Abstract
Photosynthesis is sensitive to environmental stress and must be efficiently modulated in response to abiotic stress. However, the underlying mechanisms are not well understood. Here we report that ARABIDOPSIS THALIANA HOMEOBOX 17 (ATHB17), an Arabidopsis HD-Zip transcription factor, regulated the expression of a number of photosynthesis associated nuclear genes (PhANGs) involved in the light reaction and ATSIG5 in response to abiotic stress. ATHB17 was responsive to ABA and multiple stress treatments. ATHB17-overexpressing plants displayed enhanced stress tolerance, whereas its knockout mutant was more sensitive compared to the wild type. Through RNA-seq and quantitative real-time reverse transcription PCR (qRT-PCR) analysis, we found that ATHB17 did not affect the expression of many known stress-responsive marker genes. Interestingly, we found that ATHB17 down-regulated many PhANGs and could directly modulate the expression of several PhANGs by binding to their promoters. Moreover, we identified ATSIG5, encoding a plastid sigma factor, as one of the target genes of ATHB17. Loss of ATSIG5 reduced salt tolerance while overexpression of ATSIG5 enhanced salt tolerance, similar to that of ATHB17. ATHB17 can positively modulate the expression of many plastid encoded genes (PEGs) through regulation of ATSIG5. Taken together, our results suggest that ATHB17 may play an important role in protecting plants by adjusting expression of PhANGs and PEGs in response to abiotic stresses.
Abiotic stresses such as salinity, drought, high light and unfavorable temperatures, adversely affect the growth and development of plants. Photosynthesis in chloroplasts is one of the primary processes to be affected by abiotic stress1. The effects can be direct, such as decreased CO2 diffusion caused by stomata closure2 or affecting ribulose bisphosphate carboxylase/oxygenase (Rubisco) activity3. More importantly, abiotic stress reduces the threshold intensity for the onset of photo-inhibition, and results in over excitation of the photosystems (PSs), dramatically increasing reactive oxygen species (ROS) production4,5. Therefore, rapid response of photosynthetic machinery and metabolism is key for plants to cope with the fluctuating environment6.
Chloroplasts are genetically semi-autonomous organelles that evolutionarily retain an eubacteria-type of circular genome DNA. In higher plants, the chloroplast 120–150 kb genome encodes only about 120 genes7. More required proteins are encoded by the nuclear genome and imported to play roles in the chloroplasts after translation in the cytosol8.
Chloroplast gene transcription in higher plants is performed by at least two types of RNA polymerases, plastid encoded RNA polymerase (PEP) and nuclear encoded RNA polymerase (NEP). PEP holoenzyme is a complex composed by five subunits: α, β, β′, ω (omega), σ (sigma), in which the α2ββ′ω constitutes the catalytic core, while the nuclear-encoded σ subunit recognizes the specific promoter region and initiates transcription of the core complex9.
The transcription of photosynthesis related genes in chloroplasts is mainly dependent on PEP, and the nuclear-encoded sigma factors play special roles in regulating the chloroplast transcription10,11. Since the first chloroplast sigma factor gene was isolated from red algae nuclear genome12,13, more chloroplast sigma factors (e.g. ATSIG1-6) had been identified in different plant species14,15. Most plastid encoded genes appear to be regulated by several sigma factors with overlapping functions. However, within a particular time frame during plant development, plastid genes are likely to be coordinated by a distinct sigma factor, for example, psaA and rbcL by ATSIG116, psaJ by ATSIG217, psbN by ATSIG318, ndhF by ATSIG419, psbBT, psbD, psbC, psbZ, psbA and psaAB by ATSIG517,20,21,22.
Among the six sigma factors in Arabidopsis, only ATSIG5 expression is stress inducible and phylogenetically specific23,24,25. It is induced by high light, low temperature, high salt and osmotic stress17, as well as blue light23. Beside these stresses, MpSIG5 of liverwort Marchantia polymorpha is significantly induced by reactive oxygen species (ROS) stress26. ATSIG5 regulates the repair capacity from injury to the PS II reaction center by salt stress. It does this by determining the promoter recognition specificity of PEP in plastid gene expression that activates psbD from the blue-light responsive promoter (BLRP)9,27. In addition, ATSIG5 regulates chloroplast psbD and psbA coding for the PS II core proteins D1 and D2 in response to light quality and intensity, and combines extrinsic and intrinsic signals important for adjusting nuclear and plastid gene transcription in light acclimation processes28.
Coordinating the transcription of photosynthesis associated nuclear genes (PhANGs) and plastid encoded genes (PEGs) to maintain the proper stoichiometry of nuclear encoded proteins, plastid proteins, carotenoids and chlorophylls, is critical for correct assembly of functional photoprotective and photosynthetic complexes within chloroplasts under stress conditions29. Although many abiotic stress-responsive transcription factors (TFs) have been studied, very few are known to modulate the expression of photosynthesis-related genes5.
The homeodomain leucine-zipper (HD-Zip) TFs are the most abundant group of homeobox genes in plants, which have diverse functions during plant development and stress adaptation30,31. According to their distinctive features such as gene structures, DNA-binding specificities, additional common motifs and physiological functions, HD-Zip TFs can be classified into four subfamilies30. There are 10 HD-Zip II genes in the Arabidopsis genome, which play important roles ranging from auxin response to shade avoidance. Five HD-Zip class II genes, including HOMEOBOX ARABIDOPSIS THALIANA 1 (HAT1), HAT2, HAT3, ATHB2, ATHB4, are known to respond to light quality changes32. Auxin response analyses strongly suggest that HAT1, HAT3 and ATHB4 are under the control of the phytochrome system as is ATHB233. Of the remaining class II HD-Zip members, little is known about their functions, excepting ATHB17.
ATHB17 localizes to both the cytoplasm and nuclei; the distribution is determined by its unique N-terminus. Overexpression of ATHB17 in Arabidopsis enhances chlorophyll content in the leaves, while expression of a truncated ATHB17 protein in maize increases ear weight at silking34,35. ATHB17-overexpressing Arabidopsis plants are sensitive to ABA and NaCl, whereas ATHB17 knockout mutants are insensitive to ABA and NaCl at post germination stage. However, these phenotypes are weak and the expression of ABA-responsive genes is not significantly altered in the ATHB17-overexpressing plants compared with wild-type (WT) plants36. At this stage it remains unclear if the phenotypes result from modulating ABA signaling or other mechanisms.
In contrast, in this study, we found that ATHB17 regulated the expression of PhANGs and ATSIG5 to cope with environmental stresses. ATHB17 responded to multiple abiotic stresses. Overexpression of ATHB17 enhanced plant tolerance to salt, drought and oxidative stresses, and ATHB17 knockout resulted in the opposite phenotypes. By RNA-seq analysis, we find ATHB17 repressed the expression of many PhANGs while it activated the transcription of ATSIG5. Further analysis revealed that ATHB17 directly bound to the promoter of several PhANGs and most likely regulated their expression. Moreover, ATHB17 directly bound to the ATSIG5 promoter and activated ATSIG5 expression to regulate some PEG expression to deal with environmental stress. Our study revealed another stress response pathway that modulates the photosynthesis light reaction by down regulating PhANGs and up regulating PEGs in response to multiple abiotic stresses, therefore protecting plants from photosynthesis-derived damage.
Results
ATHB17 is preferentially expressed in roots and responsive to multiple stress signals
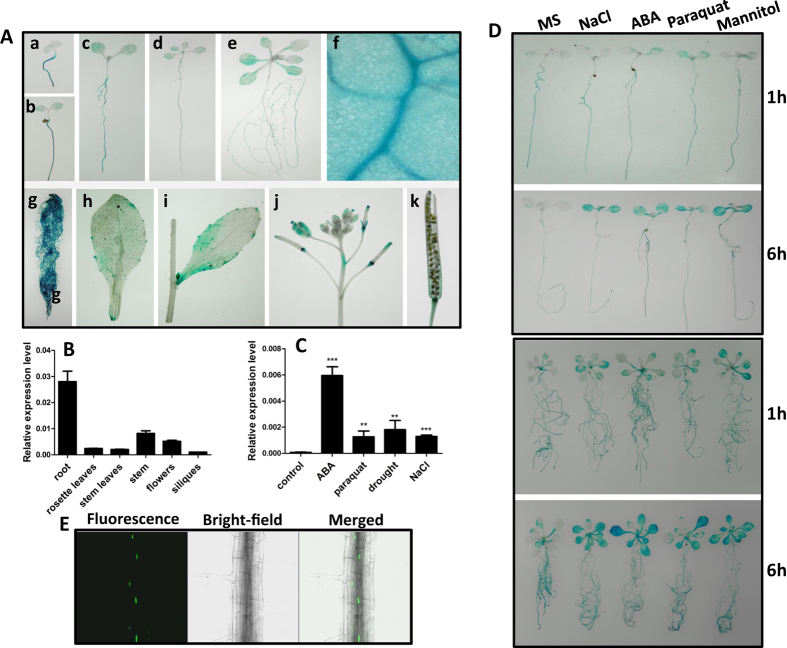
To reveal the expression pattern of ATHB17, we analyzed transgenic plants harboring the ATHB17 promoter-GUS reporter construct (pATHB17::GUS). Strong GUS activity was detected in roots of seedlings at different ages (Fig. 1A-a to A-e and A-g). ATHB17 was also expressed in rosette leaves with much higher expression levels in the leaf veins (Fig. 1A-f). However, at mature stage, ATHB17 was mainly expressed in roots (Fig. 1A-g), in agreement with the results of Park et al.36. There was only weak expression in other organs, such as rosette leaf (Fig. 1A-h), cauline leaf (Fig. 1A-i), flower and young silique (Fig. 1A-j), and mature silique (Fig. 1A-l). These results were confirmed by quantitative real-time reverse transcription PCR (qRT-PCR) analysis shown in Fig. 1B, implying a potential function of ATHB17 in roots.
Figure 1. Expression pattern of ATHB17 and response to different stress signals.
(A) The expression pattern of ATHB17 was revealed by GUS staining of pATHB17::GUS transgenic plants. GUS activity was observed in 2-day-old (a), 4-day-old (b), 7-day-old (c), 9-day-old (d), or 14-day-old (e) seedlings, leaves of 9-day-old plant (f), roots of mature plants (g), rosette leaf (h), cauline leaf (i), flower and young silique (j), old silique (k). (B) Analysis of the ATHB17 expression patterns by qRT–PCR. UBQ5 was used as an internal control. Values are mean ± SD of three replica experiments. (C) Expression levels of ATHB17 after different treatments. Induction levels of ATHB17 in 10-day-old plants by ABA (100 μM, 4 h), paraquat (5 μM, 4 h), drought (4 h), NaCl (200 mM, 4 h) were determined by qRT–PCR. Values are mean ± SD of three replica experiments (Student’s t-test *P < 0.05, **P < 0.01). (D) Effects of different stress treatments on pATHB17::GUS expression, 7-day-old or 14-day-old pATHB17::GUS transgenic seedlings grown on MS medium were transferred to MS liquid medium or MS liquid medium containing NaCl (200 mM), ABA (10 μM), paraquat (5 μM), mannitol (200 mM) for 1–6 h and then the seedlings were harvested for GUS staining for 3 h. (E) ATHB17 protein localization in the root cells of transgenic plants expressing ATHB17–GFP under the control of the ATHB17 native promoter.
Moreover, we found that ATHB17 could be significantly induced by ABA (abscisic acid), paraquat, drought, and NaCl treatments (Fig. 1C). Consistent with this result, GUS staining of the pATHB17::GUS reporter line treated with NaCl, ABA, PQ, and mannitol also showed strongly induced expression of ATHB17, especially in the leaves (Fig. 1D).
To study the localization of ATHB17 protein in plant cell, we obtained transgenic plants expressing green fluorescent protein (GFP) and ATHB17 fusion protein under the control of its own promoter (pATHB17::ATHB17::GFP). By examining GFP florescence in root of the transgenic plant, we found that ATHB17 was predominantly localized in the nucleus (Fig. 1E), which is consistent with the previous report36.
ATHB17 is a positive regulator of tolerance to multiple abiotic stresses
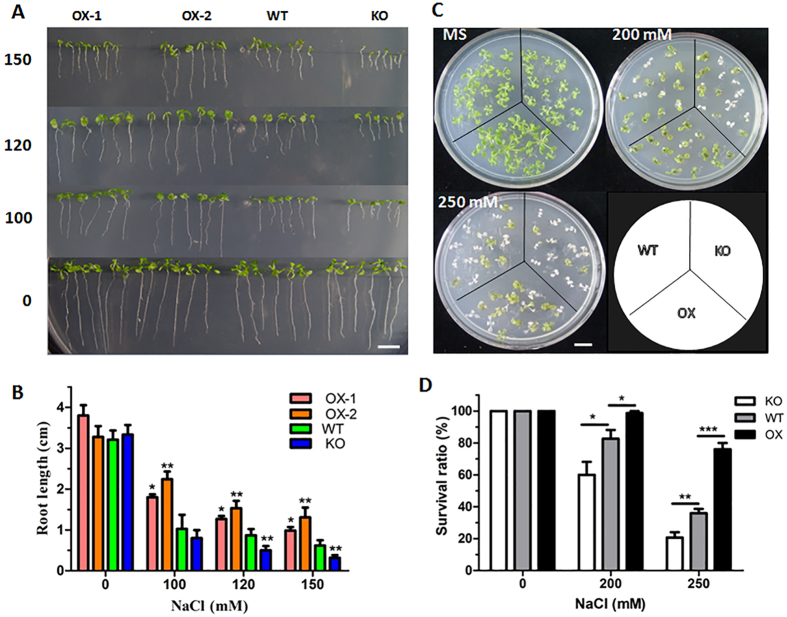
In order to uncover the functions of ATHB17, we generated 35S:: ATHB17 overexpression (OX) transgenic Arabidopsis plants, and obtained an ATHB17 knockout (KO) mutant from Arabidopsis Biological Resource Center (Supplementary Fig. S2). To test whether ATHB17 was involved in salt tolerance, we determined the NaCl sensitivity of the ATHB17 OX and KO lines. Firstly, the NaCl sensitivity at germination and seedling establishment stage was assayed. Seeds were germinated and grown on MS (Murashige and Skoog) medium containing different concentration of NaCl. The results showed that the root growth of ATHB17 OX lines was more resistant to salt stress compared with the WT plants. In contrast, the ATHB17 KO plants were more sensitive to NaCl stress with significantly reduced root growth under stress. However, there was no difference in germination or root elongation on MS medium without NaCl (Fig. 2A,B). In another salt tolerance assay, 7-day-old ATHB17 OX, KO and WT plants were transferred to MS medium or MS medium containing NaCl. After another 7 days growth, the ATHB17 OX plants showed higher survival ratio whereas the ATHB17 KO plants showed opposite phenotypes compared with the WT (Fig. 2C,D). Moreover, salt tolerance assay with older plants grown in soil showed that ATHB17 positively regulated plant survival to salt stress (Supplementary Fig. S2).
Figure 2. ATHB17 is a positive regulator for salt stress resistance.
(A,B) Salt tolerance assay of ATHB17 OX and KO Phenotypes of seedlings (A) and root length (B) of ATHB17 OX, KO and WT plants grown on medium containing indicated concentrations of NaCl. Seeds were germinated and grown vertically on MS medium or MS medium containing the indicated concentration of NaCl for 10 days. Values are mean ± SD (n = 60, *P < 0.05, **P < 0.01). Bar = 1 cm. (C) Phenotype of ATHB17 OX, KO and WT plants on MS medium with different concentrations of NaCl. Seven-day-old plants on MS medium were transferred to MS medium or MS medium containing the indicated concentrations of NaCl for 7 days. Bar = 4 cm. (D) Survival ratio of the ATHB17 OX, KO and WT plants on MS medium or MS medium with indicated concentrations of NaCl. Values are mean ± SD of three biological replicates each containing 14–16 plants per genotype (*P < 0.05, **P < 0.01, ***P < 0.001).
To confirm this, we conducted complementation analysis by expressing ATHB17 in the ATHB17 KO plants, which restored the salt tolerance of KO plants to WT level (Supplementary Fig. S3). In addition, ATHB17 was able to confer salt tolerance in tobacco at different developmental stages when overexpressed (Supplementary Fig. S4).
Since ATHB17 was responsive to multiple stresses as demonstrated above, we also analyzed the role of ATHB17 in drought stress and oxidative stress. The ATHB17 OX Arabidopsis plants showed enhanced drought tolerance while the ATHB17 KO plants were drought sensitive compared with WT plants (Supplementary Fig. S5A,B), which is consistent with the result of Park et al.36. In addition, overexpression of ATHB17 also conferred improved tolerance to paraquat (Supplementary Fig. S5C,D). These data indicate ATHB17 is a positive regulator of multiple abiotic stresses.
The expression of stress-responsive marker genes was not significantly affected by ATHB17
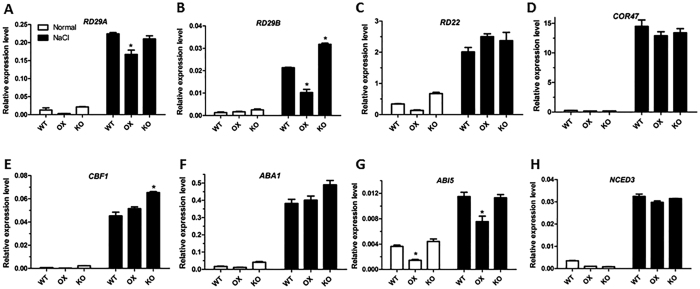
To investigate the mechanism of ATHB17 modulated stress tolerance, we compared the transcriptomes of ATHB17 OX, KO and WT plants grown on NaCl-free MS medium and MS medium containing 200 mM NaCl using RNA-seq method. Surprisingly, we found the expression of classic stress-induced marker genes, such as responsive to desiccation 29A (RD29A), RD29B, RD20, RD22, cold-regulated 47 (COR47), COR45B, CRT/DRE binding factor 1 (CBF1), salt overly sensitive 2 (SOS2) and SOS3, were not significantly influenced by ATHB17 under both normal and salt treated conditions. Expression levels of ABA signaling and synthesis pathway genes were also found not significantly changed in the ATHB17 OX and KO plants (Table 1). To validate the results of RNA-seq profiling analysis, eight stress-responsive genes were selected for qRT-PCR analysis. The results showed that the response of these genes was not significantly affected by the expression level of ATHB17 (Fig. 3). These results implicate that ATHB17-conferred stress tolerance, probably arose not through the classical stress response pathway, but through a new pathway.
Table 1. Relative expression levels of stress-responsive genes identified by RNA-seq profiling.
| Gene ID | Annotation | Normal (TPM*) |
NaCl (TPM*) |
||||
|---|---|---|---|---|---|---|---|
| WT | OX | KO | WT | OX | KO | ||
| AT5G52310 | RD29A (responsive to desiccation 29A) | 0.99 | 0.38 | 0.01 | 71.54 | 73.85 | 52.09 |
| AT5G52300 | RD29B (responsive to desiccation 29B) | 0.01 | 0.01 | 0.28 | 16.32 | 14.61 | 12.21 |
| AT5G25610 | RD22 (responsive to desiccation 22) | 36.67 | 11.53 | 44.81 | 182.8 | 280.38 | 373.55 |
| AT2G33380 | RD20 (responsive to desiccation 20) | 16.52 | 20.36 | 10.65 | 116.37 | 146.30 | 133.47 |
| AT1G20440 | COR47 (cold regulated 47) | 943.44 | 770.26 | 1405.98 | 3953.93 | 4602.60 | 5296.48 |
| AT2G42530 | COR15B (cold regulated 15B) | 0.01 | 0.01 | 0.01 | 4.12 | 3.80 | 6.10 |
| AT4G25490 | CBF1 (C-repeat/DRE binding factor 1) | 0.01 | 0.01 | 0.01 | 15.49 | 20.61 | 19.94 |
| AT4G26080 | ABI1 (abscisic acid insensitive 1) | 36.01 | 46.48 | 44.95 | 122.14 | 122.88 | 186.78 |
| AT5G57050 | ABI2 (abscisic acid insensitive 2) | 0.66 | 5.57 | 2.63 | 74.01 | 64.04 | 61.45 |
| AT2G36270 | ABI5 (abscisic acid insensitive 3) | 3.96 | 3.07 | 4.29 | 17.47 | 23.62 | 16.68 |
| AT5G67030 | ABA1 (abscisic acid deficient 1) | 49.55 | 44.95 | 75.24 | 287.96 | 282.38 | 264.50 |
| AT1G16540 | ABA3 (abscisic acid deficient 3) | 8.26 | 17.86 | 8.71 | 24.23 | 35.42 | 17.09 |
| AT3G14440 | NCED3 (9-cis-epoxycarotenoid dioxygenase 3) | 0.99 | 0.38 | 0.01 | 71.54 | 73.85 | 52.09 |
| AT5G35410 | SOS2 (salt overly sensitive 2) | 9.58 | 10.95 | 6.64 | 7.75 | 10.21 | 7.32 |
| AT5G24270 | SOS3 (salt overly sensitive 3) | 1.32 | 4.03 | 1.38 | 0.82 | 0.40 | 0.01 |
*TPM: number of transcripts per million tags.
Figure 3. qRT-PCR validation of the data of stress-responsive marker genes in the RNA-seq profiling.
About 0.1 g of 12-day old seedlings of the ATHB17 OX, KO lines and WT plants were treated with liquid MS medium containing 0 or 200 mM NaCl for 5 h. Total RNA were extracted and reverse-transcribed as templates for qRT-PCR. UBQ5 was used as an internal control. Values are mean ± SD of three independent experiments (*P < 0.05).
ATHB17 negatively regulates some PhANGs
Based on Gene Ontology (GO) term enrichment analysis of the RNA-seq profiling data, we found that the PhANGs were significantly enriched among the different expressed genes between the ATHB17 OX and ATHB17 KO plants both under normal and salt stress conditions (Supplementary Data 1 and 2). The expression of 26 of the 63 PhANGs in the Arabidopsis genome were down-regulated (>2 fold) in the ATHB17 OX compared with ATHB17 KO plants under normal or NaCl treated conditions. More specifically, under normal condition, 20 of the 26 PhANGs had reduced expression (>1.5 fold) in the ATHB17 OX plants compared with ATHB17 KO plants, with 16 of these 20 PhANGs up-regulated in the ATHB17 KO plants while down-regulated in the ATHB17 OX plants compared with WT plants (Table 2).
Table 2. Expression levels of PhANGs in ATHB17 OX, KO and WT plants under normal or NaCl treated conditions identified by RNA-seq profiling.
| Gene ID | Description | Normal (TPM*) |
NaCl (TPM*) |
||||
|---|---|---|---|---|---|---|---|
| KO | WT | OX | KO | WT | OX | ||
| AT1g29930 | CAB1 (chlorophyll a/b binding protein 1) | 2659.41 | 1921.89 | 172.11 | 5531.28 | 4153.04 | 1530.00 |
| AT1g31330 | PSAF (PS I subunit F) | 1117.89 | 912.05 | 284.09 | 1296.45 | 946.94 | 291.59 |
| AT2g34420 | LHB1B2 (light-harvesting chlorophyll protein complex II subunit B2) | 2781.39 | 2899.35 | 712.06 | 4826.90 | 4362.05 | 1421.93 |
| AT3g55330 | PPL1 (PsbP-like protein 1) | 37.48 | 14.53 | 5.57 | 29.30 | 17.14 | 9.41 |
| AT3g61470 | LHCA2 (PS I light harvesting complex gene 2) | 2269.95 | 2062.94 | 705.34 | 4063.92 | 3235.44 | 1422.93 |
| AT4g10340 | LHCB5 (light harvesting complex of PS II subunit 5) | 836.73 | 592.29 | 86.05 | 1080.78 | 707.61 | 357.03 |
| AT4g30950 | FAD6 (fatty acid desaturase 6) | 182.28 | 154.60 | 67.61 | 260.43 | 212.79 | 127.28 |
| AT5g66570 | PSBO1 (PS II oxygen-evolving complex 1) | 2749.86 | 2251.56 | 485.78 | 4219.77 | 3937.28 | 1953.87 |
| AT1g52230 | PSAH2 (PS I subunit H2) | 69.84 | 92.16 | 26.70 | 72.84 | 60.00 | 30.22 |
| AT2g34430 | LHB1B1 (light-harvesting chlorophyll protein complex II subunit B1) | 1449.82 | 1655.64 | 306.57 | 4226.69 | 3826.35 | 2079.15 |
| AT3g16140 | PSAH1 (PS I subunit H1) | 143.97 | 158.89 | 57.24 | 196.95 | 166.81 | 48.03 |
| AT1g15820 | LHCB6 (light harvesting complex of PS II subunit 6) | 1560.05 | 1324.97 | 614.10 | 1194.31 | 1474.56 | 735.88 |
| AT1g29910 | CAB3 (chlorophyll a/b binding protein 3) | 821.65 | 702.62 | 110.45 | 1632.16 | 1800.76 | 510.13 |
| AT2g39470 | PPL2 (PsbP-like protein 2) | 46.05 | 21.47 | 15.17 | 63.48 | 79.12 | 26.82 |
| AT3g47470 | LHCA4 (light-harvesting chlorophyll protein complex I subunit A4) | 1025.23 | 863.49 | 428.16 | 1665.12 | 2171.46 | 1119.13 |
| AT3g50820 | PSBO2 (PS II subunit O-2) | 146.05 | 87.21 | 46.29 | 316.18 | 454.27 | 462.50 |
| AT3g54110 | PUMP1 (plant uncoupling mitochondrial protein 1) | 87.13 | 71.68 | 40.53 | 43.95 | 49.28 | 64.84 |
| AT4g02770 | PSAD1 (PS I subunit D-1) | 2057.38 | 1748.79 | 900.69 | 1283.02 | 1341.55 | 856.36 |
| AT5g54270 | LHCB3 (light-harvesting chlorophyll B-binding protein 3) | 294.44 | 231.23 | 70.69 | 320.65 | 348.94 | 127.28 |
| AT1g61520 | LHCA3 (PS I light harvesting complex gene 3) | 2178.95 | 1902.07 | 1325.00 | 3721.29 | 2438.82 | 1668.09 |
| AT1g45474 | LHCA5 (PS I light harvesting complex gene 5) | 55.87 | 67.72 | 31.50 | 46.39 | 35.77 | 20.21 |
| AT1g55670 | PSAG (PS I subunit G) | 37.48 | 37.33 | 23.63 | 38.66 | 24.56 | 13.81 |
| AT1g67740 | PSBY (PS II BY) | 89.90 | 63.75 | 142.91 | 135.10 | 154.12 | 64.44 |
| AT1g76570 | LHCB7 (light-harvesting complex B7) | 8.44 | 7.93 | 9.03 | 24.82 | 11.21 | 10.81 |
| AT3g54890 | LHCA1 (PS I light harvesting complex gene 1) | 53.38 | 122.55 | 222.82 | 103.76 | 94.78 | 50.03 |
| AT4g28660 | PSB28 (PS II reaction centre PSB28 protein) | 33.33 | 10.90 | 20.75 | 13.43 | 10.71 | 4.00 |
*TPM: transcripts per million tags.
Under salt treatment, 21 of the 26 PhANGs were down-regulated in the ATHB17 OX plants. In contrast, 16 PhANGs had higher expression in ATHB17 KO plants while lower expression in ATHB17 OX plants compared with WT plants (Table 2). These genes mainly encode proteins in PS I complexes (PSAF, PSAH2, PSAH1, PSAD1, PSAG), light-harvesting complexes (LHCA1, LHCA2, LHCA3, LHCA4, LHCA5, LHCB3, LHCB5, LHCB6, LHCB7, LHB1B1, LHB1B2), chlorophyll a/b-binding (CAB1, CAB3) proteins, and PS II oxygen evolving complex (PSBO1, PSBO2, PPL1, PPL2).
qRT-PCR analysis was carried out to validate the results of expression profiling analysis by RNA-seq. The data in Fig. 4A showed that 11 of the 13 tested PhANGs had higher transcript levels in ATHB17 KO plants and lower expression levels in ATHB17 OX plants compared with WT plants. This result is consistent with the data of RNA-seq profiling analysis. Together, these data indicate that ATHB17 may act as a repressor of PhANGs, which agrees with its function as a repressor reported by Rice et al.35.
Figure 4. qRT-PCR analysis of the expression level of PhANGs and PEGs.
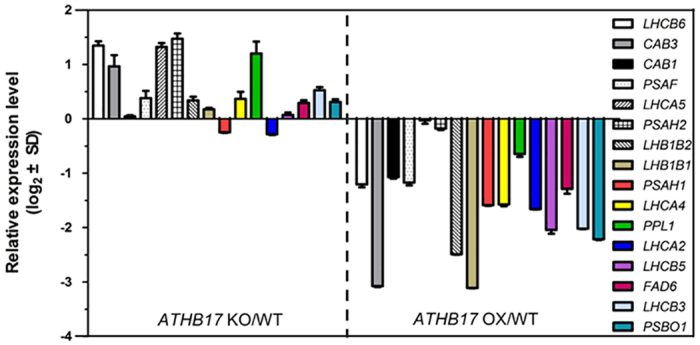
About 0.1 g 12-day-old plants grown under normal condition were used for RNA extraction. First-strand cDNA was synthesized from 1 μg total RNA with Oligo dT primer for qRT-PCR analysis. Relative transcription levels of the genes in ATHB17 OX and KO are normalized to levels in WT control (WT = 0). Values are mean ± SD of three independent experiments.
ATHB17 can directly bind to the promoters of several PhANGs
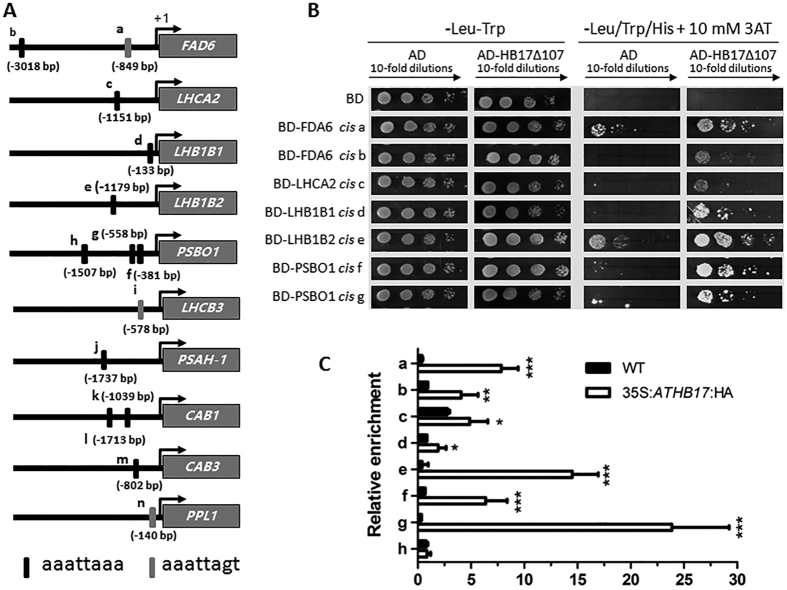
HD-ZIP transcription factors show a binding preference for variant HD-binding sequences30,37. According to our bioinformatic analysis, combined with the information from AGRIS (Arabidopsis Gene Regulatory Information Server), we selected six types of 8–9 bp sequences as potential HD binding sites. We firstly tested the binding affinities of ATHB17 to these sequences by yeast one-hybrid (Y1H) assay. We found full length ATHB17 protein had strong self-activation activity. It has been reported that ATHB17 protein lacking the first 113 amino acids (ATHB17Δ113) can still homo-dimerize and specifically recognize the sequence requirements for DNA binding35. Thus we used ATHB17 protein lacking the first 107 amino acids (ATHB17Δ107) for Y1H and electrophoretic mobility shift assay (EMSA). As results shown in Supplementary Fig. S6A, ATHB17Δ107 had no self-activation activity. Y1H assay reveled ATHB17 had strong binding affinities to these HD binding cis-elements: aaattagt, tttaattt and taaatgta (Supplementary Fig. S6B). Then we performed EMSA to confirm the binding affinities of ATHB17Δ107 to the potential ATHB17 binding cis-elements in vitro. Supplementary Fig. S6C shows that ATHB17Δ107 could directly bind to the sequences aaattagt and tttaattt, but failed to bind to taaatgta in vitro. The results of EMSA indicate that ATHB17Δ107 protein was able to directly bind to the tttaattt motif in vitro, and the binding was specific as demonstrated by competition assay using unlabelled (competitor) and non-specific probes (non-competitor) (Supplementary Fig. S6D).
Based on the above analysis, we chose aaattagt and tttaattt as the ATHB17 binding cis-elements. Promoter sequence analyses of 19 ATHB17 down-regulated PhANGs revealed that 10 of the 19 genes had at least one ATHB17 binding cis-element (Fig. 5A). To analyze whether ATHB17 could directly regulate transcription of all these 10 genes by binding to the ATHB17 cis-elements in their promoters, we performed a Y1H assay. As shown in Fig. 5B, ATHB17Δ107 could only bind to the promoters of 5 PhANGs (FDA6, LHCA2, LHB1B1, LHB1B2, PSBO1) with different binding affinities. Subsequently, chromatin immunoprecipitation (ChIP) assays using the transgenic plants expressing 35S-haemagglutinin (HA)-ATHB17 plants were conducted to validate the binding affinities in vivo. ChIP-qPCR showed that ATHB17 could directly bind to ATHB17 binding cis-element motifs in the promoters of the 5 genes screened out by Y1H. However, ATHB17 failed to bind the h fragment in PSBO1 promoter, which also had no binging affinity to ATHB17Δ107 in yeast (Fig. 5C). These results are consistent with the data of Y1H and the expression pattern of these genes in the ATHB17 OX, KO and WT plants (Table 2), indicating that ATHB17 can directly bind to the promoter of a number of PhANGs to regulate their expression.
Figure 5. ATHB17 binds to the promoters of several PhANGs.
(A) Location of ATHB17 binding cis-elements in the promoter of several PhANGs. The ATHB17 binding cis-elements are indicated with filled rectangles, above/below which the sites of the last base of the cis-elements relative to the start code are shown. (B) Y1H assay for ATHB17 binding to the 25 bp fragment containing ATHB17 binding cis-element from the promoter of five PhANGs, respectively. (C) ChIP assay. About 70–200 bp promoter fragments containing ATHB17 binding cis-element were enriched by anti-HA antibodies in ChIP–qPCR analysis. Values are mean ± SD of three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001).
ATHB17 binds to the HD binding cis-elements in the ATSIG5 promoter
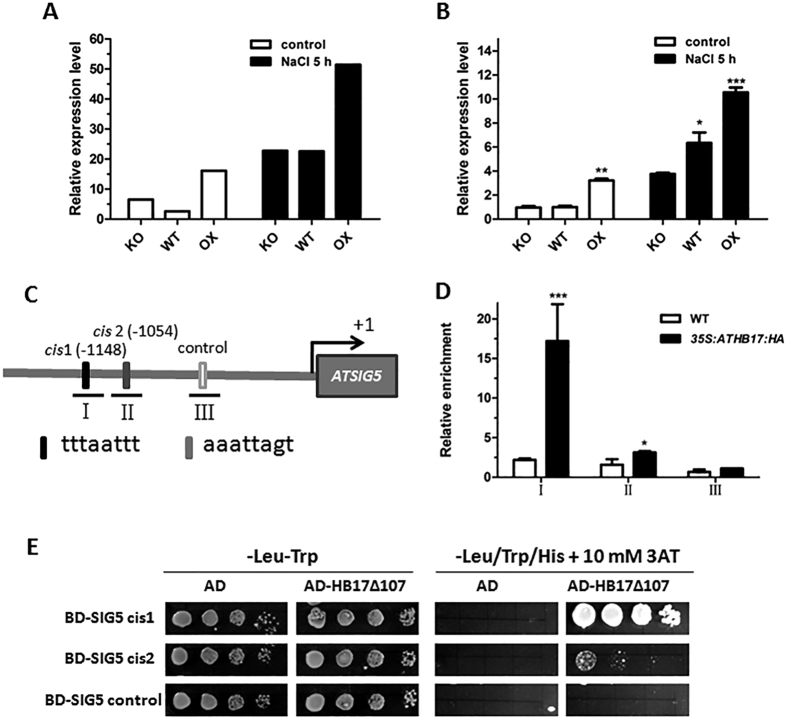
Through RNA-seq profiling analysis, we found a nuclear encoded sigma factor, ATSIG5, had higher expression in ATHB17 OX plants than in WT and ATHB17 KO plants both under normal and salt stress conditions (Fig. 6A). qRT-PCR validation showed that ATHB17 OX plants had increased ATSIG5 transcription compared with WT and ATHB17 KO plants under normal condition. After salt treatment, ATSIG5 transcript was significantly up-regulated in ATHB17 OX plants, while down-regulated in ATHB17 KO plants compared with WT plants (Fig. 6B). Promoter sequence analysis revealed two potential ATHB17 binding cis-elements in the ATSIG5 promoter: cis1 tttaattt and cis2 aaattagt located at 1148 bp and 1054 bp upstream of start codon, respectively (Fig. 6C). These data suggest that ATSIG5 may be a candidate target of ATHB17.
Figure 6. ATHB17 directly regulates the transcription of ATSIG5.
(A) Expression levels of ATSIG5 in the RNA-seq profiling data. (B) qRT-PCR validation of the expression levels of ATSIG5 in RNA-seq profiling. About 0.1 g 12-day-old plants grown on MS medium were used transferred to liquid MS medium containing 0 or 200 mM NaCl for 5 h. The plants were harvested for qRT-PCR analysis. Values are mean ± SD of three independent experiments and asterisks denote Student’s t-test significance compared with KO (*P < 0.05, **P < 0.01, ***P < 0.001). (C) The schematic illustration of the locations of ATHB17 binding cis-element in the promoters of ATSIG5 and the fragments (short lines) used in ChIP-qPCR assay. cis1 is tttaattt located 1148 bp upstream of start codon, cis2 is aaattagt located 1054 bp upstream of start codon, control is a random 8 bp sequence. (D) qPCR data from ChIP assay with antibody against HA. A fragment without ATHB17 binding cis-element (fragment III) was used as negative control. Values are mean ± SD of three replica experiments (*P < 0.05,***P < 0.001); (E) Y1H assay for ATHB17 binding to the 25 bp fragment containing ATHB17 binding cis-element from the promoter of ATSIG5. Fragment containing no ATHB17 binding cis-element was used as a negative control.
ChIP and Y1H assay was conducted to determine whether ATHB17 could directly bind to the ATHB17 binding cis-elements in the promoter of ATSIG5. The results of ChIP-qPCR showed in Fig. 6D revealed that ATHB17 was able to bind to the ATSIG5 promoter DNA fragments I and II which contained the ATHB17 binding cis-element. However, the DNA fragment III containing no ATHB17 binding cis-element was not enriched, suggesting that ATHB17 specifically bound to the ATHB17 binding cis-element motifs in the ATSIG5 promoter. Binding of ATHB17 to the ATSIG5 promoter was further tested by Y1H assay. Consistent with the ChIP assay results, ATHB17 could directly bind to both of the promoter fragments containing ATHB17 binding cis-element cis1 and cis2 in yeast cells, with much stronger binding affinities to the region containing cis1, but could not bind to the negative control promoter fragment without HD binding cis-element (Fig. 6E). These data indicate that ATSIG5 was a direct target of ATHB17.
ATHB17 regulates salt tolerance partly by modulating ATSIG5
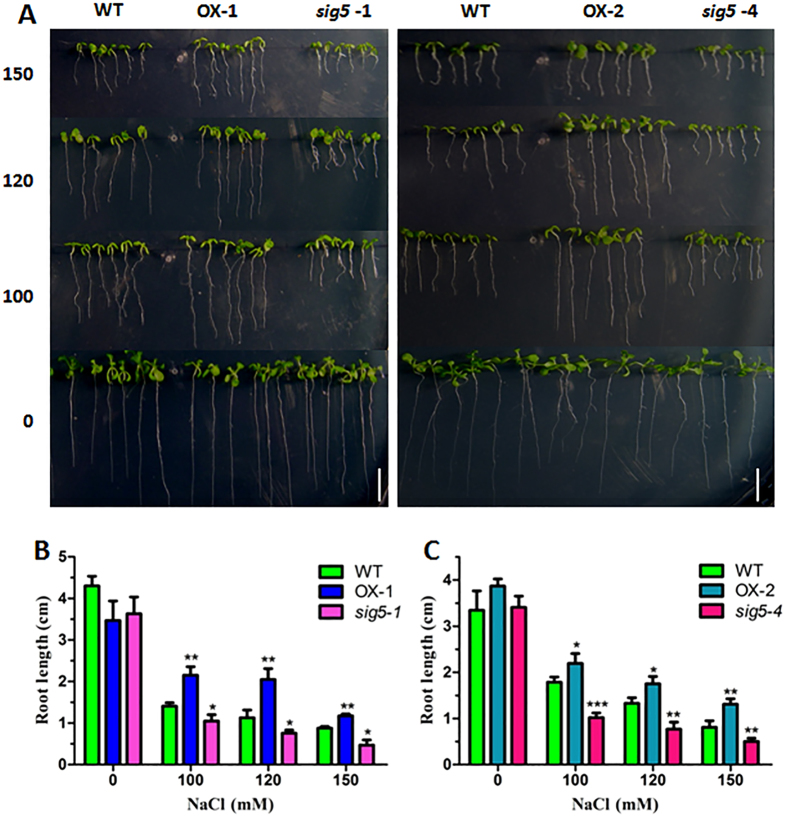
ATSIG5 was reported to respond to multiple stress signals, including salt stress. The sig5-1 mutant is hypersensitive to NaCl treatment17. To study the functions of ATSIG5, we generated 35S::ATSIG5 (OX) lines and obtained ATSIG5 knockout mutants sig5-1 and sig5-4 (Supplementary Fig. S7). After germination and growth vertically on MS or MS medium containing NaCl for 10 days, the ATSIG5 OX showed salt tolerant phenotypes with better root growth while sig5-1 and sig5-4 showed salt sensitive phenotypes compared with the WT (Fig. 7). These data agree with the results reported by Nagashima et al.17.
Figure 7. ATSIG5-overexpressing transgenic plants were more tolerant while its knockout mutants were more sensitive to salt stress.
(A) Salt tolerance assay of ATSIG5-overexpressing and knockout plants. Seeds were sowed on MS medium containing 0, 100, 120, 150 mM NaCl and grown vertically for 10 days. Bar = 1 cm; (B,C) Root length of the 10-day-old plants grown on MS medium containing indicated concentrations of NaCl. Values are mean ± SD (n = 45, *P < 0.05, **P < 0.01, ***P < 0.001).
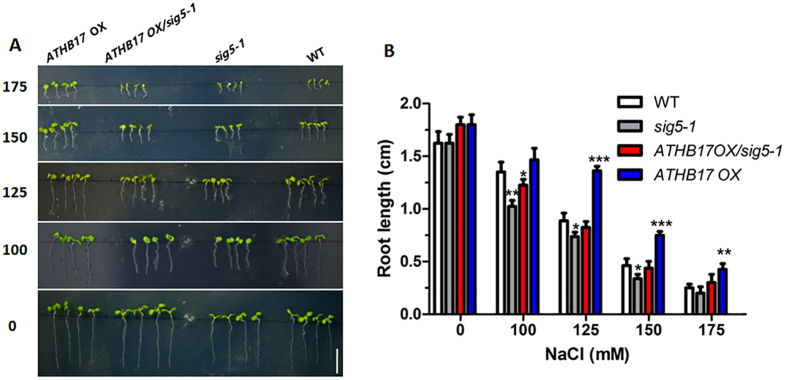
To investigate further whether ATHB17 modulated salt stress through regulating ATSIG5 expression, we introduced ATHB17 OX into sig5-1 background by crossing. The ATHB17 OX sig5-1 offspring were tested for salt tolerance. Although ATHB17 gene was overexpressed in the hybrid offspring, the ATHB17 OX sig5-1 seedlings did not show salt stress resistant phenotypes as ATHB17OX plants did but an intermediate phenotype between the two parents (Fig. 8), implying that the ATHB17-conferred stress tolerance was partially dependent on ATSIG5.
Figure 8. ATSIG5 acts downstream of ATHB17.
(A) The phenotypes of ATHB17 OX, ATHB17 OX/sig5-1, sig5-1 and WT seedlings grown on MS medium containing indicated concentrations of NaCl. Seeds were sowed on MS medium containing 0, 100, 125, 150 and 175 mM NaCl and grown vertically for 10 days. Bar = 1 cm. (B) root length of the 10-day-old plants grown on MS medium containing different concentrations of NaCl. Values are mean ± SD (n = 45, *P < 0.05, **P < 0.01, ***P < 0.001).
ATSIG5 was found to play an important role in regulating many PEGs which respond to environmental signals17,20,21. In order to study whether ATHB17 could modulate PEGs transcription through regulating ATSIG5, we investigated the expression of several downstream PEGs of ATSIG5 which encoded components of PS I and II in ATHB17 OX and KO plants by qRT-PCR. As shown in Fig. 9A, under normal condition, all the 6 PEGs tested were down-regulated in ATHB17 KO plants, while 4 were up-regulated in ATHB17 OX plants, compared with WT plants. However, after NaCl stress treatment, the expression of all these genes was reduced in ATHB17 KO plants, and increased in ATHB17 OX plants (Fig. 9B). These results indicated that ATHB17 might positively regulate PEG expression through ATSIG5.
Figure 9. The expression levels of several PEGs in ATHB17 OX and KO plants.
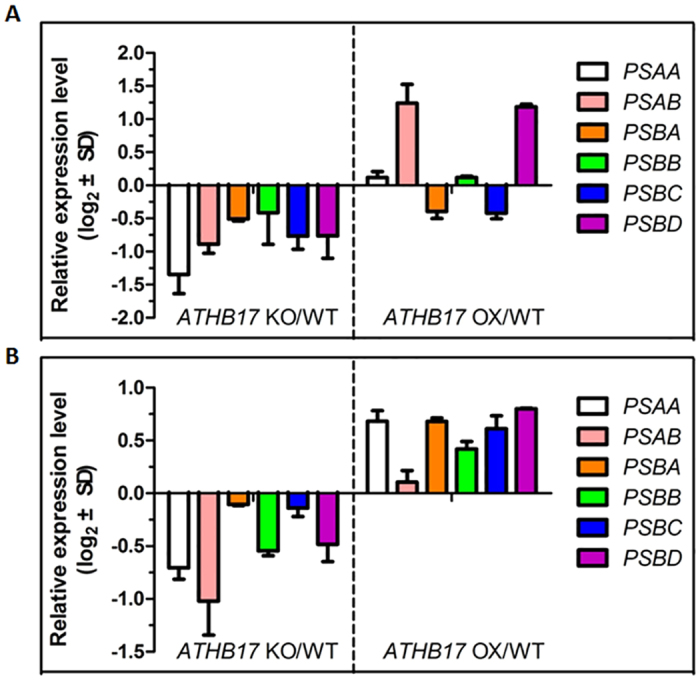
Twelve-day-old plants grown under normal conditions were transferred to liquid MS medium containing 0 or 200 mM NaCl for 6 h. The plants were harvested for RNA extraction. First-strand cDNA was synthesized from 1 μg total RNA with random primer for qRT-PCR analysis. Relative transcription levels of the genes in ATHB17 OX and KO were normalized to levels in WT control (WT = 0). Values are mean ± SD of three independent experiments. (A) PEGs expression levels in ATHB17OX, KO and WT plants treated with NaCl-free liquid MS. (B) PEG expression levels in ATHB17OX, KO and WT plants treated with liquid MS containing 200 mM NaCl.
Discussion
Previous studies on salt stress tolerance have mainly focused on the genes related to ion homeostasis, metabolites or osmo-protectants, antioxidant, hormones, ABA synthesis and signaling, and stress-responsive TFs38,39,40. Very few researches focus on the coordination of photosynthetic genes to improve plant salt tolerance. In this study, we did not find any significant difference in the expression of classic stress-induced marker genes between ATHB17 OX and KO plants, including many stress-responsive genes, ABA synthesis and signaling pathway genes, as well as SOS genes (Table 1 and Fig. 3). These results partially agree with the results by Park et al.36, who reported that ATHB17 overexpression did not affect the expression of a number of ABA-responsive genes. These findings implicate that ATHB17-confered stress tolerance may arise through a pathway different from the traditional ones.
Environmental stresses present great challenges to the normal development and growth of plants. In addition to developmental changes, chloroplasts constantly experience changing environments, thus a tight coordination between the nucleus and chloroplast is crucial to the survival of plants. These genome-coordinating mechanisms are achieved through both anterograde (nucleus to organelle) and retrograde (organelle to nucleus) signals41,42. Most of chloroplast proteins are nuclear-encoded, and the concentrations of these proteins are efficiently regulated by nuclear transcription43. TFs play important roles in the nuclear-chloroplast communication. Nuclear-encoded sigma factors regulate PEP activity to modulate the expression of different sets of genes responding to the external environmental signals44. Besides sigma factors, very few TFs have been isolated that regulate transcripts of nuclear photosynthetic genes and chloroplast genes to date. GATA-type TFs GNC and CGA1 were reported to modulate the expression of chloroplast protein genes genomes uncoupled 4 (GUN4) and HEMA145. Abscisic acid insensitive 4 (ABI4) represses the expression of photosynthetic nuclear genes, potentially acting as a master switch required for the modulation of nuclear genes in response to environmental signals and developmental cues46. Higher yield rice (HYR) is a master regulator in rice, responding to environmental stress, directly activating several photosynthesis genes by binding to their promoters47. In addition, Golden2-like 1 (GLK1) and GLK2 TFs coordinate the expression of the photosynthetic apparatus genes in Arabidopsis48.
On the other hand, the chloroplast can act as an environmental sensor. Metabolite and protein signals, such as the recently discovered dihydroxyacetone phosphate (DHAP), heme, methylerythritolcyclodiphosphate (MEcPP), 3′-phosphoadenosine 5′-phosphate (PAP), GUN1, PTM, A-type heat-shock transcription factors (HSFA1D, HSFA2 and HSFA3), and Heat Shock 90 (HSP90) produced in chloroplasts act as novel types of retrograde signals to regulate PhANGs in the nucleus42,49,50,51,52,53.
In this study, we found that ATHB17, a HD-ZIP TF, played important roles in regulating the expression of PhANGs in response to salt stress. By genetic analysis with ATHB17 knockout mutants and ATHB17-overexpressing lines, we have demonstrated that ATHB17 is a positive regulator in response to salt stress (Fig. 2). RNA-seq profiling analysis revealed that ATHB17 acted as a repressor of PhANGs. ATHB17 was reported as a transcriptional repressor containing EAR (ERF-associated amphiphilic repression)-like motif35. Consistent with this result, we found that ATHB17 repressed the transcription of 26 PhANGs under normal or salt stress conditions (Table 2 and Fig. 4), with 5 of them (FDA6, LHCA2, LHB1B1, LHB1B2 and PSBO1) directly regulated by ATHB17 (Fig. 5). ATHB17 repressed the expression of genes related to light-harvesting complexes (LHCA1, LHCA2, LHCA3, LHCA4, LHCA5, LHCB3, LHCB5, LHCB6, LHCB7, LHB1B1, LHB1B2), chlorophyll a/b-binding proteins (CAB1, CAB3), PS II oxygen evolving complex (PSBO1, PSBO2, PPL1, PPL2), thus reducing light harvesting to alleviate photo-oxidative damage. The light-harvesting protein complex together with chlorophyll captures light energy and delivers it to the PSs. Under stress conditions, light harvesting must be reduced in order to avoid over-excitation and damage of PSs. Therefore, we can speculate that decreasing light capture by ATHB17 is one of the ways to protect plants from photo-damage under stress conditions.
Furthermore, we also found that ATHB17 could directly activate the transcript of ATSIG5 (Fig. 6), a multiple-stress responsive TF which can be induced by various stresses, such as high light, salt, osmotic stress and low temperature17. Light induction of ATSIG5 expression was strongly dependent on cryptochrome1 (CRY1), and long hypocotyl 5 (HY5) which could directly bind to ATSIG5 promoter to regulate ATSIG5 expression in a light responsive manner28. However, stress and light responsiveness of ATSIG5 seemed to be independent17. It remains unclear what factors are involved in modulating ATSIG5 expression in response to other stresses. Here, we found HB17 is one of the upstream regulators of ATSIG5 in response to salt stress. We found that, similar with ATSIG5, ATHB17 is also a multiple stress-responsive TF. NaCl, mannitol, paraquat, and ABA could highly induce its expression in leaves (Fig. 1). Knockout of ATHB17 impaired the induction of ATSIG5 by salt stress (Fig. 6B). Through Y1H and ChIP assay, we found ATHB17 could directly bind to the ATSIG5 promoter to regulate its transcription (Fig. 6). Overexpression of ATSIG5 in Arabidopsis enhanced its salt tolerance, while sig5 knockout plants became salt sensitive (Fig. 7). However, when ATHB17 was overexpressed in the ATSIG5 knockout background, salt tolerance of the plants was partially impaired (Fig. 8). These results indicated that ATHB17 is one of the upstream regulators of ATSIG5 in response to salt, and ATHB17-conferred salt resistance partly depends on ATSIG5.
The activity of PS II can be inactivated by a variety of environmental stresses, which inhibit the repair of PS II rather than directly attacking it54,55. D1 and D2, which are encoded by chloroplast gene psbA and psbD, bind all the redox-active components related to electron transfer of PS II and create oxidative power to break water molecules. Therefore, D1 and D2 are the main targets of oxidative damage56,57. After degradation of the photo-damaged D1 protein, a new functional copy is inserted into the core complex following de novo synthesis, to repair of PS II in response to stresses58. ATSIG5 was not only involved in the response of plants to blue light, but also involved in protection of chloroplasts under various stress conditions through enhancing repair of the PS II reaction center17. ATSIG5 binds to the psbD promoter, which drives the expression of psbD-psbC-psbZ operon encoding core-proteins of PS II, in response to blue light and various abiotic stress17,59. ATSIG5 also activates expression of the psbA in chloroplasts20, and acts as a key mediator of circadian regulation of expression of several chloroplast genes21. Therefore, ATSIG5 is likely to combine extrinsic and intrinsic signals, which are important in adjusting plastid and nuclear gene expression, to light and environmental stress to successfully adapt plants to changing environments20. Under stress conditions, ATHB17 may coordinate the expression of PEGs through regulating ATSIG5 (Fig. 9), thus enhancing PS II reaction center repair to survive better.
Moreover, ATHB17 may be involved in balancing the stoichiometry of PS I to PS II under salt stress condition. It is well-known that PS II is the main target of oxidative damage under stress conditions, therefore, the subunit proteins of PS II are maintained with a rapid turnover rate to facilitate the repair cycle56,57. Recent studies found the PS II photo-inhibition-repair cycle can also protect PS I from irreversible damage60. Although PS II in chloroplasts undergoes a frequent repair cycle, the functional PS II is decreased, and becomes rate-limiting to photosynthesis under stress conditions. During acclimation, when either PS becomes rate-limiting to photosynthesis, expression of genes for its reaction center proteins are up-regulated, while genes for reaction center proteins of the other PS, which has surplus photochemical capacity, are simultaneously decreased61. Thus, in order to balance the stoichiometry of PS I to PS II under salt stress condition, expression of genes encoding subunits of PS II should be induced and genes encoding subunits of PS I should be simultaneously repressed. By RNA-seq profiling and qRT-PCR analyses, we found that ATHB17 repressed the expression of several nuclear genes which encoded PS I reaction center subunits (psaF, psaH1, psaH2, psaD1,psaG) (Table 2, Fig. 4), and activated the expression ATSIG5, which may up-regulate several chloroplast encoded PS II subunit genes (Fig. 9), thus benefiting the stoichiometry of PS I to PS II.
More interestingly, ATHB17 is a multiple stress responsive gene. Under normal conditions, we found that ATHB17 was primarily expressed in the root, with weak expression in the shoot, consistent with the previous report36. However, Hymus et al.34 only detected very weak ATHB17 signal in specific cells in the embryonic, QC area of primary and lateral roots, through in situ hybridization analysis. These differences may result from low sensitivity of in situ hybridization. Under stress conditions, ATHB17 was significantly induced, especially in the leaves (Fig. 1C and D). As discussed above, the induction of ATHB17 expression in the leaves may alleviate the damage to chloroplast, thus enhance plant tolerance to environmental stresses. Consistent with this hypothesis, overexpression of ATHB17 could also enhance plant tolerance to drought and oxidative stresses (Supplementary Fig. S5), indicating a mechanism similar to that of salt stress may exist in responding to these stresses. Taken together, our results imply that ATHB17 may act as an important TF regulating both PhANGs and PEGs to decrease light harvesting, enhance PS repair, and balance PS stoichiometry under stress conditions, thus improving the tolerance of plants to multiple stresses.
In conclusion, our study demonstrated that ATHB17 was an important TF in coordinating the expression of PhANGs and PEGs to cope with multiple stresses. Under stress conditions, the expression of ATHB17 was induced, especially in the photosynthetic leaves. As a TF, ATHB17 repressed the transcription of many PhANGs indirectly or directly by binding to their promoters. ATHB17 also directly activated transcription of ATSIG5, whose protein is then translocated to chloroplast as an important regulator of many chloroplast genes, such as psbA, psbB, psbC, psbD and psBT. However, it may modulate several other chloroplast genes through other unknown pathways. Overall, it eventually reduces light harvest under stress, enhances PS repair cycle and balances the stoichiometry of PS I to PS II in the chloroplast, therefore alleviating the damage to chloroplast under stress conditions and improving plant stress tolerance.
Material and Methods
Plant materials and growth conditions
Arabidopsis thaliana (Col-0) and tobacco (Nicotiana tobaccum, NC89) were used for transformation. Seeds were sterilized in 10% (w/v) bleach for 10 min, and washed five to six times with sterile water. For Arabidopsis, the seeds were first treated at 4 °C for 3 days vernalization, then sowed on MS medium. Seven-day-old Arabidopsis seedlings were transferred to soil. For tobacco, the washed seeds were directly germinated on MS medium. The growth house was controlled at 22 ± 1 °C with a 16 h a photoperiod.
Constructs and preparation of transgenic plants
To generate 35S::ATHB17 and 35S::ATSIG5 overexpression binary vectors, the ATHB17 or ATSIG5 cDNA was isolated by RT-PCR with ATHB17-attb-LP and ATHB17-attb-RP or ATSIG5-attb-LP and ATSIG5-attb-RP and cloned into pCB200462 via the GATEWAY cloning system. To analyze the ATHB17 expression pattern, a 3.0 kbp promoter fragment was amplified with the primers pATHB17-LP and pATHB17-RP and then shuttled into the vector pCB308R62. To get the native promoter-gene fusion construct, a DNA fragment containing ATHB17 promoter and coding region amplified by genomic PCR with primers ATHB17-attb-LP2 and ATHB17-attb-RP2 was cloned into pMDC110 to fuse with GFP63. For ChIP assays, ATHB17 full-length coding sequence amplified by RT–PCR using specific primers ATHB17-HA-attb-LP and ATHB17-attb-RP was inserted into pCB2004 to get pCB2004::ATHB17-HA by the GATEWAY cloning system. All the constructs were electroporated into Agrobacterium tumefaciens C58C1, which were used to transform wild type Arabidopsis plants as described64,65. All the primers used are listed in Supplementary Table 1.
Identification of ATHB17 and ATSIG5 knockout mutants and overexpression plants
The T-DNA insertion site of the SALK_095524 mutant in ATHB17 was confirmed by genomic PCR using three specific primers: SALK_095524-LP, SALK_095524-RP, LBb1.3. Similarly, the sig5-1 (SALK_049021) mutant was identified by genomic PCR with three primers: sig5-1 LP, sig5-1 RP and LBb1.3. Mutant of sig5-4 (SALK_101921) were identified by genomic PCR with three primers: sig5-4 LP, sig5-4 RP and LBb1.3. For expression analysis of the knockout mutants and overexpression transgenic plants, Quantitative RT-PCR was used with primers for the full-length coding sequence. All the primers used are listed in Supplementary Table 1.
GUS activity staining assay
GUS staining of pATHB17::GUS transgenic plants was performed as described66. Arabidopsis seedlings were soaked in staining buffer containing 1 mM 5-bromo-4-chloro-3-indoryl-β-D-glucuronide (X-Gluc, Rose Scientific Inc., Somerset, NJ, U.S.A.) overnight, then decolored using an ethanol series. Individual representative seedlings were photographed.
Hydroponic culture
20-day-old seedlings germinated on MS agar medium were used for hydroponic culture as previous described67. The root of each tobacco seedling was wrapped in a sponge strip and inserted into a hole made in thick polystyrene foam board. The foam boards were floated on MS hydroponic solution with or without NaCl. Plants were cultured at 22 °C under 16-h photoperiod. Nutrient solution was changed every 7 days.
RNA-seq
Two-week-old seedlings of ATHB17 OX, KO, and the WT control were treated with or without 200 mM NaCl in liquid MS medium, 60 rpm shaking in the air for 6 h, then total RNA isolated with TRIzol reagent (Invitrogen Inc.). RNA from 3 independent replicates were mixed by equal volume. RNA-seq was performed and analyzed by BGI (Beijing Genome Institute, Shenzhen) corporation following the protocol provided by the manufacturer. The Illumina HiSeq™ 2000 platform was used for sequencing. For annotation, all clean tags were mapped to the reference sequence and allowed no more than one nucleotide mismatch. Clean tags mapped to reference sequences from multiple genes were filtered. For gene expression analysis, the number of clean tags for each gene was calculated and then normalized to the number of transcripts per million clean tags (TPM)68.
qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen Inc.), and first-strand cDNA synthesized from 1 μg total RNA in a 20 μl reaction mixture with Prime Script RT regent kit (TAKARA BIOTECHNOLOGY CO., LTD). For nuclear encoded gene expression analysis, oligo dT primer was used for cDNA synthesis. For PEGs expression analysis, random primers was used for cDNA synthesis. The transcript levels of classic stress-related genes and other genes were examined with specific primers. The PCR was performed on ABI step-one instrument with the amplification conditions of 40 cycles of 95 °C for 1 min, 62 °C for 20 seconds and 72 °C for 30 seconds. UBQ5 was used as the internal control. The relative expression levels were calculated by the 2−ΔΔCT method. All the primers used are listed in Supplementary Table 1.
Yeast one-hybrid assay (Y1H)
A cDNA fragment encoding ATHB17Δ107 was amplified by the primers: the forward with restriction endonuclease BamH I, 5′-CGGGATCCGACCAGCTA AGGCTAGACATGAA-3′and the reverse with restriction endonuclease Xba I, 5′-GCTCTAGATCAACGATCACGCTCTTGCG-3′, and inserted into plasmid pAD-GAL4-2.1 (pAD) to get AD/ATHB17Δ107.
To get the report vectors containing HD-binding sequences, three copies of the HD-binding sequence, containing Sac I and Mlu I adaptors, were annealed and inserted into the Sac I and Mlu I sites of pHIS2. To get the report vectors containing promoter sequence of different genes, 25 bp promoter segment containing HD-binding sequence with Sac I and Mlu I adaptors was annealed and inserted into pHIS2, respectively. The constructs were confirmed by sequencing. The pAD and pHIS2 empty vector were used for negative control. AD/ATHB17Δ107 and the reporter pHIS2 containing different DNA sequence were co-transfected into yeast cells Y187 respectively. The yeast was first grown on SD-Trp-Leu medium for 3 days at 30 °C, and then transferred to SD-Trp-Leu-His medium with 10 mM or 20 mM 3-aminotriazole (3-AT, sigma) at different dilutions. The yeasts were incubated at 30 °C for 5 days and the extent of yeast growth was determined.
ChIP-PCR assay
Leaves of 10-day-old T3 homozygote of 35S::ATHB17-HA transgenic plants and anti-HA tag antibody (Cali-Bio, CB100005M) were used for pulling down the chromatin, as previously described69. After degrading the associated proteins with proteinase K, the chromatin DNA samples were treated using phenol/chloroform, then precipitated and finally eluted in 30 μl TE buffer. ChIP-qPCR was then used to verify the promoter segment of related genes using the primers listed in Supplementary Table 1.
EMSA assay
ATHB17Δ107 protein was expressed in Escherichia coli with pET28a (+) protein expression system (Novagen) and purified by Ni2+ chromatography. Three copes of complementary single-stranded HD-binding sequence were synthesized and annealed to form double-stranded DNA fragment. The DNA fragments were marked with α-32P-dCTP and gel purified as probes. Purified ATHB17Δ107 protein (200 ng) was incubated with probes for 30 min on ice. For the competition test, non-labeled probe and non-specific probes were added into the binding reaction. Each reaction was loaded on a 4.5% (w/v) native polyacrylamide gel with 0.5 × TBE buffer. The gel then exposed to X-ray film.
Statistical analysis
Statistically significant differences (P < 0.05 or P < 0.01 or P < 0.001) were computed based on the Student’s t-tests.
Additional Information
How to cite this article: Zhao, P. et al. ATHB17 enhances stress tolerance by coordinating photosynthesis associated nuclear gene and ATSIG5 expression in response to abiotic stress. Sci. Rep. 7, 45492; doi: 10.1038/srep45492 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by Ministry of Science and Technology of China (Grant # 2012CB114304 to C.-B.X), NNSFC (Grant # 30700051 to C.-B.X) and the Chinese Academy of Sciences (Grant # KSCX3–EW–N–007 to C.-B. X). We thank ABRC for offering T-DNA insertion lines.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.X. and L.Y. designed the experiments. P.Z., R.C., and L.Y. performed most of the experiments and data analyses. P.X., J.W., J.-L.M., Y.C., and C.-Z.Z. assisted in experiments and data analyses. P.Z. and L.Y. wrote the manuscript. C.X supervised the project and revised the manuscript.
References
- Munns R., James R. A. & Lauchli A. Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany 57, 1025–1043 (2006). [DOI] [PubMed] [Google Scholar]
- Flexas J., Bota J., Loreto F., Cornic G. & Sharkey T. D. Diffusive and metabolic limitations to photosynthesis under drought and salinity in C(3) plants. Plant Biology 6, 269–279 (2004). [DOI] [PubMed] [Google Scholar]
- Lawlor D. W. & Cornic G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ 25, 275–294 (2002). [DOI] [PubMed] [Google Scholar]
- Vasilikiotis C. & Melis A. Photosystem-Ii Reaction-Center Damage and Repair Cycle - Chloroplast Acclimation Strategy to Irradiance Stress. P Natl Acad Sci USA 91, 7222–7226 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo N. J. M., Lourenco T. & Oliveira M. M. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot-London 103, 609–623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri M. Z., Moumeni A. & Komatsu S. Abiotic Stresses: Insight into Gene Regulation and Protein Expression in Photosynthetic Pathways of Plants. Int J Mol Sci 16, 20392–20416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakamura Y., Kaneko T., Asamizu E. & Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA research 6, 283–290 (1999). [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., Schutze K., Fey V., Sherameti I. & Oelmuller R. Chloroplast redox control of nuclear gene expression–a new class of plastid signals in interorganellar communication. Antioxidants & redox signaling 5, 95–101 (2003). [DOI] [PubMed] [Google Scholar]
- Kanamaru K. & Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Bioscience, biotechnology, and biochemistry 68, 2215–2223 (2004). [DOI] [PubMed] [Google Scholar]
- Legen J. et al. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. The Plant journal 31, 171–188 (2002). [DOI] [PubMed] [Google Scholar]
- Krause K., Maier R. M., Kofer W., Krupinska K. & Herrmann R. G. Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Molecular & general genetics: MGG 263, 1022–1030 (2000). [DOI] [PubMed] [Google Scholar]
- Liu B. & Troxler R. F. Molecular characterization of a positively photoregulated nuclear gene for a chloroplast RNA polymerase sigma factor in Cyanidium caldarium. P Natl Acad Sci USA 93, 3313–3318 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. et al. Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science 272, 1932–1935 (1996). [DOI] [PubMed] [Google Scholar]
- Lysenko E. A. Analysis of the evolution of the family of the sig genes encoding plant sigma factors. Russ J Plant Physl+ 53, 605–614 (2006). [Google Scholar]
- Yao J., Roy-Chowdhury S. & Allison L. A. AtSig5 is an essential nucleus-encoded Arabidopsis sigma-like factor. Plant physiology 132, 739–747 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J., Turkeri H., Kolpack A. & Link G. Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription - Recent lessons from Arabidopsis thaliana. European Journal of Cell Biology 89, 940–946 (2010). [DOI] [PubMed] [Google Scholar]
- Nagashima A. et al. The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol 45, 357–368 (2004). [DOI] [PubMed] [Google Scholar]
- Zghidi W., Merendino L., Cottet A., Mache R. & Lerbs-Mache S. Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Res 35, 455–464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory J. J. et al. Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Res 33, 5991–5999 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama Y. et al. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. P Natl Acad Sci USA 101, 3304–3309 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordally Z. B. et al. Circadian Control of Chloroplast Transcription by a Nuclear-Encoded Timing Signal. Science 339, 1316–1319 (2013). [DOI] [PubMed] [Google Scholar]
- Yamburenko M. V., Zubo Y. O. & Borner T. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: repression by guanosine-3′-5′-bisdiphosphate and activation by sigma factor 5. Plant J 82, 1030–1041 (2015). [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y., Morikawa K., Shiina T. & Toyoshima Y. Blue light specific and differential expression of a plastid sigma factor, Sig5 in Arabidopsis thaliana. FEBS Lett 516, 225–228 (2002). [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant molecular biology 76, 235–249 (2011). [DOI] [PubMed] [Google Scholar]
- Lysenko E. A. Plant sigma factors and their role in plastid transcription. Plant cell reports 26, 845–859 (2007). [DOI] [PubMed] [Google Scholar]
- Kanazawa T., Ishizaki K., Kohchi T., Hanaoka M. & Tanaka K. Characterization of four nuclear-encoded plastid RNA polymerase sigma factor genes in the liverwort Marchantia polymorpha: blue-light- and multiple stress-responsive SIG5 was acquired early in the emergence of terrestrial plants. Plant Cell Physiol 54, 1736–1748 (2013). [DOI] [PubMed] [Google Scholar]
- Thum K. E., Kim M., Christopher D. A. & Mullet J. E. Cryptochrome 1, cryptochrome 2, and phytochrome a co-activate the chloroplast psbD blue light-responsive promoter. The Plant cell 13, 2747–2760 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellenthin M., Ellersiek U., Börger A. & Baier M. Expression of the Arabidopsis Sigma Factor SIG5 Is Photoreceptor and Photosynthesis Controlled. Plants 3, 359–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. & Montgomery B. L. Phytochrome-dependent coordinate control of distinct aspects of nuclear and plastid gene expression during anterograde signaling and photomorphogenesis. Front Plant Sci 5, 171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F. D., Manavella P. A., Dezar C. A. & Chan R. L. The true story of the HD-Zip family. Trends in Plant Science 12, 419–426 (2007). [DOI] [PubMed] [Google Scholar]
- Turchi L., Baima S., Morelli G. & Ruberti I. Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. Journal of Experimental Botany 66, 5043–5053 (2015). [DOI] [PubMed] [Google Scholar]
- Ciarbelli A. R. et al. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol 68, 465–478 (2008). [DOI] [PubMed] [Google Scholar]
- Kunihiro A. et al. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant & cell physiology 52, 1315–1329 (2011). [DOI] [PubMed] [Google Scholar]
- Hymus G. J. et al. Application of HB17, an Arabidopsis class II homeodomain-leucine zipper transcription factor, to regulate chloroplast number and photosynthetic capacity. J Exp Bot 64, 4479–4490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice E. A. et al. Expression of a Truncated ATHB17 Protein in Maize Increases Ear Weight at Silking. Plos One 9(4): e94238. doi 10.1371/journal.pone.0094238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. Y., Kim S. A., Lee S. J. & Kim S. Y. ATHB17 is a positive regulator of abscisic acid response during early seedling growth. Mol Cells 35, 125–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R. et al. The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60, 564–574 (2009). [DOI] [PubMed] [Google Scholar]
- Gupta B. & Huang B. R. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int J Genomics (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. et al. GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci Rep 6, 35040 doi: 10.1038/srep35040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J. D. & Chory J. Coordination of gene expression between organellar and nuclear genomes. Nature Reviews Genetics 9, 383–395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. X., Phua S. Y., Crisp P., McQuinn R. & Pogson B. J. Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annu Rev Plant Biol 67, 25–53 (2016). [DOI] [PubMed] [Google Scholar]
- Kleffmann T. et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14, 354–362 (2004). [DOI] [PubMed] [Google Scholar]
- Kanamaru K. & Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Biosci Biotech Bioch 68, 2215–2223 (2004). [DOI] [PubMed] [Google Scholar]
- Hudson D. et al. GNC and CGA1 Modulate Chlorophyll Biosynthesis and Glutamate Synthase (GLU1/Fd-GOGAT) Expression in Arabidopsis. Plos One 6 (11): e26765. doi 10.1371/journal.pone.0026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon P., Gregorio J. & Cordoba E. ABI4 and its role in chloroplast retrograde communication. Frontiers in Plant Science 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambavaram M. M. R. et al. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat Commun 5 : 5302 doi: 10.1038/ncomms6302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T. et al. GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell 21, 1109–1128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo G. M. et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J. D., Perez-Ruiz J. M. & Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21, 897–903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y. M. et al. Retrograde Signaling by the Plastidial Metabolite MEcPP Regulates Expression of Nuclear Stress-Response Genes. Cell 149, 1525–1535 (2012). [DOI] [PubMed] [Google Scholar]
- Chi W., Feng P., Ma J. & Zhang L. Metabolites and chloroplast retrograde signaling. Curr Opin Plant Biol 25, 32–38 (2015). [DOI] [PubMed] [Google Scholar]
- Jung H. S. et al. Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci USA 110, 14474–14479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y. & Allakhverdiev S. I. Photoinhibition of photosystem II under environmental stress. Bba-Bioenergetics 1767, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- Allakhverdiev S. I. et al. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiology 130, 1443–1453 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen R., Tyystjarvi E. & Aro E. M. Degradation pattern of photosystem II reaction center protein D1 in intact leaves. The major photoinhibition-induced cleavage site in D1 polypeptide is located amino terminally of the DE loop. Plant Physiol 111, 1183–1190 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Timberg R. & Ohad I. Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem 177, 403–410 (1988). [DOI] [PubMed] [Google Scholar]
- Theis J. & Schroda M. Revisiting the photosystem II repair cycle. Plant Signal Behav 11, e1218587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffer P. H. & Christopher D. A. Structure and blue-light-responsive transcription of a chloroplast psbD promoter from Arabidopsis thaliana. Plant Physiol 115, 213–222 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M., Mekala N. R. & Aro E. M. Photosystem II photoinhibition-repair cycle protects Photosystem I from irreversible damage. Biochim Biophys Acta 1837, 210–215 (2014). [DOI] [PubMed] [Google Scholar]
- Rochaix J. D. Posttranscriptional control of chloroplast gene expression. From RNA to photosynthetic complex. Plant Physiol 125, 142–144 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z. Y. et al. High-throughput binary vectors for plant gene function analysis. Journal of Integrative Plant Biology 49, 556–567 (2007). [Google Scholar]
- Curtis M. D. & Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R. B. et al. Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proc Natl Acad Sci USA 83, 2571–2575 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A. & Bevan M. W. Gus Fusions - Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher-Plants. Embo J 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. H. et al. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6, 27795; doi: 10.1038/srep27795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- t Hoen P. A. C. et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.