Abstract
Helicobacter pylori colonizes the human stomach and induces inflammation, and in some cases persistent infection can result in gastric cancer. Attachment to the gastric mucosa is the first step in establishing bacterial colonization, and outer membrane proteins (OMPs) play a pivotal role in binding to human cells. Some OMP interaction molecules are known in H. pylori, and their associated host cell responses have been gradually clarified. Many studies have demonstrated that OMPs are essential to CagA translocation into gastric cells via the Type IV secretion system of H. pylori. This review summarizes the mechanisms through which H. pylori utilizes OMPs to colonize the human stomach and how OMPs cooperate with the Type IV secretion system.
Keywords: Helicobacter pylori, outer membrane protein, pathogenesis, Type IV secretion system
1. Introduction
Membrane proteins are classified by configuration, which determines their function and location. α-Helical membrane proteins are found in almost all membranes; however, β-barrel membrane proteins are only found in the outer membranes of Gram-negative bacteria, and not in Gram-positive bacteria. In bacteria, β-barrel outer membrane proteins (OMPs) have many functions including importing nutrients and translating signals from the outside environment. With respect to human infection, OMPs play a pivotal role in colonization, the expression of pathogenic factors, and antibiotic resistance. For Acinetobacter baumannii, OMPs are essential for establishing infection, and an interaction network with β-lactamase regulates antibiotic inactivation [1,2]. Thus, the role of OMPs in bacteria has been clarified, and new mechanisms to understand the biological roles of OMPs have been provided. For Helicobacter pylori, many virulence factors have been identified, and their functions have been gradually revealed. In addition, interaction partners of OMPs have been identified and their biological functions were elucidated. Recently, the interplay between OMPs and virulence factors has been suggested. However, the dynamic functions of OMPs in H. pylori are still unknown, and investigating these aspects represents an exciting field.
H. pylori is a Gram-negative bacterium, with which half of the world’s population is infected. This species is a Class I carcinogen that colonizes the human stomach, wherein it can induce inflammatory disorders (chronic gastritis and ulceration) and malignant neoplastic diseases (mucosa-associated lymphoid tissue and gastric cancer). H. pylori has mechanisms to survive the human stomach, which is a very harsh environment in which it is difficult for bacteria to survive. As a first line of defense, urease is produced by H. pylori, which results in the generation of ammonia to neutralize the acidic condition. Subsequently, H. pylori attaches to gastric epithelial cells via OMPs. The outer membrane of H. pylori plays a pivotal role in the attachment and colonization of gastric cells. After colonization, H. pylori pathogenesis results in inflammation. H. pylori-infected patients exhibit inflammation of the gastric mucosa, which can result in metaplasia. Finally, some H. pylori-infected patients develop gastric cancer; however, this typically requires long-term, persistent infection.
Among the virulence factors of H. pylori, cytotoxin-associated gene A product (CagA), urease, γ-glutamine transferase, high temperature requirement A, and vacuolation associated gene A (VacA) were identified as secreted [3]. Specifically, CagA is injected into human gastric cells via the Type IV secretion system (T4SS) of H. pylori to induce the production of inflammatory cytokines in gastric epithelial cells [4,5,6]. Recently, many studies have suggested that the OMPs of H. pylori play a role not only in the attachment to gastric cells but also in the enhancement of CagA translocation via the T4SS. In 1997, the genome of H. pylori strain 26695 was sequenced completely, and it was suggested to encode 32 OMPs [7]. Subsequent studies revealed that sialic acid-binding adhesin (SabA), blood-group-antigen-binding adhesin (BabA), adherence-associated lipoprotein A and B (AlpA/B), outer inflammatory protein A (OipA), and Helicobacter outer membrane protein Q (HopQ) were involved in adhesion (Table 1). Therefore, OMPs are important for colonization and the establishment of inflammation in the stomach. The functions of OMPs are summarized in this review to provide an overview of how H. pylori utilizes membrane proteins to colonize the stomach, a process that results in pathogenesis. Additionally, the evidence suggesting that OMPs cooperate with the T4SS to translocate CagA into gastric cells is summarized. The biological function of CagA protein has been revealed; however, how OMPs cooperate with Type IV secretion system is unclear. Thus, this review contributes to a deeper understanding of the dynamic action between OMP and CagA translocation via the T4SS.
Table 1.
Helicobacter pylori outer membrane proteins that interact with the host cell.
| Outer Membrane Protein | Interaction Partner | Suggested Protein Function |
|---|---|---|
| BabA (HopS) | Lewis B, Globo H hexaglycosylceramide, Globo A heptaglycosylceramide |
Adhesion to host cell, enhancing translocation of CagA via the T4SS |
| SabA (HopP) | Sialyl Lewis X, Sialyl Lewis A, Lewis X | Adhesin to host cell |
| OipA (HopH) | Not known | Adhesion, induction of inflammatory cytokine production |
| HopQ | CEACAM1, 3, 5, 6 | Adhesion to host cell, translocation of CagA via the T4SS |
| AlpA/B (HopC/B) | Collagen IV, Laminin | Adhesion to ECM |
2. The Genetic Features of Helicobacter OMPs
Members of the large Helicobacter outer membrane protein (Hop) family were first characterized as OMPs in H. pylori. Five proteins (HopA–E) were isolated and these were demonstrated to function as porins for transmitting small hydrophobic molecules, nutrients, and some antibiotics by passive diffusion [8,9]. The genome of H. pylori 26695 was completely sequenced, and subsequent analysis identified 21 members of the Hop family; these genes encoded proteins that possess one domain of similarity at the N-terminal end and seven domains of similarity at their C-terminal ends [7]. A C-terminal phenylalanine residue is essential for the correct assembly of bacterial OMPs [10,11]. However, half of the Hop family proteins contain a C-terminal tyrosine. In addition, all Hop proteins possess highly similar C-termini, which contain aromatic residues. With phenylalanine at their C-terminal ends, Hop proteins are less divergent in this region, and thus the central hyper-variable domain contains the majority of the membrane-specific sequence. Indeed, for several Hop proteins with phenylalanine at the C-terminal end, the similarity of this region between paralogues within a strain is higher than that of orthologous proteins between isolates. H. pylori also contains orthologues of hop-related (Hor) protein, Helicobacter OMP family (Hof) protein, and Helicobacter outer membrane (Hom) family proteins. The functions of these proteins have not been revealed.
Most Hop proteins are predicted to fold into anti-parallel amphipathic β-sheets that organize into a β-barrel structure. Typically, β-sheet domains localize to the membrane region, and these domains are connected by short amino acid loops in the periplasmic region, whereas longer amino acid loops connect these domains in the extracellular region. It is known that the membrane domain is difficult to overexpress and purify. For H. pylori, the extracellular domain of OMP was recently purified and crystallized. However, the full length OMP has not yet been purified for H. pylori.
3. AlpA/B
AlpA and AlpB are involved in the adhesion of the pathogen to gastric epithelial cells [12]. Collagen IV and laminin, which exist in the extracellular matrix, are the suggested binding partners of BabA and BabB [13,14]. Both genes have high homology, specifically 46.7% in H. pylori 26695, and both proteins are important for adhesion to gastric cells [7]. In addition, Odenbreit et al. reported that H. pylori expresses AlpA and AlpB; however, all other OMPs (BabA, BabB, BabC, HopM, SabA, and OipA) were shown to be produced depending on clinical isolate strains [15]. This indicates that AlpA and AlpB are essential and play a main role in colonization. Subsequently, studies demonstrated that AlpA- and AlpB-mutant H. pylori strains poorly colonize the stomachs of guinea pigs, mice, and Mongolian gerbils [14,16,17]. Thus, it is clear that these factors are important for bacterial colonization. However, the related role in pathogenesis of AlpA and AlpB is controversial. Lu et al. suggested that these proteins do not affect CagA translocation via the T4SS and that they are involved in the stimulation of certain signaling pathways (MAPKs, c-Fos, c-Jun, CREB, AP-1, and NF-κB pathway), which can induce production of interleukin-8 (IL-8). Furthermore, these factors are associated with a reduction in chemokine KC and interleukin-6 in the gastric mucosa [18]. In contrast, AlpA- and AlpB-mutant H. pylori caused a more severe inflammatory reaction in Mongolian gerbils than the wild type [16]. The reason for the contradictory results might be due to differences in host animal species or H. pylori strains.
4. SabA
H. pylori adhesion via SabA is important for colonization and the induction of inflammation in the gastric mucosa. Sialyl LewisX/a glycosphingolipid (sLex and sLea) was identified as a receptor for a 70 kDa protein in H. pylori, and this was named SabA [18,19,20]. Recent reports revealed the three-dimensional structure of the extracellular domain of SabA and identified the domain responsible for binding to sLex and LewisX antigen [21]. The functional status of SabA is regulated by slipped strand mispairing, which is determined by the number of CT dinucleotide repeats in the 5′ region of the gene. Switch “on” means that SabA is functional, whereas Switch “off” means that it is non-functional [7,22,23].
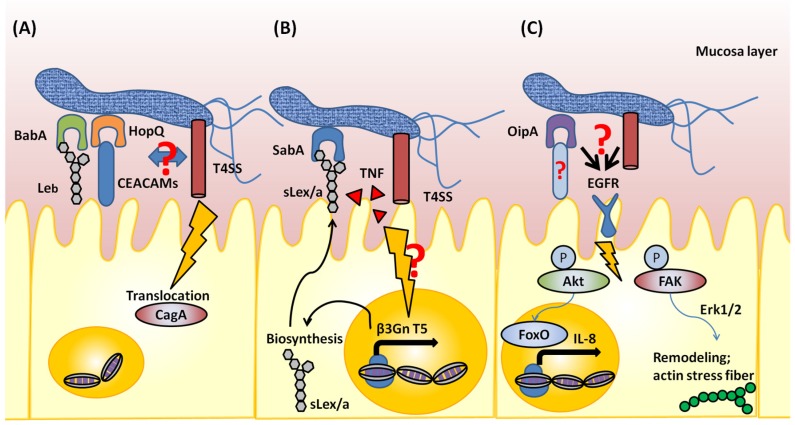
The adhesion process via SabA is dynamic and co-operates with the T4SS and TNF. In fact, sLex antigen is absent in healthy gastric mucosa, and it was previously unclear how H. pylori uses sLex antigen for adhesion. The T4SS and TNF is suggested to induce sLex expression. Marcos et al. demonstrated that cag PAI-positive H. pylori induces β3 GlcNAcT5 (β3GnT5) expression in gastric epithelial cells. β3GnT5 is a glycosyltransferase and is essential for the biosynthesis of the Lewis antigens. Furthermore, the induction of β3GnT5 is dependent on TNF, but not on IL-8. In regard to the binding capacity of H. pylori via SabA, β3GnT5 expression was shown to increase the cell adhesion rates for SabA-positive H. pylori strains [24]. In addition, acid-responsive signaling in H. pylori regulates SabA transcription [25]. These data indicate that H. pylori adaptation to the environment is a precursor of adhesion via SabA; subsequently, inflammation might support H. pylori colonization of gastric epithelial cells (Figure 1).
Figure 1.
Schematic of outer membrane protein-mediated pathogenesis. (A) BabA interacts with Lea antigen and enhances CagA translocation via the Type 4 secretion system (T4SS). HopQ interacts with CEACAMs, and is essential for CagA translocation. BabA and HopQ might interact with the T4SS in the membrane region of H. pylori, although this is unclear. (B) Absence of sLex antigen expression in the healthy stomach. H. pylori infection induces β3GnT5 expression in gastric epithelial cells and biosynthesis of the sLex antigen; sLex localizes to the membrane region of gastric epithelial cells. As a result, H. pylori can colonize by utilizing SabA, which interacts with the sLex antigen. Although the detailed mechanism is unclear, TNF and the T4SS are suggested to induce β3GnT5 expression. (C) OipA is suggested to induce phosphorylation of EGFR, leading to activation of focal adhesion kinase (FAK) and Akt-related signaling. Activated FAK induces actin stress fiber remodeling via the MAPK and Erk1/2 signaling pathway. In addition, phosphorylation of Akt can activate FoxO transcription factors and induce IL-8 production. The binding partner of OipA and whether OipA can cooperate with the T4SS are unclear.
5. BabA
BabA was identified as the first adhesin molecule of H. pylori, and this protein can bind to the Lewis B (Leb) blood group antigen [26,27]. Subsequent studies demonstrated that BabA has adapted to fucosylated blood group antigens, which are most prevalent in certain local populations [28]. In Europe and the United States, where Blood Groups A, B, and O are common, H. pylori strains can bind to Blood Groups A, B, and O Type 1 determinants (H Type 1 determinants); this is referred to as a “generalist strain.” Meanwhile, a “specialist strain” can bind to H Type 1 determinants only. A specialist strain was found in the South American native population, in which only the Blood Group O phenotype is present. Recently, the Globo H hexaglycosylceramide, e.g., the Blood Group O determinant on a Type 4 core chain, was suggested to be a novel glycosphingolipid binding partner for BabA [29]. Both strains can bind the Globo H hexaglycosylceramide; however, only the generalist strain can bind the Globo A heptaglycosylceramide, e.g., the Blood Group A determinant on a Type 4 core chain.
BabA is suggested to enhance CagA translocation into gastric cells and induce severe inflammation in the stomach (Figure 1) [30]. Triple-positive (BabA, vacA-s1 type, CagA) H. pylori can colonize a greater proportion of the gastric mucosa, and induce severe inflammation in the stomach [31]. In addition, triple-positive H. pylori-infected patients exhibit a higher incidence of intestinal metaplasia, compared to that in patients infected with double-positive (vacA-s1 type, CagA) H. pylori [32]. Recently, the extracellular domain of BabA was crystallized, and this provided new insights into the protein features. Moonens et al. identified the domain of BabA that is responsible for interaction with blood group antigens. Furthermore, N-acetylcysteine (NAC), a redox-active compound, was shown to disrupt the disulfide bond of the interaction domain, inactivate the binding properties of BabA, and thus block BabA-mediated adherence to the gastric mucosa of mice [33]. In addition, treatment of H. pylori-infected Leb-expressing mice with NAC lowers gastric mucosal neutrophil infiltration. This might provide the basis for possible H. pylori eradication therapies.
6. OipA
OipA was initially identified as a protein that induces a pro-inflammatory response based on the fact that an oipA mutant resulted in the reduced production of IL-8 in a gastric epithelial cell line [34]. In contrast, some studies have indicated that OipA plays a role in H. pylori adhesion to host cells and does not influence the production of IL-8 in gastric cells [35,36]. Whether OipA can induce IL-8 production is controversial. The functional status of OipA is regulated by slipped strand mispairing, which is the same mechanism utilized by SabA [34,35,36,37]. Many OMP family genes demonstrate a loss of one resident locus (hopM/N, babA/B/C, and sabA/B), whereas it was reported that the OipA locus was duplicated, referred to as OipA-2 [38]. OipA-2 is found in H. pylori hspEAsia and hspAmerind strains, but not in hpEurope or hspWAfrica strains, based on multi-locus sequence typing analysis.
The role of OipA as an adhesion molecule is suggested by in vitro experiments using gastric epithelial cells [34,35,39]. However, results in an animal model indicate that the adhesive action of OipA is dependent on the H. pylori strain. Akanuma and colleagues reported that the oipA-mutant H. pylori TN2 strain does not infect Mongolian gerbils [40]. In contrast, Franco and co-workers reported that the oipA-mutant H. pylori 7.13 strain infected Mongolian gerbils; however, oipA mutant strain-infected animals did not develop gastric cancer, whereas 27% of those infected with the wild-type strain did develop this condition [41]. Thus, the adhesion of OipA depends on H. pylori strain and controversial. Therefore, it is difficult to demonstrate the relationships between OipA status and molecular processes as well as clinical outcomes.
Based on epidemiological studies, functional OipA correlates with other virulence factors such as cag PAI, vacA-s1 type, and vacA-m1 type [35,42,43,44]. Therefore, it is difficult to demonstrate the direct function of OipA by using mutant H. pylori strains; however, OipA-related signaling has been reported. OipA is suggested to induce the remodeling of actin stress fibers by activating the phosphorylation of focal adhesion kinase (FAK); this leads to the activation of extracellular signal-regulated kinases 1 and 2 (Erk1/2) [45]. OipA-mediated FAK activation might be a consequence of altered epidermal growth factor receptor (EGFR) signaling. oipA mutant H. pylori could not activate EGFR-related signaling involving phosphatidylinositol 3-kinase (PI3K) [45], phosphoinositol dependent kinase-1 (PDK1), and Akt, which regulate FoxO forkhead transcription factor activity, leading to IL-8 induction [46]. Recently, EGFR/FAK signaling was shown to lead to phosphorylation of paxillin and subsequent actin remodeling (Figure 1) [47].
It is difficult to demonstrate the relationships between OipA status and clinical outcomes. However, it is clear that OipA status is associated with the presence of the other virulence factors in H. pylori. Therefore, OipA might cooperate with other virulence factors during the pathogenesis of H. pylori.
7. HopQ
HopQ is a paralog of the Hop family that includes BabA and SabA [48]. HopQ is classified into Type I and Type II alleles, and these alleles correlate with other virulence factors. Ping and Timothy reported that Type I HopQ alleles are significantly more common in cagA-positive and vacA-s1 type strains than in cagA-negative and vacA-s2 strains [49]. Furthermore, based on an epidemiological study, it was revealed that Type I HopQ alleles are more common in East Asian H. pylori strains [50]. This indicates the presence of geographic differences for the HopQ allele type, as with vacA and cagA genotypes.
HopQ was reported as a non-cagPAI-encoded co-factor of the T4SS. Belogolva et al. suggested that HopQ is essential for CagA translocation, and that deletion of HopQ reduces T4SS-dependent activation of signaling (NF-κB, MAPK signaling, and IL-8 production) [51]. Furthermore, this observation was reported in human granulocytes [52]. Recently, the carcinoembryonic antigen-related cell adhesion molecule family (CEACAMs) was defined as a group of host cell receptors for HopQ. CEACAMs have isoforms, and HopQ binds the amino-terminal IgV-like domain of human CEACAM1, CEACAM3, CEACAM5, and CEACAM6 proteins. Based on the crystal structure of the extracellular domain of HopQ, a β-hairpin insertion in the extracellular 3 + 4 helix bundle domain of HopQ is important for CEACAM binding, and this determines the type of HopQ allele. A peptide derived from this domain competitively inhibits HopQ-mediated Cag translocation, similar to that observed with genetic or antibody-mediated abrogation of HopQ function [53,54]. This suggests the possibility that CagA translocation is mediated not only by the T4SS, but also by membrane proteins (Figure 1). The CEACAM-binding site of HopQ has been found to exhibit sequence diversity. Therefore, such variations might affect the translocation of CagA via the T4SS.
8. Conclusions
H. pylori expresses many different virulence factors. The function of each virulence factor has gradually been revealed. In regard to membrane proteins, the binding partners of BabA, SabA, and HopQ were identified, and subsequently BabA and HopQ were suggested to enhance CagA translocation into gastric cells via the T4SS. In addition, many epidemiological studies have demonstrated that OMPs correlate with other virulence factors and clinical outcomes. Therefore, these proteins might have a dynamic function in cooperating with the T4SS and other factors. However, less is known about the interplay between membrane proteins and the membrane region of H. pylori. Recently, it is clear that outer membrane vesicles (OMVs) have a unique function. OMVs are involved with periplasmic and outer membrane proteins, peptideglycanes, lipopiolysaccharides, DNA, RNA, and enzymes. OMVs are also involved in a mechanism to transfer virulence factors and maintain bacteria communities. Thus, OMPs at MOVs might play an important role in communicating with bacteria. Studies that focus on local or temporal interactions among virulence factors and the other molecules are required for a deeper understanding of the dynamic pathogenesis and communication of H. pylori.
Many studies demonstrated that H. pylori utilize OMPs to colonize the human stomach. This indicates that OMPs might be a vaccine candidate. In fact, many reports suggested that OMPs might be a candidate as a vaccine based on in vitro experiments. However, there has been no report that shows that OMPs are useful as target molecules for vaccines in animal models. An animal model is essential to demonstrate the efficacy of a vaccine. Thus, this problem needs to be overcome to produce a vaccine for H. pylori infection.
Acknowledgments
We sincerely apologize to all authors whose important work is not cited in this review due to page limitations. This report was based on work supported in part by grants from the National Institutes of Health (DK62813) (Yoshio Yamaoka) and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (25293104, 26640114 and 15H02657) (Yoshio Yamaoka).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.McConnell M.J., Actis L., Pachón J. Acinetobacter baumannii: Human infections; factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013;37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 2.Wu X., Chavez J.D., Schweppe D.K., Zheng C., Weisbrod C.R., Eng J.K., Murali A., Lee S.A., Ramage E., Gallagher L.A., et al. In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat. Commun. 2016;11:13414. doi: 10.1038/ncomms13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posselt G., Backert S., Wessler S. The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun. Signal. 2013;7:77. doi: 10.1186/1478-811X-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terradot L., Waksman G. Architecture of the Helicobacter Cag-type IV secretion system. FEBS J. 2011;278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- 5.Backert S., Fronzes R., Waksman G. VirB2 and VirB5 proteins: Specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 2008;16:409–413. doi: 10.1016/j.tim.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Fischer W. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 2011;278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 7.Exner M.M., Doig P., Trust T.J., Hancock R.E. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 1995;63:1567–1572. doi: 10.1128/iai.63.4.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doig P., Exner M.M., Hancock R.E., Trust T.J. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 1995;177:5447–5452. doi: 10.1128/jb.177.19.5447-5452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struyvé M., Moons M., Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-F. [DOI] [PubMed] [Google Scholar]
- 10.De Cock H., Struyvé M., Kleerebezem M., van der Krift T. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J. Mol. Biol. 1997;269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 11.Tomb J.F., White O., Kerlavage A.R., Clayton R.A., Sutton G.G., Fleischmann R.D., Ketchum K.A., Klenk H.P., Gill S., Dougherty B.A., et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 12.Odenbreit S., Till M., Hofreuter D., Faller G., Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- 13.Trust T.J., Doig P., Emödy L., Kienle Z., Wadström T., O’Toole P. High-affinity binding of the basement membrane proteins collagen type IV and laminin to the gastric pathogen Helicobacter pylori. Infect. Immun. 1991;59:4398–4404. doi: 10.1128/iai.59.12.4398-4404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senkovich O.A., Yin J., Ekshyyan V., Conant C., Traylor J., Adegboyega P., McGee D.J., Rhoads R.E., Slepenkov S., Testerman T.L. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect. Immun. 2011;79:3106–3116. doi: 10.1128/IAI.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odenbreit S., Swoboda K., Barwig I., Ruhl S., Borén T., Koletzko S., Haas R. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect. Immun. 2009;77:3782–3790. doi: 10.1128/IAI.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H., Wu J.Y., Beswick E.J., Ohno T., Odenbreit S., Haas R., Reyes V.E., Kita M., Graham D.Y., Yamaoka Y. Functional and intracellular signaling differences associated with the Helicobacter pylori AlpAB adhesin from Western and East Asian strains. J. Biol. Chem. 2007;282:6242–6254. doi: 10.1074/jbc.M611178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Jonge R., Durrani Z., Rijpkema S.G., Kuipers E.J., van Vliet A.H., Kusters J.G. Role of the Helicobacter pylori outer-membrane proteins AlpA and AlpB in colonization of the guinea pig stomach. J. Med. Microbiol. 2004;53 Pt 5:375–379. doi: 10.1099/jmm.0.45551-0. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber S., Konradt M., Groll C., Scheid P., Hanauer G., Werling H.O., Josenhans C., Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. USA. 2004;101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aspholm M., Olfat F.O., Nordén J., Sondén B., Lundberg C., Sjöström R., Altraja S., Odenbreit S., Haas R., Wadström T., et al. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. 2006;2:e110. doi: 10.1371/journal.ppat.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahdavi J., Sondén B., Hurtig M., Olfat F.O., Forsberg L., Roche N., Angstrom J., Larsson T., Teneberg S., Karlsson K.A., et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang S.S., Nguyen S.T., Perry A.J., Day C.J., Panjikar S., Tiralongo J., Whisstock J.C., Kwok T. The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori. J. Biol. Chem. 2014;289:6332–6340. doi: 10.1074/jbc.M113.513135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alm R.A., Ling L.S., Moir D.T., King B.L., Brown E.D., Doig P.C., Smith D.R., Noonan B., Guild B.C., deJonge B.L., et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 23.Oh J.D., Kling-Bäckhed H., Giannakis M., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: Evolution during disease progression. Proc. Natl. Acad. Sci. USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcos N.T., Magalhães A., Ferreira B., Oliveira M.J., Carvalho A.S., Mendes N., Gilmartin T., Head S.R., Figueiredo C., David L., et al. Helicobacter pylori induces β3GnT5 in human gastric cell lines; modulating expression of the SabA ligand sialyl-Lewis X. J. Clin. Investig. 2008;118:2325–2336. doi: 10.1172/JCI34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodwin A.C., Weinberger D.M., Ford C.B., Nelson J.C., Snider J.D., Hall J.D., Paules C.I., Peek R.M., Jr., Forsyth M.H. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology. 2008;154 Pt 8:2231–2240. doi: 10.1099/mic.0.2007/016055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilver D., Arnqvist A., Ogren J., Frick I.M., Kersulyte D., Incecik E.T., Berg D.E., Covacci A., Engstrand L., Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 27.Borén T., Falk P., Roth K.A., Larson G., Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 28.Aspholm-Hurtig M., Dailide G., Lahmann M., Kalia A., Ilver D., Roche N., Vikström S., Sjöström R., Lindén S., Bäckström A., et al. Functional adaptation of BabA: The H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- 29.Benktander J., Ångström J., Breimer M.E., Teneberg S. Redefinition of the Carbohydrate Binding Specificity of Helicobacter pylori BabA Adhesin. J. Biol. Chem. 2012;287:31712–31724. doi: 10.1074/jbc.M112.387654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishijima N., Suzuki M., Ashida H., Ichikawa Y., Kanegae Y., Saito I., Borén T., Haas R., Sasakawa C., Mimuro H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 2011;286:25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rad R., Gerhard M., Lang R., Schöniger M., Rösch T., Schepp W., Becker I., Wagner H., Prinz C. The Helicobacter pylori blood group antigen-binding adhesin facilitates bacterial colonization and augments a nonspecific immune response. J. Immunol. 2002;168:3033–3041. doi: 10.4049/jimmunol.168.6.3033. [DOI] [PubMed] [Google Scholar]
- 32.Gerhard M., Lehn N., Neumayer N., Borén T., Rad R., Schepp W., Miehlke S., Classen M., Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moonens K., Gideonsson P., Subedi S., Bugaytsova J., Romaõ E., Mendez M., Nordén J., Fallah M., Rakhimova L., Shevtsova A., et al. Structural insights into polymorphic ABO glycan binding by Helicobacter pylori. Cell Host Microbe. 2016;19:55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaoka Y., Kwon D.H., Graham D.Y. A Mr 34,000 proinflammatory outer membrane protein (OipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dossumbekova A., Prinz C., Mages J., Lang R., Kusters J.G., Van Vliet A.H., Reindl W., Backert S., Saur D., Schmid R.M., et al. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: Genetic and functional genomic analysis of hopH gene polymorphisms. J. Infect. Dis. 2006;194:1346–1355. doi: 10.1086/508426. [DOI] [PubMed] [Google Scholar]
- 36.Odenbreit S., Kavermann H., Püls J., Haas R. CagA tyrosine phosphorylation and interleukin-8 induction by Helicobacter pylori are independent from AlpAB, HopZ and Bab group outer membrane proteins. Int. J. Med. Microbiol. 2002;292:257–266. doi: 10.1078/1438-4221-00205. [DOI] [PubMed] [Google Scholar]
- 37.Saunders N.J., Peden J.F., Hood D.W., Moxon E.R. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 38.Kawai M., Furuta Y., Yahara K., Tsuru T., Oshima K., Handa N., Takahashi N., Yoshida M., Azuma T., Hattori M., et al. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11:104. doi: 10.1186/1471-2180-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaoka Y., Kudo T., Lu H., Casola A., Brasier A.R., Graham D.Y. Role of interferon-stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology. 2004;126:1030–1043. doi: 10.1053/j.gastro.2003.12.048. [DOI] [PubMed] [Google Scholar]
- 40.Akanuma M., Maeda S., Ogura K., Mitsuno Y., Hirata Y., Ikenoue T., Otsuka M., Watanabe T., Yamaji Y., Yoshida H., et al. The evaluation of putative virulence factors of Helicobacter pylori for gastroduodenal disease by use of a short-term Mongolian gerbil infection model. J. Infect. Dis. 2002;185:341–347. doi: 10.1086/338772. [DOI] [PubMed] [Google Scholar]
- 41.Franco A.T., Johnston E., Krishna U., Yamaoka Y., Israel D.A., Nagy T.A., Wroblewski L.E., Piazuelo M.B., Correa P., Peek R.M., Jr. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaoka Y., Kikuchi S., el-Zimaity H.M., Gutierrez O., Osato M.S., Graham D.Y. Importance of Helicobacter pylori OipA in clinical presentation; gastric inflammation; and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 43.Markovska R., Boyanova L., Yordanov D., Gergova G., Mitov I. Helicobacter pylori OipA genetic diversity and its associations with both disease and cagA, vacA s, m, and i alleles among Bulgarian patients. Diagn. Microbiol. Infect. Dis. 2011;71:335–340. doi: 10.1016/j.diagmicrobio.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Ando T., Peek R.M., Pride D., Levine S.M., Takata T., Lee Y.C., Kusugami K., van der Ende A., Kuipers E.J., Kusters J.G., et al. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 2002;40:239–246. doi: 10.1128/JCM.40.1.239-246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabassam F.H., Graham D.Y., Yamaoka Y. OipA plays a role in Helicobacter pylori-induced focal adhesion kinase activation and cytoskeletal re-organization. Cell. Microbiol. 2008;10:1008–1020. doi: 10.1111/j.1462-5822.2007.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabassam F.H., Graham D.Y., Yamaoka Y. Helicobacter pylori-associated regulation of forkhead transcription factors FoxO1/3a in human gastric cells. Helicobacter. 2012;17:193–202. doi: 10.1111/j.1523-5378.2012.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabassam F.H., Graham D.Y., Yamaoka Y. Paxillin is a novel cellular target for converging Helicobacter pylori-induced cellular signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:G601–G611. doi: 10.1152/ajpgi.00375.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alm R.A., Bina J., Andrews B.M., Doig P., Hancock R.E., Trust T.J. Comparative genomics of Helicobacter pylori: Analysis of the outer membrane protein families. Infect. Immun. 2000;68:4155–4168. doi: 10.1128/IAI.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao P., Cover T.L. Two different families of hopQ alleles in Helicobacter pylori. J. Clin. Microbiol. 2002;40:4504–4511. doi: 10.1128/JCM.40.12.4504-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao P., Lee K.J., Blaser M.J., Cover T.L. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol. Lett. 2005;251:37–43. doi: 10.1016/j.femsle.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 51.Belogolova E., Bauer B., Pompaiah M., Asakura H., Brinkman V., Ertl C., Bartfeld S., Nechitaylo T.Y., Haas R., Machuy N., et al. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell. Microbiol. 2013;15:1896–1912. doi: 10.1111/cmi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Busch B., Weimer R., Woischke C., Fischer W., Haas R. Helicobacter pylori interferes with leukocyte migration via the outer membrane protein HopQ and via CagA translocation. Int. J. Med. Microbiol. 2015;305:355–364. doi: 10.1016/j.ijmm.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Javaheri A., Kruse T., Moonens K., Mejías-Luque R., Debraekeleer A., Asche C.I., Tegtmeyer N., Kalali B., Bach N.C., Sieber S.A., et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016;2:16189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- 54.Königer V., Holsten L., Harrison U., Busch B., Loell E., Zhao Q., Bonsor D.A., Roth A., Kengmo-Tchoupa A., Smith S.I., et al. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat. Microbiol. 2016;2:16188. doi: 10.1038/nmicrobiol.2016.188. [DOI] [PubMed] [Google Scholar]