Abstract
Pseudomonas aeruginosa owns a variability of virulence factors. These factors can increase bacterial pathogenicity and infection severity. Despite the importance of knowledge about them, these factors are not more characterized at level of strains derived from local food products. This study aimed to characterize the virulence potential of P. aeruginosa isolated from various animal products. Several structural and virulence genes of P. aeruginosa including lasB, exoS, algD, plcH, pilB, exoU, and nan1 were detected by polymerase chain reaction (PCR) on 204 strains of P. aeruginosa. They were isolated from bovine meat (122), fresh fish (49), and smoked fish (33). The 16S rRNA gene was detected on 91.1% of the presumptive strains as Pseudomonas. The rpoB gene showed that 99.5% of the strains were P. aeruginosa. The lasB gene (89.2%) was the most frequently detected (p < 0.05). In decreasing importance order, exoS (86.8%), algD (72.1%), plcH (72.1%), pilB (40.2%), and exoU (2.5%) were detected. The lasB gene was detected in all strains of P. aeruginosa serogroups O11 and O16. The prevalence of algD, exoS, and exoU genes in these strains varied from 51.2% to 87.4%. The simultaneous determination of serogroups and virulence factors is of interest for the efficacy of surveillance of infections associated with P. aeruginosa.
Keywords: Pseudomonas aeruginosa, PCR, virulence, serogroups, bovine meat, fresh fish, smoked fish
Introduction
Pseudomonas aeruginosa is a bacterium characterized by its high genetic plasticity and potential for adapting to various environments. The species are frequently isolated from soil and water or colonizes several anatomical sites such as plants, insects, animals, and humans [1–3]. The bacterium may be involved in food poisoning and has many virulence factors [4, 5]. As consequence, P. aeruginosa is an opportunistic pathogen, frequently implicated in nosocomial infections [6–8].
The bacterium is found in approximately 50% to 60% of the fragile or immunocompromised people [9, 10]. It is implicated in 16% of nosocomial pneumonia, 12% in urinary tract infections, 8% in surgical infections, and 10% in bloodstream infections [11]. Otherwise, there is increasing evidence implication of the species in foodborne infections [9, 12, 13]. The pathogenicity of P. aeruginosa is attributed to its ability to form biofilms and to the production of numerous membrane and extracellular virulence factors [4, 5]. Membrane factors include lipopolysaccharides, flagella, adhesion factor (pili type IV), and alginate, while extracellular factors are mainly exotoxins (exotoxin A), exo-proteases (elastase, staphylolysin, alkaline protease, protease IV), phospholipase C, chromophores, and exoenzymes S, T, and U [2, 9]. Virulence factors associated with the membrane are generally involved in colonization and chronic infection, while extracellular factors, extremely toxic, are associated with acute infection [9, 14]. P. aeruginosa can also cause lung lesions by different mechanisms. The mutation responsible for the mucoid form (muc) is associated with activation of the transcription of the alginate gene (algD). The formation of mucoid colonies of P. aeruginosa composed of alginates, involving algD genes, protects the bacterium from the host’s immune response and from antibiotics. As lactoperoxidase (LPS), alginates are involved in the adhesion of the bacterium to the respiratory epithelium [2].
Pili are multifunctional and play a crucial role in the initiation of colonization, allowing adherence of the bacteria to host epithelial surfaces [2, 15]. Pili are also important for biofilms formation [2, 16]. These genes called pil are found in different regions of bacterial chromosome [17]. The expression of these genes is controlled by the two-component systems, Pil-S and Pil-R, and the alternative sigma factor RpoN.
Exoenzyme S, encoded by the exoS gene, is an ADP-ribosyltransferase that is secreted by the type III secretion system directly into the cytosol of epithelial cells [2, 18]. Injection of exotoxin exoU is the cause of rapid death (1 to 2 h) of host cells [2]. This death is characterized by loss of the integrity of the plasma membrane, which is typical of necrosis [19]. In addition to its cytotoxic activity, exoU induces expression of the genes of inflammation [20, 21]. ExoU is 100 times more cytotoxic than exoS according to Lee et al. [2, 19, 22]. LasB elastase, a zinc metalloprotease encoded by the LasB gene, has an elastolytic activity on lung tissue, attacks eukaryotic proteins such as collagen and elastin, and destroys the structural proteins of the cell.
In addition, the phospholipids contained in pulmonary surfactants may be hydrolyzed by two phospholipases C encoded by plcH and plcN (PLC-H and PLC-N, respectively) [2, 5]. The gene called nan1 codes for a sialidase which is responsible for the adhesion of the respiratory tract [5, 18]. An extracellular neuraminidase is thought to play an important role in implantation of the bacterium, but the genetic basis of this process is still unknown [5, 18]. This variety of virulence factors contributes to the pathogenicity of P. aeruginosa. However, these virulence factors are not often studied in the strains isolated from some local food products. Those local food products and mainly products of animal origin harbor strains that may possess virulence factors and some risky zoonotic serogroups.
This study aimed to characterize the virulence factors determining the pathogenicity of P. aeruginosa strains isolated from animal products.
Materials and methods
P. aeruginosa isolates
The study has been carried out over a period between February 2015 and November 2015 in 5 districts of Abidjan (Côte d’Ivoire). A total of 204 isolates of P. aeruginosa were used in the present study. The isolates were obtained from different samples of animal products, including beef (114), fresh fish (45), and smoked fish (30). Presumptive strains of Pseudomonas were isolated on CFC (cetrimide, fucidine, cephaloridine; Oxoid, Code: 0559, England) base Pseudomonas and Cetrimide agar (Oxoid, England). P. aeruginosa biochemical identifications were done by API 20NE (bioMérieux, Marcy l’Etoile, France) and API database. For molecular identifications, Pseudomonas isolates were identified by 16S rRNA and rpoB gene confirmed that some of them were P. aeruginosa. The reference strain P. aeruginosa ATCC 27853 was used as quality control.
Detection of identification and virulence genes by polymerase chain reaction (PCR)
Extraction and purification of DNA
Template DNA was extracted from whole organisms by boiling [22]. Bacteria were harvested from an overnight broth culture (Biokar Diagnostics, BK015HA, France), suspended in 1 ml sterile Milli-Q water (Milli-Q™, Millipore Corporation, USA). A suspension of 200 μl was incubated at –20 °C for 15 min and boiled at 95 °C for 15 min. The suspension was immediately cooled at 4 °C for 10 min and then centrifuged at 14,000 rpm for 10 min to pellet the cell debris.
The DNA template was purified according to the method described by Zimmermann et al. [23]. The purity and DNA concentration of the extract were determined by spectrophotometer (Eppendorf BioPhotometer plus, USA).
Amplification of 16S rRNA, rpoB, algD, PilB, lasB, nan1, plcH, exoS, and exoU genes
Single PCR was used to detect 16S RNA and rpoB gene. PCR mixtures with a final volume of 25 μl consisted of 16 μl sterile Milli-Q water (Milli-Q™, Millipore Corporation, USA), 5 μl 5XTP, 1.5 μl MgCl2 (2 mM), 0.2 μl dNTPs (10 mM), 0.1 μl each primer (20 mM) (Integral DNA Technology, France), 0.1 μl Go tag polymerase (Promega Corporation, Madison, WI 53711-5399, USA), and 2 μl template DNA.
Multiplex PCRs were used to detect algD, PilB, lasB, nan1, plcH, exoS, and exoU. PCR mixtures with a final volume of 25 μl consisted of 15.8 μl sterile Milli-Q water (Milli-Q™, Millipore Corporation, USA), 5 μl 5XTP, 1.5 μl MgCl2 (2 mM), 0.2 μl dNTPs (10 mM), 0.1 μl each primer (20 mM) (Integral DNA Technology, France), 0.1 μl Go tag polymerase (Promega Corporation, Madison, WI 53711-5399, USA), and 2 μl DNA template. A quantity of 2 μl of sterile Milli-Q water was used for negative control, and DNA of ATCC reference strain 27853 was used for positive control.
The oligonucleotide primers used in this study and the amplification program are listed in Tables 1 and 2. Each PCR was performed using thermocycleur type T3000 Thermocycler, block type standard 3a (Biometra, Germany). The amplified DNA was separated by gel electrophoresis with 2% agarose containing 0.5 μg/ml with ethidium bromide for 30 min at 130 V, visualized under UV transillumination, and photographed (Molecular Imager Gel DocTM XR+, Bio-Rad). A molecular weight marker (TriDye™, 100 bp or 1-kb DNA Ladder, Biolabs) was used. Amplified genes were identified on the basis of fragment size shown in Tables 1 and 2.
Table 1.
Primers used for amplification of identification genes in single PCR
| Primers | Target gene | Sequence (5′-3′) | Product size (bp) | Amplification program | Annealing temperature (°C) | Source |
|---|---|---|---|---|---|---|
| 16S-F 16S-R |
16S rRNA | AGAGTTTGATCCTGGCTCAG CTACGGCTACCTTGTTACGA |
≈1351 | 94 °C, 2 min 5 × [94 °C, 45 s; 55 °C, 1 min; 72 °C, 2 min] 35 × [92 °C, 45 s; 60 °C, 45 s; 72 °C, 2 min] 72 °C, 2 min; 4 °C… |
55 60 |
[25] |
| rpoB-F rpoB-R |
rpoB | CAGTTCATGGACCAGAACAACCCG ACGCTGGTTGATGCAGGTGTTC |
≈759 | 94 °C, 3 min 30 × [94 °C, 1 min; 58 °C, 1 min; 72 °C, 2 min] 72 °C, 2 min; 4 °C… |
58 | [27] |
PCR: polymerase chain reaction; F: forward; R: reverse
Table 2.
Primers used for amplification of virulence genes in multiplex PCR
| Primers | Target gene | Sequence (5′-3′) | Product size (bp) | Amplification program | Annealing temperature (°C) | Source |
|---|---|---|---|---|---|---|
| pilB-F pilB-R |
pilB | ATG AAC GAC AGC ATC CAA CT GGG TGT TGA CGC GAA AGT CGA T |
826 | 94 °C, 5 min 35 × [94 °C, 35 s; 60 °C, 1 min; 72 °C, 1 min] 72 °C, 7 min; 4 °C… |
60 | [29] |
| LasB-F LasB-R |
LasB | GGA ATG AAC GAG GCG TTC TC GGT CCA GTA GTA GCG GTT GG |
300 | [28] | ||
| ExoS-F ExoS-R |
exoS | CTT GAA GGG ACT CGA CAA GG TTC AGG TCC GCG TAG TGA AT |
504 | [28] | ||
| algD-F algD-R |
algD | ATG CGA ATC AGC ATC TTT GGT CTA CCA GCA GAT GCC CTC GGC |
1310 | 94 °C, 5 min 35 × [94 °C, 35 s; 61 °C, 1 min; 72 °C, 1 min] 72 °C, 7 min; 4 °C… |
62 | [28] |
| plcH-F plcH-R |
plcH | GAA GCC ATG GGC TAC TTC AA AGA GTG ACG AGG AGC GGTAG |
307 | 60 | [28] | |
| nan1-F nan1-R |
nan1 | ATG AAT ACT TAT TTT GAT AT CTA AAT CCA TGC TCT GAC CC |
1317 | 94 °C, 5 min 35 × [94 °C, 35 s; 57 °C, 1 min; 72 °C, 1 min] 72 °C, 7 min; 4 °C… |
53 | [29] |
| exoU-F exoU-R |
exoU | GGG AAT ACT TTC CGG GAA GTT CGA TCT CGC TGC TAA TGT GTT |
428 | 60 | [29] |
PCR: polymerase chain reaction; F: forward; R: reverse. AlgD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; pilB, type IV fimbrial biogenesis protein PilB-encoding gene; nan1, neuraminidase-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene; lasB, elastase LasB-encoding gene; exoS, exoenzyme S-encoding gene; exoU, exo-enzyme U-encoding gene
Serotyping of P. aeruginosa isolates
The O-serotypes were determined by a slide agglutination test using four pools (OMA, OMC, OME, and OMF) and 20 monovalent antisera, O1 to O20 (SanofiDiagnostics Pasteur), according to the manufacturer’s recommendations.
Statistical analysis
The statistical analysis was carried out on the software Statistical Package for the Social Sciences (SPSS) 20.0 (IBM SPSS, Chicago, IL, United States of America). The Student’s t test, Mann–Whitney U test, Spearman’s correlation analysis, and multiple regression analysis were performed. Statistical significance was set at p < 0.05.
Results
P. aeruginosa strains detected by 16S rRNA and rpoB genes
Out of 225 presumptive isolates of Pseudomonas, 205 (91.1%) Pseudomonas were identified by 16S rRNA (Fig. 1; Table 3) and 204 (99.5%) strains were confirmed by the rpoB gene (Fig. 2; Table 3) as P. aeruginosa.
Fig. 1.
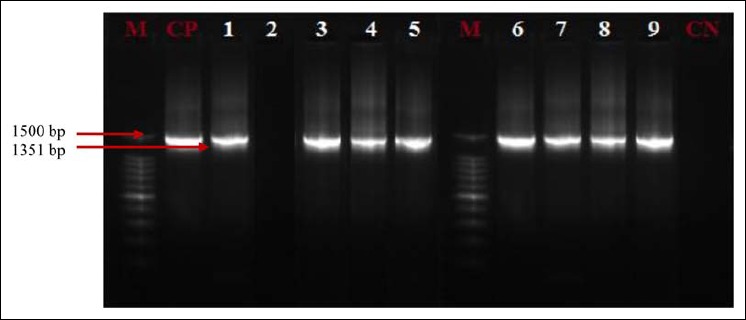
16S rRNA profiles of Pseudomonas isolates. Lanes 1, 3–9: Presence of Pseudomonas in analyzed products; lane 2: absence of Pseudomonas in analyzed products; CP: positive control (Pseudomonas aeruginosa ATCC 27853); CN: negative control; M: marker gene ruler, 100 bp (Bench Top, 100-bp DNA Ladder, Promega Corporation, USA)
Table 3.
Frequency of strains confirmed by the 16S rRNA and rpoB genes
| Genes | Number of isolates presumptive Pseudomonas N = 225 | ||
|---|---|---|---|
| Confirmed species | Effective | Percentage | |
| 16S rRNA | Pseudomonas SP | 205 | 91.1 |
| rpoB | Pseudomonas aeruginosa | 204 | 99.5 |
Fig. 2.
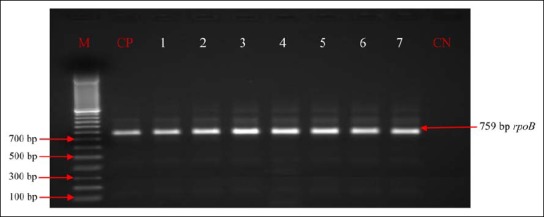
RpoB profiles of Pseudomonas aeruginosa isolates. Lanes 1–7: Presence of Pseudomonas aeruginosa in analyzed products; CP: positive control (Pseudomonas aeruginosa ATCC 27853); CN: negative control; M: marker gene ruler, 100 bp (Bench Top, 100-bp DNA Ladder, Promega Corporation, USA)
Virulence genes of P. aeruginosa
Out of seven genes researched, six virulence genes of P. aeruginosa were detected in some animal samples (line 5: lasB, exoS, algD, plcH, pilB, and exoU; Figs. 3–5). Other samples have fewer than six virulence factors (lines 2–4 and 6–13; Figs. 3–5).
Fig. 3.
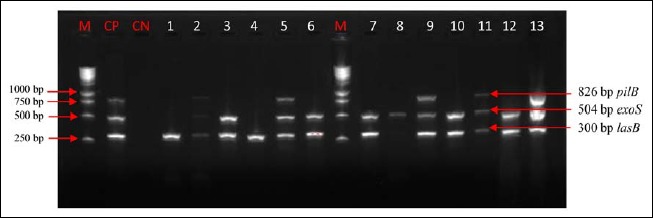
Electrophoretic profile of amplification products of the virulence genes pilB, exoS, and lasB in analyzed products. Virulence genes were present in analyzed products. Lane 1: lasB; lane 2: pilB, exoS, and lasB; lane 3: exoS and lasB; lane 4: lasB; lane 5: pilB, exoS, and lasB; lane 6: exoS and lasB; lane 7: exoS and lasB; lane 8: exoS and lasB; lane 9: pilB, exoS, and lasB; lane 10: exoS and lasB; lane 11: pilB, exoS, and lasB; lane 12: exoS and lasB; lane 13: pilB, exoS, and lasB; M: marker gene ruler, 250 bp (Bench Top, 1-kb DNA Ladder, Promega Corporation, USA); pilB, type IV fimbrial biogenesis protein PilB-encoding gene; lasB, elastase LasB-encoding gene; exoS, exoenzyme S-encoding gene
Fig. 4.
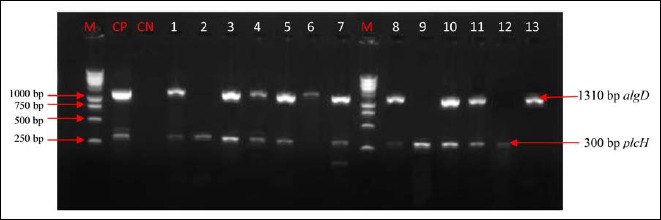
Electrophoretic profile of amplification products of the virulence genes algD and plcH in analyzed products. Virulence genes were present in analyzed products. Lane 1: algD and plcH; lane 2: plcH; lane 3: algD and plcH; lane 4: algD and plcH; lane 5: algD and plcH; lane 6: algD; lane 7: algD and plcH; lane 8: algD and plcH; lane 9: plcH; lane 10: algD and plcH; lane 11: algD and plcH; lane 12: plcH; lane 13: algD; M: marker gene ruler, 250 bp (Bench Top, 1-kb DNA Ladder, Promega Corporation, USA); algD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene
Fig. 5.
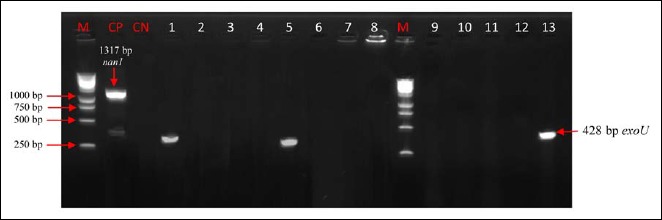
Electrophoretic profile of amplification products of the virulence gene exoU and nan1 in analyzed products. Virulence genes were present in analyzed products. Lanes 1, 5, 13: exoU; Virulence genes were absent in analyzed products; lanes 2–4, 6–12; M: marker gene ruler, 250 bp (Bench Top, 1-kb DNA Ladder, Promega Corporation, USA); algD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene. M: marker gene ruler, 250 bp (Bench Top, 1-kb DNA Ladder, Promega Corporation, USA); exoU, exoenzyme U-encoding gene
Prevalence of virulence genes
Table 4 showed the prevalence of genes detected out of 204 strains of P. aeruginosa. According to the table, lasB gene with 89.2% has been most detected following by exoS (86.8%). Genes algD and plcH had the same prevalence (72.1%). The prevalence of pilB and exoU genes was, respectively, 40.2% and 2.5%. The nan1 gene had not been detected during this study.
Table 4.
Prevalence of P. aeruginosa virulence genes isolated from animal products (n = 204)
| Virulence genes | Prevalence of Pseudomonas aeruginosa virulence genes isolated from animal products | |||||||
|---|---|---|---|---|---|---|---|---|
| Bovine meat (n = 122) | Fresh fish (n = 49) | Smoked fish (n = 33) | Total (n = 204) | |||||
| Effective (N) |
Prevalence (%) |
Effective (N) |
Prevalence (%) |
Effective (N) |
Prevalence (%) |
Effective (N) |
Prevalence (%) |
|
| lasB | 118 | 96.7 | 35 | 71.4 | 29 | 87.9 | 182 | 89.2 |
| exoS | 118 | 96.7 | 33 | 67.3 | 26 | 78.8 | 177 | 86.8 |
| algD | 91 | 74.5 | 38 | 77.5 | 18 | 54.5 | 147 | 72.1 |
| plcH | 88 | 72.1 | 37 | 75.5 | 22 | 66.7 | 147 | 72.1 |
| pilB | 52 | 42.6 | 19 | 38.8 | 11 | 33.3 | 82 | 40.2 |
| exoU | 5 | 4.1 | 0 | 0.0 | 0 | 0.0 | 5 | 2.5 |
| nan1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
AlgD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; pilB, type IV fimbrial biogenesis protein PilB-encoding gene; nan1, neuraminidase-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene; lasB, elastase LasB-encoding gene; exoS, exoenzyme S-encoding gene; exoU, exoenzyme U-encoding gene
The prevalence of virulence genes varied from 0% to 96.7%, from 0% to 87.9%, and from 0% to 77.5%, respectively, in bovine meat, smoked fish, and fresh fish (Table 4). The prevalence of lasB and exoS genes was higher in bovine meat (96.7% and 96.7%), followed by smoked fish (87.9% and 78.8%) and fresh fish (71, 4%, and 67.3%) (Fig. 6). That of algD and plcH gene was respectively higher in fresh fish (77.5% and 75.5%), followed by bovine meat (74.5% and 72.1%) and smoked fish (54.5% and 66.7%) (Fig. 6). The prevalence of the pilB gene was less than 50% in all animal products analyzed (Fig. 6). The exoU gene was detected only in strains from bovine meat.
Fig. 6.
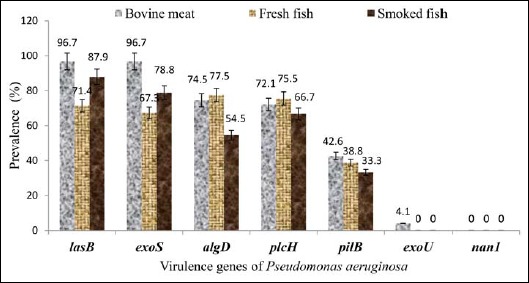
Virulence genes according to the origin of the strain. AlgD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; pilB, type IV fimbrial biogenesis protein PilB-encoding gene; nan1, neuraminidase-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene; lasB, elastase LasB-encoding gene; exoS, exoenzyme S-encoding gene; exoU, exoenzyme U-encoding gene
Diversity of P. aeruginosa serogroups
The mostly isolated serogroups were O11 with 25.5%, followed by serogroups O5 (21.1%) and O16 (20.6%). According to Table 5, non-serotyping strains and serogroups O7, O8, O9, O15, O1, O10, O12, O2, and O4 were less than 5%.
Table 5.
Serogroups of Pseudomonas aeruginosa
| Serogroups | Number of strains (n = 204) |
Prevalence (%) |
|---|---|---|
| O11 | 52 | 25.5 |
| O5 | 43 | 21.1 |
| O16 | 42 | 20.6 |
| NS | 10 | 4.9 |
| O7 | 10 | 4.9 |
| O8 | 8 | 3.9 |
| O9 | 8 | 3.9 |
| O15 | 7 | 3.4 |
| O1 | 6 | 2.9 |
| O10 | 6 | 2.9 |
| O12 | 6 | 2.9 |
| O2 | 3 | 1.5 |
| O4 | 3 | 1.5 |
NS: not serotypeable; O: serogroups
Prevalence of the virulence genes of P. aeruginosa according to serogroups
Elastase-producing strains of P. aeruginosa (lasB) belonging to serogroups O11 and O16 had a prevalence of 100% (Fig. 7). However, the same strains of serogroups O11 and O16 harbored the alginate (algD), exoenzyme (exoS), and (exoU) with a prevalence ranging from 51.2% to 87.4%. Elsewhere, genes (plcH) and (pilB) had been detected from 8.4% to 12.2% (Fig. 7). P. aeruginosa strains belonging to O5 serogroups had a prevalence ranging from 27.2% to 34.9% for strains with virulence factors. No strain of the three serogroups produced neuraminidase 1.
Fig. 7.
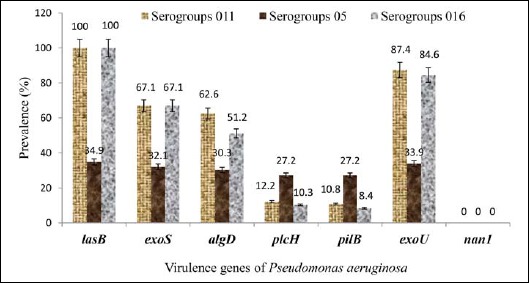
Virulence genes and serogroups of Pseudomonas aeruginosa. AlgD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; pilB, type IV fimbrial biogenesis protein PilB-encoding gene; nan1, neuraminidase-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene; lasB, elastase LasB-encoding gene; exoS, exoenzyme S-encoding gene; exoU, exoenzyme U-encoding gene
Distribution of P. aeruginosa virulence genes
According to Fig. 8, virulence factors have been distributed at 99.7% on the axes F1 and F2. The F1 axis represents 97.1%, and the F2 axis, 2.6% (Fig. 8). All the variables bovine meat, fresh fish, and smoked fish are positively represented on axis 1. This axis contrasts the lasB, exoS, algD, and plcH genes that are positively correlated with the pilB, exoU, and nan1 genes which correlation is negative. Thus, the lasB, exoS, algD and plcH genes are the most found in the three food matrices (bovine meat, fresh fish, and smoked fish).
Fig. 8.
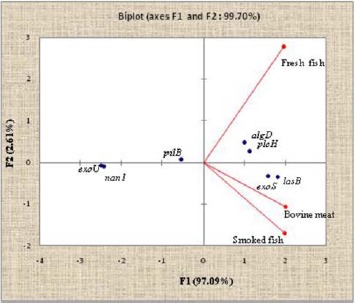
Distribution of Pseudomonas aeruginosa virulence genes isolated from animal products. The axes F1 and F2 of the ACP account for 99.7% of the inertia. The F1 axis represents 97.09% and the F2 axis 2.61%. AlgD, GDP-mannose 6-dehydrogenase AlgD (alginate)-encoding gene; pilB, type IV fimbrial biogenesis protein PilB-encoding gene; nan1, neuraminidase-encoding gene; plcH, hemolytic phospholipase C precursor-encoding gene; lasB, elastase LasB-encoding gene; exoS, exoenzyme S-encoding gene; exoU, exoenzyme U-encoding gene
Discussion
16S rRNA gene analysis revealed that 91.1% of the analyzed food products were positive for the genus Pseudomonas, whereas in 99.5% of the Pseudomonas-positive samples, the rpoB gene could be detected, indicating contamination of the products by P. aeruginosa. This result shows that the resolving power of this marker is higher than that of 16S rRNA [25]. This result also highlights the strong discriminating power of the identification method using rpoB gene and confirms the heterogeneity of the P. aeruginosa observed by many authors [25, 29]. This high molecular identification rate showed that genomic studies are needed to confirm the exact taxonomic position of P. aeruginosa. The performance of the rpoB gene on the detection of P. aeruginosa strains could also be explained by the fact that the discrimination between species very close to Pseudomonas is obtained by the analysis of the rpoB gene [25, 30].
The results also showed that the prevalence of virulence genes of P. aeruginosa predominantly detected was 89.2% for lasB and 86.8% for exoS, followed by 72.1% for algD and 72.1% for plcH. The high prevalence of elastase (89.2%) encoding the lasB (zinc metalloprotease) gene and secreted by type II secretion system indicates that this protease can be important for the pathogenesis of P. aeruginosa isolated from animal products cleaving elastin and collagen [2, 5].
Indeed, this enzyme is at the origin of a destruction of the junctions between the epithelial cells [2, 5]. This protease increases the production of IL-8 and reduces the innate immune response by cleaving proteins from surfactant, SP-A and SP-D, and protease receptors [5]. Elastase B is also able of inactivating other proteins such as IgA, IgG, and complement compounds, thus, modulating the immune response. It also inhibits the repair of injured epithelial cells by altering cell mobility [2–4].
The high prevalence of algD gene could be at the origin of the conversion of P. aeruginosa strains to a mucoid phenotype overproducing alginates [31]. This alginate prevalence indicates that isolated strains of animal products are involved in the formation of biofilms as alginates have been widely regarded as the major exopolysaccharides of the biofilm matrix [31, 32]. The overproduction of alginates protects P. aeruginosa from antibiotics and also alleviates the immune response by inhibiting complement activation, reducing polymorphonuclear chemotaxis and decreasing phagocytosis [31]. The expression of algD and lasB genes could be correlated with appearance of a mucoid phenotype. Also, alginate and elastase production could be characteristic of installation of a chronic infection in consumers of animal products [32]. These same high prevalences of these virulence genes were obtained by Mitov et al. [4].
The prevalences for the plcH and exoS genes were 72.1% and 86.8%, respectively. These prevalences showed that strains isolated were able of secreting hemolytic exoenzymes and phospholipase C, and thus, strains could be involved in pulmonary infections [33, 34]. The exoS was responsible for tissue destruction; it is implicated to pulmonary infection and can be important for bacterial dissemination [4]. This prevalence was 40.2%, 2.5%, and 0.0%, respectively, for the pilB, exoU, and nan1 gene. The prevalence of the pilB gene which codes for the formation of pili indicates that the strains are also involved in the twitching motility which allows the displacement on a solid surface, thanks to the extension and then the retraction of the pili and the mobility of the “swarming” type [16]. This prevalence shows that strains isolated from animal products could be involved in the formation of biofilm and the initiation of infections by bacteriophages whose pili is considered as a receptor.
The relatively low prevalence for exoenzyme-U could be explained by the fact that 90% of P. aeruginosa strains producing exoU are associated with serious infections, and therefore, the strains of animal origin are not extremely associated with severe infections [4, 19]. Among type III secretion proteins, exoU is the most cytotoxic gene. The exoU secretion is a marker of invasive isolates of P. aeruginosa [5, 19].
These results indicate that some of virulence factors assist bacterial establishment and colonization on the surface of the host, while others expedite invasion of numerous tissue. Some factors such as flagella, fimbriae, surface polysaccharides, and type IV pili are involved in bacterial colonization. All these elements are considered essential for attachment mechanism. P. aeruginosa has the ability to invade tissue. P. aeruginosa also produces toxins and enzymes that disrupt carnal barriers by disrupting cell membranes despite the immune system of the host.
In addition, the results of this study showed that all elastase-producing strains of P. aeruginosa (lasB) belonged to serogroups O11 and O16 with a prevalence of 100%. Recently, Le-Berre et al. showed that the O11 serogroups, elastase production, and TTSS were associated with increased lung injury in a murine model of pneumonia [35]. They found that serogroups O11 strains were significantly more virulent than nontypable strains and serotype O6.
These same strains of serogroups O11 and O16 had prevalence between 51.2% and 87.4% for the alginate (algD), exoenzyme (exoS), and (exoU) producing strains. P. aeruginosa strains belonging to O5 serogroups had a prevalence ranging from 27.2% to 34.9% for all the strains with virulence factors in this study. Neuraminidase was not produced by any strains of the three serogroups produced.
All these results could indicate that strains of O11 and O16 serogroups are more associated with virulence factors than strains belonging to O5 serogroups [36, 37].
The results of this study showed that lasB, exoS, algD, and plcH genes are the most frequently found in the three food matrices. P. aeruginosa strains isolated from animal products are able to harbor genes of resistance and virulence belonging to particular serogroups.
Conclusion
The study showed that the rpoB gene confirms the identification of P. aeruginosa. It also revealed that strains of P. aeruginosa isolated from animal products harbor distinct virulence genes. The lasB, exoS, algD, and plcH genes are the most found in the three food matrices. The study also indicated that strains of serogroups O11 and O16 were more associated with the virulence genes of P. aeruginosa isolated from animal products.
Acknowledgements
The authors thank the INRS – Institut Armand Frappier of Canada, the Pasteur Institute of Paris, and the Institut Pasteur of Cote d’Ivoire for their advice and excellent technical assistance.
Funding Statement
Funding sources: There has been no source of funding.
References
- 1.Edit K, Sándor S, Gyula D, Júlia R, Balázs K, Balázs K: Pathogenic and phylogenetic features of 2 multiresistant Pseudomonas aeruginosa strains originated from remediated sites. Int J Occup Med Environ Health 29, 503–516 (2016) [DOI] [PubMed] [Google Scholar]
- 2.Fadhil L, Al-Marzoqi AH, Zahraa MA, Shalan AA: Molecular and phenotypic study of virulence genes in a pathogenic strain of Pseudomonas aeruginosa isolated from various clinical origins by PCR: profiles of genes and toxins. Res J Pharm, Biol Chem Sci 7, 590–598 (2016) [Google Scholar]
- 3.Streeter K, Katouli M: Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect Epidemiol Med 2, 25–32 (2016) [Google Scholar]
- 4.Mitov I, Tanya S, Boyka M: Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz J Microbiol 41, 588–595 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khattab MA, Nour MS, ElSheshtawy NM: Genetic identification of Pseudomonas aeruginosa virulence genes among different isolates. J Microb Biochem Technol 7, 274–277 (2015) [Google Scholar]
- 6.Amazian K, Rossello J, Castella A, Sekkat S, Terzaki S, Dhidah L: Prevalence of nosocomial infections in 27 hospitals in the Mediterranean region. East Mediterr Health J 16, 1070–1078 (2010) [PubMed] [Google Scholar]
- 7.AL-Kadhmi NA, AL-Thwaini AN, AL-Turk WA, AL-Taif KI: Studies on the multidrug resistance to Pseudomonas aeruginosa isolated from infected wounds. Int J Curr Microbiol App Sci 5, 963–970 (2016) [Google Scholar]
- 8.Kakupa DK, Muenze PK, Byl B, Wilm MD: Study of the prevalence of nosocomial infections and associated factors in the two university hospitals of Lubumbashi, Democratic Republic of Congo. Pan Afr Med J 24, 275–280 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bricha S, Ounine K, Oulkheir S, Haloui N, Attarassi B: Virulence factors and epidemiology related to Pseudomonas aeruginosa. Tunis J Infect Dis 2, 7–14 (2009) [Google Scholar]
- 10.Minchella A, Molinari L, Alonso S, Bouziges N, Sotto A, Lavigne JP: Evolution of antimicrobial resistance against Pseudomonas aeruginosa in a French university hospital between 2002 and 2006. Pathol Biol (Paris) 58, 1–6 (2010) [DOI] [PubMed] [Google Scholar]
- 11.Vitkauskienė A, Skrodenienė E, Dambrauskienė A, Macas A, Sakalauskas R: Pseudomonas aeruginosa bacteremia: resistance to antibiotics, risk factors, and patient mortality. Medicina (Kaunas) 46, 490–495 (2010) [PubMed] [Google Scholar]
- 12.Bhattarai RR, Nawaz MA: Isolation and identification of Yersinia and Pseudomonas sp. from Australian milk and salad using 16s rDNA. Peer J Pre Prints 7, 1–7 (2015) [Google Scholar]
- 13.Virupakshaiah DBM, Hemalata VB: Molecular identification of Pseudomonas aeruginosa from food borne isolates. Int J Curr Microbiol App Sci 5, 1026–1032 (2016) [Google Scholar]
- 14.Nikbin V, Aslani M, SharafiZ Hashemipour M, Shahcheraghi F, Ebrahimipour G: Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iranian J Microbiol 4, 118–123 (2012) [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta SK, Berk RS, Masinick S, Hazlett LD: Pili and lipopolysaccharide of Pseudomonas aeruginosa bind to the glycolipid asialo GM1. Infect Immun 62, 4572–4579 (1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asikyan ML, Kus JV, Burrows LL: Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J Bacteriol 190, 7022–7034 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holloway BW, Römling U, Tümmler B: Genomic mapping of Pseudomonasaeruginosa PAO. Microbiology 140, 2907–2929 (1994) [DOI] [PubMed] [Google Scholar]
- 18.Ghanbarzadeh ZC, Khorshidi A, Firoozeh F, Aznaveh AM, Akbari H: Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol 8, e22345 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawa T, Shimizu M, Moriyama K, Wiener-Kronish JP: Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review. Critical Care 18, 1–11 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stirling FR, Cuzick A, Kelly SM, Oxley D, Evans TJ: Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol 8, 1294–1309 (2006) [DOI] [PubMed] [Google Scholar]
- 21.Saliba AM, Nascimento DO, Silva MC, Assis MC, Gayer CR, Raymond B, Coelho MG, Marques EA, Touqui L, Albano RM, Lopes UG, Paiva DD, Bozza PT, Plotkowskiet MC: Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cellular Microbiol 7, 1811–1822 (2005) [DOI] [PubMed] [Google Scholar]
- 22.Lee VT, Smith RS, Tummler BLS: Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect Immun 73, 1695–1705 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann A, Lüth J, Pauli U: Quantitative and qualitative evaluation of nine different extraction methods for nucleic acids on soya bean food samples. Z Lebensm Unters Forsch 207, 81–90 (1998) [Google Scholar]
- 24.Franzetti L, Scarpellini M: Characterization of Pseudomonas spp. isolated from foods. Annu Microbiol 57, 39–47 (2007) [Google Scholar]
- 25.Ait-Tayeb L, Ageron E, Grimont F, Grimont PAD: Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res Microbiol 156, 763–773 (2005) [DOI] [PubMed] [Google Scholar]
- 26.De Jonghe V, Coorevits A, Van Hoorde K, Messens W, Van Landschoot A, De Vos P, Heyndrickx M: Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl Environ Microbiol 77(2), 460–470 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, Quentin R: Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol 53, 73–81 (2004) [DOI] [PubMed] [Google Scholar]
- 28.Strateva T. (2008): Microbiological and molecular-genetic investigations on resistance mechanisms and virulence factors in clinical strains of Pseudomonas aeruginosa. Sofia, Bulgaria, PhD Dissertation, Medical University of Sofia, Bulgaria, p. 210 [Google Scholar]
- 29.Mehri I, Turki Y, Daly I, Rjab AB, Hassen A, Maher G: Molecular identification and assessment of genetic diversity of fluorescent Pseudomonads based on different polymerase chain reaction (PCR) methods. Afr J Microbiol Res 7, 2103–2113 (2013) [Google Scholar]
- 30.Mulet M, Lalucat J, Garcia-Valdés E: DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12, 1513–1530 (2010) [DOI] [PubMed] [Google Scholar]
- 31.Whitney J C, Whitfield GB, Marmont LS, Yip P, Neculai AM, Lobsanov Y D, Robinson H, Ohman DE, Howell PL: Dimeric c-di-GMP is required for posttranslational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem 290, 12451–12462 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O: Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35, 322–332 (2010) [DOI] [PubMed] [Google Scholar]
- 33.Barker AP, Vasil AI, Filloux A, Ball G, Wilderman PJ, Vasil ML: A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol Microbiol 53, 1089–1098 (2004) [DOI] [PubMed] [Google Scholar]
- 34.Jackson AA, Gross MJ, Daniels EF, Hampton TH, Hammons JH, Vallet-Gely I: Anr and its activation by PlcH activity in Pseudomonas aeruginosa host colonization and virulence. J Bacteriol 195, 3093–3104 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le-Berre R, Nguyen S, Nowak E, Kipnis E, Pierre M, Quenee L, Ader F, Lancel S, Courcol R, Guery BP, Faure K: Relative contribution of three main virulence factors in Pseudomonas aeruginosa pneumonia. Crit Care Med 39, 2113–2120 (2011) [DOI] [PubMed] [Google Scholar]
- 36.Lu Q, Philippe E, Charles-Edouard L, Michel W, Michael T, Emmanuelle M, Jorge G, Pierre-François L, Holger K, Bruno F, Verena G, Erkan M, Antonio P, Hedvika L, Chastre J, Rouby JJ: Pseudomonas aeruginosa serotypes in nosocomial pneumonia: prevalence and clinical outcomes. Critic Care 18, R17 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Q, Rouby JJ, Laterre PF, Eggimann P, Dugard A, Giamarellos-Bourboulis EJ, Mercier E, Garbino J, Luyt CE, Chastre J, Georgescu-Kyburz V, Rudolf MP, Gafner V, Lazar H, Koch H, Perez A, Kramer SD, Tamm M: Pharmacokinetics and safety of panobacumab: specific adjunctive immunotherapy in critical patients with nosocomial Pseudomonas aeruginosa O11 pneumonia. J Antimicrob Chemother 66, 1110–1116 (2011) [DOI] [PubMed] [Google Scholar]


