Abstract
Clozapine is an atypical antipsychotic, that is established as the treatment of choice for treatment-resistant schizophrenia (SCZ). To date, no study investigating comprehensive DNA methylation changes in SCZ patients treated with chronic clozapine has been reported. The purpose of the present study is to reveal the effects of clozapine on DNA methylation in treatment-resistant SCZ. We conducted a genome-wide DNA methylation profiling in peripheral leukocytes (485,764 CpG dinucleotides) from treatment-resistant SCZ patients treated with clozapine (n = 21) in a longitudinal study. Significant changes in DNA methylation were observed at 29,134 sites after one year of treatment with clozapine, and these genes were enriched for “cell substrate adhesion” and “cell matrix adhesion” gene ontology (GO) terms. Furthermore, DNA methylation changes in the CREBBP (CREB binding protein) gene were significantly correlated with the clinical improvements. Our findings provide insights into the action of clozapine in treatment-resistant SCZ.
Keywords: schizophrenia, DNA methylation, clozapine
1. Introduction
Schizophrenia (SCZ) is a mental disorder characterized by symptoms that include delusions, hallucinations, and disorganized speech [1]. Approximately, 30% of SCZ patients are treatment-resistant [2], and the atypical antipsychotic clozapine has become the treatment of choice in this setting [3,4]. Recently, we have demonstrated that cell adhesion molecules, which play an important role in brain development including in axonal/dendrite growth, synapse formation and plasticity [5], were potential candidates for the molecular basis of clozapine response by conducting gene expression profiling using induced pluripotent stem (iPS) cell-based technology [6]. However, the molecular and epigenetic mechanisms underlying the therapeutic efficacy of clozapine have not yet been fully elucidated.
DNA methylation is an epigenetic modification that plays a critical role in brain function [7,8]. DNA methylation mainly occurs at the 5′ position of the cytosine base followed by a guanine base that is called CpG [9]. A number of studies have demonstrated aberrant DNA methylation in SCZ [10,11,12,13,14,15,16,17,18]. Furthermore, growing evidence suggests that DNA methylation may be involved in the therapeutic efficacy of atypical antipsychotic drugs [19,20,21,22,23,24]. With respect to clozapine, two animal studies have demonstrated that acute clozapine treatment induces DNA demethylation in the promoters of specific GABAergic and glutamatergic genes [25,26]. However, to our knowledge, no study investigating comprehensive DNA methylation changes in SCZ patients treated with chronic clozapine has been reported.
In the present study, we comprehensively analyzed changes in DNA methylation in peripheral leukocytes from treatment-resistant SCZ patients treated with clozapine in a longitudinal study. Next, we examined the correlation between changes in DNA methylation in response to clozapine and clinical improvements in treatment-resistant SCZ.
2. Results
2.1. Changes in DNA Methylation in Leukocytes after Clozapine Treatment
Of the 350,142 CpG sites analyzed, significant changes in DNA methylation were observed at 29,134 sites when we compared samples from 21 patients collected before and after one year of treatment with clozapine. The top 100 CpG sites are shown in Supplementary Table 1. Of the 29,134 CpG sites showing significant differences in methylation, clozapine treatment increased DNA methylation at 13,052 sites (44.8%) and decreased DNA methylation at 16,082 sites (55.2%).
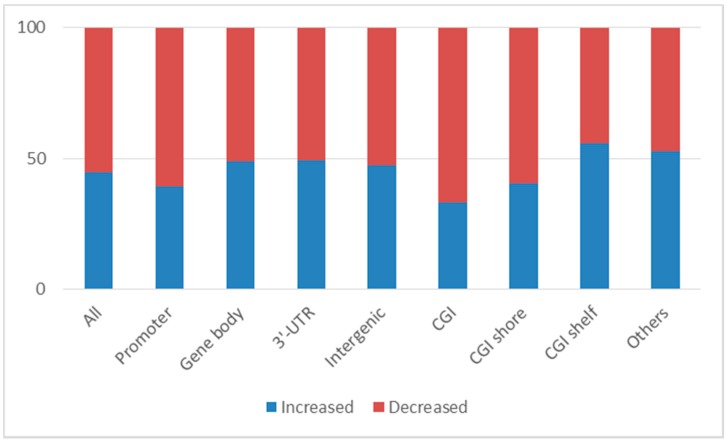
Classification of significant CpG sites based on their locations within genes revealed that 11,850 sites (40.7%) were located in promoter regions; 9479 sites (32.5%) were in gene bodies; 864 sites (3.0%) were in 3′-UTRs; the remainders were found in intergenic regions. Decreases in DNA methylation following clozapine treatment were more likely to occur in promoter regions than in other regions (60.5% in promoter regions vs. 51.6% in other regions; Fisher’s exact test p = 7.00 × 10−16; Figure 1). Classification of CpG sites based on location relative to CpG content in the genes (CpG island (CGI), CGI shore, CGI shelf, and others) revealed that 7656 sites (26.3%) were located within CGIs; 7334 sites (25.2%) were in CGI shores; and 2846 sites (9.8%) were in CGI shelves. Sites of decreased DNA methylation following clozapine treatment were more likely to occur in CGI regions than in other regions (66.9% in CGI regions versus 51.0% in other regions; Fisher’s exact test p = 3.83 × 10−36; Figure 1). Interestingly, significant CpG sites of decreased DNA methylation which were located in CGIs in promoter regions include several GABA, glutamate, and related SCZ susceptibility genes [27], such as GAD1 (glutamate decarboxylase 1), GRIN2A (glutamate ionotropic receptor NMDA type subunit 2A), GRIN2D (glutamate ionotropic receptor NMDA type subunit 2D), and GRM7 (glutamate metabotropic receptor 7).
Figure 1.
The proportions of CpG sites which showed increased or decreased DNA methylation changes after clozapine treatment. Of 29,134 significant CpG sites, clozapine caused an increased DNA methylation at 13,052 sites (44.8%) and decreased DNA methylation at 16,082 sites (55.2%). Of 29,134 significant CpG sites, 11,850 sites (40.7%) were located in the promoter regions (increased DNA methylation: 39.5%, decreased DNA methylation: 60.5%), 9479 sites (32.5%) in gene bodies (increased DNA methylation: 49.1%, decreased DNA methylation: 50.9%), and 864 sites (3.0%) in 3′-UTRs (increased DNA methylation: 47.5%, decreased DNA methylation: 52.5%). Of 29,134 CpG sites, 7656 sites (26.3%) were located in the CGIs (CpG island) (increased DNA methylation: 33.1%, decreased DNA methylation: 66.9%), 7334 sites (25.2%) in CGI shores (increased DNA methylation: 40.3%, decreased DNA methylation: 59.7%), and 2846 sites (9.8%) in CGI shelves (increased DNA methylation: 55.9%, decreased DNA methylation: 44.1%).
A list of the CpG sites located in promoter regions with average DNA methylation differences (Δβ) greater than 0.05 and paired t-test p-values less than 0.001 is shown in Table 1. Two of these CpG sites are located in the TRIM15 (tripartite motif containing 15) gene, which has been implicated in SCZ (Figure 2) [28]. Additionally, gene ontology (GO) analysis revealed that genes with DNA methylation changes following clozapine treatment were enriched for the “cell substrate adhesion” and “cell matrix adhesion” GO terms (False discovery rate (FDR) q < 0.05).
Table 1.
A list of the significant CpG sites with average Δβ > 0.05 and paired t-test p-value < 0.001 in the gene promoter regions. CGI, CpG island; UCSC, University of California Santa Cruz; * Positions refer to Genome Research Consortium human genome build 37 (GRCh37/UCSC human genome 19 (hg19).
| Probe ID | Average Beta Difference before Treatment of Clozapine | Average Beta Difference after Treatment of Clozapine | Average Beta Difference between Treatment of Clozapine | p-Value | Chromosome | Position * | UCSC RefGene Name | UCSC RefGene Group | Relation to UCSC CpG Island |
|---|---|---|---|---|---|---|---|---|---|
| cg15542713 | 0.427 | 0.496 | 0.070 | 1.1 × 10−4 | 1 | 42385581 | HIVEP3 | Promoter | CGI shore |
| cg02772121 | 0.570 | 0.624 | 0.054 | 3.2 × 10−4 | 6 | 30130881 | TRIM15 | Promoter | Others |
| cg10864200 | 0.608 | 0.557 | −0.050 | 7.3 × 10−4 | 4 | 720809 | PCGF3 | Promoter | CGI shelf |
| cg12422154 | 0.601 | 0.652 | 0.050 | 2.5 × 10−4 | 6 | 30130819 | TRIM15 | Promoter | Others |
Figure 2.
DNA methylation changes following clozapine treatment in the TRIM15 gene. Clozapine caused increased DNA methylation changes at two CpG sites (cg02772121 and cg12422154) in the promoter region of the TRIM15 gene (p = 3.2 × 10−4, and p = 2.5 × 10−4, respectively).
2.2. Correlations between Changes in DNA Methylation in Leukocytes and Clinical Outcomes
Analysis of correlations between changes in psychotic symptoms (% Positive and Negative Syndrome Scale (PANSS)) and clinical variables (clozapine dose, duration of clozapine treatment, age of onset, gender, and use of mood stabilizers) revealed no significant correlations (p > 0.05). Upon analysis of correlations between ∆β-values for the 29,134 CpG sites showing significant changes in DNA methylation after clozapine treatment and % PANSS, a CpG site associated with the CREBBP (CREB binding protein) gene, cg05151055, was the only site that was significantly correlated with changes in psychotic symptoms (FDR q < 0.05; Figure 3).
Figure 3.
The correlation between beta difference of DNA methylation changes in the CREBBP gene and % Positive and Negative Syndrome Scale (PANSS) changes. X axis represents beta difference of DNA methylation in the CREBBP gene (cg05151055). Y axis represents % PANSS changes. Each dot represents samples. Significant negative correlation between beta difference of DNA methylation in the CREBBP gene and % PANSS changes was observed (p = 2.7 × 10−7).
3. Discussion
To the best of our knowledge, this study represents the first comprehensive analysis of the effects of clozapine treatment on DNA methylation in leukocytes from treatment-resistant SCZ patients. We identified 29,134 CpG sites that showed significant changes in DNA methylation following chronic clozapine treatment. The proportion of CpG sites with decreased DNA methylation was higher than the proportion of sites with increased DNA methylation after clozapine treatment (55.2% vs. 44.8%, respectively). Consistent with this finding, we previously demonstrated that DNA hyper-methylation patterns frequently occurred in medication-free patients with SCZ, while hypo-methylation was more common in patients with SCZ treated with antipsychotics [12,14]. We also found that decreased DNA methylation following clozapine treatment was more likely to occur at CpG sites located in CGIs in gene promoter regions. This trend is consistent with results reported in a previous study of the effects of blonanserin, another atypical antipsychotic [22].
In this study, we found that genes that showed DNA methylation changes following clozapine treatment were enriched for the GO terms “cell substrate adhesion”, which is essential for cells to interact with their environments [29], and “cell matrix adhesion”, which is essential for cell migration, tissues organization, and differentiation [30]. Similarly, in a recent analysis of differential gene expression profiles in neurons from twins with treatment-resistant SCZ who had discordant responses to clozapine using pluripotent stem (iPS) cell based technology, we found that differentially expressed genes were enriched for “cell adhesion” and “biological adhesion” [6]. Cell adhesion is essential for forming tissue and neuronal connections crucial for nervous system development [31]. Animal studies suggest that dysfunction of neuronal cell adhesion molecules, which are expressed primarily in the central nervous system where they regulate synaptic signaling, may lead to impairment of memory and learning [32,33]. Furthermore, pathways related to cell adhesion have been shown to contribute to SCZ susceptibility in a genome-wide association study [34], and increased blood levels of neuronal cell adhesion molecules have been observed in SCZ [35,36]. These results suggest that changes of neuronal cell adhesion-related molecules may be implicated in the pathophysiology of SCZ, and clozapine may exert its therapeutic effects by altering DNA methylation of genes encoding these molecules.
We found that increases in DNA methylation of the CREBBP gene following clozapine treatment was significantly correlated with clinical improvements in treatment-resistant SCZ. This result suggests that epigenetic modification of the CREBBP gene in peripheral leukocytes can predict clinical responses to clozapine in treatment-resistant SCZ. CREBBP is a protein that possesses intrinsic histone acetyltransferase activity, in addition to acting as a scaffold to stabilize protein interactions within the transcription complex [37]. Pathway analyses of results from genome-wide association studies have demonstrated that this gene is associated with SCZ [38,39]. Furthermore, CREBBP variants have been associated not only with clinical symptoms, but also with cognitive phenotypes in SCZ [36,40].
Our study has several limitations that should be noted. First, our sample size was small, and studies that replicate our findings in a larger cohort are needed. Second, patients enrolled in this study had been treated with various kinds of antipsychotics prior to treatment with clozapine. Third, the dosage and duration of clozapine treatment were not uniform across the patient cohort. Fourth, it is unclear whether changes in DNA methylation following clozapine treatment are normalized to control levels due to a lack of control data. Fifth, it is also unclear whether the DNA methylation changes following the treatment are specific to clozapine or are common among other antipsychotics. Direct comparisons to previous studies that examined the effects of antipsychotics on DNA methylation are hampered by the use of different study designs and analysis of different tissues [19,20,21,22,23,24]. Further blood studies using other antipsychotics are needed to reveal differences between clozapine and other antipsychotics. Sixth, we did not investigate the relationship between DNA methylation and expression. Further functional studies, including transcriptome analysis and cell adhesion assays, are required to clarify the molecular mechanisms of clozapine action. Finally, it is important to note that the observed changes in DNA methylation after clozapine treatment were analyzed only in peripheral leukocytes, and studies in brain tissue are needed to confirm the mechanistic interpretation of our results.
4. Materials and Methods
4.1. Subjects
Twenty-one patients with SCZ (mean age: 42.1 ± 11.4 years; eight males and 13 females) were recruited from Kochi, Tokushima, and Osaka University Hospitals in Japan. All subjects were of Japanese origin and had been treated with various antipsychotic drugs before treatment with clozapine. Peripheral blood was collected twice from each patient, just before introduction of clozapine and after one year of treatment. The mean dose of clozapine was 473.8 ± 91.3 mg/day, and the mean duration of clozapine treatment was 340.8 ± 182.7 days. The psychotic symptoms of the patients were evaluated using PANSS at the time of peripheral blood sample collection. The mean PANSS score was 113.4 at baseline and 91.7 at the end of treatment, respectively. The clinical characteristics of the patients are summarized in Supplementary Table 2. SCZ was diagnosed according to DSM-IV criteria by at least two expert psychiatrists on the basis of extensive clinical interviews and a review of medical records. All patients met the criteria for treatment-resistant SCZ and clozapine administration, as described in the clozapine drug information provided in Japan [41,42]. No psychiatric comorbidities were present in any of the patients. The study protocol was approved by the institutional ethics committee of Tokushima University Graduate School (Project ID, H28-1; Date of approval, 23 June 2016), and all enrolled participants provided their signed, written, informed consent for participation.
4.2. Analysis of DNA Methylation
Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Germantown, MD, USA). Bisulfite conversion of 500 ng of genomic DNA was performed with the EZ DNA methylation kit (Zymo Research, Irvine, CA, USA). DNA methylation levels were assessed using Infinium® HumanMethylation450 BeadChips (Illumina Inc., San Diego, CA, USA), which makes it possible to examine DNA methylation status at more than 485,000 CpG sites, according to the manufacturer’s instructions, and the resulting data was analyzed using the methylation analysis module within the BeadStudio software (Illumina Inc.). The DNA methylation status of CpG sites (termed the β-value) was calculated based on the ratio of the signal from a methylated probe relative to the sum of the signal from both methylated and unmethylated probes. β values ranged from 0 (completely unmethylated) to 1 (fully methylated). For intra-chip normalization of probe intensities, colored balance and background corrections were performed for every set of 12 samples from the same chip using internal control probes. CpG sites used for statistical analyses met the following criteria: (1) β-values with detection p-values < 0.05; (2) autosomal CpGs, with no missing values in any subjects; (3) no probe single nucleotide polymorphisms (SNPs) with minor allele frequencies ≥5% in the HapMap-JPT population; (4) no probe cross-reactivity; and (5) no SNPs at CpG sites and single-base extension sites detailed in a previous paper [43]. The final data set for peripheral leukocytes included 350,142 sites.
4.3. Statistical Analysis
A paired t-test was used to assess DNA methylation changes following clozapine treatment. p-values < 0.05 were considered statistically significant. Gene ontology analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [44]. An FDR correction was applied at the 0.05 level for multiple comparisons. Pearson’s correlations were performed to assess the effects of clinical variables, including clozapine dose, duration of clozapine treatment, age of onset, gender, and use of mood stabilizers on PANSS percentage changes (% PANSS; defined as (PANSS at the end of the study − PANSS at baseline)/PANSS at baseline). A univariate linear regression model was used to examine the relationship between β-values for probes with significant DNA methylation changes following clozapine treatment and % PANSS. An FDR correction was applied at the 0.05 level for multiple comparisons.
5. Conclusions
We report a comprehensive analysis of DNA methylation changes in peripheral leukocytes from patients with treatment-resistant SCZ following clozapine treatment. We found that genes with clozapine-induced changes in DNA methylation were associated with cell substrate adhesion and cell matrix adhesion. These results provide insight into potential mechanisms of action of clozapine in treatment-resistant SCZ. Further functional studies are needed to clarify the molecular mechanisms of clozapine action.
Acknowledgments
The authors would like to thank all the volunteers who understood our study purpose and participated in this study and the physicians who helped us to collect clinical data and blood samples. The authors would also like to thank Akemi Okada for her technical assistance. This manuscript has been edited by native English-speaking experts from BioMed Proofreading LLC. This work was supported in part by Japan Agency for Medical Research and development, AMED (Hidenaga Yamamori, Masataka Kikuchi, Hitoshi Hashimoto, Ryota Hashimoto, Shusuke Numata, Takanobu Nakazawa and Tetsuro Ohmori), Grant-in-Aid for Young Scientists (B) (No.16K19768) (Makoto Kinoshita), Grant-in-Aid for Scientific Research (C) (No. 16K07222) (Masataka Kikuchi and Akihiro Nakaya), SENSHIN Medical Research Foundation (Shusuke Numata), and the JSPS Program (No. S2603) (Hitoshi Hashimoto).
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/3/632/s1.
Author Contributions
Ryota Hashimoto and Shusuke Numata designed the study; Ryota Hashimoto, Shusuke Numata and Tetsuro Ohmori managed the research; Hidehiro Umehara, Hidenaga Yamamori, Michiko Fujimoto, Makoto Kinoshita, Ryota Hashimoto, Shusuke Numata, Shinji Shimodera, Shinya Watanabe, Tetsuro Ohmori and Yuka Yasuda collected samples and acquired the data; Atsushi Tajima, Issei Imoto and Makoto Kinoshita undertook the statistical analysis; Makoto Kinoshita and Shusuke Numata wrote the draft of this paper; Akihiro Nakaya, Hitoshi Hashimoto, Ryota Hashimoto, Masataka Kikuchi, Takanobu Nakazawa and Tetsuro Ohmori helped to interpret data and edited the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gaebel W., Zielasek J. Schizophrenia in 2020: Trends in diagnosis and therapy. Psychiatry Clin. Neurosci. 2015;69:661–673. doi: 10.1111/pcn.12322. [DOI] [PubMed] [Google Scholar]
- 2.Meltzer H.Y. Treatment-resistant schizophrenia—The role of clozapine. Curr. Med. Res. Opin. 1997;14:1–20. doi: 10.1185/03007999709113338. [DOI] [PubMed] [Google Scholar]
- 3.Kane J., Honigfeld G., Singer J., Meltzer H. Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Arch. Gen. Psychiatry. 1988;45:789–796. doi: 10.1001/archpsyc.1988.01800330013001. [DOI] [PubMed] [Google Scholar]
- 4.Wahlbeck K., Cheine M., Essalim A., Adams C. Evidence of Clozapine’s Effectiveness in Schizophrenia: A Systematic Review and Meta-Analysis of Randomized Trials. Am. J. Psychiatry. 1999;156:990–999. doi: 10.1176/ajp.156.7.990. [DOI] [PubMed] [Google Scholar]
- 5.Corvin A.P. Neuronal cell adhesion genes: Key players in risk for schizophrenia, bipolar disorder and other neurodevelopmental brain disorders? Cell Adhes. Migr. 2010;4:511–514. doi: 10.4161/cam.4.4.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakazawa T., Kikuchi M., Ishikawa M., Yamamori H., Nagayasu K., Matsumoto T., Fujimoto M., Yasuda Y., Fujiwara M., Okada S., et al. Differential gene expression profiles in neurons generated from lymphoblastoid B-cell line-derived iPS cells from monozygotic twin cases with treatment-resistant schizophrenia and discordant responses to clozapine. Schizophr. Res. 2016 doi: 10.1016/j.schres.2016.10.012. in press. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Tang B., He Y., Jin P. DNA methylation dynamics in neurogenesis. Epigenomics. 2016;8:401–414. doi: 10.2217/epi.15.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tognini P., Napoli D., Pizzorusso T. Dynamic DNA methylation in the brain: A new epigenetic mark for experience-dependent plasticity. Front. Cell. Neurosci. 2015;9:331. doi: 10.3389/fncel.2015.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith Z.D., Meissner A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 10.Pidsley R., Mill J. Epigenetic studies of psychosis: Current findings, methodological approaches, and implications for postmortem research. Biol. Psychiatry. 2011;69:146–156. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Nishioka M., Bundo M., Kasai K., Iwamoto K. DNA methylation in schizophrenia: Progress and challenges of epigenetic studies. Genome Med. 2012;4:96. doi: 10.1186/gm397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinoshita M., Numata S., Tajima A., Shimodera S., Ono S., Imamura A., Iga J., Watanabe S., Kikuchi K., Kubo H., et al. DNA methylation signatures of peripheral leukocytes in schizophrenia. Neuromol. Med. 2013;15:95–101. doi: 10.1007/s12017-012-8198-6. [DOI] [PubMed] [Google Scholar]
- 13.Akbarian S. Epigenetic mechanisms in schizophrenia. Dialogues Clin. Neurosci. 2014;16:405–417. doi: 10.31887/DCNS.2014.16.3/sakbarian. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinoshita M., Numata S., Tajima A., Ohi K., Hashimoto R., Shimodera S., Imoto I., Takeda M., Ohmori T. Aberrant DNA methylation of blood in schizophrenia by adjusting for estimated cellular proportions. Neuromol. Med. 2014;16:697–703. doi: 10.1007/s12017-014-8319-5. [DOI] [PubMed] [Google Scholar]
- 15.Numata S., Ye T., Herman M., Lipska B.K. DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front. Genet. 2014;5:280. doi: 10.3389/fgene.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cariaga-Martinez A., Saiz-Ruiz J., Alelú-Paz R. From Linkage Studies to Epigenetics: What We Know and What We Need to Know in the Neurobiology of Schizophrenia. Front. Neurosci. 2016;10:202. doi: 10.3389/fnins.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montano C., Taub M.A., Jaffe A., Briem E., Feinberg J.I., Trygvadottir R., Idrizi A., Runarsson A., Berndsen B., Gur R.C., et al. Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study. JAMA Psychiatry. 2016;73:506–514. doi: 10.1001/jamapsychiatry.2016.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teroganova N., Girshkin L., Suter C.M., Green M.J. DNA methylation in peripheral tissue of schizophrenia and bipolar disorder: A systematic review. BMC Genet. 2016;17:27. doi: 10.1186/s12863-016-0332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melka M.G., Castellani C.A., Laufer B.I., Rajakumar R.N., O’Reilly R., Singh S.M. Olanzapine induced DNA methylation changes support the dopamine hypothesis of psychosis. Mol. Psychiatry. 2013;1:19. doi: 10.1186/2049-9256-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melka M.G., Laufer B.I., McDonald P., Castellani C.A., Rajakumar R.N., O’Reilly R., Singh S.M. The effects of olanzapine on genome-wide DNA methylation in the hippocampus and cerebellum. Clin. Epigenet. 2014;6:1. doi: 10.1186/1868-7083-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melka M.G., Castellani C.A., Rajakumar N., O’Reilly R., Singh S.M. Olanzapine-induced methylation alters cadherin gene families and associated pathways implicated in psychosis. BMC Neurosci. 2014;15:112. doi: 10.1186/1471-2202-15-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murata Y., Nishioka M., Bundo M., Sunaga F., Kasai K., Iwamoto K. Comprehensive DNA methylation analysis of human neuroblastoma cells treated with blonanserin. Neurosci. Lett. 2014;563:123–128. doi: 10.1016/j.neulet.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 23.Melka M.G., Rajakumar R.N., O’Reilly R., Singh S.M. Olanzapine-induced DNA methylation in the hippocampus and cerebellum in genes mapped to human 22q11 and implicated in schizophrenia. Psychiatr. Genet. 2015;25:88–94. doi: 10.1097/YPG.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 24.Sugawara H., Bundo M., Asai T., Sunaga F., Ueda J., Ishigooka J., Kasai K., Kato T., Iwamoto K. Effects of quetiapine on DNA methylation in neuroblastoma cells. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2015;56:117–121. doi: 10.1016/j.pnpbp.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Dong E., Nelson M., Grayson D.R., Costa E., Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc. Natl. Acad. Sci. USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong E., Tueting P., Matrisciano F., Grayson D.R., Guidotti A. Behavioral and molecular neuroepigenetic alterations in prenatally stressed mice: Relevance for the study of chromatin remodeling properties of antipsychotic drugs. Transl. Psychiatry. 2016;6:e711. doi: 10.1038/tp.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherlyn S.Y., Woon P.S., Liu J.J., Ong W.Y., Tsai G.C., Sim K. Genetic association studies of glutamate, GABA and related genes in schizophrenia and bipolar disorder: A decade of advance. Neurosci. Biobehav. Rev. 2010;34:958–977. doi: 10.1016/j.neubiorev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee S., Guha S., Ikeda M., Iwata N., Malhotra A.K., Pe’er I., Darvasi A., Lencz T. Excess of homozygosity in the major histocompatibility complex in schizophrenia. Hum. Mol. Genet. 2014;23:6088–6095. doi: 10.1093/hmg/ddu308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coburn L., Lopez H., Caldwell B.J., Moussa E., Yap C., Priya R., Noppe A., Roberts A.P., Lobaskin V., Yap A.S., et al. Contact inhibition of locomotion and mechanical cross-talk between cell–cell and cell–substrate adhesion determine the pattern of junctional tension in epithelial cell aggregates. Mol. Biol. Cell. 2016;27:3436–3448. doi: 10.1091/mbc.E16-04-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berrier A.L., Yamada K.M. Cell-matrix adhesion. J. Cell. Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 31.Nagappan-Chettiar S., Johnson-Venkatesh E.M., Umemori H. Activity-dependent proteolytic cleavage of cell adhesion molecules regulates excitatory synaptic development and function. Neurosci. Res. 2016 doi: 10.1016/j.neures.2016.12.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bukalo O., Fentrop N., Lee A.Y., Salmen B., Law J.W., Wotjak C.T., Schweizer M., Dityatev A., Schachner M. Conditional ablation of the neural cell adhesion molecule reduces precisionof spatial learning, long-term potentiation, and depression in the CA1subfield of mouse hippocampus. J. Neurosci. 2004;24:1565–1577. doi: 10.1523/JNEUROSCI.3298-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoenica L., Senkov O., Gerardy-Schahn R., Weinhold B., Schachner M., Dityatev A. In vivo synaptic plasticity in the dentate gyrus of mice deficient inthe neural cell adhesion molecule NCAM or its polysialic acid. Eur. J. Neurosci. 2006;23:2255–2264. doi: 10.1111/j.1460-9568.2006.04771.x. [DOI] [PubMed] [Google Scholar]
- 34.O’Dushlaine C., Kenny E., Heron E., Donohoe G., Gill M., Morris D., International Schizophrenia Consortium. Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol. Psychiatry. 2011;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 35.Piras F., Schiff M., Chiapponi C., Bossù P., Mühlenhoff M., Caltagirone C., Gerardy-Schahn R., Hildebrandt H., Spalletta G. Brain structure, cognition and negative symptoms in schizophrenia are associated with serum levels of polysialic acid-modified NCAM. Transl. Psychiatry. 2015;5:e658. doi: 10.1038/tp.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aonurm-Helm A., Jaako K., Jürgenson M., Zharkovsky A. Pharmacological approach for targeting dysfunctional brain plasticity: Focus on neural cell adhesion molecule (NCAM) Pharmacol. Res. 2016;113 Pt B:731–738. doi: 10.1016/j.phrs.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Crisafulli C., Chiesa A., Han C., Lee S.J., Shim D.S., Balzarro B., Andrisano C., Sidoti A., Patkar A.A., Pae C.U., et al. Possible influence of CREB1, CREBBP and CREM variants on diagnosis and treatment outcome in patients with schizophrenia. Neurosci. Lett. 2012;508:37–41. doi: 10.1016/j.neulet.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Jia P., Wang L., Meltzer H.Y., Zhao Z. Common variants conferring risk of schizophrenia: A pathway analysis of GWAS data. Schizophr. Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y.H., Kim J.H., Song G.G. Pathway analysis of a genome-wide association study in schizophrenia. Gene. 2013;525:107–115. doi: 10.1016/j.gene.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Need A.C., Keefe R.S., Ge D., Grossman I., Dickson S., McEvoy J.P., Goldstein D.B. Pharmacogenetics of antipsychotic response in the CATIE trial: A candidate gene analysis. Eur. J. Hum. Genet. 2009;17:946–957. doi: 10.1038/ejhg.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamori H., Hashimoto R., Ishima T., Kishi F., Yasuda Y., Ohi K., Fujimoto M., Umeda-Yano S., Ito A., Hashimoto K., et al. Plasma levels of mature brain-derived neurotrophic factor (BDNF) and matrix metalloproteinase-9 (MMP-9) in treatment-resistant schizophrenia treated with clozapine. Neurosci. Lett. 2013;556:37–41. doi: 10.1016/j.neulet.2013.09.059. [DOI] [PubMed] [Google Scholar]
- 42.Yamamori H., Hashimoto R., Ishima T., Kishi F., Yasuda Y., Ohi K., Fujimoto M., Umeda-Yano S., Ito A., Hashimoto K., et al. Changes in plasma d-serine, l-serine, and glycine levels in treatment-resistant schizophrenia before and after clozapine treatment. Neurosci. Lett. 2014;582:93–98. doi: 10.1016/j.neulet.2014.08.052. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.