Abstract
In 1987, when I became interested in the notion of antisense technology, I returned to my roots in RNA biochemistry and began work to understand how oligonucleotides behave in biological systems. Since 1989, my research has focused primarily on this topic, although I have been involved in most areas of research in antisense technology. I believe that the art of excellent science is to frame large important questions that are perhaps not immediately answerable with existing knowledge and methods, and then conceive a long-term (multiyear) research strategy that begins by answering the most pressing answerable questions on the path to the long-term goals. Then, a step-by-step research pathway that will address the strategic questions posed must be implemented, adjusting the plan as new things are learned. This is the approach we have taken at Ionis. Obviously, to create antisense technology, we have had to address a wide array of strategic questions, for example, the medicinal chemistry of oligonucleotides, manufacturing and analytical methods, pharmacokinetics and toxicology, as well as questions about the molecular pharmacology of antisense oligonucleotides (ASOs). Each of these endeavors has consumed nearly three decades of scientific effort, is still very much a work-in-progress, and has resulted in hundreds of publications. As a recipient of the Lifetime Achievement Award 2016 granted by the Oligonucleotide Therapeutic Society, in this note, my goal is to summarize the contributions of my group to the efforts to understand the molecular mechanisms of ASOs.
Keywords: : RNase H, oligonucleotide, antisense, mechanisms, RNA, DNA
Introduction
Throughout my career, my primary research interests have focused on understanding the molecular mechanisms by which drugs work, molecular pharmacology. Early in my career, I focused on the molecular mechanisms of antineoplastic drugs such as bleomycin, cisplatinum, and anthracyclines [1–3]. I then focused on more traditional receptor biological questions with a particular emphasis on leukotriene receptor signaling pathways and other G-protein-coupled receptors [4]. In 1987 when I became interested in the notion of antisense technology, I returned to my roots in RNA biochemistry and began work to understand how oligonucleotides behave in biological systems. Since 1989, my research has focused primarily on this topic, although I have been involved in most areas of research in antisense technology.
I believe that the art of excellent science is to frame large important questions that are perhaps not immediately answerable with existing knowledge and methods, and then conceive a long-term (multiyear) research strategy that begins by answering the most pressing answerable questions on the path to the long-term goals. Then, a step-by-step research pathway that will address the strategic questions posed must be implemented, adjusting the plan as new things are learned. This is the approach we have taken at Ionis Pharmaceuticals (formerly, Isis Pharmaceuticals). Obviously, to create antisense technology, we have had to address a wide array of strategic questions, for example, the medicinal chemistry of oligonucleotides, manufacturing and analytical methods, pharmacokinetics and toxicology, as well as questions about the molecular pharmacology of antisense oligonucleotides (ASOs). Each of these endeavors has consumed nearly three decades of scientific effort, is still very much a work-in-progress, and has resulted in hundreds of publications. That progress has also been chronicled in a number of books that I have edited [5–8]. (Although it is clearly time to edit a new volume on antisense technology.) As a recipient of the Lifetime Achievement Award 2016 granted by the Oligonucleotide Therapeutic Society, in this note, my goal is to summarize the contributions of my group to the efforts to understand the molecular mechanisms of ASOs.
Phases of Antisense Drug Action
The molecular events that result in the activities of ASOs can be divided into three phases: prehybridization, hybridization, and posthybridization. In my laboratory, we have attempted to address all three phases. In the prehybridization phase, an ASO must enter the cell, distribute within the cell, and achieve effective concentrations at the target RNA site(s). It must then sort through the cellular nucleic acid sequence space to hybridize to its cognate site. The concentrations of the cognate site are extremely low relative to total cellular RNA and certainly relative to conditions used in in vitro hybridization assays. Hybridization to the cognate site in the cell is a much more complex process than in a test tube and must involve interactions with proteins, such as Ago2, or other cellular components that facilitate hybridization [9]. Once the ASO is bound to its cognate site, depending on the chemical design of the ASO, a variety of events may be induced that alter the target RNA to achieve the desired pharmacological outcome [10].
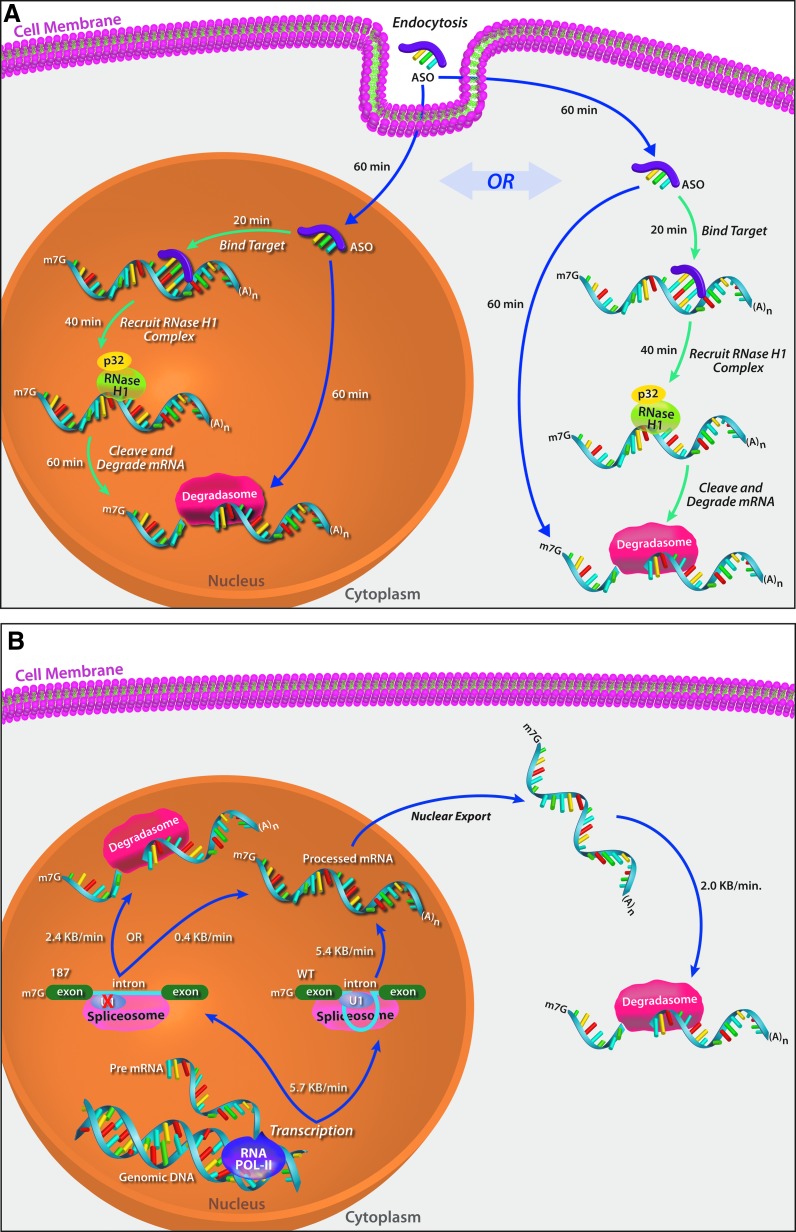
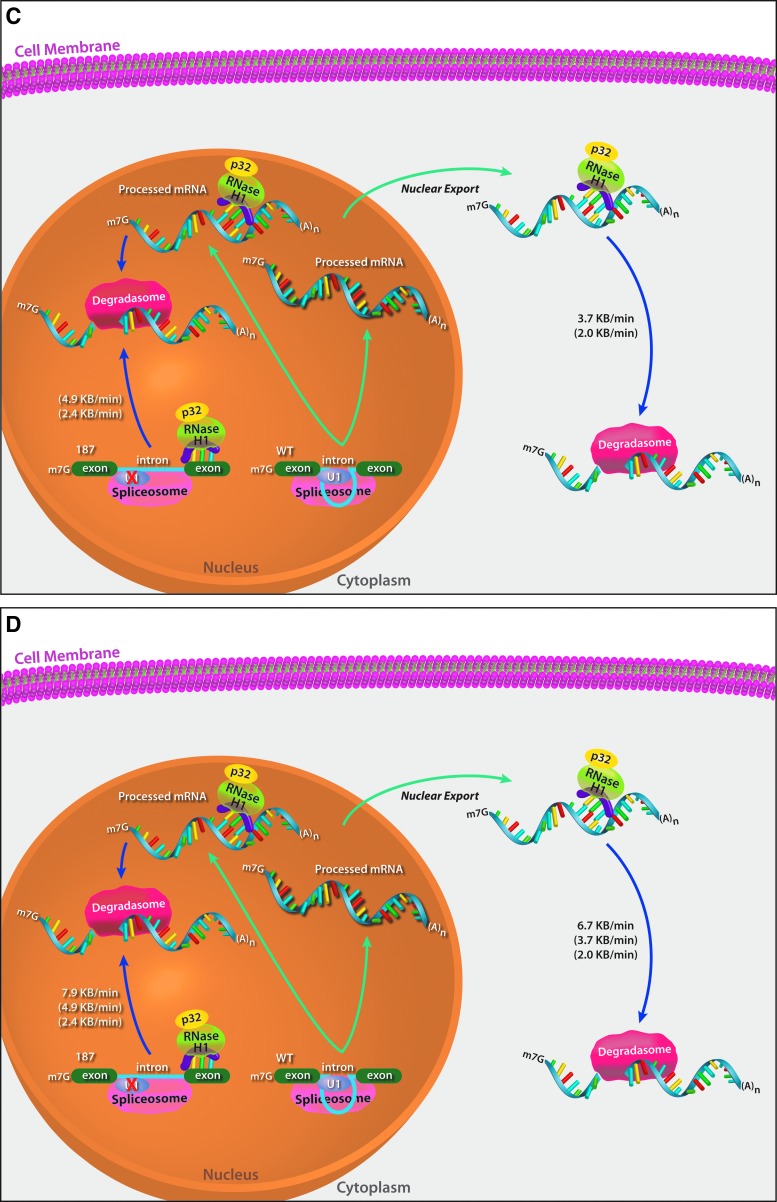
One of the more interesting features of the molecular pharmacology of ASOs is that the kinetics are remarkably slow. We have characterized the kinetics of the major steps in the molecular pharmacology of RNase H1 activating ASOs [11] showing that the onset of action occurs about two hours after transfection, with 60, 20, and 40 min required for intracellular distribution, RNA sequence searching and hybridization to the cognate site and RNase H1 recruitment and cleavage, respectively. The recruitment of RNase H1 and subsequent RNase H1-mediated cleavage of the target RNA increase the degradation rate of the target RNA by 2- to 4-fold compared with the intrinsic rate of cellular RNA degradation (2.0 or 2.4 kb/degradation rate in the cytoplasm or nucleus, respectively) (Fig. 1).
FIG. 1.
Rates of steps in RNase H1 activating ASOs activity [11]. (A) After transfection, about 60 min is required for intracellular distribution of ASOs. In both the nucleus and cytoplasm, about 20 min are required to screen nucleic acid sequence and bind to the cognate site. Approximately 40 min are then required to recruit RNase H1 and achieve measurable RNA target reduction. (B) The transcription and splicing rates for the wild-type SOD1 construct and a mutant (187) with substantially reduced splicing. (C) An effective RNase H1 activating ASO approximately doubles the intrinsic rate of cellular RNA degradation. (D) The concentration of RNAse H1 is rate limiting. Over-expression of Escherichia coli RNase H1 again doubles the rate reduction induced by an effective RNase H1 ASO. ASO, antisense oligonucleotide.
Prehybridization Events
One of our long-term goals has been to understand the events that take place before hybridization at the target sequence in RNA by the ASO. Clearly to hybridize to the “receptor” sequence, effective concentrations in the region, in which the target RNA resides, must be achieved. We began by cataloging the major intracellular proteins that bind phosphorothioate (PS) ASOs using an affinity capture method to identify the proteins in cellular homogenates involved in those interactions [12]. Somewhat surprisingly, only 58 proteins were identified that bind PS ASOs. The affinity capture method we have used would not be expected to identify proteins with low affinity for PS ASOs or proteins that are present in low concentrations. Nor is it likely that the method would identify proteins that bind only when complexed with other proteins or other cellular components. However, since we are primarily interested in bulk movements of PS ASOs in cells, this catalog of proteins likely constitutes most of the abundant proteins of interest and is certainly a good starting point.
Not surprisingly, many of the proteins that bind PS ASOs contain nucleic acid-binding domains or are chaperone proteins. However, a number of proteins that might not have been expected to bind PS ASOs were identified, for example, annexin A2 [13]. Then, by reducing and overexpressing various proteins, we identified several proteins that either reduce or enhance the potency of PS ASOs. We have characterized the mechanisms accounting for the effects on antisense activity of a number of these proteins. Some, through various mechanisms, inhibit ASO activity, for example, by competing with RNase H1 for binding to the RNA-ASO heteroduplex [14]. Others increase ASO activity by various mechanisms, for example, alteration of subcellular localization [15]. Other proteins affect ASO cellular toxicities, for example, members of the drosophila behavior human splicing (DBHS) family including, PSF and P54nrb [16]. Still others, such as HSP90 [17], have effects for which the mechanisms remains to be defined.
Next, we identified the major sites of subcellular accumulation and characterized the kinetics, with which these sites accumulate PS ASOs after transfection, free uptake, and electroporation [12,13,15,18,19]. By coupling this information with the time course of drug action of PS ASOs, we were able to associate various sites of accumulation with the onset of activity, peak effects, and duration of effects of PS ASOs. Reduction of specific proteins that caused changes in subcellular localization of PS ASOs then supported the identification of subcellular sites, at which PS ASOs are active, or inactive. For example, paraspeckles, PS bodies, P bodies, and lysosomes are all sites, in which these ASOs are inactive [12,15]. Two particularly informative examples are PS bodies and paraspeckles. PS bodies form in the nucleus when TCP1β binds PS ASOs. Reducing TCP1β resulted in reduction of PS bodies and PS ASO activity. Similarly, we showed that PS ASOs can displace a long noncoding RNA, NEAT1, and form functional paraspeckles in the nucleus, and these paraspeckle proteins inhibit PS ASO activities [18]. Thus, we have a full catalog of intracellular sites, in which ASOs accumulate over time after transfection, free uptake, and electroporation and the proteins responsible for accumulation of PS ASOs at the various sites (Fig. 2).
FIG. 2.
Subcellular distribution of PS ASOs and cleavage of target RNA by RNase H1. PS ASO distribute within the cell via interactions with specific proteins. Once the ASO has bound to its cognate site in the target RNA, RNase H1 (and P32) is recruited to degrade the target RNA. Post-RNase H1 cleavage and the fragments are processed by normal cellular RNA degradation pathways. PS, phosphorothioate.
We then applied the same methodology to characterize the major interactions of PS ASOs at the cell surface [12]. Although much of this work is still in the process of being published, we believe that the main cell surface interactions of PS ASOs are once again with proteins. We have identified new cell surface proteins that bind PS ASOs and confirmed the interactions of a number of proteins previously reported to bind to these agents [12,20,21]. We have also identified key membrane lipids that appear to affect, although indirectly, interactions with cell surface proteins (S. Wang et al., unpublished data) and shown that the decision to enter productive or nonproductive subcellular distribution pathways is likely made at the cell surface. Finally, we have begun to characterize the processes, by which PS ASOs distribute intracellularly, for example, we have shown that PS ASOs are released mainly from late endosomes and this leads to ASO activity (S. Wang et al., unpublished data).
Thus, progress over the last few years provides a detailed road map of where in the cell ASOs accumulate, how they get to those sites, and the sites that appear to be associated with ASO activity. We know that proteins determine the fate of PS ASOs at the cell surface and within the cell. We have a solid understanding of the kinetics of PS ASO distribution in cells and opportunities to enhance ASO performance by using this information in the design of new PS ASOs that more effectively take advantage of proteins that contribute to ASO activities.
Hybridization
In principle, PS ASOs of any chemistry should not be expected to hybridize to their cognate sequences in cells [9]. For most ASOs, this step remains very much a black box, as despite years of effort, proteins or lipids that might facilitate hybridization have not been identified. The single exception, however, serves as an excellent model. Ago2 binds the guide strand of double-stranded siRNA and single-strand RNAi molecules and “preorganizes” the ASO, thus, facilitating binding to the cognate site in the target RNA [9]. Thanks to the work of many laboratories, a great deal is understood about how Ago2 binds oligoribonucleotides and facilitates hybridization [9]. The research continues to identify other factors (proteins, lipids) that may facilitate the hybridization of other chemical classes of oligonucleotides. However, we do understand factors that influence both potency and specificity of the potential hybridization events. We know that RNA structure is a major determinant while only rarely do proteins that are bound to RNA targets alter ASO activity [22]. We know that the number of copies of target RNAs is irrelevant as is the RNA half-life, except for rapidly cleared RNAs such as cMyc [14,22,23]. We also know that nonpromiscuous repeats are present in many pre-mRNAs and these make excellent sites for ASO activity [24]. As we make additional progress in understanding prehybridization events, we hope that we will identify the cellular factors that enhance the hybridization of ASOs of various chemical compositions, similar to the effects of Ago2 with RNA-like ASOs [9].
Posthybridization Mechanisms
Depending on the design of the ASO, after hybridization a number of mechanisms can be exploited to degrade, disable, or modify the target RNA to achieve the desired pharmacologic effects. This has also been one of the significant research interests of my laboratory [5,10].
RNase H1
Mammalian cells contain two RNase H enzymes, H1 and H2, which cleave RNA only when it is a DNA-RNA heteroduplex [25]. As RNase H1 cleavage of target RNAs has proven to be a robust and versatile mechanism, by which DNA-like ASOs induce pharmacological effects, we have invested substantially in understanding the RNase H enzymes and how they participate in ASO activities. We showed that although RNase H2 is much more abundant than RNase H1 in mammalian cells, in intact cells, only RNase H1 participates in ASO activities [25–27]. This is probably because RNase H2 is tightly bound to chromatin while RNase H1 is present in the nucleus, cytoplasm, and mitochondria. We characterized the structure and enzymological properties of the RNase H1 [28–37] and evaluated how the enzyme is regulated. We showed that RNase H1 participates with a binding partner, a protein involved in a number of RNA metabolic activities, P32, and that in some circumstances P32 appeared to enhance the specificity of cleavage of RNase H1 [32]. These studies supported optimization of chimeric ASOs designed to activate RNase H1. To confirm that RNase H1 is the only nuclease responsible for the antisense activity of DNA-like ASOs, we constructed hepatocytes-specific RNase H1 knockout mice [38] and showed that in the absence of RNase H1, all activities were lost.
We have also been interested in understanding the biological roles of mammalian RNase H1, showing that it was essential for mitochondrial ribosomal RNA transcription [32]. Using the knockout mice system, we confirmed and extended these observations demonstrating that RNase H1 is essential for efficient R loop clearance formed during transcription of both mitochondrial and nuclear preribosomal RNA [38]. In work that is yet to be published, we have shown that RNase H1 and topoisomerase 1 are redundant in R loop clearance and that they are coordinately regulated (W. Shen et al., unpublished data).
Armed with a solid understanding of the properties of mammalian RNase H1, we have characterized the key factors that contribute to the specificity of cleavage by RNase H1 of cognate sites for DNA-like ASOs [22] and shown that RNase H1 is limiting. We have defined the rates of all the major steps in the pharmacological effects of DNA-like ASOs [11] and shown that several proteins may reduce or block the cleavage of target RNAs by competing with RNase H1 for binding to the DNA-RNA-like heteroduplex formed by the ASOs and target RNAs [12,14]. Further, as mentioned previously, we have shown that many pre-mRNAs contain nonpromiscuous repeat sequences that serve as particularly good targets for RNase H1 activating ASOs [24]. We now know that the RNA fragments produced by RNase H1 after ASO treatment are processed by the normal cellular RNA degradation pathways in both the nucleus and cytoplasm [39]. Finally, we have demonstrated that one pathway that may account for some cellular toxicities of ASOs involves off-target cleavage of sensitive sites, particularly in long pre-mRNAs [40].
These studies have contributed to steadily improving performance of DNA-like ASOs, as we have optimized ASO chemistries and designs to maximize activity and specificity by designing ASOs to interact with attractive sites in target RNAs and serve as optimal substrates for RNase H1.
Ago2-Mediated Cleavage of Target RNA
The original definition of antisense [41] was entirely agnostic as to the type of oligonucleotide, with which to hybridize to the target RNA and what might happen after binding. So, all postbinding mechanisms may be formally considered under the broad rubic, antisense, including siRNAs. In fact, Ago2 has an RNase H domain, so we have always considered Ago2-mediated RNA degradation an “antisense mechanism.” Although a large number of laboratories have contributed to a detailed understanding of the molecular mechanisms of Ago2-induced target RNA reduction [42], we have focused on the properties of Ago2-induced cleavage that may affect the pharmacological properties of both single- and double-stranded RNA-like oligonucleotides that bind to Ago2. We characterized the binding and cleavage specificities of human Ago2 [43,44] and demonstrated that siRNAs competed with microRNAs and other siRNAs for binding [45]. We showed that Dicer interacts with both single- and double-stranded RNA-like oligonucleotides [42]. To better understand the risk of off-target cleavage of RNAs by siRNAs, we showed that competition between siRNAs and microRNAs for binding to Ago2 could result in unexpected cellular effects due to changes in microRNA activities [46]. Finally, we have synthesized and stabilized single-strand RNA-like ASOs that bind to Ago2 and result in cleavage of the target RNA, thus, obviating the need for a passenger strand and simplifying the design of the siRNAs [47,48].
Other Posthybridization Mechanisms
ASOs can be designed to take advantage of other posthybridization mechanisms (for review see Ref. [10]). One mechanism of interest and of particular importance for some diseases is the use of ASOs to alter splicing, for example, spinal muscle atrophy [49]. We first identified the key features of splice sites that influence the ability of ASOs to alter splicing [50] and then optimized the design of ASOs to take advantage of this mechanism. It is also possible to design ASOs that alter the site of polyadenylation, thus affecting RNA stability [51]. In contrast, we showed that the notion of using “U1 adapters” to sequester U1 RNPs was associated with nonspecific changes in splicing that would lead to numerous undesired effects [52]. More recently, we have reported that it is possible to design ASOs that bind to upstream open reading frames [53] or translation suppressor elements (manuscript submitted) in the 5′ untranslated region of mRNAs to selectively enhance the translation of specific mRNAs, thus creating the opportunity to develop “agonist-like” ASOs. As more than 80% of human mRNAs have such elements, this presents a broad new opportunity for the technology.
Future Focus
The work in our laboratory has resulted in steady advances in understanding the molecular pharmacology of PS ASOs, and lays a strong foundation for rapid advances in the future, particularly when considered in the context of the contributions of many other laboratories. Based on our work, we are convinced that to fully understand the molecular pharmacology of PS ASOs, it is essential to understand the “nucleic acid code” in more detail. Thanks to nearly 30 years of effort, the structure–activity relationship (SAR) of ASO-nucleic acid interactions are well understood. Nevertheless, there are elements of this code that we would like to better understand. For example, we are attempting to map the structures of all cellular RNAs using recently developed next-generation sequencing methods. We also need to understand the roles and effects of post-transcriptional modifications of RNAs on ASO activities.
Our efforts to understand the proteins that bind PS ASOs have convinced us that proteins determine the fates of PS ASOs, demanding that we understand the amino acid code. Thanks to a new assay developed in our laboratory, progress of this has been gratifyingly rapid [53]. We have a preliminary grasp of the SAR of protein interactions from the ASO side and with regard to protein domains that bind PS ASOs. We have shown that PS content, the hydrophobicity of the 2′ modifications, charge, and sequence influence protein binding and different proteins display different proclivities. What is tremendously exciting is that we have demonstrated that very minor modifications in ASO design can have substantial effects on ASO–protein interactions [54]. We will continue to advance our understanding of the SAR of these interactions and, as we do, an enormous new scope for medicinal chemistry is presented suggesting that in the future we can tailor PS ASOs to interact with proteins that enhance their pharmacological property and avoid those that limit their activities or exaggerate adverse events.
Perhaps the most gratifying aspect of the progress made over these decades to me is that we now have sufficient knowledge to compose data-based testable models for essentially all events observed after PS ASO administration. This should result in more efficient basic research and more rapid conversion of the knowledge gained to advances in ASO performance that will benefit patients with a wide range of diseases. Given the breadth of the potential for antisense technology, that is a truly thrilling vision of the future of technology. Moreover, we have used much of the information we have developed directly in the identification of potential new medicines to steadily improve the performance of ASOs. With Vitravene and Kynamro having been marketed, and nusinersen (Spinraza) approaching approval, and a pipeline of 38 antisense drugs in development at Ionis, based on several chemical classes, including 2′ methoxyl ethyl, 2′cEt, and GalNAc, it is thrilling to contemplate the potential benefit to patients that antisense technology may bring in the near future.
Acknowledgments
It has “taken a village” to create antisense technology. So I accepted the lifetime achievement award on behalf of all my colleagues who contributed. Similarly, although I wrote this “anthology” of our work on the molecular mechanisms of ASOs, the work was a product of the wonderful people who have been in my research group and, of course, we have benefited from being a part of Ionis Pharmaceuticals.
I specially want to thank Xue-Hai Liang and Tim Vickers for editing the manuscript and Donna Parrett for excellent administrative support.
Author Disclosure Statement
The author is employed by Ionis Pharmaceuticals.
References
- 1.Crooke ST. and Reich SD, eds. (1980). Anthracyclines: Current Status and New Developments. Academic Press, New York, NY [Google Scholar]
- 2.Prestayko AW, Crooke ST, and Carter SK, eds. (1980). Cisplatin—Current Status and New Developments. Academic Press, New York, NY [Google Scholar]
- 3.Carter SK, Crooke ST, and Umezawa H, eds. (1978). Bleomycin: Current Status and New Developments. Academic Press, New York, NY [Google Scholar]
- 4.Crooke ST. and Wong A, eds. (1991). Lipoxygenases and Their Products. Academic Press, New York, NY [Google Scholar]
- 5.Crooke ST, ed. (2008). Antisense Drug Technology: Principles, Strategies, and Applications , Second Edition CRC Press, Baco Raton, FL [Google Scholar]
- 6.Crooke ST, ed. (2001). Antisense Drug Technology: Principles, Strategies, and Applications. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 7.Crooke ST, ed. (1998). Antisense Research and Application Handbook of Experimental Pharmacology. Springer-Verlag, Berlin, Heidelberg [Google Scholar]
- 8.Crooke ST. (1995). Therapeutic Applications of Oligonucleotides. RG Landes Co. Austin, TX [Google Scholar]
- 9.Salomon WE, Samson MJ, Moore MJ, Zamore PD. and Serebrov V. (2015). Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell 162:84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooke ST, Vickers TA, Lima WF. and Wu H. (2008). Mechanisms of antisense drug action, an introduction. In: Antisense Drug Technology: Principles, Strategies, and Applications , Second Edition Crooke ST, ed. CRC Press, Baco Raton, FL, pp. 3–46 [Google Scholar]
- 11.Vickers TA. and Crooke ST. (2015). The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acid Res 43:8955–8963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang XH, Sun H, Shen W. and Crooke ST. (2015). Identification and characterization of intracellular proteins that bind phosphorothioate-containing oligonucleotides. Nucleic Acid Res 43:2927–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Sun H, Tanowitz M, Liang XH. and Crooke ST. (2016). Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acid Res 44:7314–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickers TA. and Crooke ST. (2014). Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLoS One 9:e108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crooke ST, Wang S, Vickers TA, Shen W. and Liang XH. (2016). Cellular uptake and trafficking of antisense oligonucleotides. Nature Biotech (In press) [DOI] [PubMed] [Google Scholar]
- 16.Shen W, Liang XH, Sun H. and Crooke ST. (2015). 2′-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acid Res 43:4569–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang XH, Shen W, Sun H, Kinberger GA, Prakash TP, Nichols JG. and Crooke ST. (2016). Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2′-modifications and enhances antisense activity. Nucleic Acid Res 44:3892–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen W, Liang XH. and Crooke ST. (2014). Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structure. Nucleic Acid Res 14:8648–8662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang XH, Shen W, Sun H, Prakash TP. and Crooke ST. (2014). TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acid Res 42:7819–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juliano RL, Ming X, Carver K. and Laing B. (2014). Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther 24:101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juliano RL. and Carver K. (2015). Cellular uptake and intracellular trafficking of oligonucleotides. Adv Drug Deliv Rev 87:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima WF, Vickers TA, Nichols J, Li C. and Crooke ST. (2014). Defining the factors that contribute to on-target specificity for antisense oligonucleotides. PLos One 9:e101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miraglia L, Watt AT, Graham MJ. and Crooke ST. (2000). Variations in mRNA content have no effect on the potency of antisense oligonucleotides. Antisense Nucleic Acid Drug Dev 10:453–461 [DOI] [PubMed] [Google Scholar]
- 24.Vickers TA, Freier SM, Bui HH, Watt A. and Crooke ST. (2014). Targeting of repeated sequences unique to a gene results in significant increases antisense oligonucleotide potency. PLoS One 9:e110615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima WF, Wu H. and Crooke ST. (2008). The RNase H Mechanism. In: Antisense Drug Technology: Principles, Strategies, and Applications , Second Edition Crooke ST, ed. Baco Raton, FL, pp. 47–74 [Google Scholar]
- 26.Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST. (2004). Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J Biol Chem 279:17181–17189 [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Lima WF. and Crooke ST. (1998). Molecular cloning and expression of cDNA for human RNase H. Antisense Nucleic Acid Drug Dev 8:53–61 [DOI] [PubMed] [Google Scholar]
- 28.Lima WF, Rose JB, Nichols JG, Wu H, Migawa MT, Wyrzykiewicz TK, Siwkowski AM. and Crooke ST. (2007). Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol Pharmacol 71:83–91 [DOI] [PubMed] [Google Scholar]
- 29.Lima WF, Nichols JG, Wu H, Prakash TP, Migawa MT, Wyrzykiewicz TK, Bhat B. and Crooke ST. (2004). The structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J Biol Chem 279:36317–36326 [DOI] [PubMed] [Google Scholar]
- 30.Lima WF, Wu H, Nichols JG, Prakash TP, Ravikumar V. and Crooke ST. (2003). Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3′-DNA/5′-RNA terminus of the heteroduplex substrate. J Biol Chem 278:49860–49867 [DOI] [PubMed] [Google Scholar]
- 31.Lima WF, Wu H, Nichols JG, Manalili SM, Drader JJ, Hofstadler SA. and Crooke ST. (2003). Human RNase H1 activity is regulated by a unique redox switch formed between adjacent cysteines. J Biol Chem 278:14906–14912 [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Sun H, Liang XH, Lima WF. and Crooke ST. (2013). Human RNase H1 is associated with Protein P32 and is involved in mitrochondrial pre-rRNA processing. PLos One 8:e71006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooke ST. (2004). Antisense strategies. Curr Mol Med 4:465–487 [DOI] [PubMed] [Google Scholar]
- 34.Crooke ST. (2004). Progress in antisense technology. Annu Rev Med 55:61–95 [DOI] [PubMed] [Google Scholar]
- 35.Lima WF. and Crooke ST. (2001). Human RNase H. Methods Enzymol 341:430–440 [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Lima WF. and Crooke ST. (199). Properties of cloned and expressed Human RNase H. J Biol Chem 274:28270–28278 [DOI] [PubMed] [Google Scholar]
- 37.Lima WF, Rose JB, Nichols JG, Wu H, Migawa MT, Wyrzykiewicz TK, Vasquez G, Swayze EE. and Crooke ST. (2007). The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol Pharmacol 71:73–82 [DOI] [PubMed] [Google Scholar]
- 38.Lima WF, Murray HM, Damle SS, Hart CE, Hung G, De Hoyos CL, Liang XH. and Crooke ST. (2016). Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acid Res 44:5299–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lima WF, De Hoyos CL, Liang XH. and Crooke ST. (2016). RNA cleavage products generated by antisense oligonucleotides and siRNAs are processed by the RNA surveillance machinery. Nucleic Acid Res 44:3351–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burel S, Hart CE, Cauntay P, Hsiao J, Machemer T, Katz M, Watt A, Bui HH, Younis H, et al. (2016). Hepatotoxicity of high affinity Gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acid Res 44:2093–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamecnik PC. and Stephenson ML. (1978). Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligonucleotide. Proc Natl Acad Sci U S A 75:280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lima WF, Murray H, Nichols JG, Wu H, Sun H, Prakash TP, Berdeja AR, Gaus HJ. and Crooke ST. (2009). Human Dicer binds short single-strand and double-strand RNA with high affinity and interacts with different regions of the nucleic acids. J Biol Chem 284:2535–2548 [DOI] [PubMed] [Google Scholar]
- 43.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM. and Baker BF. (2003). Efficient reduction of target RNAs by siRNA and RNase H-dependent antisense agents: a comparative analysis. J Biol Chem 278:7108–7118 [DOI] [PubMed] [Google Scholar]
- 44.Lima WF, Wu H, Sun H, Murray HM. and Crooke ST. (2009). Binding and cleavage specificities of human Argonaute2. J Biol Chem 284:25653–25663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vickers TA, Lima WF, Nichols JG. and Crooke ST. (2007). Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res 35:6598–6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang XH, Hart CH. and Crooke ST. (2013). Transfection of siRNAs can alter miRNA levels and trigger non-specific protein degradation in mammalian cells. Biochim Biophys Acta 1829:455–468 [DOI] [PubMed] [Google Scholar]
- 47.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP. and Corey DR. (2012). Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 150:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, et al. (2012). Single-stranded siRNAs activate RNAi in animals. Cell 150:883–894, 2012 [DOI] [PubMed] [Google Scholar]
- 49.Sazani P, Graziewicz MA. and Kole R. (2008). Splice switching oligonucleotides as potential therapeutics. In: Antisense Drug Technology: Principles, Strategies, and Applications , Second Edition Crooke ST, ed. CRC Press, Baco Raton, FL, pp. 89–114 [Google Scholar]
- 50.Hodges D. and Crooke ST. (1995). Inhibition of splicing of wild-type and mutated luciferase-adenovirus pre-mRNAs by antisense oligonucleotides. Mol Pharm 48:905–918 [PubMed] [Google Scholar]
- 51.Vickers TA. and Crooke ST. (2001). siRNAs targeted to certain polyadenylation signals promote specific, RISC-independent degradation of messenger RNAs. Nucleic Acid Res 40:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vickers TA, Sabripour M. and Crooke ST. (2011). U1 Adaptors result in reduction of multiple pre-mRNA species principally by sequestering U1snRNP. Nucleic Acids Res 39:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang XH, Shen W, Sun H, Migawa MT, Vickers TA. and Crooke ST. (2016). Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat Biotech 34:875–880 [DOI] [PubMed] [Google Scholar]
- 54.Vickers TA. and Crooke ST. (2016). Development of a quantitative BRET affinity assay for nucleic acid-protein interactions. PLoS One 11:e0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]