Abstract
Progressive macular hypomelanosis (PMH) is a skin disorder that is characterized by hypopigmented macules and usually seen in young adults. The skin microbiota, in particular the bacterium Propionibacterium acnes, is suggested to play a role.
Here, we compared the P. acnes population of 24 PMH lesions from eight patients with corresponding nonlesional skin of the patients and matching control samples from eight healthy individuals using an unbiased, culture-independent next-generation sequencing approach. We also compared the P. acnes population before and after treatment with a combination of lymecycline and benzoylperoxide.
We found an association of one subtype of P. acnes, type III, with PMH. This type was predominant in all PMH lesions (73.9% of reads in average) but only detected as a minor proportion in matching control samples of healthy individuals (14.2% of reads in average). Strikingly, successful PMH treatment is able to alter the composition of the P. acnes population by substantially diminishing the proportion of P. acnes type III.
Our study suggests that P. acnes type III may play a role in the formation of PMH. Furthermore, it sheds light on substantial differences in the P. acnes phylotype distribution between the upper and lower back and abdomen in healthy individuals.
Keywords: progressive macular hypomelanosis, Propionibacterium acnes, Cutibacterium acnes, next-generation sequencing, subtype III, skin microbiota, single locus sequencing type, phylotype
Introduction
Progressive macular hypomelanosis (PMH) is characterized by symmetric nonscaly hypopigmented skin areas that are predominantly visible in the sebaceous areas of the trunk. In the lower back and abdomen, discrete lesions are distinguished while they are more confluent on the upper trunk. There is no inflammation, pain, or itching associated with PMH, but the disease can have a major psychosocial effect on patients. PMH appears to be more frequent in young women, and although the disorder has a worldwide distribution, it is most often identified in dark-skinned populations [1–3].
Several treatment modalities are used against PMH including topical benzoylperoxide 5% (BPO) and clindamycin 1% alone or in combination with ultraviolet A (UVA) or narrow band ultraviolet B (UVB) irradiation [4–7], oral lymecycline in combination with topical BPO 5% for 3 months [8], and low-dose isotretinoin for 1 month [9]; however, the ideal treatment is not yet defined.
The etiology of PMH is not known; however, several studies indicate that the Gram-positive anaerobic bacterium Propionibacterium acnes may play a pivotal role [10, 11]. Biopsy specimens from PMH lesions contained P. acnes in pilosebaceous ducts in contrast to biopsies from healthy skin [11], real-time PCR showed a significant predominance of P. acnes in lesional skin as compared to nonlesional skin [10], and red fluorescence was detected in lesions when subjected to Woods light [11]. Finally, antibacterial treatment effective against P. acnes leads to repigmentation [4, 8]. It is not known how P. acnes could induce hypopigmentation. Using microscopy, a decrease in melanin production and a change in the distribution of melanosomes with a resultant decrease in epidermal melanin was shown in PMH lesions [12, 13].
Based on multilocus sequence typing (MLST) and single-locus sequence typing (SLST) schemes and complete genome sequencing, the population of P. acnes has been shown to consist of several phylogenetic subtypes commonly designated IA1, IA2, IB, IC, II, and III [14–21]. A previous study based on bacterial cultivation has indicated that certain subtypes of P. acnes may be associated with PMH: an abundance of a specific but unidentified type of P. acnes was observed in PMH lesions that is different from P. acnes types isolated from acne lesions [22]. Recently, we cultured P. acnes isolates belonging to the otherwise uncommon type III from lesions of PMH patients and sequenced their genomes [23], and a very recent study revealed an abundance of P. acnes type III in bacterial cultures from lesional skin in 14 of 34 PMH patients [24].
In the present study, the type distribution of the entire population of P. acnes in affected and unaffected skin areas of PMH patients, including samples after treatment, and matching control samples was determined using a culture-independent next-generation sequencing (NGS)-based SLST approach. Results show a strong association of P. acnes type III with disease.
Materials and methods
Patient and control cohort, treatment regimen
Eight patients with PMH were recruited by voluntary consent in a private dermatology practice in Aalborg, Denmark. The patients were all clinically examined by a specialist in dermatology (H.B. Lomholt), and PMH was diagnosed based on the finding of clinically characteristic lesions, patient history, and a lack of Malassezia in microscopic inspections. All patients were female between 18 and 31 years of age (mean, 23.5 years) (see Supplementary Table S1 online). Five of the patients were treated in a 3-month course with oral lymecycline (300 mg, daily) combined with a daily wash using a 5% BPO washing gel, and one patient used only the 5% BPO wash (see Supplementary Table S2 online). As controls, eight healthy volunteers were recruited among students at the University of Aarhus, Denmark. They were all female between 24 and 31 years of age (mean, 26.5 years). Information regarding the study participants and treatment is summarized in Supplementary Tables S1 and S2 online.
Table S1.
Data on eight patients (P) and eight controls (C) included in the study
| Sex | Age | Sex | Age | ||
|---|---|---|---|---|---|
| P1 | Female | 19 | C1 | Female | 28 |
| P2 | Female | 24 | C2 | Female | 31 |
| P3 | Female | 18 | C3 | Female | 27 |
| P4 | Female | 22 | C4 | Female | 26 |
| P5 | Female | 24 | C5 | Female | 25 |
| P6 | Female | 31 | C6 | Female | 25 |
| P7 | Female | 29 | C7 | Female | 26 |
| P8 | Female | 21 | C8 | Female | 24 |
Table S2.
Treatment and treatment responses in six PMH patients treated with benzoylperoxide (BPO) daily washes and/or peroral lymecycline (LC) 300 mg once daily for three months
| Patient no. | Treatment | Outcome |
|---|---|---|
| P1 | BPO, LC | Good response with a few residual lesions |
| P2 | BPO, LC | Good response |
| P3 | Irregular BPO, no LC | Improved with residual lesions |
| P4 | BPO, LC | Good response |
| P7 | Irregular BPO, irregular LC | Good response with a few residual lesions |
| P8 | BPO, LC | Good response |
Sampling sites and procedure
Samples were taken in all patients from three lesional areas including the lower, middle, or upper back (depending on the position of the PMH lesions) and the abdomen, and additional samples were taken from patients in unaffected adjacent skin areas. One patient had no lesions on the abdomen and was sampled from all three positions on the back. Six of the patients were additionally sampled after treatment from the same areas as the pretreatment sample. Samples from the lower and upper back, and abdomen were taken from the eight healthy controls. In addition, samples from the forehead and the buccal cavity were taken from patients (Fig. S1). Samples for NGS-based SLST were obtained by swabbing the skin firmly for 20 s with a sterile cotton swab moistened in sampling buffer (0.1% detergent [Triton X-100] in 0.075 M phosphate buffer, pH 7.9) [25]. DNA was extracted from all collected samples, and SLST fragment amplification was performed as described previously [20]. The amplicons were then subjected to next-generation sequencing (NGS) using the pyrosequencing technology. Subsequent bioinformatics analysis included quality control and sequence read assignment to the STs of the SLST scheme (http://medbac.dk/slst/pacnes), resulting in a high-resolution phylotype analysis of the P. acnes population in each sample.
Samples for bacterial cultivation were taken with a sterile charcoal swab moistened in sampling buffer firmly scrubbed on the skin for 20 s. From each patient, two swabs were obtained from lesional skin on the back and abdomen, respectively, and, in addition, two swabs from adjacent nonlesional skin. For a semiquantitative estimation of the number of bacteria, the primary charcoal cotton swabs were streaked on tryptone-yeast-glucose (TYG) agar plates and incubated for 120 h in an anaerobic chamber (Forma Scientific Anaerobic System model 1024).
Next-generation sequencing-based single-locus sequence typing (NGS-based SLST)
Cotton swab samples from both patients and healthy controls were transferred to 1.5-ml Eppendorf tubes, and bacterial DNA was isolated and purified using PowerLyzer® PowerSoil® DNA Isolation Kit (MO BIO, Carlsbad, CA, United States) according to the manufacturer’s instruction. The isolated DNA was then subjected to amplification by PCR as described previously [20]. All primer sequences are given in Supplementary Table S3 online. All PCR reactions were made by mixture of 5 μl DNA sample, 2.5 μl AccuPrime PCR Buffer II (Invitrogen), 1.5 μl of forward primer, 1.5 μl of reverse primer (10 μM), 0.15 μl AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen), and 14.35 μl PCR-grade water into a total volume of 25 μl. All samples were amplified by an initial denaturation for 2 min at 94 °C, followed by 35 cycles of 20 s of denaturation at 94 °C, 30 s at 55 °C for annealing, and 60 s of extension at 68 °C, ending with a 5-minute step of extension at 68 °C. Three PCRs per sample were performed and verified on an agarose gel, pooled together, and purified using a NucleoSpin Extract Kit (Macherey-Nagel). The concentration of purified DNA samples was measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific). The amplicons were pooled in batches of 20 and sequenced unidirectionally from the forward primer using Roche GS FLX+ pyrosequencing technology either at Institute of Microbiology and Genetics, Georg-August University Göttingen, Göttingen, Germany or at Eurofins Genomics, Ebersberg, Germany. The data were then processed using the PyroNoise implementation in Mothur v. 1.36.1 [26]. All sequences have been deposited at NCBI with the project number PRJNA347641. The sequence reads were aligned to all the known STs in the SLST database (http://medbac.dk/slst/pacnes) using BLASTn [27] and assigned an individual ST based on a best-hit model with a cut-off value of 99.5%; anything below this threshold or reads with two or more identical best hits were discarded as unassigned reads.
Table S3.
Primers used in this study
| Primers for NGS-based SLST | |
| SLST_PA_REV | 5’-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG-CCGGCTGGCAAATGAGGCAT-3’ |
| SLST_PA_MID1 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAG-ACGAGTGCGT-CAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID2 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAG-ACGCTCGACA-CAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID3 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGAGACGCACTCCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID4 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGAGCACTGTAGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID5 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGATCAGACACGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID6 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGATATCGCGAGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID7 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGCGTGTCTCTACAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID8 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGCTCGCGTGTCCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID10 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCTCTATGCGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID11 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGTGATACGTCTCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID13 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGCATAGTAGTGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID14 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGCGAGAGATACCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID15 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGATACGACGTACAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID16 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCACGTACTACAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID17 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGCGTCTAGTACCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID18 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCTACGTAGCCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID19 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGTGTACTACTCCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID20 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGACGACTACAGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID21 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGCGTAGACTAGCAGCGGCGCTGCTAAGAACTT-3 |
| SLST_PA_MID22 | 5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGTACGAGTATGCAGCGGCGCTGCTAAGAACTT-3 |
| Primers for Sanger-sequencing-based SLST | |
| Sanger_SLST_for | 5’-CGCCATCAAGGCACCAACAA-3’ |
| Sanger_SLST_rev | 5’-ATATCGGCCCGTATTTGGGC-3’ |
Sanger sequencing-based SLST of bacterial isolates
SLST typing of P. acnes isolates has been described previously [20]. In brief, the sampled charcoal cotton swabs were streaked on tryptone-yeast-glucose (TYG) agar plates and incubated for 72 h in an anaerobic chamber (Forma Scientific Anaerobic System model 1024). This primary culture was then examined; up to 10 random single colonies resembling P. acnes were selected and individually subcultivated on new TYG agar plates for 72 h under anaerobic conditions. After growth, the P. acnes isolates were harvested and subjected to DNA extraction by a boiling procedure at 100 °C for 10 min in 0.5 ml Eppendorf PCR tubes containing 300 μl PCR-grade water. A PCR was carried out with the SLST primers (see Supplementary Table S3 online) as follows: 2 μl of the crude DNA extract was mixed with 1 μl of each forward and reverse primer (10 μM), 10 μl 5’-PRIME Hotmastermix (5 PRIME, Hamburg, Germany), and 11 μl of PCR-grade water. PCR conditions were as follows: initial denaturation of 40 s at 96 °C followed by 35 cycles of 35 s of denaturation at 96 °C, 40 s of annealing at 55 °C, and 40 s of extension at 72 °C, followed by a finale 7-minute extension step at 72 °C. The resulting PCR products were run on a 1% agarose gel to ensure quality and were then sequenced at GATC Biotech AG (Konstanz, Germany) with the forward and reverse SLST primers. The sequences were assembled and trimmed to the known SLST fragment size (484 bp) using MEGA v.6.06 [28]. Finally, the resulting sequences were assigned to STs using the SLST database (http://medbac.dk/slst/pacnes).
Statistical analysis
The proportions of P. acnes type III were compared between mean values of the three lesional sites in patients and corresponding sites in controls using the unpaired Wilcoxon rank sum test/Mann–Whitney U test. Lesional sites in patients before and after treatment were compared using the paired Wilcoxon signed rank test.
Ethics statement
The study protocol was approved by the Ethics Committee of Region North, Denmark (document N-20120050), and the study was conducted according to the principles of the declaration of Helsinki. Written informed consent was obtained from all study participants.
Results
Type III P. acnes predominates in PMH samples
SLST amplicons from 96 samples were selected for pyrosequencing. In average, 7911 sequence reads per sample were obtained and 97% of the reads could be assigned to a P. acnes sequence type (ST) (see Supplementary Table S4 online). In the whole data set, the SLST scheme distinguished 90 different STs of P. acnes.
Table S4.
Next-generation sequencing data: numbers of sequence reads and ST assignment in 96 samples
| Patient | P1 | P1 | P1 | P1 | P1 | P1 | P1 | P1 | P1 | P1 | P1 | P2 | P2 | P2 | P2 | P2 | P2 | P2 | P2 | P2 | P3 | P3 | P3 | P3 |
| Sampling spot | LB | LB | UB | UB | ABD | F | M | 2-LB | 2-UB | 2-UB | 2-ABD | LB | LB | MB | ABD | F | 2-LB | 2-MB | 2-ABD | 2-A | LB | LB | UB | UB |
| Number of reads | 6661 | 10951 | 3840 | 5336 | 10036 | 14883 | 781 | 3938 | 5059 | 690 | 6379 | 4974 | 4383 | 9758 | 3189 | 7728 | 3760 | 3046 | 2485 | 185 | 10144 | 7440 | 2064 | 5196 |
| Number of assigned reads | 6341 | 10762 | 3175 | 5250 | 9992 | 14715 | 760 | 3872 | 5002 | 685 | 6245 | 4928 | 4330 | 9697 | 3127 | 7641 | 3665 | 2972 | 2450 | 184 | 10023 | 7315 | 1999 | 5121 |
| % unassigned reads | 4.80 | 1.73 | 17.32 | 1.61 | 0.44 | 1.13 | 2.69 | 1.68 | 1.13 | 0.72 | 2.10 | 0.92 | 1.21 | 0.63 | 1.94 | 1.13 | 2.53 | 2.43 | 1.41 | 0.54 | 1.19 | 1.68 | 3.15 | 1.44 |
| Unassigned | 320 | 189 | 665 | 86 | 44 | 168 | 21 | 66 | 57 | 5 | 134 | 46 | 53 | 61 | 62 | 87 | 95 | 74 | 35 | 1 | 121 | 125 | 65 | 75 |
| A1 | 12 | 19 | 4 | 261 | 19 | 809 | 461 | 123 | 384 | 141 | 2398 | 16 | 749 | 86 | 1846 | 96 | 1535 | 1435 | 1200 | 4 | 132 | 1385 | 127 | 737 |
| A2 | 0 | 0 | 0 | 0 | 0 | 22 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| A4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| A5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| A6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 0 | 1 | 1 | 3 | 2 | 19 | 0 | 2 | 3 | 0 | 5 |
| A7 | 0 | 0 | 0 | 4 | 0 | 6 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| A8 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| A9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| A10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A13 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| A16 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A17 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| A18 | 0 | 2 | 0 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 9 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 |
| A19 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| A20 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| A21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A22 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
| B1 | 0 | 2 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 15 | 61 | 20 | 102 |
| C1 | 0 | 0 | 0 | 0 | 1 | 476 | 1 | 2 | 1 | 0 | 3 | 0 | 6 | 0 | 11 | 0 | 7 | 17 | 26 | 0 | 0 | 470 | 1 | 137 |
| C2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| C3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| C4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| D1 | 0 | 0 | 0 | 0 | 0 | 1 | 12 | 0 | 4 | 0 | 12 | 0 | 64 | 8 | 13 | 4 | 83 | 113 | 29 | 0 | 63 | 289 | 91 | 504 |
| D2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| E1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| E4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| E7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 11 | 0 | 1 | 0 | 21 | 80 | 4 | 122 |
| E8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 70 | 503 | 159 | 883 |
| F1 | 0 | 3 | 0 | 4 | 1 | 2252 | 52 | 0 | 1 | 0 | 151 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F3 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F4 | 0 | 0 | 0 | 0 | 0 | 67 | 49 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 13 | 44 | 1 | 0 | 25 | 463 | 216 | 278 |
| F5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| F6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| F7 | 0 | 8 | 0 | 12 | 21 | 6897 | 33 | 8 | 33 | 0 | 365 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F8 | 0 | 0 | 0 | 0 | 0 | 1 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| F9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| G1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 7 | 50 | 0 | 169 | 140 | 84 | 0 | 0 | 0 | 0 | 1 |
| H1 | 0 | 0 | 0 | 22 | 0 | 4158 | 1 | 18 | 21 | 0 | 1 | 0 | 161 | 3 | 155 | 90 | 718 | 633 | 518 | 0 | 0 | 1 | 1 | 14 |
| H2 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| H3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K1 | 37 | 7 | 38 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 4 | 410 | 60 | 425 | 6623 | 554 | 400 | 386 | 114 | 1 | 187 | 1 | 39 |
| K2 | 30 | 5 | 72 | 20 | 10 | 0 | 2 | 0 | 14 | 1 | 7 | 0 | 28 | 1 | 2 | 148 | 74 | 21 | 17 | 7 | 69 | 0 | 0 | 0 |
| K3 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K4 | 0 | 0 | 0 | 0 | 0 | 0 | 63 | 0 | 0 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17 |
| K5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K7 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 10 | 0 | 0 | 47 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| K8 | 800 | 300 | 620 | 2758 | 54 | 3 | 17 | 121 | 4503 | 472 | 61 | 0 | 117 | 19 | 490 | 623 | 442 | 109 | 167 | 57 | 0 | 9 | 0 | 0 |
| K9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| K10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K13 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L1 | 5288 | 10375 | 2191 | 2002 | 9822 | 7 | 33 | 11 | 4 | 31 | 3177 | 0 | 2715 | 9503 | 57 | 0 | 5 | 23 | 0 | 0 | 0 | 0 | 25 | 3 |
| 12 | 149 | 38 | 145 | 136 | 56 | 0 | 0 | 3587 | 9 | 0 | 38 | 0 | 2 | 1 | 14 | 0 | 20 | 7 | 0 | 0 | 0 | 0 | 1 | 0 |
| L3 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L5 | 25 | 2 | 71 | 6 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 4907 | 6 | 5 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | 1 | 21 |
| L6 | 0 | 1 | 0 | 14 | 1 | 1 | 7 | 0 | 0 | 0 | 5 | 1 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 9623 | 3839 | 1349 | 2245 |
| Patient | P3 | P3 | P3 | P3 | P3 | P4 | P4 | P4 | P4 | P4 | P4 | P4 | P4 | P4-2 | P5 | P5 | P5 | P5 | P5 | P6 | P6 | P6 | P6 | P6 |
| Sampling spot | ABD | F | 2-LB | 2-UB | 2-ABD | LB | LB | MB | UB | F | 2-LB | 2-UB | 2-UB | 2-UB-2 | LB | LB | ABD | CH | F | LB | LB | UB | ABD | F |
| Number of reads | 11989 | 7710 | 11831 | 16240 | 14294 | 11453 | 5424 | 16143 | 9719 | 5933 | 4381 | 3373 | 4487 | 8107 | 668 | 1858 | 12708 | 6808 | 14254 | 6239 | 4225 | 7404 | 13528 | 12788 |
| Number of assigned reads | 11816 | 7493 | 11443 | 14467 | 13916 | 11273 | 5296 | 15702 | 9234 | 5858 | 4288 | 3316 | 4418 | 8035 | 643 | 1816 | 12527 | 6613 | 13176 | 6111 | 4158 | 7122 | 13245 | 12196 |
| % unassigned reads | 1.44 | 2.81 | 3.28 | 10.92 | 2.64 | 1.57 | 2.36 | 2.73 | 4.99 | 1.26 | 2.12 | 1.69 | 1.54 | 0.89 | 3.74 | 2.26 | 1.42 | 2.86 | 7.56 | 2.05 | 1.59 | 3.81 | 2.09 | 4.63 |
| Unassigned | 173 | 217 | 388 | 1773 | 378 | 180 | 128 | 441 | 485 | 75 | 93 | 57 | 69 | 72 | 25 | 42 | 181 | 195 | 1078 | 128 | 67 | 282 | 283 | 592 |
| A1 | 266 | 908 | 5007 | 626 | 2666 | 37 | 1287 | 28 | 7 | 559 | 2601 | 29 | 1802 | 104 | 6 | 184 | 29 | 1262 | 324 | 397 | 967 | 3463 | 908 | 1883 |
| A2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| A3 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| A4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 8 | 1 |
| A6 | 1 | 2 | 16 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 11 | 0 | 0 | 1 | 0 | 3 | 0 | 1 | 3 | 19 | 3 | 0 |
| A7 | 0 | 1 | 2 | 1 | 1 | 0 | 9 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 4 | 4 | 0 |
| A8 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 2 |
| A9 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A11 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| A12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| A14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A15 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| A16 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| A18 | 1 | 9 | 3 | 0 | 3 | 0 | 7 | 0 | 0 | 0 | 1 | 1 | 2 | 3 | 0 | 0 | 0 | 12 | 0 | 1 | 1 | 2 | 2 | 3 |
| A19 | 2 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| A20 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 12 | 1 | 0 |
| A21 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| A22 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 0 |
| A23 | 0 | 0 | 2 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| B1 | 7 | 41 | 187 | 1019 | 65 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 4 | 0 | 0 | 1 | 20 | 89 | 2 |
| C1 | 19 | 35 | 174 | 47 | 29 | 0 | 13 | 1 | 1 | 10 | 22 | 5 | 10 | 1 | 1 | 0 | 2 | 42 | 154 | 0 | 5 | 25 | 330 | 244 |
| C2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 313 | 0 | 216 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C3 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 |
| C4 | 0 | 0 | 2 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| D1 | 40 | 32 | 1148 | 1711 | 177 | 28 | 269 | 99 | 1432 | 121 | 88 | 464 | 416 | 818 | 105 | 618 | 9 | 1583 | 424 | 475 | 285 | 1603 | 918 | 4209 |
| D2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| E1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 21 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E3 | 0 | 0 | 2 | 27 | 2 | 3 | 1 | 0 | 3 | 1 | 13 | 103 | 42 | 177 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| E4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E7 | 38 | 38 | 115 | 849 | 65 | 93 | 544 | 221 | 1617 | 128 | 308 | 2705 | 1137 | 6920 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| E8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E9 | 16 | 1359 | 1785 | 5413 | 772 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 |
| F2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F4 | 134 | 790 | 337 | 882 | 1062 | 28 | 1532 | 0 | 2 | 1122 | 163 | 3 | 426 | 2 | 121 | 408 | 20 | 326 | 1708 | 2 | 261 | 13 | 25 | 603 |
| F5 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F6 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F8 | 10 | 40 | 2 | 0 | 0 | 3 | 10 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 47 | 0 | 5 | 0 | 0 | 1 |
| F9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F10 | 0 | 1 | 1 | 0 | 86 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| G1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| H1 | 10 | 14 | 12 | 2 | 213 | 0 | 0 | 0 | 0 | 3861 | 415 | 2 | 105 | 0 | 272 | 123 | 576 | 659 | 9462 | 0 | 59 | 11 | 0 | 5195 |
| H2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 3 |
| H3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| H4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| H5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 21 |
| K1 | 9 | 3 | 20 | 0 | 75 | 0 | 4 | 0 | 0 | 4 | 16 | 0 | 11 | 0 | 15 | 126 | 51 | 72 | 389 | 0 | 176 | 0 | 3 | 0 |
| K2 | 0 | 788 | 20 | 7 | 87 | 0 | 0 | 0 | 0 | 12 | 58 | 0 | 23 | 0 | 0 | 1 | 0 | 2 | 631 | 0 | 0 | 0 | 1 | 0 |
| K3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| K5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K8 | 4 | 97 | 6 | 0 | 41 | 8 | 0 | 0 | 0 | 0 | 100 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| K13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L1 | 17 | 2 | 31 | 393 | 68 | 4 | 19 | 16 | 36 | 0 | 57 | 0 | 38 | 0 | 116 | 348 | 11721 | 2628 | 1 | 5217 | 2372 | 1661 | 10929 | 23 |
| 12 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 81 | 0 | 71 | 0 | 0 | 0 | 8 | 0 | 0 | 3 | 2 | 0 | 1 | 0 |
| L3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L5 | 2 | 0 | 7 | 3 | 2 | 1 | 2 | 6 | 4 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 107 | 9 | 0 | 7 | 13 | 260 | 6 | 0 |
| L6 | 11240 | 3330 | 2554 | 3459 | 8482 | 11051 | 1582 | 15328 | 6109 | 23 | 5 | 2 | 55 | 0 | 1 | 1 | 3 | 1 | 1 | 2 | 0 | 4 | 12 | 0 |
| Patient | P6 | P7 | P7 | P7 | P7 | P7 | P7 | P7 | P7-2 | P7-2 | P8 | P8 | P8 | P8 | P8 | P8-2 | P8-2 | P8-2 | C1 | C1 | C1 | C1 | C2 | C2 |
| Sampling spot | M | LB | LB | MB | ABD | M | 2-LB | 2-LB | 2-MB | 2-ABD | LB | LB | ABD | F | M | 2-LB | 2-UB | 2-ABD | LB | LB | UB | ABD | LB | LB |
| Number of reads | 2655 | 4321 | 290 | 10068 | 8766 | 3223 | 1864 | 5707 | 9899 | 13630 | 7437 | 5757 | 10330 | 8430 | 1366 | 3912 | 10423 | 2636 | 11079 | 14191 | 9044 | 13576 | 9422 | 9230 |
| Number of assigned reads | 2612 | 3943 | 285 | 9934 | 8671 | 3179 | 1811 | 5627 | 9314 | 13499 | 7364 | 5667 | 10194 | 8187 | 1348 | 3860 | 10240 | 2609 | 10891 | 13926 | 8920 | 13378 | 9225 | 9033 |
| % unassigned reads | 1.62 | 8.75 | 1.72 | 1.33 | 1.08 | 1.37 | 2.84 | 1.40 | 5.91 | 0.96 | 0.98 | 1.56 | 1.32 | 2.88 | 1.32 | 1.33 | 1.76 | 1.02 | 1.70 | 1.87 | 1.37 | 1.46 | 2.09 | 2.13 |
| Unassigned | 43 | 378 | 5 | 134 | 95 | 44 | 53 | 80 | 585 | 131 | 73 | 90 | 136 | 243 | 18 | 52 | 183 | 27 | 188 | 265 | 124 | 198 | 197 | 197 |
| A1 | 1757 | 2 | 0 | 53 | 45 | 687 | 78 | 1592 | 38 | 469 | 78 | 2702 | 680 | 1291 | 1130 | 2770 | 8093 | 2023 | 5468 | 12157 | 247 | 10963 | 2877 | 4718 |
| A2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 4 | 0 | 1 | 4 | 6 | 0 | 4 | 1 | 0 | 0 | 2 |
| A3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 33 | 57 | 7 | 0 | 2 |
| A4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 2 | 3 | 1 | 0 | 0 |
| A5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 4 | 0 | 0 | 32 | 12 |
| A6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 7 | 0 | 6 | 7 | 13 | 0 | 4 | 18 | 1 | 27 | 25 | 0 | 9 | 12 | 5 |
| A7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 13 | 0 | 2 | 17 | 0 | 18 | 9 | 10 |
| A8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 4 | 1 | 1 | 0 | 2 | 3 | 1 |
| A9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | 1 | 8 | 0 |
| A10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A11 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 3 | 0 | 1 | 2 | 0 |
| A12 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| A13 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 0 |
| A14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A15 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 4 | 0 | 0 |
| A16 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 0 | 4 | 0 | 5 |
| A17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 4 | 4 | 0 | 0 | 1 | 0 |
| A18 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 6 | 0 | 5 | 2 | 13 | 1 | 4 | 15 | 1 | 18 | 30 | 0 | 31 | 9 | 4 |
| A19 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 5 | 0 | 2 | 0 | 2 |
| A20 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 5 | 0 | 1 | 24 | 0 | 4 | 2 | 3 |
| A21 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 20 | 1 | 1 | 27 | 0 | 1 | 0 | 1 |
| A22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 3 | 2 | 10 |
| A23 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 18 | 1 | 4 | 17 | 0 | 1 | 3 | 20 |
| B1 | 0 | 0 | 0 | 1 | 0 | 6 | 4 | 0 | 0 | 14 | 2 | 17 | 4 | 6 | 34 | 2 | 0 | 0 | 763 | 44 | 82 | 12 | 1 | 3 |
| C1 | 0 | 12 | 0 | 0 | 0 | 0 | 1 | 9 | 4 | 31 | 6 | 224 | 42 | 75 | 0 | 175 | 371 | 89 | 62 | 63 | 8 | 415 | 14 | 23 |
| C2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| C3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 1 | 1 | 0 | 13 | 0 | 1 |
| C4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D1 | 77 | 355 | 1 | 1 | 31 | 171 | 131 | 963 | 492 | 212 | 3 | 418 | 118 | 1324 | 0 | 486 | 665 | 251 | 1644 | 1057 | 8501 | 1332 | 5384 | 3034 |
| D2 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 2 | 2 |
| D3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 4 | 0 | 29 | 1 |
| E1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| E3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| E4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E7 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 8 | 45 | 0 | 1 | 7 | 0 | 10 | 10 | 38 |
| E8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F1 | 0 | 0 | 0 | 1 | 0 | 14 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 120 | 45 | 0 | 82 | 304 | 109 |
| F2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F4 | 111 | 11 | 0 | 3 | 32 | 384 | 34 | 522 | 51 | 197 | 0 | 3 | 1 | 0 | 13 | 3 | 6 | 0 | 7 | 5 | 0 | 1 | 261 | 380 |
| F5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| F7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F8 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 1 |
| G1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 190 | 486 |
| H1 | 192 | 300 | 3 | 39 | 158 | 1765 | 391 | 2263 | 6553 | 931 | 0 | 65 | 32 | 772 | 54 | 61 | 102 | 17 | 0 | 4 | 0 | 0 | 23 | 8 |
| H2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 66 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H5 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 9 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K1 | 60 | 2342 | 0 | 33 | 2 | 49 | 10 | 212 | 2156 | 9 | 4 | 14 | 34 | 35 | 38 | 178 | 396 | 163 | 148 | 270 | 12 | 352 | 8 | 7 |
| K2 | 0 | 124 | 0 | 0 | 7 | 2 | 2 | 2 | 0 | 21 | 452 | 242 | 30 | 3969 | 21 | 89 | 319 | 11 | 1 | 1 | 0 | 18 | 8 | 0 |
| K3 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55 | 0 | 32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 344 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K7 | 0 | 1 | 0 | 6 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 13 |
| K8 | 348 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 11 | 0 | 7 | 47 | 1 | 6 | 1 | 10 | 3 | 0 | 9 | 3 | 35 |
| K9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K11 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 8 | 0 | 1 | 0 | 2 | 0 | 0 |
| K13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L1 | 24 | 765 | 273 | 9760 | 8360 | 53 | 1146 | 26 | 7 | 11578 | 0 | 0 | 1 | 208 | 0 | 0 | 0 | 11 | 2513 | 3 | 0 | 5 | 0 | 1 |
| 12 | 1 | 0 | 1 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 6795 | 1874 | 9241 | 3 | 7 | 66 | 109 | 19 | 1 | 57 | 0 | 69 | 22 | 93 |
| L3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L5 | 0 | 25 | 4 | 1 | 1 | 0 | 5 | 0 | 0 | 5 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 |
| L6 | 0 | 0 | 1 | 28 | 28 | 1 | 3 | 0 | 1 | 10 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 52 | 4 | 1 | 0 | 0 | 0 |
| Patient | C2 | C2 | C3 | C3 | C3 | C3 | C4 | C4 | C4 | C4 | C5 | C5 | C5 | C6 | C6 | C6 | C6 | C7 | C7 | C7 | C8 | C8 | C8 | C8 |
| Sampling spot | UB | ABD | LB | LB | UB | ABD | LB | LB | UB | ABD | LB | LB | ABD | LB | LB | UB | ABD | LB | LB | ABD | LB | LB | UB | ABD |
| Number of reads | 10716 | 11233 | 13394 | 8265 | 19242 | 16172 | 12935 | 5910 | 13972 | 4298 | 7483 | 4560 | 14148 | 13528 | 3374 | 12825 | 6836 | 9334 | 4230 | 5383 | 5438 | 7202 | 16129 | 11094 |
| Number of assigned reads | 10559 | 10936 | 12988 | 7949 | 17996 | 15895 | 12806 | 5843 | 13558 | 4250 | 7313 | 4494 | 13645 | 13391 | 3303 | 12406 | 6710 | 9060 | 4083 | 5236 | 5267 | 7065 | 15614 | 10816 |
| % unassigned reads | 1.47 | 2.64 | 3.03 | 3.82 | 6.48 | 1.71 | 1.00 | 1.13 | 2.96 | 1.12 | 2.27 | 1.45 | 3.56 | 1.01 | 2.10 | 3.27 | 1.84 | 2.94 | 3.48 | 2.73 | 3.14 | 1.90 | 3.19 | 2.51 |
| Unassigned | 157 | 297 | 406 | 316 | 1246 | 277 | 129 | 67 | 414 | 48 | 170 | 66 | 503 | 137 | 71 | 419 | 126 | 274 | 147 | 147 | 171 | 137 | 515 | 278 |
| A1 | 9314 | 5265 | 2052 | 2611 | 8947 | 14187 | 282 | 1235 | 5191 | 1353 | 2035 | 892 | 1101 | 446 | 805 | 690 | 2054 | 1094 | 704 | 1033 | 1633 | 1755 | 13979 | 2008 |
| A2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 5 |
| A3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| A4 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A5 | 4 | 0 | 3 | 0 | 1 | 2 | 0 | 15 | 7 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| A6 | 12 | 13 | 6 | 6 | 17 | 10 | 1 | 5 | 8 | 3 | 16 | 3 | 0 | 0 | 2 | 1 | 19 | 6 | 2 | 0 | 3 | 3 | 8 | 0 |
| A7 | 4 | 25 | 2 | 1 | 6 | 30 | 0 | 2 | 5 | 0 | 0 | 0 | 1 | 0 | 1 | 6 | 3 | 2 | 2 | 1 | 3 | 3 | 12 | 1 |
| A8 | 9 | 5 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 2 | 3 | 1 | 0 | 0 | 0 | 6 | 0 | 2 | 0 | 0 | 4 | 0 | 12 | 0 |
| A9 | 1 | 1 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| A10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A11 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| A12 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| A13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| A14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A15 | 1 | 10 | 0 | 1 | 1 | 4 | 0 | 6 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 |
| A16 | 5 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 |
| A17 | 1 | 1 | 1 | 2 | 2 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 |
| A18 | 42 | 8 | 7 | 15 | 4 | 20 | 1 | 1 | 9 | 2 | 1 | 5 | 0 | 0 | 0 | 0 | 4 | 1 | 4 | 3 | 5 | 1 | 10 | 5 |
| A19 | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 10 | 1 |
| A20 | 3 | 1 | 1 | 0 | 0 | 5 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 2 | 1 | 0 | 3 | 1 | 0 | 5 | 0 |
| A21 | 0 | 0 | 0 | 1 | 0 | 22 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 2 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| A22 | 0 | 4 | 0 | 6 | 15 | 0 | 0 | 1 | 2 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 1 | 0 |
| A23 | 14 | 6 | 0 | 4 | 5 | 6 | 0 | 0 | 17 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 12 | 1 |
| B1 | 7 | 2 | 7 | 0 | 0 | 1 | 2 | 9 | 103 | 9 | 1 | 0 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 8 |
| C1 | 55 | 35 | 12 | 73 | 1425 | 377 | 3 | 16 | 9 | 5 | 20 | 42 | 29 | 0 | 7 | 6 | 51 | 124 | 16 | 1 | 77 | 334 | 482 | 215 |
| C2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C3 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 |
| C4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D1 | 813 | 4237 | 668 | 2640 | 6895 | 505 | 10 | 2 | 0 | 60 | 1761 | 1571 | 3364 | 95 | 447 | 6424 | 914 | 1101 | 553 | 69 | 586 | 785 | 55 | 1567 |
| D2 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| D3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 2 |
| E1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E3 | 0 | 0 | 1 | 4 | 67 | 1 | 3 | 36 | 36 | 38 | 4 | 1 | 6 | 2 | 16 | 29 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| E4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E7 | 16 | 21 | 1 | 45 | 440 | 108 | 197 | 1324 | 1886 | 1175 | 119 | 53 | 363 | 44 | 639 | 2697 | 356 | 26 | 30 | 23 | 0 | 0 | 1 | 3 |
| E8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| F1 | 33 | 106 | 48 | 135 | 1 | 3 | 3 | 20 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| F2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F4 | 30 | 609 | 1 | 3 | 160 | 3 | 0 | 82 | 2086 | 0 | 0 | 0 | 0 | 0 | 15 | 4 | 31 | 0 | 0 | 0 | 0 | 1 | 0 | 15 |
| F5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F10 | 0 | 42 | 0 | 0 | 0 | 0 | 0 | 0 | 244 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| G1 | 145 | 388 | 6 | 2 | 0 | 12 | 0 | 9 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 2 | 0 | 0 |
| H1 | 0 | 0 | 8 | 13 | 0 | 48 | 1 | 17 | 2 | 6 | 40 | 22 | 20 | 14 | 12 | 3 | 207 | 1490 | 300 | 622 | 158 | 292 | 20 | 923 |
| H2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| H3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| H4 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 13 | 53 | 0 | 0 | 0 | 0 |
| H5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 |
| K1 | 0 | 2 | 3 | 10 | 1 | 204 | 1 | 10 | 0 | 0 | 416 | 300 | 56 | 9 | 17 | 12 | 649 | 106 | 1075 | 1920 | 1860 | 2450 | 259 | 3862 |
| K2 | 0 | 0 | 4620 | 172 | 2 | 77 | 0 | 0 | 0 | 0 | 683 | 393 | 3790 | 12671 | 1184 | 2497 | 1646 | 4063 | 995 | 1060 | 34 | 40 | 8 | 64 |
| K3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 26 | 0 | 0 | 0 | 52 | 53 | 431 | 30 |
| K5 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 26 | 0 | 6 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 12 | 2 | 0 | 1 | 0 | 0 |
| K6 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| K7 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 9 |
| K8 | 2 | 20 | 5527 | 2164 | 2 | 225 | 0 | 16 | 0 | 6 | 10 | 1 | 3 | 32 | 29 | 12 | 296 | 19 | 22 | 35 | 48 | 84 | 13 | 874 |
| K9 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| K10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K11 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| K12 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 7 | 8 | 0 | 0 | 0 | 0 |
| K13 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| K14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| L1 | 24 | 1 | 3 | 31 | 2 | 5 | 12275 | 2992 | 3930 | 1572 | 357 | 188 | 875 | 71 | 116 | 0 | 126 | 35 | 32 | 105 | 0 | 0 | 0 | 0 |
| 12 | 19 | 109 | 0 | 1 | 0 | 1 | 3 | 1 | 0 | 1 | 27 | 26 | 11 | 0 | 0 | 0 | 0 | 20 | 89 | 158 | 48 | 17 | 1 | 26 |
| L3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1688 | 891 | 3859 | 0 | 0 | 0 | 0 | 576 | 208 | 128 | 0 | 0 | 0 | 0 |
| L4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L5 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 9 | 0 | 108 | 87 | 88 | 0 | 1 | 0 | 283 | 1 | 0 | 0 | 737 | 1235 | 251 | 1182 |
| L6 | 0 | 0 | 2 | 1 | 0 | 0 | 13 | 2 | 6 | 4 | 1 | 0 | 48 | 1 | 4 | 0 | 5 | 371 | 4 | 5 | 0 | 2 | 0 | 1 |
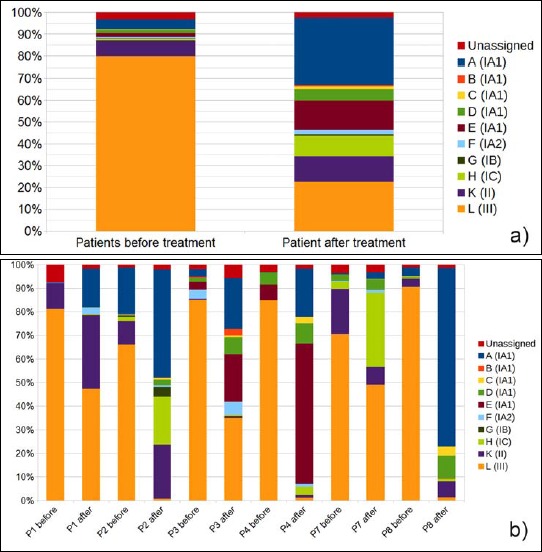
The results of the P. acnes ST distribution in PMH lesions from eight patients and eight matching healthy controls are shown in (Fig. 1a,b). A clear distinction was found when comparing PMH lesional samples to matching controls: P. acnes type III (the corresponding ST is designated “L”) was the dominating phylotype in PMH lesions (Fig. 1a); on average, 73.9% of the reads belonged to this type. In contrast, in the matching control samples, only 14.2% of the reads could be assigned to P. acnes type III (p value 7.8 × 10-5). Instead, control samples contained a higher proportion of type IA1 (STs “A” to “E”) strains (56.8%) and type II (ST “K”) strains (22.3%), while these were found in much lower proportions in PMH lesions: type IA1 (12.8%) and type II (5.6%). Individual variation among the patients and controls was observed: the proportion of type III in the total P. acnes population varied between 50% and 91% in PMH lesions and between 0% and 53% in control samples (Fig. 1b).
Fig. 1.
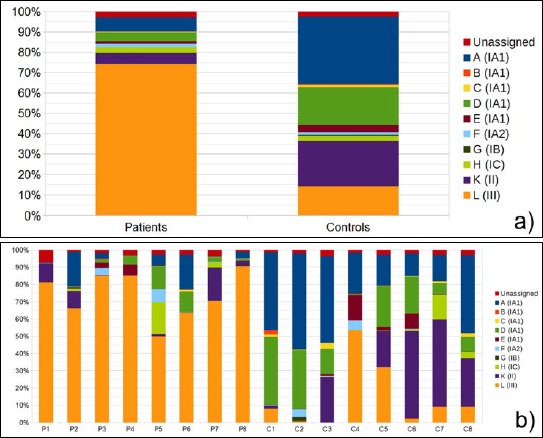
Comparison of P. acnes ST distribution in PMH samples and controls based on next-generation sequencing data. a) Average of P. acnes ST proportions in patient versus controls samples. b) Each column represents ST proportions as an average of the three sampling sites (upper back, lower back, and abdomen). Data are given for eight patients, P1 to P8, and eight controls, C1 to C8. Each ST (A to L) is given a color as indicated, and the corresponding P. acnes subtype is given in brackets
We wanted to confirm these findings with a culture-dependent technique: swab samples taken from PMH lesions and healthy controls were cultivated anaerobically. Up to 10 P. acnes colonies per sample were randomly selected from the agar plates; for each isolate, the classical SLST assignment by Sanger sequencing of the PCR-amplified SLST fragment was carried out. In average, 39.6% of the P. acnes colonies were type III in lesion samples, in contrast to only 12.5 % in controls (see Supplementary Fig. S2 online).
Fig. S2.
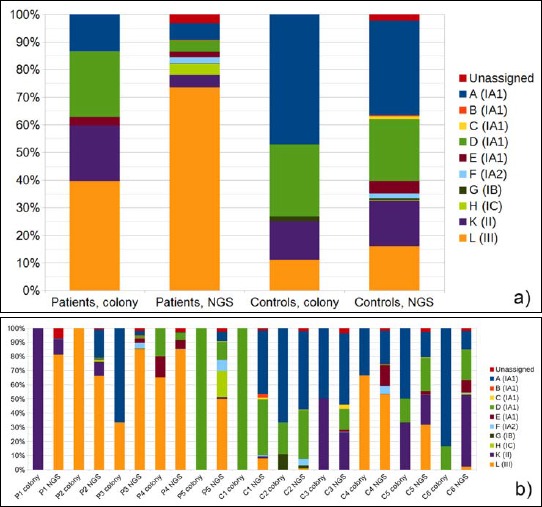
Comparison of two methods of P. acnes ST distribution analysis: cultivation and subsequent SLST of up to 10 colonies per lesion versus next-generation sequencing (NGS)-based SLST. a) Average ST proportions in patient and control samples analysed by culture versus NGS. b) Culture versus NGS determination of ST proportions shown for individual patients and controls. Each ST type A to L is given a color as indicated, and the corresponding P. acnes subtype is given in brackets
P. acnes type III in PMH patients is enriched in PMH regions and adjacent skin areas but is rarely found at other body sites
A recent study [29] suggested that the phylotype distribution of P. acnes is relatively uniform and stable throughout the body. Thus, we wanted to investigate if PMH patients are also predominantly colonized by P. acnes type III at different body sites other than the PMH lesions. Samples from different locations including the lower and upper back, the forehead and the buccal mucosa were analyzed from patients and controls.
Type III was rarely detected on the forehead or in the buccal mucosa of patients (Fig. 2a). Most patients had a mixture of different P. acnes types on the forehead, in particular, strains of the phylotypes IA2, IC, and II. Only one patient was found to have a significant proportion of type III P. acnes on the forehead. On the buccal mucosa, type IA1 was predominant in most patients.
Fig. 2.
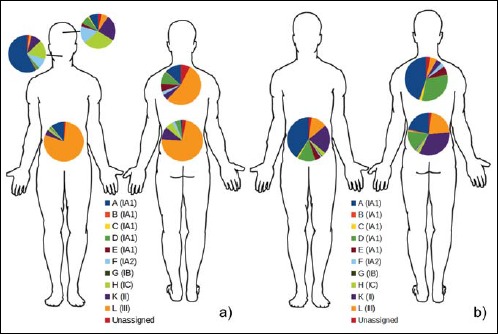
Average P. acnes ST distribution shown for different body sites in patients and controls. a) P. acnes ST distribution in patient PMH lesions on lower back, upper back, and abdomen, and unaffected skin on the forehead and buccal mucosa. b) ST distribution in controls at sites corresponding to patient lesions on lower back, upper back, and abdomen. Each ST (A to L) is given a color as indicated, and the corresponding P. acnes subtype is given in brackets
Looking at the P. acnes phylotype distribution of patients on nonlesional skin sites, a dominance of type III strains was detected, albeit at a lesser extent than on lesional skin, 53% versus 74%, respectively (Fig. 3).
Fig. 3.
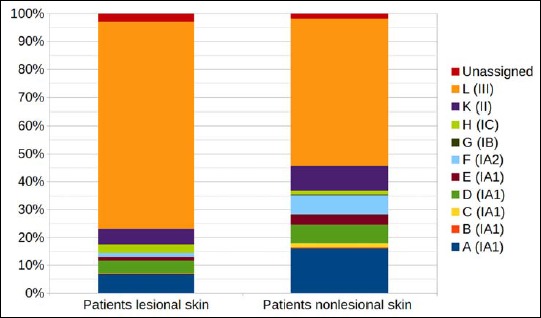
Comparison of average P. acnes ST distribution in PMH patients in lesional versus nonlesional skin based on next-generation sequencing data. Each ST (A to L) is given a color as indicated, and the corresponding P. acnes phylobtype is given in brackets
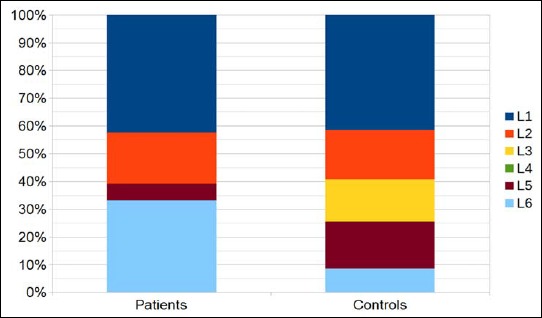
Healthy individuals were analyzed as well. Interestingly, in average, we could detect a larger proportion of type III strains (21.7%) on the lower back skin compared to the upper back skin (5.0%), indicating that the lower back is the preferred habitat for type III strains (Fig. 2b). The dominant P. acnes type on the lower back skin of healthy controls was type II (33.5%), a type that was rarely detected on the upper back (4.0%). The dominating type of the upper back and the abdomen was type IA1 (44.7% and 40.6%, respectively).
PMH treatment alters the P. acnes population and diminishes the proportion of type III strains
Next, we wanted to investigate how the treatment of PMH with a combination of lymecycline and BPO might alter the P. acnes type distribution. Six patients were treated for 3 months, and samples were taken before and after treatment from the same skin location. Information about the treatment regimen and the response for each patient is given in Supplementary Table S2 online. Representative images of the back skin of two patients who responded well to the treatment are shown (Fig. 4).
Fig. 4.
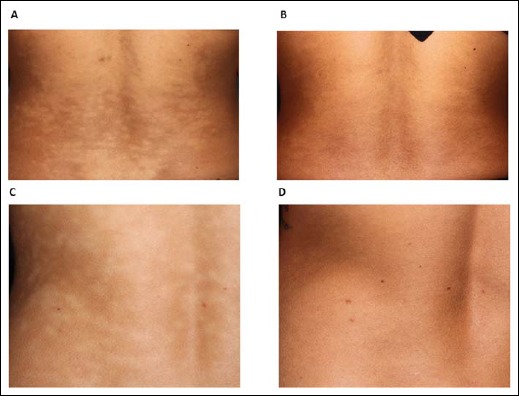
Clinical PMH treatment responses. Lesional skin on the back before and after treatment with lymecycline 300 mg and BPO daily washes for 3 months shown for patient P2 (A and B) and patient P4 (C and D)
We could detect a striking reduction of the proportion of type III P. acnes from an average of 80% before treatment to 22% after treatment (p = 0.015) (Fig. 5a). In all six patients, the type III proportion was diminished after treatment in PMH-affected skin sites. In three patients (P2, P4, and P8), the type III population was almost completely eradicated after treatment (Fig. 5b). Interestingly, these three patients showed a particular good response to the treatment with almost no remaining PMH lesions. The type distribution after treatment resembled in average the one detected in controls samples (Fig. 1b). In contrast, in patients with a less good treatment response, a substantial type III population could be still detected (Fig. 5b).
Fig. 5.
Comparison of P. acnes ST distribution in PMH patients before and after treatment based on next-generation sequencing data. a) Average P. acnes ST proportions in patient samples before and after treatment. b) Each column represents ST proportions as an average of the three sampling sites (upper back, lower back, and abdomen). Data are given for six patients before and after treatment. Each ST type A to L is given a color as indicated, and the corresponding P. acnes subtype is given in brackets
Existence of a specific type III linage associated with PMH lesions?
Since type III strains were also detected, albeit fewer, in healthy samples, in particular on the lower back skin, we wanted to investigate if PMH-associated type III strains belong to a specific lineage that is different from health-associated type III strains. Our SLST scheme can differentiate six STs within the type III lineage. The analyses showed that PMH-associated type III P. acnes belong predominantly to the STs L1 (56%) and L6 (25%); the latter ST was detected at lower rates among health-associated type III P. acnes (9%) (Fig. 6). Overall, our data do not reveal a clear-cut difference between the type III populations of healthy and PMH-affected individuals.
Fig. 6.
Comparison of average ST distribution among P. acnes subtype III strains in PMH patients versus controls based on next-generation sequencing data. Each type III ST (L1 to L6) is given a color as indicated
Discussion
We report the hitherto most detailed data on the distribution of P. acnes subtypes on multiple skin sites on eight PMH patients and eight healthy controls, using a highly discriminative NGS-based SLST approach. The study revealed an association of one particular type of P. acnes, the phylotype III, with the skin disorder progressive macular hypomelanosis. Moreover, an indication that subtype III is involved in the PMH disease pathogenesis was revealed by the comparison of patient samples before and after treatment: therapy with lymecycline and BPO, both highly active against P. acnes, led to a diminished proportion of type III, which was paralleled by the disappearance of clinical PMH lesions.
The applied NGS-based SLST approach gave an average of 7911 reads per sample with 97% of reads assigned to known P. acnes STs. This provided a robust basis for a high-resolution estimate of the type distribution in each sample. Though this technique is regarded as less biased than culture-dependent techniques, a PCR bias due to disproportional amplification of certain sequences cannot be ruled out. Therefore, up to 10 randomly selected colonies were cultured from samples and analyzed by traditional Sanger sequencing for comparison. Importantly, the dominance of type III strains in PMH lesions as compared to controls was consistent in both techniques.
P. acnes subtype III was first reported as a new phylogenetic type in 2008 based on four strains isolated from spinal intervertebral disc material [17]. In addition, subtype III isolates were recently detected in surgically excised lumbar disc herniations from 5 of 24 patients [30]. Subtype III bacterial cells differ from other P. acnes types in showing a long filamentous morphology reminiscent of Propionibacterium propionicum. Subtype III differs also in biochemical tests, matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) spectra, and genetic markers; thus, it was recently proposed as a new subspecies with the name P. acnes subsp. elongatum [31]. This subtype has rarely been cultured from healthy controls, opportunistic infections, or acne patients [14, 16, 18, 32]. In contrast, the present study showed a striking predominance of subtype III in PMH lesional samples from the lower, middle, and upper back, and abdomen. This corroborates the previous finding of a unique, unidentified P. acnes type in PMH patients [22], and the recent report that type III strains were cultured from lesions of 14 of 34 PMH patients [24].
The NGS-based SLST approach provided no data on the presence of other bacterial species or the total bacterial numbers but only on the proportional distribution of P. acnes subtypes. Previous studies have found a highly significant increase in P. acnes numbers in PMH lesions [11, 24]. In accordance, we could also confirm that P. acnes is more abundant in lesions compared to nonlesional skin: a semiquantitative analysis revealed that all PMH samples from patients had a higher colony-forming unit count as compared to adjacent nonlesional skin (data not shown). As in other studies, no Malassezia fungus was detected and only few bacteria of other species, mainly Staphylococcus epidermidis, were found.
Our study revealed a high proportion (74%) of P. acnes type III in lesional skin and also a relatively high proportion (53%) of type III isolates in adjacent nonlesional skin of the patients. In contrast, Barnard et al. cultured P. acnes from nonlesional skin in only one of 34 patient samples, indicative of low P. acnes numbers in nonlesional skin. Therefore, the type distribution at normal skin sites could not be assessed in their study [24]. In addition, the difference may reflect different sampling and sample processing approaches in the two studies. Barnard et al. cultivated bacteria from a homogenized 4-mm punch skin biopsy sample, whereas we used surface skin swabs from a circular area of approximately 1.5 cm in diameter. We noticed that it is difficult to completely separate lesions from adjacent nonlesional skin sites as lesions may be small and plenty and not well defined. This may explain the relatively high proportion of type III strains in nonlesional skin of patients in the present study. Normal controls harbored only a minor proportion of P. acnes type III on corresponding skin sites.
The SLST scheme can distinguish six different STs among type III strains and four of these were detected in PMH lesions with no clear differences in their relative proportions in patients and controls. Interestingly, a regional difference in prevalent type III clones was suggested, based on comparisons of PMH isolates from Europe and Brazil [24].
The mechanism leading to macular hypomelanosis is not known, but ultrastructural studies have shown less melanized and aggregated melanosomes instead of single mature melanosomes transferred from melanocytes to keratinocytes [12]. The association with P. acnes type III suggests that a type III-specific factor could be involved. Comprehensive comparison of type III genomes to P. acnes genomes of other subtypes has identified several genomic regions specific to type III genomes encoding functions such as type II secretion system, ABC transporter, inositol transport/modification, gyrase, integrase, transposase, oligopeptide transport, and processing of sugars/amino acids [24, 33, 34]. In addition, some genes are absent from type III genomes but present in all other types including hyaluronate lyase, magnesium-chelatase, iron transporter, bacteriocin, 3-isopropylmalate dehydrogenase, maltose transporter, and periplasmic binding protein. It has to be 2investigated in the future if and which factors of P. acnes type III are important in PMH pathogenesis.
At present, there is a major interest in defining the normal human skin microbiome as it is considered important for maintaining healthy skin. Several metagenomic studies have described the skin microbiome on species level, but it has become increasingly clear that subtypes within species may play important roles. Most previous P. acnes studies have shown a predominance of type IA1 followed by type II and IB among skin isolates derived from the face or upper back [14, 16, 18]. The present study is the most detailed study on P. acnes subtype distribution on different body sites. It was shown that subtype III is not normally dominant in skin areas of healthy skin; it was rarely found on the face and in the oral cavity. On healthy skin, type III is more frequent at the lower back and abdomen, together with type II, relative to other body sites.
The present study has some limitations. The number of patients and controls is low, even though many samples were analyzed from each person. In general, nonlesional skin was sampled lateral to the lesional area and, therefore, not on exactly corresponding skin areas. After treatment, faint residual lesions in three patients were detectable, even though the clinical response was satisfactory. We do not know if the bacteria most important in the disease process reside on the skin or in the follicles and the surface swab technique employed may have missed bacteria residing in follicles. Furthermore, detailed data on the total number of bacteria at each site and the presence and proportions of other microorganisms besides P. acnes were not obtained in this study.
In conclusion, the present study showed that P. acnes phylotype III is associated with PMH lesions and, therefore, may play a role in the disease pathogenesis. In the normal human microbiome, P. acnes subtype III appears to constitute only a minor portion, mainly residing on the lower back and abdomen, among the P. acnes population. The findings open for future studies of specific type III traits to further define the role of the bacterium and increase our understanding of the PMH disease pathogenesis.
Fig. S1.
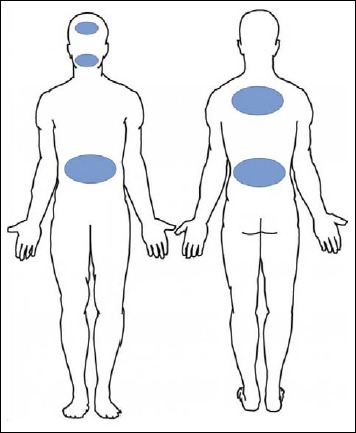
Sampling sites in patients and controls. Patients were sampled from lesional skin on lower back, upper back, and abdomen and adjacent non-affected skin. In addition, unaffected skin of the forehead and buccal mucosa were sampled. After treatment and in healthy controls samples were obtained from areas corresponding to lesions
Acknowledgements
The authors thank Andrea Thürmer, University of Göttingen Genomics Laboratory, Göttingen, Germany, for NGS sequencing support.
Funding Statement
Funding sources: The work was funded in part by the Danish Medical Research council (DFF-1331-00241) to H.B. (http://ufm.dk/) and by Fonden for Faglig Udvikling af Speciallægepraksis to H.B.L. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Footnotes
Abbreviations:
PMH, progressive macular hypomelanosis; SLST, single-locus sequencing typing; ST, sequence type; NGS, next-generation sequencing; BPO, benzoylperoxide
References
- 1.Guillet G, Helenon R, Guillet MH, Gauthier Y, Menard N: Progressive and confluent hypomelanosis of the melanodermic metis. Ann Dermatol Venereol 119, 19–24 (1992) [PubMed] [Google Scholar]
- 2.Guillet G, Helenon R, Gauthier Y, Surleve-Bazeille JE, Plantin P, Sassolas B: Progressive macular hypomelanosis of the trunk: primary acquired hypopigmentation. J Cutan Pathol 15, 286–289 (1988) [DOI] [PubMed] [Google Scholar]
- 3.Relyveld GN, Menke HE, Westerhof W: Progressive macular hypomelanosis: an overview. Am J Clin Dermatol 8, 13–19 (2007) [DOI] [PubMed] [Google Scholar]
- 4.Relyveld GN, Kingswijk MM, Reitsma JB, Menke HE, Bos JD, Westerhof W: Benzoyl peroxide/clindamycin/UVA is more effective than fluticasone/UVA in progressive macular hypomelanosis: a randomized study. J Am Acad Dermatol 55, 836–843 (2006) [DOI] [PubMed] [Google Scholar]
- 5.Santos JB, Almeida OL, Silva LM, Barreto ER: Efficacy of topical combination of benzoyl peroxide 5% and clindamycin 1% for the treatment of progressive macular hypomelanosis: a randomized, doubleblind, placebo-controlled trial. An Bras Dermatol 86, 50–54 (2011) [DOI] [PubMed] [Google Scholar]
- 6.Sim JH, Lee DJ, Lee JS, Kim YC: Comparison of the clinical efficacy of NBUVB and NBUVB with benzoyl peroxide/clindamycin in progressive macular hypomelanosis. J Eur Acad Dermatol Venereol 25, 1318–1323 (2011) [DOI] [PubMed] [Google Scholar]
- 7.Kim MB, Kim GW, Cho HH, Park HJ, Kim HS, Kim SH, Kim BS, Ko HC: Narrowband UVB treatment of progressive macular hypomelanosis. J Am Acad Dermatol 66, 598–605 (2012) [DOI] [PubMed] [Google Scholar]
- 8.Cavalcanti SM, Querino MC, Magalhaes V, Franca ER, Magalhaes M, Alencar E: The use of lymecycline and benzoyl peroxide for the treatment of progressive macular hypomelanosis: a prospective study. An Bras Dermatol 86, 813–814 (2011) [DOI] [PubMed] [Google Scholar]
- 9.Kim YJ, Lee DY, Lee JY, Yoon TY: Progressive macular hypomelanosis showing excellent response to oral isotretinoin. J Dermatol 39, 937–938 (2012) [DOI] [PubMed] [Google Scholar]
- 10.Cavalcanti SM, de Franca ER, Lins AK, Magalhaes M, de Alencar ER, Magalhaes V: Investigation of Propionibacterium acnes in progressive macular hypomelanosis using real-time PCR and culture. Int J Dermatol 50, 1347–1352 (2011) [DOI] [PubMed] [Google Scholar]
- 11.Westerhof W, Relyveld GN, Kingswijk MM, de Man P, Menke HE: Propionibacterium acnes and the pathogenesis of progressive macular hypomelanosis. Arch Dermatol 140, 210 –214 (2004) [DOI] [PubMed] [Google Scholar]
- 12.Relyveld GN, Dingemans KP, Menke HE, Bos JD, Westerhof W: Ultrastructural findings in progressive macular hypomelanosis indicate decreased melanin production. J Eur Acad Dermatol Venereol 22, 568–574 (2008) [DOI] [PubMed] [Google Scholar]
- 13.Wu XG, Xu AE, Song XZ, Zheng JH, Wang P, Shen H: Clinical, pathologic, and ultrastructural studies of progressive macular hypomelanosis. Int J Dermatol 49, 1127–1132 (2010) [DOI] [PubMed] [Google Scholar]
- 14.Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, Modlin RL, Miller JF, Sodergren E, Craft N, Weinstock GM, Li H: Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol 133, 2152–2160 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilian M, Scholz CF, Lomholt HB: Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes. J Clin Microbiol 50, 1158–1165 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomholt HB, Kilian M: Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One 5, e12277 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDowell A, Perry AL, Lambert PA, Patrick S: A new phylogenetic group of Propionibacterium acnes. J Med Microbiol 57, 218–224 (2008) [DOI] [PubMed] [Google Scholar]
- 18.McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, Nagy I, Lambert PA, Patrick S: A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology 157, 1990–2003 (2011) [DOI] [PubMed] [Google Scholar]
- 19.McDowell A, Barnard E, Nagy I, Gao A, Tomida S, Li H, Eady A, Cove J, Nord CE, Patrick S: An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic’, ‘commensal’ and antibiotic resistant strains. PLoS One 7, e41480 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholz CF, Jensen A, Lomholt HB, Bruggemann H, Kilian M: A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo. PLoS One 9, e104199 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Champer J, Garban H, Kim J: Typing of Propionibacterium acnes: a review of methods and comparative analysis. Br J Dermatol 172, 1204–1209 (2015) [DOI] [PubMed] [Google Scholar]
- 22.Relyveld GN, Westerhof W, Woudenberg J, Kingswijk M, Langenberg M, Vandenbroucke-Grauls CM, Bos JD, Savelkoul PH: Progressive macular hypomelanosis is associated with a putative Propionibacterium species. J Invest Dermatol 130, 1182–1184 (2010) [DOI] [PubMed] [Google Scholar]
- 23.Petersen R, Lomholt HB, Scholz CF, Bruggemann H: Draft genome sequences of two Propionibacterium acnes strains isolated from progressive macular hypomelanosis lesions of human skin. Genome Announc 3, (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnard E, Liu J, Yankova E, Cavalcanti SM, Magalhaes M, Li H, Patrick S, McDowell A: Strains of the Propionibacterium acnes type III lineage are associated with the skin condition progressive macular hypomelanosis. Sci Rep 6, 31968 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson P, Kligman AM: A new method for the quantitative investigation of cutaneous bacteria. J Invest Dermatol 45, 498–503 (1965) [DOI] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF: Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL: BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S: MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA: Temporal stability of the human skin microbiome. Cell 165, 854 – 866 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rollason J, McDowell A, Albert HB, Barnard E, Worthington T, Hilton AC, Vernallis A, Patrick S, Elliott T, Lambert P: Genotypic and antimicrobial characterisation of Propionibacterium acnes isolates from surgically excised lumbar disc herniations. Biomed Res Int 2013, 530382 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekio I, Culak R, Misra R, Gaulton T, Fang M, Sakamoto M, Ohkuma M, Oshima K, Hattori M, Klenk HP, Rajendram D, Gharbia SE, Shah HN: Dissecting the taxonomic heterogeneity within Propionibacterium acnes: proposal for Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes subsp. elongatum subsp. nov. Int J Syst Evol Microbiol 65, 4776–4787 (2015) [DOI] [PubMed] [Google Scholar]
- 32.Lomholt HB, Kilian M: Clonality and anatomic distribution on the skin of antibiotic resistant and sensitive Propionibacterium acnes. Acta Derm Venereol 94, 534–538 (2014) [DOI] [PubMed] [Google Scholar]
- 33.Scholz CF, Bruggemann H, Lomholt HB, Tettelin H, Kilian M: Genome stability of Propionibacterium acnes: a comprehensive study of indels and homopolymeric tracts. Sci Rep 6, 20662 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, Li H: Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio 4, e00003–00013 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]


