Abstract
The ex vivo challenge assay is being increasingly used as an efficacy endpoint during early human clinical trials of HIV prevention treatments. There is no standard methodology for the ex vivo challenge assay, although the use of different data collection methods and analytical parameters may impact results and reduce the comparability of findings between trials. In this analysis, we describe the impact of data imputation methods, kit type, testing schedule and tissue type on variability, statistical power, and ex vivo HIV growth kinetics. Data were p24 antigen (pg/ml) measurements collected from clinical trials of candidate microbicides where rectal (n = 502), cervical (n = 88), and vaginal (n = 110) tissues were challenged with HIV-1BaL ex vivo. Imputation of missing data using a nonlinear mixed effect model was found to provide an improved fit compared to imputation using half the limit of detection. The rectal virus growth period was found to be earlier and of a relatively shorter duration than the growth period for cervical and vaginal tissue types. On average, only four rectal tissue challenge assays in each treatment and control group would be needed to find a one log difference in p24 to be significant (alpha = 0.05), but a larger sample size was predicted to be needed for either cervical (n = 21) or vaginal (n = 10) tissue comparisons. Overall, the results indicated that improvements could be made in the design and analysis of the ex vivo challenge assay to provide a more standardized and powerful assay to compare efficacy of microbicide products.
Keywords: : HIV prevention, bioinformatics, drug discovery, HIV
Introduction
Hiv infects predominantly through the mucosal tissue following sexual intercourse; therefore, the female genital tract (vaginal and cervical) as well as the rectal mucosae have been extensively studied. Consequently, HIV prevention products aim to prevent the sexual transmission of HIV-1. HIV-1 infection of human genital and rectal tract tissues biopsied from individuals following an in vivo HIV-1 prevention regimen has been used as a measure of drug efficacy and is referred to as the “ex vivo challenge assay.” In the ex vivo challenge assay, fresh tissue samples are infected with HIV-1; then, after a washout, HIV-1 growth is monitored in the tissue supernatant for up to 21 days postinfection. Virus levels in the tissue supernatant are tested every 1–4 days, and low or no HIV-1 growth indicates treatment efficacy. There is currently no standard methodology for the ex vivo challenge assay, although this assay is increasingly being used as an exploratory endpoint in phase 1 and 2 clinical trials of candidate microbicides.1–6
HIV-1 infection can be measured with HIV-1 RNA and DNA by real-time polymerase chain reaction7–9 and fixing the tissue for immunohistochemistry to detect p24 expressing cells.10 The p24 antigen release assay has been the most commonly used viral endpoint to measure ex vivo HIV-1 growth in rectal,11 cervical, and vaginal tissue.10 The purpose of this article is to present an interlaboratory retrospective analysis of ex vivo challenge p24 antigen release data to help improve and standardize ex vivo challenge assay methodology to allow for comparisons of treatment efficacy across laboratories, studies, and tissue types.
Analytical measurements, such as the p24 antigen assay, have a limit of detection (LOD) that is the lowest concentration that can be determined to be statistically different from a blank measurement. For some assays, a lower limit of quantification (LLOQ) is used as the lowest concentration measured with precision and accuracy. Values below the LOD or LLOQ are referred to as “left censored,” that is when a measurement below a lower limit is made, the concentration is unknown and reported as either missing or zero. It has been shown that the logic underlying not reporting left-censored values is flawed as distortions in values above the LOD can be worse than those below the LOD.12 In addition, prediction of values below the LLOQ can provide more information than the mere statement that the value is less than the LLOQ.13 Indeed, in the ex vivo challenge assay, left-censored values of low or no virus growth indicate successful virus suppression. As virus growth measurements are log-normally distributed,14 virus growth data are often log transformed to use standard, parametric statistical methods, and therefore, a data imputation method is needed to retain such instances of HIV suppression. Statistical methods that account for left-censored data have been developed particularly for measurement of environmental contaminants15 and virological research.16 Four imputation methods, including simple substitution and more complex model-based approaches,17 were compared here with the goal of finding an optimum method for both ease of application and model fit for the ex vivo challenge assay.
In the ex vivo challenge assay, the frequency and sequence of testing days are not standardized but chosen by the laboratory to capture the time period of likely virus growth. Choice of the frequency and duration of testing days can be based on scientific and logistical factors, especially if the assay is run within the context of a multi-site clinical trial. Ideally, fresh tissue is started in culture shortly after (e.g., 1–2 h) biopsy collection,18 placing the burden of supernatant collection on a local laboratory team. Minimizing the number of testing time points and/or duration of the assay would increase the feasibility of running this assay. In this large retrospective analysis, virus growth kinetics were compared across laboratories and tissue types to determine the active virus growth periods in rectal, vaginal, and cervical tissues during the ex vivo challenge assay.
The statistical power of an experiment is the likelihood that a study will detect a statistically significant effect when there is a true difference. Statistical power is driven by the size of the difference to be detected and the variability in the data. The statistical power of a treatment versus placebo/control comparison using the ex vivo challenge assay is affected by the virus endpoint, number of testing days collected, and variability inherent in the tissue and assay methods used. Variability across different p24 endpoints has been compared. A cross-sectional index calculated from a growth curve reflective of the virus growth achieved in an assay (SOFT), a cumulative p24 endpoint (i.e., sum of all p24 measurements across the duration of the assay), and p24 on specific days were found to provide less measurement variability than the AUC (i.e., area under the virus growth curve) and slope of the virus growth curve.2,14 The cumulative p24 endpoint is now widely used as a readily calculable measurement that captures the total virus growth achieved in an assay.19–23 The number of testing days used will have a direct impact on the value of the cumulative p24 endpoint as more frequent testing over longer assay durations will likely increase cumulative p24 measurements. In addition, variability in the cumulative p24 endpoint may not be equivalent across different tissue types or laboratory methods. The expected difference in cumulative p24 between a treatment and a control condition and the variability in these measurements will have a direct impact on statistical power. Statistical power was compared here for rectal, cervical, and vaginal data to determine the number of tissue samples that would be needed, per treatment group, to find a one log10 difference in virus growth to be statistically significant.
Imputation methods, virus growth kinetics, and statistical power were compared using a multistudy data set compiled from clinical trials of HIV microbicides where p24 measurements were collected during the ex vivo challenge assay.3–6,10,18,24 Only those tissue samples collected from nontreated subjects at baseline or following a placebo treatment were used to provide a large homogeneous data set of nondrug-treated ex vivo human tissues infected with HIV-1BaL at ∼104 TCID50 (50% Tissue Culture Infective Dose; a measure of infectious virus titer).
Materials and Methods
Data Sources and Inclusion Criteria
Data were p24 antigen (pg/ml) measurements collected from early phase clinical trials of candidate microbicides. The p24 antigen assay was used to measure HIV-1BaL concentration postinfection of biopsy tissues (rectal, vaginal, and cervical) during the ex vivo challenge assay. The first ex vivo challenge phase 1 experiment compared two infectious doses of HIV-1 BaL, 102 and 104 TCID50: 100% of biopsies were infected at baseline with the higher titer, whereas only about 60% were infected with the lower titer.3 All data in this study were from tissue samples infected with 104 TCID50. Data were included in this study if the p24 measurements were from the following: (1) a rectal, vaginal, or cervical human tissue biopsy following ex vivo infection with ∼104 TCID50 HIV-1BaL, (2) a phase 1 or 2 clinical trial testing a candidate oral PrEP or microbicide product with in vivo treatment followed by ex vivo challenge assay, (3) fresh (not frozen) tissue samples, and (4) a baseline, placebo, or no treatment condition.
Ex vivo challenge assay
The assay methodologies used for the p24 data sets have been reported in detail elsewhere3,6,10 and are summarized here in Tables 1 and 2. Generally, the ex vivo challenge assays used tissue biopsies collected from human subjects by endoscopy (rectal biopsies) or direct biopsy (cervical or vaginal biopsies). Tissue samples were placed in a medium and transported to a laboratory, and, within 1–3 h following incubation or on ice overnight, samples were infected with HIV-1BaL at ∼104 TCID50 and followed in culture for up to 21 days. During the culture period, supernatants for p24 quantification were collected every 1–4 days. Interleukin-2 (50 U/ml; Roche Life Sciences, Indianapolis, IN) was added to the culture medium for data sets V1, V2, C1, C2, and C4. Concentrations of p24 in the supernatant were quantified using a range of ELISA kits (Table 1) where the LOD or LLOQ was provided by the laboratory or was the lowest nonzero p24 measurement for that study. Successive p24 concentrations (pg/ml), at each time point, were added to calculate cumulative p24 (pg/ml).1 The cumulative p24 endpoint was not intended to be a measure of total p24 in the assay but, instead, a measure of the accumulation of successive p24 concentrations.23,25–27
Table 1.
Ex Vivo Challenge Assay Methods for Rectal (R1–R8), Cervical (C1–C4), and Vaginal (V1–V5) Data Sets: Treatments, Kits, and p24 Testing Days
| Data set | No. of ex vivo samplesa(donors) | Treatment(s) | p24 Kit | p24 Testing days (% missing) |
|---|---|---|---|---|
| R1 | 56 (24) | Baseline, placebo | National Cancer Instituteb | 1 (93), 4 (86), 7 (34), 11 (4), 14 (0) |
| R2 | 112 (48) | Baseline, placebo | National Cancer Institute | 1 (54), 4 (54), 7 (6), 11 (1), 14 (0) |
| R3 | 9 (3) | Baseline | National Cancer Institute | 4 (100), 7 (44), 11 (0), 14 (0) |
| R4 | 30 (10) | Baseline | National Cancer Institute | 4 (90), 7 (40), 11 (10), 14 (0) |
| R5 | 127 (32) | Baseline | Alliancec | 3 (89), 7 (52), 14 (24) |
| R6 | 14 (4) | Baseline | ZeptoMetrixd | 3 (0), 7 (0), 11 (0), 15 (0) |
| R7 | 144 (24) | Placebo | National Cancer Institute | 1 (99), 4 (97), 7 (51), 11 (13), 14 (6) |
| R8 | 10 (5) | Baseline | National Cancer Institute | 4 (40), 7 (0), 10 (0), 14 (0) |
| C1 | 28 (28) | Baseline | Alliance | 4 (0), 7 (0), 11 (0), 14 (0), 17 (0), 21 (0) |
| C2 | 30 (30) | Placebo | AlphaLISAb | 4 (0), 7 (0), 11 (0), 14 (0), 17 (0), 21 (0) |
| C3 | 24 (24) | Baseline | Alliance | 3 (0), 7 (0), 10 (0), 14 (0) |
| C4 | 6 (6) | Placebo | Alliance | 4 (0), 7 (0), 11 (0), 14 (0), 17 (0), 21 (0) |
| V1 | 29 (28) | Baseline | Alliance | 4 (0), 7 (0), 11 (0), 14 (0), 17 (0), 21 (0) |
| V2 | 30 (30) | Placebo | AlphaLISA | 4 (0), 7 (0), 11 (0), 14 (0), 17 (0), 21 (0) |
| V3 | 24 (24) | Baseline | Alliance | 3 (0), 7 (0), 10 (0), 14 (0) |
| V4 | 19 (19) | Baseline | Alliance | 7 (0), 14 (0), 21 (0) |
| V5 | 8 (2) | Baseline | ZeptoMetrix | 3 (0), 7 (0), 11 (0), 15 (0) |
All cultures were nonpolarized. Tissue setup within 1 h with the exception of V4, which was on ice overnight.
700 total tissue samples.
Frederick National Laboratory for Cancer Research, Frederick, MD.
Perkin Elmer, Waltham, MA.
Zeptometrix, Buffalo, NY.
Table 2.
Ex Vivo Challenge Assay Methods for Rectal (R1–R8), Cervical (C1–C4), and Vaginal (V1–V5) Data Sets: Supernatant Volume, HIV-1 BaL Source, Tissue and Forceps Details
| Data set | Supernatant Vol (μl) | HIV-1 BaL source | Mean wt/size (pre/post) | Forceps brand; Manufacturer | Forceps size, mm |
|---|---|---|---|---|---|
| R1a | 400 | NIH, Catalog No. 510 | 20–30 mg (pre) | Radial Jaw 4; Boston Scientific | 3.8 |
| R2a | 400 | NIH, Catalog No. 510 | 20–30 mg (pre) | Radial Jaw 4; Boston Scientific | 3.8 |
| R3a | 500 | Advanced | 15–20 mg (pre) | Radial Jaw 4; Boston Scientific | 2.8 |
| R4a | 500 | Advanced | 15–20 mg (pre) | Radial Jaw 4; Boston Scientific | 2.8 |
| R5a | 500 | Advanced | 15–20 mg (pre) | Radial Jaw 4; Boston Scientific | 2.8 |
| R6a | 200 | NIH | 3 × 3 × 1 mm (pre) | Sarratt; Stericom | 4 |
| R7a | 400 | NIH, Catalog No. 510 | 20–30 mg (pre) | Radial Jaw 4; Boston Scientific | 3.8 |
| R8b | 500 | NIH, Catalog No. 510 | 20–30 mg (pre) | Radial Jaw 4; Boston Scientific | 3.8 |
| C142 | 700 | Advanced | 9–30 mg (post) | Tischler; BD | 2.3 × 4.2 |
| C242 | 700 | Advanced | 9–30 mg (post) | Tischler; BD | 2.3 × 4.2 |
| C3a | 500 | Advanced | 15–20 mg (pre) | Radial Jaw 4; Boston Scientific | 2.8 |
| C442 | 700 | Advanced | 4–13 mg (post) | Tischler; BD | 2.3 × 4.2 |
| V142 | 700 | Advanced | 11–26 mg (post) | Tischler; BD | 2.3 × 4.2 |
| V242 | 700 | Advanced | 11–26 mg (post) | Tischler; BD | 2.3 × 4.2 |
| V3a | 500 | Advanced | 15–20 mg (pre) | Radial Jaw 4; Boston Scientific | 2.8 |
| V443 | 100 | NIH | 30 mg (pre) | Tischler; BD | 3 × 5 |
| V5a | 100 | NIH | 3 × 3 × 1 mm (pre) | Sarratt; Stericom | 4 |
HIV-1 BaL Sources: NIH = NIH AIDS Research and Reference Reagent Program, Bethesda, MD (www.aidsreagent.org/) Advanced = Advanced Biotechnologies, Inc. (https://abionline.com/).
TCID50 titration methods were:
Titrated on activated peripheral blood mononuclear cells. Calculated by Reed-Muench Formula.
Statistical analyses
A total of 17 data sets were integrated using a “transform and recode” process28 where all p24 measurements were transformed into pg/ml units, nontreated samples were defined as either placebo or baseline depending on the study design, assays from the same study but performed at different sites were coded as different data sets, and data were selected only for those ex vivo tissue samples infected with ∼104 TCID50 HIV-1BaL.
Three developmental (A–C) and one commonly used imputation method (“Z”17,29) were used to impute missing, zero, or p24 measurements below the LOD or LLOQ. The commonly used method (“Z”) imputes missing values with either ½LLOQ or ½LOD. The rationale for this imputation is that data below a lower cutoff (i.e., LLOQ or LOD) will have a normal distribution where the mean of the data falls halfway between the cutoff and zero. All developmental models (A–C) included a random effect to account for within-subject repeatability. The first developmental imputation method (“A”) used a nonlinear mixed effect model30–32 so that missing or zero results were imputed with predicted values from the model. Nonlinear mixed effect models have been used to impute left-censored values for longitudinal HIV measures of HIV-infected patients where RNA levels often drop below the LOD with highly active antiretroviral treatments.29 The second developmental imputation method (“B”) took advantage of the virus growth kinetics during active infection where virus replication classically follows a nonlinear s-shaped curve with gradual increase in virus before a vigorous growth phase followed by low or no additional growth.14 In method B, imputation was performed using a nonlinear quadratic fit across days of culture and when enough detectable measurements (>3) were available for curve fitting, followed by method A for those tissue cultures that did not provide enough data to model virus growth curve. The third developmental method (“C”) combined methods A and B in an iterative manner, randomly choosing single data values for imputation with method A after all possible data had been imputed with method B, thus reducing bias introduced into the nonlinear mixed effect model by the order that values were imputed. To assess each method for variability, a summary measure called the sums of squares for imputation (SSI) was used. This is the sum of the squared differences between the measurements (imputed and detectible) and the predicted values from the model fit where a low SSI indicates a better fit.
A nonlinear growth curve [Eq. (1)] was fit to each data set, and the second derivative of each curve33 was used to identify the lower and upper inflection points as the beginning and end of the virus growth period. The virus growth periods were compared across tissue types and data sets.
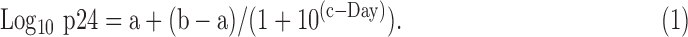 |
The number of tissue samples needed, per treatment group, to find a one log10 difference in cumulative p24 to be significant at alpha = 0.05, with 80% power by t-test, was determined for each data set. Using this approach, a statistically significant one-log reduction in cumulative p24 was found in the ex vivo challenge assay following in vivo use of UC781 2.5% gel, a candidate microbicide3 in the first phase 1 pharmacodynamic study of ex vivo efficacy, and Tenofovir 1% gel,6 in a subsequent phase 1 pharmacodynamic study.
All statistical analyses were performed with SAS® v9.3 (Cary, NC). The SAS program written to perform and compare imputation methods A, B, C, and Z is included in Supplementary Data (Supplementary Data are available online at www.liebertpub.com/aid).
Results
HIV-1BaL p24 antigen measurements were collected from a total of 700 tissue samples (Table 1) where 1–3 biopsy samples, per donor, were entered into the analyses. Data sets were coded for each laboratory and study according to tissue type: rectal (R1–R8), cervical (C1–C4), and vaginal (V1–V5).
Comparison of p24 imputation methods
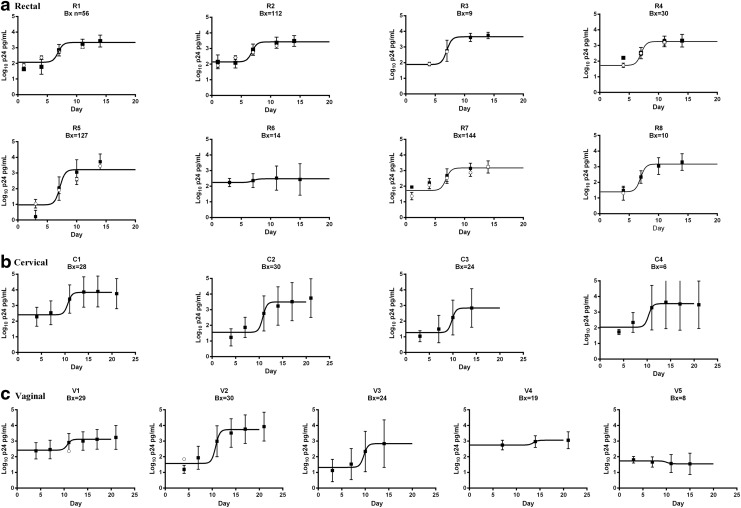
There was a pattern to the missing data where a greater proportion was found for the rectal data sets and at the earlier p24 testing time points (Table 1). Imputation methods A, B, C, and Z were compared for data sets where at least 10 biopsies for ex vivo challenge assays were used, data were left censored or missing, and the assays included at least four p24 testing days, as the models were noncalculable when these requirements were not met. There were seven rectal data sets with missing or left-censored data (R1–R5, R7, and R8; Table 1), and of these data sets, R5 and R8 only collected p24 measurements on three testing days and R3 data set comprised only nine tissue samples (Table 1). Following imputations, data were compared across methods A, B, C, and Z for sets R1, R2, R4, and R7 (Table 3). High SSI values indicated a poorer fit and more variability for imputation method Z compared to methods A–C for bolded values in Table 3. A significant difference between the data sets was found when methods A, B, and C were compared to method Z (p < .0001; Table 3). These results indicate that imputation using methods A–C provided both improved model fit and a different result outcome when compared to imputation method Z. Method A was the preferred method as imputations using this method could be readily calculated in commonly available software. Missing data were imputed with method A for the remainder of the analyses, and the imputed data (open squares; Fig. 1) were mostly within mean ± 1 SD of the nonimputed data (filled squares, error bars; Fig. 1).
Table 3.
Comparison of Imputation Methods for Missing or Nondetected p24 Measurements
| Data imputation methoda | |||||
|---|---|---|---|---|---|
| Data set | Parameter | A | B | C | Z |
| R1b | Difference to Z | <0.0001 | <0.0001 | <0.0001 | NA |
| SSIc | 76 | 91 | 89 | 516 | |
| R2b | Difference to Z | <0.0001 | <0.0001 | <0.0001 | NA |
| SSI | 325 | 434 | 434 | 1223 | |
| R4b | Difference to Z | <0.0001 | <0.0001 | <0.0001 | NA |
| SSI | 41 | 41 | 50 | 173 | |
| R7d | Difference to Z | <0.0001 | <0.0001 | <0.0001 | NA |
| SSI | 208 | 259 | 405 | 1863 | |
Imputation Methods A = missing values were imputed with those predicted from a nonlinear mixed effect model, and B = missing values were imputed with a nonlinear quadratic fit of virus growth across days of culture for assays with >3 testing time points and a nonlinear mixed effect model for assays with ≤3 testing time points. C = missing values were imputed with a combination of methods A and B in an iterative manner, and Z = missing values were imputed with ½LOD.
Differences for A versus B, A versus C, and B versus C nonsignificant.
SSI = sum of squares for imputation. This was the sum of the squared differences between the measurements (imputed and detectible) and the predicted values from the model fit where a low SSI indicates a better fit.
Differences for A versus B and A versus C significant at p < .05, B versus C nonsignificant.
LOD, limit of detection.
High SSI values indicated a poorer fit and more variability for imputation method Z compared to methods A–C for bolded values.
FIG. 1.
Tissue virus growth in ex vivo challenge assays for rectal (a), cervical (b), and vaginal (c) data sets. Log10 p24 means and SD are indicated for nonimputed (filled squares, ±1 SD) and imputed (open circles) data. Missing data were imputed using a nonlinear mixed effect model (Method “A”). The solid black line indicates a nonlinear growth curve model fit to each data set.
Virus growth in the ex vivo challenge assay
Virus growth, as measured by the p24 assay, was modeled with a nonlinear virus growth curve for each data set (Fig. 1). There was considerable similarity in rectal virus growth curves across laboratories and studies where active virus growth reached a p24 level of around 3 log10 (Fig. 1a) and variability (vertical bars) was relatively low across the eight rectal data sets evaluated. Cervical virus growth reached a p24 level between 3 log10 and 4 log10 (Fig. 1b), and variability was markedly greater for the cervical compared to the rectal data sets (i.e., longer error bars). Vaginal tissue virus growth was variable both within and between data sets where virus growth reached between 1 log10 and 4 log10 p24 (Fig. 1c).
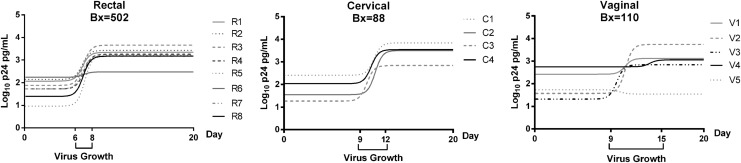
The active virus growth period was defined as the time span between the lower and upper inflection points of each curve (“Virus Growth”; Fig. 2) where the first and last inflection points are indicated as the virus growth duration on the x-axis. The virus growth period for rectal tissues was earlier (days 6–8) and of a relatively shorter duration (3 days total) compared to the cervical (days 9–12) and vaginal (days 9–15; Fig. 2) tissues.
FIG. 2.
Nonlinear growth curve models for rectal, cervical, and vaginal data sets. The first and last inflection points of the nonlinear curves, per tissue type, are indicated on the x-axis as the beginning and end of the virus growth period.
Statistical power of the ex vivo challenge assay
The ex vivo cervical, rectal, and vaginal tissue protocols used various time points for up to 21 days postinfection, several ELISA kits (Table 1), and other within-laboratory potential sources of variability that were not measured but could nonetheless affect statistical power. For example, cumulative p24 would be expected to be higher when more testing time points were used: log10 mean cumulative p24 ranged from 4.13 to 4.57 pg/ml among cervical tissue experiments using the same six time points (C1, C2, and C4; Table 4) but dropped to 3.06 log10 mean cumulative p24 when only four time points were used (C3; Table 4). There was a trend for variability (i.e., log10 SDs) to be lower for the rectal data sets (0.19–0.54) compared to the cervical (0.85–1.30) and vaginal data sets (0.48–1.21; Table 4). The p24 kits used for each data set are listed in Table 1, and there was some evidence for potential differences in p24 results due to kit type: the V1 data set using the Perkin Elmer Alliance kit had a log10 mean cumulative p24 of 3.91 (95% CI 3.7–4.1), but, for the same testing days, the V2 data using the AlphaLISA kit had a log10 mean cumulative p24 of 4.31 pg/ml (95% CI 4–4.6; Table 4). As cumulative p24 could be affected by the number of days and interval of days of virus collection, the duration of the assay, tissue type, p24 kit, and possibly other assay-related factors, a power analysis was run per data set (“N per Group*”; Table 4). Statistical power was determined for comparisons of cumulative p24 between a nondrug-treated condition (placebo and baseline) and an anticipated one log10 change in p24 following an in vivo drug treatment. The number of ex vivo challenge assays that would need to be performed to find a one log10 difference in p24 to be statistically significant was averaged across rectal, vaginal, and cervical tissue data sets (average N per Group in Table 4). On average, only four rectal tissue challenge assays in each treatment and control group would be needed to find a one log10 difference in p24 following a drug treatment when using rectal tissue in the ex vivo challenge assay. This relatively low number of tissue samples needed for 80% power was due to the low variability in the rectal tissue virus growth data (0.19–0.54 log10 SD; Table 4). A larger sample of vaginal ex vivo tissues (n = 10; Table 4) would be needed to find this same one log10 difference in cumulative p24 to be different between a treatment and control condition. The cervical tissue data were the most variable with log10 SDs ranging between 0.85 and 1.3 (Table 4) and this was reflected in the relatively larger number of cervical ex vivo tissue samples (n = 21) that would be needed to find this same one log10 difference to be statistically significant.
Table 4.
Summary Statistics and Power Analysis Results for p24 (pg/ml) Rectal, Cervical, and Vaginal Data Sets
| Tissue | Data set | p24 Sampling days | Biopsy (donor) | Cumulative log10 p24 (pg/mL) | SD | 95% CI | N per groupa |
|---|---|---|---|---|---|---|---|
| Rectal | R1 | 1, 4, 7, 11 and 14 | 56 (24) | 3.77 | 0.27 | 3.7–3.8 | 3 |
| R2 | 1, 4, 7, 11 and 14 | 112 (48) | 3.85 | 0.29 | 3.8–3.9 | 3 | |
| R3 | 4, 7, 11 and 14 | 9 (3) | 4.04 | 0.19 | 3.9–4.2 | 3 | |
| R4 | 4, 7, 11 and 14 | 30 (10) | 3.67 | 0.28 | 3.6–3.8 | 3 | |
| R5 | 3, 7 and 14 | 127 (32) | 3.77 | 0.47 | 3.7–3.9 | 5 | |
| R6 | 3, 7, 11 and 15 | 14 (4) | 3.17 | 0.54 | 2.9–3.5 | 6 | |
| R7 | 1, 4, 7, 11 and 14 | 144 (24) | 3.57 | 0.32 | 3.5–3.6 | 3 | |
| R8 | 4, 7, 10 and 14 | 10 (5) | 3.54 | 0.50 | 3.2–3.9 | 6 | |
| Average | 3.67 | 4 | |||||
| Cervical | C1 | 4, 7, 11, 14, 17 and 21 | 28 (28) | 4.57 | 0.85 | 4.2–4.9 | 13 |
| C2 | 4, 7, 11, 14, 17 and 21 | 30 (30) | 4.13 | 1.15 | 3.7–4.6 | 22 | |
| C3 | 3, 7, 10 and 14 | 24 (24) | 3.06 | 1.08 | 2.6–3.5 | 20 | |
| C4 | 4, 7, 11, 14, 17 and 21 | 6 (6) | 4.32 | 1.30 | 2.9–5.7 | 28 | |
| Average | 4.02 | 21 | |||||
| Vaginal | V1 | 4, 7, 11, 14, 17 and 21 | 29 (28) | 3.91 | 0.48 | 3.7–4.1 | 5 |
| V2 | 4, 7, 11, 14, 17 and 21 | 30 (30) | 4.31 | 0.88 | 4–4.6 | 14 | |
| V3 | 3, 7, 10 and 14 | 24 (24) | 3.18 | 1.21 | 2.7–3.7 | 24 | |
| V4 | 7, 14 and 21 | 19 (19) | 3.48 | 0.40 | 3.3–3.7 | 4 | |
| V5 | 3, 7, 11 and 15 | 8 (2) | 2.32 | 0.43 | 2–2.7 | 5 | |
| Average | 3.44 | 10 |
The number of ex vivo samples needed to provide 80% statistical power to detect a one log10 difference in cumulative p24 between treatment groups at alpha = 0.05.
Discussion
The results of this retrospective analysis provide evidence to support a number of practical guidelines for conducting ex vivo challenge assays related to choice of (1) imputation methods, (2) testing days, and (3) number of tissue samples to be used.
All three novel imputation methods tested here provided an improved model fit compared to the ubiquitous ½LOD type methods. There was a pattern to the missing data, occurring only in the rectal data sets and mostly at the early time points where low or no virus growth had yet occurred. A model-based approach has been recommended for such “missing not at random” data,34 especially when 50% or more data are missing.35 Replacement of nondetected measurements with ½LOD is considered acceptable when less than 15% data are missing.35 Clearly, practical concerns will play an important role when choosing a data imputation method as ease of computation is important for any method that is to be routinely used by a scientific team. The nonlinear mixed effect model was considered the simplest of the novel imputation methods tested, where imputations could be made with commonly available software (please see free imputation software at www.alphastatconsult.com//). Although the imputation methods were tested with p24 assay data sets, the nonlinear mixed effect model here could equally be used to impute missing data collected from other HIV-1 strains or biomedical analytical procedures, such as RNA, cytokine, and chemokine quantification.
Periods of active virus growth were found to vary across tissue types with the rectal tissue providing an early short growth period, cervical tissue providing a later growth period, and vaginal tissue providing a later and longer growth period. The transformation zone is the area between the ectocervix and endocervix, where the epithelium changes from stratified squamous to columnar. While there are resident dendritic cells, macrophage, natural killer cells, and lymphocytes throughout the female genital tract, the transformation zone typically has the highest concentration of immune cells.36 In contrast, the gastrointestinal mucosa contains the majority of the body's CD4+ lymphocyte population and likely represents the largest reservoir of HIV and site of viral replication.37 Rectal subepithelial stromal tissues are densely populated with organized lymphoid tissue, dendritic cells, macrophages, and T cells, all susceptible to HIV infection, whereas female genital tract tissue is less well defined with a higher density of immune cells and cervical columnar epithelial cells that produce mucus and antimicrobial proteins.38 Differences have been found between rectal and genital tissue types in efflux transporter mRNA where OAT1 protein was detected in 100% of rectal tissues but not female genital tissues.39 Such anatomical and functional differences between rectal and female genital sites could account for the differences found here in HIV kinetics, where the rectal tissue displayed rapid viral replication with less variable kinetics compared to cervical and vaginal tissue. Changes in the woman's menstrual cycle, contraception method, and other cervicovaginal factors could impact infectivity in the ex vivo challenge assay.40,41 The finding of differences in ex vivo HIV growth support the standardization of p24 testing days per tissue type to allow direct comparisons in cumulative p24 to be made across studies and testing sites. Defining the active virus growth period and maximum levels of growth obtained using such a large body of data for each tissue type will allow research teams to choose and standardize the p24 testing days to provide a strong and reliable p24 signal in the baseline or placebo conditions to compare to expected HIV suppression in the active drug treatment arms.
Cervical and vaginal tissue assays present more challenges to the researcher due to greater variability in virus replication (cumulative p24) and kinetics. This variability in kinetics for the cervical and most markedly the vaginal tissue may hamper efforts to shorten the duration of this assay with active growth found out to 15 days, although sampling time points beyond 15 days would not be predicted to capture any significant additional growth. This study demonstrates that the variability in the tissue ex vivo virus growth can have a direct impact on the power of placebo-controlled microbicide trials using this exploratory efficacy endpoint. The ex vivo challenge assay has been an exploratory endpoint in phase 1 and 2 clinical trials powered for the primary endpoints of pharmacokinetics, safety, and acceptability, resulting in the inclusion of predominantly males as rectal microbicide products were tested.3,4,6 As the female participants in these studies usually provided both genital and rectal tissue, there can be over twice as many rectal tissue ex vivo data compared to cervical and vaginal tissue, per study. In addition, there are practical limitations to the number of mucosal biopsies that can be collected. Using flexible sigmoidoscopy, it is possible to collect 20–30 mucosal biopsies, and, consequently, there are no limitations on the numbers of biopsies that can be used for the ex vivo challenge assay. In contrast, typically only 2–4 cervical or vaginal biopsies are collected. This problem is exacerbated by competing needs for biopsies that include measurement of drug concentration in mucosal biopsies. The finding that more vaginal and cervical data are needed to provide the equivalent statistical power as the rectal data is the exact opposite of the balance of ex vivo data that have been generated in recent studies. Rectal data sets are, therefore, more likely to be powered to find significant differences between treatment and control conditions, and, conversely, a lack of significant treatment effects in cervical and vaginal tissue may be due to insufficient statistical power.
A difficulty that was not addressed in this article was the effect of the various treatment regimens on the kinetics of virus growth, as only nontreated (baseline or placebo) data were used in the analysis. This decision was based on the need to more fully understand how the untreated virus grows in the different tissue models so that suppression of growth, as would be expected from an efficacious microbicide, could be identified in the nontreated condition. The findings of this analysis are specific to the HIV-1BaL virus type used in this retrospective analysis of phase 1 and 2 clinical trials of HIV prevention treatments. Although other virus isolates, for example, Transmitter/Founder isolates and HIV-1 variants, have been found to be equally infective in ex vivo cervical tissue,23 they have not been used for the ex vivo challenge assay as of yet. In addition, as assay parameters were not independently varied but a result of the choices made by each scientific team, factors other than those included here (e.g., testing days, p24 kits, tissue type, treatment) may have affected the p24 results in ways that were not discovered during this retrospective analysis.
A key feature of this article is to demonstrate the utility of a retrospective analysis of data from multiple trials to improve assay methodology and statistical power in treatment comparisons. Overall, results indicated that improvements could be made in the design and analysis of the ex vivo challenge assay to provide a more standardized and powerful tool to compare efficacy of oral PrEP and microbicide products designed to suppress HIV-1BaL infection.
Supplementary Material
Acknowledgments
The authors thank Julie Elliott, Louise A. Ouattara, Kevin Uranker, Natalia Olejniczak, and Jarret Engstrom for performing the ex vivo challenge assays and providing technical details of the assays for this article. This analysis was supported by a subcontract with Advanced BioScience Laboratories, Inc., Rockville, MD through a NIH/NIAID/DAIDS contract: “Comprehensive Resources for HIV Microbicides and Biomedical Prevention” (no. HHSN272201000001C). The authors would like to acknowledge the support of the following grants: NIH U19 CHARM-01 (U19AI082637), Bill and Melinda Gates Foundation (OPP1045325), NIH U19 MDP (no. U19AI06061), UCLAs NIH CFAR Mucosal Immunology and Seed Grant Cores (no. AI28697) and CTRC (no. UL1TR000124), NIH FAME (U19 AI082639), an intra-agency agreement between the NIH/NIAID/DAIDS, USAID/OHA (NIAID Y1-AI-1756-01), CONRAD (GPO-A-00-08-00005-00), and ViiV contract WS2184024. The Microbicide Trials Network is funded by the National Institute of Allergy and Infectious Diseases (UM1AI068633, UM1AI068615, UM1AI106707), with cofunding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Institute of Mental Health, all components of the U.S. National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Finally, the authors would like to thank all of the clinical trial participants and the clinical team members, without whom this work would not have been possible.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Richardson-Harman N, Hendrix CW, Bumpus NN, Mauck C, Cranston RD, Yang K, et al. : Correlation between compartmental tenofovir concentrations and an ex vivo rectal biopsy model of tissue infectibility in the RMP-02/MTN-006 phase 1 study. PLoS One 2014;9:e111507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson-Harman N, Mauck C, McGowan I, Anton P: Dose-response relationship between tissue concentrations of UC781 and explant infectibility with HIV type 1 in the RMP-01 rectal safety study. AIDS Res Hum Retroviruses 2012;28:1422–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anton PA, Saunders T, Elliott J, Khanukhova E, Dennis R, Adler A, et al. : First phase 1 double-blind, placebo-controlled, randomized rectal microbicide trial using UC781 gel with a novel index of ex vivo efficacy. PLoS One 2011;6:e23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGowan I, Cranston RD, Duffill K, Siegel A, Engstrom JC, Nikiforov A, et al. : A phase 1 randomized, open label, rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of three formulations of tenofovir 1% gel (the CHARM-01 study). PLoS One 2015;10:e0125363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen BA, Panther L, Marzinke MA, Hendrix CW, Hoesley CJ, van der Straten A, et al. : Phase 1 safety, pharmacokinetics, and pharmacodynamics of dapivirine and maraviroc vaginal rings: A Double-Blind Randomized Trial. J Acquir Immune Defic Syndr 2015;70:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anton PA, Cranston RD, Kashuba A, Hendrix CW, Bumpus NN, Richardson-Harman N, et al. : RMP-02/MTN-006: A phase 1 rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of tenofovir 1% gel compared with oral tenofovir disoproxil fumarate. AIDS Res Hum Retroviruses 2012;28:1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicol MR, Emerson CW, Prince HM, Nelson JA, Fedoriw Y, Sykes C, et al. : Models for predicting effective HIV chemoprevention in women. J Acquir Immune Defic Syndr 2015;68:369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher PS, Elliott J, Grivel JC, Margolis L, Anton P, McGowan I, et al. : Ex vivo culture of human colorectal tissue for the evaluation of candidate microbicides. AIDS 2006;20:1237–1245 [DOI] [PubMed] [Google Scholar]
- 9.Janocko L, Althouse AD, Brand RM, Cranston RD, McGowan I: The molecular characterization of intestinal explant HIV infection using polymerase chain reaction-based techniques. AIDS Res Hum Retroviruses 2015;31:981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dezzutti CS, Uranker K, Bunge KE, Richardson-Harman N, Macio I, Hillier SL: HIV-1 infection of female genital tract tissue for use in prevention studies. J Acquir Immune Defic Syndr 2013;63:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti L, Barry SM, Hope TJ, Shattock RJ: HIV-1 infection of human penile explant tissue and protection by candidate microbicides. AIDS 2009;23:319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y, Harel O, Little RJ: How well quantified is the limit of quantification? Epidemiology 2010;21 Suppl 4:S10–S16 [DOI] [PubMed] [Google Scholar]
- 13.Succop PA, Clark S, Chen M, Galke W: Imputation of data values that are less than a detection limit. J Occup Environ Hyg 2004;1:436–441 [DOI] [PubMed] [Google Scholar]
- 14.Richardson-Harman N, Lackman-Smith C, Fletcher PS, Anton PA, Bremer JW, Dezzutti CS, et al. : Multisite comparison of anti-human immunodeficiency virus microbicide activity in explant assays using a novel endpoint analysis. J Clin Microbiol 2009;47:3530–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slymen DJ, de Peyster A, Donohoe RR: Hypothesis testing with values below detection limit in environmental studies. Environ Sci Technol 1994;28:898–902 [DOI] [PubMed] [Google Scholar]
- 16.Moulton LH, Halsey NA: A mixture model with detection limits for regression analyses of antibody response to vaccine. Biometrics 1995;51:1570–1578 [PubMed] [Google Scholar]
- 17.Krishnamoorthy K, Mallick A, Mathew T: Model-based imputation approach for data analysis in the presence of non-detects. Ann Occup Hyg 2009;53:249–263 [DOI] [PubMed] [Google Scholar]
- 18.Dezzutti CS, Richardson-Harman N, Rohan LC, Marzinke M, Hoesley MD, et al. : Impact of cryopreservation on pharmacodynamic correlations following use of vaginal rings containing dapivirine and/or maraviroc. Medicine 2016;95:e4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinnadurai R, Rajan D, Munch J, Kirchhoff F: Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J Virol 2007;81:6563–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobos Jimenez V, Booiman T, de Taeye SW, van Dort KA, Rits MA, Hamann J, et al. : Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons. Sci Rep 2012;2:763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geuenich S, Kaderali L, Allespach I, Sertel S, Keppler OT: Biological signature characteristics of primary isolates from human immunodeficiency virus type 1 group O in ex vivo human tonsil histocultures. J Virol 2009;83:10494–10503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heigele A, Camerini D, van't Wout AB, Kirchhoff F: Viremic long-term nonprogressive HIV-1 infection is not associated with abnormalities in known Nef functions. Retrovirology 2014;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merbah M, Arakelyan A, Edmonds T, Ochsenbauer C, Kappes JC, Shattock RJ, et al. : HIV-1 expressing the envelopes of transmitted/founder or control/reference viruses have similar infection patterns of CD4 T-cells in human cervical tissue ex vivo. PLoS One 2012;7:e50839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurman AR, Chandra N, Yousefieh N, Zalenskaya Z, Kimble T, Asin S, Rollenhagen C, Anderson S, Herold B, Mesquita PMM, Richardson-Harman N, Cunningham T, Schwartz J, Doncel G: Comparison of follicular and luteal phase mucosal markers of HIV susceptibility in healthy women. AIDS Res Hum Retroviruses 2016;32:547–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckstein DA, Penn ML, Korin YD, Scripture-Adams DD, Zack JA, Kreisberg JF, et al. : HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 2001;15:671–682 [DOI] [PubMed] [Google Scholar]
- 26.Kmiec D, Iyer SS, Sturzel CM, Sauter D, Hahn BH, Kirchhoff F: Vpu-mediated counteraction of tetherin is a major determinant of HIV-1 interferon resistance. MBio 2016;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Yorke E, Hancock G, Clutton G, Sande N, Angus B, et al. : Improved quantification of HIV-1-infected CD4+ T cells using an optimised method of intracellular HIV-1 gag p24 antigen detection. J Immunol Methods 2013;391:174–178 [DOI] [PubMed] [Google Scholar]
- 28.Carrig MM, Manrique-Vallier D, Ranby KW, Reiter JP, Hoyle RH: A nonparametric, multiple imputation-based method for the retrospective integration of data sets. Multivariate Behav Res 2015;50:383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacqmin-Gadda H, Thiebaut R, Chene G, Commenges D: Analysis of left-censored longitudinal data with application to viral load in HIV infection. Biostatistics 2000;1:355–368 [DOI] [PubMed] [Google Scholar]
- 30.Vonesh EFaC VM: Linear and Nonlinear Models for the Analysis of Repeated Measurements. New York, Marcel Dekker, 1997 [Google Scholar]
- 31.Davidian MaG DM: Nonlinear Models for Repeated Measurement Data. New York, Chapman & Hal, 1995 [Google Scholar]
- 32.Wu H, Wu L: Identification of significant host factors for HIV dynamics modelled by non-linear mixed-effects models. Stat Med 2002;21:753–771 [DOI] [PubMed] [Google Scholar]
- 33.Taylor TH, Jr.: Finding usable portion of sigmoid curve. U.S. Patent 7,469,186, December 23, 2008
- 34.Graham JW: Missing data analysis: Making it work in the real world. Annu Rev Psychol 2009;60:549–576 [DOI] [PubMed] [Google Scholar]
- 35.Environmental Protection Agency: Guidance for Data Quality Assessment. (Edited by Office of Environmental Information). Washington DC, Environmental Protection Agency, 2000, pp. 153–165 [Google Scholar]
- 36.Pudney J, Quayle AJ, Anderson DJ: Immunological microenvironments in the human vagina and cervix: Mediators of cellular immunity are concentrated in the cervical transformation zone. Biol Reprod 2005;73:1253–1263 [DOI] [PubMed] [Google Scholar]
- 37.Mowat AM, Viney JL: The anatomical basis of intestinal immunity. Immunol Rev 1997;156:145–166 [DOI] [PubMed] [Google Scholar]
- 38.McGowan I, Dezzutti C: Rectal microbicide development. Curr Top Microbiol Immunol 2014;383:117–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicol MR, Fedoriw Y, Mathews M, Prince HM, Patterson KB, Geller E, et al. : Expression of six drug transporters in vaginal, cervical, and colorectal tissues: Implications for drug disposition in HIV prevention. J Clin Pharmacol 2014;54:574–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thurman AR, Chandra N, Yousefieh N, Zalenskaya I, Kimble T, Asin S, et al. : Comparison of follicular and luteal phase mucosal markers of HIV susceptibility in healthy women. AIDS Res Hum Retroviruses 2016;32:547–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurman AR: Comparison of mucosal markers of HIV susceptibility in healthy premenopausal versus postmenopausal women. J Acquir Immune Defic Syndr (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed LJ, Muench H: A simple method of estimating fifty per cent endpoints. Am J Hygiene 1938;27:493–497 [Google Scholar]
- 43.Kaerber G: Beitrag zur Kollektiven Behandlung Pharmakologischer Reihenversuche. Arch Exp Path Pharma 1931;162:480–487 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.