactonase/LRE-like region |
| AN5608 | 2.4 | Conserved hypothetical protein | Carboxylesterase |
| AN1043 | 2.3 | Conserved hypothetical protein | Glycosyl hydrolases family 43 |
| AN2779 | 2.2 | Dipeptidase | Amidohydrolase family |
| AN2388 | 2.2 | Beta-1,4-endoglucanase | Not found |
| AN1826 | 2.1 | Metallo-beta-lactamase domain-containing protein | Metallo-beta-lactamase superfamily |
| AN2424 | 2.1 | Beta-N-acetylhexosaminidase | Glycosyl hydrolase family 20, catalytic domain |
| AN7911 | 2.1 | Conserved hypothetical protein | Amidohydrolase |
| AN2777 | 2.0 | Fumarylacetoacetate hydrolase | Fumarylacetoacetate (FAA) hydrolase family, alpha/beta hydrolase fold |
| UNCLASSIFIED | |||
| AN2587 | 8.7 | Conserved hypothetical protein | CFEM domain |
| AN11810 | 3.7 | Putative GNAT-type acetyltransferase | Not found |
| AN8980 | 3.4 | AlcM, conserved hypothetical protein | Not found |
| AN8092 | 3.3 | Conserved hypothetical protein | pfs domain Potential Cdc28p substrate |
| AN0180 | 3.0 | Putative Enoyl-CoA hydratase | Enoyl-CoA hydratase/isomerase family |
| AN7539 | 3.0 | Hydrophobin | Fungal hydrophobin |
| AN8913 | 3.0 | Predicted glycosylphosphatidylinositol (GPI)-anchored protein | Not found |
| AN8614 | 2.7 | Conserved hypothetical protein | Cupin domain |
| AN9354 | 2.7 | Putative transcription factor | NmrA-like family Transcription factor |
| AN2954 | 2.6 | Extracellular serine-rich protein | Not found |
| AN1982 | 2.5 | Cell cycle regulatory protein, putative | WD domain |
| AN5273 | 2.5 | Hydroxymethylglutaryl-CoA lyase | Enoyl-CoA hydratase/isomerase family,HMGL-like |
| AN8468 | 2.2 | Wd-repeat protein | NACHT domain, WD domain |
| AN9297 | 2.2 | FAD binding, oleate hydratase activity and role in fatty acid metabolic process | Not found |
| AN8622 | 2.2 | Phosphodiesterase/alkaline phosphatase D | PhoD-like phosphatase |
| AN1046 | 2.2 | Chitin synthase D | Chitin synthase |
| AN2022 | 2.2 | Putative heterokaryon incompatibility protein | Not found |
| AN11080 | 2.1 | DMATS type aromatic prenyltransferase | Tryptophan dimethylallyltransferase, arom_pren_DMATS: aromatic prenyltransferase, DMATS type |
| AN3730 | 2.1 | 1,3-beta-glucanosyltransferase | X8 domain, Glycolipid anchored surface protein (GAS1) |
| AN0017 | 2.1 | Conserved hypothetical protein | CutC family |
| AN2623 | 2.1 | Acyl-coenzyme A:6-aminopenicillanic-acid-acyltransferase | Not found |
| AN5833 | 2.1 | Acyl-CoA synthetase | AMP-binding enzyme, Domain of unknown function (DUF3448) |
| AN7279 | 2.1 | GPI anchored protein poly(beta-D-mannuronate) lyase activity | Not found |
| AN2346 | 2.1 | Conserved hypothetical protein | Pfs domain |
| AN10302 | 2.1 | Hypothetical protein | NACHT domain |
| AN3159 | 2.0 | Conserved hypothetical protein | RasGEF domain |
| AN8368 | 2.0 | Conserved hypothetical protein | Spherulation-specific family 4 |
| AN2778 | 2.0 | Conserved hypothetical protein | Cytochrome b5-like Heme/Steroid binding domain |
| HYPOTHETICAL PROTEINS WITH NO IDENTIFIED DOMAINS | |||
| AN2676 | 4.9 | Conserved hypothetical protein | Not found |
| AN7203 | 3.5 | Conserved hypothetical protein | Not found |
| AN8369 | 3.4 | Predicted protein | Not found |
| AN11341 | 3.2 | Predicted protein | Not found |
| AN10086 | 3.1 | DUF1446 domain-containing protein | Protein of unknown function (DUF1446) |
| AN9331 | 3.1 | Conserved hypothetical protein | Not found |
| AN6797 | 3.1 | Conserved hypothetical protein | Not found |
| AN4122 | 3.1 | Conserved hypothetical protein | Protein of unknown function |
| AN2322 | 3.0 | Conserved hypothetical protein | Not found |
| AN10360 | 2.9 | Conserved hypothetical protein | Not found |
| AN9323 | 2.9 | Conserved hypothetical protein | Not found |
| AN7261 | 2.8 | Conserved hypothetical protein | Not found |
| AN5190 | 2.8 | Conserved hypothetical protein | Not found |
| AN2700 | 2.8 | Conserved hypothetical protein | Not found |
| AN0972 | 2.7 | Conserved hypothetical protein | Not found |
| AN2750 | 2.7 | Conserved hypothetical protein | Not found |
| AN1928 | 2.7 | Conserved hypothetical protein | Not found |
| AN12341 | 2.7 | Hypothetical protein | Not found |
| AN1042 | 2.7 | Conserved hypothetical protein | Not found |
| AN10326 | 2.6 | Predicted protein | Not found |
| AN7333 | 2.6 | Conserved hypothetical protein | Not found |
| AN12061 | 2.6 | Conserved hypothetical protein | Not found |
| AN8240 | 2.6 | Conserved hypothetical protein | Not found |
| AN7985 | 2.5 | Conserved hypothetical protein | Not found |
| AN11290 | 2.5 | Conserved hypothetical protein | Not found |
| AN7270 | 2.5 | Conserved hypothetical protein | Not found |
| AN4554 | 2.5 | Conserved hypothetical protein | Not found |
| AN11809 | 2.5 | Conserved hypothetical protein | Not found |
| AN2041 | 2.4 | Conserved hypothetical protein | Not found |
| AN10048 | 2.4 | Conserved hypothetical protein | Not found |
| AN1925 | 2.4 | Conserved hypothetical protein | Not found |
| AN8994 | 2.4 | Conserved hypothetical protein | Not found |
| AN11281 | 2.4 | Conserved hypothetical protein | Not found |
| AN12238 | 2.3 | Conserved hypothetical protein | Not found |
| AN10320 | 2.3 | Conserved hypothetical protein | Not found |
| AN9521 | 2.2 | Conserved hypothetical protein | Not found |
| AN7124 | 2.2 | Conserved hypothetical protein | Not found |
| AN4319 | 2.2 | Conserved hypothetical protein | Not found |
| AN0005 | 2.2 | Hypothetical protein | Not found |
| AN4136 | 2.2 | Conserved hypothetical protein | Not found |
| AN11585 | 2.2 | Conserved hypothetical protein | Not found |
| AN7976 | 2.1 | Conserved hypothetical protein | Not found |
| AN11872 | 2.1 | Predicted protein | Not found |
| AN8148 | 2.1 | Conserved hypothetical protein | Not found |
| AN6648 | 2.1 | Conserved hypothetical protein | Not found |
| AN11777 | 2.1 | Predicted protein | Not found |
| AN11865 | 2.1 | Conserved hypothetical protein | Not found |
| AN0481 | 2.1 | Conserved hypothetical protein | Not found |
| AN8008 | 2.1 | Conserved hypothetical protein | Not found |
| AN1649 | 2.1 | Conserved hypothetical protein | Not found |
| AN7099 | 2.1 | Conserved hypothetical protein | Not found |
| AN8740 | 2.1 | Conserved hypothetical protein | Not found |
| AN4642 | 2.0 | Conserved hypothetical protein | Not found |
| AN0539 | 2.0 | Conserved hypothetical protein | Not found |
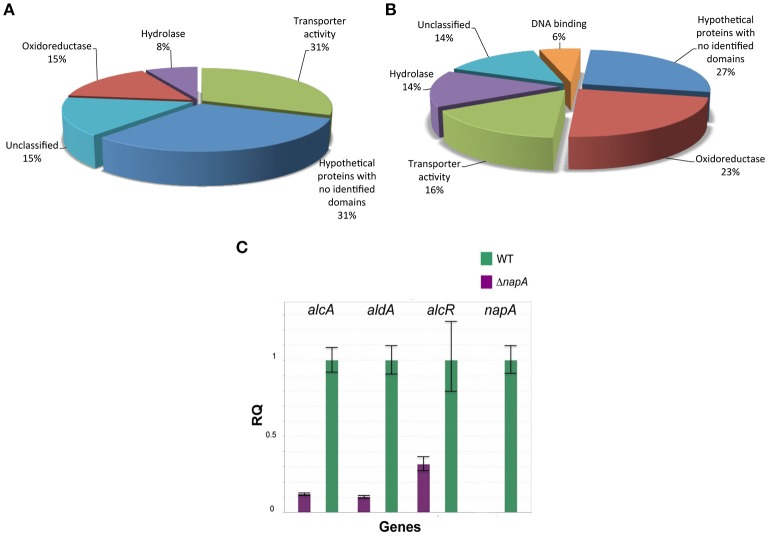
Our results show that the number of genes negatively regulated by NapA is small (13), compared with the number of genes positively regulated by NapA (201). Among genes most highly repressed by NapA, there is the putative HET (heterokaryon incompatibility) containing domain protein AN7834, the cysteine-rich secreted protein AN7836 and the putative ABC transporter AN7839, which seems to represent the only NapA-regulated ABC-type transporter. In addition, this group includes genes for 3 additional putative transporters (MSF type), 2 oxidoreductases, 1 hydrolase, 1 acetyltransferase, 1 protein with a nucleoside phosphorylase domain and 2 proteins with no identifiable domains (Figure 7A, Table 1).
Figure 7.
NapA is required for gene regulation in asexual spores. Transcriptomic analysis reveals that NapA is needed for the negative regulation of at least 13 genes (A) and the positive regulation of at least 201 genes (B) in conidia. (C) Confirmation of NapA-dependent expression of ethanol utilization genes during conidiation by qPCR. Genes alcA, aldA and alcR were used to confirm transcriptome results from conidia. Gene expression was normalized to H2B histone transcript levels. Data are mean values of three independent biological replicates. Bars represent the standard deviation (SD).
Among the 201 genes positively regulated by NapA (Figure 7B, Table 2), 12 encode putative (TFs) not yet characterized in A. nidulans, most of them belonging to the Zn(2)-Cys(6) DNA-binding domain family. Remarkably, 46 genes encode different types of oxidoreductases, including multiple monooxygenases, dehydrogenases, several members of the P450 cytochrome drug detoxifying enzyme family and the catalase peroxidase CatD/CpeA (AN7388). Other genes encode proteins with lipase, peptidase and other hydrolytic activities. 32 genes encode putative membrane proteins involved in the transport of sugars, drugs, amino acids, metals or other metabolites, 21 of which are members of the major facilitator superfamily (MSF). 29 genes encode proteins with predicted or confirmed hydrolytic enzyme activity. Notably, several of these are related to the hydrolysis of plant cell-wall complex carbohydrates, such as putative cutinase AN10346, alpha-amylase AN3402, endo-beta-1,6-glucanase AN3777, xylanase AN3613, endo-polygalacturonase AN6656, xylosidase AN7275, beta-1,4-endoglucanase AN2388, while xylose inducible alpha-galactosidase aglC/AN8138, alpha-xylosidase agdD/AN7505, putative beta-glucosidase bglH/AN3903, putative alpha-amylase amyC/AN4507 were detected as NapA regulated with logFC values between 2 and 1.5 (Table 2 and Table S4). Other hydrolases like putative beta-lactamases AN5422 and AN1826 might be related to antibiotic degradation.
Twenty eight genes encode proteins that have recognizable domains but did not belong to the previous categories. Among these, AN2587 shows the highest LogFC value (8.7), and encodes a putative membrane protein with a cysteine-rich CFEM domain, recently shown to be involved in Fe3+ heme acquisition (Nasser et al., 2016). This and the fact NapA also regulates ferric reductase AN7981, putative heme binding protein AN2778 and NRPS SidC (AN0607, siderophore biosynthesis; Table S4) genes supports a NapA role in iron acquisition. Also notable is NapA regulation of several genes involved in the biosynthesis of secondary metabolites (Tables 2 and Table S4), such as sterigmatocystin (stcU/AN7806), penicillin (ipnA/AN2622), monodictyphenone and prenyl xanthones (Bok et al., 2009; Sanchez et al., 2011; Andersen et al., 2013) (xptB/AN12402, mdpB/AN10049, mdpC/AN0146 and mdpD/AN0147), all with clear antimicrobial activity. Other genes are involved in the biosynthesis of asperfuranone (afoE/AN1034) or unknown metabolites (PKS AN11191). In contrast, NapA has been reported to repress the production of secondary metabolites in A. parasiticus (Reverberi et al., 2008) and A. nidulans (Yin et al., 2013) during growing conditions, suggesting that NapA regulates secondary metabolism in opposite ways during growth and conidiation. Finally, NapA regulates at least 54 genes encoding hypothetical proteins with no recognizable domains (Figure 7B, Table 2).
In summary, our transcriptomic results support a model in which during conidiation NapA regulates functions associated with the successful germination of conidia in natural environments. In the case of A. nidulans saprophytic life style this would include a large group of enzymes to degrade plant complex carbohydrates, proteins to transport the corresponding derived sugars and enzymes needed to transform them into acetyl-CoA. NapA also regulates the production of secondary metabolites, such as penicillin, which would prevent growth from competing organisms, as well as enzymes and transporters to detoxify drugs produced by competitors. In the same line, NapA regulates enzymes and proteins involved in iron scavenging, such as those involved in siderophore biosynthesis, iron reduction and transport, and heme acquisition. As such functions are also important during pathogenic interactions, we propose that these and not NapA ROS-detoxification roles, might be relevant for NapA critical virulence roles (Molina and Kahmann, 2007; Guo et al., 2011; Huang et al., 2011), specially when conidia mediate the infection process.
NapA is required for alc gene expression and the utilization of ethanol, arabinose, and fructose as sole carbon sources
It called our attention that the mRNA levels of genes alcR (AN8978; Table S4), alcA (AN8979), alcU (AN8982; Table S4) and aldA (AN0554), all members of the well-studied ethanol system (Fillinger and Felenbok, 1996), were clearly reduced in ΔnapA conidia. The ethanol system is composed by genes alcP, alcR, alcO, alcA, alcM, alcS, and alcU, all clustered in chromosome VII and aldA located in chromosome VIII. alc genes share the same regulation, being strongly induced by the physiological inducer acetaldehyde and the non-physiological inducer 2-butanone (Flipphi et al., 2002). They are also subject to strict control by the transcriptional activator AlcR and are repressed by glucose via the CreA repressor (Fillinger and Felenbok, 1996). However, only AlcR, alcohol dehydrogenase I AlcA, and aldehyde dehydrogenase AldA are required for ethanol utilization, and the specific function of the other alc genes is unknown. Yet, AlcS is a membrane protein with homology to acetate transporters that is nevertheless dispensable for growth on ethanol, acetaldehyde or acetate (Flipphi et al., 2006). Therefore, we decided to confirm alcR, alcA and aldA expression results by using qPCR. Results in Figure 7C show that indeed, the accumulation of the corresponding mRNAs in conidia depends on NapA.
To test the physiological significance of NapA-mediated alc gene regulation, we examined the ability of ΔnapA mutants to grow on ethanol as sole carbon source. As control we included a creAd204 mutant, which shows derepressed expression of alcA in the presence of glucose (Shroff et al., 1996). Since AlcA transforms allyl alcohol (AA) into the toxic compound acrolein, this creA mutant is unable to grow on glucose plus AA (Figures 8A,B). Notably, ΔnapA conidia and mycelia were unable to grow on ethanol, supporting NapA function in proper alcA expression. However, the ΔnapA mutant was also unable to grow on glucose plus AA (Figures 8A,B). AA/acrolein causes oxidative stress and induces Yap1 activation in S. cerevisiae (Kwolek-Mirek et al., 2009; Golla et al., 2015) and therefore it might result particularly toxic for the ΔnapA mutant. To test this, we generated ΔalcA and ΔnapA ΔalcA mutants and examined them for ethanol utilization and AA resistance. As seen in Figure 8C, the ΔalcA mutant is not sensitive to H2O2, is unable to grow on ethanol and is fully resistant to AA. In contrast, a ΔnapA ΔalcA mutant is sensitive to H2O2, unable to grow on ethanol and more sensitive to AA than the ΔalcA mutant, supporting the idea that ΔnapA mutants are per se more sensitive to AA. The failure to grow in 1% ethanol was not remediated by the presence of the antioxidant N-acetylcysteine (5 mM), suggesting that such failure is related to the reduced expression of the alc genes. However, it cannot be excluded that this might be also related to an increased ethanol sensitivity of the ΔnapA mutants. In summary, these results show that NapA is required for expression of the alc regulon during asexual development and for growth on ethanol, adding a new level of regulation to this well know pathway.
Figure 8.
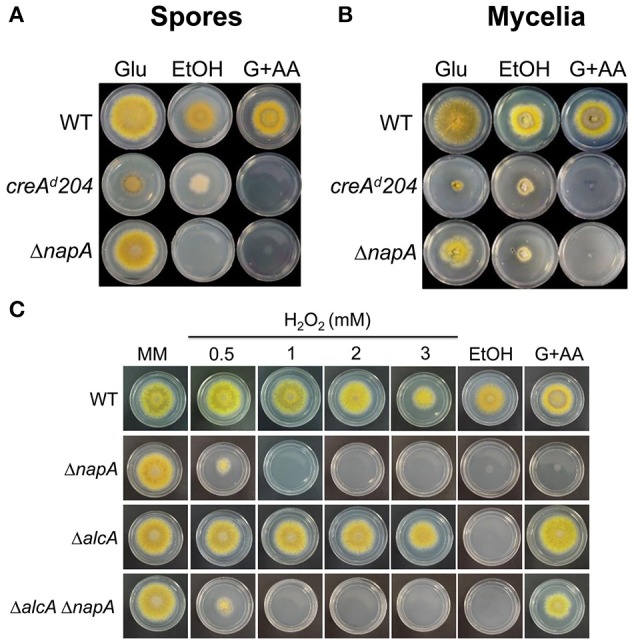
NapA is required for the ethanol utilization and for resistance to Allyl alcohol. (A) Conidia (1 × 103) from strains CLK43 (WT), CFL7 (ΔnapA) and MH440 (creAd204) were inoculated on supplemented MM plates containing either 1% glucose, 1% ethanol or 1% glucose plus 5 mM Allyl alcohol (AA) and were incubated at 37°C for 4 days. (B) Mycelial plugs cut from the growing edge of 5-day colonies from the strains in (A) were transferred to the indicated media and incubated at 37°C for 4 days. (C) Conidia (1 × 103) from strains CLK43 (WT), CFL7 (ΔnapA), CAM17 (ΔalcA), and CAM18 (ΔalcA ΔnapA) were inoculated on supplemented MM plates containing either H2O2 at the indicated concentrations or 1% glucose (MM), 1% ethanol or 1% glucose plus 5 mM allylic alcohol (AA) and incubated at 37°C for 4 days.
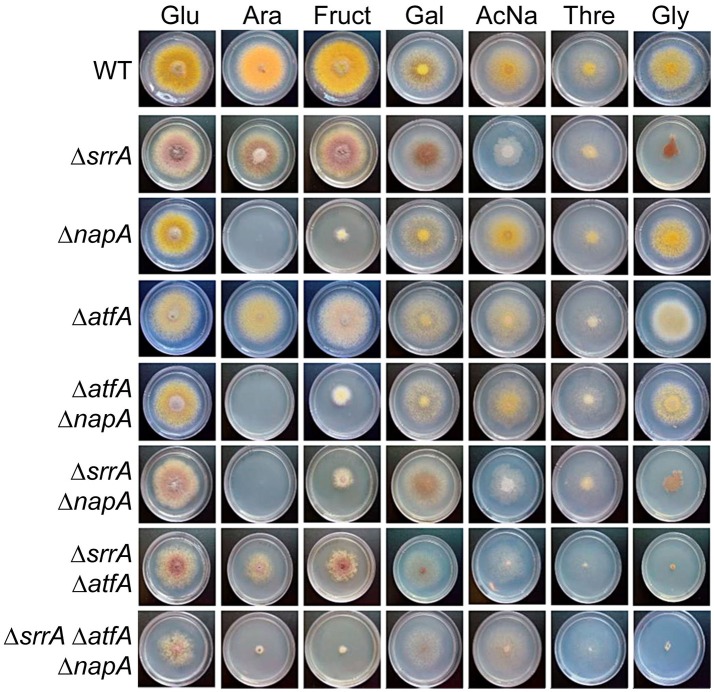
These results prompted us to determine if NapA and TFs SrrA and AtfA were also required for conidia germination and growth on ethanol or other alternative carbon sources. As seen in Figure 9, NapA was also required for arabinose and fructose utilization, while SrrA was partially required for glycerol utilization and AtfA was dispensable for the utilization of all carbon sources tested. These results indicate that NapA plays an important role in the utilization of carbon sources alternative to glucose.
Figure 9.
NapA is also necessary for the utilization of arabinose and fructose as sole carbon sources. Conidia (1 × 103) from strains CLK43 (WT), COSsrrA3 (ΔsrrA), CFL7 (ΔnapA), TFLΔatfA-04 (ΔatfA), CAM7 (ΔatfA ΔnapA), CAM6 (ΔsrrA ΔnapA), CAM8 (ΔsrrA ΔatfA), and CAM9 (ΔsrrA ΔatfA ΔnapA) were inoculated on supplemented MM plates containing either glucose (Glu), arabinose (Ara), fructose (Fruct), galactose (Gal), sodium acetate (AcNa), threonine (Thre) or 1% glycerol (Gly) as sole carbon sources and incubated at 37°C during 4 days.
NapA is localized in nuclei during growth in ethanol or glucose starvation
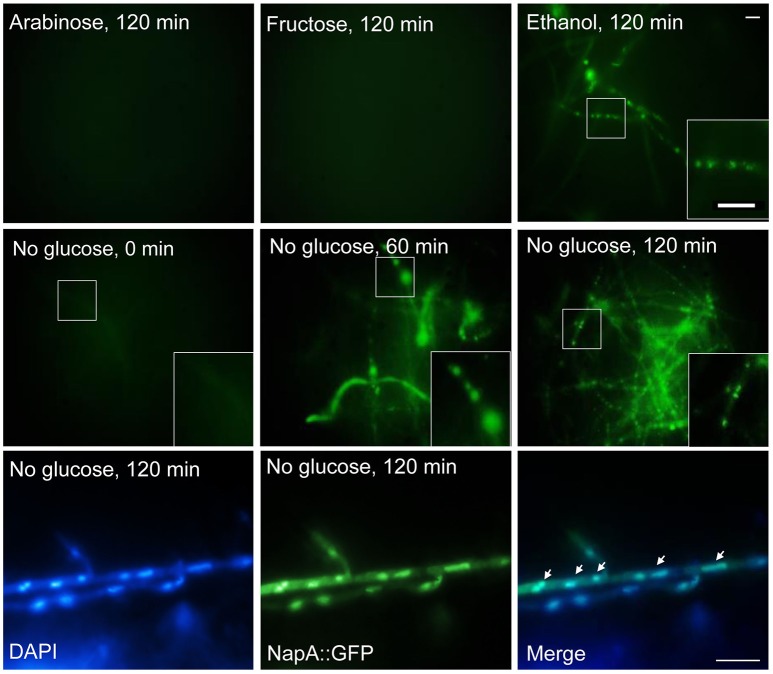
Having found that NapA is necessary for proper growth in arabinose and fructose, we explored NapA localization during growth in poor carbon sources, as well as during glucose starvation. Results in Figure 10 show that as it occurs during growth in glucose, NapA expression is very low during growth in arabinose and fructose as sole carbon sources. In contrast, NapA::GFP expression was induced during growth in ethanol, as well as during glucose starvation, and NapA::GFP nuclear localization was detected between 60 and 120 min after shifting to ethanol or glucose lacking media. In addition, we found that a osmotic stress treatment with 1 M NaCl is able to induce NapA nuclear localization under normal glucose growth conditions (Figure S7), despite the fact that ΔnapA mutants are not sensitive to osmotic stress.
Figure 10.
Glucose starvation induces NapA nuclear localization. NapA does not accumulate in nuclei during growth in arabinose or fructose as sole carbon sources but it does accumulate in nuclei during glucose starvation. Conidia from strain CAM20 (NapA::GFP) were grown for 18 h in MM at 37°C and then mycelia was shifted to MM with or without glucose or with fructose or glycerol as carbon source for indicated times (0–120 min). Mycelial samples were observed in vivo and photographed every 60 min using Epifluorescence microscopy. Lower panel shows nuclei (DAPI) and NapA::GFP fluorescence in mycelia starved for glucose during 120 min, fixed and photographed. Larger square areas in each picture show enlargements of the areas indicated by smaller squares.
Discussion
AtfA, SrrA, and NapA play non-redundant and differential roles in the antioxidant response
At least three pathways, mediated by TFs, AtfA, SrrA and NapA, are involved in fungal responses to oxidative and other types of stress and we have studied them here in the same organism. We have reported that four catalases present in A. nidulans display differential regulation during growth, stress and development (Navarro et al., 1996; Navarro and Aguirre, 1998; Kawasaki and Aguirre, 2001). Notably, catalase CatA is found only in asexual and sexual spores, despite the fact that catA mRNA accumulates under many stress conditions (Navarro and Aguirre, 1998; Lara-Rojas et al., 2011). Our results here show that TFs, AtfA, SrrA and NapA are essential components in such catalase gene regulation. NapA and SrrA are both required for CatB induction by H2O2, only AtfA is required for CatA expression in conidia and none of them is required for CatB induction during late stationary phase of growth, while CatD/CpeA activity levels are influenced by SrrA. The phenotypic analysis of oxidative stress resistance using ΔnapA, ΔsrrA, and ΔatfA single, double and triple mutants also showed that TFs NapA, SrrA, and AtfA play differential roles in oxidative stress resistance.
The functions of Ap1-like TFs in different fungi show both similarities and differences. For example and in contrast to our results, yapA deletion in symbiotic fungus Epichloë festucae causes sensitivity to H2O2 and t-BOOH in conidia but not in mycelia and YapA, instead of AtfA, is required for expression of the spore-specific catalase CatA (Cartwright and Scott, 2013). Neurospora crassa mutants lacking the NapA homolog NcAp-1 were reported as showing no sensitivity to osmotic stress and only a slight sensitivity to H2O2 (Takahashi et al., 2010), although recent work showed that mutants were sensitive to osmotic stress, cadmium and H2O2 (Tian et al., 2011). In Magnaporthe oryzae inactivation of NapA homolog MoAP1 causes only a mild sensitivity to H2O2 (Guo et al., 2011), while Ustilago maydis yap1 null mutants are sensitive to H2O2 and their virulence is significantly reduced (Molina and Kahmann, 2007). A. fumigatus, yap1 null mutants are also sensitive to H2O2 and menadione but not affected in pathogenicity (Lessing et al., 2007).
Like in the case of Yap1 and Pap1, the activation of NapA homologs by H2O2 results in nuclear accumulation in all filamentous fungi where this has been studied, and in U. maydis two Yap1 conserved cysteines (Cys-399 and Cys-407) were shown to be crucial for both, nuclear accumulation and functionality (Molina and Kahmann, 2007). An additional level of redox regulation has been proposed in A. nidulans, where the CCAAT-binding complex (CBC) represses napA expression under low ROS levels, while high ROS levels result in oxidation of two cysteines in HapC CBC subunit, and the transcriptional activation of napA (Thön et al., 2010). We found that in addition to H2O2 and menadione, osmotic stress (Figure S7), glucose starvation stress and growth on ethanol also induced NapA nuclear localization, supporting the idea that different types of stress can all lead to oxidative stress (Hansberg and Aguirre, 1990).
GpxA, TpxA, and TpxB peroxiredoxin function in the antioxidant response and conidiation
We analyzed the role of Gpx3 peroxiredoxin homolog GpxA and Tpx1 homologs TpxA and TpxB in NapA antioxidant and developmental functions. We found that none of these peroxiredoxins was required for H2O2 or menadione resistance and therefore are unnecessary for NapA activation. This is consistent with results in E. festucae, where peroxiredoxins Gpx3 (GpxA) and Tpx1 (TpxA) were not needed for YapA H2O2-induced nuclear accumulation (Cartwright and Scott, 2013). On the contrary, M. oryzae mutants lacking Gpx3 functional homolog MoHYR1 are sensitive to H2O2, fail to express several genes related to the antioxidant response and show reduced virulence. However, initial data showing that MoYap1 mutants are no affected in pathogenicity indicates that MoHYR1 functions in virulence are not mediated by MoYap1 (Huang et al., 2011). In A. fumigatus AfYap1 also accumulates in the nuclei in response to H2O2 and notably, GpxA homolog AspF3 and TpxA require AfYap1 for its H2O2-mediated induction (Lessing et al., 2007). These and our results suggest that NapA might be required for H2O2-induced expression of gpxA, tpxA and tpxB. A minor direct or indirect role for these peroxiredoxins in H2O2 detoxification is indicated by the fact that in a ΔnapA background, the simultaneous inactivation of Gpx3, TpxA, and TpxB resulted in increased sensitivity to H2O2. Our results suggest that TpxA and NapA regulate conidiation through the same pathway. One possibility is that NapA mediates TpxA induction during conidiation, although we did not detect tpxA mRNA in our transcriptomic experiments, and TpxA could in turn regulate other activities needed for full sporulation.
NapA regulates development
We reported that the regulated production of ROS is essential for fungal sexual development (Lara-Ortíz et al., 2003; Cano-Domínguez et al., 2008), a finding demonstrated in several fungi (Malagnac et al., 2004; Siegmund et al., 2013; Dirschnabel et al., 2014). In addition to their roles in the antioxidant response, AtfA and SrrA play different roles in gene regulation during development. AtfA mediates SakA roles in sexual development and conidial viability (Kawasaki et al., 2002; Lara-Rojas et al., 2011), while SrrA is required for normal conidiation and conidial viability (Vargas-Perez et al., 2007). The results reported here uncovered a novel developmental role for NapA, by showing that NapA represses sexual development and is needed for full conidiation, as well as for the accumulation of multiple mRNAs in conidia. The requirement for increased ROS levels during sexual development is consistent with the fact that ΔnapA mutants show an increased production of fruiting bodies, as NapA is required to maintain low ROS levels and to express genes involved in the biosynthesis of cleistothecial melanin, a well-known antioxidant.
The lower conidiation observed in ΔnapA mutants might be related to their lower expression of gmcA gene (AN8547), encoding a putative glucose-methanol-choline oxidoreductase required for early stages of conidiophore development (Etxebeste et al., 2012). The fact that GmcA ortholog AFUA_3G01580 is also induced by H2O2 in an AfYap1-dependent manner in A. fumigatus (Lessing et al., 2007) suggests that gmcA regulation by NapA is conserved, at least in the Aspergilli. Moreover, NapA roles in development might be conserved in fungi. In M. oryzae MoAP1 deletion causes only mild sensitivity to H2O2 but mutants show a drastic reduction in formation of aerial mycelium and conidiation. Notably, the same phenotypes are observed in mutants affected in MoAP1 regulated genes MGG_01230 and MGG_15157, encoding succinic semialdehyde dehydrogenase MoSsadh and acetyltransferase MoAct, respectively (Guo et al., 2011), which however we did not detect as NapA-dependent. In the dimorphic fungus Talaromyces marneffei, yapA mutants are sensitive to H2O2 and menadione, show decreased radial growth, produce conidiophores with fewer phialides and conidia, and conidia show decreased germination rates, while yeast cells fail to undergo binary fission (Dankai et al., 2016). Overall, the developmental roles of TFs long associated only with ROS detoxification support the role of ROS as developmental signals (Hansberg and Aguirre, 1990; Aguirre et al., 2005).
Genes regulated by NapA during asexual development
A comparison between Ap1-like dependent regulons under H2O2 stress shows common themes in different fungi. DNA microarray analysis of yap1-dependent genes in U. maydis identified 221 down regulated genes with a fold change >1.5, that included genes for ROS decomposing enzymes and enzymes involved in biosynthesis of low molecular weight antioxidants and NADPH generation (Molina and Kahmann, 2007). Similarly, in Cochliobulus heterostrophus CHAP1-dependent genes included genes for thioredoxin reductase, γ-glutamyl cysteine synthetase, glutathione reductase, glutathione synthetase and thioredoxin (Lev et al., 2005). In B. cinerea, genes for catalaseC, thioredoxin reductase, glutaredoxin, glutathione-S-transferase1, thioredoxin and a hypothetical glutathione-S-transferase were found to be Bap1-dependent (Temme and Tudzynski, 2009) and a similar pattern was observed in A. fumigatus (Lessing et al., 2007).
Overall, such relatively conserved H2O2-induced Ap1-mediated gene expression patterns are different from the one we observe during conidiation. Indeed, we do not find genes involved in major NADPH or GSH generation pathways. Instead, we find a large number of genes involved in drug efflux and detoxification, including several genes for enzymes with putative cytochrome P450 activity, as well as other oxidases. Notably, Yap1 (Lee et al., 1999) and Pap1 (Chen et al., 2008) regulate genes coding for efflux pumps and dehydrogenases that seem necessary for defense against multiple drugs. In fact, pap1 was first identified as a gene whose overexpression conferred resistance to drugs like brefeldin A, staurosporine or caffeine (Toda et al., 1991). Furthermore, Calvo et al. (2012) have shown that in response to H2O2 Pap1 requires the transcription factor Prr1 to activate the antioxidant but not the drug tolerance genes, providing a possible mechanism to explain the major role that NapA shows in the regulation of drug tolerance genes during conidiation. Indeed, the possibility that NapA plays overlapping but different functions during oxidative stress, conidiation and sexual development deserves further research.
Among the NapA-dependent genes, we also identified two members of the cupin superfamily, which includes metal-dependent and independent enzymes, as well as catalytically inactive proteins associated with abiotic stress and quiescent structures. AN8614 encodes a single-domain 153 amino-acid cupin showing similarity to germins and oxalate oxidases (Dunwell et al., 2000), some of which surprisingly also show superoxide dismutase activity (Woo et al., 2000). AN8368 encodes a 305 amino-acid protein identified as a spherulin 4-like protein. We detected that AN8368 is an ortholog of AN2952, and A. fumigatus and A. clavatus Sph3, which correspond to a glycoside hydrolase essential for the biosynthesis of the exopolysaccharide galactosaminogalactan (Bamford et al., 2015).
The role of NapA in carbon catabolism
Conidia and mycelia from ΔnapA mutants are unable to grow on ethanol and arabinose and show reduced growth on fructose, as sole carbon sources. A derepression of alcA during conidiation was observed before in conidiation defective mutants carrying alleles of the affected gene fused to the alcA promoter (Arratia-Quijada et al., 2012). The lack of growth on ethanol can be at least partially explained by the fact that NapA is required for full expression of alcR, alcA, and aldA genes during conidiation and presumably also during mycelial growth. Since NapA is needed for normal expression of AlcR and this TF is required for its own transcription, it seems likely that AlcR mediates the regulation of alcP, alcA, alcM, alcS, and aldA exerted by NapA. In this scenario alcR full expression might require CreA derepression, activation by NapA, as well as AlcR autoinduction. The fact that alcR promoter contains putative Ap1 binding sites is consistent with this interpretation. It seems unlikely that CreA represses napA expression because NapA antioxidant function must be required during normal glucose metabolism. In addition, creAd204 mutants do not show increased resistance to H2O2 and qPCR analysis did not show increased napA mRNA levels in conidia from the creAd204 mutant, as compared to wild type conidia (not shown). Three lines of evidence indicate that ethanol utilization represents a condition in which NapA antioxidant role becomes more important. First, NapA is required for full expression of most alc genes during conidiation. Second, ethanol growth induces napA expression and NapA nuclear localization. Third, NapA is needed for growth in ethanol. Evidence indicates that ethanol toxicity is mediated by AlcA product acetaldehyde, which itself can cause oxidative stress. Interestingly, it has been considered unlikely that ethanol catabolism alone requires such a high expression levels and subtle regulation of the alc genes (Flipphi et al., 2006). We have added another regulation layer that might help to understand the physiological significance of this complex regulation.
NapA requirement to utilize arabinose and fructose might be explained at different levels. First, NapA is necessary for transcript accumulation of the xylitol/sorbitol dehydrogenase (AN2666) in conidia and possibly in mycelia. AN2666 is 52% identical to A. niger enzyme XdhA, which participates in conversation of xylitol to D-xylulose during arabinose metabolism (de Groot et al., 2007). Xylitol/sorbitol dehydrogenase also shows 77% identity to SdhA, an enzyme involved in reversible transformation between fructose and sorbitol (Koivistoinen et al., 2012). This suggests that AN2666 participates in fructose assimilation and is essential for arabinose assimilation. In addition, arabinose and/or fructose transport might be compromised in ΔnapA mutants, considering that NapA regulates several genes encoding proteins with transport activity. Among these, AN10891 and AN2665 encode proteins that are 28 and 26% identical to B. cinerea fructose specific transporter FRT1 (Doehlemann et al., 2005). Second, arabinose and fructose metabolism share some regulatory steps. Within the Aspergilli transcriptional activators AraR, GalR and XilR regulate the metabolic conversion of L-arabinose, D-galactose and D-xylose, respectively. A recent report shows that both AraR and XlnR regulate the pentose catabolism genes, as well as the oxido-reductive D-galactose catabolic pathway (Kowalczyk et al., 2015). However, it is interesting that mutants lacking both AraR and XlnR still show a detectable growth on arabinose, indicating the participation of additional regulatory mechanisms. Also interesting is that A. niger XlnR is somehow related to ROS metabolism, as xlnR null mutants are sensitive to oxidative stress and show a qualitative increase in ROS levels (Raulo et al., 2016). Third, NapA homologs are necessary for expression of enzymes involved in the generation of cellular reducing power (NADPH, GSH and thioredoxins). Although this was not detected during conidiation, during growth A. nidulans ΔnapA mutants show a decreased GSH/GSSG ratio and a 1.6 fold decrease in total glutathione content when compared to a WT strain (Thön et al., 2010). This is consistent with the fact that ΔnapA mutants are very sensitive to AA/acrolein, known to cause GSH depletion (Kwolek-Mirek et al., 2009). Such a decrease in reducing power might provide additional difficulties in catabolizing carbon sources that are more demanding on these resources. Indeed, ethanol and arabinose catabolism results in an imbalance of redox cofactors (Seiboth and Metz, 2011).
A role for NapA homologs in carbon metabolism appears conserved in fungi. In S. cerevisiae the involvement of Yap1 in responding to carbon stress was proposed after it was recovered in two-hybrid assays as interactor of the Sip2 subunit of the Snf1 kinase, needed for adaptation to glucose limitation, and showing that media shifting from glucose to glycerol or to glucose lacking media induced Yap1 nuclear accumulation (Wiatrowski and Carlson, 2003). Likewise, in S. pombe glucose but not nitrogen starvation induces Pap1 nuclear localization in a process that depends on the stress MAPK Spc1/Sty1 (Madrid et al., 2004). However, a direct involvement of Yap1 or NapA in carbon utilization was not demonstrated in these cases.
In nature, fungal spores germinate on environments containing diverse, often-poor carbon sources as well as drugs produced by competing organisms or that are detritus-derived. In addition, conidia contain chemicals and toxins derived form secondary metabolism as well as auto-inhibitors of germination. Our results support the proposal that oxidative stress is produced during conidiation and that NapA plays a crucial role in the regulation of multiple genes during this process. Although the rates of conidia germination are not decreased in ΔnapA mutants under laboratory conditions, the set of genes regulated by NapA during conidiation suggest that they might be important for spores to germinate in natural heterogeneous environments.
Author contributions
JA: Designed experiments, wrote the MS, and obtained funding. AM, FL and OS: Performed and designed experiments, contributed to MS writing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by CONACYT grants CB-2014-01-238492, Investigación en Fronteras de la Ciencia 2015-I-319 and PAPIIT-UNAM IN208916. AM received a Doctoral fellowship from CONACYT and this work is part of her Ph.D. Thesis, as student in the program Maestría y Doctorado en Ciencias Bioquímicas from Universidad Nacional Autónoma de México. We thank IFC-UNAM Molecular Biology, Imagenology and Computer Units. We also thank Ricardo Grande, Veronica Jimenez-Jacinto and Alejandro Sanchez-Flores for sequencing and bioinformatics support as part of the “Unidad de Secuenciación Masiva y Bioinformática” of the “Laboratorio Nacional de Apoyo Tecnológico a las Ciencias Genómicas,” CONACyT #260481, at Instituto de Biotecnología/UNAM.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00516/full#supplementary-material
References
- Aguirre J., Ríos-Momberg M., Hewitt D., Hansberg W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13, 111–118. 10.1016/j.tim.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Andersen M. R., Nielsen J. B., Klitgaard A., Petersen L. M., Zachariasen M., Hansen T. J., et al. (2013). Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. U.S.A. 110, E99–E107. 10.1073/pnas.1205532110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo-Bazan L., Fernández-Martinez J., de los Ríos V. M., Etxebeste O., Albar J. P., Penalva M. A., et al. (2008). NapA and NapB are the Aspergillus nidulans Nap/SET family members and NapB is a nuclear protein specifically interacting with importin alpha. Fungal Genet. Biol. 45, 278–291. 10.1016/j.fgb.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Arratia-Quijada J., Sánchez O., Scazzocchio C., Aguirre J. (2012). FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 11, 1132–1142. 10.1128/EC.00101-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y., Hagiwara D., Yamashino T., Mizuno T. (2007). Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71, 1800–1803. 10.1271/bbb.70133 [DOI] [PubMed] [Google Scholar]
- Bamford N. C., Snarr B. D., Gravelat F. N., Little D. J., Lee M. J., Zacharias C. A., et al. (2015). Sph3 Is a Glycoside hydrolase required for the biosynthesis of galactosaminogalactan in Aspergillus fumigatus. J. Biol. Chem. 290, 27438–27450. 10.1074/jbc.M115.679050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Ni M., Bok J. W., Helmstaedt K., Valerius O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. 10.1126/science.1155888 [DOI] [PubMed] [Google Scholar]
- Bok J. W., Chiang Y. M., Szewczyk E., Reyes-Dominguez Y., Davidson A. D., Sanchez J. F., et al. (2009). Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 5, 462–464. 10.1038/nchembio.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck V., Quinn J., Soto Pino T., Martin H., Saldanha J., Makino K., et al. (2001). Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 12, 407–419. 10.1091/mbc.12.2.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo I. A., Garcia P., Ayte J., Hidalgo E. (2012). The transcription factors Pap1 and Prr1 collaborate to activate antioxidant, but not drug tolerance, genes in response to H2O2. Nucleic Acids Res. 40, 4816–4824. 10.1093/nar/gks141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Domínguez N., Alvarez-Delfín K., Hansberg W., Aguirre J. (2008). The NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot. Cell 7, 1352–1361. 10.1128/EC.00137-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright G. M., Scott B. (2013). Redox regulation of an AP-1-like transcription factor, YapA, in the fungal symbiont Epichloe festucae. Eukaryot. Cell 12, 1335–1348. 10.1128/EC.00129-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira G. C., Arnaud M. B., Inglis D. O., Skrzypek M. S., Binkley G., Simison M., et al. (2014). The aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 42, D705–D710. 10.1093/nar/gkt1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. R., Wilkinson C. R. M., Watt S., Penkett C. J., Toone W. M., Jones N., et al. (2008). Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell 19, 308–317. 10.1091/mbc.E07-08-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankai W., Pongpom M., Youngchim S., Cooper C. R., Jr., Vanittanakom N. (2016). The yapA encodes bZIP transcription factor involved in stress tolerance in pathogenic fungus Talaromyces marneffei. PLoS ONE 11:e0163778. 10.1371/journal.pone.0163778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G., Russell P. (1997). Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol. Cell. Biol. 17, 3356–3363. 10.1128/MCB.17.6.3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G., Shiozaki K., Russell P. (1996). Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 16, 2870–2877. 10.1128/MCB.16.6.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot M. J. L., van den Dool C., Wösten H. A. B., Levisson M., vanKuyk P. A., Ruijter G. J. G., et al. (2007). Regulation of pentose catabolic pathway genes of Aspergillus niger. Food Technol. Biotechnol. 45, 134–138. [Google Scholar]
- Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481. 10.1016/S0092-8674(02)01048-6 [DOI] [PubMed] [Google Scholar]
- de Nadal E., Ammerer G., Posas F. (2011). Controlling gene expression in response to stress. Nat. Rev. Genet. 12, 833–845. 10.1038/nrg3055 [DOI] [PubMed] [Google Scholar]
- Dirschnabel D. E., Nowrousian M., Cano-Dominguez N., Aguirre J., Teichert I., Kück U. (2014). New insights into the roles of NADPH oxidases in sexual development and ascospore germination in Sordaria macrospora. Genetics 196, 729–744. 10.1534/genetics.113.159368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Molitor F., Hahn M. (2005). Molecular and functional characterization of a fructose specific transporter from the gray mold fungus Botrytis cinerea. Fungal Genet. Biol. 42, 601–610. 10.1016/j.fgb.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Dunwell J. M., Khuri S., Gane P. J. (2000). Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179. 10.1128/MMBR.64.1.153-179.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebeste O., Herrero-García E., Cortese M. S., Garzia A., Oiartzabal-Arano E., de los Ríos V., et al. (2012). GmcA is a putative glucose-methanol-choline oxidoreductase required for the induction of asexual development in Aspergillus nidulans. PLoS ONE 7:292. 10.1371/journal.pone.0040292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger S., Felenbok B. (1996). A newly identified gene cluster in Aspergillus nidulans comprises five novel genes localized in the alc region that are controlled both by the specific transactivator AlcR and the general carbon-catabolite repressor CreA. Mol. Microbiol. 20, 475–488. 10.1046/j.1365-2958.1996.5301061.x [DOI] [PubMed] [Google Scholar]
- Flipphi M., Kocialkowska J., Felenbok B. (2002). Characteristics of physiological inducers of the ethanol utilization (alc) pathway in Aspergillus nidulans. Biochem. J. 364, 25–31. 10.1042/bj3640025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flipphi M., Robellet X., Dequier E., Leschelle X., Felenbok B., Vélot C. (2006). Functional analysis of alcS, a gene of the alc cluster in Aspergillus nidulans. Fungal Genet. Biol. 43, 247–260. 10.1016/j.fgb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Furukawa K., Hoshi Y., Maeda T., Nakajima T., Abe K. (2005). Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56, 1246–1261. 10.1111/j.1365-2958.2005.04605.x [DOI] [PubMed] [Google Scholar]
- Golla U., Bandi G., Tomar R. S. (2015). Molecular cytotoxicity mechanisms of allyl alcohol (acrolein) in budding yeast. Chem. Res. Toxicol. 28, 1246–1264. 10.1021/acs.chemrestox.5b00071 [DOI] [PubMed] [Google Scholar]
- Guo M., Chen Y., Du Y., Dong Y., Guo W., Zhai S., et al. (2011). The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7:e1001302. 10.1371/journal.ppat.1001302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Asano Y., Marui J., Yoshimi A., Mizuno T., Abe K. (2009). Transcriptional profiling for Aspergillusnidulans HogA MAPK signaling pathway in response to fludioxonil and osmotic stress. Fungal Genet. Biol. 46, 868–878. 10.1016/j.fgb.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Hagiwara D., Matsubayashi Y., Marui J., Furukawa K., Yamashino T., Kanamaru K., et al. (2007). Characterization of the NikA histidine kinase implicated in the phosphorelay signal transduction of Aspergillus nidulans, with special reference to fungicide responses. Biosci. Biotechnol. Biochem. 71, 844–847. 10.1271/bbb.70051 [DOI] [PubMed] [Google Scholar]
- Han K. H., Park J. S., Chae K. S., Han D. M. (2010). Simple identification of veA1 mutation in Aspergillus nidulans. J. Microbiol. 48, 885–887. 10.1007/s12275-010-0506-y [DOI] [PubMed] [Google Scholar]
- Han K. H., Prade R. A. (2002). Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43, 1065–1078. 10.1046/j.1365-2958.2002.02774.x [DOI] [PubMed] [Google Scholar]
- Hansberg W., Aguirre J. (1990). Hyperoxidant states cause microbial cell differentiation by cell isolation from dioxygen. J. Theor. Biol. 142, 201–221. 10.1016/S0022-5193(05)80222-X [DOI] [PubMed] [Google Scholar]
- Hill T. W., Käfer E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium stock solution. Fungal Genet. Newsl. 48, 20–21. 10.4148/1941-4765.1173 [DOI] [Google Scholar]
- Huang K., Czymmek K. J., Caplan J. L., Sweigard J. A., Donofrio N. M. (2011). HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7:e1001335. 10.1371/journal.ppat.1001335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer E. (1965). Origins of translocations in Aspergillus nidulans. Genetics 52, 217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki L., Aguirre J. (2001). Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 183, 1434–1440. 10.1128/JB.183.4.1434-1440.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki L., Sánchez O., Shiozaki K., Aguirre J. (2002). SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45, 1153–1163. 10.1046/j.1365-2958.2002.03087.x [DOI] [PubMed] [Google Scholar]
- Kawasaki L., Wysong D., Diamond R., Aguirre J. (1997). Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 179, 3284–3292. 10.1128/jb.179.10.3284-3292.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Han K., Kim K., Han D., Jahng K., Chae K. (2002). The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37, 72–80. 10.1016/S1087-1845(02)00029-4 [DOI] [PubMed] [Google Scholar]
- Koivistoinen O. M., Richard P., Penttilä M., Ruohonen L., Mojzita D. (2012). Sorbitol dehydrogenase of Aspergillus niger, SdhA, is part of the oxido-reductive D-galactose pathway and essential for D-sorbitol catabolism. FEBS Lett. 586, 378–383. 10.1016/j.febslet.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Kowalczyk J. E., Gruben B. S., Battaglia E., Wiebenga A., Majoor E., de Vries R. P. (2015). Genetic Interaction of Aspergillus nidulans galR, xlnR and araR in regulating D-Galactose and L-Arabinose release and catabolism gene expression. PLoS ONE 10:43200. 10.1371/journal.pone.0143200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwolek-Mirek M., Bednarska S., Bartosz G., Bilinski T. (2009). Acrolein toxicity involves oxidative stress caused by glutathione depletion in the yeast Saccharomyces cerevisiae. Cell Biol. Toxicol. 25, 363–378. 10.1007/s10565-008-9090-x [DOI] [PubMed] [Google Scholar]
- Lara-Ortíz T., Riveros-Rosas H., Aguirre J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50, 1241–1255. 10.1046/j.1365-2958.2003.03800.x [DOI] [PubMed] [Google Scholar]
- Lara-Rojas F., Sánchez O., Kawasaki L., Aguirre J. (2011). Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 80, 436–454. 10.1111/j.1365-2958.2011.07581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Godon C., Lagniel G., Spector D., Garin J., Labarre J., et al. (1999). Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040–16046. 10.1074/jbc.274.23.16040 [DOI] [PubMed] [Google Scholar]
- Lessing F., Kniemeyer O., Wozniok I., Loeffler J., Kurzai O., Haertl A., et al. (2007). The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell 6, 2290–2302. 10.1128/EC.00267-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Hadar R., Amedeo P., Baker S. E., Yoder O. C., Horwitz B. A. (2005). Activation of an AP1-like transcription factor of the maize pathogen Cochliobolus heterostrophus in response to oxidative stress and plant signals. Eukaryot. Cell 4, 443–454. 10.1128/EC.4.2.443-454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid M., Soto T., Franco A., Paredes V., Vicente J., Hidalgo E., et al. (2004). A cooperative role for Atf1 and Pap1 in the detoxification of the oxidative stress induced by glucose deprivation in Schizosaccharomyces pombe. J. Biol. Chem. 279, 41594–41602. 10.1074/jbc.M405509200 [DOI] [PubMed] [Google Scholar]
- Malagnac F., Lalucque H., Lepère G., Silar P. (2004). Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41, 982–997. 10.1016/j.fgb.2004.07.008 [DOI] [PubMed] [Google Scholar]
- Millar J. B., Buck V., Wilkinson M. G. (1995). Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 9, 2117–2130. 10.1101/gad.9.17.2117 [DOI] [PubMed] [Google Scholar]
- Molina L., Kahmann R. (2007). An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19, 2293–2309. 10.1105/tpc.107.052332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye-Rowley W. S., Harshman K. D., Parker C. S. (1989). Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3, 283–292. 10.1101/gad.3.3.283 [DOI] [PubMed] [Google Scholar]
- Nasser L., Weissman Z., Pinsky M., Amartely H., Dvir H., Kornitzer D. (2016). Structural basis of haem-iron acquisition by fungal pathogens. Nat. Microbiol. 1:16156. 10.1038/nmicrobiol.2016.156 [DOI] [PubMed] [Google Scholar]
- Navarro R. E., Aguirre J. (1998). Posttranscriptional control mediates cell type-specific localization of catalase a during Aspergillus nidulans development. J. Bacteriol. 180, 5733–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro R. E., Stringer M. A., Hansberg W., Timberlake W. E., Aguirre J. (1996). catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr. Genet. 29, 352–359. 10.1007/s002940050056 [DOI] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., Murray S. L., et al. (2006). A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172, 1557–1566. 10.1534/genetics.105.052563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. N., Lee A., Place W., Shiozaki K. (2000). Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 11, 1169–1181. 10.1091/mbc.11.4.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. N., Shiozaki K. (1999). Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 13, 1653–1663. 10.1101/gad.13.13.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y. T., Ahn C. S., Kim J. G., Ro H. S., Lee C. W., Kim J. W. (2010). Proteomic analysis of early phase of conidia germination in Aspergillus nidulans. Fungal Genet. Biol. 47, 246–253. 10.1016/j.fgb.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Qiao J., Kontoyiannis D. P., Calderone R., Li D., Ma Y., Wan Z., et al. (2008). Afyap1, encoding a bZip transcriptional factor of Aspergillus fumigatus, contributes to oxidative stress response but is not essential to the virulence of this pathogen in mice immunosuppressed by cyclophosphamide and triamcinolone. Med. Mycol. 46, 773–782. 10.1080/13693780802054215 [DOI] [PubMed] [Google Scholar]
- Quinn J., Malakasi P., Smith D. A., Cheetham J., Buck V., Millar J. B., et al. (2011). Two-component mediated peroxide sensing and signal transduction in fission yeast. Antioxid. Redox Signal. 15, 153–165. 10.1089/ars.2010.3345 [DOI] [PubMed] [Google Scholar]
- Raulo R., Kokolski M., Archer D. B. (2016). The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. Amb Express 6:5. 10.1186/s13568-016-0177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi M., Zjalic S., Ricelli A., Punelli F., Camera E., Fabbri C., et al. (2008). Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: a role for the ApyapA gene. Eukaryot. Cell 7, 988–1000. 10.1128/EC.00228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. G. (2016). Overview on Peroxiredoxin. Mol. Cells 39, 1–5. 10.14348/molcells.2016.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J. F., Entwistle R., Hung J. H., Yaegashi J., Jain S., Chiang Y. M., et al. (2011). Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J. Am. Chem. Soc. 133, 4010–4017. 10.1021/ja1096682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez O., Aguirre J. (1996). Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet. Newslett. 43, 48–51. 10.4148/1941-4765.1317 [DOI] [Google Scholar]
- Sánchez O., Navarro R. E., Aguirre J. (1998). Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 258, 89–94. 10.1007/s004380050710 [DOI] [PubMed] [Google Scholar]
- Scherer M., Wei H. J., Liese R., Fischer R. (2002). Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell 1, 725–735. 10.1128/EC.1.5.725-735.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth B., Metz B. (2011). Fungal arabinan and L-arabinose metabolism. Appl. Microbiol. Biotechnol. 89, 1665–1673. 10.1007/s00253-010-3071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. (1995). Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 378, 739–743. 10.1038/378739a0 [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. (1996). Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10, 2276–2288. 10.1101/gad.10.18.2276 [DOI] [PubMed] [Google Scholar]
- Shroff R. A., Lockington R. A., Kelly J. M. (1996). Analysis of mutations in the creA gene involved in carbon catabolite repression in Aspergillus nidulans. Can. J. Microbiol. 42, 950–959. 10.1139/m96-122 [DOI] [PubMed] [Google Scholar]
- Siegmund U., Heller J., van Kan J. A., Tudzynski P. (2013). The NADPH oxidase complexes in Botrytis cinerea: evidence for a close association with the ER and the tetraspanin Pls1. PLoS ONE 8:e55879 10.1371/annotation/84c258cf-4f98-43d8-a4f0-7032cba36f22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana T., Okazaki S., Murayama A., Naganuma A., Nomoto A., Kuge S. (2009). A major peroxiredoxin-induced activation of yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J. Biol. Chem. 284, 4464–4472. 10.1074/jbc.M807583200 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Yamashita K., Shiozawa A., Ichiishi A., Fukumori F., Fujimura M. (2010). An AP-1-like transcription factor, NAP-1, regulates expression of the glutathione S-transferase and NADH:flavin oxidoreductase genes in Neurospora crassa. Biosci. Biotechnol. Biochem. 74, 746–752. 10.1271/bbb.90790 [DOI] [PubMed] [Google Scholar]
- Temme N., Tudzynski P. (2009). Does botrytis cinerea Ignore H2O2-induced oxidative stress during infection? Characterization of botrytis activator protein 1. Mol. Plant Microbe Interact. 22, 987–998. 10.1094/MPMI-22-8-0987 [DOI] [PubMed] [Google Scholar]
- Thön M., Al Abdallah Q., Hortschansky P., Scharf D. H., Eisendle M., Haas H., et al. (2010). The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 38, 1098–1113. 10.1093/nar/gkp1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Li J., Glass N. L. (2011). Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 157, 747–759. 10.1099/mic.0.045468-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Shimanuki M., Yanagida M. (1991). Fission yeast genes that confer resistance to staurosporine encode an Ap-1-like transcription factor and a protein-kinase related to the mammalian Erk1/Map2 and budding yeast fus3 and Kss1 Kinases. Genes Dev. 5, 60–73. 10.1101/gad.5.1.60 [DOI] [PubMed] [Google Scholar]
- Vargas-Perez I., Sanchez O., Kawasaki L., Georgellis D., Aguirre J. (2007). Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 6, 1570–1583. 10.1128/EC.00085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos A. P., Castillo E. A., Biteau B., Nicot C., Ayté J., Toledano M. B., et al. (2005). A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 8875–8880. 10.1073/pnas.0503251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos A. P., Castillo E. A., Jones N., Ayté J., Hidalgo E. (2004). Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 52, 1427–1435. 10.1111/j.1365-2958.2004.04065.x [DOI] [PubMed] [Google Scholar]
- Wiatrowski H. A., Carlson M. (2003). Yap1 accumulates in the nucleus in response to carbon stress in Saccharomyces cerevisiae. Eukaryot. Cell 2, 19–26. 10.1128/EC.2.1.19-26.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo E. J., Dunwell J. M., Goodenough P. W., Marvier A. C., Pickersgill R. W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7, 1036–1040. 10.1038/80954 [DOI] [PubMed] [Google Scholar]
- Yin W. B., Reinke A. W., Szilágyi M., Emri T., Chiang Y. M., Keating A. E., et al. (2013). bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 159, 77–88. 10.1099/mic.0.063370-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Hamari Z., Han K. H., Seo J. A., Reyes-Dominguez Y., Scazzocchio C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.