Abstract
The cytotoxicity of the venom of 25 species of Old World elapid snake was tested and compared with the morphological and behavioural adaptations of hooding and spitting. We determined that, contrary to previous assumptions, the venoms of spitting species are not consistently more cytotoxic than those of closely related non-spitting species. While this correlation between spitting and non-spitting was found among African cobras, it was not present among Asian cobras. On the other hand, a consistent positive correlation was observed between cytotoxicity and utilisation of the defensive hooding display that cobras are famous for. Hooding and spitting are widely regarded as defensive adaptations, but it has hitherto been uncertain whether cytotoxicity serves a defensive purpose or is somehow useful in prey subjugation. The results of this study suggest that cytotoxicity evolved primarily as a defensive innovation and that it has co-evolved twice alongside hooding behavior: once in the Hemachatus + Naja and again independently in the king cobras (Ophiophagus). There was a significant increase of cytotoxicity in the Asian Naja linked to the evolution of bold aposematic hood markings, reinforcing the link between hooding and the evolution of defensive cytotoxic venoms. In parallel, lineages with increased cytotoxicity but lacking bold hood patterns evolved aposematic markers in the form of high contrast body banding. The results also indicate that, secondary to the evolution of venom rich in cytotoxins, spitting has evolved three times independently: once within the African Naja, once within the Asian Naja, and once in the Hemachatus genus. The evolution of cytotoxic venom thus appears to facilitate the evolution of defensive spitting behaviour. In contrast, a secondary loss of cytotoxicity and reduction of the hood occurred in the water cobra Naja annulata, which possesses streamlined neurotoxic venom similar to that of other aquatic elapid snakes (e.g., hydrophiine sea snakes). The results of this study make an important contribution to our growing understanding of the selection pressures shaping the evolution of snake venom and its constituent toxins. The data also aid in elucidating the relationship between these selection pressures and the medical impact of human snakebite in the developing world, as cytotoxic cobras cause considerable morbidity including loss-of-function injuries that result in economic and social burdens in the tropics of Asia and sub-Saharan Africa.
Keywords: cytotoxin, cobra, Hemachatus, Naja, Ophiophagus, Elapidae, evolution, antipredator defense
1. Introduction
Members of the front-fanged venomous snake family Elapidae are present throughout the tropical and temperate parts of the Old and New World. These snakes are responsible for many clinically significant bites to humans including fatalities. Species with potently cytotoxic venom, particularly members of the genus Naja (cobras) in Africa and tropical Asia, are major causes of loss-of-function injuries, which may render survivors unable to work [1,2,3]. Little reliable data exists for the incidence of morbidity following snakebite in developing countries, but it is predicted that the morbidity and mortality burden is considerable [4]. The permanent sequelae resulting from bites by cytotoxic species of snake can result in lifelong disabilities that render victims unable to perform manual labour. As snakebite is a disease that primarily affects the poor [5], and as young males are at highest risk [6,7], cytotoxic envenomations generate a considerable socioeconomic burden in developing countries. Understanding the evolutionary pressure that shapes the activity of snake venoms may aid attempts to address the snakebite crisis in sub-Saharan Africa and southern Asia both by providing valuable insights into the relationship of snake behaviour to venom activity and by assisting the development of effective and affordable snakebite therapeutics. Currently, the evolution of elapid snake venom, including its connection to behavioural and morphological patterns, is poorly understood. Specifically, little is known concerning the selection pressures that resulted in the evolution of potent cytotoxicity within the elapid snake family, particularly among cobras. Why snakes from a family typified by venoms rich in potent neurotoxins, which are devastatingly effective in prey subjugation [8,9], should evolve less potent cytotoxins and why this should have occurred primarily in one clade (cobras and their close relatives), are intriguing and important questions for evolutionary toxinology to address.
Elapidae is a large and diverse family which underwent a major radiation at the end of the Eocene period [10], spreading over much of Africa and southern Asia and ultimately reaching Australasia approximately 25 million years ago [11]. These fast moving snakes typically rely on cryptic colouration to remain undetected by predators during periods of rest. However, some lineages (Naja and closely related genera) have evolved defensive behaviour characterised by the extension of “hoods”, formed when elongate ribs in the neck are spread, which often reveal bright colours and intricate patterns [12]. This morphological and behavioural adaptation also calls for a “plan B”—a defensive strategy in case the display fails [13,14]. Typically, plan B involves painful defensive bites, but the ability to spit venom is believed, on the basis of variations in morphology and associated behaviour, to have evolved three times independently within the Naja + Hemachatus clade [13,15].
Most prior research has focussed on the spitting behavior itself, treating it as a peculiar oddity in nature without investigating associated evolutionary trends (e.g., venom composition) [16], and it has been described at the morphological [17,18], mechanical [12,19,20] and behavioural levels [13,21]. Hooding is a distinctive characteristic of the genera Hemachatus, Naja and Ophiophagus and members of these genera are found in Africa and Asia [22]. Since Ophiophagus is not closely related to Naja and Hemachatus [15,17], hooding has likely evolved on at least two separate occasions. Previous studies have investigated the morpho-kinetic action of hooding [16] or simply observed that hooding is a form of defensive reaction [12] that presumably functions as an aposematic signal or intimidation display by giving the appearance of being larger.
Myriad toxins are responsible for the cytotoxicity (cell death) of snake venoms. The tissue destruction that often results from envenomation by Naja spp. is caused by a specialised class of 3-finger toxins (3FTx)—the cytotoxins or “cardiotoxins” [1]. In Ophiophagus envenomations, on the other hand, L-amino acid oxidase is responsible [23]. Despite the iconic nature of the species involved, the connection between the hooding and spitting behaviours and the cytotoxicity of the venoms has not been previously investigated. Indeed, hooding behaviour has puzzled scientists for more than 200 years [14,24]. Thus, this project aimed to test the relationship between these three character states with a view to understanding how this suite of defensive adaptations has coevolved.
2. Results and Discussion
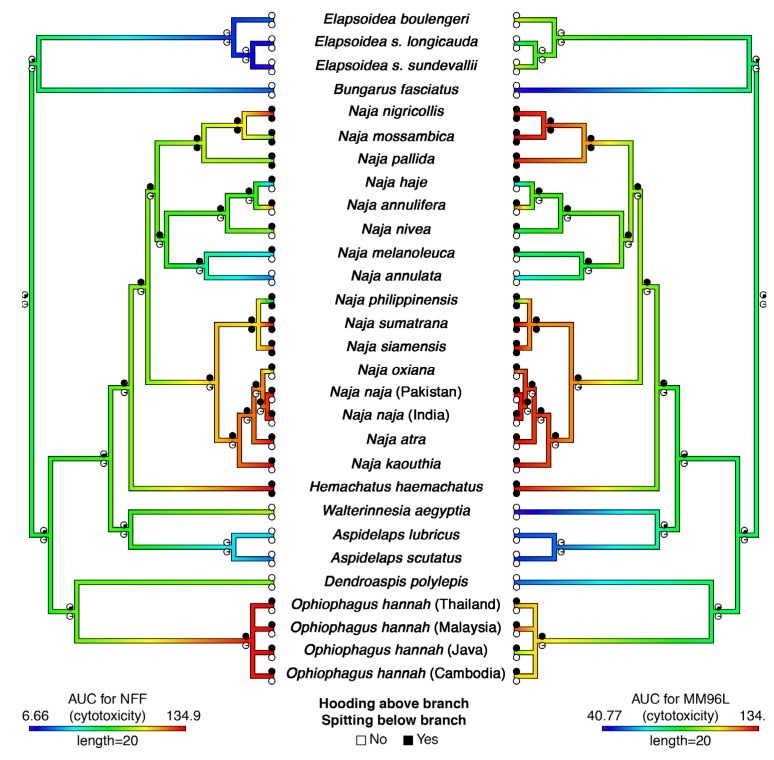
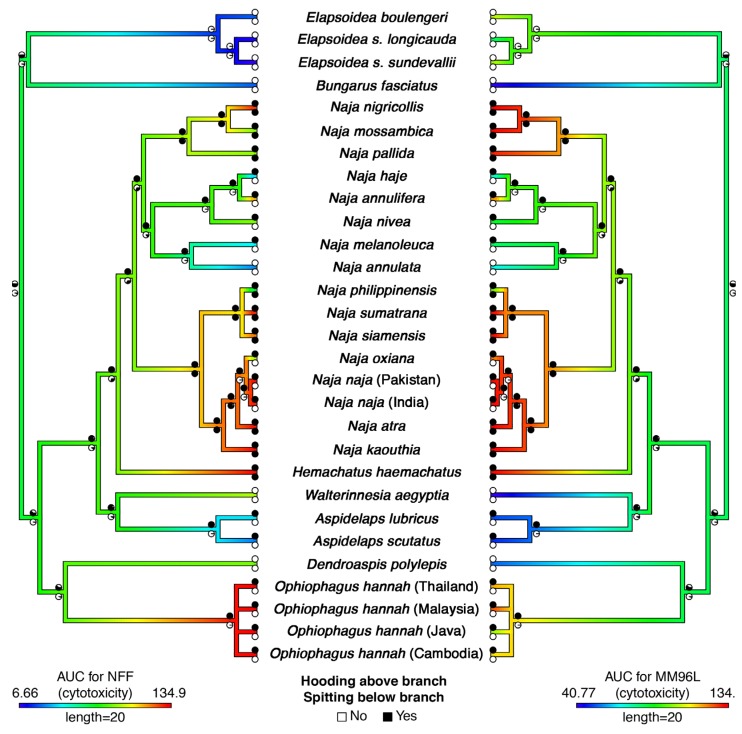
In order to account for variance by cell type, we tested the cytotoxic effects of crude venoms on one healthy-type cell line and one cancerous cell line and looked for congruence in effect between the two cell types. Colorimetric MTT testing revealed substantial variation between non-hooding and hooding species in relative toxicities across cell types and concentrations (Figure 1 and Supplementary Tables) (raw data in Supplementary Tables). Cytotoxicity of Naja venom appears to have increased on several separate occasions (Figure 1 and Supplementary Tables). Increased cytotoxicity was also observed in Hemachatus and Ophiophagus. Notably, increased cytotoxicity is not estimated to have occurred concurrently with hooding, but rather to have evolved subsequent to the evolution of hooding behavior (Figure 1 and Supplementary Tables). Exceptional hooding behavior itself is estimated to have evolved twice, once on the branch leading to Ophiophagus and once at the base of the clade Naja + Hemachatus (Figure 1). However, if weakly hooding species such as Aspidelaps are classified as ‘hooding’, we find modest evidence for an earlier single origin of hooding ability in clade comprising Aspidelaps, Hemachatus, Naja and Walterinnesia (Figure 2). According to this interpretation, Walterinnesia would have lost the propensity to hood, presumably as a result of their occupation of a nocturnal niche in which visual defensive displays are less effective.
Figure 1.
Ancestral state reconstructions of hooding, spitting, and cytotoxicity; based on ambiguous Aspidelaps species coded as non-hooding as well as the ambiguous N. atra and N. kaouthia also coded as non-spitting. Reconstruction over branches represents the AUC (area under the curve) for the non-transformed (NFF) cell line (left) and the melanoma (MM96L) cancer cell line (right), where warmer colours represent higher cytotoxicity against cell lines (raw data in Supplementary Tables). Pie charts are the same on both trees and represent estimates of ancestral states for hooding (above branch) and spitting (below branch) where black and white represent the trait being present or absent respectively. States at tips represent the data collected. Phylogeny follows Lee (2016) [11].
Figure 2.
Ancestral state reconstructions of hooding, spitting, and cytotoxicity; based on ambiguous Aspidelaps species coded as hooding as well as the ambiguous spitters N. atra and N. kaouthia also coded as spitting. Reconstruction over branches represents the AUC for the non-transformed NFF cell line (left) and the melanoma (MM96L) cancer line (right) where warmer colours represent higher cytotoxicity against that cell line (raw data in Supplementary Tables). Pie charts are the same on both trees and represent estimates of ancestral states for hooding (above branch) and spitting (below branch) where black and white represent the trait being present or absent respectively. States at tips represent the data collected. Phylogeny follows Lee (2016) [11].
Based on our ancestral state reconstructions, we found evidence that spitting behavior has evolved on three occasions (Figure 1), as predicted and based on morphological and behavioural adaptations [13,15]. To our knowledge, this is the first attempt to formally estimate the number of origins of spitting and confirms earlier suggestions by Wuster et al. (2007): one origin in Hemachatus, one in Asian Naja, and one in African Naja [25]. We found a difference in the placement of origins of spitting behavior in the Asian Naja clade based on how N. atra and N. kaouthia are coded (Figure 1 and Figure 2). These species are not considered ‘true’ spitters as they do not possess specialised morphological adaptations and do not commonly spit, however, both have been recorded to spit venom at a substantial distance on rare occasions [26,27]. When focused only on the highly specialised spitters, we find that spitting evolved at the base of the lineage of highly adapted Asian Naja (represented in this study by N. philippinensis, N. siamensis and N. sumatrana) (Figure 1). Alternatively, when including the weakly adapted cases (Figure 2), we find an origin of spitting at the base of the Asian Naja clade followed by a loss in the N. naja + N. oxiana clade Although counterintuitive at first glance, this scenario would interpret the limited spitting ability of N. atra and N. kaouthia as a vestigial relic of spitting ancestors and would also explain the increased cytotoxicity across the whole Asian Naja clade (Figure 2). Specialised morphological adaptations for spitting may also be a more recent innovation in the Asian Naja than in their African counterparts, as they are known to be less accurate spitters and the trait seems less fixed given the (at least) two species (N. atra and N. kaouthia) that contain both spitting and non-spitting individuals [28]. Nevertheless, more complete data regarding spitting behavior in these “quasi-spitters” would be required to rigorously verify this suggested scenario. Taken together, however, the evidence uncovered in the present study suggests that the evolution of hooding and defensive cytotoxic venom in turn produced the selection pressures that facilitated the evolution of spitting.
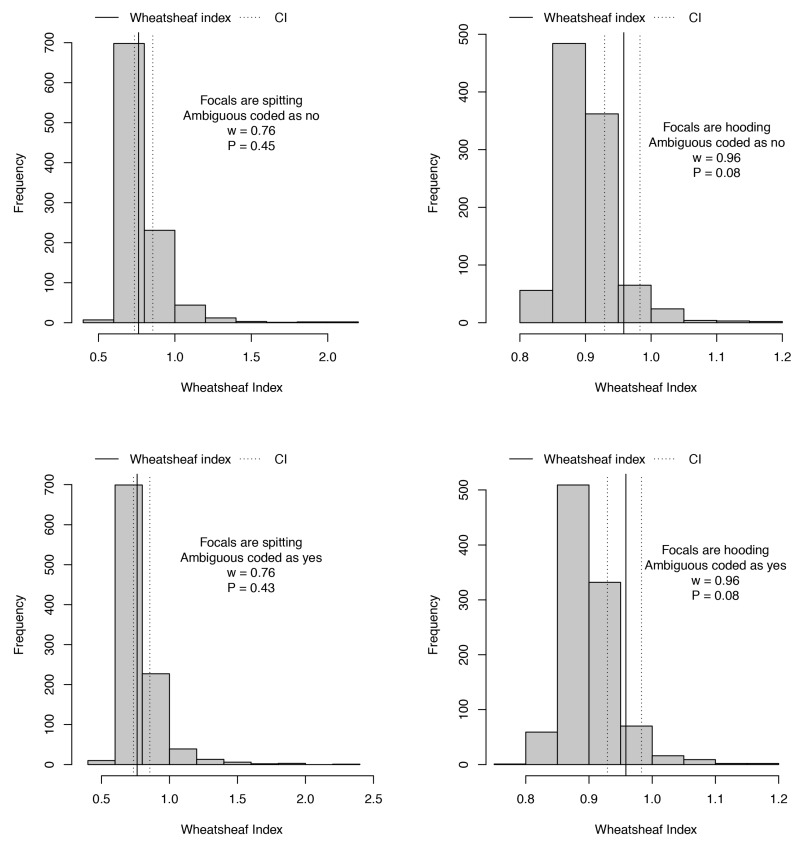
In further statistical investigations of the relative importance of hooding versus spitting in the evolution of cytotoxicity, we found that the best supported pGLS model for toxicity to both NFF and MM96L cell lines included hooding but not spitting (NFF line, ambiguous coded no: t1,27 = 3.80, p = 0.0008; MM96L line, ambiguous coded no: t1,27 = 4.70, p = 0.0001; MM96L line, ambiguous coded yes: t1,27 = 2.63, p = 0.014). These models provide complementary and consistent support for hooding being more important than spitting in the evolution of cytotoxicity. Finally, we also applied the Wheatsheaf index [29], which controls for undue weight given to close relatives and also for the possible distributions of traits over a given tree, to ask whether convergence in cytotoxicity was stronger for spitting or hooding species. Although neither trait was significantly associated with exceptionally strong convergence across the tree (Figure 3), hooding was associated with stronger convergence in cytotoxicity than was spitting (wspitting = 0.76, p = 0.44; whooding = 0.96, p = 0.08). Therefore, although it is difficult to make robust evolutionary inferences about suites of traits when each has very few origins, the consistent picture arising from the several statistical analyses utilised suggests that, in the evolution of this group of snakes, hooding is more strongly associated with cytotoxicity than spitting. Our results also display a clear link between the evolution of strong hood or body patterns, as part of enhanced visual displays, and parallel increases in cytoxicity.
Figure 3.
Analyses of strength of convergence in cytotoxicity in association with spitting or hooding behaviour, under both coding schemes for ambiguous species. Histograms show that the null distribution represented expected strengths of convergence in each analysis, with solid and dotted lines overlaid to show calculated Wheatsheaf index (w) and its 95% confidence interval respectively. Wheatsheaf index is presented alongside the p-value for exceptionally strong convergence in each case. Note that alternative codings make a little difference to results but convergence in cytotoxicity is slightly stronger (with a higher Wheatsheaf index) when associated with hooding than spitting behaviour.
The most recent common ancestor of Naja and Hemachatus appears to have possessed moderately cytotoxic venom (Figure 1) and morphologically may have resembled today’s Naja haje, which is drab in colour, displaying little or no patterning when hooding (Figure 4A,B). These snakes strike nervously and flee when possible [30]. In both cell types tested the African non-spitting Naja such as N. haje were higher in cytotoxicity than non-hooding snakes but lower than all other Naja, especially those with aposematic markings (Figure 1 and Supplementary Tables). This pattern was consistent with that seen in another cytotoxicity studies which compared N. haje with Naja mossambica and Naja nigricollis [31]. The Asian non-spitting Naja have evolved broader hoods along with aposematic hood markings (Figure 4C,D) and are notorious for standing their ground when faced with a threat. These snakes were significantly more toxic to both cell types than African non-spitting Naja (Figure 1 and Supplementary Tables). Basal African spitting cobras such as Naja nubiae (Figure 4E) and Naja ashei (Figure 4F) resemble N. haje in being drably coloured with non-descript patterning, if any. Similar, possibly plesiomorphic, colour and patterning is seen in the non-spitting Naja oxiana and spitting Naja philippinensis Asian species, both of which are nested within clades of aposematically marked species. N. oxiana is sister to the ornate N. naja while N. philippinensis is related to the brightly banded species N. siamensis. In both cases, the non-aposematically marked state is accompanied by a decrease in cytotoxicity (Figure 1 and Supplementary Tables). It is notable that the desert dwelling population of N. naja from Pakistan, which becomes melanistic as adults (Figure 4I) but displays the aposematic hood markings as juveniles and subadults, does not show a decreased cytotoxicity relative to the aposematic hood marked population studied from India.
Figure 4.
Relative degree of aposematic hood marking between (A,B) African (Naja haje) and (C,D) Asian (Naja naja) cobras with their higher levels of cytotoxicity;. Similar patterning to N. haje are seen in the the basally coloured African spitting cobras lacking aposematic marking like N. haje (E) N. nubiae and (F) N. ashei. Convergent reversal from aposematic markings to the basal drab coloured state accompanied by a lowering of cytotoxicity has occurred in (G) the Asian non-spitting cobra N. oxiana and (H) the spitting cobra N. phillipinensis. The Pakistan Sindh desert population of N. naja displays the aposematic hood marking as juveniles and subadults but not (I) as adults, without any loss of cytotoxicity. Photos: (A,B) Arno Naude; (C,D) Gowri Mallapur; (E) HG Hjim; (F) Anothony Childs; (G,H) Randy Ciuros; (I) Bryan Fry.
N. annulifera was the only African non-spitting cobra with an increase of cytotoxicity (Figure 1 and Supplementary Tables). This species is also unique amongst African non-spitting Naja in being brightly banded (Figure 5A). The evolution of aposematic body banding occurred convergently in four other lineages: Hemachatus haemachatus (Figure 5B), N. nigricincta (Figure 5C), N. siamensis (Figure 5D), and O. hannah (Figure 5E). In N. siamensis the hood markings are obscured by solid black colouring suggesting (at least for this species) an inverse relationship between aposematic hood and body markings—non-spitting Asian cobras often have aposematic hood marks (Figure 3D). Unlike African spitting cobras, the Asian spitting cobras do not display a dramatic rise in cytotoxicity to either cell type relative to aposematically marked non-spitting Asian Naja. However, as stated above, the aposematic N. siamensis displayed higher toxicity than N. philippinensis (Figure 1 and Supplementary Tables).
Figure 5.
Convergent aposematic banding in the strongly cytotoxic species (A) Naja annulifera; (B) Hemachatus haemachatus; (C) Naja nigricincta; (D) Naja siamensis; and (E) Ophiophagus hannah; Photos by (A,D) Randy Ciuros; (B) Giuseppe Mazza; (C,E) Tom Charlton.
Within the African Naja spitting cobras, N. katiensis (Figure 6A), which is a relatively dull red, has less cytotoxic venom than the scarlet N. pallida (Figure 6B) [32]. While N. mossambica (Figure 6C) and N. nigricollis (Figure 6D) both exhibit bright red aposematic markings on their necks, there is a notable difference in the location and colour of the markings. N. nigricincta represents a aposematically black and white banded derivation within this aposematically red coloured clade (Figure 6C). In both cell types the N. nigricollis venom was the most potent, which is congruent with documented effects on human bite victims. Similarly (and apparently convergently), some populations of O. hannah (e.g., those from Malaysia) in which adults are golden in colour with subtle reticulated patterns instead of aposematically banded, snakes exhibit vibrant orange aposematic colouring on the front side of the hood analogous to the red markings of the African spitting cobras (Figure 6E). The Malaysian population was also the most cytotoxic of the Ophiophagus populations tested (Figure 1 and Supplementary Tables).
Figure 6.
Aposematic hood colouring in the African spitting cobras such as (A) Naja katiensis; (B) Naja pallida; (C) Naja mossambica; (D) Naja nigricollis; and (E) convergently in the adult colouring in the Malaysian population of Ophiophagus hannah (the most cytotoxic O. hannah population). Photos (A) Stephen Spawls; (B) Wikimedia Commons; (C,D) Randy Ciuros; (E) Kevin Messenger.
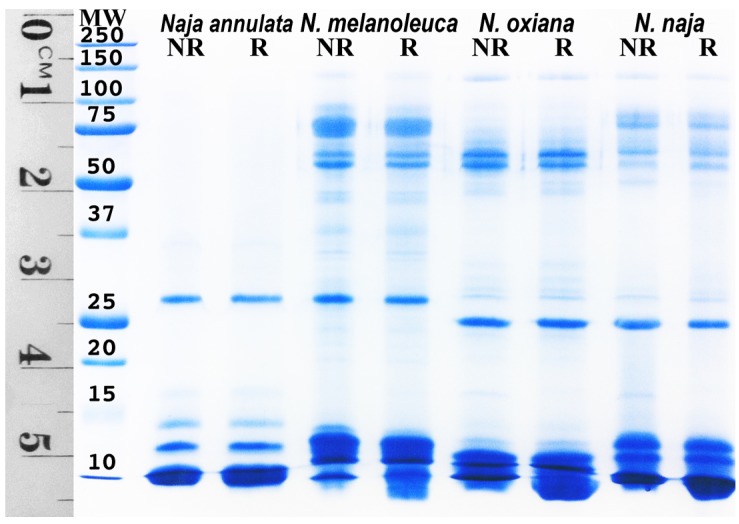
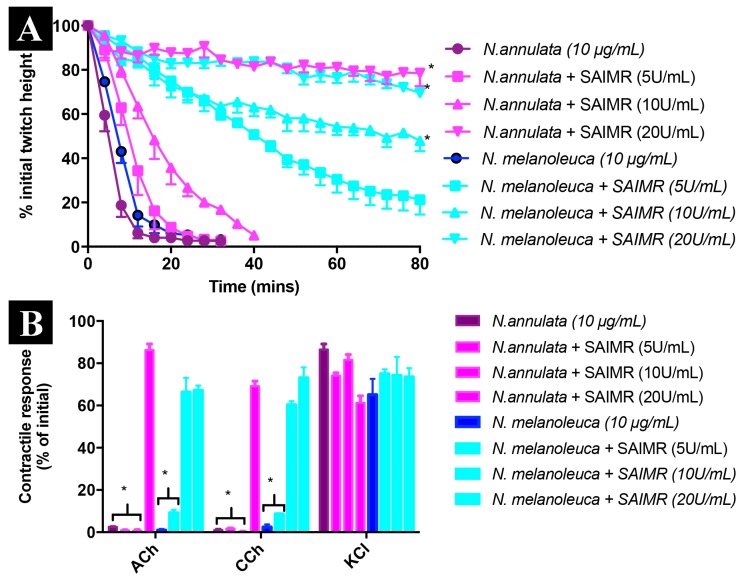
In contrast to the evolution of aposematic body banding, the banding of the water cobra N. annulata likely serves a camouflage purpose in the aquatic niche they occupy, similar to that selected for in other aquatic lineages from sea snakes to fish (Figure 7). As well as for the loss of aposematic hood markings, there would be a selection pressure in this environment for the shortening of the elongate ribs of the hood in order to increase streamlining and allow great neck flexibility for swimming and hunting fish in crevices. In both cell types tested N. annulata was significantly less cyototoxic than all other Naja (Figure 1 and Supplementary Tables). The venom of this species has also undergone a proteomic streamlining (relative to that of other Naja—Figure 8) in a manner analogous to the evolution of simplified venoms in other aquatic elapid snakes—sea kraits and sea snakes [33,34]. The absence of cytotoxins in the venom of this species is thus linked to the behavioural changes associated with occupying a new ecological niche, that of an aquatic snake that spends most if not all of its time in or around a body of water and has undergone a shift to a primarily piscivorous diet. The easy escape mechanism of fleeing into the water [35], an almost unique adaptation within Naja, may be the reason for the lack of cytotoxins in the venom of this species. The loss of strong cytotoxic activity and reduction of hooding as a defensive behaviour are both derived character states of N. annulata and this shift is reflected in the degree of relative neurotoxicity, with N. annulata being more neurotoxic than its closest relative N. melanoleuca (Figure 9A) with t90s of 10 min and 15 min respectively. The neurotoxic action was mediated by alpha-neurotoxins (Figure 9B), which are retained in N. melanoleuca. Thus, as the latter species is used in the antivenom immunizing mixture, cross-reactivity occurred with the N. annulata alpha-neurotoxins (Figure 9A).
Figure 7.
Disruptive camouflage patterning in aquatic snakes (A,B) Naja annulata which also has a secondarily extremely reduced hood and also secondarily lost its cytotoxicity; (C) Laticauda colubrina; (D) Emydocephalus annulatus; (E) Eunectes murinus and fish (F) Cichla orinocensis; (G) Salmo trutta. Photos (A) Markus Oulehla; (B,D) Wikimedia Commons; (C) Jan Messersmith; (E) Rhett A. Butler; (F) Ivan Mikolji; (G) Phil Skinner.
Figure 8.
1D SDS-PAGE mini-gel showing the streamlining of the aquatic cobra species Naja annulata in comparison to the closest relative N. melanoleuca, and the Asian non-spitting species N. oxiana and N. naja. Running conditions: NR = non-reduced, R = reduced.
Figure 9.
Neurotoxic effects of Naja annulata and Naja melanoleuca alone (10 µg/mL) and in the presence of SAIMR polyvalent antivenom (SAIMR PAV; 5, 10, 20 U/mL; n = 3) on (A) indirect twitches (B) responses to exogenous agonists the chick biventer cervicis preparation. * p < 0.05, significantly different to venom alone.
The banding and body marking of Aspidelaps is convergent with that of other nocturnal, semi-fossorial reptiles and likely functions as camouflage, not as an aposematic marker (Figure 10). While A. lubricus is significantly less cytotoxic than even N. haje, it is still notably more cytotoxic than A. scutatus (Figure 1 and Supplementary Tables). Consistent with this, if caught out in the open during the day, the orange and black banding of A. lubricus may well serve an ancillary (and perhaps epiphenomenal) aposematic function, just as aposematically banded diurnal species would benefit from the banding as camouflage during nocturnal activity periods. This “dual-purpose” of banding suggests a possible evolutionary mechanism for the origin of aposematic banding in snakes. Extrapolating from these evolutionary patterns and from the low cytotoxicity of the narrow-hooding, semi-fossorial Aspidelaps species (Figure 1 and Supplementary Tables), we hypothesise that venom of the burrowing cobra Naja multifasciata (not investigated in the present study) is likely to possess reduced cytotoxicity as it also has secondarily reduced its hood and defensive displays parallel to the evolution of a fossorial lifestyle. Similarly, the tree cobra N. goldii (not investigated in this study) also has a secondary reduction of hood and defensive displays parallel to the evolution of an arboreal lifestyle and so we likewise hypothesise a reduced cytotoxicity for its venom.
Figure 10.
Disruptive camouflage patterning the nocturnal semi-fossorial reptiles (A) Aspidelaps scutatus; (B) Aspidelaps lubricus; (C) Chionactis occipitalis; (D) Eremiascincus fasciolatus; and (E) the hypervariable Brachyurophis genus (counting from the left starting with the top row) B. approximans (1–9), B. fasciolatus (10–15) and B. semifasciatus (16–20). Photos (A) Tyrone Ping; (B) Tony Phelps; (C) Richard Cazares; (D) Wikimedia Commons; (E) Brian Bush.
Mapping these changes over the phylogenetic tree allows for a ready visualisation of this complex interplay (Figure 1). It should also be noted that while the evolution of the upright hooding display is convergent between Hemachatus + Naja and Ophiophagus, so is the evolution of gross cytotoxicity, with the specific cytotoxin types used differing between these two clades. The Naja + Hemachatus clade utilise derived 3FTx peptides (cytotoxins or “cardiotoxins”) [1], whereas Ophiophagus utilises L-amino acid oxidase (LAAO) [31] enzymes in the same functional role. Thus there is convergence between the two groups in upright hooding displays being associated with defensive cytotoxic function, and as they have evolved the function independently the underlying chemical mechanisms are not homologous. Hemachatus + Naja venoms have a high concentration of the cytotoxic 3FTx unique to this clade [36,37,38,39,40,41,42,43,44,45]. Similarly O. hannah venoms have the highest concentration of L-amino acid oxidase of any snake venom and also the most derived forms of venom L-amino acid oxidase [23,46,47,48,49,50,51]. Taken together these results are strongly indicative of a co-evolutionary relationship between hooding behaviour and cytotoxic venom.
Cytotoxic 3FTx make very little contribution to the lethal effects of cobra venom, having LD50s ranging from 3.8 to 9.7 mg/kg [52,53]. In comparison alpha-neurotoxic 3FTx (also present in the venoms of cobras) have intravenous LD50s of 0.07–0.2 mg/kg [34]. Cytotoxic 3FTx are thus likely to be considerably less effective in prey subjugation than their neurotoxic peptide ancestors. This, along with the demonstration in the present study that cytotoxicity is associated with an obvious defensive adaptation (hooding behaviour), makes a strong case that their evolution has been shaped by the defensive deployment of venom. Similar results are available for the LAAO enzymes responsible for the cytotoxicity of O. hannah venom [54], with this enzyme class having an unimpressive LD50 of 5 mg/kg [49]. Low toxicity and pain-inducing activities are properties of defensive toxins observed in other lineages [55]. The use of venom in defense by snakes has been a contentious subject, given that loss or reduction of the venom system has been observed in some species subsequent to a transition to defenseless prey [56,57,58,59,60]. It has been suggested, however, that maintenance of the venom system for a predatory function might facilitate defensive deployment [61]. Certainly, all species in the present study utilise their venom in predation, but to our knowledge this is the first study to demonstrate strong evidence of the evolution of certain snake venom components (toxins) driven largely by a defensive function. This is congruent with the streamlining of the venom (Figure 8) and rise in relative neurotoxicity (Figure 9) paralleling the secondary loss of cytotoxicity of in the aquatic N. annulata, with these changes accompanied by a loss of the long hood ribs (Figure 7A).
It has long been clear that spitting cobras possess morphological and behavioural traits that evolved for defensive purposes [28], but the relationship between these traits and venom composition has been uncertain. As cytotoxins are the primary toxins implicated in the ocular irritation and damage caused by spat Naja venom, our results suggest a clear evolutionary trajectory in which the evolution of hooding as a confrontational defensive behaviour likely led to selection for a damaging, pain-inducing venom as a result of its new major role, which then facilitated the evolution of specialised mechanisms of long-distance venom delivery in three independent lineages. Reliance on bold defensive displays likely increases the frequency with which snake species are forced to deploy their venom defensively, thus driving the selection of venom that causes painful local damage to potential predators. Both the confrontational nature of the hooding display and its upright posture may be crucial in the evolution of spitting behaviour, which could explain why only snakes that adopt this defensive posture have evolved spitting, i.e., it is necessary for a snake to be looking forward and upwards at incoming predators for them to be able to spit venom effectively. As to why spitting behavior has never evolved in Ophiophagus despite the potent cytotoxicity providing the selection pressure of venom “worth spitting”, it may be that the very large, globular enzymes underlying this function in Ophiophagus venom are less efficiently absorbed in the eyes than are the very small peptides that underpin the cytotoxic function in Naja. Thus, it is clear that a strong co-evolutionary relationship exists between three of the character states that define cobras as a group: exceptional hooding displays (which have evolved twice within the clade), spitting behaviour/associated morphology (evolved three times within the clade) and increases in cytotoxic activity linked to the evolution of aposematic markings. Nevertheless, it remains a mystery why hooding (at least to the extent seen in cobras) is relatively rare in snakes and therefore what factors initiate the above scenario for evolution of this defensive suite.
To summarise, in Naja there is extensive variation in hooding (Figure 4), aposematic markings (Figure 4, Figure 5 and Figure 6) and camouflage (Figure 7 and Figure 10) and these variables are linked to patterns in relative cytotoxicity (Figure 1 and Supplementary Tables), venom complexity (Figure 8) and/or changes in neurotoxicity (Figure 9). In contrast to the variation within Naja a high level of defensive cytotoxicity is preserved across the full range of Ophiophagus venoms examined in this study, as are the morphological and behavioural attributes of hooding in this species. It is notable, however, that the Malaysian population with bright orange aposematic hood colouration in adult snakes (Figure 6E) is also the most cytotoxic population. This relative conservation of L-amino acid driven defensive cytotoxicity is in contrast to documented variation among O. hannah populations in the relative presence and concentration of toxins used in predation [62]. Thus, as discussed above, this species provides additional evidence that hooding is associated with defensive cytotoxic activity even in the face of substantial variation in the toxins selected for use in predation.
Overall, our results suggest that cytotoxicity evolved first with hooding. Hooding precedes several independent increases in cytotoxicity in the Naja + Hemachatus clade and evolved concurrently with increased cytotoxicity in Ophiophagus (Figure 1 and Supplementary Tables). No spitting accompanies the increase in cytotoxicity in N. annulifera. However, spitting is clearly closely associated with a secondary increase in cytotoxicity in Hemachatus and African Naja. The Asian Naja could either have evolved spitting at the same time as increased cytotoxicity or possibly afterwards, depending on whether weakly spitting N. atra and N. kauothia specimens are deemed informative. The inverse relationship between classes of aposematic marking (ornate hood patterns or flashes of red on the hood versus bold body banding) is an intriguing aspect, but in either case these aposematic markings are linked to further rises in cytotoxicity.
Given the significant human impact of the cytotoxic venoms of Naja sp. in both sub-Saharan Africa and southern Asia, the results of this study are also evidence of the mutually enlightening relationship between evolutionary toxinology and clinical toxinology. Understanding the evolutionary selection pressures resulting in the evolution of cytotoxic venom can help us understand the association between snake behaviour and the impact of snakebite, and could also be utilised to develop next-generation snakebite therapeutics, which are particularly desirable for the treatment of local damage inflicted by cytotoxins, as this common, high-impact, result of envenomation is poorly treated by currently available antivenoms [63]. In addition, the investigation of novel cytotoxins may provide lead compounds for the use in anticancer drug design and development.
3. Materials and Methods
3.1. Species Studied
- Non-hooding species
- Bungarus fasciatus
- Dendroaspis polylepis
- Elapsoidea boulengerii, Elapsoidea sundevali longicauda, Elapsoidea sundevali sundevali
- Walterinnesia aegyptia
- Naja annulata
- Narrow hooding species
- Aspidelaps lubricus, Aspidelaps scutatus (nar)
- Hooding species
- Hemachatus haemachatus
- African Naja
- Non-spitters = N. annulata, N. annulifera, N. haje, N. melanoleuca, N. nivea
- Spitters = N. mossambica, N. nigricollis, N. pallida
- Asian Naja
- Non-spitters = N. atra, N. kaouthia, N. naja, N. oxiana (Note that N. atra and N. kaouthia are slightly ambiguous as they are not considered as true spitting cobras, and lack the highly specialised adaptations of the other species I this category, but have been documented to spit significantly under strong duress [27,28]
- Spitters = N. philippinensis, N. siamensis, N. sumatrana
- Ophiophagus hannah clade (Four geographically different samples from O. hannah from Cambodia, Malaysia, Thailand, East Java).
Hooding and spitting were categorized based on experience with these animals combined with literature sources. For the comparative analyses described below, two separate datasets were used when investigating either hooding or spitting. One wherein ambiguous species were considered to lack the trait and another wherein ambiguous species were considered to have the trait. Ambiguous species for hooding were Aspidelaps lubricus and A. scutatus and those for spitting were Naja atra and N. kaouthia, such that the number of species affected by alternative coding was small but it still allowed us to ensure robustness of results to differing interpretations.
3.2. Venom Preparation
Venoms were obtained in lypholized form and prepared by resuspension in deionized water (dH2O), centrifugation at 4000 RCF, and supernatant passed through a 0.45 µm Millipore (Brisbane, QLD, Australia) syringe driven filter unit. Protein concentrations were determined using a NanoDrop 2000 UV-Vis Spectrophotometer (Thermofisher, Sydney, NSW, Australia) at an absorbance of 280 nm.
3.3. Cell Lines and Cell Culture
The effect of each snake venom was assessed on human neonatal foreskin fibroblast (NFF) and malignant melanoma (MM96L) cell lines, supplied by QIMR Berghofer Medical Research institute. Venom mediated cytotoxicity is often responsible for the degradation and destruction of skin and connective tissue, therefore the chosen cell lines were deemed appropriate. Cell lines were maintained in RPMI medium supplemented with 1% penicillin-streptomycin and foetal calf serum (FCS), 10% FCS for NFF and 5% FCS for MM96L. Cells were split 24 h prior to the experiment (for up to 25 passages for MM96L and 10 passages for NFF) using 0.25% trypsin, and seeded in 96-well flat bottom plates at a density of 5000 and 2500 cells/well for NFF and MM96L cells, respectively. Plates were incubated overnight at 37 °C in a 5% CO2-95% humidified environment prior to treatment.
3.4. MTT Assays
Cell viability was evaluated using colorimetric MTT (Thiazolyl Blue Tetrazolium Bromide; Sigma-Aldrich M5655, Sydney, NSW, Australia) assays. Venom was added to cells at 5 µg, 1 µg, 0.5 µg, and 0.1 µg protein amounts and followed by a 48-h incubation period. MTT was added at a concentration of 5 mg/mL per well. An amount of 0.1% sodium dodecyl sulphate (SDS) was used as a positive control to achieve 100% toxicity, and the protocol was followed according to the manufacturer’s description. The absorbance was read at 570 nm on the PowerWave XS2 plate-reader (Bio-Tek Instruments, Winooski, VT, USA), using Gen5 software. Three independent experiments were conducted with a minimum of three replicates per treatment. Cell viability readings were normalized as a percent of untreated control cells, and viability expressed as a percentage of toxicity ± standard error of the mean (SEM). The relationship between venom dose and cytotoxic response was calculated via area under the curve (AUC) analysis, using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA) (Supplementary Tables).
3.5. Phylogenetic Comparative Analyses
A phylogeny was assembled using [11] as a starting point as this is currently the most comprehensive time-calibrated phylogeny available for elapids. This tree included all species within the current dataset except Aspidelaps lubricus, Elapsoidea boulengeri (and the two E. sundevallii ssp.), Naja oxiana, N. philippinensis, and the localities of N. naja and Ophiophagus hannah. Consequently, these species were added to the tree (by directly manipulating the newick-formatted file) at positions which were deemed plausible to give a final working phylogeny. A. lubricus and E. boulengeri were added as sister species to their congeners with a divergence of 5 mya as this is approximately representative of the divergence of many sister species in this tree. For E. sundevallii ssp. and O. hannah localities, we created a split and a polytomy (respectively) at 2.5 mya for the same reasons (half the 5 mya to reflect the smaller divergence within species). N. philippinensis was included as a polytomy with N. sumatrana and N. siamensis because the relationships in this clade are currently very uncertain, however due to the general similarity of N. oxiana and N. naja, we added the former as a sister species to the latter at the midpoint of the divergence between N. naja and N. atra (sister groups before the inclusion of N. oxiana). For the two N. naja populations, we added those as a split diverging midway from the divergence from N. oxiana. The resulting phylogeny is shown in Figure 1 and was used for all further analyses conducted in R v3.2.5 [64] using the ape package [65] plus others introduced for particular analyses.
First, the phylogenetic signal of all traits used in the following analyses was assessed to examine to what extent variation in the traits reflected evolutionary history. For binary traits (hooding and spitting), we estimated Fritz and Purvis’ D [66] in the R package caper [67], testing for significant signal using 5000 permutations. For continuous traits (cytotoxicity measured as AUC for each cell line), we estimated Blomberg et al.’s K [68] in the R package phytools [69], and similarly tested for significance using 5000 simulations. In all cases, detectable phylogenetic signal was found (p < 0.0002 for spitting, however ambiguous species were coded and also for hooding when ambiguous species coded as not hooded; p = 0.001 for hooding when ambiguous species were coded as having hoods and also for cytotoxicity to the NFF cell line; p = 0.0002 for cytotoxicity to the MM96L cell line), emphasizing the need to analyze the data in a comparative framework.
Ancestral states were estimated and reconstructed over the tree in order to investigate the evolutionary history of the traits and consequently their relation to one another over time. For binary traits (spitting and hooding), we used the rerooting method [70] in phytools [69] as this method can handle polytomies when reconstructing categorical traits. The underlying model used was an equal rates model for both traits, chosen after log-likelihood comparison with a different rates model. The continuous traits (cytotoxicity on each cell line measured as AUC) were reconstructed by maximum likelihood in the contMap function in phytools [69].
To compare the strength of convergence in cytotoxicity in association with hooding vs. spitting, we calculated the Wheatsheaf index [29] for cytotoxicity as a two-dimensional trait (i.e., combining both cell lines) with either hooding or spitting as the focal trait. We were mainly interested in comparing the index between the two categorical traits but also tested for a signal of extremely strong convergence using 1000 bootstrap replicates to generate a null distribution. These analyses were conducted in the R package windex [71].
As a final estimate of the relative importance of hooding and spitting in the evolution of cytotoxicity, we fit a series of pGLS models in caper [67]. The same strategy was used for each of four datasets: the two coding versions for ambiguous species and AUC for the two cell lines. For each dataset the model set consisted of four models with cytotoxicity as the response variable and either hooding, spitting, both, or neither (intercept-only) as explanatory variables. Models were then compared with Akaike information criterion (AIC) and where the best model was not the null (intercept-only) model the output was checked to ensure that the explanatory variable had a significant effect before being finally accepted as the best model.
3.6. SDS-PAGE
One-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (1D SDS- PAGE) was performed as per previously described methods [72] to compare N. annulata, N. melanoleuca, N. oxiana and N. naja venoms.
3.7. Neurotoxicity Assays
The in vitro neurotoxicity of venoms was tested using the chick biventer cervices nerve-muscle preparation as described previously [73]. This procedure was approved by the Monash Animal Research Platform (MARP) Animal Ethics Committee, Monash University, Australia MARP/2014/97 (approved in December 2014).
Acknowledgments
Timothy N.W. Jackson, Ivan Koludarov and Renan Castro Santana were supported by University of Queensland PhD scholarships.
Supplementary Materials
The following are available online at www.mdpi.com/2072-6651/9/3/103/s1, Table S1: NFF venom cytotoxicity, Table S2: MM96L venom cytotoxicity.
Author Contributions
B.G.F. conceived and designed the experiments; N.P., M.P.I., S.A.A., I.H., H.v.d.P., J.T.-A-P., R.v.d.B., H.M.I.K., F.J.V., A.N., M.S., L.J., N.D., J.M. and B.G.F. contributed reagents/materials/analysis tools; N.P., T.N.W.J., M.P.I., K.A., R.P., D.C.Y., S.A.A., S.P.B., I.K., J.D., B.S., A.A., R.C.S., M.J., W.C.H., J.M. and B.G.F. performed the experiments, analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ismail M., al-Bekairi A.M., el-Bedaiwy A.M., Abd-el Salam M.A. The ocular effects of spitting cobras: II. Evidence that cardiotoxins are responsible for the corneal opacification syndrome. J. Toxicol. Clin. Toxicol. 1993;31:45–62. doi: 10.3109/15563659309000373. [DOI] [PubMed] [Google Scholar]
- 2.Ismail M., al-Bekairi A.M., el-Bedaiwy A.M., Abd-el Salam M.A. The ocular effects of spitting cobras: I. The ringhals cobra (Hemachatus haemachatus) venom-induced corneal opacification syndrome. J. Toxicol. Clin. Toxicol. 1993;31:31–41. doi: 10.3109/15563659309000372. [DOI] [PubMed] [Google Scholar]
- 3.Kasturiratne A., Wickremasinghe A.R., de Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D.G., de Silva H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5:e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez J.M., Williams D., Fan H.W., Warrell D.A. Snakebite envenoming from a global perspective: Towards an integrated approach. Toxicon. 2010;56:1223–1235. doi: 10.1016/j.toxicon.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 5.Harrison R.A., Hargreaves A., Wagstaff S.C., Faragher B., Lalloo D.G. Snake envenoming: A disease of poverty. PLoS Negl. Trop. Dis. 2009;3:e569. doi: 10.1371/journal.pntd.0000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chippaux J.P. Estimate of the burden of snakebites in sub-Saharan Africa: A meta-analytic approach. Toxicon. 2011;57:586–599. doi: 10.1016/j.toxicon.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Joshi H.S., Mahmood S.E., Joshi M.C., Shaifali I., Srivastava P.C. Clinico-epidemiological profile of snake bite cases in Western Nepal. TAF Prev. Med. Bull. 2012;11:57–62. doi: 10.5455/pmb.20111023012020. [DOI] [Google Scholar]
- 8.Fry B.G., Wuster W., Kini R.M., Brusic V., Khan A., Venkataraman D., Rooney A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 2003;57:110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- 9.Sunagar K., Jackson T., Undheim E., Ali S., Antunes A., Fry B. Three-fingered RAVERs: Rapid accumulation of variations in exposed residues of snake venom toxins. Toxins. 2013;5:2172–2208. doi: 10.3390/toxins5112172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly C.M.R., Barker N.P., Villet M.H., Broadley D.G. Phylogeny, biogeography and classification of the snake superfamily Elapoidea: A rapid radiation in the late Eocene. Cladistics. 2009;25:38–63. doi: 10.1111/j.1096-0031.2008.00237.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee M.S., Sanders K.L., King B., Palci A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae) R. Soc. Open Sci. 2016;3:150277. doi: 10.1098/rsos.150277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthe R.A., de Pury S., Bleckmann H., Westhoff G. Spitting cobras adjust their venom distribution to target distance. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2009;195:753–757. doi: 10.1007/s00359-009-0451-6. [DOI] [PubMed] [Google Scholar]
- 13.Berthe R.A., Westhoff G., Bleckmann H. Potential targets aimed at by spitting cobras when deterring predators from attacking. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2013;199:335–340. doi: 10.1007/s00359-013-0796-8. [DOI] [PubMed] [Google Scholar]
- 14.Russell P., Home E. Remarks on the voluntary expansion of the skin of the neck in the Cobra de Capello or hooded snake of the East Indies. With a description of the structures of the parts which perform that office. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1804;94:346–352. doi: 10.1098/rstl.1804.0015. [DOI] [Google Scholar]
- 15.Young B.A., Boetig M., Westhoff G. Functional bases of the spatial dispersal of venom during cobra “spitting”. Physiol. Biochem. Zool. 2009;82:80–89. doi: 10.1086/595589. [DOI] [PubMed] [Google Scholar]
- 16.Young B.A., Kardong K.V. The functional morphology of hooding in cobras. J. Exp. Biol. 2010;213:1521–1528. doi: 10.1242/jeb.034447. [DOI] [PubMed] [Google Scholar]
- 17.Young B.A., Dunlap K., Koenig K., Singer M. The buccal buckle: The functional morphology of venom spitting in cobras. J. Exp. Biol. 2004;207:3483–3494. doi: 10.1242/jeb.01170. [DOI] [PubMed] [Google Scholar]
- 18.Young B.A., Herzog F., Friedel P., Rammensee S., Bausch A., van Hemmen J.L. Tears of venom: Hydrodynamics of reptilian envenomation. Phys. Rev. Lett. 2011;106:198103. doi: 10.1103/PhysRevLett.106.198103. [DOI] [PubMed] [Google Scholar]
- 19.Cascardi J., Young B.A., Husic H.D., Sherma J. Protein variation in the venom spat by the red spitting cobra, Naja pallida (Reptilia: Serpentes) Toxicon. 1999;37:1271–1279. doi: 10.1016/S0041-0101(98)00264-5. [DOI] [PubMed] [Google Scholar]
- 20.Triep M., Hess D., Chaves H., Brucker C., Balmert A., Westhoff G., Bleckmann H. 3D flow in the venom channel of a spitting cobra: Do the ridges in the fangs act as fluid guide vanes? PLoS ONE. 2013;8:e61548. doi: 10.1371/journal.pone.0061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westhoff G., Tzschatzsch K., Bleckmann H. The spitting behavior of two species of spitting cobras. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2005;191:873–881. doi: 10.1007/s00359-005-0010-8. [DOI] [PubMed] [Google Scholar]
- 22.Wallach V., Wüster W., Broadley D.G. In praise of subgenera: Taxonomic status of cobras of the genus Naja Laurenti (Serpentes: Elapidae) Zootaxa. 2009;2236:26–36. [Google Scholar]
- 23.Ahn M.Y., Lee B.M., Kim Y.S. Cytotoxicity and L-amino acid oxidase activity of animal venoms. Arch. Pharm. Res. 1997;20:13–16. doi: 10.1007/BF02974035. [DOI] [PubMed] [Google Scholar]
- 24.Jara M., Pincheira-Donoso D. The neck flattening defensive behaviour in snakes: First record of hooding in the South American colubrid genus Philodryas. Anim. Biol. 2015;65:73–79. doi: 10.1163/15707563-00002459. [DOI] [Google Scholar]
- 25.Wuster W., Crookes S., Ineich I., Mane Y., Pook C.E., Trape J.F., Broadley D.G. The phylogeny of cobras inferred from mitochondrial DNA sequences: Evolution of venom spitting and the phylogeography of the African spitting cobras (Serpentes: Elapidae: Naja nigricollis complex) Mol. Phylogenet. Evol. 2007;45:437–453. doi: 10.1016/j.ympev.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 26.Wüster W. The cobras of the genus Naja in India. Hamadryad. 1998;23:15–32. [Google Scholar]
- 27.Chan S. A Field Guide to the Venomous Land Snakes of Hong Kong. Cosmos Books Ltd.; Hong Kong, China: 2006. [Google Scholar]
- 28.Wüster W., Thorpe R.S. Dentitional phenomena in cobras revisited: Spitting and fang structure in the Asiatic species of Naja (Serpentes: Elapidae) Herpetologica. 1992;48:424–434. [Google Scholar]
- 29.Arbuckle K., Bennett C.M., Speed M.P. A simple measure of the strength of convergent evolution. Methods Ecol. Evol. 2014;5:685–693. doi: 10.1111/2041-210X.12195. [DOI] [Google Scholar]
- 30.Marais J. A Complete Guide to the Snakes of Southern Africa. Struik Nature; Cape Town, South Africa: 2004. [Google Scholar]
- 31.Kalam Y., Isbister G.K., Mirtschin P., Hodgson W.C., Konstantakopoulos N. Validation of a cell-based assay to differentiate between the cytotoxic effects of elapid snake venoms. J. Pharmacol. Toxicol. Methods. 2011;63:137–142. doi: 10.1016/j.vascn.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Mendez I., Gutierrez J.M., Angulo Y., Calvete J.J., Lomonte B. Comparative study of the cytolytic activity of snake venoms from African spitting cobras (Naja spp., Elapidae) and its neutralization by a polyspecific antivenom. Toxicon. 2011;58:558–564. doi: 10.1016/j.toxicon.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Calvete J.J., Ghezellou P., Paiva O., Matainaho T., Ghassempour A., Goudarzi H., Kraus F., Sanz L., Williams D.J. Snake venomics of two poorly known Hydrophiinae: Comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J. Proteom. 2012;75:4091–4101. doi: 10.1016/j.jprot.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 34.Fry B.G., Wuster W., Ramjan S.F.R., Jackson T., Martelli P., Kini R.M. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun. Mass Spectrom. 2003;17:2047–2062. doi: 10.1002/rcm.1148. [DOI] [PubMed] [Google Scholar]
- 35.Spawls S., Branch B. Dangerous Snakes of Africa. Blandford Press; London, UK: 1995. [Google Scholar]
- 36.Dutta S., Chanda A., Kalita B., Islam T., Patra A., Mukherjee A.K. Proteomic analysis to unravel the complex venom proteome of eastern India Naja naja: Correlation of venom composition with its biochemical and pharmacological properties. J. Proteom. 2017;156:29–39. doi: 10.1016/j.jprot.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Huang H.W., Liu B.S., Chien K.Y., Chiang L.C., Huang S.Y., Sung W.C., Wu W.G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015;128:92–104. doi: 10.1016/j.jprot.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y., Li Y., Lee W., Xu X., Zhang Y., Zhao R., Wang W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011;12:1. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauridsen L.P., Laustsen A.H., Lomonte B., Gutierrez J.M. Exploring the venom of the forest cobra snake: Toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteom. 2017;150:98–108. doi: 10.1016/j.jprot.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 40.Laustsen A.H., Gutierrez J.M., Lohse B., Rasmussen A.R., Fernandez J., Milbo C., Lomonte B. Snake venomics of monocled cobra (Naja kaouthia) and investigation of human IgG response against venom toxins. Toxicon. 2015;99:23–35. doi: 10.1016/j.toxicon.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Petras D., Sanz L., Segura A., Herrera M., Villalta M., Solano D., Vargas M., Leon G., Warrell D.A., Theakston R.D., et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the pan-African EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J. Proteome Res. 2011;10:1266–1280. doi: 10.1021/pr101040f. [DOI] [PubMed] [Google Scholar]
- 42.Sintiprungrat K., Watcharatanyatip K., Senevirathne W.D., Chaisuriya P., Chokchaichamnankit D., Srisomsap C., Ratanabanangkoon K. A comparative study of venomics of Naja naja from India and Sri Lanka, clinical manifestations and antivenomics of an Indian polyspecific antivenom. J. Proteom. 2016;132:131–143. doi: 10.1016/j.jprot.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki-Matsubara M., Athauda S.B., Suzuki Y., Matsubara K., Moriyama A. Comparison of the primary structures, cytotoxicities, and affinities to phospholipids of five kinds of cytotoxins from the venom of Indian cobra, Naja naja. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2016;179:158–164. doi: 10.1016/j.cbpc.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J. Proteom. 2015;120:105–125. doi: 10.1016/j.jprot.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Yap M.K., Fung S.Y., Tan K.Y., Tan N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra) Acta Trop. 2014;133:15–25. doi: 10.1016/j.actatropica.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Fung S.Y., Lee M.L., Tan N.H. Molecular mechanism of cell death induced by king cobra (Ophiophagus hannah) venom L-amino acid oxidase. Toxicon. 2015;96:38–45. doi: 10.1016/j.toxicon.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Tan C.H., Tan K.Y., Fung S.Y., Tan N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah) BMC Genom. 2015;16:687. doi: 10.1186/s12864-015-1828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan N.H., Hj M.N. Enzymatic and toxic properties of Ophiophagus hannah (king cobra) venom and venom fractions. Toxicon. 1989;27:689–695. doi: 10.1016/0041-0101(89)90020-2. [DOI] [PubMed] [Google Scholar]
- 49.Tan N.H., Saifuddin M.N. Isolation and characterization of an unusual form of L-amino acid oxidase from King cobra (Ophiophagus hannah) venom. Biochem. Int. 1989;19:937–944. [PubMed] [Google Scholar]
- 50.Tan N.H., Saifuddin M.N. Isolation and characterization of a hemorrhagin from the venom of Ophiophagus hannah (king cobra) Toxicon. 1990;28:385–392. doi: 10.1016/0041-0101(90)90076-J. [DOI] [PubMed] [Google Scholar]
- 51.Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J., Kerkkamp H.M., Vos R.A., Guerreiro I., Calvete J.J., et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joubert F.J. Snake venom toxins. The amino-acid sequences of three toxins (9B, 11 and 12A) from Hemachatus haemachatus (Ringhals) venom. Eur. J. Biochem. 1977;74:387–396. doi: 10.1111/j.1432-1033.1977.tb11403.x. [DOI] [PubMed] [Google Scholar]
- 53.Rong H., Li Y., Lou X.H., Zhang X., Gao Y.X., Teng M.K., Niu L.W. Purification, partial characterization, crystallization and preliminary X-ray diffraction of a novel cardiotoxin-like basic protein from Naja naja atra (South Anhui) venom. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:130–134. doi: 10.1107/S1744309107002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahn M.Y., Lee B.M., Kim Y.S. Characterization and cytotoxicity of L-amino acid oxidase from the venom of king cobra (Ophiophagus hannah) Int. J. Biochem. Cell Biol. 1997;29:911–919. doi: 10.1016/S1357-2725(97)00024-1. [DOI] [PubMed] [Google Scholar]
- 55.Casewell N.R., Wuster W., Vonk F.J., Harrison R.A., Fry B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 56.Fry B.G., Casewell N.R., Wuster W., Vidal N., Young B., Jackson T.N. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon. 2012;60:434–448. doi: 10.1016/j.toxicon.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Fry B.G., Sunagar K., Casewell N.R., Kochva E., Roelants K., Scheib H., Wüster W., Vidal N., Young B., Burbrink F., et al. The origin and evolution of the Toxicofera reptile venom system. In: Fry B.G., editor. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press; New York, NY, USA: 2015. pp. 1–31. [Google Scholar]
- 58.Fry B.G., Undheim E.A., Ali S.A., Jackson T.N., Debono J., Scheib H., Ruder T., Morgenstern D., Cadwallader L., Whitehead D., et al. Squeezers and leaf-cutters: Differential diversification and degeneration of the venom system in toxicoferan reptiles. Mol. Cell. Proteom. 2013;12:1881–1899. doi: 10.1074/mcp.M112.023143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M., Fry B.G., Kini R.M. Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii) J. Mol. Evol. 2005;60:81–89. doi: 10.1007/s00239-004-0138-0. [DOI] [PubMed] [Google Scholar]
- 60.Li M., Fry B.G., Kini R.M. Putting the brakes on snake venom evolution: The unique molecular evolutionary patterns of Aipysuras eydouxii (Marbled sea snake) phospholipase A(2) toxins. Mol. Biol. Evol. 2005;22:934–941. doi: 10.1093/molbev/msi077. [DOI] [PubMed] [Google Scholar]
- 61.Jackson T.N., Fry B.G. A tricky trait: Applying the fruits of the “function debate” in the philosophy of biology to the “venom debate” in the science of toxinology. Toxins. 2016;8:263. doi: 10.3390/toxins8090263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang H.C., Tsai T.S., Tsai I.H. Functional proteomic approach to discover geographic variations of king cobra venoms from Southeast Asia and China. J. Proteom. 2013;89:141–153. doi: 10.1016/j.jprot.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 63.Williams D.J., Gutierrez J.M., Calvete J.J., Wuster W., Ratanabanangkoon K., Paiva O., Brown N.I., Casewell N.R., Harrison R.A., Rowley P.D., et al. Ending the drought: New strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteom. 2011;74:1735–1767. doi: 10.1016/j.jprot.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 64.R-Core-Team, R . A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [(accessed on 27 January 2017)]. Available online: https://www.R-project.org/ [Google Scholar]
- 65.Paradis E., Claude J., Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 66.Fritz S.A., Purvis A. Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits. Conserv. Biol. 2010;24:1042–1051. doi: 10.1111/j.1523-1739.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 67.Orme D., Freckleton R., Thomas G., Petzoldt T., Fritz S., Isaac N., Pearse W. Caper: Comparative Analyses of Phylogenetics and Evolution in R. 2015. [(accessed on 1 February 2017)]. R Package Version 0.5.2. Available online: https://CRAN.R-project.org/package=caper.
- 68.Blomberg S.P., Garland T., Ives A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 69.Revell L.J. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 70.Yang Z., Kumar S., Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arbuckle K., Minter A. Windex: Analyzing convergent evolution using the Wheatsheaf index in R. Evol. Bioinform. 2015;11:11–14. doi: 10.4137/EBO.S20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ali S.A., Jackson T.N., Casewell N.R., Low D.H., Rossi S., Baumann K., Fathinia B., Visser J., Nouwens A., Hendrikx I., et al. Extreme venom variation in Middle Eastern vipers: A proteomics comparison of Eristicophis macmahonii, Pseudocerastes fieldi and Pseudocerastes persicus. J. Proteom. 2015;116:106–113. doi: 10.1016/j.jprot.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 73.Ali S.A., Yang D.C., Jackson T.N., Undheim E.A., Koludarov I., Wood K., Jones A., Hodgson W.C., McCarthy S., Ruder T., et al. Venom proteomic characterization and relative antivenom neutralization of two medically important Pakistani elapid snakes (Bungarus sindanus and Naja naja) J. Proteom. 2013;89:15–23. doi: 10.1016/j.jprot.2013.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.