Abstract
The apicomplexan parasite Sarcocystis neurona causes equine protozoal myeloencephalitis (EPM), a degenerative neurological disease of horses. Due to its host range expansion, S. neurona is an emerging threat that requires close monitoring. In apicomplexans, protein kinases (PKs) have been implicated in a myriad of critical functions, such as host cell invasion, cell cycle progression and host immune response evasion. Here, we used various bioinformatics methods to define the kinome of S. neurona and phylogenetic relatedness of its PKs to other apicomplexans. We identified 97 putative PKs clustering within the various eukaryotic kinase groups. Although containing the universally-conserved PKA (AGC group), S. neurona kinome was devoid of PKB and PKC. Moreover, the kinome contains the six-conserved apicomplexan CDPKs (CAMK group). Several OPK atypical kinases, including ROPKs 19A, 27, 30, 33, 35 and 37 were identified. Notably, S. neurona is devoid of the virulence-associated ROPKs 5, 6, 18 and 38, as well as the Alpha and RIO kinases. Two out of the three S. neurona CK1 enzymes had high sequence similarities to Toxoplasma gondii TgCK1-α and TgCK1-β and the Plasmodium PfCK1. Further experimental studies on the S. neurona putative PKs identified in this study are required to validate the functional roles of the PKs and to understand their involvement in mechanisms that regulate various cellular processes and host-parasite interactions. Given the essentiality of apicomplexan PKs in the survival of apicomplexans, the current study offers a platform for future development of novel therapeutics for EPM, for instance via application of PK inhibitors to block parasite invasion and development in their host.
Keywords: Sarcocystis neurona, EPM, apicomplexans, phylogeny, homology modeling
1. Introduction
Equine protozoal myeloencephalitis (EPM) is an infectious, progressive, degenerative neurological disease of horses caused by the apicomplexan parasite, Sarcocystis neurona [1]. To complete its life cycle, this heteroxenous parasite requires a reservoir host (i.e., opossums; Didelphis virginiana, Didelphis albiventris) and an aberrant (horses) or intermediate host (cats, skunks, raccoons and sea otters) [2]. Opossums become infected upon ingestion of sarcocysts containing hundreds of bradyzoites. The bradyzoites undergo gametogony and sporulate into mature oocysts that are then shed in the feces. After ingestion by the intermediate or aberrant hosts, the oocysts transform into the environmentally-resistant sporozoites that chronically parasitize the neural and inflammatory cells of the host’s central nervous system (CNS). Clinical EPM symptoms depend on the part of the CNS that is parasitized and in general results in abnormal gait, dysphagia and muscle atrophy in affected horses [3].
The intracellular nature of S. neurona and its ability to evade the host’s immune surveillance [4] makes EPM treatment expensive, lengthy and challenging. Traditionally, clinical treatment of EPM involved inhibitors of folate synthesis and metabolism (sulfonamides/pyrimethamine combination) over a prolonged period [5]. More recently, triazines derivatives (diclazuril, ponazuril) that target the parasite’s apicoplast [6], nitazoxanide, a pyruvate:ferredoxin oxidoreductase analogue that inhibits the parasite’s anaerobic metabolism [7], and anti-inflammatory agents and immune stimulants [8] have been used with variable success in eliminating clinical signs. Despite the availability of these drugs, EPM treatment is complicated by the emergence of drug-resistance (due to intermittent or periodic treatments), cost of therapies and drug toxicity and infection relapses due to re-growth of residual parasites after the treatment regimes [2]. As such, the discovery and development of novel therapeutics for EPM is imperative.
To successfully invade the host cells, apicomplexans utilize three specialized exocytic organelles (micronemes, rhoptries and dense-granules) [9]. The microneme is used for host cell recognition, binding, penetration and gliding along the cytoskeletal structures. Rhoptry proteins are discharged into the host cell during parasite internalization and are crucial in the formation of the parasitophorous vacuoles (PVs). Developing zoites contain non-pedunculated condensing vesicles that synthesize and package inactive rhoptry proteins, which are proteolytically activated when the rhoptry contents are condensed [10]. The PVs facilitate parasite development by allowing nutrient transport from the host cell and by blocking lysosomal fusion, which would otherwise kill the parasites [11]. Upon internalization, zoites use the dense-granules to remodel the PVs into functionally-active organelles.
The proliferation and differentiation of apicomplexans are influenced by protein kinases (PKs) that are involved in the invasion and modification of host cell structure and function. Generally, PKs can be classified into the conventional (“typical”) eukaryotic PK (ePK) and “atypical” PK (aPK) superfamilies [12,13,14]. Based on the controlled vocabulary of Hanks et al.’s [12,15] classification scheme, there are eight ePK families. These include PKs A, G and C (AGCs), calmodulin/calcium-dependent PKs (CAMKs), CMGC (including cyclin-dependent kinases (CDKs), mitogen-activated protein kinases (MAP kinases), glycogen synthase kinases (GSK) and CDK-like kinases), casein kinase 1 (CK1), “sterile-phenotype” kinases (STEs), receptor guanylate cyclase (RGC), tyrosine kinases (TKs), tyrosine kinase-like kinases (TKLs) and the “other PKs” sub-family (OPKs) [16]. The aPK superfamily consist of the Alpha-kinases, pyruvate dehydrogenase kinases (PDHK), phosphatidylinositol 3-kinase-related kinases (PIKK) and right open reading frame (RIO) kinases [17]. Although generally lacking or having limited sequence similarity to the ePKs and constituting small families in all organisms, some aPKs are homologous to catalytically-active PKs [12].
Several kinomes have been characterized in various organisms [14], including yeast, fruit fly, roundworms and human [18]. In Apicomplexans, the kinome of the malaria parasite, Plasmodium falciparum, was initially reported to contain 85 typical ePKs, which clustered into several groups including five of the major ePKs (i.e., CK1, TKL, CMGC, CAMK and AGC), but was devoid of STEs and TKs [18]. Subsequent studies on the Plasmodium kinome resulted in the identification of more PKs and PK-like proteins, adding up to 99 PKs [19,20]. However, despite the diverse repertoire of the Plasmodium ePKs, reverse genetics studies revealed that over 30% of the kinases are nonessential for the parasite’s asexual blood-stage development; only three of the 12 ePKs required for Plasmodium transmission in vivo have been conclusively demonstrated to be essential for the parasite’s asexual development [21]. Kinomes of a dozen other apicomplexan species have been reported, notable of which are Toxoplasma, Cryptosporidium and Babesia species [17]. Talevich et al. [22] recently classified ePKs into 17 genomes in Apicomplexa (Coccidia, Piroplasmida and Haemosporida species). The Rhoptry kinases (ROPKs) and pseudokinases in some coccidian genomes (Toxoplasma gondii, Neospora caninum, Eimeria tenella and portions of S. neurona) have been recently catalogued into 42 subfamilies [23]. Overall, at least 65 orthologous PK groups amongst the 12 kinomes described in the apicomplexans are shared with other alveolates and/or metazoans [17,24].
Each of the PK families has vital roles in parasite’s survival. For instance, PfPK-B (AGC family), PfTKL3 (TKL family) and four of the seven CDPKs (CAMK family) are required by Plasmodium parasites to complete their asexual cycle [25,26,27]. In a recent study, deletion of TgCK1α (CK1 family) resulted in defective replication of T. gondii in vitro [28]. The P. falciparum CKL and SRPK1 (CMGC family) complement each other in the regulation of mRNA splicing [29]. Since apicomplexans lack typical MAPK cascades, the STEs are not well studied. However, in the parasites that do have the MAPK pathways, STEs are essential for parasite growth (e.g., in the human parasites, Schistosoma mansoni [30]). Further, for parasites without the conventional MAPK cascades, PKs may activate the signaling pathways, for instance the Plasmodium Pfnek-1/3 [31,32]. It should however be noted that the activation of Plasmodium MAPK via Pfnek-1/3-mediated phosphorylation has only been demonstrated in vitro; there is no sufficient evidence of MAPK signaling in vivo in the parasite. Finally, some of the notable OPKs include aurora kinases, rhoptry kinases (ROPKs) and parasite-specific eukaryotic initiation factor-2 (elF2) kinases (elF2K), which are important in parasite virulence and differentiation [23,33,34].
Here, we used a genome-wide approach to define the kinome of S. neurona and determined the relatedness of the putative PKs to those reported in other apicomplexans. Defining the S. neurona kinome is not only important in providing insights into the parasite biology, but also identification of potential novel drug targets that can be used to clear chronic S. neurona infections and reduce parasite survival.
2. Results
2.1. Sarcocystis neurona Encodes 97 Putative Kinases
To date, at least 15 apicomplexan genomes (coccidians, gregarines, hemosporidians and piroplasmids) have either been fully sequenced or partially annotated [24]. In the current study, we conducted an exhaustive genome-wide search of the newly-sequenced S. neurona genome [35], and identified 97 putative PKs (Table 1). The identified PKs contained the characteristic PK (IPR000719) or PK-like (IPR011009) domains and three conserved amino acids constituting the catalytic triad (Lys30, Asp125, Asp143). The PKs had sizes ranging between 152 and 6544 amino acids and relative molecular weights of between 15.94 and 671.51 kDa. The majority of the PKs had an isoelectric point (pI) greater than 7.0, implying that the PKs have low turnover rates, since in general, acidic proteins are thought to be degraded more rapidly than neutral or basic proteins [36].
Table 1.
Description of the 97 putative protein kinases (PKs) identified in the kinome of Sarcocystis neurona. The putative PKs could be classified into eight groups. The amino acid coordinates of the conserved PK domains in the protein sequences and the PK homologies to other apicomplexan PKs are shown in Columns 7–12.
| Description of the Putative Protein Kinases (PKs) in the Genome of S. neurona | Description of Protein Kinase (PK) Homologies (BLASTp) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID a | Sequence Annotations; Description b | Family; (Subfamily) c | Length (aa) | pI | MW (kDa) | PK Domain Coordinates | Sequence Name; (Apicomplexan) | Bit Score | E-Value | Identity (%) | Accession Number |
| 1. Kinase Group AGC (Protein kinases A (PKA), G (PKG) and C (PKC) families) | |||||||||||
| SRCN_1312 | AGC kinase | 3-phosphoinositide dependent PK-1 (PDK1) | 903 | 5.52 | 101.13 | 137–481 | PDPK; (T. gondii RUB) | 417 | 6.00 × 10−135 | 59 | KFG61374.1 |
| SRCN_3339 | AGC kinase | PKA | 1428 | 8.85 | 147.52 | 1102–1417 | Putative AGC kinase; (N. caninum L) | 611 | 0.0 | 81 | CEL65574.1 |
| SRCN_4249 | AGC kinase | Nuclear dbf2-related (NDR) | 152 | 8.91 | 17.47 | 6–141 | Putative AGC kinase; (N. caninum L) | 219 | 2.00 × 10−68 | 76 | XP_003883757.1 |
| SRCN_4518 | PK G AGC kinase family member PKG | Ciliate-E2 | 425 | 5.83 | 48.78 | 97–399 | AGC kinase TgPKG1; (T. gondii ME49) | 830 | 0.0 | 92 | EPR61116.1 |
| SRCN_3990 | cAMP-dependent kinase | CAMKL; (MELK) | 1907 | 9.55 | 217.55 | 782–1634 | cAMP-dependent protein kinase (T. gondii VEG) | 75.9 | 1 × 10−12 | 30 | ESS31194.1 |
| SRCN_5165 | cAMP-dependent PK, catalytic chain | PKA | 343 | 8.99 | 39.36 | 20–338 | AGC kinase; (T. gondii ARI) | 425 | 3.00 × 10−150 | 90 | KYF43224.1 |
| SRCN_4913 | Putative PK | PKD | 2330 | 6.42 | 244.64 | 1093–1737 | Putative PK; (E. tenella) | 107 | 9.00 × 10−22 | 60 | XP_013228294.1 |
| SRCN_5430 | AGC kinase | Ribosomal protein S6 Kinases (RSK; (p70)) | 1378 | 5.54 | 139.28 | 824–1344 | AGC kinase; (T. gondii MAS) | 223 | 3.00 × 10−60 | 59 | KFH07588.1 |
| SRCN_5610 | cAMP-dependent PK, catalytic chain | PKA | 333 | 9.00 | 37.96 | 12–318 | cAMP-dependent PK, catalytic subunit; (T. gondii ME49) | 641 | 0.0 | 92 | XP_002366464.1 |
| 2. Kinase Group calcium (Ca2+)-/calmodulin-regulated kinases (CAMK) | |||||||||||
| SRCN_1071 | Ca2+-dependent kinase | CAMK1 | 1495 | 9.47 | 152.66 | 1085–1401 | Putative PK; (T. gondii VEG) | 188 | 1.00 × 10−47 | 67 | ESS31884.1 |
| SRCN_2032 | Putative PK | Ciliate-C1 | 297 | 6.22 | 33.26 | 15–297 | PK; (H. hammondi) | 424 | 1.00 × 10−146 | 68 | XP_008882026.1 |
| SRCN_2165 | Ca2+-dependent kinase CDPK2B | CDPK | 692 | 7.33 | 75.65 | 101–401 | Ca2+-dependent PK CDPK2A; (T. gondii ARI) | 674 | 0.0 | 90 | KYF44522.1 |
| SRCN_2257 | Histone kinase | CAMKL; (AMP-activated protein kinase (AMPK)) | 1800 | 8.98 | 187.71 | 1159–1448 | Putative CAM kinase, SNF1 family; (E. acervulina) | 377 | 7.00 × 10−105 | 64 | XP_013252246.1 |
| SRCN_2544 | CAM SNF1 AMK1 family | CAMKL;(AMPK-regulated kinase novel kinase (NUAK)) | 333 | 5.71 | 37.85 | 62–333 | CAM kinase, SNF1/AMK1 family ToxPK1; (N. caninum L) | 480 | 9.00 × 10−167 | 79 | XP_003882065.1 |
| SRCN_2937 | Ca2+-signaling kinase MARK | CAMKL; (microtubule affinity regulating kinase (MARK)) | 278 | 9.62 | 31.29 | 1–250 | Putative Ca2+ signaling PK MARK; (T. gondii GT1) | 356 | 5.00 × 10−124 | 72 | EPR59053.1 |
| SRCN_3011 | Calmodulin-dependent PK (CAM) CDPK6 | CDPK | 1435 | 9.25 | 154.44 | 1238–1435 | Cdpk kinase domain; (T. gondii) | 179 | 6.00 × 10−49 | 75 | 3IS5_A |
| SRCN_3314 | A Chain crystal Structure of TgCDPK1 with inhibitor bound | CDPK1 | 519 | 5.99 | 58.89 | 37–335 | Calmodulin-domain PK 1; (T. gondii) | 536 | 0.0 | 97 | 3MA6_A |
| SRCN_3583 | Ca2+-dependent kinase CDPK5 | CDPK | 454 | 6.09 | 50.38 | 35–308 | Ca2+-dependent PK CDPK5; (T. gondii ARI) | 776 | 0.0 | 89 | KYF43137.1 |
| SRCN_3701 | Ca2+-dependent kinase CDPK3 | CDPK | 560 | 5.91 | 62.07 | 77–362 | Ca2+-dependent Kinase; (T. gondii) | 493 | 1.00 × 10−172 | 87 | 3DXN_A |
| SRCN_4076 | CAM CDPK family | CDPK | 1701 | 5.93 | 181.42 | 1079–1663 | CAM kinase, CDPK family; (H. hammondi) | 239 | 2.00 × 10−62 | 69 | XP_008884897.1 |
| SRCN_4093 | PK | CAMKL; (AMP-activated protein kinase (AMPK)) | 1155 | 9.02 | 118.33 | 1–261 | Putative atypical MEK-related kinase; (N. caninum L) | 222 | 1.00 × 10−59 | 70 | XP_003880869.1 |
| SRCN_4390 | Ca2+-dependent kinase CDPK2 | CDPK | 790 | 6.24 | 85.47 | 280–556 | Ca2+-dependent PK, related; (N. caninum L) | 1130 | 0.0 | 74 | XP_003884321.1 |
| SRCN_4815 | Histone kinase (partial) | CAMKL; (AMP-activated protein kinase (AMPK)) | 711 | 6.53 | 75.98 | 1–314 | SNF1-related PK catalytic-α KIN10, 5 AMP-activated PK; (N. caninum L) | 568 | 0.0 | 48 | CEL67550.1 |
| SRCN_5227 | CAM CDPK CDPK8-like | CDPK | 2748 | 8.98 | 282.12 | 796–885 | Putative CAM kinase, CDPK family; (N. caninum L) | 117 | 3.00 × 10−24 | 59 | XP_003881901.1 |
| SRCN_5410 | Calmodulin-dependent PK (CAM-SNF1 family) | CAMK1 | 467 | 8.94 | 52.04 | 168–446 | CAM kinase, SNF1 family; (H. hammondi) | 431 | 5.00 × 10−134 | 78 | XP_008883430.1 |
| SRCN_5812 | Ca2+-dependent kinase CDPK9 | CDPK | 760 | 8.37 | 84.23 | 254–573 | Ca2+-dependent PK CDPK9; (H. hammondi) | 1139 | 0.0 | 81 | XP_008889286.1 |
| SRCN_5948 | Ca2+-dependent kinase CDPK8 | CDPK | 3298 | 7.11 | 345.85 | 208–860 | EF-hand domain-containing protein; (T. gondii ME49) | 114 | 1.00 × 10−23 | 58 | XP_002368547.1 |
| SRCN_6597 | Ca2+ dependent kinase CDPK7 | CAMK1 | 1374 | 9.09 | 138.28 | 365–623 | PK-PH domain-containing protein; (T. gondii ME49) | 813 | 0.0 | 80 | XP_002366487.1 |
| SRCN_6606 | Ca2+-dependent kinase CDPK4 | CDPK | 1632 | 9.42 | 170.93 | 813–1236 | Ca2+-dependent PK; (T. gondii) | 731 | 0.0 | 58 | CAD32376.2 |
| 3. Kinase Group casein kinase 1 (cell kinase 1) | |||||||||||
| SRCN_3445 | Casein kinase I | CK1-D | 323 | 9.34 | 37.65 | 6–290 | Casein kinase 1; (T. gondii ME49) | 603 | 0.0 | 94 | XP_002366683.1 |
| SRCN_4587 | Casein kinase I | CK1-D | 137 | 7.78 | 15.94 | 1–137 | Casein kinase I; (H. hammondi) | 226 | 3.00 × 10−71 | 81 | XP_008883809.1 |
| SRCN_4645 | Casein kinase I | CK1-D | 229 | 9.51 | 25.66 | 38–229 | Casein kinase I; (T. gondii GAB2-2007-GAL-DOM2) | 259 | 1.00 × 10−86 | 79 | KFG42638.1 |
| 4. Kinase Group CMGC (including cyclin-dependent kinases, mitogen-activated PKs, glycogen synthase kinases and CDK-like kinases) | |||||||||||
| SRCN_1104 | Cyclin-dependent kinase family 5 | Ca2+-dependent PK-L (CDKL) | 372 | 9.23 | 42.71 | 1–318 | Cyclin-dependent kinase family 5 protein; (H. hammondi) | 490 | 2.00 × 10−173 | 76 | XP_008884207.1 |
| SRCN_1236 | Cell-cycle-associated kinase (SRPK) | Serine-arginine rich PK (SRPK) | 2911 | 5.37 | 302.90 | 713–1837 | PK; (T. gondii ME49) | 476 | 9.00 × 10−141 | 76 | XP_002369401.1 |
| SRCN_1479 | CMGC Lammer | CLK | 748 | 10.23 | 79.09 | 485–748 | Cell-cycle-associated PK CLK; (T. gondii FOU) | 288 | 2.00 × 10−81 | 74 | KFG33061.1 |
| SRCN_1611 | CMGC Dual-specificity tyrosine-regulated kinase (DYRK) | DYRK; (DYRKP) | 1504 | 5.86 | 160.34 | 551–1498 | Cell-cycle-associated PK DYRK; (T. gondii VEG) | 223 | 2.00 × 10−57 | 63 | ESS33160.1 |
| SRCN_1731 | Cell-cycle-associated kinase GSK | Glycogen synthase kinase (GSK) | 219 | 6.59 | 24.39 | 1–175 | Cell-cycle-associated PK GSK; (H. hammondi) | 330 | 2.00 × 10−112 | 82 | XP_008887193.1 |
| SRCN_1732 | Cell-cycle-associated kinase GSK | Glycogen synthase kinase (GSK) | 203 | 10.78 | 20.72 | 86–203 | CMGC kinase, GSK family TgPK3; (E. brunetti) | 114 | 1.00 × 10−28 | 91 | CDJ46527.1 |
| SRCN_2759 | Cell-cycle-associated kinase partial | Ca2+-dependent PK (CDK); (CRK7) | 1122 | 6.14 | 118.67 | 541–1122 | Cell-cycle-associated PK CDK; (T. gondii VAND) | 118 | 1.00 × 10−25 | 79 | KFH12036.1 |
| SRCN_2845 | CMGC DYRK PRP4 kinase | DYRK; (PRP4) | 1665 | 9.76 | 177.08 | 1267–1596 | Putative PK (CLK3); (P. malariae) | 330 | 1.00 × 10−102 | 69 | SBS85334.1 |
| SRCN_3891 | CMGC kinase | DYRK; (DYRK2) | 674 | 8.85 | 73.85 | 399–674 | Putative CMGC kinase; (T. gondii ME49) | 80.1 | 6.00 × 10−14 | 67 | EPT25192.1 |
| SRCN_4209 | CMGC MAPK family (ERK) MAPK-1 | Mitogen-activated PK (MAPK); (ERK)) | 2361 | 6.73 | 247.71 | 94–754 | CMGC, MAPK/ (ERK) TgMAPK-1; (E. brunetti) | 137 | 1.00 × 10−30 | 74 | CDJ49492.1 |
| SRCN_4674 | Cyclin-dependent kinase | Ca2+-dependent PK (CDK); (CDK7) | 138 | 7.80 | 15.50 | 1–138 | Cyclin-dependent kinase; (T. gondii GT1) | 108 | 7.00 × 10−27 | 58 | EPR60430.1 |
| SRCN_4801 | Cell-cycle-associated kinase | Ca2+-dependent PK (CDK); (CDK5) | 300 | 6.08 | 34.33 | 1–289 | CMGC kinase, CDK family TgPK2; (N. caninum L) | 576 | 0.0 | 91 | XP_003885801.1 |
| SRCN_5365 | Cell-cycle-associated kinase MAPK | Mitogen-activated PK (MAPK; (ERK)) | 417 | 6.77 | 48.32 | 7–363 | Cell-cycle-associated PK MAPK; (H. hammondi) | 823 | 0.0 | 93 | XP_008886907.1 |
| SRCN_6346 | Cell-cycle-associated kinase CDK | Ca2+-dependent PK (CDK); (CDK5) | 690 | 9.55 | 80.90 | 208–603 | Putative cell-cycle-associated PK CDK; (T. gondii ARI) | 390 | 2.00 × 10−122 | 87 | KYF45878.1 |
| SRCN_6427 | CMGC CK2 kinase | Cell Kinase 2 (CK2) | 1395 | 10.29 | 144.86 | 885–1356 | CMGC kinase, CK2 family; (T. gondii MAS) | 241 | 6.00 × 10−73 | 98 | KFH07655.1 |
| SRCN_6472 | Cell-cycle-associated kinase ERK7 | Mitogen-activated PK (MAPK; (ERK)) | 983 | 9.28 | 104.95 | 7–317 | Cell-cycle-associated PK ERK7; (T. gondii ARI) | 647 | 0.0 | 81 | KYF46268.1 |
| SRCN_761 | Cell-cycle-associated kinase | Ca2+-dependent PK (CDK); (CDK7) | 577 | 9.34 | 58.39 | 144–490 | Cell-cycle-associated PK; (H. hammondi) | 283 | 3.00 × 10−88 | 68 | XP_008882409.1 |
| SRCN_895 | Cell-cycle-associated kinase | Ca2+-dependent PK (CDK); (CDK10) | 340 | 8.93 | 38.57 | 1–307 | Cell-cycle-associated PK; (T. gondii ARI) | 234 | 6.00 × 10−75 | 76 | KYF44017.1 |
| SRCN_977 | Cell-cycle-associated kinase CDK | Ca2+-dependent PK (CDK); (PITSLRE/CDK11) | 1502 | 7.38 | 156.35 | 1114–1429 | Cell-cycle-associated PK CDK; (T. gondii p89) | 454 | 3.00 × 10−135 | 92 | KFG28420.1 |
| 5. Kinase Group ‘Other’ (OPK; i.e., kinases with conventional PK (ePK) domains that do not fit into any of the other major groups of kinases) | |||||||||||
| SRCN_108 | Unc-51-like autophagy activating kinase 1 (ULK1) | ULK | 343 | 7.13 | 38.90 | 1-223 | ULK kinase; (T. gondii VAND) | 376 | 2.00 × 10−130 | 75 | KFH07419.1 |
| SRCN_1606 | eIF2 kinase IF2K-C | PEK; (general control nonderepressible 2 (GCN2)) | 4034 | 8.98 | 406.57 | 1235–2178 | eIF2 kinase IF2K-C; (T. gondii VAND) | 259 | 4.00 × 10−67 | 35 | KFH07289.1 |
| SRCN_2076 | Rhoptry kinase family ROP30 | Conserved hypothetical protein | 1276 | 9.18 | 134.73 | 812–1260 | ROP30 (T. gondii VEG) | 230 | 3.00 × 10−63 | 53 | CEL76436.1 |
| SRCN_2123 | Rhoptry kinase family ROP35 | PLK; (PLK-Unclassified) | 291 | 9.30 | 33.50 | 53–265 | ROP35; (T. gondii RUB) | 207 | 2.00 × 10−61 | 43 | KFG59037.1 |
| SRCN_3216 | Rhoptry kinase family ROP32 | CAMK-Unique | 523 | 7.08 | 57.00 | 214–520 | Putative PK; (T. gondii VAND) | 167 | 1.00 × 10−42 | 30 | KFH00232.1 |
| SRCN_2183 | Rhoptry kinase family ROP35 | Aurora-like | 226 | 6.36 | 25.84 | 1–212 | ROP35; (T. gondii VEG) | 198 | 8.00 × 10−59 | 48 | ESS33297.1 |
| SRCN_2271 | Putative PK (incomplete catalytic triad) | NimA (Never in mitosis gene A)-related Kinase (NEK) | 1463 | 9.02 | 157.18 | 437–1145 | Putative PK; (N. caninum L) | 327 | 5.00 × 10−90 | 68 | XP_003881849.1 |
| SRCN_2403 | Aurora kinase (incomplete catalytic triad) | PLK; (SAK/Plk4) | 778 | 9.79 | 79.92 | 492–778 | Putative Aurora kinase; (N. caninum L) | 127 | 3.00 × 10−28 | 44 | XP_003880644.1 |
| SRCN_2630 | NimA related kinase (NEK) family protein | NEK | 351 | 8.70 | 38.38 | 1–336 | NEK kinase; (T. gondii ME49) | 242 | 8.00 × 10−75 | 52 | XP_018638598.1 |
| SRCN_286 | Wee kinase | Inhibitory regulator of the RAS-cAMP (IRA1) kinase suppressor (IKS) | 1019 | 6.20 | 106.67 | 598–959 | Wee kinase; (H. hammondi) | 445 | 5.00 × 10−141 | 58 | XP_008882669.1 |
| SRCN_3075 | Tyrosine kinase-like (TKL) protein | Numb-associated kinase (NAK) | 1571 | 8.41 | 164.18 | 16–500 | TKL; (T. gondii TgCatPRC2) | 138 | 1.00 × 10−32 | 73 | KYK64203.1 |
| SRCN_3142 | PIK3R4 kinase-related | Aurora | 997 | 8.72 | 106.54 | 548–899 | Putative PIK3R4 kinase-related protein; (N. caninum L) | 449 | 2.00 × 10−137 | 60 | XP_003885774.1 |
| SRCN_3151 | NimA related kinase (NEK) family protein | NEK | 3186 | 7.96 | 318.69 | 352–656 | NEK kinase; (T. gondii VEG) | 468 | 7.00 × 10−131 | 73 | CEL78174.1 |
| SRCN_3247 | Rhoptry kinase family ROP27 | Ciliate-D | 345 | 8.94 | 38.81 | 23–325 | ROP27; (T. gondii p89) | 163 | 4.00 × 10−43 | 31 | KFG37427.1 |
| SRCN_3417 | Aurora kinase | Aurora | 438 | 7.65 | 48.49 | 14–289 | Aurora kinase; (T. gondii TgCatPRC2) | 490 | 3.00 × 10−155 | 76 | KYK63669.1 |
| SRCN_3444 | Unc-51-like Autophagy activating kinase 1 (ULK1) | ULK | 406 | 6.52 | 44.69 | 12–406 | ULK kinase; (T. gondii RUB) | 232 | 3.00 × 10−71 | 61 | KFG59767.1 |
| SRCN_3669 | CMGC kinase | ULK | 1803 | 8.41 | 189.65 | 736–1200 | Putative CMGC kinase; (N. caninum L) | 624 | 0.0 | 62 | CEL65030.1 |
| SRCN_4410 | Rhoptry kinase family ROP35 | PKA-like | 204 | 9.44 | 23.50 | 1–166 | ROP35; (H. hammondi) | 107 | 1.00E × 10−25 | 39 | XP_008885989.1 |
| SRCN_4503 | eIF2 kinase IF2K-B | PEK; (general control nonderepressible 2 (GCN2)) | 158 | 5.76 | 17.59 | 1–158 | eIF2 kinase IF2K-B (T. gondii TgCatPRC2) | 149 | 5.00 × 10−41 | 74 | KYK69938.1 |
| SRCN_4528 | NimA related kinase (NEK) family protein | NEK | 187 | 8.20 | 21.23 | 1–186 | NEK kinase; (H. hammondi) | 177 | 4.00 × 10−54 | 64 | XP_008885186.1 |
| SRCN_2404 | Aurora kinase (incomplete catalytic triad) | Serum and glucocorticoid induced Kinase (SGK) | 295 | 8.81 | 31.42 | 1–249 | Putative Aurora kinase; (N. caninum L) | 126 | 7.00 × 10−31 | 43 | CEL65223.1 |
| SRCN_5653 | PEK kinase | Aurora | 626 | 8.27 | 60.75 | 513–626 | PEK kinase (T. gondii TgCatPRC2) | 251 | 1.00 × 10−76 | 60 | KYK62422.1 |
| SRCN_5943 | NIMA-related kinase NIMA1 | NEK | 2842 | 9.04 | 295.44 | 73–383 | NIMA-related PK NIMA1; (T. gondii MAS) | 486 | 2.00 × 10−140 | 67 | KFH05809.1 |
| SRCN_6157 | Unc-51-like autophagy activating kinase 1 (ULK1) | ULK | 2420 | 9.38 | 250.58 | 1380–1672 | ULK kinase (T. gondii ME49) | 99.8 | 3 × 10−21 | 38 | XP_018635814.1 |
| SRCN_6184 | Myosin-light-chain kinase | Ciliate-E2-Unclassified | 478 | 5.42 | 53.65 | 177–474 | ROP19A (T. gondii ME49) | 127 | 3.00 × 10−30 | 27 | XP_018637476.1 |
| SRCN_6572 | Tyrosine kinase-like (TKL) | ULK | 622 | 6.13 | 68.80 | 1–345 | TKL; (T. gondii VAND) | 181 | 1.00 × 10−46 | 74 | KFH00338.1 |
| SRCN_6812 | PK | ULK | 199 | 6.74 | 22.72 | 1–183 | PK; (H. hammondi) | 172 | 3.00 × 10−48 | 53 | XP_008887491.1 |
| SRCN_7083 | Rhoptry kinase family ROP35 | PKA-like | 262 | 9.62 | 30.26 | 1–242 | ROP35; (H. hammondi) | 127 | 6.00 × 10−32 | 39 | XP_008885989.1 |
| SRCN_4310 | Rhoptry kinase family ROP33 | Kinase Homologous to SPS1/STE20 (KHS) | 1591 | 9.85 | 169.63 | 1265–1578 | ROP33; (H. hammondi) | 306 | 2.00 × 10−87 | 39 | XP_008887632.1 |
| SRCN_7082 | Rhoptry kinase family ROP33 | Kinase Homologous to SPS1/STE20 (KHS) | 403 | 9.59 | 45.92 | 77–390 | ROP33 (T. gondii p89) | 277 | 3.00 × 10−89 | 40 | KFG45248.1 |
| SRCN_7084 | Rhoptry kinase family ROP37 | Ribosomal protein S6 Kinases (RSK; (RSK)) | 339 | 5.41 | 38.09 | 19–334 | ROP37; (N. caninum L) | 144 | 1.00 × 10−36 | 36 | CEL64242.1 |
| 6. Kinase Group “Sterile” serine/threonine kinase, or sterile-phenotype kinases (STE) | |||||||||||
| SRCN_1328 | Serine threonine kinase | Conserved hypothetical protein | 1461 | 9.29 | 158.74 | 559–671 | Hypothetical protein, conserved; (E. maxima) | 88.6 | 5.00 × 10−16 | 68 | XP_013335801.1 |
| SRCN_5172 | “Sterile” serine/threonine kinase (STE) | Mammalian Sterile 20-like (MST)) | 6552 | 6.14 | 671.51 | 3410–4122 | STE kinase; (T. gondii TgCatPRC2) | 412 | 1.00 × 10−114 | 54 | KYK71951.1 |
| 7. Kinase Group Tyrosine Kinase-Like (TKL) | |||||||||||
| SRCN_1435 | Tyrosine kinase-like (TKL) | Mixed lineage kinase (MLK); (Leucine Zipper-bearing Kinase (LZK)) | 3064 | 8.05 | 306.74 | 2540–3060 | Tyrosine kinase-like (TKL) protein; (N. caninum L) | 278 | 3.00 × 10−73 | 72 | CEL64955.1 |
| SRCN_1571 | Tyrosine kinase-like (TKL) | Microtubule-associated S/T kinase (MAST) | 550 | 9.76 | 59.91 | 135–501 | Conserved hypothetical protein; (E. praecox) | 76.3 | 6.00 × 10−13 | 53 | CDI87140.1 |
| SRCN_3466 | Tyrosine kinase-like (TKL) | TKL-Unique | 3002 | 9.87 | 320.97 | 2342–2997 | Tyrosine kinase-like (TKL) protein; (H. hammondi) | 202 | 3.00 × 10−50 | 65 | XP_008887506.1 |
| SRCN_3928 | Tyrosine kinase-like (TKL) | LISK - LIMK (LIM kinase) and TESK (Testicular protein Kinase); (DD1) | 5842 | 8.78 | 608.80 | 3639–4268 | Tyrosine kinase-like (TKL) protein; (T. gondii TgCatPRC2) | 216 | 2.00 × 10−61 | 79 | KYK63216.1 |
| SRCN_4277 | Kinase domain-containing protein | TKL-ciliate1 | 2256 | 8.43 | 240.25 | 1570–2256 | Tyrosine kinase-like (TKL) protein; (N. caninum L) | 204 | 8.00 × 10−51 | 61 | CEL67693.1 |
| SRCN_811 | Tyrosine kinase-like (TKL) | TKL-Unique | 1099 | 9.18 | 119.26 | 814–1083 | Putative tyrosine kinase-like (TKL) protein; (E. acervulina) | 403 | 6.00 × 10−125 | 59 | XP_013252162.1 |
| 8. Kinase Group Atypical (aPKs) | |||||||||||
| SRCN_3601 | Atypical MEK-related kinase | Muscle-associated kinase TRIO | 950 | 7.20 | 103.14 | 381–850 | Atypical MEK-related kinase; (T. gondii GT1) | 171 | 1.00 × 10−42 | 32 | EPR62774.1 |
| SRCN_5962 | Atypical MEK-related kinase | Rho-associated protein kinase (ROCK)-like | 805 | 5.01 | 87.71 | 525–805 | Atypical MEK-related kinase; (H. hammondi) | 127 | 4.00 × 10−29 | 56 | XP_008884362.1 |
| SRCN_3988 | Phosphatidylinositol 3-/4-kinase (PI3K) | Atypical/PIKK/ATM | 4251 | 5.95 | 440.5 | 3671–3772 | PI3K, ; (H. hammondi) | 268 | 1 × 10−69 | 55 | XP_008886631.1 |
| SRCN_6465 | Phosphatidylinositol 3-/4-kinase (PI3K) | Atypical PIKK/ATM | 207 | 5.2 | 23.24 | 5-122 | Phosphatidylinositol 4-kinase, partial; (T. gondii p89) | 272 | 4 × 10−93 | 96 | KFG28404.1 |
| SRCN_1259 | Phosphatidylinositol 3-/4-kinase (PI3K) | Atypical/PIKK/FRAP | 1362 | 9.56 | 142.85 | 1140–1311 | PI3K; (T. gondii MAS) | 317 | 3 × 10−94 | 76 | KFH10008.1 |
| SRCN_6464 | Phosphatidylinositol 3-/4-kinase (PI3K) | No hits found | 2108 | 8.95 | 209.68 | 1626–1722 | Phosphatidylinositol 3-4-kinase; (T. gondii p89) | 244 | 1 × 10−63 | 66 | KFG28409.1 |
| SRCN_1743 | Pyruvate dehydrogenase kinase | Atypical/PDHK/BCKDK | 930 | 6.50 | 101.41 | 291–426 | PDHK, isoenzyme-2; (N. caninum L) | 458 | 6 × 10−146 | 45 | CEL70411.1 |
a The protein sequences and their corresponding identified were obtained from the Toxoplasma Genomics Resource database (Release 28; Version May 2016) [42]; b the descriptions of the protein sequence are based on BLASTp annotations using Blast2GO (see the text for details); c the kinase classification is based on BLASTp on the kinase database.
Assignment of S. neurona PK groups was accomplished by sequence clustering using Blast2GO [37] and by BLASTp searches in the Kinase database [14]. Out of the eleven known PK groups [12,13,14], S. neurona PKs segregated into the AGC (n = 9), CAMK (n = 20), CK1 (n = 3), CMGC (n = 19), STE (n = 2), TKL (n = 6), aPK (n = 7) and OPK (n = 31) (Table 1). Apart from the 31 OPKs that do not fit into the major kinase groups, the CAMK and CMGC groups, whose members are essential for the parasite’s host cell invasion [38] and differentiation (via cell-cycle regulation) [39], respectively, had the highest number of PKs, underlying the importance of these processes in the parasite. Unlike in some parasites, such as P. falciparum that lack STEs [18], S. neurona contains STE PKs.
2.1.1. The AGC Group
The numbers of apicomplexan AGCs range from four (in Babesia bovis) to 15 (in T. gondii) [17]. Based on our Blast2GO annotations and BLASTp homology searches against the kinome database, five out of the nine S. neurona AGCs (SRCN_3339, SRCN_3990, SRCN_5165, SRCN_5610 and SRCN_1312) were homologs to the universally-conserved PKAs that are found in N. caninum and T. gondii (see Table 1). The PKAs are essential for the completion of schizogony (asexual reproduction) in Plasmodium parasites [40]. Further, S. neurona contains a putative PKG (SRCN_4518), which shows high homology (92%) to the T. gondii TgPKG1 (Table 1); PKGs are essential in apicomplexans [41].
2.1.2. The CAMK Group
CAMKs form the second-largest apicomplexan PKs (after OPKs). Apicomplexan kinomes constitute varying numbers of CAMKs, which range from seven (in B. bovis) to 29 (in T. gondii) [17]. The most important CAMK family is the CDPK, which appeared to constitute almost 50% of S. neurona putative CAMKs (see Table 1). In terms of homologies, the S. neurona kinome contained orthologs to the T. gondii CDPK1 (SCRN_3314), CDPK2B (SCRN_2165), CDPK3 (SCRN_3701), CDPK4 (SCRN_6606), CDPK5 (SCRN_3583), CDPK6 (SCRN_3011), CDPK7 (SCRN_6597) and CDPK8 (SCRN_5948). Other CDPK orthologs were to the N. caninum CDPK2 (SCRN_4390) and Hammondia hammondi CDPK9 (SCRN_5812) (Table 1). Inhibition of TgCDPK1 has been shown to disrupt the motility, host cell invasion and egress of T. gondii [43]. Owing to the absence of mammalian CDPK homologs, the identification of a relatively large number of CDPK homologs in S. neurona could be utilized in the rational design of anti-parasitic therapeutics.
2.1.3. The CK1 Group
It is notable that S. neurona putatively encodes for three CK1 enzymes. Apart from T. gondii and some alveolates (e.g., Cryptosporidium hominis and Cryptosporidium parvum, important causative agents of diarrhea in children), which have three and two CK1 enzymes, respectively, most apicomplexans possess a single CK1 enzyme [17]. Two of the three S. neurona putative CK1 (SRCN_3445 and SRCN_4645) showed high sequence similarity (>90%) to the T. gondii TME49_040640 (TgCK1-α) and TGME49_089320 (TgCK1-β), respectively (Table 1). Inhibition of CK1 showed potential for anti-parasitic interventions in T. gondii [44]. CK1 is critical for the asexual proliferation of the Plasmodium parasites and is expressed in all of the life-cycle stages of the parasite [45]. Three putative S. neurona CK1 had significant sequence similarity to the P. falciparum PfCK1, i.e., 74% (SRCN_3445), 65% (SRCN_4587) and 56% (SRCN_4645) (data not shown).
2.1.4. The CMGC Group
The CMGC is the largest PK group in apicomplexans; CMGC numbers range from 15 in B. bovis to 23 in Plasmodium vivax [17], which is within the range we identified in the S. neurona kinome in our study (i.e., 19 CMGCs; see Table 1). Notable of these were the two GSK homologs (SRCN_1731 and SRCN_1732). This finding is similar to what has been observed in Plasmodium parasites in which two GSK-3 enzymes have been reported, both of which are essential for the parasite [46]. Homology searches showed considerable sequence similarity (51% and 41% for SRCN_1731 and SRCN_1732, respectively) to the PfGSK-3 enzymes (data not shown). Notably, eight of the 19 CMGCs in S. neurona were CDKs, including CDK7 (SRCN_4674, SRCN_2759 and SRCN_761), CDK10 (SRCN_895) and CDK11 (SRCN_977). Available data show that CDKs are essential in P. falciparum [24]. We also identified two putative MAPK homologs (SRCN_4209 and SRCN_5365) and ERK7 (SRCN_6472) (see Table 1), a result that is comparable to the two MAPKs in the kinome of P. falciparum [17].
2.1.5. The OPK Group
The apicomplexan-specific OPKs are a tight cluster of PKs without clear relation to any of the other major PK groups. Notable of these are ROPKs, which have high sequence divergence and have been thought to be largely restricted to T. gondii [47], which has a total of 34 members spread in over 40 distinct sub-families [23]. Although their diversification in apicomplexans is poorly understood, some ROPKs are key virulence factors in T. gondii [23]. At least nine putative ROPKs could be identified in S. neurona, including ROPK19A (SRCN_6184), ROP27 (SRCN_3247), ROP30 (SRCN_2076), ROP33 (SRCN_7082 and SRCN_7086), ROP35 (SRCN_2183, SRCN_2123, SRCN_7083 and SRCN_4410) and ROP37 (SRCN_7084), implying that the ROPKs are not restricted to T. gondii. Although largely presumed to be inactive, ROPKs are implicated in the regulation of the host transcription [47], and their presence in S. neurona may support the hypothesis that the ROPKs have a unique activation mechanisms in their regulatory functions that facilitate apicomplexan pathogenesis [24,48]. Other notable OPKs included two parasite-specific eukaryotic initiation factor-2 (elF2) kinases (elF2K-C (SRCN_1606) and elF2K-B (SRCN_4503)), four NEKs (SRCN_4528, SRCN_2630, SRCN_286 and SRCN_3151) and four ULKs (SRCN_3444, SRCN_3669, SRCN_6812 and SRCN_6157) (Table 1). The elF2Ks are conserved in apicomplexans and are important for the induction of parasite differentiation into the bradyzoites cysts, which are clinically important [34].
2.1.6. The STE Group
The STEs are poorly represented in apicomplexans, and although most apicomplexans have one or two STE genes per genome, some parasites, such as C. parvum, are reported to harbor up to six STEs [17,20]. Our results suggest that S. neurona has at least one putative STE (Table 1). STEs are thought to function in MAPK pathway cascades despite the fact that this pathway is absent in apicomplexans. The small repertoire of apicomplexan STEs is in contrast to that reported in other parasites, such as trypanosomatids, in which these enzymes regulate the length of the flagella [49].
2.1.7. The TKL Group
Apicomplexans harbor a maximum of seven TKL-coding genes, which makes it notable that we identified six putative TKLs in S. neurona (Table 1). Reverse genetics studies have demonstrated that some of the conserved TKLs, for instance PfTKL3, are essential for the asexual Plasmodium proliferation [27], thereby a potential drug target. Two of the six S. neurona putative TKLs had considerable sequence similarities to the Plasmodium TKLs, including SRCN_3466 (36% similar to Plasmodiuim malariae TKL1) and SCRN_1435 (49% similar to Plasmodium ovale TKL3) (data not shown).
2.1.8. The aPK Group
The aPKs have been detected in apicomplexan parasites, such as P. falciparum [17,18] and T. gondii, which has at least four genes thought to encode these enzymes, the products of which are hypothesized to be part of the ovoid mitochondrial cytoplasmic (OMC) complex [50], a composite assembly of organelles observed only in growing tachyzoites of T. gondii. An exhaustive search of the S. neurona proteome revealed four putative PIKKs (SRCN_3988, SRCN_6464, SRCN_6465, SRCN_1259) and one PDHK (SRCN_1743) (Table 1). Whereas PIKKs have been identified in at least 12 apicomplexan kinomes, PDHK seem to have been identified only in the T. gondii kinome [17]. Our analyses of the putative S. neurona PKs did not yield any homologs of the Alpha and RIO kinases, implying that these PKs are absent from the kinome of this parasite; RIO kinases have been reported in P. falciparum [17,18], as well as in the kinomes of other apicomplexans including C. parvum, T. gondii and B. bovis [17].
2.2. Evolution of S. neurona Protein Kinases
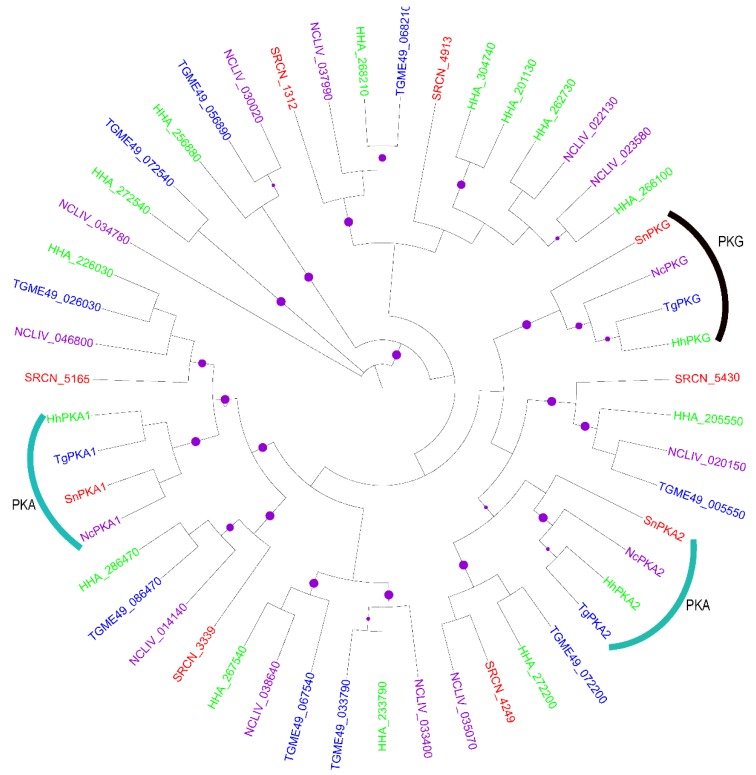
We investigated the evolutionary relationships among the various S. neurona PK groups and their homologs in related apicomplexans. Our analysis revealed valuable insights into the biology of these organisms. The kinome of S. neurona is comprised of slightly fewer AGCs (n = 9) compared to the kinomes of T. gondii (n = 11), N. caninum (n = 13) and H. hammondi (n = 15). In general, the phylogenetic clustering of the S. neurona AGCs mirrored the homologies of these enzymes to those of the three apicomplexans used in this study (Figure 1; compare with Table 1). Sequence analysis of S. neurona AGCs revealed significant divergence with only ~30% sequence similarity amongst members of this group. Two S. neurona AGCs SRCN_5610 (SnPKA1) and SRCN_3990 (SnPKA2) clearly cluster with T. gondii PKAs TGME49_028420 and TGME49_015670 [51] (Figure 1). Moreover, SRCN_5610 shares high (~60%) full length sequence identity with its ortholog, TgPKA1. It is also notable that the single putative PKG (SRCN_4518) distinctly clustered with its T. gondii ortholog, TGME49_111360 (TgPKG) (Figure 1). It has recently been shown that P. falciparum PKG acts as a signaling hub that plays a central role in a number of core parasite processes [52].
Figure 1.
Mid-point rooted maximum likelihood (ML) phylogenetic tree of apicomplexan AGCs. The terminal branches are color-coded for AGCs in the kinomes of Sarcocystis neurona (SRCN; red), Toxoplasma gondii, ME-49 strain (TGME49; blue), Hammondia hammondi (HHA; green) and Neospora caninum, Liverpool strain (NCLIV; purple). A solid purple circle on a branch indicates bootstrap support greater than 70. The phylogenetic tree was inferred from a multiple sequence alignment using PhyML with the Le and Gascuel (LG) amino acid substitution model and the gamma model of substitution rate heterogeneity. The tree image was rendered with iTOL.
In addition to the kinase domain, SnPKA1, SRCN_3339, SRCN_5165 and SRCN_4518 possess the AGC-kinase C-terminal domain, which contains two of the three conserved phosphorylation sites in AGCs (data not shown). These conserved sites serve as phosphorylation-regulated switches in the control of both intra- and inter-molecular interactions [53]. Like T. gondii, S. neurona lacks PKB and PKC. However, S. neurona contains a putative phosphoinositide-dependent kinase-1, PDPK1 (SRCN_1312), that clusters with the T. gondii PDPK1 (TGME49_268210) [51].
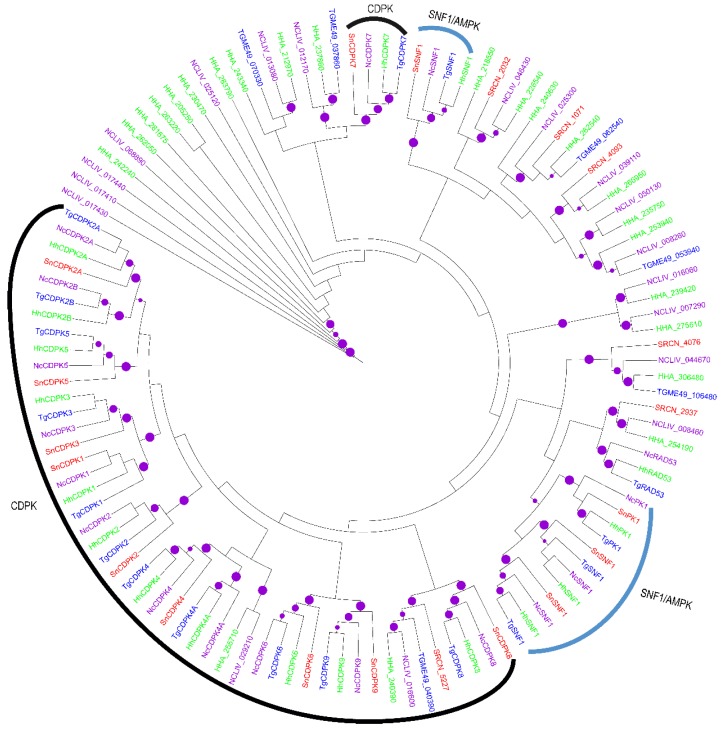
Despite the absence of PKC in S. neurona, CAMK family members were identified, which perhaps underscores the importance of Ca2+ regulation in this apicomplexan parasite. The majority of the identified S. neurona CAMKs segregated with their orthologs in T. gondii, N. caninum and H. hammondi in clades with robust bootstraps (Figure 2), thus validating the annotation of the CAMKs. Amongst the CAMKs, SRCN_2544 clustered with T. gondii PK1 (TGME049_243500) of the AMPK/SNF1 sub-family. There were also three additional SNF1 members in S. neurona (SRCN_5410, SRCN_4815 and SRCN_2257), which clustered with T. gondii TGME49_315190, TGME49_233905 and TGME49_291050, respectively.
Figure 2.
Mid-point rooted ML phylogenetic tree of apicomplexan CAMKs. The terminal branches are color-coded for AGCs in the kinomes of S. neurona (SRCN; red), T. gondii, ME-49 strain (TGME49; blue), H. hammondi (HHA; green) and N. caninum, Liverpool strain (NCLIV; purple). A solid purple circle on a branch indicates bootstrap support greater than 70. The phylogenetic tree was inferred from a multiple sequence alignment using PhyML with the LG amino acid substitution model and the gamma model of substitution rate heterogeneity. The tree image was rendered with iTOL.
Based on the clustering with T. gondii CDPK orthologs, 10 S. neurona CDPKs, including CDPK1 (SRCN_3314), CDPK2 (SRCN_4390), CDPK2A (SRCN_2165), CDPK3 (SRCN_3701), CDPK4 (SRCN_6606), CDPK5 (SRCN_3583), CDPK6 (SRCN_3011), CDPK7 (SRCN_6597), CDPK8 (SRCN_5948) and CDPK9 (SRCN_5812), were identified (Figure 2). This result implies that S. neurona has potentially lost at least two CDPKs (compared to the 12 CDPK that have been reported in T. gondii [26]). The possible loss notwithstanding, S. neurona contained the six CDPKs that are expressed and are well-conserved in most apicomplexans (i.e., CDPK1, CDPK3, CDPK4, CDPK5, CDPK6 and CDPK7) [54]. Sequence analysis revealed that, like in other apicomplexans, all identified S. neurona CDPKs except SnCDPK7 contained both a kinase domain and a Ca2+-binding domain known as the EF-hand domain [26]. Like its T. gondii ortholog, TgCDPK7) SnCDPK7 contains a pleckstrin-homology (PH) domain just upstream of its PK domain [54]. The domain architecture in CDPKs is such that kinase activity is stimulated upon Ca2+-binding. Putative additional S. neurona CDPKs include SRCN_5227, which falls within the CDPK cluster and segregates with TGME49_040390 that is annotated as a CDPK and SRCN_4076 that clusters with TGME49_106480, also annotated as a CDPK (Figure 2).
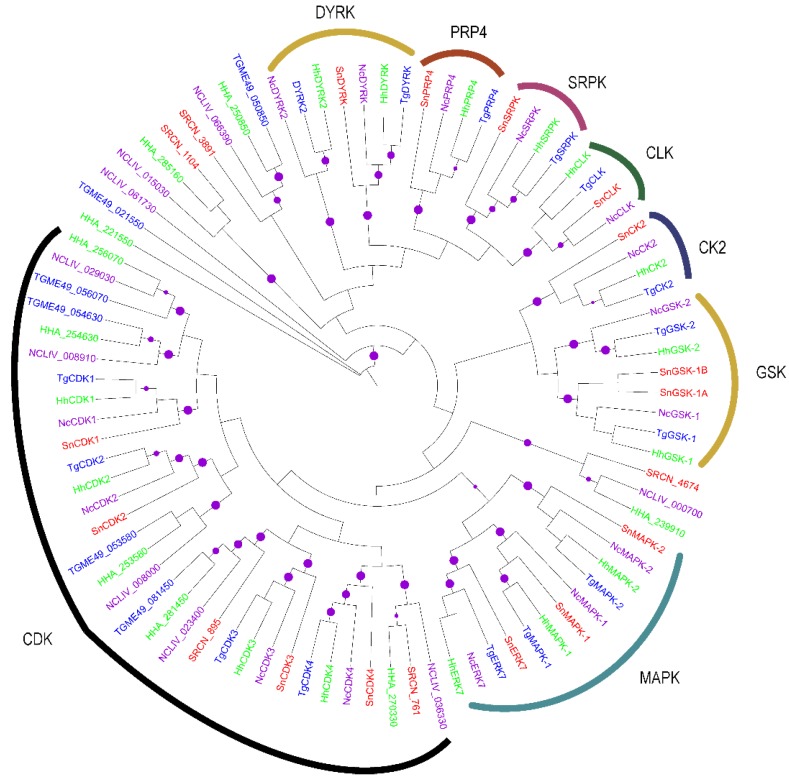
The majority of the putative CMGCs identified in S. neurona clustered with robust bootstrap support values with the conserved CMGCs in T. gondii, N. caninum and H. hammondi (Figure 3). Based on the segregation of the CMGC kinases, four S. neurona CDK1 (SRCN_4801), CDK2 (SRCN_2759), CDK3 (SRCN_977) and CDK4 (SRCN_6346) were identified (Figure 3). CDKs are amongst the main molecular switches that regulate cell cycle progression in apicomplexan parasites [55]. Additional S. neurona CMGC kinases identified include SRPK (SRCN_1236), CLK (SRCN_1479), PRP4 (SRCN_2845), DYRK (SRCN_1611), GSK-1A (SRCN_1731), GSK-1B (SRCN_1732), CK2 (SRCN_6427), ERK7 (SRCN_6472), MAPK-2 (SRCN_5365) and MAPK-1 (SRCN_4209), all of which fall in orthologous clades with robust bootstrap values. SRPK, CLK and PRP4 kinases most likely have crucial roles in parasite survival given their involvement in cycle-regulatory regulation [56,57,58], potentially via alternative mRNA splicing. Inhibition of PfCLK-mediated SR protein phosphorylation impaired blood stage replication and malaria transmission in Plasmodium [59]. The DYRK is implicated in a myriad of cell cycle functions that make this PK, hence an attractive drug target [60]. Other drug targets include MAPKs, which regulate diverse cellular functions, such as tissue morphogenesis, cytoskeletal rearrangements, proliferation, differentiation, survival, immune responses and adaptation/stress-responses [61]. The S. neurona putative MAPK-1 (SRCN_4209) ortholog in T. gondii (TgMAPK-1) is a virulence factor that alters IFN-γ-mediated control of Toxoplasma tachyzoite proliferation by manipulating IFN-γ-mediated nitric oxide synthase (iNOS) and NO generation [62].
Figure 3.
Mid-point rooted ML phylogenetic tree of apicomplexan CMGCs. The terminal branches are color-coded for AGCs in the kinomes of S. neurona (SRCN; red), T. gondii, ME-49 strain (TGME49; blue), H. hammondi (HHA; green) and N. caninum, Liverpool strain (NCLIV; purple). A solid purple circle on a branch indicates bootstrap support greater than 70. The phylogenetic tree was inferred from a multiple sequence alignment using PhyML with the LG amino acid substitution model and the gamma model of substitution rate heterogeneity. The tree image was rendered with iTOL.
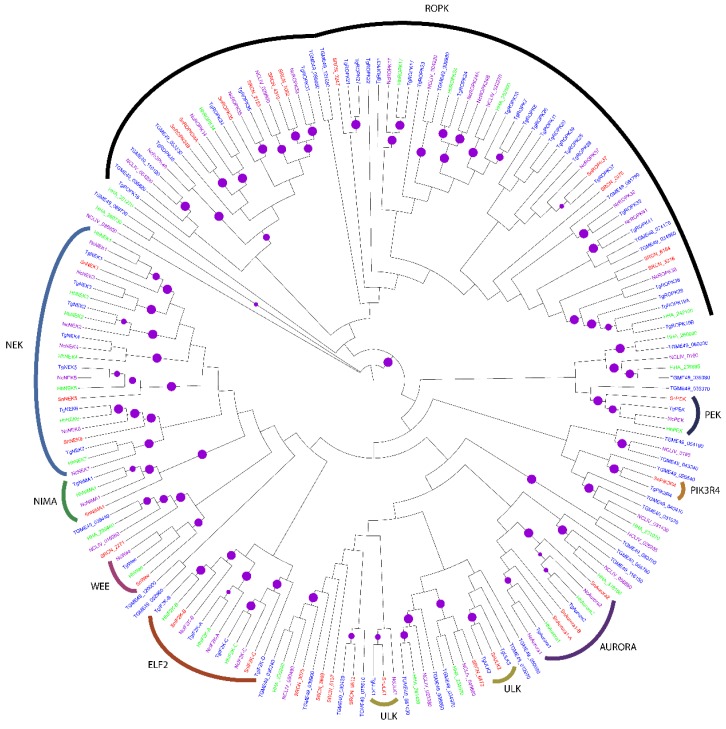
In the OPK family, SRCN_4528, SRCN_2630 and SRCN_3151 are putative NEKs given their clustering with T. gondii NEK kinases TgNEK1 (TGME49_319700), TgNEK6 (TGME49_294260) and TgNEK5 (TGME49_018400), respectively (Figure 4). S. neurona has two putative ULK kinases SRCN_108 (SnULK2) and SRCN_3444 (SnULK1) that cluster with their T. gondii orthologs TgULK1 (TGME49_235750) and TgULK2 (TGME49_240630), respectively). S. neurona contains three putative Aurora kinases; SRCN_2404 (SnAurora1-A), SRCN_2403 (SnAurora1-B) and SRCN_3417 (SnAurora2) that cluster with T. gondii aurora kinases, TgAurora1 (TGME49_118770) and TgAurora2 (TGME49_003010) (Figure 4). S. neurona also contains a putative Wee kinase, SnWee (SRCN_286) that clusters with T. gondii Wee kinase, TgWee (TGME49_273690), as well as a NIMA kinase, SnNIMA1 (SRCN_5943), that clusters with TgNIMA1 (TGME49_292140). NIMA-related kinases are implicated in cell cycle control. Like its apicomplexan relatives T. gondii and P. falciparum, S. neurona contains tyrosine-kinase-like (TKL) kinase, SRCN_6572, that clusters with TGME49_234970. Moreover, S. neurona contains two elF2 kinases, SnIF2K-B (SRCN_4503) and SnIF2K-C (SRCN_1606) (Figure 4). SRCN_4503 shows 74% sequence similarity to the T. gondii TgCatPRC2 [IF2K-B] (Table 1).
Figure 4.
Mid-point rooted ML phylogenetic tree of apicomplexan OPKs. The terminal branches are color-coded for AGCs in the kinomes of S. neurona (SRCN; red), T. gondii, ME-49 strain (TGME49; blue), H. hammondi (HHA; green) and N. caninum, Liverpool strain (NCLIV; purple). A solid purple circle on a branch indicates bootstrap support greater than 70. The phylogenetic tree was inferred from a multiple sequence alignment using PhyML with the LG amino acid substitution model and the gamma model of substitution rate heterogeneity. The tree image was rendered with iTOL.
The clustering of putative S. neurona ROPKs was also notable. For instance, SRCN_7084 (SnROPK37) clustered with T. gondii TgROPK37 (TGME49_094560), implying that it is a ROPK37 (Figure 4). SRCN_7082 and SRCN_4310 cluster with T. gondii TgROPK33 (TGME49_001130), suggesting that they are isoforms of ROPK33. Annotations in Table 1 indicate that SRCN_4310 is a putative ROP33 (39% sequence similarity to H. hammondi ROP33). Moreover, SRCN_2183 (SnROPK35) clusters with TgROPK35 (TGME49_104740). Other putative S. neurona ROPKs include SRCN_7083 (SnROPK34A) and SRCN_4410 (SnROPK34B) that cluster with TgROP34 (TGME49_040090) (Figure 4). SnROPK34A and SnROPK34B are potentially duplicated forms of TgROPK34.
Moreover, SRCN_3142 (SnPIK3R4) segregates with T. gondii TgPIK3R4 (TGME49_018550). NEKs are involved in cell cycle regulation, while Aurora kinases play pivotal roles in endodyogeny, duplication rate and parasite virulence [33]. Taken together, the presence of a variety of ROPKs in S. neurona is interesting given the fact that in T. gondii, ROPKs are key virulence factors [63].
3. Discussion
The kinomes of apicomplexans range from 35 PKs (in B. bovis) to 135 PKs (in T. gondii) [24]. We identified a total of 97 putative PKs in the kinome of S. neurona, compared to the PKs reported in the kinomes of P. falciparum (n = 99), T. gondii (n = 135), N. caninum (n = 130) and H. hammondi (n = 124) [17,19]. Although the total number of S. neurona PKs appeared markedly reduced compared to that of its close coccidian relatives (T. gondii and N. caninum [23]), taken as a percentage of total genome size, the proportion of S. neurona PKs is comparable to the 2% observed in humans [13] and other coccidians [23]. The contraction of the S. neurona kinome could be attributed to genome compaction, which occasionally offsets lineage-specific expansions of specific gene families. Notably, genome contraction is a common mode of genomic evolution in intracellular parasites, including apicomplexans [64,65]. As such, the evolution of PKs may be in tandem to the overall genomic adaptive strategies of these parasites.
Using a hierarchical scheme based on the major PK groups, the S. neurona kinases could be classified and phylogenetically clustered into the various PK families. A complement of nine putative AGC kinases was identified in S. neurona, which is reduced compared with that of T. gondii, N. caninum and H. hammondi. Despite this potential gene loss, seven of the nine AGCs (SRCN_5165, SRCN_5610, SRCN_3339, SRCN_4249, SRCN_3990, SRCN_5430 and SRCN_1312) had orthologs in T. gondii, N. caninum and H. hammondi. In agreement with the observation that PKA is conserved in apicomplexans [23], two PKAs (SRCN_5610 and SRCN_3990) were identified in S. neurona. In T. gondii, increases in cytosolic cAMP levels activate PKA to trigger the developmental switch from the rapidly proliferating tachyzoites to the quiescent bradyzoites [66]. Additionally, two other S. neurona AGCs (SRCN_5165 and SRCN_3339) were putative PKAs given that they contained the characteristic GxGxxG motif found in PKA [51]. Notably, based on orthology, S. neurona contains a single putative PKG (SRCN_4518) that distinctly clustered with T. gondii PKG (TGME49_111360).
In apicomplexans, CAMKs modulate the intracellular Ca2+ concentration, which in turn regulates vital processes, such as host-cell invasion, protein secretion and parasite differentiation. We identified four potential AMPK/SNF1 family members (SRCN_2544, SRCN_5410, SRCN_4815 and SRCN_2257). The AMP-activated PK cascade acts as a metabolic sensor that monitors cellular AMP and ATP levels and is activated by an elevation of the AMP:ATP ratio. Further, we identified 12 putative CDPKs in the S. neurona, including CDPK1 (SRCN_3314), CDPK2 (SRCN_4390), CDPK2A (SRCN_2165), CDPK3 (SRCN_3701), CDPK4 (SRCN_6606), CDPK5 (SRCN_3583), CDPK6 (SRCN_3011), CDPK7 (SRCN_6597), CDPK8 (SRCN_5948), CDPK9 (SRCN_5812), SRCN_5227 and SRCN_4076. Compared to the 12 CDPKs reported in T. gondii [26], it appears that S. neurona had all six well-conserved apicomplexan CDPKs (CDPK1, CDPK3, CDPK4, CDPK5, CDPK6 and CDPK7), which provide a link between Ca2+ signaling and parasite differentiation, motility, invasion and egress [54]. In T. gondii, downregulation of CDPK1 interfered with parasite motility, host cell invasion and egress [43], while disruption of CDPK3 caused defective parasite egress [67]. Further, the essentiality of CDPK6 and CDPK7 in T. gondii has recently been demonstrated [68]. Indeed, TgCDPK1 has been targeted for the development of new drugs for toxoplasmosis [69]. Sequence analysis revealed that, similar to other apicomplexans, all identified S. neurona CDPKs except CDPK7 (SRCN_6597) contain both a PK domain and an EF-hand (Ca2+-binding) domain [26]. Similar to its T. gondii ortholog, TGME49_028750 (TgCDPK7), the S. neurona CDPK7 (SRCN_6597) contains a pleckstrin-homology (PH) domain just upstream of its PK domain [54]; the domain architecture is such that kinase activity is stimulated upon Ca2+ binding. Moreover, our phylogeny provided clues of possible gene duplications giving rise to SRCN_3990 and SRCN_3011, as well as SRCN_4093 and SRCN_1071. Interestingly, based on phylogenetic analysis, S. neurona probably contains four (SRCN_5227, SRCN_1071, SRCN_4093 and SRCN_3011) species-specific CAMKs.
The CMGCs, comprising CDKs, MAPKs, GSKs and CLKs, coordinate a wide range of cellular functions in different species. By both annotations and phylogenetic analyses, we identified four putative CDKs sub-family members; CDK5 (SRCN_4801 and SRCN_6346), CDK7 (SRCN_2759, SRCN_4674 and SRCN_761), CDK10 (SRCN_895) and CDK11 (SRCN_977). The finding of CDKs in S. neurona suggests that this parasite’s cell cycle regulation could be CDK-dependent and perhaps similar to that of higher eukaryotes [70]. The identification of three putative MAPKs (SRCN_4209, SRCN_6472 and SRCN_5365) in S. neurona points to the existence of MAPK regulated transduction pathway(s) in this pathogen. Similar to its T. gondii ortholog (TgMAPK1), which is a p38α MAPK homolog [71], SRCN_4209 may be potentially involved in parasite proliferation/stage differentiation, stress response and manipulation of the host immunity to enhance virulence. On the other hand, SRCN_6472 and SRCN_5365 may augment the roles of SRCN_4209 in the parasite. In T. gondii, MAPK1/ERK7 is involved in intracellular proliferation [72]. We also identified two putative GSKs (SRCN_1731 and SRCN_1732). A genome-wide gene knockout approach in P. falciparum demonstrated PfGSK-3 to be critical for schizogony of the parasite [19]. Other S. neurona CMGC kinases identified include CLK (SRCN_1479), PRP4 (SRCN_2845), DYRK (SRCN_1611) CK2 (SRCN_6427) and SRPK (SRCN_1236).
ROPKs are secreted by T. gondii into the host cell and play roles in adhesion, motility and manipulation of immune responses [73]. We identified 11 putative ROPK sub-family members in the S. neurona kinome, i.e., ROPK37 (SRCN_7084), ROPK35 (SRCN_2183), ROPK33A (SRCN_7082), ROPK33B (SRCN_4310), ROPK34A (SRCN_7083), ROPK34B (SRCN_4410), SRCN_6184, SRCN_3216, SRCN_2076, SRCN_3247 and SRCN_2123. It has been recently demonstrated that T. gondii ROP21 and ROP27 play a role in a constitutive pathway based on their localization in the PV and cyst matrix [74]. However, the S. neurona putative ROPs 21 and 27 could not be clearly delineated in our clustering. Moreover, T. gondii ROP35 has been shown to play a crucial role in chronic infection [75]. Although the S. neurona genome is more than twice the size of other coccidians whose genomes were sequenced so far (e.g., Toxoplasma and Neospora), it has a considerably reduced number of ROPKs that nevertheless may have vital roles in the parasite’s virulence. Specifically, S. neurona is devoid of ROP5, ROP16, ROP18 and ROP38, which have been shown to confer virulence and alter the host’s cellular signaling pathways [72]. Putatively therefore, S. neurona ROPKs may have multiple roles in the survival of the parasite. In the search of drug targets against S. neurona, the reduced ROPKs with possible multiple roles and absent in the vertebrate host are thus attractive candidates.
4. Conclusions and Future Perspectives
The kinome of S. neurona contains members of the major classes of PKs, including AGC, CMGC, GSK, CAMK, CK, TKL, aPKs and several PKs in the OPK family. Similar to other apicomplexans, S. neurona kinome is devoid of PKC, the TKs, Alpha kinases, as well as RIO kinases. Further, the S. neurona kinome harbors two putative MAPK homologs, a finding that is similar to some apicomplexans, such as P. falciparum. S. neurona kinome also lacks some of the ROPKs that have been implicated in the virulence of T. gondii. Given the central roles played by PKs in the regulation of the host-parasite interactions and in the facilitation of the parasite proliferation and differentiation, delineation of the S. neurona kinome offers a platform for future development of efficacious drugs for EPM, for instance via parasite transmission blocking vaccine against the parasites (specific inhibition of the parasite’s PKs). This approach is made possible by the differences between parasite and host PK homologs [76]. Zhang et al. [77] reviewed the applications and the progress made in the targeting of specific PKs as antimalarial drugs against Plasmodium parasites. Proof of principle of this approach has been demonstrated by the inhibition of human PKs using chemical ligands to treat cancers and other diseases [78,79]. Recently, Ojo et al. [80] provided evidence that PKs can be targeted for rationally-designed drugs that can potently inhibit the growth of S. neurona. The technology is available and approved for therapeutic intervention, thus offering a unique prospect of repurposing chemical ligands to manage S. neurona infections [81]. It is however important to note that experimental validations are required to validate the S. neurona putative PKs to facilitate the development of anti-parasitic interventions. A potential approach is the application of genetically-encoded sensors to identify inhibitors of important parasite signaling pathways.
5. Materials and Methods
5.1. Genome-Wide Identification of Putative S. neurona PKs
The predicted S. neurona proteome was downloaded from the Toxoplasma Genomics Resource database (Release 28; Version May 2016) [42]. A hidden Markov model (HMM) profile of signature PK domains obtained from the Kinomer database v 1.0 [82] was used to search for S. neurona kinases using HMMER v 3.1b2 [83]. The sequences having PK domain (IPR011009) or PK-like domain (IPR000719) were considered as putative kinases. Annotation of the putative kinase sequences was performed by BLASTp search against the non-redundant (nr)-NCBI protein and UniProtKB/Swiss-Prot databases at an e-value of ≤10−6. The identified S. neurona putative PKs were subsequently classified by BLASTp interrogations into the KinBase [84]. Gene ontology (GO) mapping was performed using Blast2GO v 4.0.7 [37]. The molecular weight (Mw) and isoelectric point (pI) were obtained using the ExPASy compute pI/Mw tool [85]. Motifs analysis was performed with the MEME Suite v 4.11.2 [86]. The parameters were as follows: number of repetitions, any; maximum numbers of motifs, 30; and the optimum motif widths, between 6 and 200 residues.
5.2. Phylogenetic Analysis
Phylogenetic trees were constructed to decipher the orthologous and paralogous relationships of S. neurona kinases. Protein kinase domains from putative S. neurona kinase groups were extracted and aligned with protein kinase domains from their homologs in T. gondii [42], H. hammondi and N. caninum using MUSCLE [87]. The alignments were subsequently manually edited in Jalview [88] for curation of alignment to remove uncertain regions due to gaps and poor alignment. Phylogenetic reconstruction was undertaken using the maximum likelihood program PhyML 3.0 [89] and RAxML v 8.0 [90] and the Bayesian inference program MrBAYES v 3.2 [91]. For PhyML, the LG substitution model was selected assuming an estimated proportion of invariant sites and four gamma-distributed rate categories to account for rate heterogeneity across sites. The gamma shape parameter was estimated directly from the data. The robustness of internal branches was evaluated using 100 bootstraps. MrBayes was run for 5,000,000 generations with two runs and four chains in parallel and a burn-in of 25%. Obtained trees were rendered with the Interactive Tree of Life server (iTOL) [92].
Acknowledgments
The authors acknowledge Peter Waweru and Rosaline Macharia of the University of Nairobi for insightful discussions. Egerton University funded the open access publication of this paper.
Author Contributions
Edwin Kimathi Murungi developed the concept. Edwin Kimathi Murungi and Henry Muriuki kariithi designed and performed the experiments, analyzed the data and wrote the paper. The authors approved the final version of the manuscript.
Conflicts of Interest
The authors declare that there is no conflict of interest in this work.
References
- 1.Dubey J.P., Lindsay D.S., Saville W.J., Reed S.M., Granstrom D.E., Speer C.A. A review of Sarcocystis neurona and equine protozoal myeloencephalitis (EPM) Vet. Parasitol. 2001;95:89–131. doi: 10.1016/S0304-4017(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 2.Reed S.M., Furr M., Howe D.K., Johnson A.L., MacKay R.J., Morrow J.K., Pusterla N., Witonsky S. Equine protozoal myeloencephalitis: An updated consensus statement with a focus on parasite biology, diagnosis, treatment, and prevention. J. Vet. Intern. Med. 2016;30:491–502. doi: 10.1111/jvim.13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubey J.P., Howe D.K., Furr M., Saville W.J., Marsh A.E., Reed S.M., Grigg M.E. An update on Sarcocystis neurona infections in animals and equine protozoal myeloencephalitis (EPM) Vet. Parasitol. 2015;209:1–42. doi: 10.1016/j.vetpar.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe D.K., MacKay R.J., Reed S.M. Equine protozoal myeloencephalitis. Vet. Clin. N. Am. Equine Pract. 2014;30:659–675. doi: 10.1016/j.cveq.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Colahan P.T., Bailey J.E., Cheeksa J.P., Jones G.L., Yangc M. Effect of sulfadiazine and pyrimethamine on selected physiologic and performance parameters in athletically conditioned thoroughbred horses during an incremental exercise stress test. Vet. Ther. 2002;3:49–63. [PubMed] [Google Scholar]
- 6.McClure S.R., Palma K.G. Treatment of equine protozoal myeloencephalitis with nitazoxanide. J. Equine Vet. Sci. 1999;19:639–641. doi: 10.1016/S0737-0806(06)82197-0. [DOI] [Google Scholar]
- 7.Bernard W.V., Beech J. Neurological examination and neurological conditions causing gait deficits. In: Ross M.W., Dyson S.J., editors. Diagnosis and Management of Lameness in the Horse. 2nd ed. Elsevier Saunders; St. Louis, MO, USA: 2003. pp. 135–145. [Google Scholar]
- 8.Warschauer B.A., Sondhof A. Equine protozoal myeloencephalitis. Iowa State Univ. Vet. 1998;60:Article 10. [Google Scholar]
- 9.Dubremetz J.F., Garcia-Reguet N., Conseil V., Fourmaux M.N. Apical organelles and host-cell invasion by Apicomplexa. Int. J. Parasitol. 1998;28:1007–1013. doi: 10.1016/S0020-7519(98)00076-9. [DOI] [PubMed] [Google Scholar]
- 10.Sadak A., Taghy Z., Fortier B., Dubremetz J.F. Characterization of a family of rhoptry proteins of Toxoplasma gondii. Mol. Biochem. Parasitol. 1988;29:203–211. doi: 10.1016/0166-6851(88)90075-8. [DOI] [PubMed] [Google Scholar]
- 11.Cesbron-Delauw M.F., Gendrin C., Travier L., Ruffiot P., Mercier C. Apicomplexa in mammalian cells: Trafficking to the parasitophorous vacuole. Traffic. 2008;9:657–664. doi: 10.1111/j.1600-0854.2008.00728.x. [DOI] [PubMed] [Google Scholar]
- 12.Hanks S.K., Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 13.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 14.Manning G., Plowman G.D., Hunter T., Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002;27:514–520. doi: 10.1016/S0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 15.Hanks S.K., Quinn A.M., Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 16.Miranda-Saavedra D., Barton G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins Struct. Funct. Bioinform. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- 17.Miranda-Saavedra D., Gabaldón T., Barton G.J., Langsley G., Doerig C. The kinomes of apicomplexan parasites. Microbes Infect. 2012;14:796–810. doi: 10.1016/j.micinf.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Ward P., Equinet L., Packer J., Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: The kinome of a divergent eukaryote. BMC Genom. 2004 doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan N., Krupa A. A genomic perspective of protein kinases in Plasmodium falciparum. Proteins Struct. Funct. Bioinform. 2005;58:180–189. doi: 10.1002/prot.20278. [DOI] [PubMed] [Google Scholar]
- 20.Talevich E., Tobin A.B., Kannan N., Doerig C. An evolutionary perspective on the kinome of malaria parasites. Philos. Trans. R. Soc. B. 2012;367:2607–2618. doi: 10.1098/rstb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tewari R., Straschil U., Bateman A., Böhme U., Cherevach I., Gong P., Pain A., Billker O. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talevich E., Mirza A., Kannan N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol. Biol. 2011;11:321. doi: 10.1186/1471-2148-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talevich E., Kannan N. Structural and evolutionary adaptation of rhoptry kinases and pseudokinases, a family of coccidian virulence factors. BMC Evol. Biol. 2013;13:117. doi: 10.1186/1471-2148-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talevich E., Kannan N., Miranda-Saavedra D. Computational analysis of apicomplexan kinomes. In: Doerig C., Späth G., Wiese M., editors. Protein Phosphorylation in Parasites: Novel Targets for Antiparasitic Intervention. 5th ed. Wiley-Blackwell; Weinheim, Germany: 2014. pp. 1–36. [Google Scholar]
- 25.Kumar A., Vaid A., Syin C., Sharma P. PfPKB, a novel protein kinase B-like enzyme from Plasmodium falciparum I. Identification, characterization, and possible role in parasite development. J. Biol. Chem. 2004;279:24255–24264. doi: 10.1074/jbc.M312855200. [DOI] [PubMed] [Google Scholar]
- 26.Billker O., Lourido S., Sibley L.D. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe. 2009;5:612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdi A., Eschenlauer S., Reininger L., Doerig C. SAM domain-dependent activity of PfTKL3, an essential tyrosine kinase-like kinase of the human malaria parasite Plasmodium falciparum. Cell. Mol. Life Sci. 2010;67:3355–3369. doi: 10.1007/s00018-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Wang S., Wang W., Gu Y., Liu H., Wei F., Liu Q. Targeted disruption of CK1a in Toxoplasma gondii increases acute virulence in mice. Eur. J. Protistol. 2016;56:90–101. doi: 10.1016/j.ejop.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal S., Kern S., Halbert J., Przyborski J.M., Baumeister S., Dandekar T., Doerig C., Pradel G. Two nucleus-localized CDK-like kinases with crucial roles for malaria parasite erythrocytic replication are involved in phosphorylation of splicing factor. J. Cell. Biochem. 2011;112:1295–1310. doi: 10.1002/jcb.23034. [DOI] [PubMed] [Google Scholar]
- 30.Andrade L.F., Nahum L.A., Avelar L.G., Silva L.L., Zerlotini A., Ruiz J.C., Oliveira G. Eukaryotic protein kinases (ePKs) of the helminth parasite Schistosoma mansoni. BMC Genom. 2011;12:215. doi: 10.1186/1471-2164-12-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low H., Lye Y.M., Sim T.S. Pfnek3 functions as an atypical MAPKK in Plasmodium falciparum. Biochem. Biophys. Res. Commun. 2007;361:439–444. doi: 10.1016/j.bbrc.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 32.Dorin D., Le Roch K., Sallicandro P., Alano P., Parzy D., Poullet P., Meijer L., Doerig C. Pfnek-1, a NIMA-related kinase from the human malaria parasite Plasmodium falciparum. Eur. J. Biochem. 2001;268:2600–2608. doi: 10.1046/j.1432-1327.2001.02151.x. [DOI] [PubMed] [Google Scholar]
- 33.Berry L., Chen C.-T., Reininger L., Carvalho T.G., El Hajj H., Morlon-Guyot J., Bordat Y., Lebrun M., Gubbels M.-J., Doerig C. The conserved apicomplexan Aurora kinase TgArk3 is involved in endodyogeny, duplication rate and parasite virulence. Cell. Microbiol. 2016;18:1106–1120. doi: 10.1111/cmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan W.J., Narasimhan J., Bhatti M.M. Parasite-specific eIF2 (eukaryotic initiation factor-2) kinase required for stress-induced translation control. Biochem. J. 2004;380:523–531. doi: 10.1042/bj20040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blazejewski T., Nursimulu N., Pszenny V., Dangoudoubiyam S., Namasivayam S., Chiasson M.A., Chessman K., Tonkin M., Swapna L.S., Hung S.S., et al. Systems-based analysis of the Sarcocystis neurona genome identifies pathways that contribute to a heteroxenous life cycle. mBio. 2015;6:e02445–14. doi: 10.1128/mBio.02445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dice J.F., Goldberg A.L. Relationship between in vivo degradative rates and isoelectric points of proteins. Proc. Natl. Acad. Sci. USA. 1975;72:3893–3897. doi: 10.1073/pnas.72.10.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 38.Kato K., Sugi T., Takemae H., Takano R., Gong H., Ishiwa A., Horimoto T., Akashi H. Characterization of a Toxoplasma gondii calcium calmodulin-dependent protein kinase homolog. Parasites Vectors. 2016;9:405. doi: 10.1186/s13071-016-1676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan F., Tang J., Qin C., Kim K. Cyclin-dependent kinase TPK2 is a critical cell cycle regulator in Toxoplasma gondii. Mol. Microbiol. 2002;45:321–332. doi: 10.1046/j.1365-2958.2002.03026.x. [DOI] [PubMed] [Google Scholar]
- 40.Solyakov L., Halbert J., Alam M.M., Semblat J.P., Dorin-Semblat D., Reininger L., Bottrill A.R., Mistry S., Abdi A., Fennell C. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat. Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- 41.Gurnett A.M., Liberator P.A., Dulski P.M., Salowe S.P., Donald R.G., Anderson J.W., Wiltsie J., Diaz C.A., Harris G., Chang B. Purification and molecular characterization of cGMP-dependent protein kinase from apicomplexan parasites a novel chemotherapeutic target. J. Biol. Chem. 2002;277:15913–15922. doi: 10.1074/jbc.M108393200. [DOI] [PubMed] [Google Scholar]
- 42.Kissinger J.C., Gajria B., Li L., Paulsen I.T., Roos D.S. ToxoDB: Accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lourido S., Shuman J., Zhang C., Shokat K.M., Hui R., Sibley L.D. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010;465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donald R.G., Zhong T., Meijer L., Liberator P.A. Characterization of two T. gondii CK1 isoforms. Mol. Biochem. Parasitol. 2005;141:15–27. doi: 10.1016/j.molbiopara.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Dorin-Semblat D., marta-Gatsi C., Hamelin R., Armand F., Carvalho T.G., Moniatte M., Doerig C. Malaria parasite-Infected erythrocytes secrete PfCK1, the Plasmodium homologue of the pleiotropic protein kinase casein kinase 1. PLoS ONE. 2015;10:e0139591. doi: 10.1371/journal.pone.0139591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masch A., Kunick C. Selective inhibitors of Plasmodium falciparum glycogen synthase-3 (PfGSK-3): New antimalarial agents? Biochim. Biophys. Acta Proteins Proteom. 2015;1854:1644–1649. doi: 10.1016/j.bbapap.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Peixoto L., Chen F., Harb O.S., Davis P.H., Beiting D.P., Brownback C.S., Ouloguem D., Roos D.S. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host. Microbe. 2010;8:208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibley L.D., Qiu W., Fentress S., Taylor S.J., Khan A., Hui R. Forward genetics in Toxoplasma gondii reveals a family of rhoptry kinases that mediates pathogenesis. Eukaryot. Cell. 2009;8:1085–1093. doi: 10.1128/EC.00107-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erdmann M., Scholz A., Melzer I.M., Schmetz C., Wiese M. Interacting protein kinases involved in the regulation of flagellar length. Mol. Biol. Cell. 2006;17:2035–2045. doi: 10.1091/mbc.E05-10-0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kõhler S. Multi-membrane-bound structures of Apicomplexa: II. the ovoid mitochondrial cytoplasmic (OMC) complex of Toxoplasma gondii tachyzoites. Parasitol. Res. 2006;98:355–369. doi: 10.1007/s00436-005-0066-y. [DOI] [PubMed] [Google Scholar]
- 51.Artz J.D., Wernimont A.K., lali-Hassani A., Zhao Y., Amani M., Lin Y.H., Senisterra G., Wasney G.A., Fedorov O., King O. The Cryptosporidium parvum kinome. BMC Genom. 2011;12:478. doi: 10.1186/1471-2164-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alam M.M., Solyakov L., Bottrill A.R., Flueck C., Siddiqui F.A., Singh S., Mistry S., Viskaduraki M., Lee K., Hopp C.S. Phosphoproteomics reveals malaria parasite Protein Kinase G as a signalling hub regulating egress and invasion. Nat. Commun. 2015 doi: 10.1038/ncomms8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 54.Morlon-Guyot J., Berry L., Chen C.-T., Gubbels M.-J., Lebrun M., Daher W. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell. Microbiol. 2014;16:95–114. doi: 10.1111/cmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwanaga T., Sugi T., Kobayashi K., Takemae H., Gong H., Ishiwa A., Murakoshi F., Recuenco F.C., Horimoto T., Akashi H. Characterization of Plasmodium falciparum cdc2-related kinase and the effects of a CDK inhibitor on the parasites in erythrocytic schizogony. Parasitol. Int. 2013;62:423–430. doi: 10.1016/j.parint.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Aubol B.E., Adams J.A. Recruiting a silent partner for activation of the protein kinase SRPK1. Biochemistry. 2014;53:4625–4634. doi: 10.1021/bi500483m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eckert D., Andrée N., Razanau A., Zock-Emmenthal S., Lützelberger M., Plath S., Schmidt H., Guerra-Moreno A., Cozzuto L., Ayté J. Prp4 kinase grants the license to splice: Control of weak splice sites during spliceosome activation. PLoS Genet. 2016;12:e1005768. doi: 10.1371/journal.pgen.1005768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gazarini M.L., Garcia C.R.S. Interruption of the blood-stage cycle of the malaria parasite, Plasmodium chabaudi, by protein tyrosine kinase inhibitors. Braz. J. Med. Biol. Res. 2003;36:1465–1469. doi: 10.1590/S0100-879X2003001100003. [DOI] [PubMed] [Google Scholar]
- 59.Kern S., Agarwal S., Huber K., Gehring A.P., Strõdke B., Wirth C.C., Brügl T., Abodo L.O., Dandekar T., Doerig C. Inhibition of the SR protein-phosphorylating CLK kinases of Plasmodium falciparum impairs blood stage replication and malaria transmission. PLoS ONE. 2014;9:e105732. doi: 10.1371/journal.pone.0105732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rüben K., Wurzlbauer A., Walte A., Sippl W., Bracher F., Becker W. Selectivity profiling and biological activity of novel ß-carbolines as potent and selective DYRK1 kinase inhibitors. PLoS ONE. 2015;10:e0132453. doi: 10.1371/journal.pone.0132453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brumlik M.J., Pandeswara S., Ludwig S.M., Murthy K., Curiel T.J. Parasite mitogen-activated protein kinases as drug discovery targets to treat human protozoan pathogens. J. Signal Transduct. 2011 doi: 10.1155/2011/971968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brumlik M.J., Pandeswara S., Ludwig S.M., Jeansonne D.P., Lacey M.R., Murthy K., Daniel B.J., Wang R.F., Thibodeaux S.R., Church K.M. TgMAPK1 is a Toxoplasma gondii MAP kinase that hijacks host MKK3 signals to regulate virulence and interferon-g-mediated nitric oxide production. Exp. Parasitol. 2013;134:389–399. doi: 10.1016/j.exppara.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox B.A., Rommereim L.M., Guevara R.B., Falla A., Triana M.A.H., Sun Y., Bzik D.J. The Toxoplasma gondii rhoptry kinome is essential for chronic infection. mBio. 2016;7:e00193–16. doi: 10.1128/mBio.00193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lawrence J.G., Hendrix R.W., Casjens S. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 2001;9:535–540. doi: 10.1016/S0966-842X(01)02198-9. [DOI] [PubMed] [Google Scholar]
- 65.Templeton T.J., Iyer L.M., Anantharaman V., Enomoto S., Abrahante J.E., Subramanian G.M., Hoffman S.L., Abrahamsen M.S., Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugi T., Ma Y.F., Tomita T., Murakoshi F., Eaton M.S., Yakubu R., Han B., Tu V., Kato K., Kawazu S.I. Toxoplasma gondii cyclic AMP-dependent protein kinase subunit 3 is involved in the switch from tachyzoite to bradyzoite development. mBio. 2016;7:e00755–16. doi: 10.1128/mBio.00755-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaji R.Y., Johnson D.E., Treeck M., Wang M., Hudmon A., Arrizabalaga G. Phosphorylation of a myosin motor by TgCDPK3 facilitates rapid Initiation of motility during Toxoplasma gondii egress. PLoS Pathog. 2015;11:e1005268. doi: 10.1371/journal.ppat.1005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Long S., Wang Q., Sibley L.D. Analysis of noncanonical calcium-dependent protein kinases in Toxoplasma gondii by targeted gene deletion using CRISPR/Cas9. Infect. Immun. 2016;84:1262–1273. doi: 10.1128/IAI.01173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ojo K.K., Larson E.T., Keyloun K.R., Castaneda L.J., DeRocher A.E., Inampudi K.K., Kim J.E., Arakaki T.L., Murphy R.C., Zhang L. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat. Struct. Mol. Biol. 2010;17:602–607. doi: 10.1038/nsmb.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 71.Cao L., Wang Z., Wang S., Li J., Wang X., Wei F., Liu Q. Deletion of mitogen-activated protein kinase 1 inhibits development and growth of Toxoplasma gondii. Parasitol. Res. 2016;115:797–805. doi: 10.1007/s00436-015-4807-2. [DOI] [PubMed] [Google Scholar]
- 72.Li Z.-Y., Wang Z.-D., Huang S.-Y., Zhu X.-Q., Liu Q. TgERK7 is involved in the intracellular proliferation of Toxoplasma gondii. Parasitol. Res. 2016;115:3419–3424. doi: 10.1007/s00436-016-5103-5. [DOI] [PubMed] [Google Scholar]
- 73.Behnke M.S., Fentress S.J., Mashayekhi M., Li L.X., Taylor G.A., Sibley L.D. The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog. 2012;8:e1002992. doi: 10.1371/journal.ppat.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones N.G., Wang Q., Sibley L.D. Secreted protein kinases regulate cyst burden during chronic toxoplasmosis. Cell. Microbiol. 2016 doi: 10.1111/cmi.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knoll L.J. Functional analysis of the rhoptry kinome during chronic Toxoplasma gondii infection. mBio. 2016;7:e00842–16. doi: 10.1128/mBio.00842-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doerig C. Protein kinases as targets for anti-parasitic chemotherapy. Biochim. Biophys. Acta Proteins Proteom. 2004;1697:155–168. doi: 10.1016/j.bbapap.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 77.Zhang M., Chavchich M., Waters C. Targeting protein kinases in the malaria parasite: Update of an antimalarial drug target. Curr. Top. Med. Chem. 2012;12:456–472. doi: 10.2174/156802612799362922. [DOI] [PubMed] [Google Scholar]
- 78.Cohen P. The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/S0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 79.Eglen R.M., Reisine T. The current status of drug discovery against the human kinome. Assay Drug Dev. Technol. 2009;7:22–43. doi: 10.1089/adt.2008.164. [DOI] [PubMed] [Google Scholar]
- 80.Ojo K.K., Dangoudoubiyam S., Verma S.K., Scheele S., DeRocher A.E., Yeargan M., Choi R., Smith T.R., Rivas K.L., Hulverson M.A. Selective inhibition of Sarcocystis neurona calcium-dependent protein kinase 1 for equine protozoal myeloencephalitis therapy. Int. J. Parasitol. 2016;46:871–880. doi: 10.1016/j.ijpara.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dissous C., Grevelding C.G. Piggy-backing the concept of cancer drugs for schistosomiasis treatment: A tangible perspective? Trends Parasitol. 2011;27:59–66. doi: 10.1016/j.pt.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Martin D.M., Miranda-Saavedra D., Barton G.J. Kinomer v. 1.0: A database of systematically classified eukaryotic protein kinases. Nucleic Acids Res. 2009;37:D244–D250. doi: 10.1093/nar/gkn834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 84.Sundarsanam S., Bingham J., Manning G., Charydczak G., Chen M.J. Kinase.com—Genomics, Evolution and Function of Protein Kinases. 1999. [(accessed on 30 January 2017)]. Available online: www.kinase.com.
- 85.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. 1st ed. Humana Press; Totowa, NJ, USA: 2005. pp. 571–607. [Google Scholar]
- 86.Bailey T.L., Bodén M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2: A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 90.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ronquist F., Teslenko M., van der M.P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Letunic I., Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]