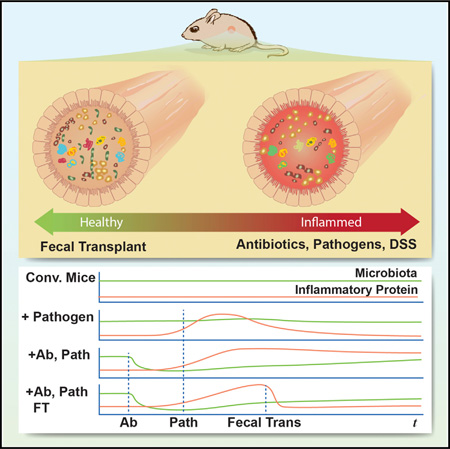
SUMMARY
Improved understanding of the interplay between host and microbes stands to illuminate new avenues for disease diagnosis, treatment, and prevention. Here, we provide a high-resolution view of the dynamics between host and gut microbiota during antibiotic-induced intestinal microbiota depletion, opportunistic Salmonella typhimurium and Clostridium difficile pathogenesis, and recovery from these perturbed states in a mouse model. Hostcentric proteome and microbial community profiles provide a nuanced longitudinal view, revealing the interdependence between host and microbiota in evolving dysbioses. Time- and condition-specific molecular and microbial signatures are evident and clearly distinguished from pathogen-independent inflammatory fingerprints. Our data reveal that mice recovering from antibiotic treatment or C. difficile infection retain lingering signatures of inflammation, despite compositional normalization of the microbiota, and host responses could be rapidly and durably relieved through fecal transplant. These experiments demonstrate insights that emerge from the combination of these orthogonal, untargeted approaches to the gastrointestinal ecosystem.
Graphical abstract
INTRODUCTION
Antibiotic-associated diarrhea occurs in 5%–25% of patients treated with antibiotics and causes thousands of deaths and billions of dollars of additional costs to the health care industry each year (Bergogne-Bérézin, 2000). The general mechanism behind the pathogenesis of these diseases is well understood: broad-spectrum antibiotics deplete the commensal gastrointestinal (GI) microbiota and allow pathogens to colonize and proliferate (Högenauer et al., 1998). Once established in GI niches, pathogens can harm the host through multiple mechanisms. The antibiotic-associated pathogenic bacteria Salmonella tyhpimurium and Clostridium difficile cause GI dysbioses through two distinct mechanisms. Salmonella is an intracellular pathogen, invading intestinal cells via a type-3 secretion system (Ohl and Miller, 2001) and promoting intestinal permeability. In contrast, C. difficile remains extracellular but, instead, secretes toxin proteins that are endocytosed by intestinal epithelia, causing apoptotic cell death (Nam et al., 2010).
Despite these explicit mechanisms of infection, the systems-wide effects on the host and microbiota within the GI tract are poorly understood. Global microbial responses to, and recovery from, antibiotics have been clearly demonstrated using unbiased community profiling approaches (Dethlefsen and Relman, 2011), but without a similarly objective metric to measure host responses, a comprehensive view of the ecosystem has remained out of reach. Anti-microbial peptides (Cash et al., 2006), immunoglobulins (Suzuki and Fagarasan, 2008), and mucin proteins (Martens et al., 2008) all respond to and directly interact with the microbiota, protecting the host from commensal or pathogen outgrowth. The longitudinal and long-term effects of microbiota disturbance, pathogen invasion, intestinal inflammation, and recovery on these and other yet-to-be-discovered interactions remain unknown.
The intestinal microbiota is inherently variable across individuals and between biological states. This variability complicates efforts to draw direct and meaningful comparisons between any two states using genomic technologies (Costello et al., 2009). Conversely, the host genome is essentially invariant over experimental time frames and is generally better defined than microbial metagenomes. Furthermore, proteins expressed by the single host genome stand to directly reflect how changing conditions impact the host and its influence on the microbiome. Thus, focused attention on intestinally expressed host proteins promote robust and readily interpretable comparisons across experimental states. Toward this end, we recently developed an untargeted, mass-spectrometry-based proteomics approach to measure GI host responses to the microbiota (Lichtman et al., 2013). By applying a “host-centric” strategy of stool proteomics to multiple mouse and human microbial states, we showed that the relative abundances of hundreds of host-derived fecal proteins serve as a sensitive fingerprint of gut homeostasis from the perspective of the host. With this method, we also evaluated how wide-ranging changes to the microbiota impacts host physiology across distinct GI tract regions (Lichtman et al., 2015).
In the present study, we applied the “host-centric proteomics of stool” method to fecal specimens collected from mice in a time-course fashion. This experimental model provided a non-invasive avenue for learning how antibiotic disruption impacts host biology and promotes pathogenesis. Our analysis provides orthogonal insights into antibiotic-associated diseases learned from combined host- and microbe-centric analyses. Multi-dimensional host protein profiles provided a high-resolution view of gut perturbation and the dynamics of recovery. Considered in combination with microbial enumeration, they led to the following principal conclusions: (1) an antibiotic cocktail followed by clindamycin treatment induced a far more durable disruption to the host proteome than streptomycin, despite promoting similar global perturbations to the microbiota; (2) the host proteome reflects clear signatures of sub-pathological Salmonella colonization; (3) condition-specific signatures of host inflammation, whether pathogen dependent or independent, are apparent from the host proteome; and (4) inflammatory signatures in host responses persist after pathogen clearance and microbiota recovery yet can be immediately normalized by fecal transplantation. Through these investigations, we demonstrate that the host is an essential and dynamic component in modulating changes to highly complex microbiome systems.
RESULTS
To examine the effects of antibiotics and antibiotic-associated infection on the host-microbiota dynamic, we initiated two distinct infection models in mice, Salmonella typhimurium (Salmonella) and Clostridium difficile (C. difficile) (Figure S1). In these established models, streptomycin promotes the expansion of Salmonella, causing a terminal infection; a cocktail of antibiotics followed by a single dose of clindamycin allows for the emergence of C. difficile, from which the host can recover (Ng et al., 2013). We tracked the microbiota and host proteome of individual mice from their conventional state, through antibiotic treatment, pathogen infection, and, in the case of C. difficile, recovery aided by fecal transplant. In total, we assessed the microbiota and host proteome of 37 animals distributed across 11 conditions and 16 time points. The coincident dynamics of over 100 microbial taxa (operational taxonomic units; OTUs) and 1,425 host proteins allowed us to directly interrogate interdependencies between the host and microbiota.
Differential Antibiotic Recovery Dynamics between Host Proteome and Microbiota
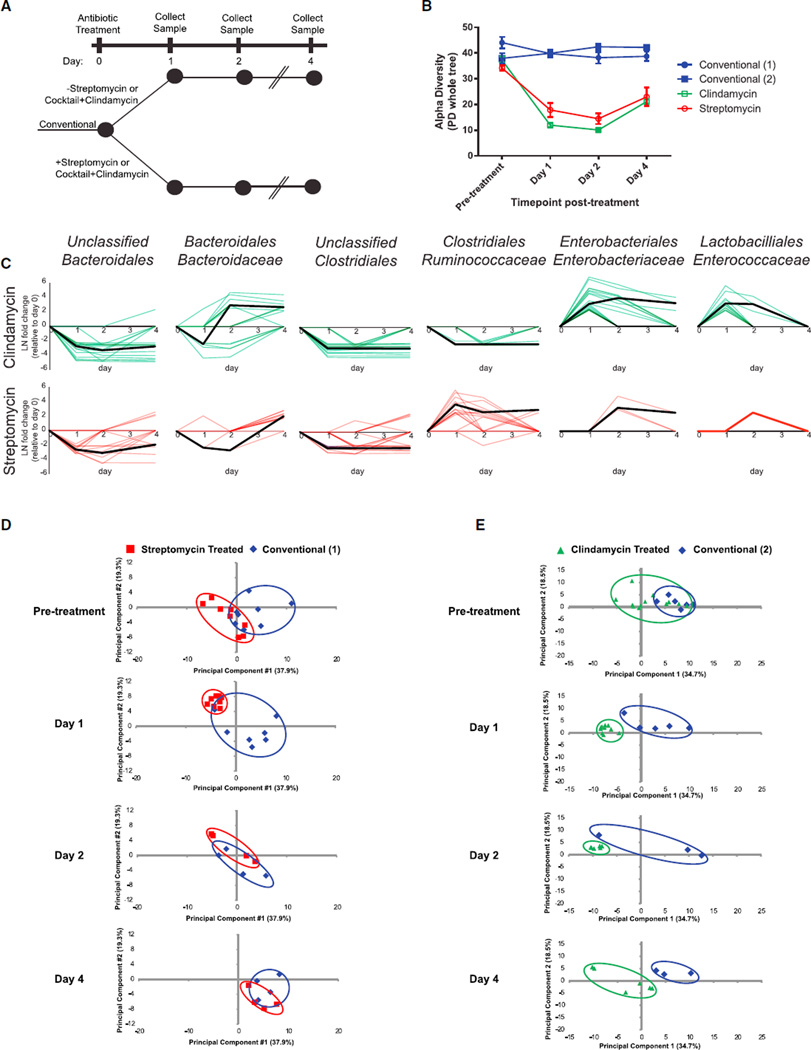
Antibiotics are a necessary step toward initiating many enteric diseases, but even broad-spectrum antibiotics such as streptomycin and clindamycin can have differing effects on microbiota subpopulations (Sekirov et al., 2008). Therefore, we sought to first compare the extent to which these antibiotic treatments caused lasting changes to host and microbiota in ways that could promote pathogen colonization, as measured by 16S rRNA microbial community profiling and host-centric proteomics. Conventional mice were treated with either streptomycin or a combination of an antibiotic cocktail and clindamycin (see Experimental Procedures), and fecal samples were collected for 4 days after treatment (Figure 1A). As expected, alpha diversity, a metric for the complexity of the microbial community, decreased by more than 2-fold in both antibiotic-treated conditions (Figure 1B). In both cases, the diversity of the microbiota remained 2-fold decreased by the fourth day after treatment (Figure 1B), and the microbiota composition remained perturbed as measured by principal-component analysis (PCA) (Figures S2A and S2B). Of 123 significantly changing (adjusted p < 0.01) OTUs grouped by microbial family (Table S1), the two taxa most negatively affected by both antibiotic treatments were undefined members of the Bacteroidales (average, 35-fold) and undefined members of the Clostridiales (average, 19-fold) (Figure 1C). Other taxa were differentially affected by the two antibiotics: 1 day following treatment, streptomycin-treated mice exhibited an average 32-fold increase in Ruminococcaceae, whereas mice treated with clindamycin exhibited a bloom in Enterobaceriaceae (115-fold) and Enterococcaceae (32-fold) (Figure 1C). By 4 days post-treatment, the increases in Ruminococcaceae, Enterobacterieacea, and Enterococcaceae had resolved, returning to their original relative abundances, while the unclassified Bacteroidales and Clostridiales groups remained significantly decreased (Figure 1C). In both treatment conditions, members of the Bacteroidaceae family bloomed on day 4 post-treatment (Figure 1C). These data confirm that several types of bacteria are able to survive antibiotic treatment, which alters their apparent relative abundances over short timescales. Our findings show that these two broad-spectrum antibiotics made similar global alterations to the microbiota but with idiosyncratic opportunistic outgrowths of individual taxa. Next, we examined whether these changes to the microbiota were associated with changes in the host protein expression.
Figure 1. Antibiotic Treatment Elicits Distinct Effects on the Host and Microbiota.
(A) Conventional mice were dosed with one of two antibiotic treatments: streptomycin or a 2-day course of an antibiotic cocktail followed by a single dose of clindamycin. Antibiotic- and vehicle-treated mice were followed for 4 days, and stool samples were collected for 16S rRNA analysis and host-centric proteomics.
(B) Alpha diversity at each time point (mean ± SEM) measured by the PD_whole_tree metric from 16S rRNA sequencing data. Conventional (1) refers to the control mice from the streptomycin experiment, whereas Conventional (2) refers to the control mice from the clindamycin treatment.
(C) OTUs that are significantly affected by antibiotic treatment, grouped by microbial family. Briefly, DESeq was used to identify significantly changing OTUs and their respective fold changes (natural log, LN) when comparing a particular time point with the same mice recorded at day 0. For each comparison, each significantly changed OTU at any time point was plotted as a colored line individual point. Zero values are plotted for insignificantly or unchanged OTUs and time points. Black lines represent the median value of OTU fold changes that significantly deviated from day-0 levels.
(D and E) Principal-component analysis was conducted on all 1,425 host proteins identified across all samples in the (D) streptomycin and (E) clindamycin experiments and plotted by time point. Diminishing numbers of mice exist because mice were put into different arms of the general experimental paradigm (Figure S1).
Host fecal proteomes provided us with an orthogonal dimension for evaluating antibiotic-induced perturbations. We identified an average of 6,616 spectra mapping to 326 proteins per fecal sample collected from each mouse at each time point and treatment condition. Proteins were quantitatively compared between each condition using normalized spectral counts, which span 4 orders of magnitude of dynamic range. These data exhibited distinct temporal dynamics from the microbiota and served as state-specific fingerprints used for PCA. The day following each antibiotic treatment, host proteomes coalesced into a single state, distinct from untreated animals (Figures 1D and 1E). Throughout the 4-day time course, host proteomes measured from clindamycin-treated mice retained proteomic profiles that were distinct from those of untreated mice and consistent with the pronounced perturbation observed on days 1 and 2 (Figure 1E). These dynamics mimic the effects of clindamycin on microbiota complexity (Figure 1B). Conversely, streptomycin-treated mice returned to a proteome state indistinguishable from that of the conventional mice by day 4, even though the microbiota diversity remained depleted (Figures 1B and 1D). The observation that only streptomycin-treated host proteomes recovered to a conventional state while the microbiota remained perturbed led us to investigate the specific proteins that influenced these global PCA signatures.
We examined the proteins that significantly changed within mice before, and 1 day after, antibiotic treatment (false discovery rate [FDR] < 0.05, fold change > 1 ln [natural log]). Although similar numbers of proteins met these criteria from both antibiotic-treatment groups (Figures S2C and S2D), only six (12.5%) overlapped between datasets: two were more abundant in both treatments (transthyretin, chymotrypsin-like elastase family member 2A); two were downregulated in both treatments (chymotrypsinogen B, phospholipase A2); and two were downregulated in streptomycin-treated mice but were upregulated in clindamycin-treated mice (myosin-1a, cytochrome c) (Figure S2E). Phospholipase A2 has known anti-Gram-positive activity (Harwig et al., 1995). Thus, under both treatment conditions in which Gram-positive Clostridiales levels were low, phospholipase A2 would be expected to be downregulated. Clindamycin treatment appears to upregulate a variety of peptidases, as well as proteins involved in actin polymerization. Meanwhile, streptomycin treatment significantly upregulated immunoglobulin-related proteins (i.e., immunoglobulin heavy-chain [IGH] and immunoglobulin kappa locus [IGK]) (p = 8e-3) while decreasing the abundance of proteins involved in T cell activation (DPP4, PIP homolog). Collectively, these data clearly reveal that different antibiotics have distinct, multi-faceted effects on the host intestine. These effects manifest on timescales that correspond with vulnerability to intestinal pathogens.
Dual Host-Microbiota Analyses Identify Unique Signatures of Pathogen-Induced Inflammatory States
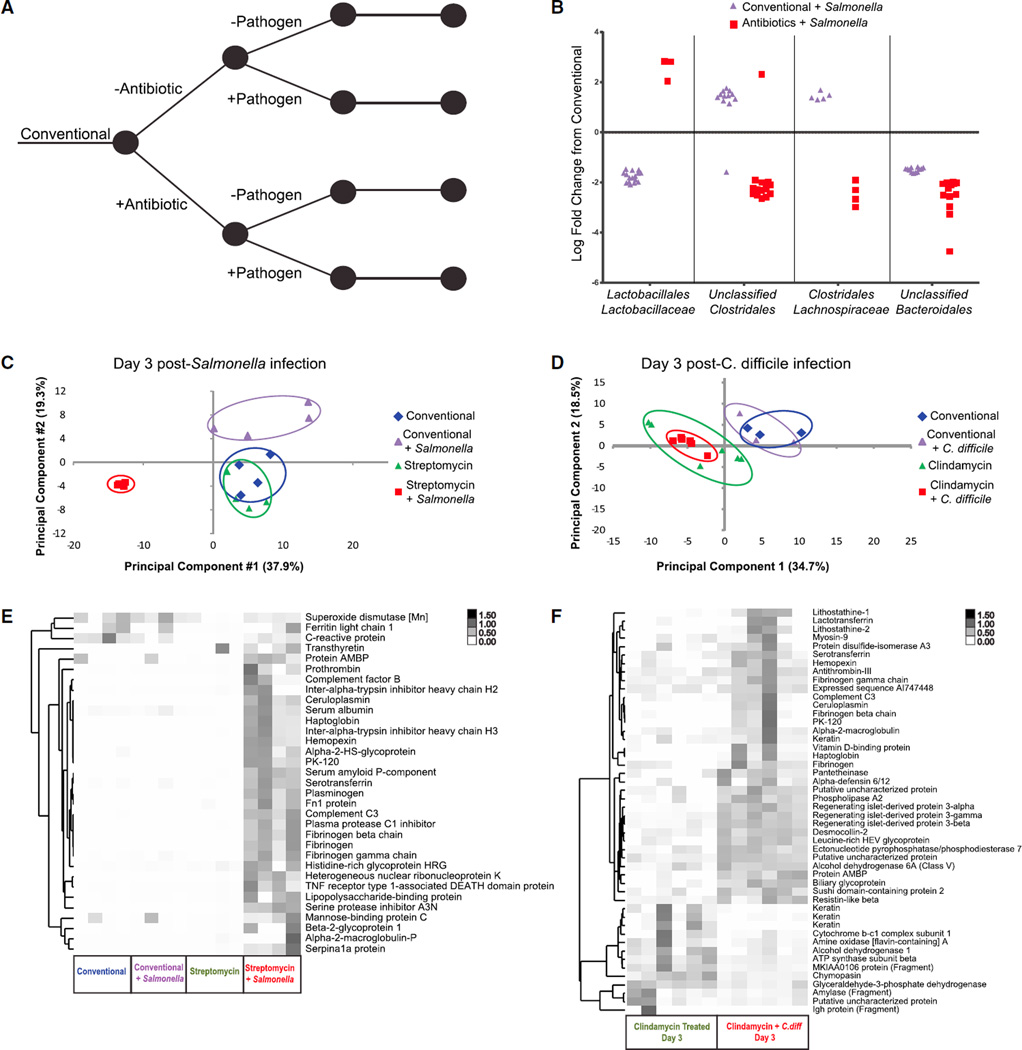
Given the clear effects that antibiotics have on both the microbiota and host, we set out to measure the extent to which pathogens further derange the commensal microbiota as well as host protein expression. Mice treated with antibiotics or vehicle control were subsequently infected with or without pathogen via intragastric gavage (Figure 2A). In both C. difficle and Salmonella treatment groups, mice that were administered antibiotics prior to infection experienced a bloom in the abundance of the respective pathogen. Conversely, pathogen loads remained below detection levels in mice that were inoculated with either pathogen without prior antibiotic treatment, as measured by qPCR (Figures S3A and S3B). Relatively few commensal microbial taxa demonstrated altered abundances in mice inoculated with Salmonella without streptomycin (Figures 2B and S3C; Table S2), whereas no significant changes were evident in mice exposed to C. difficile without prior clindamycin treatment (data not shown). To further test whether these pathogens were able to colonize recipient mice in the absence of prior antibiotic treatment, we performed selective plating from the same stool specimens used in parallel microbe enumeration and proteomic assays. These experiments confirmed extremely low-level Salmonella infection (<104 colony-forming units [CFU] per gram) in mice exposed to the pathogen without antibiotics. No such signal was detected in negative controls or antibiotic-free animals infected with C. difficile.
Figure 2. The Effect of Pathogen Infection on the Host and Microbiota.
(A) Conventional mice were treated with antibiotic (as in Figure 1), 1 day prior to infection with Salmonella (streptomycin treated) or C. difficile (clindamycin treated).
(B) OTUs from four taxonomic groups that are significantly affected by Salmonella infection, grouped by microbial family. DESeq was used to identify significantly changing OTUs and their respective fold changes when comparing infected mice with the same mice in the starting conventional state. For each comparison, each significant OTU was plotted as an individual point. The entire list of significantly changing OTUs can be found in Table S2.
(C and D) Principal-component analysis of (C) Salmonella-infected mice and (D) C. difficile-infected mice and the relevant controls at 3 days post-infection.
(E) Cluster analysis of acute-phase inflammation-induced protein abundances, measured from all mice in the Salmonella experiment on day 3 post-infection.
(F) Cluster analysis of protein abundances determined to be significantly changed (FDR < 0.05, > 1 loge fold change) between clindamycin-treated control mice and clindamycin-treated, C. difficile-infected mice at 3 days post-infection.
Antibiotic-treated, Salmonella-infected mice developed a microbiota that differed from antibiotic-only-treated mice at the same time point. Specifically, whereas streptomycin treatment promoted increases in the Lachnospiraceae and Bacteroidaceae relative abundances (7-fold and 9-fold, respectively; Figure 1C; Table S1), the expansion of these taxa was abrogated by Salmonella infection. Alternatively, Enterobacteriaceae family members were increased in mice with antibiotic-associated infection, ranging from 7- to 367-fold increases (Figure 1B). These data suggest that members of this family, which includes Salmonella, may benefit from niches Salmonella made available as the pathogen achieved greater abundances and induced severe disease, as seen in previous studies (Stecher et al., 2007). These data confirm that discrete signatures of microbiota dysbiosis depend on the levels at which a pathogen colonizes the host.
Considering that Salmonella exposure with and without prior antibiotic treatment induced measurable changes to the microbiota, we investigated whether the same specific microbial taxa responded to these two experimental perturbations and found substantial differences between them. Notably, members of the Clostridales increased in abundance by 5.7-fold in mice infected with Salmonella without streptomycin, whereas they decreased in abundance by 16-fold with antibiotic-associated infection (Figure 2B). Similarly, members of the Lactobacillales decreased in abundance by 12-fold in the mice exposed to Salmonella alone, whereas they increased by 5.5-fold with antibiotic-associated infection (Figure 2B). These data further support a model in which Salmonella infection with prior antibiotic exposure creates a distinct perturbation state relative to infection without antibiotics.
The streptomycin-associated Salmonella infection model used here is particularly severe; the infection becomes systemic, and mice succumb to the disease around 5 days post-infection. From the perspective of the host fecal proteome, the antibiotic-associated Salmonella infection resulted in a highly specific and easily distinguished state (Figure 2C). We identified 225 proteins demonstrating significantly changed fecal levels between this severe perturbation and untreated control mice (Table S3). Acute-phase inflammatory response proteins were highly represented among these induced proteins (14 of the 225; Figure 2E) and included complement C3 and alpha-2-macroglobulin. Meanwhile, the pancreatic enzymes (chymotrypsin-like elastase, carboxypeptidase B1) that were upregulated in the conventional Salmonella infection were significantly downregulated during the antibiotic-associated infection.
It should be expected that the severe antibiotic-associated Salmonella infection model resulted in well-distinguished host protein profiles. However, given that Salmonella numbers were below the level of qPCR detection in infected mice that did not receive antibiotics, it would be reasonable to expect that host proteomic signatures would not be sufficiently sensitive to register differences between these animals and untreated controls. However, by the third day after infection, animals infected with Salmonella without antibiotics presented protein signatures distinct from both untreated and antibiotic-treated controls (Figure 2C). We conclude that host protein profiles were able to sensitively reflect very slight, pathogen-induced shifts in the intestinal microbiome.
As with the microbial taxa described earlier, proteins measured from the host stool were differentially represented between mice that were exposed to Salmonella without prior streptomycin treatment, versus untreated conventional mice (Figure S3D). For example, 3 days post-infection with Salmonella, antibiotic-free mice were found to have upregulated regenerating islet-derived protein 3γ (REG3γ) and pancreatic protease expression, whereas aminopeptidase and brush border proteins were downregulated (p < 0.05; Figure S3D). Furthermore, the large discrepancy that we measured between the host proteomes of mice inoculated to Salmonella with or without prior antibiotic treatment suggests completely distinct host responses to these paradigms of Salmonella infection. Thus, the combination of both host and microbiota measurements provides a highly sensitive measure of infection. To further evaluate this approach, we next investigated the less severe C. difficile infection model for microbial and proteomic signs of dysbiosis.
Consistent with our microbiota measurements, no global proteomic changes were evident from mice infected with C. difficile without clindamycin, providing evidence that, unlike Salmonella, this pathogen did not colonize the gut in the absence of antibiotics (Figure 2D). With qPCR, we verified that C. difficile abundance spikes the first day after antibiotic-associated infection and is undetectable 5 days post-infection (Ng et al., 2013) (Figure S3A). Given the gross pathological impact of this antibiotic-associated infection (Ng et al., 2013), we expected to identify a large increase in inflammatory and anti-microbial proteins commensurate with pathogen levels. We found increased levels of such proteins from infected mice, including serotransferrin, complement C3, and REG3γ. However, we were surprised that their observation lagged 2 days behind emergence of the pathogen perturbation (Figures S3E–S3G). In total, 48 proteins were significantly changed by the antibiotic-associated C. difficile infection on day 3 when compared to antibiotic-treated controls (Figure 2F). Having defined two different antibiotic-associated pathogen-induced disease states from a host protein perspective, we set out to determine whether the host-centric proteomic profiles we generated were microbe specific or simply described a continuum of a universal inflammation response.
Specific and Common Molecular Signatures of Host-Derived Inflammatory States
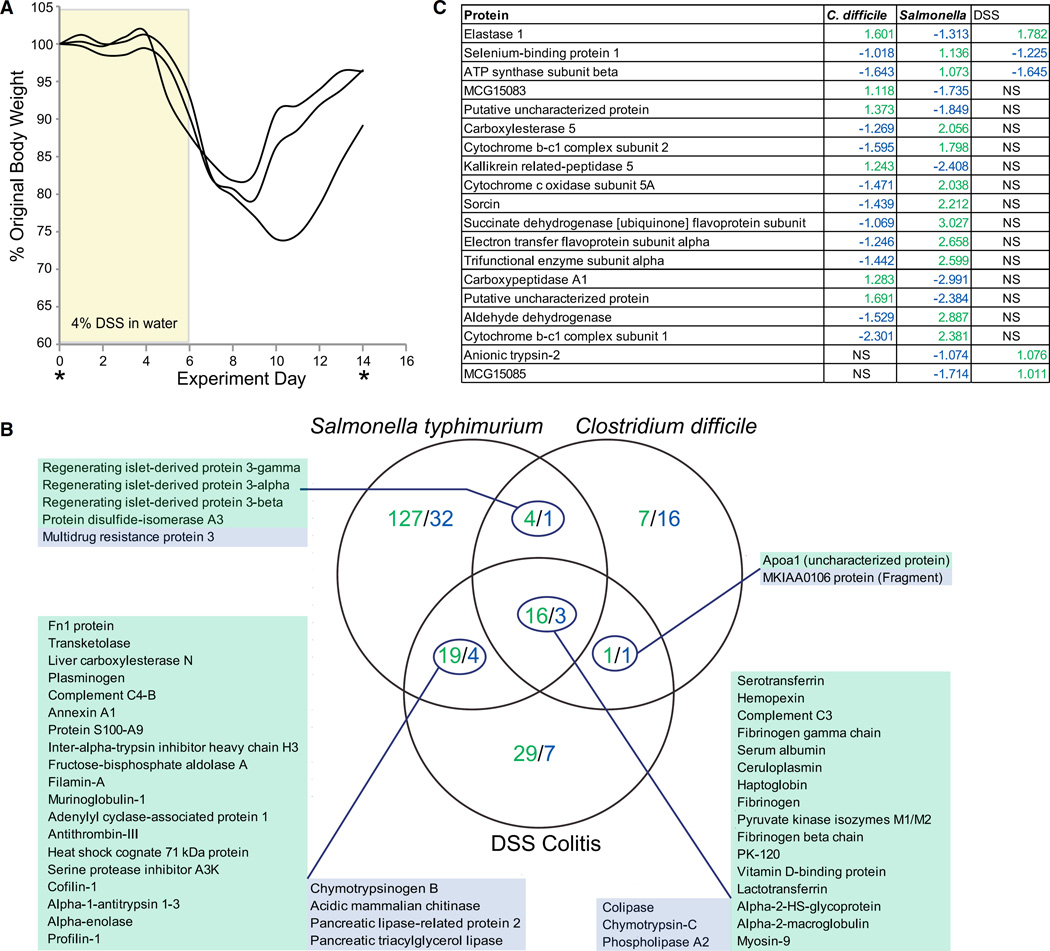
To test the microbe specificity of host-centric proteome responses, we analyzed stool collected from mice experiencing chemically induced intestinal inflammation. Mice were treated with dextran sodium sulfate (DSS) for 6 days (4% in drinking water) and then allowed to recover for 8 days with normal drinking water (Figure 3A). These conditions have been widely used to model chronic colitis (Cooper et al., 1993). Fecal samples were collected prior to treatment and at the end of recovery; 85 proteins were found to significantly differ in their abundances when the two states were compared (Table S4).
Figure 3. Host Proteins Differentiate Inflammatory Conditions.
(A) Experimental model for DSS colitis. Mice were treated with 4% DSS in drinking water for 6 days followed by 8 days on normal drinking water. Samples were taken prior to and at the end of the experiment.
(B) Venn diagram comparing the proteins significantly different (FDR < 0.05, > 1 ln fold change) in abundance when comparing the peak of inflammation with the same mice at their original conventional state. Green indicates upregulated in inflammation, Blue indicates downregulated in inflammation. The degree of overlap in proteins differentially represented between any two conditions, or between all three, was significantly less than would be expected by chance: Salmonella and C. difficile: p = 3e−46; Salmonella and DSS: p = 3e−29; C. difficile and DSS: 5e–22; all three: 3e–33 (chi-square test). Expected numbers of overlapping proteins were calculated from the degree of overlap between all proteins identified in the indicated experimental conditions and the number of proteins found to significantly change the indicated experimental conditions.
(C) Proteins that were identified as significant in more than one inflammatory condition but that were changing in expression in alternate directions and, therefore, did not fit in a particular region of the Venn diagram. NS, not significant.
Protein levels measured at the height of all three inflammation states were compared to protein levels measured from the same mice at the starting conventional state using QSpec (Choi et al., 2008). The resulting values were filtered to include only proteins that significantly (FDR < 0.05) changed by at least 1 nl. The Salmonella infection was the most substantial perturbation, resulting in the greatest number of differentially represented proteins (Figure 3B). The proteins induced across all three inflammation models included many acute-phase response proteins, known to be general markers of inflammation. Only three proteins significantly decreased in all three types of inflammation; all were associated with pancreatic enzymes (colipase, chymotrypsin-C, phospholipase A2). The greatest overlap between any two inflammatory states was observed between Salmonella and DSS, which included the induction of several other acute-phase response proteins and decreased representation of other pancreatic enzymes (Figure 3B). Many of these proteins were similarly regulated in C. difficile-mediated inflammation but did not achieve statistical significance. Likewise, the decrease in the abundance of pancreatic enzymes across all three inflammatory states is consistent with diminished pancreatic function seen in patients suffering from inflammatory bowel disease (Angelini et al., 1988). These data suggest that host-centric proteomics reflects overall disease severity, showing greater similarities between Salmonella and DSS as compared to C. difficile.
Host-centric proteomics also linked the two microbial inflammation models. All three members of the REG3 family of anti-microbial, c-type lectins were identified from Salmonella and C. difficile infections, and not DSS (Figure 3B). These peptides are produced in response to Toll-like receptor stimulation by pathogen-associated molecular patterns (Abreu, 2010). Eighteen proteins were significantly regulated in two or more conditions but in alternate directions (Figure 3C). These proteins are particularly interesting from a diagnostic perspective, as they most greatly distinguish the inflammatory states considered here.
The proteins we identified as being shared across two or more of the experimental models examined here were significantly less than expected (p < 5e-22, chi-square test; Figure 3B). This observation supports the notion that, while “core” inflammation response proteins can be found in the host fecal proteome, they are, nevertheless, a minor component. The majority of inflammation-induced proteins we found were restricted to only one experimental condition. These findings suggest that host-centric proteomic profiles can sensitively distinguish multiple inflammatory conditions in the gut. Salmonella infection uniquely upregulated proteins related to pigment granules and cell death, supporting the induction of apoptosis pathways (Walker et al., 1988). Meanwhile, DSS-induced inflammation led to increased high-density lipoproteins, which were shown to have anti-inflammatory effects in colitis models (De Nardo et al., 2014).
Fecal Transplant Promotes Immediate Host Recovery in a Microbiota-Independent Manner
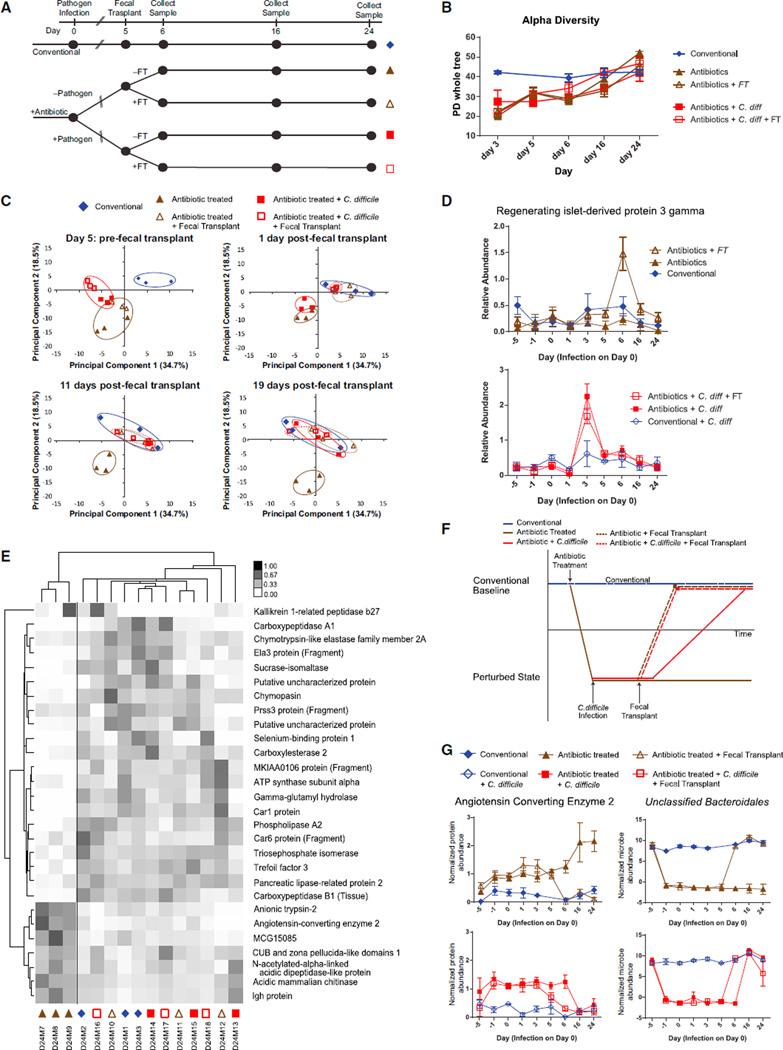
Fecal transplants have been shown to induce dramatic and rapid recovery from dysbiotic states, notably with respect to recalcitrant C. difficile infections (Brandt et al., 2012). Work with defined microbial communities suggest that these benefits are derived by changing community structure (Lawley et al., 2012), but the extent to which the host might also mediate a response to the transplant is not well understood. Given the severity of the antibiotic and inflammatory perturbations we have described thus far, we expected that recovery profiles of the host and microbiota would be linked over time and that a fecal transplant could speed the rates at which both recover. Because antibiotic-associated Salmonella infections were terminal, transient C. difficile infections gave us the opportunity to measure recovery dynamics over longer, 24-day time courses and to further understand the persistent distortion to the host proteome we observed 3 days after clindamycin treatment—even without subsequent C. difficile infection (Figure 2D). C. difficile-infected mice were treated with a fecal transplant or PBS-control gavage 5 days post-infection and then monitored for 19 days. These mice were compared to similarly treated animals that received antibiotics without subsequent C. difficile inoculation (Figure 4A).
Figure 4. Fecal Transplant Aids in the Recovery of the Host in a Microbiota-Independent Fashion.
(A) Clindamycin-treated mice were gavaged with C. difficile or PBS control. Fecal transplants from conventional mice were given to both infected and uninfected mice and all were tracked for 19 more days.
(B) Alpha diversity of mice in each treatment group, starting at 2 days prior to fecal transplant as measured by the PD_whole_tree metric (mean ± SEM).
(C) Principal-component analysis of host proteins, plotted by time point.
(D) Expression profile of REG3γ throughout the course of the entire experiment (mean ± SEM).
(E) Cluster analysis of the proteins that significantly differentiate the antibiotic-treated mice at day 24 from the rest. Symbols under the column refer to the treatment group as indicated in (C).
(F) Model of host recovery as described by the principal-component analysis.
(G) Expression profile of angiotensin-converting enzyme 2 (normalized spectral counts) and an OTU from the Bacteroidales (normalized OTU read counts) throughout the course of the experiment (mean ± SEM).
See also Figures S4 and S5 and Tables S5, S6, and S7.
Prior to receiving fecal transplants on day 5 following C. difficile infection, microbiota diversity of all clindamycin-treated mice had begun to approach that of conventional mice but was still significantly lower (p < 0.05). Interestingly, neither the prior C. difficile infection nor the subsequent fecal transplant had a noticeable effect on the microbiota diversity: all four groups exhibited similar recovery profiles (Figure 4B). By the 11th day following fecal transplant, the microbiota diversity of all four groups had recovered to a point at which they were indistinguishable from conventional mice (p > 0.05) (Figure 4B), but, importantly, several OTUs remained perturbed (Figure S4; Table S5).
The dynamics of the host proteome were found to contrast sharply with microbiota recovery profiles. The host remained in a perturbed state (distinguishable by the first two principal components) through day 5 post-infection. Remarkably, just 1 day after fecal transplant in both conditions (clindamycin with or without C. difficile), all host proteomes returned to a state indistinguishable from that of conventional, untreated animals (Figure 4C). Notably, uninfected mice that received fecal transplants exhibited a spike in anti-microbial protein REG3γ at this early time point, whereas we did not observe a similar, concurrent spike after fecal transplant in the infected mice. We did, however, note a pronounced REG3γ induction immediately after the C. difficile inoculation (Figure 4D; Table S6).
These immediate responses to fecal transplant led us to ask whether the gut proteomes of the remaining perturbed mice would ever return to the unperturbed state. The proteome of C. difficile-infected mice that did not receive fecal transplant returned to a conventional state by the 16th day post-infection. Consistent with prior observations (Buffie et al., 2012), antibiotic-treated mice that never received fecal transplants or C. difficile remained perturbed at all recorded time points until the end of the experiment, 25 days after antibiotic treatment (Figure 4C). By contrast, the two treatments that promoted eventual normalization (fecal transplant or C. difficile infection; Figure 4C) were preceded by a transient but robust inflammatory response, marked by a spike in REG3γ abundance. No such induction was observed in the antibiotic-only treatment group (Figure 4D). Further comparisons between these recovered and non-recovered states could provide insight into the processes required for re-establishing baseline homeostasis. Toward this end, we identified a small set of proteolytic enzymes that discriminated between them (Figure 4E). Three of the seven downregulated proteins in this group have, or are associated with, trypsin-like protease activity, while many of the upregulated proteins have elastase or chymotrypsin activity. This suggested to us that the repertoire of host-secreted proteolytic enzymes could contribute to re-establishing a stable microbiota or at least be further diagnostic of its perturbation.
Lastly, we sought to identify specific host-microbe interactions that may help to describe the unique dynamics of this recovery. We sought specific microbes or proteins that exhibited the same dynamics as seen in the proteome-wide analyses: specifically, (1) a return to a conventional state by day 6 post-infection (day 1 post-fecal transplant) for mice that received a fecal transplant; (2) a return to a conventional state by day 16 post-infection for infected mice not given a fecal transplant; and (3) no observed recovery for antibiotic-treated mice (Figure 4F). Angiotensin-converting enzyme 2 and an unclassified OTU from the order Bacteroidales matched these dynamics (Figure 4G). Of similar interest was a member of the Bacteroides, which did not appear to have a measurable effect on the host but appeared to be permanently excluded from recovery in the un-treated C. difficile infection (Figure S5; Table S7). Thus, our data suggest a connection between specific microbes with specific host responses through combined, untargeted microbiota and host proteome analyses.
DISCUSSION
Antibiotic-associated infections are commonly understood to arise from the outgrowth of pathogenic microbes into newly vacated ecological niches within the GI tract. While the gross effect of these types of infection—intestinal inflammation—involve the induction of host response pathways, the variety of ways in which pro-inflammatory states are initially recognized by the host and the routes by which, and extent to which, healthy states are restored are poorly understood. They are difficult to infer from measured shifts in microbial populations alone and may be impractical to measure directly from intestinal tissue biopsies. Combining microbe enumeration with host-centric proteomics, as demonstrated here, opens a direct avenue for discriminating different inflammation states based on molecular and ecological factors.
We note several surprising outcomes of this multi-omic study. In the model of Salmonella infection, we observed that streptomycin treatment introduced a clear incongruity between the microbiota and host proteome (Figures 1B and 1D). One possible explanation for this observation is that microbial enumeration by 16S rRNA sequencing is more sensitive than our proteome assays. However, even though Salmonella remained below the level of detection of DNA-based assays upon infection without prior antibiotic treatment (Figure S2), we found a global host response to Salmonella as measured from the proteome (Figure 2C). We conclude that, while these assays reflect one another to some extent (Lichtman et al., 2013), they measure distinct and complementary aspects of the intestinal microbiome; therefore, both are needed. We show here that, at least in some respects, the host proteome more sensitively reports nuanced perturbations. For instance, the decrease in the expression of phospholipase A2, a known Gram-positive antibacterial factor, after both antibiotic treatments could be a response by the host to the depletion of Gram-positive microbes such as the Clostridiales (Harwig et al., 1995). Moreover, the general perturbed microbiota state induced by streptomycin alone initiated far less of a host response than the very low-grade Salmonella infection (<104 CFU/g) without antibiotics. This finding supports the notion that pathogen surveillance within the GI tract is both highly sensitive and discriminating (Kufer et al., 2006; Macdonald and Monteleone, 2005).
In the C. difficile model, we show that antibiotic treatment prevented the full recovery of the host proteome, even after microbiota recovery. Interestingly, recovery of the host correlated with the abundance of one unclassified family member of the Bacteroidales: restoration of both appeared to be dependent on exogenous supplementation of the microbiome. Fecal transplant promoted conventional proteome signatures (Figure 4C) and levels of this OTU (Figure 4G) just 1 day after administration (with or without prior C. difficile exposure). C. difficile exposure without subsequent fecal transplant was also sufficient for restoring both proteome and this OTU to conventional states, but in a delayed fashion: Both were synchronized in their recovery by day 16 following pathogen exposure. Without the use of both microbiota and host-focused analyses, these dynamics would remain undetected.
Lastly, we were able to further demonstrate the utility of the host-centric method by identifying proteins that uniquely differentiated inflammatory states from Salmonella, C. difficile, and a chemically induced model of chronic colitis. This experiment was important in demonstrating that inflammation alone was insufficient for producing many of the antimicrobial responses observed under both Salmonella and C. difficile models. The identification of upregulated Reg3 proteins only under these conditions is strong evidence that this method accurately reports host responses to pathogens rather than non-specific signs of inflammation-induced exposure to otherwise commensal microbes. The general decrease in pancreatic enzymes such as transthyretin and chymotrypsin-like elastase (Figure S2) during all three types of inflammation could indicate a microbiota-based effect on pancreatic function. In support of this notion, the microbiota has been previously shown to modulate adaptive and innate immune system pathways in pancreatic disease. One model of antibiotic-induced pancreatitis demonstrated that commensal microbes could contribute to disease symptoms in a NOD1-dependent fashion (Tsuji et al., 2012). Furthermore, both mouse models of diabetes (Sun et al., 2015) and diabetic patients (Brown et al., 2011; de Goffau et al., 2013) demonstrated altered pancreatic output in response to lower levels of butyrate-producing microbes. Extending the untargeted, host-centric proteomic analysis presented here to a broader array of experimental conditions in mice and humans stands to better define the dynamic interactions between microbes and the pancreas in maintaining intestinal homeostasis or promoting disease.
Host-centric proteomics greatly impacts our understanding of the clinically meaningful diseases studied here. Previously, we did not know that antibiotic treatments could have such lasting effects on the host, beyond the recovery of the microbiota, nor did we know that fecal transplantation could have microbiota-independent host effects.
This work highlights the importance of non-invasive, time-course studies of the gut microbiota. Litter and cage effects that could otherwise derail such an unbiased “omics”-based investigation are evident in the results but mitigated by tracking individual mice over the course of an experiment. Comparing a perturbed state to a conventional state of the same mice diminishes the biological variability and emphasizes the proteins or microbes most important to that particular state. Conducting both of these analyses from the same fecal pellet also aids in eliminating known temporal variability (Costello et al., 2009).
We envision that the host-microbiota-monitoring approach described here can be further optimized in several ways. Since next-generation sequencing and mass spectrometry are concentration-dependent techniques, any biases toward more abundant species or proteins will be mitigated as these methods’ sensitivity continue to improve. Likewise, improvements to the quantitative accuracy of both methods will enhance the ability to distinguish small but meaningful changes in abundance. Nevertheless, statistical methodologies have been developed to address current challenges these types of data (Anders and Huber, 2010; Choi et al., 2008); the results presented here speak to the utility of combining these two methods. We also recognize the utility of these data in answering many questions that we have not pursued and, therefore, have made the data publicly available.
These host-based inflammatory fingerprints have obvious utility for the diagnosis of GI diseases in clinical settings. By extending the methods we have demonstrated in mouse models to patients, we expect that host-centric proteomics of stool will open new insights into the myriad microbial, dietary, genetic, and environmental triggers of inflammatory conditions in the gut. Disease-specific proteins identified with this approach can be further applied toward developing targeted therapeutics and companion diagnostics to these specific causes. Because these analyses are untargeted, the approach we describe here can extend beyond innate inflammation to include metabolic diseases, obesity, and cancer.
Multi-omic, non-invasive, time-course experiments become even more critical as studies move to include human patients for whom biological variability increases and individuals are limited. These techniques, combined with the exciting developments in metabolomics, will enable new dimensions for understanding of the complex ecosystem that exists in our GI tract. It is only with this understanding that we will succeed in developing novel therapeutics and diagnostics for the growing number of microbiota-associated diseases.
EXPERIMENTAL PROCEDURES
Mouse Husbandry
All experiments were conducted according to the Administrative Panel on Laboratory Animal Care (A-PLAC), the Stanford University Institutional Animal Care and Use Committee (IACUC).
Salmonella typhimurium Model
Conventionally raised, female, Swiss-Webster mice (6–10 weeks old) were separated into four cages of four mice each, and tails were marked to identify each mouse. Two groups of mice were administered 20 mg streptomycin by oral gavage, while the other two groups received vehicle control. After 24 hr, one group of streptomycin-treated mice and one group of untreated mice received 108 CFUs/ml Salmonella typhimurium via oral gavage from overnight cultures. Mice were separated into different isolators dependent on Salmonella status in order to prevent accidental contamination. Mice were monitored for 3 days post-infection and sacrificed on day 3 when antibiotic+/Salmonella+ mice became too sick. Fecal samples were taken before antibiotic treatment, before Salmonella administration, and at days 1 and 3 post-infection. See also Figure S1 for the treatment timeline.
Clostridium difficile Model
Conventionally raised, male, Swiss-Webster mice (6–10 weeks old) were separated into six cages of three mice each, and tails were marked to identify each mouse. Four groups received an antibiotic cocktail in their water consisting of kanamycin (0.4 g/l), gentamycin (0.035 g/l), colistin (0.057 g/l), metronidazole (0.215 g/l), and vancomycin (0.045 g/l) for 2 days. These mice were then placed back onto normal water for 2 days prior to a single dose of 1 mg clindamycin via oral gavage. 24 hr post-clindamycin treatment, two groups of antibiotic-treated mice and one group of untreated mice were administered 108 CFUs of Clostridium difficile via oral gavage from overnight cultures. Mice were separated into isolators based on C. difficile status in order to prevent cross-contamination by pathogen spores. 5 days post-infection, fecal transplants were conducted on mice from two groups, antibiotic+/C. difficile+ and antibiotic+/C. difficile−. Fecal samples from the fully conventional mice were taken fresh and suspended in PBS, and 200 µl were administered via oral gavage. Fecal samples were collected from all mice prior to antibiotic cocktail, prior to clindamycin treatment, prior to infection, and at days 1, 3, 5 (day of fecal transplant), 6, 7, 10, 16, and 24 post-infection. All mice were sacrificed on day 24 post-infection. See also Figure S1 for the treatment timeline.
DSS Inflammation Model
In order to obtain a chronic inflammation that has been compared to inflammatory bowel disease, mice were treated with DSS over several days and then allowed to recover. While they recovered physically, chronic GI inflammation persisted. Five conventionally raised, Swiss-Webster mice were put onto 4% (w/v) DSS in their drinking water for 6 days, and their weight was monitored daily. Two mice (with the greatest and least weight loss) were sacrificed, and the other three mice were placed back onto drinking water. Weight was measured daily for 8 additional days, as mice recovered from an acute colitis to a more chronic one. Mice were tracked via tail mark, and fecal samples were taken at the beginning and end of the experiment. Due to the traumatic nature of the DSS treatment, collecting fecal samples was not possible during the acute inflammation.
Host-centric Proteomics
We conducted fecal proteomics as previously described (Lichtman et al., 2013), with the following notable changes. Instead of analyzing all four fractions (20%, 40%, 60%, and 100% acetonitrile) from the C4 solid-phase extraction chromatography, only the 40% and 60% fractions were digested and analyzed. Each sample was only analyzed with no technical replicates on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). Furthermore, the msConvert program (v3.0.45) was used to generate peak lists from the original data, and spectra were assigned to peptides using the SEQUEST (v28.12) (Eng et al., 1994) algorithm. Spectra were queried against a composite “targetdecoy” sequence database consisting of the mouse proteome (UniProt, downloaded October 30, 2012) and reversed decoy versions of the same sequences (Elias and Gygi, 2007). SEQUEST search parameters included using a semi-specific enzyme specificity, 50 ppm precursor mass tolerance, 1-Da fragment ion mass tolerance, static carbamidomethylation of cysteines, differential oxidation of methionines, and differential deamidation of glutamines and asparagines. Each mass spectrometry run was filtered to a 1% peptide FDR, and the experiment-wide FDR was filtered to 5% using a linear discriminant analysis (Huttlin et al., 2010). All raw data are available on the PRIDE repository (Vizcaíno et al., 2013), with the dataset identifiers PXD003152, PXD003167, PXD003168, PXD003169, PXD003170, and PXD003171.
Spectral counts were normalized as a percentage of the total counts for a given fecal sample. Fold changes and statistical tests were performed using the QSpec software package (Choi et al., 2008). Centroid clustering was conducted on normalized spectral counts after normalizing to the abundance of each protein using Cluster (v3.0) and visualized using Treeview (v1.1.6r4). Principal-component analysis was conducted on normalized spectral counts using the princomp function in MATLAB (v2012a). Proteins were identified as part of particular pathways using Ingenuity Pathway Analysis, and Gene Ontology enrichments were calculated with DAVID (Huang et al., 2009a, 2009b).
16S rRNA Microbial Community Profiling
Fecal DNA was isolated (MoBio PowerFecal Kit) from 1 µl of stool, and amplicons of the V4 region of the bacterial 16S rRNA gene were made using bar-coded forward primers (515f, 806R). Samples were sequenced at the Medical Genome Facility, Mayo Clinic, using the MiSeq platform (Illumina). Sequence quality was evaluated with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and a dynamic trimmer (Cox et al., 2010) was used to remove segments of sequences of poorer quality. Paired-ends were then joined using fastq-join (Aronesty, 2013) and de-multiplexed using QIIME (Caporaso et al., 2010). OTUs were picked against the Greengenes 13.8 release database (http://greengenes.secondgenome.com), using a 97% similarity threshold. In order to capture novel sequences, those sequences that would not cluster were clustered de novo. OTUs were considered if they were represented by at least 0.001% of all sequences (Caporaso et al., 2011). Raw sequence data are available upon request.
Alpha diversity was calculated with QIIME (Qualitative Insights into Microbial Energy) using the PD_whole_tree method. For significance testing, OTUs were removed if they were not present with at least three counts across at least four individual samples. Significance testing was conducted using DESeq software (v1.4.5) (Anders and Huber, 2010) in R (v3.1.1) and filtered to an adjusted p value of <0.01.
qPCR
Total genomic DNA, extracted as described earlier, was diluted to 5 ng/µl by picoGreen assay. Primers to the Salmonella typhimurium ttrA gene and the Clostridium difficile CD2344 gene were used to assay microbial abundance via real-time qPCR.
Supplementary Material
Highlights.
Clindamycin induces lasting effects on both microbiota and host proteome
The host proteome displays clear signatures of Salmonella and Clostridium infections
Pathogen-mediated host responses are distinct from responses to chemical colitis
Fecal transplant induces faster host proteome recovery than microbiota recovery
In Brief.
Lichtman et al. compared longitudinal mouse models of antibiotic-associated inflammation. By concurrently measuring gut microbes and secreted host proteins with 16S rRNA sequencing and mass spectrometry, they found dynamic, yet distinct, microbe and proteome profiles. Inflammation-regulated proteases, antimicrobial proteins, and immunoglobulins marked multiple pathways actively shaping host responses to dysbiosis.
Acknowledgments
We would like to acknowledge all members of the J.E.E. and J.L.S. labs for assistance and discussions throughout this work, especially Steven Higginbottom for his assistance with animal husbandry; Angela Marcobal, Liz Stanley, and Drew Hryckowian for laboratory support; and Carlos Gonzalez for graphical assistance. We would also like to thank the Polk lab for discussions regarding the DSS model, the Monack lab for reagents, and Susan Holmes for discussions regarding statistical analyses. The work was supported by grants from the NIH (R01-DK085025 to J.L.S.), from the National Science Foundation (DGE-114747 to J.S.L.), and from the Bill and Melinda Gates Foundation (Stanford Human Systems Immunology Center to J.E.E.).
Footnotes
ACCESSION NUMBERS
The accession numbers for all raw data (Vizcaíno et al., 2013) are PRIDE: PXD003152 and PRIDE: PXD003167–PXD003171.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.01.009.
AUTHOR CONTRIBUTIONS
J.S.L., J.L.S., and J.E.E., designed the experiments. J.S.L., J.A.F., and K.M.N. conducted the antibiotic-associated disease experiments. J.S.L. conducted all of the sample preparation, sequencing preparation, and mass spectrometry analyses. J.S.L. and S.A.S. analyzed the data, and all authors contributed to preparing the manuscript.
REFERENCES
- Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini G, Cavallini G, Bovo P, Brocco G, Castagnini A, Lavarini E, Merigo F, Tallon N, Scuro LA. Pancreatic function in chronic inflammatory bowel disease. Int. J. Pancreatol. 1988;3:185–193. doi: 10.1007/BF02798930. [DOI] [PubMed] [Google Scholar]
- Aronesty E. Comparison of sequencing utility programs. Open Bioinform. J. 2013;7:1–8. [Google Scholar]
- Bergogne-Bérézin E. Treatment and prevention of antibiotic associated diarrhea. Int. J. Antimicrob. Agents. 2000;16:521–526. doi: 10.1016/s0924-8579(00)00293-4. [DOI] [PubMed] [Google Scholar]
- Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, Stollman N, Rohlke F, Surawicz C. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 2012;107:1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, Orivuori L, Hakala S, Welling GW, Harmsen HJ, Vaarala O. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nardo D, Labzin LI, Kono H, Seki R, Schmidt SV, Beyer M, Xu D, Zimmer S, Lahrmann C, Schildberg FA, et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 2014;15:152–160. doi: 10.1038/ni.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Harwig SS, Tan L, Qu XD, Cho Y, Eisenhauer PB, Lehrer RI. Bactericidal properties of murine intestinal phospholipase A2J. Clin. Invest. 1995;95:603–610. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högenauer C, Hammer HF, Krejs GJ, Reisinger EC. Mechanisms and management of antibiotic-associated diarrhea. Clin. Infect. Dis. 1998;27:702–710. doi: 10.1086/514958. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Banks DJ, Philpott DJ. Innate immune sensing of microbes by Nod proteins. Ann. N Y Acad. Sci. 2006;1072:19–27. doi: 10.1196/annals.1326.020. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JS, Marcobal A, Sonnenburg JL, Elias JE. Hostcentric proteomics of stool: a novel strategy focused on intestinal responses to the gut microbiota. Mol. Cell. Proteomics. 2013;12:3310–3318. doi: 10.1074/mcp.M113.029967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JS, Sonnenburg JL, Elias JE. Monitoring host responses to the gut microbiota. ISME J. 2015;9:1908–1915. doi: 10.1038/ismej.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HJ, Kang JK, Kim S-K, Ahn KJ, Seok H, Park SJ, Chang JS, Pothoulakis C, Lamont JT, Kim H. Clostridium difficile toxin A decreases acetylation of tubulin, leading to microtubule depolymerization through activation of histone deacetylase 6, and this mediates acute inflammation. J. Biol. Chem. 2010;285:32888–32896. doi: 10.1074/jbc.M110.162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 2001;52:259–274. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Furio L, Mecheri R, van der Does AM, Lundeberg E, Saveanu L, Chen Y, van Endert P, Agerberth B, Diana J. Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. 2015;43:304–317. doi: 10.1016/j.immuni.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Fagarasan S. How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol. 2008;29:523–531. doi: 10.1016/j.it.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Watanabe T, Kudo M, Arai H, Strober W, Chiba T. Sensing of commensal organisms by the intracellular sensor NOD1 mediates experimental pancreatitis. Immunity. 2012;37:326–338. doi: 10.1016/j.immuni.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker NI, Bennett RE, Axelsen RA. Melanosis coli. A consequence of anthraquinone-induced apoptosis of colonic epithelial cells. Am. J. Pathol. 1988;131:465–476. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.