Abstract
The (neo-) lacto series glycosphingolipids (nsGSLs) comprise of glycan epitopes that are present as blood group antigens, act as primary receptors for human pathogens and are also increasingly associated with malignant diseases. Beta-1, 3-N-acetyl-glucosaminyl-transferase 5 (B3GNT5) is suggested as the key glycosyltransferase for the biosynthesis of nsGSLs. In this study, we investigated the impact of CRISPR-Cas9 -mediated gene disruption of B3GNT5 (∆B3GNT5) on the expression of glycosphingolipids and N-glycoproteins by utilizing immunostaining and glycomics-based PGC-UHPLC-ESI-QTOF-MS/MS profiling. ∆B3GNT5 cells lost nsGSL expression coinciding with reduction of α2-6 sialylation on N-glycoproteins. In contrast, disruption of B4GALNT1, a glycosyltransferase for ganglio series GSLs did not affect α2-6 sialylation on N-glycoproteins. We further profiled all known α2-6 sialyltransferase-encoding genes and showed that the loss of α2-6 sialylation is due to silencing of ST6GAL1 expression in ∆B3GNT5 cells. These results demonstrate that nsGSLs are part of a complex network affecting N-glycosylation in ovarian cancer cells.
Glycosphingolipids (GSLs) have been shown to be essential in a wide variety of biological events - such as cell signalling, modification of insulin and EGF-receptor activities, and modulation of Notch ligand activity in Drosophila1. They also interact with well-known proteins such as EGFR2, TGFβ1R3, and VEGFR4 in various malignancies. GSLs are usually divided into two major families, known as galactosylated or glucosylated ceramides. The latter (glucosylceramide-related glycosphingolipids) is further divided into three major classes based on the action of specific glycosyltransferases; globo- (A4GALT), ganglio- (B4GALNT1 and ST3GAL5), and (neo-) lacto (B3GNT5) -series. Additional elongation of lipid-linked carbohydrate chains is determined by the intracellular localization, which is usually embedded in the endomembrane system and the regulation of specific glycosyltransferases within the GSL-glycan biosynthetic pathway5.
The B3GNT5 gene encodes the glycosyltransferase β-1,3-N-acetylglucosaminyl transferase 5, which attaches N-acetylglucosamine (GlcNAc) to lactosylceramide (Galβ1-4Glcβ1-1Ceramide) resulting in the precursor lactotriaosylceramide (Lc3, GlcNAcβ1-3Galβ1-4Glcβ1-1Ceramide) for synthesis of lacto (Type 1) and neolacto-series (Type 2) GSLs6 (Fig. 1A). This enzyme, together with its associated glycosidic product (Lc3), plays a role in human malignant diseases, embryonic development and cell differentiation. Specifically, Lc3 was shown to be elevated on the cell surface of human pro-myelocytic leukemia HL60 cells6,7. Moreover, it has also been suggested as a differentiation-associated GSL in the bone marrow of acute myeloid leukemia patients with corresponding elevated B3GNT5 expression8. In mice experiments, the current data on the potential function of B3GNT5 seems rather controversial, possibly due to the use of a multicellular organism and different mutation-generating techniques, thereby limiting biological interpretations regarding its role9,10,11. Other than Lc3, it is also known that B3GNT5 is the key enzyme for several GSL structures associated with human diseases such as sialyl-Lewis x12 and human blood groups ABO and P [based on the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/)]. Nevertheless, the biological function of B3GNT5-mediated GSLs is rather limited in the context of cancer and remains to be explained.
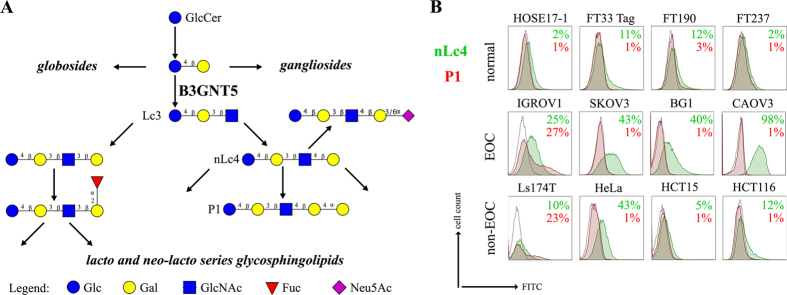
Figure 1. The heterogeneous expression of (neo-) lacto series glycosphingolipids on normal and cancer cell lines.
(A) Depiction of the three major glycosphingolipid series - globo, ganglio, and (neo-) lacto series glycosphingolipids. B3GNT5 attaches GlcNAc to the LacCer synthesizing Lc3, the precursor of all nsGSLs. Glycosidic linkages are displayed next to CFG notated monosaccharides. (B) Representative histograms of flow cytometry data on normal and cancer cell lines stained for paragloboside (nLc4, green) and P1 (red). Grey histogram depicts the negative control. Values within each plot show the mean out of three independent experiments for nLc4 (green) and P1 (red).
In this study, we successfully created a site-specific and heritable B3GNT5 knockout in human cancer cell lines. By utilizing the CRISPR Cas9 technology, we established an experimental tool for studying the function of B3GNT5-mediated GSLs, namely the entire (neo-) lacto series (nsGSL). In addition, we also performed a glycomics profiling using mass spectrometry to evaluate the effects of this GSL gene knockout on the entire glycome repertoire of membrane proteins and lipids. The specific glycan alterations described in this study are consistent in two ovarian cancer cell lines and seem to be specific for B3GNT5. It is envisioned that this gene-editing technology will serve as a useful platform to facilitate the downstream investigation of B3GNT5 and its regulation of both GSL and protein glycosylation in cancer development and progression.
Results
(Neo-) lacto- series glycosphingolipids are expressed on cancer cells
As part of our initiative to comprehensively characterize nsGSLs, we have recently reported the presence of paragloboside (nLc4, precursor of P1) and P1 pentasaccharide in tumor specimens and immortal ovarian cancer cells using two complementary methods; PGC-LC-ESI-MS/MS and flow cytometry13,14,15. In this study, we extended the profiling of nsGSLs into three distinct groups; Normal (HOSE17-1, FT33-Tag, FT190 and FT237 which were suggested as a potential origin of epithelial ovarian cancer16,17), Ovarian (IGROV1, SKOV3, BG1, and CAOV3), and Non-ovarian cancer cell lines (Ls174T, HeLa, HCT15, and HCT116). The flow cytometry data revealed a generally lower expression of nsGSLs in normal cells (nLc4 2–12% and P1 1–3%), while all four of the ovarian cancer cell lines displayed elevated expression for nLc4 (25–98%). A distinct expression of nLc4 (5–43%) was observed in non–ovarian derived cancer cells (Fig. 1B). P1 expression was observed only in IGROV1 (27%) and Ls174T (23%) cell lines (Fig. 1B). Based on their nsGSL expression levels, IGROV1 was selected for genome editing to establish a heritable and site-specific B3GNT5 knockout cell line (∆B3GNT5), which was then selected to study the influence of nsGSLs on the glycome repertoire.
Genome editing of B3GNT5 for depletion of nsGSLs and validation using flow cytometry
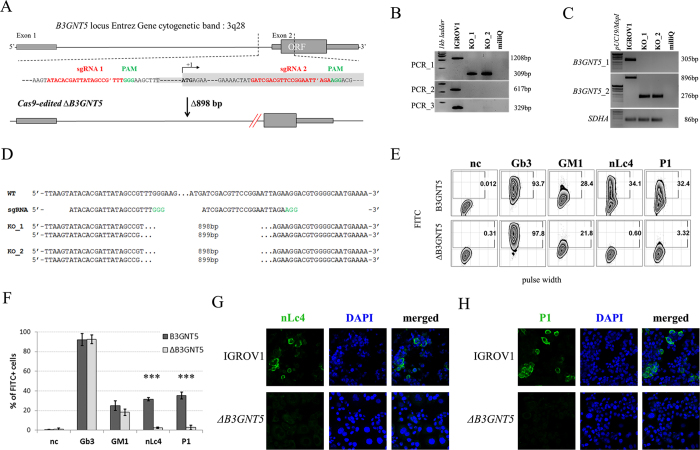
B3GNT5 is the key glycosyltransferase involved in synthesis of nsGSLs6,18. We utilized the genome editing technology, CRISPR-Cas9, to homozygously delete an 898 bp genomic region, including the translation start site of B3GNT5 (Fig. 2A). The plasmid pSpCas9(BB)-2A-GFP encoding two specific sgRNA sequences (Supplemental Table S1), was transiently transfected into IGROV1 and tested after 72 h incubation for Cas9 activity, in which an additional band at 309 bp (∆B3GNT5) observed below 1208 bp (wildtype B3GNT5) indicates an active genome editing. Next, Cas9-active cell pools were subjected to single cell sorting (Supplemental Fig. S1) and incubated until further genotyping of single cell clones. The presence of homozygous ∆B3GNT5 was verified by three independent PCRs, which showed the additional band at 309 bp in knockout (PCR_1) and no visible band at 617 bp and 329 bp, respectively for wildtype B3GNT5 (PCR_2 and PCR_3) (Fig. 2B). We identified two homozygous B3GNT5-deleted clones from a total of 320 clones profiled (0.625% efficiency). Despite equal mRNA expression of B3GNT5 transcripts (B3GNT5_1), a truncated transcript length was equally expressed in ∆B3GNT5 cells as compared to wildtype (B3GNT5_2, Fig. 2C). The homozygous deletion was confirmed by Sanger DNA sequencing, showing knockout cells with alleles in varying lengths; (indel) 898 bp and 899 bp (Fig. 2D). In regards to potential off-target effects, both sgRNAs used for genome- editing did not show off-target effects (Supplemental Fig. S2).
Figure 2. Deletion of B3GNT5 leads to depletion of nLc4 and P1.
(A) B3GNT5-editing strategy using two different sgRNAs (red) contain PAM sequence (green) deleting a genomic region up- and downstream of the transcription start site (+1) including a part of the open reading frame (ORF). In silico analysis revealed a deletion of 898 bp. (B) Representative genotyping PCR for characterization of single cell clones. PCR numbers indicate the use of different primer pairs listed in experimental procedures. (C) Semi-quantitative PCR with two different primer pairs (B3GNT5_1 and B3GNT5_2) on wildtype (pIGROV1) and two representative knockout clones. SDHA was used as a reference gene. (D) DNA sequencing results for wildtype (WT), and selected biallelically deleted knockout clones (KO_1 and KO_2). (E) Representative counter plots for presence of Gb3, GM1, nLc4 and P1 before (wildtype B3GNT5) and after gene disruption of B3GNT5 in IGROV1 cells (∆B3GNT5). Percentage of GSL-positive cells (FITC) is shown in each plot. Negative control (n.c.). (F) Box and Whisker plots representing CRISPR Cas9-mediated changes in expression of individual GSLs (percentage of FITC-positive cells, ordinate) for ∆B3GNT5 and parental cells (abscissa). ***p < 0.001 was obtained by two-way ANOVA. Data are representative out of four independent experiments. (G,H) Confocal fluorescence images displaying presence and absence of nLc4 (green, G, 40x magnification) and P1 (green, H) in B3GNT5 wildtype (B3GNT5) and knockout (∆B3GNT5) cells, respectively. Cells are counterstained with DAPI (blue). Data are represented as mean ± s.d.
The established ∆B3GNT5 cell line was further investigated for GSL expression using flow cytometry, in which the human anti-P1 IgM P3NIL100 antibody, previously validated by printed glycan array, was utilized to detect the binding specificities to P1 epitope on these cells (Supplemental Fig. S3)14. The GSL pathways affected by the genetic disruption of B3GNT5 is hypothesized (according to the scheme presented in Fig. 1A) and the binding results were in full concordance with the expression levels for nLc4 and P1 (p < 0.001, Fig. 2E and F). In conclusion, the absence of nLc4 and P1 epitopes confirmed our hypothesis that the nsGSLs were depleted in these cells, whereas ganglio- and globo- series GSLs were not affected by B3GNT5-editing (Fig. 2E and F). In addition, the absence of nLc4 and P1 in ∆B3GNT5 cells was confirmed by confocal fluorescence microscopy (Fig. 2G and H).
Validation of altered GSL-glycans in ∆B3GNT5 cells by PGC-UHPLC-ESI-QTOF-MS/MS
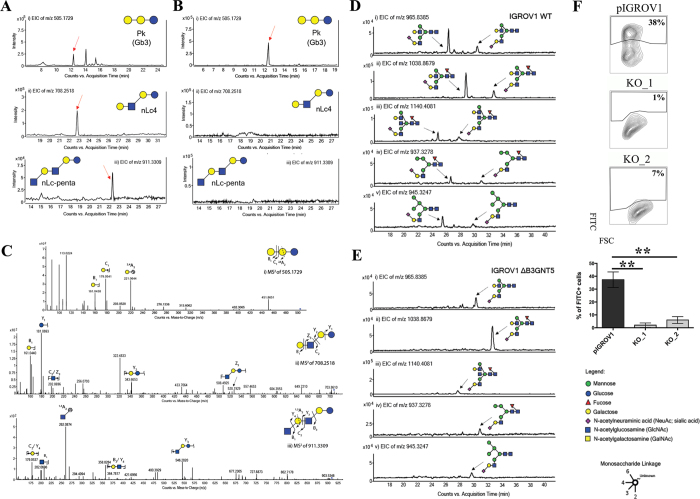
We also utilized mass spectrometry (MS) to investigate the expression of cell surface glycans, which were subsequently altered due to the genome- editing of B3GNT5 in IGROV1 cells. This glycomics-based approach was performed to confirm the absence of nsGSLs due to genetic disruption of B3GNT5. Glycans were enzymatically released from extracted GSLs of parental and ∆B3GNT5 IGROV1 cells and analyzed using negative mode UHPLC-ESI-QTOF-MS/MS. The assignment of glycan structures was facilitated using diagnostic MS2 fragment ions previously described for the analysis of N-glycans released from glycoproteins as well as GSL-derived glycans using negative mode LC-ESI-MS/MS13,19.
The MS profiling revealed three neutral GSLs comprising of globo (Gb3)-, paragloboside (nLc4) as well as neolactopentaosylceramide (nLc-penta). All three neutral GSL species differed in chromatographic retention time, as well as MS2 fragmentation patterns facilitating the structural assignment of the GSL-glycans based on the fragment ions arising from various glycosidic and cross-ring cleavages. Table 1 shows the list of GSL-glycans and their relative intensities that were observed in both cell lines. The globo series glycosphingolipid, Gb3, was detected as [M-H]1− m/z 505.17291− in wildtype and ∆B3GNT5 cells and was shown to elute at 12.16 min (Fig. 3A(i) and B(i)). The representative MS2 spectra of the precursor ion at m/z 505.17291− (Fig. 3C(i)) showed prominent glycosidic- type fragment ions (B1 at m/z 161.04381−, C1 at m/z 179.05411−) corresponding to the Gal-Gal-Glc trisaccharide sequence. The presence of the 4-linked terminal Gal to the inner (Gal-Glc) disaccharide was further confirmed by the characteristic cross ring fragment ion corresponding to 2,4A2 at m/z 221.06441−. The second glycan structure, paragloboside (nLc4), detected as [M-H]1− m/z 708.25181− was found to be present only in the wildtype IGROV1 cell line but not in the ∆B3GNT5 cell line. This glycan was shown to elute at 22.60 mins (Fig. 3A(ii)) and the MS2 spectra comprised of a mixture of B and Y ions (B1 at m/z 161.04401−, Y1 at m/z 181.06931−, Y2 at m/z 343.66531−) as well as C and Z ions (Z3 at m/z 528.19291−, C2/Z3 at m/z 202.06961−) that corresponded to the tetrasaccharide sequence, Hex-HexNAc-Gal-Glc (Fig. 3C(ii)). The absence of the cross ring cleavage ions characteristic of 4-linked GalNAc corresponding to 0,2A3 and 2,4A3 at m/z 484.16721− and m/z 424.14601−, respectively, further confirmed the presence of the 3-linked GlcNAc residue to the inner Gal-Glc of the tetrasaccharide. The late elution time observed for this glycan structure has been reported in our previous study demonstrating a similar retention time for nLc4 identified from IGROV1 cell lines using PGC-LC-ESI-IT-MS/MS (20). We also identified a third glycan structure at [M-H]1− m/z 911.33091− in wildtype IGROV1 which was shown to elute at 22.10 min (Fig. 3A(iii) and B(iii)). The MS2 spectra comprised of mainly B-, C- and Y-type fragment ions (B1 at m/z 202.06961−, C2/Y4 at m/z 179.05371−, B3/Y4 at m/z 364.78171− and Y3 at m/z 546.20201−) that corresponded to the tentative pentasaccharide sequence, HexNAc-Gal-HexNAc-Gal-Glc corresponding to a poly-LacNAc-type pentaosylceramide (Fig. 3C(iii)). The presence of the 1,3A2 at m/z 262.08741− further confirmed the terminal GlcNAc linked to the internal tetrasaccharide. However, this structure was not observed in the MS spectra of the ∆B3GNT5 cell line. Whilst we were able to confirm the absence of nLc4 in the ∆B3GNT5 cell line, the presence of the P1 epitope was not readily detected in both the IGROV1 and ∆B3GNT5 cells analyzed in this study. This could be due to the low expression of the P1 epitope, coupled with the use of different MS instrumentation in this study (as compared to our previous study using PGC-LC-ESI-IT-MS/MS that was performed on P1 – enriched cell populations of IGROV113). Moreover, this study used a larger PGC column inner diameter on a conventional LC platform performed which resulted in less sensitivity as opposed to the microflow LC platform performed previously using PGC-ESI-IT-MS/MS. Apart from the neutral GSLs, two gangliosides were also detected in the MS spectra of both the wildtype and ∆B3GNT5 cells. Both GM3 and GM2 were detected as [M-H]1− m/z 634.21421− and [M-H]1− m/z 837.29261−, respectively, while the third ganglioside, GM1, at [M-H]1− m/z 999.35481−, did not appear to be present in both the cell lines (Table 1). These results indicate that gangliosides remained unaffected by the genome-editing of B3GNT5.
Table 1. Proposed GSL-glycan structures detected on the glycolipid membranes of ovarian cancer cells, IGROV1 (wildtype) and ∆B3GNT5 cells.
| Type | No | Glycan Mass [M-H]- | Glycan Structures | PGC-ESI-QTOF-MS/MS |
PGC-ESI-IT-MS/MS | |
|---|---|---|---|---|---|---|
| IGROV1 | B3GNT5-edited IGROV1 | P1-enriched IGROV1 (Anugraham M, et al, 2015) | ||||
| Neutral GSL | 1 | 505.2 (Pk) |

|
6.34 ± 0.31 | 4.72 ± 2.38 | 0.00 |
| 2 | 546.3 |
|
0.00 | 0.00 | 0.00 | |
| 3a | 708.3 (Globo) |
|
0.00 | 0.00 | 0.00 | |
| 3b | 708.3 (Asialo-GM1) |
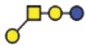
|
0.00 | 0.00 | 0.00 | |
| 3c | 708.3 (Neo-lacto) |
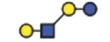
|
4.52 ± 1.58 | 0.00 | 7.64 | |
| 4 | 870.3 (P1) |
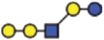
|
0.00 | 0.00 | 2.43 | |
| 5 | 911.3 |
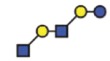
|
1.25 ± 0.48 | 0.00 | Trace | |
| Sialylated GSL | 6 | 634.2 (GM3) |
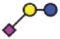
|
14.18 ± 0.61 | 23.40 ± 4.32 | 12.86 |
| 7 | 837.3 (GM2) |
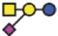
|
73.71 ± 2.40 | 71.88 ± 8.13 | 73.11 | |
| 8a | 999.3 (GM1) |
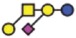
|
0.00 | 0.00 | 0.68 | |
| 8b | 999.3 (α2-3 sialyl Paragloboside) |
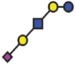
|
0.00 | 0.00 | 2.15 | |
| 8c | 999.3 (LSTc) |
|
0.00 | 0.00 | 1.14 | |
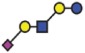
Structures were assigned based on MS/MS fragmentation (where possible) and known biological GSL synthetic pathway constraints. All structures were depicted according to the CFG (Consortium of Functional Glycomics) notation with linkage placement. Specific linkages corresponding to Gal-GlcNAc (Type 1/Type 2) lactosamine linkages are also indicated (where possible). Values represent mean relative ion intensities ± s.d. of three replicates based on their extracted ion chromatograms (EIC). GSL-glycan structures derived from P1-enriched IGROV1 cell line previously analyzed with PGC-ESI-IT-MS (20) are also shown for comparative profiling using two different MS platforms. Gb3 (Pk).
 Glucose
Glucose  Galactose
Galactose  N-acetylglucosamine (GlcNAc)
N-acetylglucosamine (GlcNAc)  N-acetylgalactosamine (GalNAc)
N-acetylgalactosamine (GalNAc)  N-acetylneruaminic acid (NeuAc)
N-acetylneruaminic acid (NeuAc)
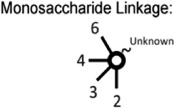
Figure 3. LC-MS profiling of neutral GSLs and monosialylated N-glycans extracted from wildtype IGROV1 and genome-edited B3GNT5 cell lines.
The extracted ion chromatograms (EIC) obtained from wildtype IGROV1 (A) and genome-edited B3GNT5 (B) cell lines are represented for Gb3 [m/z 505.17291−Galα1-4Galβ1-4Glcβ1] (i), nLc4 [m/z 708.25181−:Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ1] (ii) and nLc-pentasaccharide [m/z 911.33091−:GlcNAcβ1-3-Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ1] (iii). (C) MS2 spectrum of the precursor ion at m/z 505.17291−, m/z 708.25181− and m/z 911.33091− derived from Gb3 (i), nLc4 (ii) and nLc-penta (iii), respectively. PGC- LC allows for the separation of α2-6 and α2-3 sialylated N-glycans based on retention time. (D) The EICs obtained from the wildtype IGROV 1 depict three major monosialylated complex N-glycans at m/z 965.83852− [(Neu5Ac)1(Gal)2(GlcNAc)2+ (Man)3(GlcNAc)2] (i), m/z 1038.86792− [(Neu5Ac)1 (Gal)2(GlcNAc)2(Fuc)1+ (Man)3(GlcNAc)2] (ii) and m/z 1140.40812− [(Neu5Ac)1(Gal)2 (GlcNAc)3(Fuc)1 + (Man)3(GlcNAc)2] (iii) and two monosialylated hybrid N-glycans at m/z 937.32782− [(Neu5Ac)1(Gal)1(GlcNAc)1(Man)1 + (Man)3(GlcNAc)2] (iv) and m/z 945.32472− [(Neu5Ac)1(Gal)1(GlcNAc)1(Man)2 + (Man)3(GlcNAc)2] (v) which display α2–3 and α2–6 sialylated isomers at separate retention times. (E) The EICs of genome-edited B3GNT5 cells depicts the loss of the α2–6 sialylated isomer for all five of the above mentioned monosialylated complex [m/z 965.83852− (i), m/z 1038.86792−(ii), m/z 1140.40812− (iii)] and hybrid [m/z 937.32782− (iv), m/z 945.32472− (v)] N-glycans. (F) SNA staining confirmed reduction of α2–6 sialylation in ∆B3GNT5 cells. Representative counter plot and barograph summarizing three independent experiments on parental IGROV1 cells (pIGROV1) and ∆B3GNT5 cells (KO_1 and KO_2); **p-value < 0.01. Data are represented as mean ± s.d.
Isomeric N-glycan profiling reveals concomitant loss of α 2-6 sialylation on membrane proteins in B3GNT5-mediated depletion of nsGSL
In addition to the profiling of the GSLs, we also compared the N-glycan profiles of the wildtype and ∆B3GNT5 cell lines to gain an insight into the glycan structural changes, which could potentially be affected as a result of the GSL-related B3GNT5 genome- editing. We have previously reported an increased relative abundance of α2-6 sialylation as compared to α2-3 sialylation on membrane N-glycans of IGROV119. The presence of these isomeric structures in N-glycans can be differentiated based on their retention time on the porous graphitized carbon (PGC)-LC column, whereby glycans bearing the α2-6 sialic acid isomer are less strongly retained on the column and thus elute much earlier as compared to the α2-3 sialylated N-glycans. Likewise, in this study, a similar glycan elution pattern was observed in the wildtype IGROV1 cell lines confirming our previous results (which used another mass spectrometry setup), in which all five N-glycans comprising of the sialylated complex (m/z 965.83852−, m/z 1038.86792−, m/z 1140.40812−) (Fig. 3D(i–iii)) and hybrid (m/z 937.32782− and m/z 945.32472−) (Fig. 3D(iv–v)) glycans were found to contain α2-6 and α2-3 isomeric structures which eluted at separate retention times. Surprisingly, we noted that in comparison to the wildtype IGROV1, the genome- edited ∆B3GNT5 cells were shown to display only the α2-3 sialylated glycans but not the α2-6 sialylated N-glycan isomer for all five of the N-glycan masses described above (Fig. 3E(i–v)). We also profiled the membrane protein O-glycans of both cell lines to see if the difference in the sialylation pattern could be attributed to a global glycosylation change across all glycan types. However, we observed no change in the O-glycan sialylation profiles, further demonstrating that the preferential α2-6 sialylation was specific to only the N-glycosylation pathway, as a result of the B3GNT5 genome- editing. The N- and O-glycans identified in both cell lines and their corresponding relative intensities are shown in Table 2.
Table 2. Proposed N- and O-glycan structures detected on the membrane proteins of IGROV1 (wildtype) and ∆B3GNT5 cells.
| Type | No | Glycan Mass [M-H]− | [M-2H]2− | Glycan Structures | PGC-ESI-QTOF-MS/MS |
PGC-ESI-IT-MS/MS | |
|---|---|---|---|---|---|---|---|
| IGROV1 | B3GNT5-edited IGROV1 | IGROV1 (Anugraham M, et al, 2014) | |||||
| N -glycans -High Mannose | 1 | 1235.4 | 617.2 |
|
4.15 ± 0.11 | 5.54 ± 0.16 | 0.62 ± 0.06 |
| 2 | 1397.6 | 698.3 |
|
5.31 ± 0.24 | 4.88 ± 0.64 | 7.91 ± 0.15 | |
| 3 | 1559.6 | 779.3 |
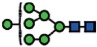
|
9.13 ± 0.27 | 9.48 ± 1.21 | 13.45 ± 1.35 | |
| 4 | 1721.6 | 860.3 |
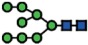
|
19.19 ± 1.15 | 20.85 ± 1.57 | 16.00 ± 3.73 | |
| 5 | 1883.8 | 941.4 |
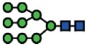
|
23.48 ± 2.02 | 22.29 ± 0.79 | 24.57 ± 6.02 | |
| 6 | 2045.6 | 1022.3 |
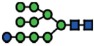
|
1.88 ± 0.21 | 1.73 ± 0.52 | 1.43 ± 0.61 | |
| N -glycans -Hybrid | 7 | 1567.6 | 783.3 |
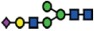
|
0.00 | 0.00 | 0.19 ± 0.03 |
| 8 | 1584.6 | 791.8 |
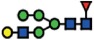
|
0.00 | 0.00 | 0.34 ± 0.11 | |
| 9 | 1600.6 | 799.8 |
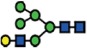
|
0.63 ± 0.01 | 0.83 ± 0.12 | 1.04 ± 0.30 | |
| 10 | 1713.6 | 856.3 |
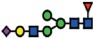
|
0.00 | 0.00 | 1.76 ± 0.21 | |
| 11 | 1729.6 | 864.3 |
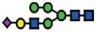
|
0.00 | 0.00 | 0.68 ± 0.20 | |
| 12 | 1746.6 | 872.8 |
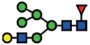
|
0.00 | 0.00 | 0.20 ± 0.00 | |
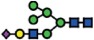
| 13a | 1875.6 | 937.3 (α2-6) |
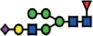
|
0.65 ± 0.03 | 0.00 | 0.20 ± 0.25 | |
| 13b | 1875.6 | 937.3 (α2-3) |
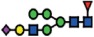
|
0.42 ± 0.05 | 0.74 ± 0.07 | 0.41 ± 0.20 | |
| 14a | 1891.6 | 945.3 (α2-6) |
|
1.34 ± 0.09 | 0.00 | 0.47 ± 0.22 | |
| 14b | 1891.6 | 945.3 (α2-3) |
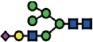
|
0.67 ± 0.05 | 1.33 ± 0.17 | 0.43 ± 0.08 | |
| N -glycans -Complex Neutral Hybrid | 15 | 1463.6 | 731.2 |
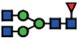
|
1.25 ± 0.20 | 1.36 ± 0.35 | 0.74 ± 0.03 |
| 16 | 1625.6 | 812.3 |
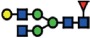
|
0.00 | 0.14 ± 0.25 | 0.64 ± 0.14 | |
| 17 | 1641.6 | 820.3 |
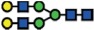
|
0.62 ± 0.01 | 1.16 ± 0.18 | 1.59 ± 0.62 | |
| 18 | 1666.4 | 832.8 |
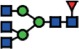
|
0.00 | 0.00 | 1.22 ± 0.40 | |
| 19 | 1682.6 | 840.8 |
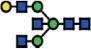
|
0.00 | 0.00 | 0.12 ± 0.05 | |
| 20 | 1771.8 | 885.4 |
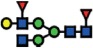
|
0.00 | 0.00 | 0.00 | |
| 21 | 1787.6 | 893.3 |
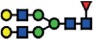
|
1.99 ± 0.05 | 3.48 ± 0.47 | 3.50 ± 0.22 | |
| 22 | 1812.8 | 905.9 |
|
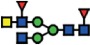
0.00 | 0.00 | 0.24 ± 0.08 | |
| 23 | 1828.8 | 913.9 |
|
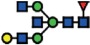
0.00 | 1.30 ± 0.34 | 0.66 ± 0.12 | |
| 24 | 1844.8 | 921.9 |
|
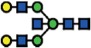
1.25 ± 0.06 | 2.01 ± 0.09 | 1.37 ± 0.19 | |
| 25 | 1869.8 | 934.4 |
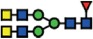
|
0.00 | 0.19 ± 0.33 | 0.51 ± 0.20 | |
| 26 | 1933.6 | 966.3 |
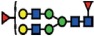
|
1.23 ± 0.78 | 1.15 ± 0.13 | 0.00 | |
| 27 | 1974.8 | 986.9 |
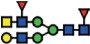
|
0.18 ± 0.31 | 0.00 | 0.79 ± 0.28 | |
| 28 | 1990.8 | 994.9 |
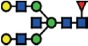
|
6.29 ± 1.36 | 8.23 ± 0.73 | 2.62 ± 0.60 | |
| 29 | 2006.8 | 1002.9 |
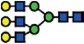
|
1.84 ± 0.39 | 1.75 ± 0.61 | 0.31 ± 0.15 | |
| 30 | 2015.8 | 1007.4 |
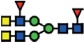
|
0.00 | 0.00 | 0.14 ± 0.06 | |
| 31 | 2079.8 | 1039.4 |
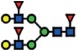
|
0.00 | 0.00 | 0.00 | |
| 32 | 2120.8 | 1059.9 |
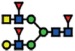
|
0.00 | 0.00 | 0.56 ± 0.19 | |
| 33 | 2136.6 | 1067.8 |
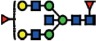
|
2.32 ± 0.27 | 3.44 ± 0.35 | 0.31 ± 0.15 | |
| 34 | 2152.8 | 1075.9 |
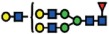
|
0.77 ± 0.40 | 1.47 ± 0.95 | 1.18 ± 0.16 | |
| 35 | 2162 | 1080.5 |
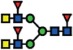
|
1.22 ± 0.22 | 0.25 ± 0.15 | 0.39 ± 0.09 | |
| 36 | 2356 | 1177.5 |
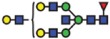
|
0.84 ± 0.07 | 1.13 ± 0.14 | 0.23 ± 0.02 | |
| 37 | 2518 | 1258.5 |
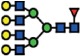
|
1.88 ± 0.24 | 1.20 ± 0.22 | 0.37 ± 0.10 | |
| N -glycans -Complex Sialylated Hybrid Complex Sialylated | 38 | 1916.6 | 957.8 |
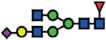
|
0.00 | 0.00 | 0.21 ± 0.06 |
| 39a | 1932.8 | 965.9 (α2-6) |
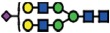
|
1.41 ± 0.11 | 0.00 | 1.38 ± 0.38 | |
| 39b | 1932.8 | 965.9 (α2-3) |
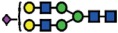
|
0.86 ± 0.05 | 1.28 + 0.24 | 1.86 ± 0.49 | |
| 40a | 2078.8 | 1038.9 (α2-6) |
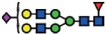
|
4.96 ± 0.35 | 0.00 | 1.91 ± 0.23 | |
| 40b | 2078.8 | 1038.9 (α2-3) |
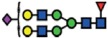
|
1.68 ± 0.16 | 3.94 ± 0.27 | 3.23 ± 0.14 | |
| 41 | 2119.8 | 1059.4 |
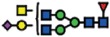
|
0.00 | 0.00 | 0.36 ± 0.15 | |
| 42 | 2223.8 | 1111.4 |
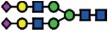
|
0.00 | 0.00 | 0.41 ± 0.07 | |
| 43 | 2265.8 | 1132.4 |
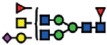
|
0.00 | 0.00 | 0.06 ± 0.02 | |
| 44a | 2282.0 | 1140.5 (α2-6) |
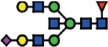
|
1.35 ± 0.11 | 0.00 | 0.35 ± 0.14 | |
| 44b | 2282.0 | 1140.5 (α2-3) |
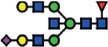
|
1.20 ± 0.34 | 2.30 ± 0.06 | 0.30 ± 0.17 | |
| 45 | 2297.8 | 1148.4 |
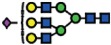
|
0.00 | 0.00 | 0.23 + 0.02 | |
| 46 | 2370.0 | 1184.5 |
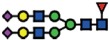
|
0.00 | 0.00 | 1.33 ± 0.36 | |
| 47 | 2411.0 | 1205 |
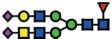
|
0.00 | 0.00 | 0.06 ± 0.02 | |
| 48 | 2444.0 | 1221.5 |
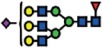
|
1.30 + 0.46 | 0.00 | 0.65 ± 0.07 | |
| 49 | 2589.0 | 1294.0 |
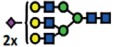
|
0.00 | 0.00 | 0.03 ± 0.00 | |
| 50 | 2735.0 | 1367.0 |
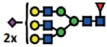
|
0.00 | 0.00 | 0.03 ± 0.00 | |
| 51 | 3026.0 | 1512.5 |
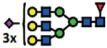
|
0.00 | 0.00 | 0.19 ± 0.08 | |
| O-glycans | 1a | 675.3 | NA |
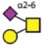
|
9.10 ± 2.32 | 13.08 ± 2.43 | NA |
| 1b | 675.3 | NA |
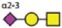
|
31.55 ± 4.19 | 27.59 ± 2.21 | NA | |
| 2 | 966.3 | NA |
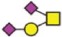
|
52.11 ± 2.60 | 52.25 ± 0.59 | NA | |
| 3 | 749.3 | NA |
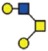
|
0.00 | 0.00 | NA | |
| 4 | 1040.5 | NA |
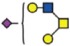
|
7.24 ± 1.26 | 7.08 ± 0.44 | NA | |
| 5 | 1331.5 | NA |
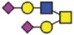
|
0.00 | 0.00 | NA | |
N- and O-glycan structures were separated by PGC-UHPLC-ESI-QTOF and their structures were assigned based on MS/MS fragmentation (where possible), retention time differences and biological pathway constraints. Structures were depicted according to the Consortium of Functional Glycomics (CFG) notation with linkages (α2, 3 and α2, 6) indicated for sialic acid (where known). Values represent mean relative ion intensities ± SD of three replicates based on their extracted ion chromatograms (EIC). N-glycan structures derived from IGROV1 cell line previously analyzed with PGC-ESI-IT-MS13,19 are also shown for comparative profiling using two different MS platforms.
 Galactose
Galactose  Mannose
Mannose  Fucose
Fucose  N-acetylglucosamine (GlcNAc)
N-acetylglucosamine (GlcNAc)  N-acetylgalactosamine(GalNAc)
N-acetylgalactosamine(GalNAc)  N-acetylneruaminic acid (NeuAc; sialic acid).
N-acetylneruaminic acid (NeuAc; sialic acid).
In order to confirm the reduction of α2-6 sialylation on membrane glycoproteins, we performed a staining using Sambucus nigra agglutinin lectin (SNA), which has been shown to bind preferentially to sialic acid attached to terminal galactose in via α2-6 linkage20. The parental IGROV1 revealed positive staining (37% ± 6% FITC+) for α2-6Neu5Ac whereas for both ∆B3GNT5 cells, the expression of α2-6 –linked Neu5Ac was significantly reduced (KO_1 = 2% ± 1.7% and KO_2 = 6% ± 2.6% FITC+) (p < 0.01; Fig. 3F), thereby confirming the loss of membrane protein α2-6 sialylation.
α2-6 sialylation is not affected in B4GALNT1 ganglioside-depleted IGROV1 cells
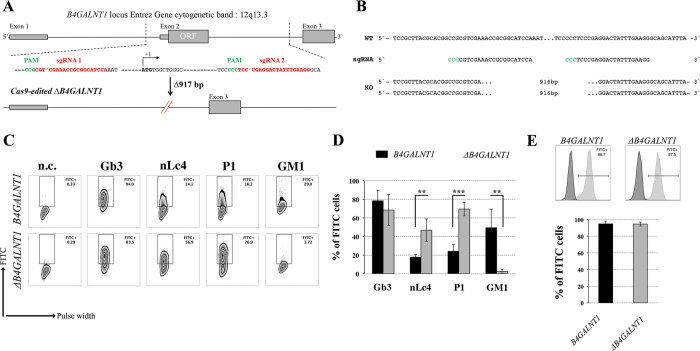
Our current data demonstrate that the deletion of B3GNT5 and consequently, the loss of nsGSLs, have unexpectedly led to the observation that α2-6 sialylation is reduced on N-glycosylated proteins. To investigate whether this effect can be also observed when other GSL series are genome-edited, we utilized the CRISPR-Cas9 to homozygously delete the B4GALNT1 gene encoding for the beta-1,4-N-acetyl-galactosaminyltransferase-1 glycosyltransferase involved in the synthesis of gangliosides. IGROV1 cells were transfected with two sgRNA Cas9-encoding constructs to delete exon 2 of B4GALNT1, resulting in a 917 bp deletion (Fig. 4A). Clones identified as homozygously edited B4GALNT1 knockout cells (ΔB4GALNT1) were confirmed by DNA sequencing based on the deletion of 916 bp (Fig. 4B). Following measurement of GSL expression, as expected, we observed a significant reduction for GM1 (p < 0.01) and no change in the expression of globoside Gb3 (Fig. 4C and D). In line with a previous report showing appearance of nLc4 after sialidase treatment in ganglioside-depleted mice21, we found that nLc4 and P1 are significantly elevated in ΔB4GALNT1 cells (p < 0.01, Fig. 4D). In regards to α2-6 sialylation, we did not observe a difference in SNA staining to IGROV1 cells compared to ΔB4GALNT1 cells (p > 0.05, Fig. 4E). Taken together, CRISPR-Cas9 mediated disruption of B4GALNT1 leading to loss of gangliosides did not subsequently alter α2-6 sialylation on glycosylated proteins in contrast to that observed for ΔB3GNT5 cells.
Figure 4. Loss of gangliosides by B4GALNT1-editing does not affect α2–6 sialylation in IGROV1 cells.
(A) Depiction of CRISPR-Cas9-mediated B4GALNT1 editing in IGROV1 cells using two different sgRNAs [red; PAM sequence (green)]. In silico analysis revealed a deletion of 917 bp including translation start site located at exon 2. (B) Verification of ∆B4GALNT1 cells by DNA sequencing. (C) Representative flow cytometry zebra blot for parental IGROV1 and ∆B4GALNT1 cells. (D) Bar chart summarizing three independent flow cytometry experiments. (E) SNA-staining remains unaffected in ∆B4GALNT1 cells compared to IGROV1 (B4GALNT1). Data are represented as mean ± s.d.
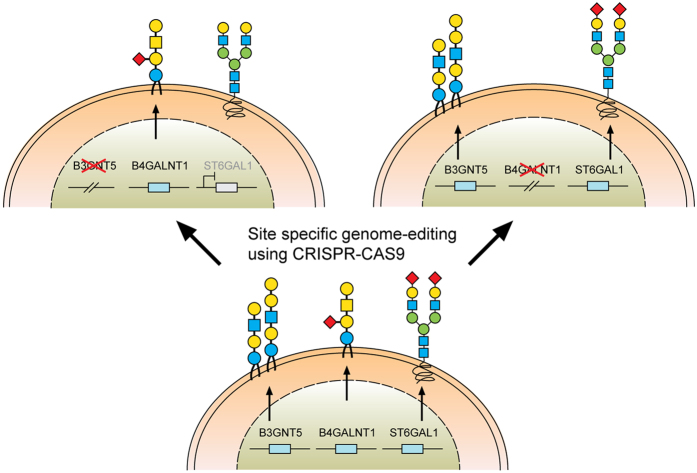
Loss of α2-6 sialylation is a result of silenced ST6GAL1 expression in ∆B3GNT5 cells
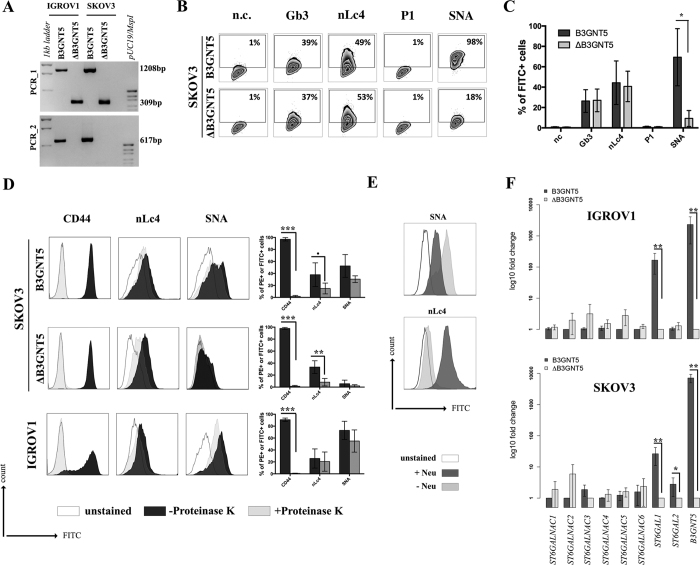
The reduction of α2-6 sialylation in ∆B3GNT5 IGROV1 cells, prompted us to investigate whether this observation is constrained to a specific cell line or reproducible in another ovarian cancer cell line. Thus, we applied the genome-editing strategy as reported for IGROV1 (Figs 2A and 5A) in SKOV3 cells, which were chosen based on their expression of nLc4 and α2-6 sialic acid (Figs 1B and 5B). In contrast, Ls174T being positive for nLc4 and P1 did not show α2-6 sialylation and was therefore not selected for genome- editing (Supplemental Fig. S4A). Analysis of the GSL expression in parental SKOV3 and corresponding ∆B3GNT5 cells revealed only marginal changes for Gb3 and nLc4, whereas SNA staining was significantly reduced (p = 0.0232). This confirms what was previously observed in ∆B3GNT5 IGROV1 cells, and further indicates that the disruption of B3GNT5 leads to a reduction in α2-6 sialylation and may be a cell line-independent phenomenon (Fig. 5B and C).
Figure 5. The reduction of α2-6 sialylation occurs in two ∆B3GNT5 ovarian cancer cell lines.
(A) Representative genotyping PCR for ∆B3GNT5 SKOV3 ovarian cancer cells. PCR_1, wildtype-specific PCR; PCR_2, deletion specific PCR. (B) Representative zebra plot for GSL expression in SKOV3 wildtype and ∆B3GNT5 cells. (C) Quantified data from three independent flow cytometry experiments. (D) The epitope for anti-nLc4 antibodies is present on glycoproteins and glycosphingolipids in SKOV3 and IGROV1 cells, respectively. Proteinase K treatment on SKOV3, SKOV3 ∆B3GNT5 and IGROV1 cells following staining for CD44, nLc4 and α2-6 neuraminic acid (with SNA). Representative histogram for unstained (white), proteinase K untreated (dark gray) and proteinase K treated cells (light gray). Bar chart summarizes three independent experiments. ***p < 0.001, **p < 0.01, *p-value < 0.05,p-value < 0.1. (E) Histogram for IGROV1 cells treated with neuraminidase and stained for α2-6 neuraminic acid (with SNA) and nLc4; unstained (white), neuraminidase treated (dark gray) and untreated cells (light gray). (F) Relative expression of genes encoding glycosyltransferase known to attach α2-6 neuraminic acid to glycoproteins and glycosphingolipids. Bar chart shows the mean and standard deviation of three independent experiments of target genes (ST6GALNAC1-6, ST6GAL1, ST6GAL2, and B3GNT5) in wildtype (IGROV1 and SKOV3) and corresponding ∆B3GNT5 cells. Data are represented as mean ± s.d.
It is interesting to note, however, that we did not observe any reduction of nLc4 in SKOV3 ∆B3GNT5 cells (Fig. 5C). Thus, we hypothesized that besides GSLs, the antibody may also recognize the terminal nLc epitope (Galβ1-4GlcNAc) on N- and O-glycoproteins carrying Type II LacNAc (Galβ1-4GlcNAc) terminated antennas and therefore, the binding levels in SKOV3 ΔB3GNT5 cells remained unchanged. To analyze whether the antibody-binding epitope is also carried on cell surface proteins, we treated SKOV3, SKOV3 ΔB3GNT5, and IGROV1 cells with Proteinase K and performed staining for nLc4, SNA, and CD44. The latter was used as a control antigen (sensitive to Proteinase K treatment). As shown in Fig. 5D, cells subjected to Proteinase K treatment showed a complete loss of CD44 epitope, an ubiquitously expressed membrane protein, indicating that proteins on the cell surface are fully digested. In contrast, SKOV3 and SKOV3 ΔB3GNT5 cells showed reduction of nLc4 staining after Proteinase K treatment whereas IGROV1 cells remained positive after nLc4 staining. This suggests that the nLc4 epitope was potentially present on cell surface proteins on SKOV3 as well as Ls174T cells (Supplemental Fig. S4B). In regards to α2-6 sialylation, Proteinase K treated cells showed only marginal reduction in SNA staining (Fig. 5D). Finally, IGROV1 cells (no reduction upon Proteinase K treatment for nLc4) were also treated with a broad-specificity neuraminidase to remove α2-6/2-3-linked Neu5Ac, resulting in the reduction of SNA staining (Fig. 5E). This observation corroborated with a further increase in nLc4 staining, indicating that nLc4 on GSLs is terminated with Neu5Ac to form sialyl- 3 or 6- paragloboside GSLs in IGROV1 cells and is further exposed on the cell surface upon de-sialylation. Taken together, our results provide evidence that apart from nLc4 expression on GSLs, the anti-nLc4 antibody epitope is also preferentially expressed on glycoproteins in SKOV3 cells, as indicated by the sustained presence of nLc4 in SKOV3 ΔB3GNT5 cells and terminal nLc4 in GSLs may further be modified to form sialo-paraglobosides, as observed in IGROV1 cells.
Next, we investigated whether mRNA expression of genes encoding sialyltransferases involved in the synthesis of α2-6-sialoglycans is altered upon deletion of B3GNT5. RT-qPCR was established in concordance with MIQE guidelines22 and glyco-gene expression was normalized to the geometric mean of three independent reference genes (Supplemental Table S2). We profiled ST6GALNAC1-6 and ST6GAL1-2 in parental IGROV1 and SKOV3 as well as their corresponding ∆B3GNT5 cells. Apart from the expected change in B3GNT5 mRNA levels, ST6GAL1 expression was found to be significantly decreased in both ∆B3GNT5 cells of up to 166-fold for IGROV1 (Fig. 5F). The expression of the remaining sialyltransferases encoding genes examined was not significantly altered in the wildtype and ∆B3GNT5 cells, except for ST6GAL2 in SKOV3 cells (Fig. 5F). In addition, ST6GAL1 expression in parental and IGROV1 ∆B4GALNT1 cells revealed equal gene expression levels (Supplemental Fig. S5), corroborating with no changes observed in SNA staining. In conclusion, these results indicate that the disruption of B3GNT5 and the corresponding loss of α2-6 sialylation on N-glycans appear due to the preferential silencing of ST6GAL1 gene expression in both ovarian cancer cell lines, IGROV1 and SKOV3 (Fig. 6).
Figure 6. A model summarizing the interaction of specific glycolipid synthases on glycosphingolipids and N-glycosylation of proteins.
Discussion
The advent of novel nuclease-based precision genome- editing techniques is starting to revolutionize the field of glycobiology, enabling stable gene editing and potentially unravelling the roles of glyco-related genes and their corresponding glycosyltransferases in mammalian cell lines. This technology was first demonstrated by Henrik Clausen and his colleagues, through the introduction of the ‘Simple Cell’ strategy, to study the role of truncated O-glycans in malignant diseases23,24, specificity of anti-glycopeptide antibodies25, profile and map the human O-GlcNAc glycoproteome26,27, and produce novel antibodies to defined glycopeptides28. Whilst a majority of this work is based on O-glycans, there is very limited information on the use of genome- editing for glycosphingolipids (GSLs) and in particular, the nsGSLs. Here, we utilized CRISPR-Cas9 nuclease genome- editing to explore the role of B3GNT5, the key glycosyltransferase responsible for the synthesis of precursor Lc3 that is extended to form the lacto (Type 1) and neo-lacto (Type II) series GSLs6. For the first time, using a combination of genomic-level editing and cell surface membrane protein and GSL-glycan profiling, we provide valuable insights into the complex interplay regulating the expression of linkage-specific α2-6 sialo-N-glycans on proteins as a consequence of GSL disruption. We demonstrate that by mutating B3GNT5, the depletion of nsGSLs consequently affects α2-6 sialylation on N-glycoproteins, not only at the cellular level, but also through the silencing of the corresponding ST6GAL1 gene.
As evidenced in this study, the genomic deletion of B3GNT5 resulted in the loss of the (neo-) lacto paragloboside (nLc4, Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ1-1Ceramide) as well as P1 (Galα1-4Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ1-1Ceramide), a pentasaccharide moiety of which has not been annotated in the KEGG database so far. We recently reported the presence of the P1 glycan in tissue samples14 as well as on P1-enriched IGROV1 ovarian cancer cell lines13, serving as a cell-recognition molecule through its binding to naturally occurring anti-glycan antibodies14,29. The presence of nLc4 and P1 was detected on IGROV1 cells using flow cytometry, in which P1 was detected using P3NIL100 antibody, a well-validated monoclonal IgM antibody shown to specifically recognize the chemically synthesized P1 trisaccharide (Galα1-4Galβ1-4GlcNAcβ-), as indicated by glycan array profiling performed in this study. Both the flow cytometry and high-resolution ESI-QTOF-MS and MS/MS profiles confirmed that the levels of nLc4 were reduced, while the globo (Gb3) and ganglio (GM3, GM2 and GM1)- series GSL glycans remained unaffected in ∆B3GNT5 cells. The presence of P1, however, was not detected on the parental IGROV1 cells by the mass spectrometry approach applied, possibly due to the low detection levels of P1 on non-P1 enriched IGROV1 cells used in this study, as well as the use of a different mass spectrometric platform. Nevertheless, we also observed that apart from IGROV1, the colon cancer cell line, Ls174T, showed detectable levels of P1 providing further evidence that P1 is not exclusively present on erythrocytes30,31,32. The results obtained herein support that the pentasaccharide P1 is indeed exclusively present as an nsGSL-glycan on membrane glycolipids and not on glycoproteins.
To date, it remains unclear if nLc4- and P1- GSL glycans are functionally relevant molecules in diseased states such as cancer and infection or if they are ubiquitously expressed. In regards to pathogenic infections, it has been previously shown that Helicobacter pylori induces B3GNT5 expression and that the ABO(H)/Lewis blood group antigens expressed in H. pylori- infected individuals act as receptors for BabA, thereby facilitating colonization of the gastric niche33. Likewise, the Shiga toxin from urinary tract infection-causing enterohemorrhagic E. coli was shown to recognize the terminal epitope of P1, Galα1-4Galβ1-4 following internalization of the toxin by receptor-mediated endocytosis34. Another study also showed that the Shiga-like toxin binds to P1 trisaccharide on core 2 O-glycoproteins in overexpressed CHO cells35. These findings indicate that this toxin may recognize P1 pentasaccharide. It is evident that the successful B3GNT5 deletion performed in this study may be further applied to investigate the lectin binding specificities (e.g. Shiga toxin) in other human cancer cell lines expressing Gb3, nLc4 and P1.
Perhaps, the most intriguing finding from this study is the unexpected loss of α2-6 sialylation observed using mass spectrometry in ∆B3GNT5 cells which was further substantiated by SNA lectin staining on cultured cells and reduced ST6GAL1 mRNA expression. We have previously shown in two separate studies, respectively, that ovarian cancer cells, IGROV1 and SKOV3, have higher levels of glycoprotein α2-6 sialylation as compared to non-cancer ovarian epithelial cells19 and nLc4 and P1 are both expressed on IGROV1 and ovarian cancer tissue-derived GSL-glycans13. Hence, the reduction of α2-6 sialylation as a direct consequence of the B3GNT5 deletion in ovarian cancer cells is indicative of their complex associations within the glycan-processing pathway. A recent publication reported a similar finding in the context of the rare autosomal recessive salt and pepper syndrome36. In this study, a homozygous transition mutation in the gene encoding sialyltransferase ST3GAL5 (GM3 synthase) was described. More importantly, in addition to the reduced GSL complexity, the comprehensive glycomics analysis performed using mass spectrometry also revealed altered N- and O-glycans on proteins due to the point mutation in the ST3GAL536. In another study on respiratory chain disorders, the authors observed significant increases of Gb3 and Gb4 and a decrease of LacCer in fibroblasts36,37. Similarly, the appearance of unexpected gangliosides in homozygously deleted ST3GAL5 murine primary embryonic fibroblast cells was also reported38. Despite the limited number of publications, our current study provides another example on how a loss of a specific glycosyltransferase interferes with glycomic changes, as observed in the reduction of α2-6 sialylation on N-glycoproteins, thereby recognizing the contribution of altered GSL expression in cell development and function. It is not known whether the loss of α2-6 sialylation is thought to provide a compensation phenotype for the depleted nsGSLs in these cells or enables them to survive despite the glycan changes. It is also unclear as to how the ST6GAL1 is regulated in the context of their translocation within the Golgi membrane or their accessibility to the membrane proteins. It will be useful to further characterize membrane proteins bearing these sialylated N-glycans and observe corresponding changes in the proteome profiles of B3GNT5 –edited ovarian cancer cells.
Of note, despite similar observations in two independent ovarian cancer cell lines and due to our carefully designed sgRNAs in a way to minimize the probability of off-target activity; we cannot exclude unintended changes in the genome of our genome- edited ovarian cancer cell lines. Here, an unbiased approach would be necessary to exclude mutations anywhere in the genome39. Therefore, the most reliable method would be whole genome sequencing40. A direct in situ breaks labeling, enrichment on streptavidin, and next-generation sequencing also referring to BLESS41 as well as linear amplification-mediated high-throughput genome-wide translocation sequencing (HTGTS)42 can also be applied as less expensive alternatives to track genomic alterations caused by off target Cas9-activity.
In conclusion, the investigation of CRISPR-Cas9-mediated deletion of glycosyltransferase in ovarian cancer cells, coupled with the use of MS-based glycomics profiling, serve as a useful model for future studies to elucidate the biological roles of nsGSLs, an important group of GSLs which may be involved in specific cell physiology functions. More importantly, the prominent changes in N-glycoprotein sialylation detected in the B3GNT5 genome- edited cells are reflective of global alterations in one or more regulatory components essential for glycan biosynthesis, thereby warranting further research efforts into the complex and overlapping roles of glycosyltransferases involved in the downstream glycan biosynthesis pathways.
Methods
Cell culture
Cell lines (HOSE17.1, IGROV1, SKOV3, BG1, CAOV3, Ls174T, HeLa, HCT15, and HCT116) were grown in RPMI1640 media supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 μg/ml) (Sigma-Aldrich, Buchs, Switzerland). Fallopian tube cell lines FT33 Tag, FT190, and FT237 (kind gifts by Dr. Drapkin) were cultured in DMEM F12/50 without HEPES supplemented with 2% (v/v) Ultroser (USG, Pall Corporation, USA) and penicillin/streptomycin. All cell lines were cultured at 37 °C in a 95% humidified atmosphere containing 5% CO2. Cell lines were short tandem repeat (STR) profiled and routinely tested for mycoplasma infection43.
Antibodies and reagents
Primary antibodies to detect the cell surface-associated GSLs were applied as described previously15. Briefly, the following primary antibodies were utilized; rat IgM anti-Pk (Gb3) antibody (ABDserotec, Germany); mouse IgM anti-nLc4 (paragloboside) antibody [clone 1B2 kindly provided by Prof. Mandel and Prof. Clausen, Denmark]44; human IgM anti-P1 antibody (Millipore, Germany), and Cholera toxin B subunit (Sigma-Aldrich, Switzerland) to detect GM1. All secondary antibodies (mouse anti-rat IgM antibody conjugated to biotin, rat anti-mouse IgM conjugated to biotin and mouse anti-human IgM conjugated to biotin) were obtained from BD Pharmingen. Cell surface GSL expression was visualized with FITC conjugated to streptavidin (BD Pharmingen). Non-viable cells were positively stained with 7-amino-actinomycin D (BD Pharmingen). Isotype controls for each primary antibody used included the following: purified rat IgM (BD Pharmingen), purified mouse IgM and chrompure human IgM (Jackson ImmunoResearch, USA). Alpha 2-6 sialylation on the cell surface was investigated using Sambucus nigra lectin (SNA) conjugated to biotin (Vector Laboratories, Reactolab, Servion, Switzerland). Lectin staining was visualized with streptavidin conjugated to FITC. Anti human CD44-PE antibody was purchased from Miltenyi Biotec (Bergisch Gladbach, Germany) for use as a positive control in Proteinase K treatment.
CRISPR-Cas9 sgRNA design and construction
The design of sgRNAs was carried out using the online program available from Zhang’s laboratory45. This program provides additional information about the quality of sgRNA by giving it a score value of up to 100 and additionally, the number and sites of potential off targets. Designed and processed sgRNAs to edit either B3GNT5 or B4GALNT1 (Supplemental Table S1) were cloned into pSpCas9(BB)-2A-GFP (addgene PX458) via BsbI restriction site using T4 DNA Ligase (Promega, Dübendorf, Switzerland).
IGROV1 and SKOV3 cells were transfected using ViaFect Transfection reagent (Promega, Dübendorf, Switzerland). A total of 4–5 × 105 cells were seeded into 6-well plates, cultured for 24 h and transfected with 2.5 μg of pSpCas9(BB)-2A-GFP. Cells were harvested after 72 h and subjected to single cell sorting.
Flow cytometry-based single-cell sorting
IGROV1 transfected with pSpCas9(BB)-2A-GFP targeting either B3GNT5 or B4GALNT1 were grown for 72 h, washed twice in PBS and harvested using non-enzymatic cell dissociation buffer (Sigma-Aldrich, Buchs, Switzerland). Cells were then resuspended in RPMI containing 10% FCS before single cell sorting was performed on a BD FACS Vantage SE DiVa Cell Sorter (BD Biosciences). Cells sorted for single DAPI− and GFP+ cells were seeded into 96-well flat-bottom plates with pre-warmed RPMI containing 10% FCS. Plates were incubated for 2 to 3 weeks following transfer to 48-well plates, prior to genomic DNA isolation for genotyping PCR to characterize single cell clones.
Genotyping PCR
Selected clones were characterized by three to four PCRs using appropriate primer pairs (Supplemental Table S1). PCR was performed using 1x GoTaq Green Master Mix (Promega, Dübendorf, Switzerland), 200 nM primer (Supplemental Table S1) and 30 ng gDNA and carried out on a Biometra Professional Trio cycler. PCR was performed under the following conditions: initial denaturation at 94 °C for 1 min followed by 33 cycles consisting of denaturation at 94 °C for 15 sec, annealing at 59 °C for 15 sec, and elongation at 72 °C for 15–45 sec depending on the amplicon length. The PCR products were separated on a 1.7% agarose gel.
Sanger DNA Sequencing
PCR products were cloned into pGEM®-T Easy Vector System (Promega) according to the manufacturer’s protocol. DNA sequencing was performed by Source Bioscience Life Sciences (Berlin, Germany). Samples were shipped at a concentration of 100 ng/μl with specific primer (if required) concentration of 3.2 pg/μl.
Off target screening
Potential off target sites were amplified (Supplemental Table S1), cloned into pGEM®-T Easy Vector System (Promega) and sequenced. A total of three colonies per clone were selected for sequencing.
Reverse transcription semi-quantitative PCR (RT-qPCR) using total RNA
Total RNA was extracted from 80% confluent 6-well plates seeded initially with 1 × 105 cells. Cells were then washed twice with sterile PBS and total RNA was extracted using the ReliaPrep RNA Cell Miniprep System (Promega, Dübendorf, Switzerland). RNA was eluted in 60 μl RNase free water and RNA concentration was measured using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Roskilde, Denmark).
Total RNA (500 ng) was reverse-transcribed using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories, Zurich, Switzerland) in a total volume of 10 μl according to the manufacturer’s instructions. RT-qPCR was performed on target and reference genes (HSPCB, SDHA, and YWHAZ) in 10 μl reactions containing 10 ng cDNA (initial total RNA), 400 nM forward and reverse primer (Supplemental Table S2), nuclease free water and 1x GoTaq® qPCR Master Mix with low ROX as the reference dye (Promega, Dübendorf, Switzerland) on a ViiA™ 7 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Reinach, Switzerland). Quantitative PCR was performed on three independent experiments, in triplicates, and analyzed as recently described19,46. Each quantitative PCR was further established with a series of six 10-fold dilutions to calculate the efficacy of each reaction and to determine the optimal range of initial mRNA concentration for analysis (Supplemental Table S2).
Flow cytometry
Immuno staining and flow cytometry was performed as previously described14,15. Biotinylated SNA lectin (1:500) containing 10 mM CaCl2 was applied for detection of α2-6 neuraminic acid using flow cytometry (BD Accuri™ C6, BD Bioscience).
Membrane protein extraction and N- and O-glycan release of parental and ∆B3GNT5 IGROV1 cell lines
Membrane protein extraction and glycan release were carried out as previously described19. Briefly, approximately 2 × 107 IGROV1 and ∆B3GNT5 IGROV1 cells were washed with PBS and homogenized in lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA and protease inhibitor, pH 7.4). Unlysed cells were pelleted through centrifugation and cellular membranes in the supernatant was subjected to ultracentrifugation at 120, 000 g prior to Triton X-114 phase partitioning of membrane proteins. Membrane proteins were acetone-precipitated, solubilized in 8 M urea and spotted onto polyvinylidene diflouride (PVDF) membrane spots placed in a 96-well microtiter plate. Protein spots were treated with PNGase F enzyme and released N-glycans were treated with 100 mM ammonium acetate (pH 5.0) and reduced to alditols with 2 M NaBH4 in 50 mM KOH. For O-glycans, the remaining PVDF spots in the 96- well microtiter plate were further subjected to reductive β-elimination by treatment with 0.5 M NaBH4 in 50 mM KOH to chemically release the O-glycans. Both the released N- and O-glycans were desalted by cation exchange chromatography as previously described and re-suspended in 15 μl of MilliQ water prior to mass spectrometry analysis.
GSL extraction and enzymatic release of GSL-glycans from cell lines
Lipid extraction and release of GSL-glycans were performed as previously described13. Briefly, GSLs were extracted from approximately 2 × 107 parental and ∆B3GNT5 IGROV1 cells using a modified Folch extraction method. Cell pellets were treated with chloroform: methanol (2:1) and subjected to centrifugation. The supernatants were evaporated to dryness and crude GSL fractions were re-dissolved in 50 μl of chloroform: methanol (2:1). Approximately 10 μl of GSL standard (neutral glycosphingolipids: LacCer, Gb3 and Gb4) and 50 μl of extracted GSLs were spotted onto the PVDF membrane and treated with 4 mU (2 μl) of Endoglycoceramidase II. GSL-glycans released from the immobilized GSL spots were reduced to alditols with 0.5 M NaBH4 in 50 mM KOH and desalted by cation exchange chromatography as previously described for N- and O- glycans.
PGC-UHPLC-ESI-QTOF-MS/MS profiling of released GSL- and N-and O-glycan alditols
The separation of glycan alditols was performed using an Agilent UHPLC modular system consisting of a degasser, binary pump, auto-sampler, thermostat and column oven (Agilent Series 1290, Agilent Technologies, Germany). The conventional flow LC system was connected to a mass spectrometer (MS) (Agilent Series 6540 Q-TOF, Agilent Technologies, USA) operating in negative mode and coupled to a jet stream electrospray ion source (ESI) as previously described for N- and O-linked glycans47. Briefly with some slight modifications for GSL-derived glycans, an aliquot of 14 μL of sample was injected onto the Hypercarb porous graphitized carbon (PGC) analytical column (ID2.1 × 150 mm column; 3 μm particle size) (Thermo Fisher Scientific, United Kingdom). The column compartment was maintained at 40 °C. The mobile phase was made up of 10 mM ammonium bicarbonate (A) and 9:1 acetonitrile/water with 10 mM ammonium bicarbonate (B). The chromatographic separation was held with the initial mobile phase composition (1% B; 0–5 mins), followed by a segmented linear gradient to 25% B (5–40 mins), 50% B (40–42 mins) and a fast step at 42.5 mins to 80% B, with a hold for 1.5 mins. The ESI source was operated with the following parameter settings: nebulizer pressure (35 psig), nozzle voltage (0 V), sheath gas flow (11 L/min), sheath gas temperature (375 °C), drying gas flow (8 L/min), drying gas temperature (250 °C), capillary voltage (4000 V) and fragmentor voltage (175 V). High-resolution MS scan (m/z 100–1700 at 1.5 Hz) and MS/MS product scan (m/z 50–1700 at 3 Hz) were acquired with information-dependent acquisition (IDA) with static exclusion range m/z 100–300. An automated mass calibration performed by delivering constantly two reference masses (m/z 119.0363 and m/z 981.9956) on the end plate of the ESI source to obtain the highest mass accuracy during the measurement. Data analysis was performed with MassHunter B.06.00 (Agilent Technologies, USA). The monosaccharide compositions of monoisotopic masses were determined using GlycoMod tool (http://web.expasy.org/glycomod/) with a mass error of ±0.5 Da. The proposed glycan structures were manually assigned and interpreted based on MS/MS fragmentation patterns of known GSL-derived glycans13 and annotated using GlycoWorkBench 2.1 software48. In addition, UniCarb KB, a web-based LC-MS/MS database49, was also utilized to confirm fragmentation and retention time of sialylated N-glycans previously reported to contain sialylated isomers19. Blank samples, fetuin glycoprotein with isomeric glycan compositions and reference glycolipid standards which served as quality control for sample preparation were randomly measured within each sequence run and checked for background contamination.
Confocal fluorescence microscopy
Cells were grown on polylysine glass slides attached to an 8-well chamber, fixed with 4% para-formaldehyde for 15 min and blocked with 5% (w/v) BSA fraction V (Sigma) dissolved in PBS for 1 h. Cells were then stained with either anti-P1 IgM15 (1:5 diluted with incubation buffer (1% BSA in PBS)) or anti-nLc4 (paragloboside) antibody (1:5) for overnight at 4 °C. Following extensive washing corresponding secondary antibodies (Biotin-anti-human IgM (1:100), Biotin- anti-mouse IgM (1:100)) were added to each chamber and incubated for 2 h at 4 °C. Afterwards, cells were washed with PBS and incubated with Streptavidin-FITC (1:200) for 1 h at 4 °C and counterstained with DAPI (Cell Signaling Technology). Fluorescence images were taken with a Zeiss LSM 710 confocal microscope (Zeiss, Feldbach, Switzerland).
Printed Glycan Array
The printed glycan array was performed as previously described with modifications29,50,51. Briefly, glycan arrays were prepared by covalent attachment of amino-spacered synthetic glycans to N-hydroxysuccinimide-activated Schott-Nexterion slides. Chemically synthesized carbohydrates with 95–98% purity were used for printing and obtained from Lectinity Holdings (Moscow, Russia) and the Consortium for Functional Glycomics (CFG). The slides were washed for 15 min with 0.1 М PBS (0.01 М Na2HPO4, 0.01 M NaH2PO4, 0.138 M NaCl, and 0.0027 M KCl, pH 7.4) containing 0.1% Tween-20. Anti-P1 antibodies were diluted 1:50 in PBS containing 1% BSA. The slides with antibody solution were shaken for 1 h and incubated under relative humidity and temperature of 80% and 37 °C, respectively. The slides were washed with PBS and labeled with secondary antibodies consisting of goat anti-human IgM conjugated with Alexa647 (Invitrogen, USA), which was diluted with PBS (1:250). After incubation, the slides were washed with PBS containing 0.001% Tween 20 and subsequently with bi-distilled water and the fluorescence intensity was measured using a confocal ScanArray Gx scanner (PerkinElmer, USA) with 5 μm resolution. The obtained data were processed using ProScanArray Express 4.0 software.
Proteinase K treatment
Proteinase K treatment was carried out as previously described52. Briefly, single-cell suspension of 2 × 106 IGROV1, Ls174T, SKOV3 and B3GNT5 - edited SKOV3 cells were treated with PBS containing 100 μg/mL proteinase K enzyme (Promega) for 1 h at 37 °C with intermittent shaking. At the same cell density the control cells were also resuspended in PBS without proteinase K. The enzymatic reaction was stopped by adding phenyl methyl sulfonyl fluoride (PMSF) at a concentration of 1 mM/mL and incubated on ice for 10 min. Cells were then fixed with 1% PFA on ice for 15 min and washed twice with 5 mL PBS and pelleted at 1200 rpm for 7 min at 4 °C. Finally, fixed cells were stained with SNA, nLc4 or anti-human CD44 antibody for flow cytometric analysis as previously described.
Neuraminidase treatment
To remove the cell surface neuraminic acids, neuraminidase treatment was performed with α2,3-6, -8,- neuraminidase (New England BioLabs, UK) according to manufacturer’s instructions. In brief, IGROV1 cells (1 × 106) were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min, and then washed with PBS twice. Cells were resuspended in 200 μl of glycobuffer (1x) containing 25 Units of the enzyme and incubated at 37 °C for 2 h. Cells were washed with PBS and followed by SNA staining and flow cytometry.
Statistical analysis
Experiments were performed in triplicates and comparisons were statistically evaluated with two-tailed Student’s t-test. P values of <0.05 were considered statistically significant (***p < 0.001, **p < 0.01, *p-value < 0.05, p-value < 0.1.).
Additional Information
How to cite this article: Alam, S. et al. Altered (neo-) lacto series glycolipid biosynthesis impairs a2-6 sialylation on N-glycoproteins in ovarian cancer cells. Sci. Rep. 7, 45367; doi: 10.1038/srep45367 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are grateful to the Flow Cytometry Facility (Danny Labes and Emmanuel Traunecker) and Microscopy Facility (Michael Abanto and Beat Erne) for providing all necessary support. We would also like to acknowledge the following people for their various contributions to this publication: Viola Heinzelmann-Schwarz, Christian Berchthold, Andreas Schötzau, and Tatiana Pochechueva. We also thank Jolene Molley for final proofreading the manuscript. This work was supported by Novartis Foundation for Biomedical Research (13B093), Krebsliga beider Basel (14-2015), Swiss Cancer League (KLS-3841-02-2016), and FreeNovation 2016 provided by Novartis all to FJ, and the Department of Biomedicine, University Hospital Basel and University of Basel. Work of NVB was supported by the Russian Science Foundation project no. 14-50-00131.
Footnotes
The authors declare no competing financial interests.
Author Contributions F.J., S.A., M.A., R.S.K., K.W., and Y.G. designed research; S.A., M.A., Y.L.H., K.W., Y.G., T.H., N.K., and F.J. performed experiments; S.A., M.A., Y.L.H., K.W., Y.G., T.H., N.K. and F.J. analyzed data; N.V.B., G.S., T.H. provided samples, reagents and analytical tools; S.A., M.A., Y.L.H. and F.J. wrote the paper. All authors reviewed and approved the manuscript.
References
- Widmer M. et al. Performance of native and recombinant antigens for diagnosis of Helicobacter pylori infection. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 18, 823–826 (1999). [DOI] [PubMed] [Google Scholar]
- Coskun U., Grzybek M., Drechsel D. & Simons K. Regulation of human EGF receptor by lipids. Proceedings of the National Academy of Sciences of the United States of America 108, 9044–9048, doi: 10.1073/pnas.1105666108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J. et al. Ganglioside GM3 participates in the TGF`beta1-induced epithelial-mesenchymal transition of human lens epithelial cells. The Biochemical journal 449, 241–251, doi: 10.1042/BJ20120189 (2013). [DOI] [PubMed] [Google Scholar]
- Chung T. W. et al. Ganglioside GM3 inhibits VEGF/VEGFR-2-mediated angiogenesis: direct interaction of GM3 with VEGFR-2. Glycobiology 19, 229–239, doi: 10.1093/glycob/cwn114 (2009). [DOI] [PubMed] [Google Scholar]
- D’Angelo G., Capasso S., Sticco L. & Russo D. Glycosphingolipids: synthesis and functions. The FEBS journal 280, 6338–6353, doi: 10.1111/febs.12559 (2013). [DOI] [PubMed] [Google Scholar]
- Togayachi A. et al. Molecular cloning and characterization of UDP-GlcNAc:lactosylceramide beta 1,3-N-acetylglucosaminyltransferase (beta 3Gn-T5), an essential enzyme for the expression of HNK-1 and Lewis X epitopes on glycolipids. The Journal of biological chemistry 276, 22032–22040, doi: 10.1074/jbc.M011369200 (2001). [DOI] [PubMed] [Google Scholar]
- Nozaki H. et al. Production and characterization of monoclonal antibodies specific to lactotriaosylceramide. Glycobiology 20, 1631–1642, doi: 10.1093/glycob/cwq117 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. High expression of lactotriaosylceramide, a differentiation-associated glycosphingolipid, in the bone marrow of acute myeloid leukemia patients. Glycobiology 22, 930–938, doi: 10.1093/glycob/cws061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oss C. J. In Immunochemistry(eds van Oss C. J. & van Regenmortel M. H. V.) (Marcel Dekker, Inc., 1994). [Google Scholar]
- Onda M., Ariga K. & Kunitake T. Activity and stability of glucose oxidase in molecular films assembled alternately with polyions. Journal of bioscience and bioengineering 87, 69–75 (1999). [DOI] [PubMed] [Google Scholar]
- Brank A. S., Eritja R., Garcia R. G., Marquez V. E. & Christman J. K. Inhibition of HhaI DNA (Cytosine-C5) methyltransferase by oligodeoxyribonucleotides containing 5-aza-2′-deoxycytidine: examination of the intertwined roles of co-factor, target, transition state structure and enzyme conformation. Journal of molecular biology 323, 53–67 (2002). [DOI] [PubMed] [Google Scholar]
- Holmes E. H., Ostrander G. K., Clausen H. & Graem N. Oncofetal expression of Lex carbohydrate antigens in human colonic adenocarcinomas. Regulation through type 2 core chain synthesis rather than fucosylation. The Journal of biological chemistry 262, 11331–11338 (1987). [PubMed] [Google Scholar]
- Anugraham M., Everest-Dass A. V., Jacob F. & Packer N. H. A platform for the structural characterization of glycans enzymatically released from glycosphingolipids extracted from tissue and cells. Rapid communications in mass spectrometry: RCM 29, 545–561, doi: 10.1002/rcm.7130 (2015). [DOI] [PubMed] [Google Scholar]
- Jacob F. et al. The glycosphingolipid P(1) is an ovarian cancer-associated carbohydrate antigen involved in migration. British journal of cancer 111, 1634–1645, doi: 10.1038/bjc.2014.455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S., Fedier A., Kohler R. S. & Jacob F. Glucosylceramide synthase inhibitors differentially affect expression of glycosphingolipids. Glycobiology 25, 351–356, doi: 10.1093/glycob/cwu187 (2015). [DOI] [PubMed] [Google Scholar]
- Flesken-Nikitin A. et al. Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche. Nature 495, 241–245, doi: 10.1038/nature11979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perets R. et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer cell 24, 751–765, doi: 10.1016/j.ccr.2013.10.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henion T. R., Zhou D., Wolfer D. P., Jungalwala F. B. & Hennet T. Cloning of a mouse beta 1,3 N-acetylglucosaminyltransferase GlcNAc(beta 1,3)Gal(beta 1,4)Glc-ceramide synthase gene encoding the key regulator of lacto-series glycolipid biosynthesis. The Journal of biological chemistry 276, 30261–30269, doi: 10.1074/jbc.M102979200 (2001). [DOI] [PubMed] [Google Scholar]
- Anugraham M. et al. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: glycan structures reflect gene expression and DNA methylation status. Molecular & cellular proteomics: MCP 13, 2213–2232, doi: 10.1074/mcp.M113.037085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerieux F. et al. Determination of the sialic acid linkage specificity of sialidases using lectins in a solid phase assay. Analytical biochemistry 211, 200–204 (1993). [DOI] [PubMed] [Google Scholar]
- Yamashita T. et al. Interruption of ganglioside synthesis produces central nervous system degeneration and altered axon-glial interactions. Proceedings of the National Academy of Sciences of the United States of America 102, 2725–2730, doi: 10.1073/pnas.0407785102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry 55, 611–622, doi: 10.1373/clinchem.2008.112797 (2009). [DOI] [PubMed] [Google Scholar]
- Radhakrishnan P. et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proceedings of the National Academy of Sciences of the United States of America 111, E4066–4075, doi: 10.1073/pnas.1406619111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo S. et al. Detection of glyco-mucin profiles improves specificity of MUC16 and MUC1 biomarkers in ovarian serous tumours. Molecular oncology 9, 503–512, doi: 10.1016/j.molonc.2014.10.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Silva L. et al. Characterization of binding epitopes of CA125 monoclonal antibodies. Journal of proteome research 13, 3349–3359, doi: 10.1021/pr500215g (2014). [DOI] [PubMed] [Google Scholar]
- Steentoft C. et al. Precision genome editing: a small revolution for glycobiology. Glycobiology 24, 663–680, doi: 10.1093/glycob/cwu046 (2014). [DOI] [PubMed] [Google Scholar]
- Steentoft C. et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. The EMBO journal 32, 1478–1488, doi: 10.1038/emboj.2013.79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Silva L. et al. A novel monoclonal antibody to a defined peptide epitope in MUC16. Glycobiology 25, 1172–1182, doi: 10.1093/glycob/cwv056 (2015). [DOI] [PubMed] [Google Scholar]
- Jacob F. et al. Serum antiglycan antibody detection of nonmucinous ovarian cancers by using a printed glycan array. International journal of cancer. Journal international du cancer 130, 138–146, doi: 10.1002/ijc.26002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki M., Fong J., Ledeen R. & Marcus D. M. Structure of the human erythrocyte blood group P1 glycosphingolipid. Biochemistry 14, 4831–4837 (1975). [DOI] [PubMed] [Google Scholar]
- Thuresson B., Westman J. S. & Olsson M. L. Identification of a novel A4GALT exon reveals the genetic basis of the P1/P2 histo-blood groups. Blood 117, 678–687, doi: 10.1182/blood-2010-08-301333 (2011). [DOI] [PubMed] [Google Scholar]
- Dunstan R. A., Simpson M. B. & Rosse W. F. Presence of P blood group antigens on human platelets. Am J Clin Pathol 83, 731–735 (1985). [DOI] [PubMed] [Google Scholar]
- Magalhaes A. et al. Helicobacter pylori chronic infection and mucosal inflammation switches the human gastric glycosylation pathways. Biochimica et biophysica acta 1852, 1928–1939, doi: 10.1016/j.bbadis.2015.07.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H. et al. Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry 37, 1777–1788, doi: 10.1021/bi971806n (1998). [DOI] [PubMed] [Google Scholar]
- Maria Cherian R. et al. Shiga-like toxin binds with high avidity to multivalent O-linked blood group P1 determinants on mucin-type fusion proteins. Glycobiology 24, 26–38, doi: 10.1093/glycob/cwt086 (2014). [DOI] [PubMed] [Google Scholar]
- Boccuto L. et al. A mutation in a ganglioside biosynthetic enzyme, ST3GAL5, results in salt & pepper syndrome, a neurocutaneous disorder with altered glycolipid and glycoprotein glycosylation. Human molecular genetics 23, 418–433, doi: 10.1093/hmg/ddt434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragaki K. et al. Refractory epilepsy and mitochondrial dysfunction due to GM3 synthase deficiency. European journal of human genetics: EJHG 21, 528–534, doi: 10.1038/ejhg.2012.202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchuk N. A. et al. Alteration of ganglioside synthesis by GM3 synthase knockout in murine embryonic fibroblasts. Biochimica et biophysica acta 1771, 1226–1234, doi: 10.1016/j.bbalip.2007.05.008 (2007). [DOI] [PubMed] [Google Scholar]
- Zischewski J., Fischer R. & Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv, doi: 10.1016/j.biotechadv.2016.12.003 (2016). [DOI] [PubMed] [Google Scholar]
- Veres A. et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30, doi: 10.1016/j.stem.2014.04.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nature methods 10, 361–365, doi: 10.1038/nmeth.2408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nature biotechnology 33, 179–186, doi: 10.1038/nbt.3101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphoff C. C. & Drexler H. G. Detection of mycoplasma contaminations. Methods in molecular biology 290, 13–23 (2005). [DOI] [PubMed] [Google Scholar]
- Young W. W. Jr., Portoukalian J. & Hakomori S. Two monoclonal anticarbohydrate antibodies directed to glycosphingolipids with a lacto-N-glycosyl type II chain. The Journal of biological chemistry 256, 10967–10972 (1981). [PubMed] [Google Scholar]
- Ran F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols 8, 2281–2308, doi: 10.1038/nprot.2013.143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. et al. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PloS one 8, e59180, doi: 10.1371/journal.pone.0059180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R. S. et al. Epigenetic activation of MGAT3 and corresponding bisecting GlcNAc shortens the survival of cancer patients. Oncotarget 7, 51674–51686, doi: 10.18632/oncotarget.10543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerell D. et al. Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool. Methods in molecular biology 1273, 3–15, doi: 10.1007/978-1-4939-2343-4_1 (2015). [DOI] [PubMed] [Google Scholar]
- Campbell M. P. et al. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic acids research 42, D215–221, doi: 10.1093/nar/gkt1128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovin N. et al. Repertoire of human natural anti-glycan immunoglobulins. Do we have auto-antibodies? Biochimica et biophysica acta 1820, 1373–1382 (2012). [DOI] [PubMed] [Google Scholar]
- Huflejt M. E. et al. Anti-carbohydrate antibodies of normal sera: findings, surprises and challenges. Molecular immunology 46, 3037–3049 (2009). [DOI] [PubMed] [Google Scholar]
- Sivasubramaniyan K. et al. Expression of stage-specific embryonic antigen-4 (SSEA-4) defines spontaneous loss of epithelial phenotype in human solid tumor cells. Glycobiology 25, 902–917, doi: 10.1093/glycob/cwv032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.