Abstract
Activation of synovial fibroblasts (SF) contributes to rheumatoid arthritis (RA) by damaging synovial membranes and generating inflammatory cytokines that recruit immune cells to the joint. In this paper we profile cytokine secretion by primary human SF from normal and RA donors and show that SF activation by TNFα, IL–1α, and Poly(I:C) causes secretion of multiple cytokines found at high levels in RA synovial fluids. We use interaction multi-linear regression to quantify therapeutic and counter–therapeutic drug effects across activators and patient donors and find that the ability of drugs to block SF activation is strongly dependent on the identity of the activating cytokine. (5z)–7–oxozeaenol (5ZO), a pre–clinical drug whose primary target is transforming growth factor β–associated kinase 1 (TAK1), is more effective at blocking SF activation across all contexts than the approved drug tofacitinib, arguing for development of molecules similar to 5ZO as RA therapeutics.
The initiating events of rheumatoid arthritis (RA), a chronic inflammatory disease that causes progressive joint destruction, are not fully understood but it is clear that abnormal adaptive and innate immunity are involved.1 Macrophages, T cells and other immune cell types infiltrate joints, resulting in swelling of the synovial membrane and causing pain and disability.2 Cytokines such as TNFα and IL–6, which are involved in cell–cell communication among immune cells and resident synovial fibroblasts, are key mediators of RA whereas drugs that inhibit these cytokines are leading RA therapies.3,4 Despite a growing number of such drugs, 40% of patients fail to fully respond to therapy5 and many experience periods of disease remission followed by flare–ups and progression. This emphasizes the need to better understand drug response and resistance and to identify new and potentially more effective therapies. Recent FDA approval for RA of the small molecule Janus kinase (JAK) inhibitor tofacitinib (Xeljanz®; see Supplementary Table 1 for a list of abbreviations and synonyms) demonstrates the therapeutic potential of small molecule drugs targeting signaling kinases that regulate inflammatory cytokine production. However, the failure in clinical trials of p38 MAP kinase inhibitors also designed to block cytokine production illustrates the difficulty of finding therapeutically efficacious modulators of inflammation.6,7
Molecular analysis of RA has concentrated on infiltrating immune cells but evidence is accumulating that synovial fibroblasts, which maintain the synovial membrane and produce lubricating molecules such as hyaluronan, play a key role in disease pathogenesis.8,9 SF are found at the leading edge of joint erosion where they adopt an activated phenotype involving secretion of inflammatory cytokines and recruitment of immune cells.8,10 SF from RA patients (RA SF) can invade and degrade human cartilage in immune deficient murine models11,12 and RA SF retain an activated phenotype in ex vivo culture for several weeks before eventually becoming quiescent.13 The activated phenotype can be regenerated by treating cells with inflammatory cytokines such as IL–1 or TNF.13
To better understand how SF respond to and shape the microenvironment of the inflamed synovium and how this might be interrupted therapeutically we exposed cells harvested from normal and RA patients to disease–relevant cytokines and then used cytokine profiling to monitor SF activation in the presence and absence of drugs that inhibit signal transduction kinases (See Figure 1a–c). We studied pre–clinical tool compounds as well as tofacitinib and a p38 inhibitor that failed in clinical trials (PH–797804). We also measured cytokine levels in RA synovial fluids to compare profiles of cytokines secreted by SF with the microenvironment of an arthritic joint. Data were analyzed in their entirety using a regression method (interaction multi–linear regression; iMLR) that leverages the multivariate perturbational structure of the data to quantify the statistical significance and effect size of cytokine–drug responses across donor sample, drug class and activating ligand.
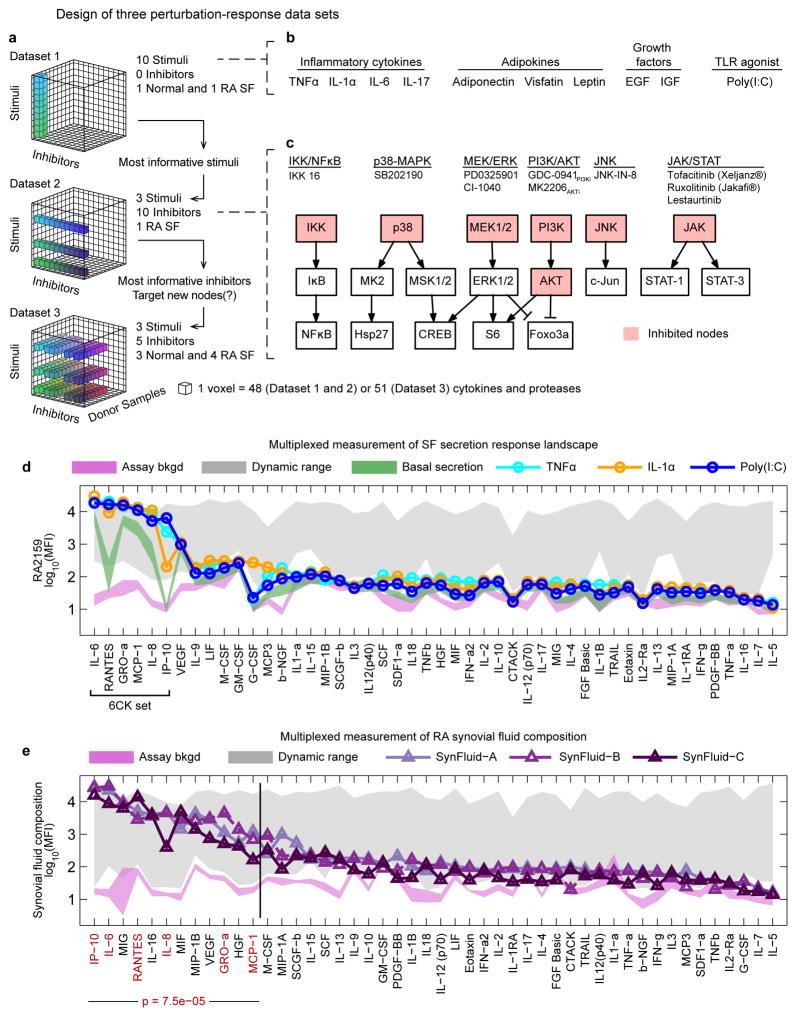
Figure 1. Experimental strategy exploring SF activation and composition of RA synovial fluids.
(a–c) Multivariate experimental design involving three successive datasets to assay basal and induced SF cytokine secretion across multiple activating ligands, small molecule drugs, and SF donor samples. Dataset 1: cytokine secretion induced in a single normal or RA SF donor by 10 stimuli; Dataset 2: evaluation of the effects of 10 kinase inhibitors on cytokine secretion induced by the top three stimuli from Dataset 1, evaluated on one RA SF sample; Dataset 3: evaluation of donor–to–donor variability for five kinase inhibitors and three stimulatory ligands across three normal or four RA SF samples. (d) Selected secretion profiles for RA2159 cells from Dataset 1 representing the three most active stimuli (profiles for all ligands are available in Supplementary Fig. 1). Magenta shaded region is the mean assay background ±2 standard deviations (S.D.) for each measured cytokine and green region is basal secretion from unstimulated SF ±2 S.D.. Gray region reflects upper and lower bounds of each cytokine assay (the dynamic range) as determined by standard curves for each measured cytokine. The “6CK set” comprises six ligands that were strongly induced by TNFα, IL–1α, and Poly(I:C). (e) Cytokine profiles in synovial fluids from three RA patients. The 6CK set (red text) is significantly enriched in the top 25% of cytokines with the highest signal in RA synovial fluids (p=7.5 × 10−5 by hypergeometric test). Normal synovial fluid was unavailable for profiling due to challenges in collecting such material from healthy individuals.46
We found that SF from both normal and RA donors are similarly activated by TNFα, IL–1α or poly(I:C) (a TLR3 agonist that mimics viral infection) such that they secrete a subset of the most abundant pro–inflammatory cytokines present in the synovial fluid of RA patients. Multiple kinase inhibitors, including tofacitinib, partially block SF activation but the magnitude of this effect is highly dependent on the identity of the activating ligand. The TAK1 inhibitor (5z)–7–oxozeaenol (5ZO) is unique among compounds studied in that it blocks induced cytokine secretion in multiple patient samples regardless of activating ligand. This argues in favor of further development of drugs related to 5ZO for treatment of RA and illustrates the potential of cue–signal–response studies14 to discriminate between potentially therapeutic and counter–therapeutic drug activities..
RESULTS
Cytokine profiles for synovial fibroblasts and synovial fluid
To screen for factors that activate and inhibit cytokine secretion by SF in culture we collected three datasets: Dataset 1 (DS1) analyzed 10 activating ligands in two patient samples to find the most potent stimuli, DS2 analyzed 10 drugs in a single patient sample, and DS3 (the largest dataset) systematically explored variation across activators and kinase inhibitors identified as most significant in DS1 and DS2 using cultures of 7 normal and RA SF (Fig. 1a; primary SF information is available in Supplementary Table 2). In DS1, SF from a normal and an RA human donor were exposed to 10 growth factors, cytokines or TLR agonists relevant to RA or SF biology (Fig. 1b; see also Supplementary Table 3 for additional details on stimuli). After 18 hr the levels of 48 cytokines present in culture supernatants were measured using commercial multiplexed bead–based antibody assays (DS1 comprised ~6 × 103 data points). These, and all subsequent data, have been formatted to NIH Library of Integrated Network–based Cellular Signatures (LINCS) standards to facilitate follow–on analysis by others.
We observed that TNFα, IL–1α, and Poly(I:C) were the most active ligands on normal and RA SF and induced secretion of multiple cytokines, among which six were produced at >500 pg/mL (IL–6, MCP–1, IL–8, GROα, RANTES, and IP–10; hereafter the “6CK set”; Fig. 1d and Supplementary Results, Supplementary Figs. 1 and 2a–f). These pro–inflammatory cytokines are involved in chemotaxis by innate or adaptive immune cells15–23 (see Supplementary Table 4) and are present at elevated levels in the serum or plasma of RA patients24–26. Secretion of several less abundant inflammatory cytokines was also observed, including the monocyte attractant MCP–3/CCL7,27 the neutrophil mitogen G–CSF,28 and the T cell/macrophage activator MIF29,30 (Fig. 1d and Supplementary Fig. 2g–h). VEGF, an angiogenic factor implicated in RA,31 was present at high levels in conditioned media from SF, but was not further induced by activating cytokines (Fig. 1d and Supplementary Fig. 1).
To compare factors secreted by cultured SF treated with TNFα, IL–1α, or Poly(I:C) to factors present in the microenvironment of a diseased joint we assayed 48 cytokines in synovial fluids from three RA patients (Supplementary Table 5). We found that members of the 6CK set were significantly enriched in the top quartile of cytokines in RA synovial fluids (Fig. 1e red text, p=7.5 × 10−5 by a hypergeometric test) and that the full cytokine profile of activated SF significantly correlated with the cytokine profile of RA synovial fluids (controlling for weak positive correlation between conditioned medium from unactivated SF and synovial fluid; Supplementary Fig. 3). In contrast, exposure of SF to the seven other stimuli in DS1 did not yield a pattern of secretion that was correlated with synovial fluid cytokine profiles (Supplementary Fig. 3). These data suggest that SF activated by TNFα, IL–1α, and Poly(I:C) play a substantial role in shaping the inflammatory microenvironment of an RA joint and may be a primary source of multiple molecules known to a play role in disease.
Effects of anti–inflammatory drugs vary with stimulatory context
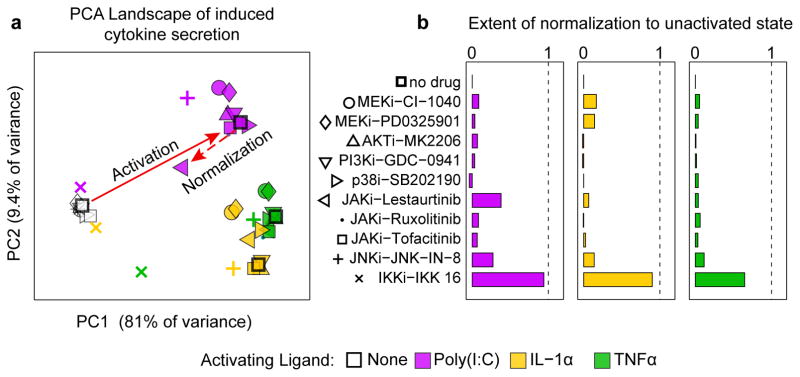
To identify small molecule kinase inhibitors that block SF activation we exposed a single patient sample to TNFα, IL–1α, or Poly(I:C) in the presence of one of 10 small molecule drugs and then measured the levels of 48 cytokines (DS2; ~5500 data points, Fig. 1a,c); for simplicity we refer to these drugs by their primary targets but most kinase inhibitors are active against multiple proteins32 (see Supplementary Table 6). Using principal component analysis (PCA) to reduce the dimensionality of the data we found that the maximal dimension of variation (principal component 1; PC1) separated basal and activated conditions whereas PC2 differentiated among the three stimuli (Fig. 2a). Effects of the drugs can be visualized by comparing positions in the PCA landscape corresponding to basal, activated and activated plus drug–treated conditions: kinase inhibitors shifted activated cells back towards the basal state, thereby “normalizing” their secretory profile. The magnitude of the activation and normalization (illustrated by arrows for Poly(I:C) and Poly(I:C)+lestaurtinib; Fig. 2a) provides a simple metric of drug effect across a multivariate SF phenotype (Fig 2b).
Figure 2. Multivariate analysis of small molecule kinase inhibitors on induced cytokine secretion.
(a) Principal component analysis of cytokine profiles differentiate secretion profiles for basal conditions (open data points) and TNFα–, IL–1α, or Poly(I:C)–activated SFs (colored data points). Solid red arrows illustrates activation by Poly(I:C)– and dotted arrow depicts the normalizing effect of lestaurtinib. (b) Ability of kinase inhibitors to prevent SF activation and restore the cytokine secretion profile to the pre–stimulus state. A value of 0 indicates no effect of the inhibitor and a value of 1 corresponds to complete normalization.
We found that tofacitinib, a JAK inhibitor approved for RA, and ruxolitinib, which is approved for myelofibrosis (and is a structural analog of baricitinib, which is currently being evaluated clinically for use in RA) resulted in normalization of ~8% of the activated cytokine profile (a normalization coefficient of 0.08). In contrast, the investigational compound lestaurtinib a multi–targeted tyrosine kinase inhibitor32 that blocks JAK2 had a normalization coefficient of 0.38 with Poly(I:C) as activator (Fig. 2b) but only 0.04 to 0.06 with TNFα or IL–1α. IKK 16, a semi–selective inhibitor of the IKK1/2 kinases that regulate the NFκB transcription factor exhibited a normalization factor of ~0.9 with IL–1α and Poly(I:C), as activators but only 0.65 with TNFα (Fig. 2b). IKK 16 has an off–target effect on the JNK pathway (Supplementary Fig. 5a) but the JNK inhibitor JNK–IN–833 had a relatively low normalization coefficient that varied with stimuli (0.11–0.27) suggesting that JNK inhibition is not the key factor in IKK 16 activity (Fig. 2b). From these data we conclude that the effect of kinase inhibitors on cytokine secretion by SF varies dramatically with activating ligand even though, once activated, SF produce a common set of cytokines (Fig. 1d). Moreover, approved and investigational JAK inhibitors only partially block SF activation, but multi–targeted kinase inhibitors such as lestaurtinib and IKK 16 are significantly more effective.
iMLR accurately quantifies context-dependent drug effects
To study the effects of donor–to–donor variability on SF activation and drug response, we collected data from four RA and three normal donor cell cultures exposed to all pairwise combinations of TNFα, IL–1α, or Poly(I:C) and five kinases inhibitors across four biological repeats (DS3 comprised ~5 × 104 data points, included data on three matrix metalloproteinases – MMP–1 to –3; the correlation between replicate experiments was r ≥0.95; Supplementary Fig. 4). We included three drugs from DS2 (JNK–IN–8, IKK16, tofacitinib), a p38 inhibitor tested in Phase II trials for RA (PH–797804) and 5ZO, a preclinical compound whose nominal target is the MAP kinase kinase kinase (MAP3K) TAK1.34,35 TAK1 regulates multiple MAPK pathways and its importance in inflammation became clear during the course of the current study.36 All drugs were used at a concentration that resulted in 95% inhibition of the primary target in SF (IC95, Supplementary Fig. 5), but below a concentration that caused cell death (Supplementary Fig. 6). Analysis of the data showed that cytokine profiles of activated SF again correlated with the profiles of RA synovial fluid and that inhibitors counter-acted the effects of the activating ligands (Supplementary Fig. 7).
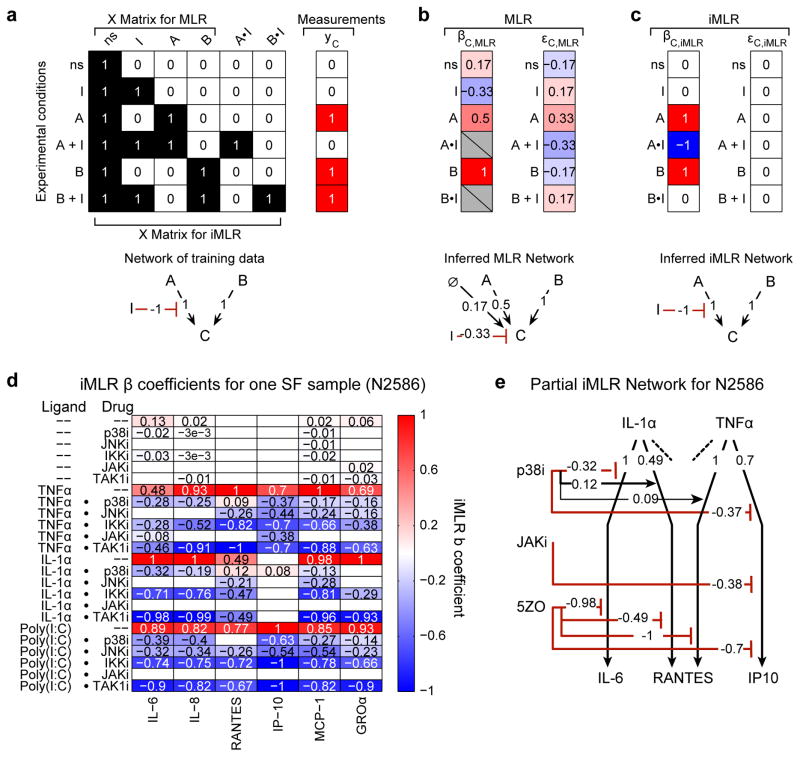
To account for variability among SF samples we used iMLR to identify statistically significant associations between drug treatment and response while accounting for context (activating ligand and patient donor).37–39 In conventional MLR the effects of activating stimuli and kinase inhibitors (the independent variables in matrix X) on measured cytokine levels (dependent variables in column vector y) are solved as y = X β+ε, where β is a vector of regression coefficients and ε is a vector of residuals. In iMLR X includes an interaction term for each pairwise combination of independent variables37 (e.g. for each activating cytokine and kinase inhibitor). Use of iMLR is illustrated with synthetic data in which cytokines A and B provoke response C and the activity of A but not B is blocked by inhibitor I (Fig. 3a). Matrix X describes an experimental design in which cells are treated with A or B in the presence or absence of I, and measurements y are made on the state of C (Fig. 3a). The network computed by MLR is depicted as a node–edge graph with β coefficients mapped to edge weights (Fig. 3b bottom). In this toy model A and B are inferred by MLR to up–regulate C (with edge weights A→C = 0.5 and B→C = 1.0) and I is inhibitory (I→C = −0.33) but error is high (εC = −0.17 to 0.33) and the topology of the network is wrong: C is incorrectly postulated to be active in the absence of stimulus and I to act directly on C (Fig. 3b). In contrast, iMLR infers an error–free network (for noise–free synthetic data) and correctly assigns edge weights of 1 to A→C and B→C and of −1 to A•I→C while eliminating the no–stimulus activity and context–independent effect of I on C (NS→C= 0 and I→C = 0; Fig. 3c). The superior performance of iMLR arises because interaction terms explicitly encode the perturbational design of the experiment.
Figure 3. Interaction–based multiple linear regression (iMLR) to analyze context dependencies in perturbation–response data.
(a) Analysis of a ‘toy’ model and synthetic data to illustrate the advantages of iMLR over MLR for analyzing perturbation–rich data. Design matrix (X) and synthetic measurements (yc) for an experiment evaluating the effect of A, B, and inhibitor I on output C; synthetic data corresponds to the network depicted below (red interactions are inhibiting and black are activating). The no-stimulus (ns) condition is mathematically represented as a column of ones. The X matrix for iMLR explicitly encodes the possibility that inhibitor I can act in a context–dependent manner on A (or B) by including the dot product of the A and I (or B and I) columns (A•I and B•I). (b–c) β coefficients (βC,MLR or βC,iMLR), model residuals (εC,MLR or εC,iMLR), and node–edge graph for networks inferred by MLR (b) or iMLR (c). (d) iMLR results for SF sample N2586 from Dataset 3; β coefficients for 6CK set cytokines (columns) and stimulus, inhibitor, and context–dependent inhibitor effects (rows). Only statistically significant effects are shown. Coefficients were scaled to column maximum of 1. (e) Node–edge graph of β coefficients for two stimulating ligands and three drugs as measured by induced IL–6, RANTES and IP10. The absence of an interaction from a drug to a ligand response indicates the absence of a significant effect; each activating ligand has additional interactions that, for simplicity, are not depicted.
Quantifying drug action across multiple donors and activators
Application of iMLR to DS3 generated one network for each donor cell sample; we assessed the statistical significance of these networks using a multi–modeling framework that merged p–value tests with Akaike and Bayesian information criteria (Supplementary Table 7, Supplementary Fig. 8–11, and Supplementary Data 1–14). We found that nearly all (407/425) of the effects of kinase inhibitors on 6CK cytokine levels were inhibitory and many of these were context–sensitive. For example the p38 inhibitor PH–797804 reduced the levels of IL–1α induced secretion of the 6CK cytokine IL–6 in in N2586 cells (IL–1α • p38i→IL–6 = −0.32) but it increased the levels IL–1α or TNFα–induced RANTES (IL–1α • p38i→RANTES = +0.12 and TNFα • p38i→RANTES = +0.09) (Fig. 3d–e). In contrast, 5ZO blocked induced secretion of IL–6 and RANTES regardless of activating ligand. In general, more drugs were effective at normalizing secretion with Poly(I:C) as an activating ligand than with TNFα or IL–1α.
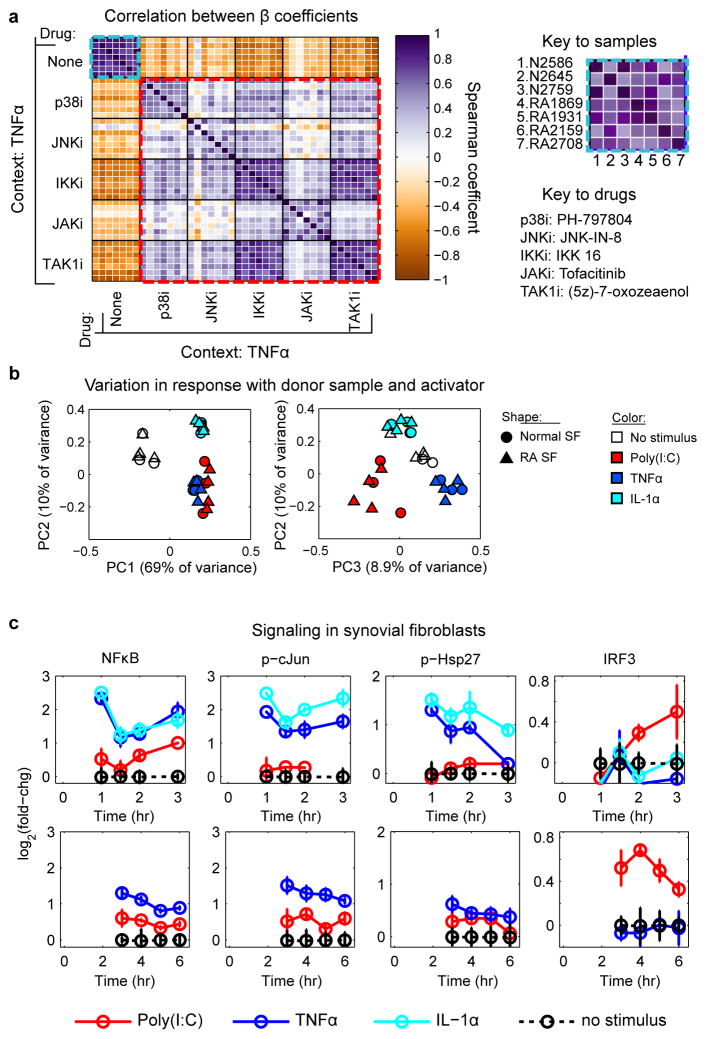
To quantify variability in drug response with donor sample and stimulatory context, we calculated Spearman correlations for iMLR coefficients compiled across all measured cytokines and conditions (Supplementary Fig. 12 and 13). We found that basal secretion profiles were correlated across all seven SF samples including between normal and RA SF; the same was true of secretion profiles for activated cells (Fig 4a and Supplementary Fig. 13). Moreover, the way SF were activated and not the identity of the donor was the primary source of variation in drug response. Consistent with these results iMLR coefficients compiled across stimulatory contexts and donors clustered in PC space by stimulatory context rather than by RA vs normal SF (Fig. 4b). Such variability reflects differences in kinase signaling by toll–like vs. cytokine receptors.40,41 Indeed, in SF we found that Poly(I:C) strongly activates IRF3 and induces low but sustained NFκB, JNK, and p38 signaling, whereas TNFα and IL-1α induce strong and rapid activation of the NFκB, JNK, and p38 pathways, but do not activate IRF3 (Fig. 4c).
Figure 4. Dependence of SF activation on the identify of the activating ligand.
(a) Spearman correlation matrix of β coefficients for different drugs and donor samples under conditions of TNFα stimulation. Each individual entry is the correlation between β coefficients across the full cytokine/protease profile, and the heavy black gridlines distinguish 7 × 7 matrices for the seven SF donor samples shown in the figure key. The light blue boxes denote the correlation of stimulated secretion across the seven SF samples, and red boxes denote the correlation between drug effects. (b) iMLR β coefficients compiled across stimuli and SF donor samples cluster according to stimulatory context in PCA space. (c) Activity of IKK/NFκB, JNK, and IKKε/TBK1–IRF3 pathways in SF sample RA2159 following 1–3 hr (top) or 3–6 hr (bottom) of cytokine stimulation.
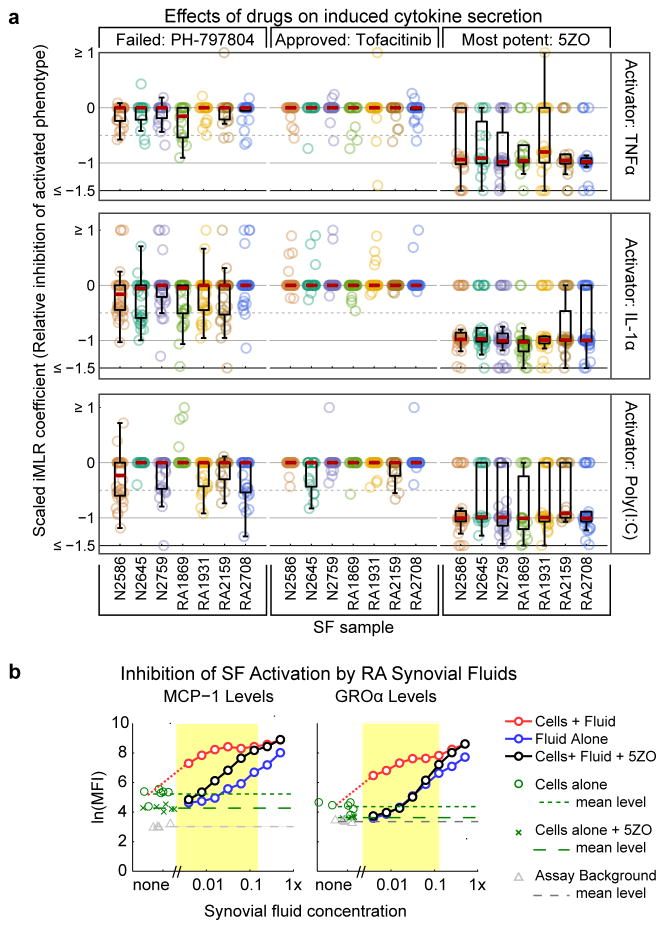
To determine the magnitude of drug–induced renormalization of cytokine secretion (effect size) we scaled iMLR coefficients between 0 (no effect) and −1 (full inhibition). We then compared the JAK inhibitor tofacitinib, an approved treatment for RA, p38 inhibitor PH–797804, a failed clinical candidate, and 5ZO and found that the latter compound was substantially more effective across conditions and donor samples tested (Fig. 5a and Supplementary Fig. 14). We also found that 5ZO could normalize SF activated by synovial fluids obtained from three different RA patients, as assayed by inhibition of induced GROα and MCP–1 secretion (Fig. 5b and Supplementary Fig. 15; the other 6CK cytokines were too abundant in synovial fluid for their induction to be scored). We conclude that 5ZO is a promising lead compound for blocking activation of SF by cytokines present in inflamed joints.
Figure 5. 5ZO normalizes SF activation across multiple contexts.
(a) Distribution of β coefficients for three kinase inhibitors across the activated cytokines profiles with values scaled so that −1 represents complete normalization to basal conditions and 0 represents no response to drug. Red dashes represents the median value of the distribution and boxes denote 25th and 75th percentile. (b) Effect of 5ZO on SF activated by RA synovial fluid. Blue line shows the level of MCP–1 (left) or GROα (right) in serially diluted synovial fluids alone. The red line depicts cytokine levels in serially diluted synovial fluids following 18 hr incubation with SF and black lines depict the same experiment performed in the presence of 0.6 μM 5ZO. Yellow region highlights the dilution range over which secretion of MCP–1 or GROα by SF is activated by synovial fluid and inhibited by 5ZO. Induction of other 6CK cytokines could not be assayed because they were too abundant in synovial fluid.
Identifying potentially counter–therapeutic drug activities
Systematic cytokine profiling followed by iMLR also provided a means to identify potentially adverse or counter–therapeutic drug effects. We observed that exposure of SF to PH–797804 increased secretion of RANTES in multiple donor samples activated by IL–1α or TNFα (Supplementary Fig. 16a). The magnitude of this effect varied with donor and stimulatory context, making it difficult to detect using conventional approaches. JNK–IN–8 elevated secretion of MMP–1 in cells activated by IL–1α and TNFα (Supplementary Fig. 16b). With 5ZO we observed potentiation of FGF–2 secretion with TNFα, but not IL–1α or Poly(I:C) as activators (Supplementary Fig. 16c); the FGF–2 growth factor has been implicated in synovial hyperplasia31 making its upregulation undesirable. However, exposure of activated SF to NG25, a structurally distinct TAK1 inhibitor, did not promote FGF–2 production (Supplementary Fig. 16d–e) showing that not all TAK1 inhibitors have this activity.
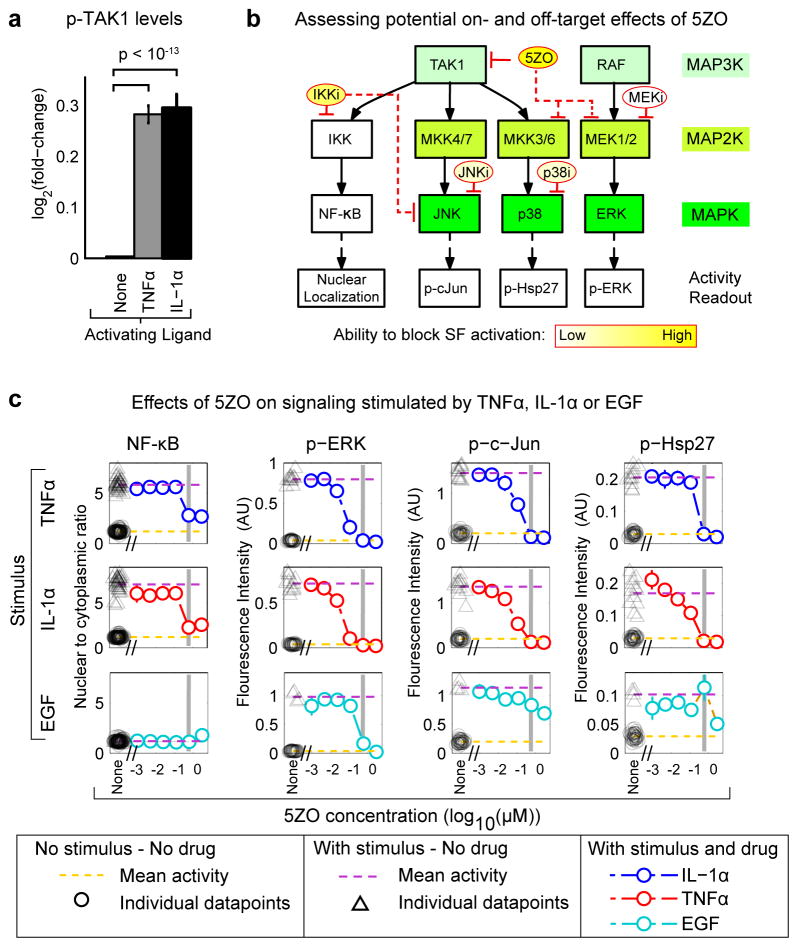
The 5ZO target TAK1 is a central mediator of SF activation
TNFα and IL–1α induce phosphorylation of TAK1 in SF (Fig. 6a and Supplementary Fig. 17). The nominal target of 5ZO is TAK1, which lies upstream of NFκB, p38 and JNK (Fig. 6b), but pre–clinical compounds are rarely mono–selective and known 5ZO targets include MEK1/2, which regulates ERK, and MKK6, which regulates p38 (Fig. 6b; International Centre for Kinase Profiling; http://www.kinase-screen.mrc.ac.uk/screening-compounds/349381). In SF, we observed that 5ZO reduced activation of NFκB, JNK, p38, and MEK by TNFα and IL–1α in a dose–dependent manner (Fig. 6c). When we assayed EGF–induced signaling, which is not believed to involve TAK1, MEK emerged as the primary off–target activity of 5ZO at the concentrations used in DS3 (gray line in Fig. 6c). However, highly selective MEK inhibitors had little or no effect on cytokine production by activated SF, nor did inhibitors of other potential 5ZO targets or of kinases lying downstream of TAK1 (e.g. p38i, JNKi; Fig. 6b). Thus, both the known off–target effects of 5ZO and targeting individual pathways downstream of TAK1 do not explain 5ZO’s anti–inflammatory activities in SF, suggesting inhibition of multiple TAK1-dependent kinase cascades are necessary to block SF activation. Moreover the TAK1 inhibitor, NG25, which has non–overlapping off–target pharmacology from that of 5ZO42, also normalized cytokine secretion by activated SF (Supplementary Fig. 16d). We therefore conclude that TAK1 is a central mediator of SF activation by TNFα, IL–1α, Poly(I:C) and RA synovial fluid. It is possible, however, that partial inhibition of multiple signaling kinases increases the effectiveness of 5ZO across donors and activators.
Figure 6. Inferring the likely targets of 5ZO based on experiments in SF and current understanding of the drug.
(a) Phosphorylation of TAK1 on activating residues (Thr184/187) 10 min after addition of 100 ng/mL TNFα or IL–1α to SF samples RA2159 in the presence of 50 nM calyculin, a serine/threonine protein phosphatase inhibitor. Error bars are SEM for detection of p–TAK1 in biological duplicate using two p–TAK1 antibodies, each at four different dilutions across three separate days (48 replicates for each stimulus; see Supplementary Fig. 17). (b) Schematic of TAK1 and MAPK signaling showing potential on–target (solid red lines) and off–target (dotted red lines) drug effects, as well as potency in inhibiting SF activation (encoded by the intensity of the yellow boxes). Analysis of DS2 and DS3 shows that secondary targets such as MEK and components of the p38 cascade have a small effects on cytokine secretion by SF relative to 5ZO. (c) Activity of signaling cascades in cells stimulated for 30 min with 100 ng/ml TNFα, IL–1α or EGF in the presence and absence of 5ZO. Activation of NFκB was scored by nuclear translocation of NFκB p65 and activation of MEK/ERK, JNK and p38 kinases by the readouts shown in panel b.
DISCUSSION
In this paper we use cytokine profiling and statistical modeling by iMLR to study the effects of small molecule drugs on the activation of SF from RA and normal donors. Among ten activating ligands examined, the most potent were TNFα and IL–1α, known mediators of RA that have been targeted therapeutically, and Poly(I:C), a TLR3 agonist designed to mimic viral infection during RA flare–ups. The profile of cytokines secreted by activated SF is rich in immune cell activators and chemoattractants and significantly correlates with the cytokine profile of synovial fluid from RA patients. These data are consistent with a growing body of evidence that (i) SF play a substantial role in shaping the inflammatory RA environment, (ii) that non–RA cells can be “activated” to an RA–like phenotype, and (iii) that this phenotype is similar (at the level of cytokine secretion) across multiple activators (Supplementary Fig. 18). Our findings are consistent with previous data showing that cultured RA SF retain a memory of the inflamed RA synovium and can alternate between quiescent and activated states in response to the addition or withdrawal of inflammatory factors.13
The persistence of inflammation in RA is thought to involve positive feedback whereby activated SF recruit immune cells to the joint, causing both cell types to shape and respond to the microenvironment in a self–sustaining manner.9,43 Cellular memory in the context of SF may be due in part to the induction of autocrine loops and factors secreted by other cell types, such as TNF which is produced primarily by immune cells. The similarity in cytokine secretion by activated SF from both normal and RA donors suggests that a self–sustaining microenvironment may be more important to disease that any intrinsic aspect of SF dysregulation. However, the number of donor samples in the current paper (n = 7) is too small for us to have detected subtle differences between normal and RA cells.
A striking result from our studies is that inhibitors of signal transduction kinases involved in cytokine production affect SF in a manner that strongly depends on how the cells are activated but much less strongly on the identity of the donor sample. We find that kinase inhibitors effective at blocking induced inflammatory cytokine secretion when Poly(I:C) is used as an activator are less effective when TNFα or IL–1α are the activators but the converse was also observed in a few cases. Context–dependent drug effects present a potential complication in the discovery of broadly active therapeutic drugs but they also represent an opportunity. We speculate that patient–to–patient differences in the efficacy of RA drugs may arise in part from differences in the way the disease (or flare–up) was induced, by viruses or other factors for example. This implies that selecting therapy based on the identity of the disease inducer might be therapeutically beneficial. The magnitude of context–dependent drug activities in RA also suggests a need to substantially improve our understanding of the complex signaling networks that regulate inflammatory cytokine production in SF and other cells in joints; such an analysis would guide further drug discovery and potentially yield useful drug response biomarkers.
Among the compounds we analyzed, 5ZO, a drug whose nominal target is the MAP3K TAK1, emerged as the most broadly effective inhibitor across donor samples and SF activators. Under virtually all conditions examined, 5ZO almost completely reverted the spectrum of cytokines expressed by activated SF. To the extent we could measure it, 5ZO also reversed activation of SF by RA synovial fluid. In contrast, the FDA approved JAK inhibitor tofacitinib was substantially less effective. These data argue for development of drugs with biological activities similar to those of 5ZO. TAK1 appears to be a primary target for this compound in SF but it will be important to consider the possibility that polypharmacology may be involved in its effectiveness.44 Moreover, potentially counter–therapeutic effects such as 5ZO–potentiated secretion of basic FGF–2 need to be engineered out.
The experimental and analytical framework we describe here for perturbational profiling45 should be applicable to other diseases characterized by multifactorial interactions between cells and their microenvironment. The approach is not geared towards high–throughput drug discovery, but rather for validation and optimization of lead compounds. Given the cost and difficulty of medicinal chemistry campaigns, adding a set of multi–dimensional assays across multiple disease modulators and donor samples is almost certainly feasible. An obvious next step in the case of RA is to analyze SF–immune cell interaction in the presence and absence of a greater diversity of activating molecules and a diversity library built around 5ZO.
ONLINE METHODS
Antibodies and reagents
Tumor necrosis factor–α (TNFα; cat. no. 300–01A), interleukin–1α (IL–1α; cat. no. 200–01A), interleukin–6 (IL–6; cat. no. 200–06), interleukin–17A (IL–17A; cat. no. 200–17), epidermal growth factor (EGF; cat. no. AF–100–15), insulin–like growth factor (IGF; cat. no. 100–11), adiponectin (cat. no. 450–24), leptin (cat. no. 300–27), and visfatin (cat. no. 130–09) were purchased from PeproTech. Poly(I:C) (cat. no. tlrl–picw) was purchased from InvivoGen. Chemical inhibitors from the following sources were dissolved in dimethyl sulfoxide (DMSO) at stock concentrations of 10 mM: IKK–1/2 inhibitor IKK–16 (cat. no. 2539) and TAK1 inhibitor (5z)–7–oxozeaenol (cat. no. 3604) were purchased from Tocris Bioscience; P38 inhibitors PH–797804 (cat. no. S2726) and SB202190 (cat. no. S1077), MEK inhibitors CI–1040 (also referred to as PD184352; cat. no. S1020) and PD0325901 (cat. no. S1036), and JAK inhibitors tofacitinib (also referred to as CP–690550; cat. no. S2789) and ruxolitinib (also referred to as INCB018424; cat. no. S1378) were purchased from Selleck Chemicals; JAK inhibitor lestaurtinib (also referred to as CEP–701; cat. no. 6307) was purchased from LC Labs; PI3K inhibitor GDC–0941 (cat. no. HY10358) and AKT inhibitor MK2206 (cat. no. HY–50094) were purchased from Haoyuan Chemexpress; JNK inhibitor JNK–IN–8 was provided by Nathanael Gray Lab, Dana Farber Cancer Institute. Phospho–p44/42 MAPK (ERK1/2)T202/Y204 (cat. no. 4370), phospho–HSP27S82, phospho–c–JunS73 (cat. no. 3270), phospho–STAT1Y701 (cat. no. 9167), phospho–STAT3Y705 (cat. no. 9145), phospho–TAK1T184/187 (cat. no. 4531), and phospho–TAK1T187 (cat. no. 4536) were purchased from Cell Signaling Technology; NFκB p65 (cat. no. sc–8008) was purchased from Santa Cruz Biotechnology. RA synovial fluids were purchased from Analytical Biological Services, Inc (see Supplementary Table 5 for details).
Tissue culture
Normal primary human synovial fibroblasts (SF) (human fibroblast–like synovioctyes (HFLS), cat. no. 408–05a) and RA SF (HFLS–RA, cat. no. 408RA–05a) were purchased from Cell Applications, Inc (see Supplementary Table 2 for details). The nomenclature for donor–derived SF samples references specific lot number (e.g. N2586 is Cell Applications HFLS lot 2586, RA2159 is Cell Applications HFLS–RA lot 2159, etc). Cells were cultured according to the supplier’s recommendations using Synoviocyte Growth Medium (Cell Applications, Inc. cat. no. 415–500) as full growth medium. Synoviocyte Basal Medium (Cell Applications, Inc. cat. no. 414–500) was used for serum starvation prior to experimental treatments. Cells were provided at passage 2 and all experiments were conducted on cells at passage 3 – 6, in accordance with published recommendations.1
Secretion response cell seeding and treatments
SF were seeded (1000 cells/well) into 384–well plates (Costar cat. no. 3712) for secretion experiments. Following ~24 hr incubation in full growth medium at 37 °C and 5% CO2 cells were starved in basal medium overnight (~16 hr) followed by an additional starvation step in basal medium that started ~4 hr prior to exposure to stimulatory factors. For Datasets 2 and 3, which included kinase inhibitors, cells were pretreated with drugs or a DMSO–only control for 3 hr prior to stimulation with cytokines. Inhibitors (described with reference to their nominal primary targets, see Supplementary Table 6) were used at the following concentrations: JNK–IN–8 (JNKi): 3 μM; PH–797804 (p38i): 0.6 μM; SB202190 (p38i): 1 μM; IKK 16 (IKKi): 2 μM; PD0325901 (MEKi): 0.1 μM; CI–1040 (MEKi): 1 μM; MK2206 (AKTi): 1 μM; GDC–0941 (PI3Ki): 1 μM; lestaurtinib (JAKi): 0.3 μM; tofacitinib (Xeljanz®, JAKi): 0.3 μM; ruxolitinib (JAKafi ®, JAKi): 0.3 μM; (5z)–7–oxozeaenol (TAK1i): 0.6 μM. All stimuli were used at a final concentration of 100 ng/mL except for Poly(I:C) (2 μg/mL), adiponectin (5 μg/mL), leptin (1 μg/mL), and visfatin (1 μg/mL), at a final volume of 50 μL/well. To determine a suitable concentration of inhibitory drug for these experiments we first evaluated the effects of a drug on a proximal downstream signaling target (e.g. p–cJun for the JNK inhibitor in cells stimulated with TNFα or IL–1α) and selected a concentration near the IC90 to achieve good target inhibition at the lowest possible dose (see Supplementary Fig. 5). We also monitored inhibitor cytotoxicity (Supplementary Fig. 6) to ensure that these concentrations did not induce significant cell death. Following 18 hr of stimulation, supernatants were recovered and clarified by 10 min centrifugation at 2000 RPM. Supernatants from two adjacent wells (e.g. well A1 and A2, which comprised biological replicates) were pooled for subsequent analysis by Luminex cytokine profiling. Downstream statistical analyses considered data from pooled supernatants as a single replicate. For Datasets 2 and 3 combinations of each stimulus and inhibitor were performed in biological duplicate and all other conditions (stimulus in absence of inhibitor, inhibitor in absence of stimulus, and unstimulated and uninhibited) were performed in at least biological quadruplicate. Pooled supernatants were adjusted with 1x phosphate buffered saline (PBS), 5% bovine serum albumin (BSA) to contain 0.25% final concentration BSA, clarified again by centrifugation, aliquoted and stored at −80 °C. For Dataset 3, the full experiment was repeated on separate days.
Secretion response measurement by Luminex assays
Multiplexed bead–based assays were analyzed on a Flexmap 3D using xPONENT software (Luminex Corp.) and reagents purchased from Bio–Rad and R&D Systems. Levels of secreted cytokines were measured using two sets of Bio–Rad detection panels: group I 27–plex (Bio–Rad cat. no. M500KCAF0Y) and group II 21–plex human cytokines (Bio–Rad cat. no. MF0005KMII) panels. Levels of MMP–1, –2, and –3 were measured using Luminex Peformance Human MMP Panel (R&D Systems cat. nos. LMP901B, LMP901C, and LMP513B). Supernatants were either diluted 1:3 with 1xPBS, 0.05% BSA, 0.05% Tween–20 (for Bio–Rad cytokines kits) or diluted 1:5 with Calibrator Diluent RD5–37 buffer (for R&D Systems MMP Panel, buffer is from Human MMP Base Kit cat. no. LMP000B) and assayed according to the supplier’s instructions alongside a 10–point standard serial four–fold dilution series (for Bio–Rad cytokines kits) or three–fold dilution series (for R&D Systems MMP kit; according to each manufacturers instructions) to determine dynamic range and infer the concentrations of each analyte.
Concentrations of cytokines in clarified supernatants were calculated using median fluorescent intensity (MFI) values and a five–parameter logistic regression curve derived from parallel measurement of serially–diluted standards. All downstream analyses used median fluorescent intensity (MFI) derived from measurement of Luminex bead intensity distributions, which is the manufacturer’s recommended metric for Luminex analysis. Upper and lower detection range of the standard curve was determined individually for each measured analyte following curve fitting by imposing a series of heuristics. For example, we considered as reliable only measurements in which the lower end of the standard curve was at least 30% higher than the assay background (by MFI) and increasing concentrations along the standard curve increased MFI signal by at least 30%.
We observed some cross–reactivity between ligand stimuli and Luminex cytokine profiling assays. For example, spiked ligand controls revealed that the same concentration of TNFα that we used in the SF activation experiments (100 ng/mL) resulted in a cross–reactive signal in unrelated Luminex kit components (see Supplementary Fig. 1). Such cross–reactivity was generally low, but could be consequential for non–6CK set analytes present at low levels (Supplementary Fig. 1). To control for cross–reactivity we measured multiple replicates of spiking controls for each stimulus at the same concentration as used to generate Datasets 1, 2, and 3. These spiked ligand controls were performed on the same Luminex assay plates as experimental samples (supernatants from activated SF). To determine the background value associated with each cytokine assay, multiple replicates of Luminex beads incubated with “mock” supernatant samples (basal media with 0.25% BSA diluted 1:3 with 1xPBS, 0.05% BSA, 0.05% Tween–20) were also included on each Luminex assay plate and were processed in an identical manner to the experimental samples.
Signaling response seeding and treatments
For signaling experiments, SF were seeded at 600 cells/well into 384–well plates and analyzed as described above for cytokine secretion experiments: following ~24 incubation in full growth medium cells were starved in basal medium overnight (~16 hr) followed by an additional starvation step in basal medium of ~4 hr duration prior to exposure to stimulatory factors (the second starvation step helps to reduce the levels of autocrine factors). Cells were pretreated with a six–point serial five–fold dilution series of inhibitors (3 μm to ~1 nM) or DMSO control for ~3 hr prior to stimulation with 100 ng/mL TNFα, IL–1α, IL–6, or EGF at a final volume of 50 μL/well; DMSO was maintained at a nominal dilution of 1:3333 for the full dilution series (which is equivalent to dilution of the 10 mM inhibitor stocks in DMSO to 3 μM maximum concentration).
Immunofluorescent microscopy and analysis
Plates were processed by adapting previously described 96–well plate protocols2,3 to 384–well plates. Briefly, supernatants were aspirated and cells were fixed for 10 min at 25 °C in 25 μL of 2% paraformaldehyde. After fixation, all subsequent steps were performed on a rocking platform. Fixed cells were washed 3x for 5 min at 25 °C using 60 μL of 1x PBS with 0.1% Tween–20 (PBST), permeabilized with 40 μL of 100% methanol for 10 min at 25 °C, washed 3x with PBST, and blocked with 60 μL of Odyssey Blocking Buffer (OBB; LI–COR) for 1 hr at 25 °C. Experiments detecting phospho–TAK1 gave low signal–to–noise and we thus made two adjustments to the protocol above: cells were fixed in PFA for 30 min (we observed that longer fixation increases signal detection for cytoplasmic proteins, but also decreases signal from nuclear proteins and transcription factors, which is why our standard protocol utilizes 10 min fixation), and permeabilized with ice cold methanol at 4 °C; all other steps were identical. Cells were stained with 25 μL of primary antibodies diluted 1:200 (p–STAT1 and p–STAT3) or 1:400 (p–ERK, p–CJUN, p–HSP27, and NFκB) in OBB, plates were sealed with foil sealing film (Bio–Rad cat. no. MSF 1001) and incubated overnight at 4 °C. Cells were washed 3x with PBST and stained with 25 μL appropriate secondary antibodies diluted 1:2000 in OBB and incubated 1 hr at 25 °C. Cells were washed 1x in PBST and then 1x in PBS and incubated with 0.25 μg/mL Hoechst–33342 (Invitrogen cat. no. H3570) and 1:1000 dilution of Whole Cell Stain Blue (Thermo Fisher cat. no. 8403202) in PBS for 30 min at 25 °C. Cells were washed 2x in PBS, sealed in foil sealing film and imaged using a 10× objective on an Operetta high–throughput microscope using Harmony software (PerkinElmer, Inc.).
We extracted quantitative data from the immunofluorescent microscopy images using Columbus software (PerkinElmer, Inc.). Briefly, Hoechst and Whole Cell Stain Blue signals were used to segment the nuclear and cellular boundaries to define nuclear, cytoplasmic, and whole cell regions. ‘Ring regions’ were defined for each individual cell and used for local background correction. The ring regions comprised the portion of the circumference of an individual cell that was devoid of neighboring segmented cells, and comprised the region 5 – 12 pixels beyond the cell boundary as defined by the segmentation algorithm. Signals for nuclear, cytoplasmic, and whole cell regions were normalized by the local background as quantified from cell–specific ring regions. This normalization strategy allowed for local background correction and decreased effects of artifacts such as non–flat illumination, bubbles, and dust. For NFκB nuclear/cytoplasmic ratios were calculated for each cell. Single–cell intensities were extracted for the appropriate cellular region, for example nuclear intensity was used for phosphorylated transcription factors such as p–c–Jun, p–STAT1, and p–STAT3, whole cell or cytoplasmic intensity was used for p–ERK1/2 and p–HSP27, and nuclear/cytoplasmic ratio was used for NFκB. Median values of single cell distributions within a given well were used for analyses of stimulus and inhibitor effects.
Secretion response of SF to RA synovial fluids
SF were prepared as described above: cells were seeded into 384–well plates, incubated overnight in full growth medium, serum starved overnight in basal medium, and basal medium refreshed ~4hr prior to stimulation. Due to extremely limited quantities of the RA synovial fluid samples, synovial fluids were serially diluted and dilution series data was used in favor of experimental replicates. Two separate eight–point 1:2 – 1:256 serial two–fold dilutions of each RA synovial fluid were prepared in basal medium: one containing DMSO control and one containing (5z)–7–oxozeaenol at 0.6 μM final concentration. Cells were incubated with 40 μL/well of the RA synovial fluid serial dilutions ((5z)–7–oxozeaenol) for 18 hr at 37 °C and 5% CO2. Supernatants were recovered as previously described: supernatants were clarified by centrifugation, adjacent wells (consisting of biological replicates) were pooled and adjusted to 0.25% BSA, supernatants were clarified again by centrifugation, aliquoted and stored at −80 °C. For analysis of cytokine profiles by Luminex, supernatants were diluted 1:3 with 1x PBS, 0.05% BSA, 0.05% Tween–20, 225 μg/mL Heteroblock (Omega Biologicals; final Heteroblock concentration 150 μg/mL) and processed according to the supplier’s protocol. Heteroblock blocks potential non–specific signal amplification by rheumatoid factor or other heterophilic antibodies in ELISA–type assays of RA samples.4 Serial two–fold dilutions (1:2 – 1:256) of RA synovial fluids (without exposure to SF) were also included on the same luminex analysis plate to allow assessment of cytokine levels in each synovial fluid both before and after incubation with SF.
DATA PROCESSING AND STATISTICAL METHODS
Analysis of basal and induced cytokine secretion by SF and of RA synovial fluid composition
We quantified the relationship between the supernatant cytokine profile following exposure to various stimuli and the cytokine profiles in RA synovial fluids by calculating the partial Pearson correlation between the supernatant and RA synovial fluid profiles when controlling for the variance explained in each profile by basal secretion levels. Briefly, cross–reactivity for each stimulating ligand against the Luminex analytes (as determined by spiking controls described above) was subtracted from the secretion response data. In cases in which the cross–reactive signal was higher than the stimulated supernatant measurement (likely resulting from consumption or degradation of the stimulating ligand by SF or loss of the ligand by adsorption to tissue culture plates), values were thresholded to the corresponding basal secretion level to minimize impact on the analysis. The natural-log of MFI data was then computed (ln(MFI)) and assay background (as determined by the assay controls described above) was subtracted. Conditions in which the measured cytokine was also the stimulating factor (e.g. stimulation with TNFα and then measurement of TNFα as part of the Bio–Rad 27–plex cytokines kit) data for these individual cytokines were filtered to a value of not–a–number (NaN), thereby excluding the data from subsequent analysis. The processed data was then analyzed using the Matlab (The Mathworks, Inc) partialcorr function and the ‘pairwise’ option (to exclude NaN values). The partialcorr function calculates the correlations between matrix X (in this case the cytokine profiles (rows) for three RA synovial fluids (columns)) and matrix Y (cytokine profiles (rows)) of SF supernatants following exposure to various stimuli (columns)) by controlling for variance explained in both X and Y by a third matrix Z (supernatant cytokine profiles of unstimulated SF). This analysis was conducted individually for each SF sample to individually control for each basal secretion profile. False discovery rate (FDR) for multiple hypothesis testing was controlled using the Benjamini–Hochberg method.5
Interaction–based multiple linear regression
Ligand–drug interaction landscapes were solved for each experimental replicate and SF donor sample in Dataset 3. Experimental design matrix describing meta data (stimuli and inhibitor treatments) was constructed for each SF donor sample with a value of 1 indicating the presence of a given stimulus or inhibitor, and a 0 the absence of the ligand or drug. Rows corresponded to experimental conditions and columns corresponded to experimental variables (stimuli and inhibitors). We then added four sets of columns to parameterize assay controls and stimulus–inhibitor interaction terms: (1) a column consisting of all 1s was added to represent the assay background, which underlies all measurements (inclusion of this column requires collection of data for the assay background, which we describe in the Luminex assay methods above); (2) a column containing a value of 1 for any cell supernatants (and a value of 0 for assay controls such as spiked ligand controls and assay background controls) was added to represent basal secretion for the given SF donor sample, which underlies all stimulated supernatant measurements; (3) a set of three columns was added to describe assay conditions containing spiked-ligand controls: TNFα, IL–1α, and Poly(I:C), respectively; and (4) interaction terms were enumerated for all stimulus+inhibitor combination conditions by taking the dot product of the respective stimulus and inhibitor columns; see “Xmat” spreadsheets in Supplementary Data 1–7 for X matrices used for each SF donor sample in Dataset 3. For the assay background and spiked ligand controls (items (1) and (3)) require the assay controls as described above in the Luminex assay methods (We included these controls on the same Luminex assay plates as the SF supernatants and processed them identically to the supernatants): item (1) requires data for Luminex beads for each analyte to enable quantification of assay background, and item (3) requires spiked ligand controls (e.g. TNFα, IL–1α, or Poly(I:C)) diluted into basal media at the same concentration as the stimulating ligand) to allow assessment of cross–reactivity of the stimulating ligand against the luminex analytes. Item (1) is also critical for the iMLR analysis because it defines the intercept in the iMLR model and enables detection of cytokines above the assay background for unstimulated SF (i.e. basally secreted cytokines).
We solved for β coefficients individually for each SF sample using the MFI for each analyte as the dependent variable and the experimental design matrix constructed as described above as the predictor matrix. We calculated 10 different MLR solutions (forward or backward stepwise regression using p–value tests, Akaike’s information criteria (AIC), or Bayesian information criteria (BIC), MLR in the absence of stepwise regression, etc; see Supplementary Table 7 for details). For p–value–based solutions we used p<0.05 as the significance cut–off for individual β coefficients. For p–value based solutions using forward selection or backward removal, model parameters were sequentially added or removed (for forward addition or backward removal, respectively) until the addition or removal of a parameter did not achieve p<0.05 for the any β coefficient. For stepwise forward selection or backward removal using information criteria (e.g. AIC or BIC) we sequentially added or removed parameters (for forward addition or backward removal, respectively) that result in the greatest decrease in the relevant information criteria value. The feature selection process was stopped when addition or removal (as appropriate) of an additional parameter did not result in a decrease of AIC or BIC. We then calculated AIC or BIC weights6 for all models considered within the given stepwise process (e.g. all forward selection models for a given analyte evaluated using BIC) and used a cumulative weight cut–off of 99% to determine the final model for the given MLR solution framework. This resulted in 10 different iMLR models. β coefficients were averaged across these 10 models; model solutions in which the β coefficient was not determined to be significant were excluded from the averaging step. We calculated confidence weights representing the fraction of the multi–model solutions inferring a significant effect for the given β coefficient. This yielded two results: (1) the β coefficient for a given effect averaged across models where the effect is deemed statistically significant, and (2) a confidence weight reflecting the fraction of our multi–modeling solutions in which the effect was scored as statistically significant. Influence matrices were constructed by signing the confidence weights (+ or −) according to the sign of the β coefficient. Influence values for β coefficients with a value of zero (e.g. 10 out of the 10 multi–modeling frameworks assign a β value of 0) were defined to be 0.
β coefficients calculated above for each stimulus were filtered using β coefficients of the spiked ligand controls. β coefficients for the stimulatory effect that were ≤1.25–fold of the β coefficient for the spiked ligand control against a given luminex analyte were regarded as reflective of a cross–reactive signal and filtered to a value of 0 for the given analyte. β coefficients for stimulatory effects that were >1.25–fold of the spiked ligand control were filtered by subtracting the value of the β coefficient of the spiked ligand control (representing the cross–reactive signal of the stimulatory factor to the luminex analyte) from the β coefficient of the stimulus effect. Cases in which the measured species were the same as the stimulating species (e.g. stimulation of SF with TNFα and measurement of TNFα with the Bio–Rad 27–plex cytokines kit) were filtered to not–a–number (NaN), as such measurements cannot discriminate detection whether an analyte is secreted by SF or simply due its addition as an SF stimulus (measurement of TNFα in other stimulatory contexts are not affected by such confounding factors and were left unchanged).
Dataset 3 comprises two identical experiments performed on separate days. The MLR analysis described above was performed independently on each experimental replicate. β coefficients were taken as significant and repeatable if they were nonzero and had the same sign in both of the experimental replicates. β coefficients and influence matrices from the two experimental replicates that passed this threshold were merged by averaging. This retains both the matrix reflecting the relative effect size (the β coefficients) and the relative confidence in this value (via the fraction of the iMLR solutions that deem the given effect is statistically significant). This multi–modeling framework enabled increased identification of potential effects through statistically rigorous methods, while flagging the effects that are only detected by a fraction of our modeling solutions as being of lower confidence.
Correlation of stimuli and inhibitor effects across SF samples
For correlational analysis of inhibitor effects across contexts and donor samples, β coefficients that had been merged across experimental replicates were compiled across all donor samples and measured analytes. β coefficients were scaled ‘column–wise’ to a maximum absolute value of 1 for each column and pairwise Spearman (rank) correlation was calculated with the Matlab corr function and the “pairwise” option (to accommodate NaN entries).
Distribution of context–dependent inhibitor effects
To evaluate the distribution of inhibitor effects across contexts and secreted cytokines, inhibitor β coefficients for each stimulus–donor context were extracted into individual matrices. For example, for donor sample N2586 separate matrices comprising β coefficients for inhibitor effects (rows) across secreted cytokines (columns) were separately compiled for IL–1α, TNFα, or Poly(I:C) stimulation. Matrices for each stimulatory context included cytokines that were stimulated by IL–1α, TNFα, or Poly(I:C), respectively, as determined by the iMLR framework above, in order to exclude inhibitor effects that are zero by virtue of the cytokine not being induced by the stimulatory factor in the given SF sample. Each donor–stimulus matrix was individually scaled such that the stimulated level for each secreted cytokine equaled 1. As such, the scaled inhibitor β coefficient represents the fractional inhibition of the secreted cytokine for each stimulatory context in the given SF sample.
Supplementary Material
Acknowledgments
We thank S. Rudnicki and the HMS ICCB for automation assistance; B. Joughin, D. Clarke, M. Morris, J. Copeland, G. Nabozni, D. Klatte, and S. Pullen for helpful discussions; and N. Gray and T. Zhang at DFCI for providing JNK–IN–8. This work was supported by the NIH LINCS grant U54HL127365 to P.K.S., P50 GM107618 to P.K.S., a sponsored research agreement dated 8/1/2009 from Boehringer Ingelheim Inc. to P.K.S. and D.A.L., and NIH NRSA Fellowship 5F32AR062931 to D.S.J.
Footnotes
Author Contributions
D.S.J, performed the experiments and computational analyses, analyzed the results, and wrote and edited the paper; A.P.J. performed the experiments; D.A.L. and P.K.S. analyzed the results and wrote and edited the paper; J.M.B. and J.L.S. made key intellectual contributions.
Competing Financial Interests
J.M.B. and J.L.S. were employees at Boehringer Ingelheim Pharmaceuticals Inc. during the course of the studies; D.A.L and P.K.S. were consultants to Boehringer Ingelheim Pharmaceuticals Inc.
References
- 1.Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. The Lancet. 2010;376:1094–1108. doi: 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- 2.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 3.Taylor PC, Feldmann M. Anti-TNF biologic agents: still the therapy of choice for rheumatoid arthritis. Nat Rev Rheumatol. 2009;5:578–582. doi: 10.1038/nrrheum.2009.181. [DOI] [PubMed] [Google Scholar]
- 4.Emery P, et al. IL-6 receptor inhibition with tocilizumab improves treatment outcomes in patients with rheumatoid arthritis refractory to anti-tumour necrosis factor biologicals: results from a 24-week multicentre randomised placebo-controlled trial. Annals of the Rheumatic Diseases. 2008;67:1516–1523. doi: 10.1136/ard.2008.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanczyk J, Ospelt C, Gay S. Is there a future for small molecule drugs in the treatment of rheumatic diseases? Curr Opin Rheumatol. 2008;20:257–262. doi: 10.1097/BOR.0b013e3282fa13ee. [DOI] [PubMed] [Google Scholar]
- 6.Lindstrom TM, Robinson WH. A Multitude of Kinases-Which are the Best Targets in Treating Rheumatoid Arthritis? Rheumatic Disease Clinics of NA. 2010;36:367–383. doi: 10.1016/j.rdc.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur JSC, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Imm. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 8.Neumann E, Lefèvre S, Zimmermann B, Gay S, Müller-Ladner U. Rheumatoid arthritis progression mediated by activated synovial fibroblasts. Trends in Molecular Medicine. 2010;16:458–468. doi: 10.1016/j.molmed.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Bottini N, Firestein GS. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat Rev Rheumatol. 2013;9:24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noss EH, Brenner MB. The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunological Reviews. 2008;223:252–270. doi: 10.1111/j.1600-065X.2008.00648.x. [DOI] [PubMed] [Google Scholar]
- 11.Lefèvre S, et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nature Medicine. 2009:1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller-Ladner U, et al. Synovial fibroblasts of patients with rheumatoid arthritis attach to and invade normal human cartilage when engrafted into SCID mice. Am J Pathol. 1996;149:1607–1615. [PMC free article] [PubMed] [Google Scholar]
- 13.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunological Reviews. 2009;233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janes KA, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 15.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta - Mol Cell Rep. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 16.Baggiolini M, Clark-Lewis I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 1992;307:97–101. doi: 10.1016/0014-5793(92)80909-z. [DOI] [PubMed] [Google Scholar]
- 17.Moser B, Clark-Lewis I, Zwahlen R, Baggiolini M. Neutrophil-activating properties of the melanoma growth-stimulatory activity. J Exp Med. 1990;171:1797–1802. doi: 10.1084/jem.171.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher C, Clark-Lewis I, Baggiolini M, Moser B. High- and low-affinity binding of GRO alpha and neutrophil-activating peptide 2 to interleukin 8 receptors on human neutrophils. Proceedings of the National Academy of Sciences. 1992;89:10542–10546. doi: 10.1073/pnas.89.21.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carr MW, Roth SJ, Luther E, Rose SS, Springer TA. Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proceedings of the National Academy of Sciences. 1994;91:3652–3656. doi: 10.1073/pnas.91.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu LL, Warren MK, Rose WL, Gong W, Wang JM. Human recombinant monocyte chemotactic protein and other C-C chemokines bind and induce directional migration of dendritic cells in vitro. Journal of Leukocyte Biology. 1996;60:365–371. doi: 10.1002/jlb.60.3.365. [DOI] [PubMed] [Google Scholar]
- 21.Maghazachi AA, Al-Aoukaty A, Schall TJ. CC chemokines induce the generation of killer cells from CD56+ cells. Eur J Immunol. 1996;26:315–319. doi: 10.1002/eji.1830260207. [DOI] [PubMed] [Google Scholar]
- 22.Conti P, DiGioacchino M. MCP-1 and RANTES are mediators of acute and chronic inflammation. Allergy Asthma Proc. 2001;22:133–137. doi: 10.2500/108854101778148737. [DOI] [PubMed] [Google Scholar]
- 23.Taub DD, et al. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177:1809–1814. doi: 10.1084/jem.177.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jager W, Prakken BJ, Bijlsma JWJ, Kuis W, Rijkers GT. Improved multiplex immunoassay performance in human plasma and synovial fluid following removal of interfering heterophilic antibodies. J Immunol Methods. 2005;300:124–135. doi: 10.1016/j.jim.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Ellingsen T, Buus A, Stengaard-Pedersen K. Plasma monocyte chemoattractant protein 1 is a marker for joint inflammation in rheumatoid arthritis. J Rheumatol. 2001;28:41–46. [PubMed] [Google Scholar]
- 26.Boiardi L, et al. Relationship between serum RANTES levels and radiological progression in rheumatoid arthritis patients treated with methotrexate. Clin Exp Rheumatol. 1999;17:419–425. [PubMed] [Google Scholar]
- 27.Opdenakker G, Froyen G, Fiten P, Proost P, Van Damme J. Human monocyte chemotactic protein-3 (MCP-3): molecular cloning of the cDNA and comparison with other chemokines. Biochem Biophys Res Commun. 1993;191:535–542. doi: 10.1006/bbrc.1993.1251. [DOI] [PubMed] [Google Scholar]
- 28.Souza LM, et al. Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science. 1986;232:61–65. doi: 10.1126/science.2420009. [DOI] [PubMed] [Google Scholar]
- 29.Bacher M, et al. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proceedings of the National Academy of Sciences. 1996;93:7849–7854. doi: 10.1073/pnas.93.15.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malemud CJ. Growth hormone, VEGF and FGF: involvement in rheumatoid arthritis. Clin Chim Acta. 2007;375:10–19. doi: 10.1016/j.cca.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 32.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nature Biotechnology. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 33.Zhang T, et al. Discovery of Potent and Selective Covalent Inhibitors of JNK. Chemistry & Biology. 2012;19:140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ninomiya-Tsuji J, et al. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai H. Targeting of TAK1 in inflammatory disorders and cancer. Trends in Pharmacological Sciences. 2012;33:522–530. doi: 10.1016/j.tips.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Sakurai H. Targeting of TAK1 in inflammatory disorders and cancer. Trends in Pharmacological Sciences. 2012;33:522–530. doi: 10.1016/j.tips.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Cohen J. Multiple regression as a general data-analytic system. Psychological Bulletin. 1968;70:426–443. [Google Scholar]
- 38.Jaccard J, Wan CK, Turrisi R. The Detection and Interpretation of Interaction Effects Between Continuous Variables in Multiple Regression. Multivariate Behavioral Research. 2010;25:467–478. doi: 10.1207/s15327906mbr2504_4. [DOI] [PubMed] [Google Scholar]
- 39.Jaccard J, Turrisi R. Interaction Effects in Multiple Regression. SAGE Publications, Inc; 2003. [Google Scholar]
- 40.Takeda K, Akira S. TLR signaling pathways. Seminars in Immunology. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Qin J. Modulation of Toll-interleukin 1 receptor mediated signaling. J Mol Med. 2005;83:258–266. doi: 10.1007/s00109-004-0622-4. [DOI] [PubMed] [Google Scholar]
- 42.Tan L, et al. Discovery of Type II Inhibitors of TGFβ-Activated Kinase 1 (TAK1) and Mitogen-Activated Protein Kinase Kinase Kinase Kinase 2 (MAP4K2) J Med Chem. 2015;58:183–196. doi: 10.1021/jm500480k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filer A. The fibroblast as a therapeutic target in rheumatoid arthritis. Current Opinion in Pharmacology. 2013;13:413–419. doi: 10.1016/j.coph.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw SY, et al. Perturbational profiling of nanomaterial biologic activity. Proc Natl Acad Sci USA. 2008;105:7387–7392. doi: 10.1073/pnas.0802878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahle P, et al. Determination of cytokines in synovial fluids: correlation with diagnosis and histomorphological characteristics of synovial tissue. Annals of the Rheumatic Diseases. 1992;51:731–734. doi: 10.1136/ard.51.6.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods References
- 1.Rosengren S, Boyle DL, Firestein GS. Acquisition, culture, and phenotyping of synovial fibroblasts. Methods Mol Med. 2007;135:365–375. doi: 10.1007/978-1-59745-401-8_24. [DOI] [PubMed] [Google Scholar]
- 2.Millard BL, Niepel M, Menden MP, Muhlich JL, Sorger PK. Adaptive informatics for multifactorial and high-content biological data. Nat Meth. 2011;8:487–492. doi: 10.1038/nmeth.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niepel M, et al. Profiles of Basal and Stimulated Receptor Signaling Networks Predict Drug Response in Breast Cancer Lines. Science Signaling. 2013;6:ra84–ra84. doi: 10.1126/scisignal.2004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd DJ, et al. Erroneous augmentation of multiplex assay measurements in patients with rheumatoid arthritis due to heterophilic binding by serum rheumatoid factor. Arthritis & Rheumatism. 2011;63:894–903. doi: 10.1002/art.30213. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995 doi: 10.2307/2346101. [DOI] [Google Scholar]
- 6.Burnham KP, Anderson DR. Model Selection and Multimodel Inference. Springer Science & Business Media; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.