Abstract
Mutations within the chromosome 9 open reading frame 72 (c9orf72) gene are associated with both familial amyotrophic lateral sclerosis and frontotemporal dementia. The mutation leads to an expanded GGGGCC hexanucleotide repeat within the first intron of c9orf72 and an expanded CCCCGG repeat within a corresponding antisense transcript. Both the mutant intronic and antisense RNAs have been implicated in disease. We have previously reported that duplex RNAs complementary to the repeats can recognize disease-causing RNA and block detection of nuclear foci formed by the mutant transcripts. Here, we test the hypothesis that inhibition can also be achieved by single-stranded silencing RNAs (ss-siRNAs). ss-siRNAs are single-stranded antisense oligonucleotides (ASOs) that function through RNAi interference (RNAi) to silence gene expression. ss-siRNAs can block the expanded repeats within both intronic RNA and the antisense transcripts. Inhibition is more potent than by analogous duplex RNAs. Our data suggest that the potent effects on foci are caused by a combination of mechanisms including RNAi and direct binding of the ss-siRNA to the target transcripts. These findings reinforce the suggestion that ss-siRNAs combine the favorable properties of duplex RNA and single-stranded ASOs.
Keywords: : RNA interference, single-strand silencing RNA, c9orf72, antisense oligonucleotide
Introduction
Progress toward capturing the potential of RNA interference (RNAi) as a mechanism for delivering therapeutic nucleic acids has been slowed by difficulties delivering sufficient amounts of duplex RNA to target tissues [1]. One approach to improving delivery uses galactose conjugates [2] that target the asialoglycoprotein receptor, a protein that is highly expressed on the surface of hepatocytes. An alternate approach to therapeutic gene silencing involves single-stranded antisense oligonucleotides (ASOs) [3]. One advantage for ASOs relative to duplex RNA is that they require only one nucleic acid strand rather that two. The ability to function with just one strand simplifies synthesis and ASOs can be delivered to tissues after administration in saline. While conjugation with galactose can improve delivery of ASOs to the liver [4], it is not necessary.
One option for increasing the value of the RNAi mechanism for therapy is to combine the strengths of the duplex RNA and ASO approaches. This goal can be achieved using single-stranded silencing RNAs (ss-siRNAs) [5–15]. ss-siRNAs are chemically modified single-stranded oligonucleotides that silence gene expression through RNAi in cell culture and animals.
ss-siRNAs are a useful scientific tool and an intriguing starting point for therapeutic discovery. Decisions regarding preclinical development of ss-siRNAs, however, must confront the fact that competing ASO and duplex RNA technologies have been optimized for many years. By contrast, much more fundamental research is needed to understand the potential of ss-siRNAs as an approach for drug discovery. This research will involve medicinal chemistry aimed at improving the characteristics of ss-siRNAs [13,14] and studies that test the boundaries of ss-siRNA function on varied gene targets.
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are devastating disorders that lack treatment options. Recently, an expanded hexanucleotide repeat has been found to be implicated in one-third of familial ALS and one-quarter of familial FTD, making it the single largest genetic factor known to contribute to these diseases [16,17]. For mRNA transcribed in the sense direction, the repeat is composed of GGGGCC hexanucleotides and is located in intron 1 (Fig. 1A). There is also an antisense transcript with a CCCCGG hexanucleotide repeat that has been implicated as contributing to disease [18].
FIG. 1.
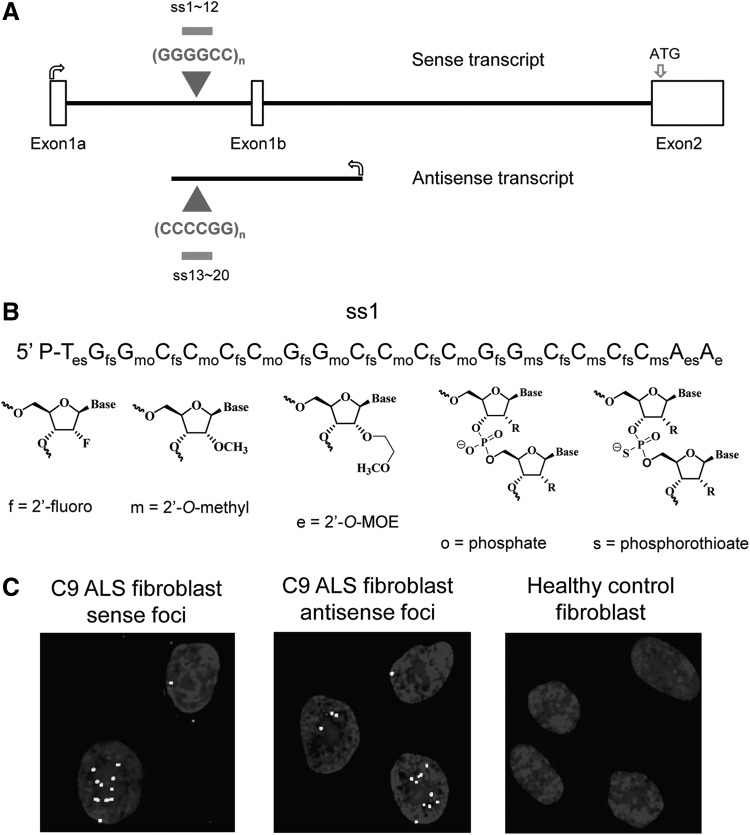
Experimental outline. (A) Diagram of c9orf72 sense and antisense transcripts containing the repeat region. (B) Example of ss-siRNA sequence and chemical modifications used to synthesize ss-siRNAs. P indicates phosphate. (C) Examples of FISH detection of c9orf72 intronic sense foci [with (CCCCGG)4-Cy3 probe] or antisense foci [with (GGGGCC)3.3-Cy3 probe] in patient-derived or healthy control fibroblast cells [with (CCCCGG)4-Cy3 probe]. FISH, fluorescent in situ hybridization.
We had previously demonstrated that duplex RNAs complementary to the hexanucleotide repeat could block both the sense and antisense transcripts, causing a substantial reduction in nuclear RNA foci [19]. We had also previously observed that ss-siRNAs could inhibit expression of genes (Huntington, atrophin-1, and ataxin-3) containing expanded CAG trinucleotide repeats [7,8,14,15]. Our goal here is to test the hypothesis that ss-siRNAs can also block the sense and antisense repeats within the c9orf72 gene.
Achieving this goal is challenging because, in contrast to our earlier use of ss-siRNAs to target genes containing expanded CAG repeats within mature mRNA to block gene translation [7,8,10,11], successful targeting c9orf72 requires (1) recognition of a hexanucleotide; (2) recognition of a sequence that is entirely composed of cytosine and guanine; (3) recognition of a target within an intron or an antisense transcript; and (4) recognition of a target within the nucleus. Here, we show that ss-siRNAs inhibit the detection of foci formed by mutant c9orf72 sense and antisense transcripts. Inhibition by ss-siRNAs was more potent than inhibition by analogous duplex RNAs and occurs through both RNAi and RNAi-independent mechanisms.
Materials and Methods
Cell culture and siRNA transfection
Mutant c9orf72 patient-derived fibroblast cells (provided by Dr. John Ravits of UCSD) were maintained at 37°C and 5% CO2 in Minimal Essential Media Eagle (MEM) (M4655; Sigma) supplemented with 15% heat inactivated fetal bovine serum (Sigma) and 0.5% MEM nonessential amino acids (Sigma). ss-siRNAs were synthesized at Ionis Pharmaceutics (Carlsbad, CA) and reconstituted in nuclease-free water. ss-siRNAs were transfected into patient-derived fibroblast cells with lipid RNAiMAX (Life Technologies) as previously described [19]. Cells were plated at a density of 80,000 per well of a six-well plate 48 h before transfection and subsequent quantitative polymerase chain reaction (qPCR) analysis. Cells were typically harvested 2 days after transfection.
qPCR analysis
c9orf72 expression was analyzed by qPCR on a 7500 real-time PCR system (Applied Biosystems) using iTaq SYBR Green Supermix (Bio-rad). Data were normalized relative to levels of GAPDH mRNA. Primers specific for c9orf72 mRNA all three variants are as follows: F 5′-AGAAGGCACAGAGAGAATGGAA-3′; R 5′-TCATCATCATTGAGTACTGTATCAGC-3′. Primers for c9orf72 intron1: F 5′-ACGCCTGCACAATTTCAGCCCAA-3′; R 5′-CAAGTCTGTGTCATCTCGGAGCTG-3′. Primers for GAPDH: F 5′-GTCATCAATGGAAATCCCATCAC-3′; R 5′-TTCTCCATGGTGGTGAAGAC-3′.
RNA FISH and imaging
RNA fluorescent in situ hybridization (FISH) was performed as described by the probe manufacturer (Biosearch Technologies) with minor modification. Fibroblast cells were plated at a density of 10,000/well into Lab-Tek 8-well chambered cover glass slides. After 1 day, siRNA/lipid complex were added at 50 nM final concentration. Forty-eight hours after transfection, cells were fixed with 4% formaldehyde in 1× phosphate-buffered saline (PBS) and permeabilized in 70% ethanol at 4°C overnight. Cells were washed with wash buffer (10% formamide in 2× saline-sodium citrate buffer [SSC]) for 5 min, and then incubated with prehybridization buffer (40% formamide in 2× SSC) at 60°C for 20 min. A (CCCCGG)4-Cy3 DNA probe or a (GGGGCC)3.3-Cy3 probe in hybridization buffer (100 mg/mL dextran sulfate and 40% formamide in 2× SSC) was added. The slide was placed in a humidified chamber and incubated in the dark at 37°C overnight. On the next day, cells were washed twice with wash buffer at 37°C, and then stained with mounting media with DAPI (H-1500; Vector Labs).
Cells were imaged at 60× magnification using a Widefield Deltavision microscope. Images were processed by blind deconvolution with AutoQuant X3. Visualization of RNA foci were made using ImageJ. For quantification, at least 20 pictures were taken from randomly chosen microscopic fields, containing 100–300 cells for each treatment. Counting of foci was performed by different investigators. All data were generated by at least three independent experiments.
Results
ss-siRNA design
For duplex RNAs, the passenger strand protects the guide strand, allowing it to be incorporated into the RNA induced silencing complex. Single-stranded unmodified RNAs are too unstable to be efficient silencing agents and ss-siRNAs are chemically modified to balance being stable inside cells while maintaining activity through RNAi. Chemical modifications include 2′-O-methyl (2′-O-me), 2′-fluoro (2′-F), and 2′-O-methoxyethyl (2′-MOE) nucleosides and phosphodiester or phosphorothioate internucleotide linkages (Fig. 1B) [4,5]. ss-siRNAs were designed to be complementary to either c9orf72 GGGGCC sense intronic RNA (ss-siRNA sequences listed in Fig. 2A) or the CCCCGG c9orf72 antisense transcript (ss-siRNA sequences listed in Fig. 3A).
FIG. 2.
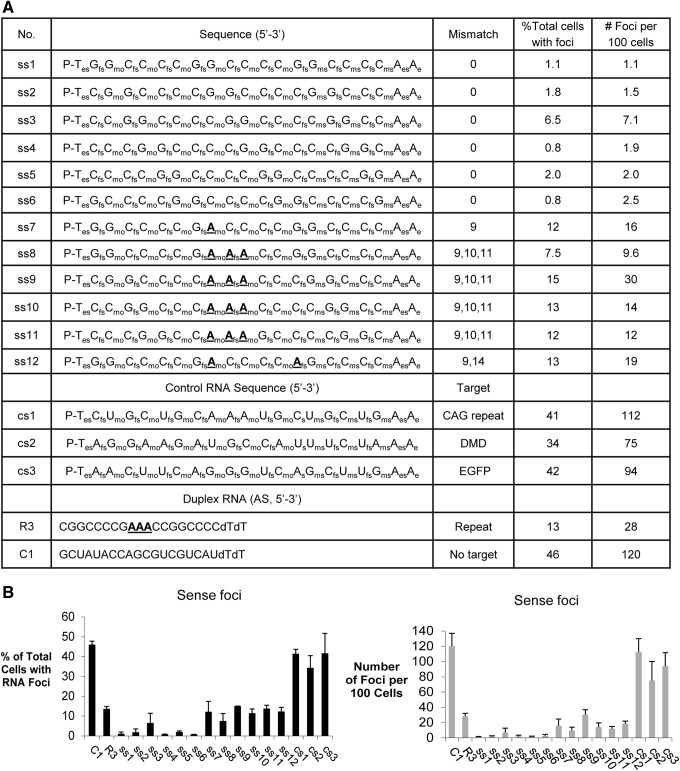
Inhibition of c9orf72 intronic sense foci by ss-siRNAs. (A) List of ss-siRNAs that are complementary to the expanded GGGGCC repeat and controls. A mismatched base is showed in bold face and underlined. (B) Quantitation of microscopy showing the percentage of cells with RNA foci and the number of foci per 100 cells. Fifty nanomolar of ss-siRNAs were transfected into c9orf72 patient-derived fibroblast cells.
FIG. 3.
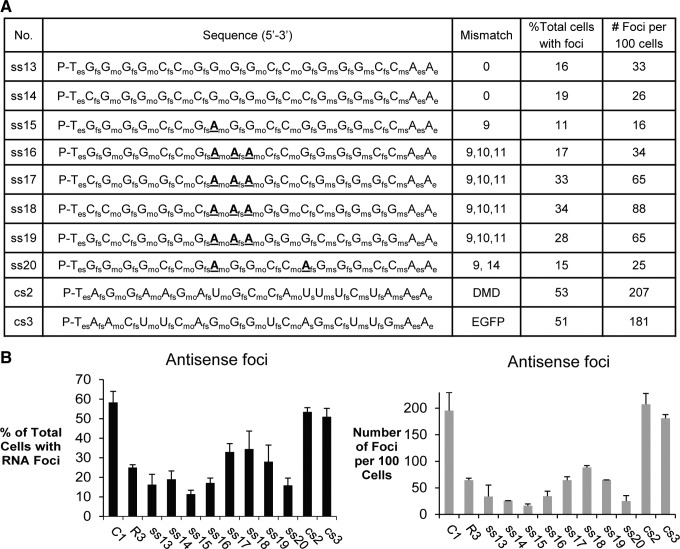
Inhibition of c9orf72 antisense foci by ss-siRNAs. (A) List of ss-siRNAs that are complementary to the expanded CCCCGG repeat and controls. (B) Quantitation of microscopy showing the percentage of cells with RNA foci and the number of foci per 100 cells. ss-siRNAs were transfected into patient-derived fibroblast cells with lipid RNAiMAX (Life Technologies) as previously described [19].
For ss-siRNAs ss7 to ss12, we introduced adenosine bases at central positions (Fig. 2A). Full complementarity between a target RNA sequence and small RNA is necessary for cleavage of target sequences by the RNAi factor argonaute [20]. We had previously observed that duplex RNAs containing central mismatches within the small RNA guide strand relative to trinucleotide [21] or hexanucleotide [19] repeat targets retain the ability to efficiently recognize mutant expanded repeat transcripts. Duplex RNAs that were fully complementarity were either not allele-selective for inhibition of genes with expanded CAG repeats (21) or were inactive toward blocking the foci formed by mutant c9orf72 (19).
Recognition of c9orf72 intronic transcript
We evaluated recognition of the expanded repeats within c9orf72 intronic RNA and the antisense transcript by measuring the ability of ss-siRNAs to block foci formation. Assays were performed in a patient-derived fibroblast cell line containing a mutant expanded repeat region. ss-siRNAs were delivered into cells by transfection using cationic lipid.
The mutant c9orf72 transcripts, both intronic and antisense, form foci that can be detected by FISH and appear in cell nuclei (Fig. 1C). For each compound, it was necessary to visualize and count several hundred cells to ensure sufficient statistical power for analysis. We determined the number of cells with detectable foci and the number of foci per hundred cells. Our laboratory has recently observed that the number of foci within cells correlates closely with the number of c9orf72 mutant expanded repeat transcripts in either the sense or antisense orientation (J. Liu, unpublished). These data suggest that each focus detected by FISH is a single RNA molecule. This conclusion was drawn from two different methods of quantifying RNA per droplet digital PCR and qPCR and comparing RNA copy number to the average number of foci detected within each cell by FISH.
We first tested ss-siRNAs targeting the GGGGCC expanded repeat with mutant c9orf72 intron 1. The first base of an ss-siRNA can begin recognizing the hexanucleotide repeat in six different registers. Therefore, we tested six different fully complementary ss-siRNAs (ss1–ss6). Each ss-siRNA was more effective (in terms of reducing the number of cells with foci and the number of foci per 100 cells) than negative control duplex RNA C1, which was inactive, and mismatch-containing RNA R3, a benchmark used in our earlier study [21] (Fig. 2B) (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat).
To evaluate the impact of disrupting the potential for target cleavage, we tested ss-siRNAs that contained 1–3 central mismatches relative to the expanded repeat target (ss7–ss12) (Fig. 2B). These mismatched ss-siRNAs were as active as mismatched duplex R3 but were less active than the fully complementary ss-siRNAs. Negative control ss-siRNAs cs1, cs2, and cs3 that lacked complementarity to target did not affect detection of foci.
Recognition of c9orf72 antisense transcript
We also examined the potential for ss-siRNAs to recognize the antisense transcript containing the CCCCGG expansion (Fig. 3A). Mismatched duplex R3 was again used as a benchmark because, since it has two strands, it targets the antisense transcript and the sense transcript [19]. Our results for the c9orf72 antisense transcript were similar to those we observed for the sense transcript. Both full-complementary and mismatch-containing ss-siRNAs reduced the total number of cells with foci and the number of foci per 100 cells (Fig. 3B). One difference between fully complementary and mismatch-containing duplex RNAs was that the fully complementary ss-siRNAs were more active than the ss-siRNAs containing central mismatches.
Mechanism of ss-siRNAs
ss-siRNAs are single-stranded oligonucleotides composed entirely of chemically modified bases. The design for chemical modifications was chosen to stabilize ss-siRNAs while also permitting them to operate through an RNAi mechanism. However, these modified bases are also used in ASOs that do not operate through an RNAi mechanism. It is possible, therefore, that RNAi may not be the only mechanism involved in gene silencing by ss-siRNAs. Direct recognition of target RNAs may also occur through a mechanism that is independent of RNAi factors and this recognition may contribute to the activity of ss-siRNAs. To test this hypothesis, we tested ss-siRNAs that were disabled with regards to function through RNAi.
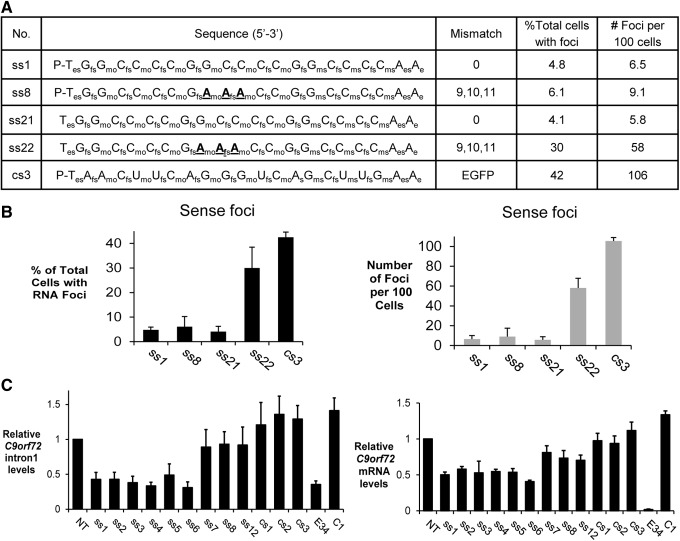
A 5′-phosphate is required for ss-siRNAs that contain a 5′-terminal methoxyethyl base to function through an RNA mechanism [5]. To test the hypothesis that multiple mechanisms contribute to blocking c9orf72 foci, we synthesized single-stranded RNAs ss21 and ss22 lacking a 5′ phosphate (Fig. 4A). These ss-siRNAs are analogs of active compounds ss1 and ss8 (Fig. 2). We first tested ss21, a single-stranded oligonucleotide that was fully complementary to intronic GGGGCC and lacked a 5′ phosphate. ss21 was as potent as analogous ss-siRNA ss1 that possessed an unblocked 5′ terminus (Fig. 4B). This result demonstrates that anti-GGGGCC ss-siRNAs can act though a non-RNAi antisense mechanism.
FIG. 4.
Mechanism of inhibition of c9orf72 intronic sense and antisense foci. Inhibition of c9orf72 intronic sense foci by ss-siRNAs. (A) List of ss-siRNAs. (B) Quantitation of microscopy showing the percentage of cells with sense RNA foci and the number of foci per 100 cells. (C) Quantitative polymerase chain reaction showing levels of sense intron 1 and mRNA after treating with 50 nM of ss-siRNAs. E34 is a duplex RNA targeting the c9orf72 coding region. ss-siRNAs were transfected into patient-derived fibroblast cells with lipid RNAiMAX (Life Technologies) as previously described [19].
Next, we tested ss-siRNA ss22, an oligonucleotide with three mismatches relative to target and lacking a 5′-phosphate. ss22 was much less active than its unblocked counterpart ss8, both in terms of the total number of cells with foci and the number of foci per cell (Fig. 4B). When mismatches are present, they destabilize interactions with the target. This destabilization may prevent efficient recognition in the absence of assistance by RNAi factors. These data show that a mismatched ss-siRNA that can function through RNAi is active, whereas the analogous mismatched oligonucleotide that is disabled for RNAi is not active. We conclude that involvement of RNAi factors like argonaute may be indispensable for maximal activity by ss-siRNAs that are not fully complementary to target.
Fully complementary duplex RNAs have the capacity to induce cleavage of target RNAs while RNAs that contain central mismatches do not [20]. We measured levels of c9orf72 intronic or mRNA after addition of ss-siRNAs (Fig. 4C). Consistent with the expectation that fully complementary RNAs would induce cleavage of targets, we observed that ss-siRNAs ss1–ss6 reduced levels of both transcripts while the mismatch-containing RNAs did not induce a significant reduction. For fully complementary ss-siRNAs, the reduction in RNA levels was ∼50%. We have previously observed this partial reduction with other repeat-containing RNA targets [12]. The partial reduction in RNA levels may reflect an inefficient cleavage mechanism when highly structured RNAs are involved. It is also possible that cleavage is less efficient because the c9orf72 intronic target is nuclear rather than cytoplasmic [22]. We note that reduction of foci was over 90% while reduction of RNA was ∼50%. This difference is consistent with a mechanism that reduced observation of foci is primarily due to blocking the repetitive RNA target rather than reducing RNA levels.
Discussion
Expansion of the GGGGCC/CCCCGG hexanucleotide repeat within intron 1 of the c9orf72 gene is a major cause of familial FTD/ALS [16–18]. Both mutant sense intronic RNA and the overlapping antisense transcript have been implicated in disease, either directly through binding to proteins or indirectly by encoding toxic dipeptides. Compounds that block expression or interfere with activity of these mutant RNAs would have the potential to treat both ALS and FTD. We have identified ss-siRNAs that can recognize and block either the mutant intronic transcript or the antisense transcript. Inhibition by fully complementary, but not mismatched, ss-siRNAs is superior in potency to inhibition by analogous duplex RNAs.
When duplex RNA is used to silence gene expression, the guide strand must dissociate from the passenger strand. For fully complementary duplexes targeting GGGGCC or CCCCGG hexanucleotide repeats, dissociation is likely to be inefficient because of exceptionally high C/G base-pairing. Inefficient dissociation of the guide strand from the passenger strand probably explains the inactivity of fully complementary duplexes reported in our previous study (21). ss-siRNAs, by definition, require no dissociation step because they are single-stranded. We find that fully complementary anti-GGGGCC or anti-CCCCGG ss-siRNAs readily recognize the c9orf72 intronic sense or antisense targets (Figs. 2 and 3). These data demonstrate that using ss-siRNAs can lead to more active compounds by eliminating any potential for the passenger strand to obstruct binding to target.
Inhibition by fully complementary oligonucleotide ss21 can be achieved when the 5′ terminus is blocked and function through RNAi is not possible (Fig. 4). This result demonstrates that ss-siRNAs can operate through a mechanism that is independent of RNAi. It is likely that this independent mechanism involves binding of oligonucleotide directly to the repeat sequence, analogous to mechanism of binding by standard ASOs. This mechanism, however, is not the primary mode of action when mismatches are introduced, the 5′-blocked mismatched RNA ss22 is inactive. From these observations, we deduce that anti-GGGGCC and anti-CCCCGG ss-siRNAs can operate by two different mechanisms, one RNAi dependent and the other RNAi independent. Neither standard ASOs nor duplex RNAs can function through both mechanisms, and the ability to combine effects may lead to greater potencies than analogous duplex RNAs.
In a previous study we examined the ability of ss-siRNAs to modulate expression of ataxin-3 by targeting an expanded CAG repeat [8]. In that report we observed allele-selective inhibition of mutant ataxin-3 expression that was dependent on RNAi. We also observed RNAi-independent production of an alternatively spliced protein isoform. Multiple mechanisms are likely to be a common feature of ss-siRNAs and are factors that should always be considered whenever ss-siRNAs are employed.
Conclusions
Our data provide another example of the activity of ss-siRNAs against challenging disease-related targets. ss-siRNAs have been successful as allele-selective inhibitors for expression of mutant huntingtin, ataxin-3, and atrophin-1. ss-siRNAs also regulate alternative splicing [12] and transcription [9]. These reports suggest that ss-siRNAs are a broadly useful platform for controlling gene expression in the laboratory. They can function against both nuclear and cytoplasmic targets.
For inhibition of c9orf72 sense and antisense transcripts, the potency was higher than the analogous duplex RNA. Because ss-siRNAs can combine function through RNAi and RNAi-independent mechanisms, they may have the potential to achieve better potencies for other targets in the future studies. Our current data support the hypothesis that ss-siRNAs are a promising approach that merits research to develop new applications.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM R35118103) and the Robert A. Welch Foundation (I-1244).
Author Disclosure Statement
T.P.P. and F.R. are employees of Ionis Pharmaceuticals. The other authors report no conflicts of interest.
References
- 1.Juliano RL. (2016). The delivery of therapeutic oligonucleotides. Nucleic Acids Res 44:6518–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair JK, Willoughby JLS, Chan A, Charisse K, Alam MD, Wang Q, Hoekstra M, Kandasamy P, Kel'in AV, et al. (2014). Multivalent N-Acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136:16958–16961 [DOI] [PubMed] [Google Scholar]
- 3.Geary RS, Norris D. and Bennett CF. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87:46–51 [DOI] [PubMed] [Google Scholar]
- 4.Prakash TP, Graham MJ, Yu J, Carty R, Low A, Chappell A, Schmidt K, Zhao C, Aghajan M, et al. (2014). Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-aceyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res 42:8796–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, et al. (2012). Single-stranded siRNAs activate RNAi in animals. Cell 150:883–894 [DOI] [PubMed] [Google Scholar]
- 6.Chorn G, Klein-McDowell M, Zhao L, Saunders MA, Flanagan WM, Willingham AT. and Lim LP. (2012). Single-stranded microRNA mimics. RNA 18:1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze E, Lima W, Crooke ST, Prakash T. and Corey DR. (2012). Single-stranded RNAs that function through RNAi are potent and allele-selective inhibitors of huntingtin expression. Cell 150:95–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Yu D, Aiba Y, Pendergraff H, Swayze EE, Lima WF, Prakash TP. and Corey DR. (2013). ss-siRNAs allele-selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res 41:9570–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui M, Prakash T. and Corey DR. (2013). Transcriptional silencing by promoter-targeted single-stranded RNAs. ACS Chem Biol 8:122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J, Lui J, Yu D, Aiba Y, Lee S, Pendergraff H, Boubaker J, Arates JW, Lagier-Tourenne C, et al. (2014). Exploring the effect of sequence length and composition on allele-selective inhibition of human huntingtin expression by single-stranded silencing RNAs. Nucleic Acid Ther 24:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, Liu J, Narayannair KJ, Lackey JG, Kuchimanchi S, Rajeev KG, Manoharan M, Swayze EE, Lima WF, et al. (2014). Allele-selective inhibition of mutant atrophin-1 expression by duplex and single-stranded RNAs. Biochemistry 53:4510–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Hu J, Hicks JA, Prakash TP. and Corey DR. (2015). Modulation of splicing by single-stranded silencing RNAs. Nucleic Acid Ther 25:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendergraff HM, Debacker AJ. and Watts JK. (2016). Single-stranded silencing RNAs: hit rate and chemical modification. Nucleic Acid Ther 26:216–222 [DOI] [PubMed] [Google Scholar]
- 14.Chang W, Pei Y, Guidry EN, Zewge D, Parish CA, Shere EC, DiMuzio J, Zhang H, South VJ, et al. (2016). Systematic chemical modifications of single stranded siRNAs significantly improved DTNNBI mRNA silencing. Bioorg Med Chem Lett 26:4513–4517 [DOI] [PubMed] [Google Scholar]
- 15.Matsui M, Prakash TP. and Corey DR. (2016). Argonaute 2-dependent regulation of gene expression by single-stranded miRNA mimics. Mol Ther 24:946–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, et al. (2012). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromsome 9p21-linked ALS-FTD. Neuron 72:257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendron TF, Bieniek KF, Zhang Y-J, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, et al. (2013). Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 53:839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J, Liu J, Liande L, Gagnon KT. and Corey DR. (2015). Targeted recognition of GGGGCC/CCCCGG repeats at the C9orf72 locus by duplex RNA. Chem Biol 22:1505–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Juranek S, Li H, Sheng G, Tuschl T. and Patel DJ. (2008). Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Liu J. and Corey DR. (2010). Allele-selectivity by switching to an miRNA-like RNAi mechanism. Chem Biol 17:1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennox KA. and Behlke MA. (2016). Cellular localization of long noncoding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44:863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.