Abstract
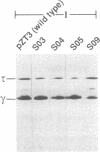
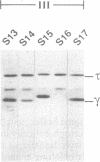
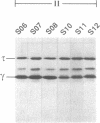
The dnaX gene (previously called dnaZX) of Escherichia coli has only one open reading frame for a 71-kDa polypeptide from which two distinct DNA polymerase III holoenzyme subunits, tau (71 kDa) and gamma (47 kDa), are produced. To determine how the gamma subunit is generated, we examined the influence of mutations in the dnaX gene on the pattern of tau and gamma production in overproducing cells. Important structural elements in dnaX mRNA include a stretch of six adenines (nucleotides 1425-1430), a stable hairpin structure (nucleotides 1437-1466), and a UGA stop codon in a -1 frame (nucleotides 1434-1436) between the stretch of adenines and the hairpin structure. Disruption of this stop codon generates a slightly larger gamma subunit, indicative of the use of a -1 stop codon farther downstream (nucleotides 1470-1472). These results suggest that a -1 frameshift during translation allows the use of this UGA codon to terminate translation of the gamma polypeptide. The amino acid composition, sequence, and mass spectra of a C-terminal peptide from mild digestion of the purified gamma protein with endoproteinase Lys-C confirms that this frameshift occurs at either of the two lysine codons in the region of the adenine stretch. Remarkable features of this frameshifting are its high frequency (i.e., about 80% in an overproducing cell) and the striking structural similarity to the frameshifting signal responsible for expression of the pol and pro genes in many retroviruses.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley K. J., Hayashi M. Role of premature translational termination in the regulation of expression of the phi X174 lysis gene. J Mol Biol. 1987 Dec 20;198(4):599–607. doi: 10.1016/0022-2836(87)90203-8. [DOI] [PubMed] [Google Scholar]
- Clare J. J., Belcourt M., Farabaugh P. J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988 Sep 5;203(1):75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- De Pauw E., Pelzer G., Dao Viet D., Marien J. On the influence of hydrophobicity in the SIMS spectra of amino acids in glycerol matrix. Biochem Biophys Res Commun. 1984 Aug 30;123(1):27–32. doi: 10.1016/0006-291x(84)90375-9. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaira M., Biswas S. B., Kornberg A. The dnaX gene encodes the DNA polymerase III holoenzyme tau subunit, precursor of the gamma subunit, the dnaZ gene product. Mol Gen Genet. 1983;192(1-2):80–86. doi: 10.1007/BF00327650. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Kanda P., Kennedy R. C., Walker J. R. Relation of the Escherichia coli dnaX gene to its two products--the tau and gamma subunits of DNA polymerase III holoenzyme. Nucleic Acids Res. 1987 Oct 12;15(19):7663–7675. doi: 10.1093/nar/15.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Maki S., Kornberg A. DNA Polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J Biol Chem. 1988 May 15;263(14):6570–6578. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. I. Purification and distinctive functions of subunits tau and gamma, the dnaZX gene products. J Biol Chem. 1988 May 15;263(14):6547–6554. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J Biol Chem. 1988 May 15;263(14):6561–6569. [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- Mullin D. A., Woldringh C. L., Henson J. M., Walker J. R. Cloning of the Escherichia coli dnaZX region and identification of its products. Mol Gen Genet. 1983;192(1-2):73–79. doi: 10.1007/BF00327649. [DOI] [PubMed] [Google Scholar]
- O'Donnell M. E. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1987 Dec 5;262(34):16558–16565. [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. ATP interactions of the tau and gamma subunits of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989 Oct 25;264(30):17790–17795. [PubMed] [Google Scholar]
- Tsurushita N., Maki H., Korn L. J. Site-directed mutagenesis with Escherichia coli DNA polymerase III holoenzyme. Gene. 1988;62(1):135–139. doi: 10.1016/0378-1119(88)90587-2. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Witten J. L., Schaffer M. H., O'Shea M., Cook J. C., Hemling M. E., Rinehart K. L., Jr Structures of two cockroach neuropeptides assigned by fast atom bombardment mass spectrometry. Biochem Biophys Res Commun. 1984 Oct 30;124(2):350–358. doi: 10.1016/0006-291x(84)91560-2. [DOI] [PubMed] [Google Scholar]
- Yin K. C., Blinkowa A., Walker J. R. Nucleotide sequence of the Escherichia coli replication gene dnaZX. Nucleic Acids Res. 1986 Aug 26;14(16):6541–6549. doi: 10.1093/nar/14.16.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K. C., Blinkowa A., Walker J. R. Nucleotide sequence of the Escherichia coli replication gene dnaZX. Nucleic Acids Res. 1986 Aug 26;14(16):6541–6549. doi: 10.1093/nar/14.16.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]