Abstract
The on-going generation of HIV-1 intersubtype recombination has led to new circulating recombinant forms (CRFs) and unique recombinant forms (URFs) in Asia. In this study, we evaluated whether previously reported URFs were actually CRFs. All available complete or near full-length HIV-1 URF sequences from Asia were retrieved from the HIV Los Alamos National Laboratory Sequence database, and phylogenetic, transmission cluster, and bootscan analyses were performed using MEGA 6.0, Cluster Picker 1.2.1, and SimPlot3.5.1. According to the criterion of new CRFs, two new HIV-1 CRFs (CRF87_cpx and CRF88_BC) were identified from these available URFs. CRF87_cpx comprised HIV-1 subtypes B, C, and CRF01_AE, and CRF88_BC comprised subtypes B and C. HIV Blast and bootscan analysis revealed that besides the three representative strains, there were two additional CRF87_cpx strains. Furthermore, we defined seven dominant URFs (dURF01-dURF07), each of which contained two strains sharing same recombination map and can be used as sequence references to facilitate the finding of new potential CRFs in future. These results will benefit the molecular epidemiological investigation of HIV-1 in Asia.
Keywords: : HIV-1, subtype, URF, CRF, Asia, cluster
The human immunodeficiency virus (HIV)-1 has high genetic diversity and recombination rate.1 There are four groups of HIV-1 (M, N, O, and P), and the M group has caused the global epidemic of HIV/AIDS. There are 11 well-defined subtypes (A1, A2, B, C, D, F1, F2, G, H, J, and K) and many recombinant forms (www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html). The complicated genetic diversity of HIV-1 is a great challenge for vaccine development and antiretroviral therapy (ART) use.1,2
HIV-1 B, C, and CRF01_AE are the major subtypes circulating in Asia, and the cocirculation of these subtypes has led to the generation of some circulating recombinant forms (CRFs) (e.g., CRF07_BC, CRF08_BC, CRF15_01B, and CRF33_01B), as well as a large number of unique recombinant forms (URFs).3 In particular, diverse HIV-1 subtypes and CRFs (e.g., B, C, CRF01_AE, CRF07_BC, and CRF08_BC) are cocirculating among injection drug users (IDUs) and other high-risk groups in the China–Myanmar border region, an important area involved in drug trafficking and HIV-1 transmission. This has resulted in a very high proportion of HIV-1 recombinants,4–9 and this area has become a “hot spot” region for the generation of various HIV-1 recombinants,4,6,7 compared to other countries and regions.3
It remains unclear, however, if many of the previously identified URFs are actually CRFs. The accepted definition of an HIV-1 CRF is a recombinant virus that has been isolated from at least three epidemiologically unlinked individuals, while a URF is defined as a recombinant virus that has been found only in one individual.10 The possible reason that many viral infections were labeled as URF, instead of CRF, is because most of the reported URFs were identified through partial genomic sequences, rather than by full-length or near full-length genomic (NFLG) sequences.4,6,7,11 In addition, most of the URFs were reported by different research groups. Therefore, we hypothesized that there were some new CRFs that were misclassified as URFs in the publically available Los Alamos National Laboratories HIV sequence database (LANL, www.hiv.lanl.gov). In this study, we searched for potential new CRFs among reported URFs by systematically reanalyzing all available NFLG sequences of URFs identified in Asia.
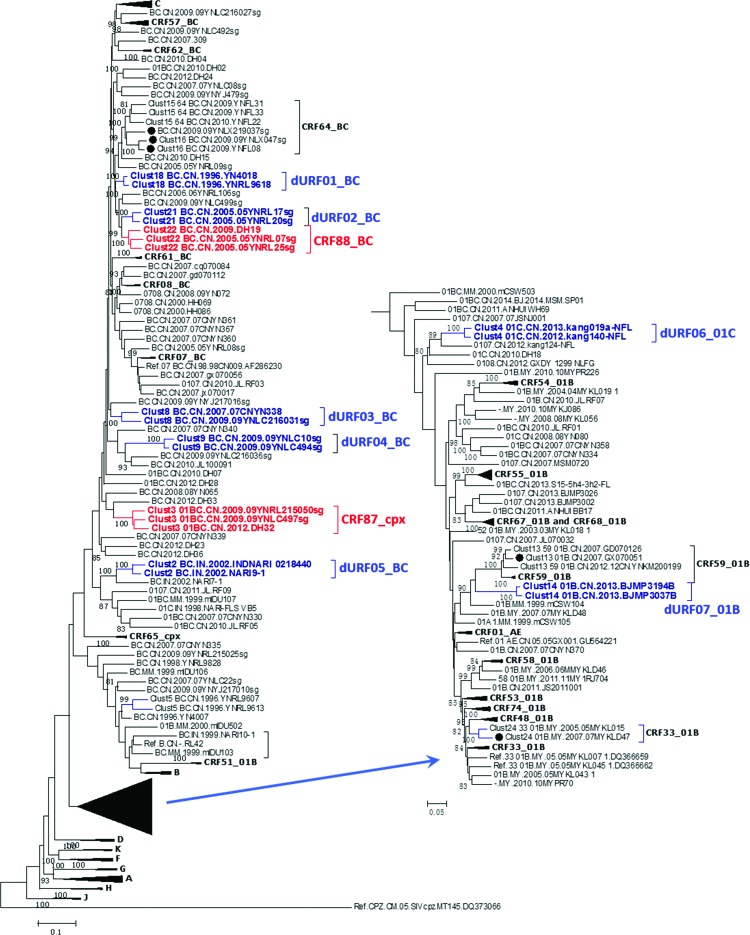
All full-length and/or NFLG HIV-1 recombinant URFs sequences from Southeast Asia were downloaded from HIV LANL database in September, 2015. A total of 172 NFLG sequences (accession numbers in the Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid) were obtained. These sequences included 14 from India, 29 from Malaysia, 7 from Myanmar, and 122 from China. These sequences were aligned with HIV-1 subtype reference sequences using MUSCLE implemented in MEGA 6.0, and a Maximum Likelihood (ML) tree was constructed using MEGA 6.0 under General Time Reversible model. Since a CRF is defined as having at least three NFLG sequences that are isolated from epidemiologically unlinked individuals and share same mosaic patterns,10 putative CRF candidates should cluster together in the ML phylogenetic tree.
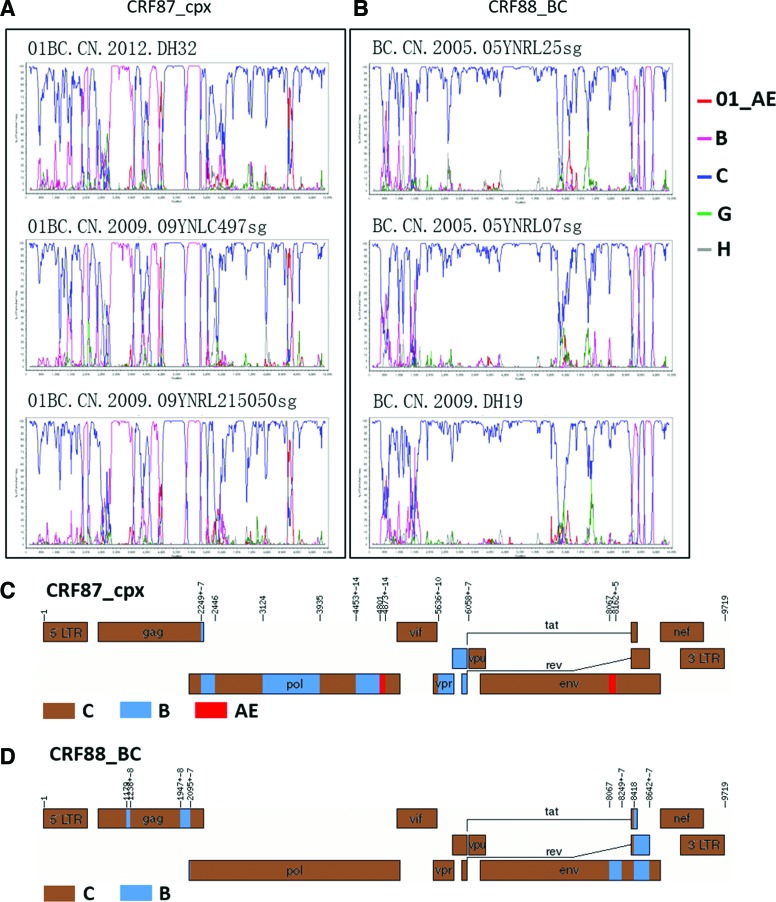
To screen for potential candidates of CRFs, clusters formed by the downloaded URFs were analyzed using Cluster Picker 1.2.112 and 11 clusters were identified (Fig. 1). Two clusters (clusters 3 and 22) contained three sequences, suggesting that they were potentially new HIV-1 CRFs. To determine whether they shared same recombination patterns, bootscanning and similarity plot analyses were performed using SimPlot v.3.5.1.13 Comparison of the bootscan plots showed that three URFs (01BC.CN.2012.DH32, 01BC.CN.2009.09YNLC497 sg, and 01BC.CN.2009.09YNLC215050 sg) in cluster 3 shared same recombination patterns (Fig. 2A). Because the URFs in cluster 3 comprised segments of CRF01_AE, B, and C, they were defined as CRF87_cpx. Specifically, CRF87_cpx was divided into two CRF 01AE, four B, and six C segments by 11 breakpoints, as follows: C (1-2248 nt), B (2249-2445 nt), C (2446-3123 nt), B (3124-3934 nt), C (3935-4452 nt), B (4453-4800 nt), CRF01AE (4801-4872 nt), C (4873-5635 nt), B (5636-6057 nt), C (6058-8066 nt), CRF01AE (8067-8161 nt), and C (8162-9719 nt), using HXB2 as a reference (Fig. 2C). In cluster 22, three URFs BC.CN.2005.05YNRL25 sg, BC.CN.2005.05YNRL07 sg, and BC.CN.2009.DH19 shared the same recombination patterns (Fig. 2B). Thus, they were designated as CRF88_BC, which consisted of three B and four C segments, as follows: C (1-1178 nt), B (1179-1237 nt), C (1238-1946 nt), B (1947-2094 nt), C (2095-8066 nt), B (8067-8248 nt), C (8249-8417 nt), B (8418-8641 nt), and C (8642-9719 nt), using HXB2 as a reference (Fig. 2D).
FIG. 1.
Phylogenetic and transmission cluster analyses of NFLG sequences of URFs available in Asia. NFLG, near full-length genome; URF, unique recombinant form. Color images available online at www.liebertpub.com/aid
FIG. 2.
Two new CRFs identified in this study. (A) and (B) bootscan plots of three representative strains of CRF87_cpx and CRF88_BC, respectively. (C) and (D) Mosaic maps of CRF87_cpx and CRF88_BC, respectively. The parameters of the SimPlot bootscan analysis were window size, 300 bps; step size, 20bps; and GapStrip on. The schematic maps were generated by Recombinant HIV-1 Drawing Tool available at the LANL web site. Breakpoint positions were numbered according to HXB2. Color images available online at www.liebertpub.com/aid
Since often the pol and vif-env are the hot-spot regions for HIV-1 recombination,2,6 to determine whether other URFs in the region also belonged to the two newly identified CRFs, we performed HIV BLAST using the pol, gag, and vif-env sequences of each representative strain of both CRF87_cpx and CRF88_BC as query. To first screen sequences with highest similarity to the query sequence, the distance trees of all hit sequences were constructed with the query sequences. The hit sequences that clustered with the query sequences were then subjected to further bootscan analyses. Two pol sequences from strains 1452 and 803 were found to cluster with the three representative sequences (01BC.CN.2012.DH32, 01BC.CN.2009.09YNLC497 sg, and 01BC.CN.2009.09YNLC215050 sg) of CRF87_cpx (Supplementary Fig. S1). Bootscan analyses showed that the pol sequences of both strains 1452 and 803 shared identical breakpoints in HIV-1 pol region with 01BC.CN.2012.DH32, 01BC.CN.2009.09YNLC497 sg, and 01BC.CN.2009.09YNLC215050 sg (Supplementary Fig. S2). These results suggest that strains 1452 and 803 might also belong to CRF87_cpx. For CRF88_BC, no hit sequence was found to cluster with the representative sequences, implying a limited prevalence.
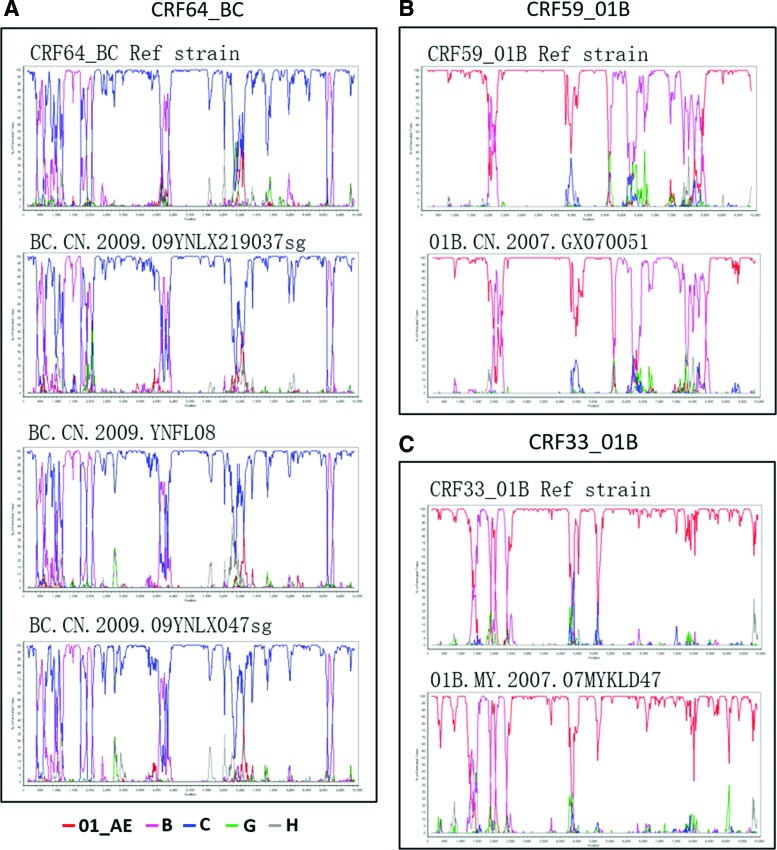
Apart from the two newly identified CRFs, we also found that three URFs (BC.CN.2009.09YNLX219037 sg, BC.CN.2009.YNFL08, and BC.CN.2009.09YNLX047 sg) closely clustered with the strains of CRF64_BC (Fig. 1). Bootscan analysis further showed that they shared identical recombination patterns with the representative strains of CRF64_01B (Fig. 3A). These results indicate that they were CRF64_01B, rather than URFs. Similarly, previously identified URF 01B.CN.2007.GX070051 and 01B.MY.2007.07MYKLD47 closely clustered with CRF59_01B and CRF33_01B, respectively, and shared a recombination pattern with the latter (Fig. 3B), indicating that 01B.CN.2007.GX070051 belonged to CRF59_01B and 01B.MY.2007.07MYKLD47 belonged to CRF33_01B (Fig. 3C). Therefore, the sequence information of the five previously identified URFs should be updated in the HIV LANL sequence database accordingly.
FIG. 3.
Bootscan analyses of four URFs that should be updated as known CRFs. (A), (B), and (C) bootscan plots of suspected CRF64_BC, CRF59_01B, and CRF33_01B strains, respectively. The suspected CRF64_BC strains contained BC.CN.2009.09YNLX219037 sg, BC.CN.2009.YNFL08, and BC.CN.2009.09YNLX047 sg. The suspected CRF59_BC strain was 01B.CN.2007.GX070051, and the CRF33_01B was 01B. MY. 2007. 07MYKLD47. Color images available online at www.liebertpub.com/aid
The cocirculation of various HIV-1 subtypes and their quick transmission across risk groups (e.g., IDUs and sexual contact) and geographic regions have facilitated the generation of HIV recombinants, both CRF and URF.3,11,14–16 An increasing number of HIV URFs was reported in Asia (especially in the China–Myanmar border region) in recent years.3 However, systematic comparison of mosaic patterns with all available URFs has not been performed often in some studies, which limits the identification of new CRFs. In this study, we showed that cluster and bootscan analyses can be used to better classify CRF versus URF in public sequence databases. Seven clusters containing two URFs sharing identical recombination patterns were found in this study (Supplementary Fig. S3). They represent the potential CRFs, given that one or more new HIV-1 sequences have been found to share the same recombination pattern to one of them. We propose that they should be defined as “dominant” URF (dURF01 to dURF07) (Table 1) in the field, and we recommend others to use them as sequence references to facilitate the finding of new potential CRFs in future. In addition, we caution that the reporting of new HIV-1 URFs should require at least NFLG sequence data and rigorous systematic phylogenetic and bootscan analyses that would include available subtypes, CRFs, and URFs, including these dURFs.
Table 1.
The Dominant Unique Recombinant Forms Identified in This Study
| dURFs | Representative strains | Accession number |
|---|---|---|
| dURF01_BC | BC.CN.1996.YN4018 | KF250380 |
| BC.CN.1996.YNRL9618 | AY967807 | |
| dURF02_BC | BC.CN.2005.05YNRL17 sg | KC899006 |
| BC.CN.2005.05YNRL20 sg | KC898978 | |
| dURF03_BC | BC.CN.2007.07CNYN338 | KF835524 |
| BC.CN.2009.09YNLC216031 sg | KC898987 | |
| dURF04_BC | BC.CN.2009.09YNLC10 sg | KC898984 |
| BC.CN.2009.09YNLC494 sg | KC898990 | |
| dURF05_BC | BC.IN.2002.INDNARI 0218440 | EU000514 |
| BC.IN.2002.NARI9-1 | EU000506 | |
| dURF06_01C | 01C.CN.2013.kang019a-NFL | KJ778895 |
| 01C.CN.2012.kang140-NFL | KJ778896 | |
| dURF07_01B | 01B.CN.2013.BJMP3194B | KP418806 |
| 01B.CN.2013.BJMP3037B | KP418805 |
The NFLG sequences of the seven dURFs are available in Supplementary Data 2.
NFLG, near full-length genome; URF, unique recombinant form.
All strains of both CRF87_cpx and CRF88_BC were isolated from Yunnan, indicating a local prevalence of both CRFs. Together with CRF65_cpx and CRF78_cpx identified in Yunnan in 2014 and 2016, respectively,17,18 there have now been found four new CRFs occurring in Yunnan in recent years. This complexity of HIV molecular epidemiology in the region is likely because Yunnan, China, borders Myanmar, Laos, and Vietnam, and contains important channels for illicit drug trafficking and HIV-1 transmission.16 The use of illicit drugs being a risk factor for the observed CRFs is supported by the observations that all new CRFs described in this study were collected from IDUs, except one CRF87_cpx strain (01BC.CN.2012.DH32) that was isolated from a man reporting sexual risk. Together, these data suggest that the region continues to be an active source of HIV genetic diversity and spread.4,6,7 Whether new CRF will result in larger spread needs close epidemiological monitoring.
In summary, we identified two new HIV-1 CRFs (CRF87_cpx and CRF88_BC) from available URFs in Asia and defined seven dominant URFs (dURF01-dURF07), each of which contained two strains sharing same recombination patterns. In addition, we updated three previously defined URFs as CRF64_BC, CRF59_01B and CRF33_01B, respectively. Altogether, these results will benefit the molecular epidemiological investigation of HIV-1 in Asia.
Sequences Data
The NLFG sequences of CRF87_cpx and CRF88_BC are available in the Supplementary Data 1, and the NFLG sequences of the seven dURFs are available in the Supplementary Data 2.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (U1302224, 81271888, 81271892 and 81601802) and National Institute of Health (MH083552). The funders had no role in study design, the collection, analysis, and interpretation of data, the writing of the report, and the decision to submit the article for publication.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Zanini F, Brodin J, Thebo L, et al. : Population genomics of intrapatient HIV-1 evolution. Elife 2015;4: pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramirez BC, Simon-Loriere E, Galetto R, Negroni M: Implications of recombination for HIV diversity. Virus Res 2008;134:64–73 [DOI] [PubMed] [Google Scholar]
- 3.Phanuphak N, Lo YR, Shao Y, et al. : HIV Epidemic in Asia: Implications for HIV vaccine and other prevention trials. AIDS Res Hum Retroviruses 2015;31:1060–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang R, Xia X, Kusagawa S, Zhang C, Ben K, Takebe Y: On-going generation of multiple forms of HIV-1 intersubtype recombinants in the Yunnan Province of China. AIDS 2002;16:1401–1407 [DOI] [PubMed] [Google Scholar]
- 5.Takebe Y, Motomura K, Tatsumi M, Lwin HH, Zaw M, Kusagawa S: High prevalence of diverse forms of HIV-1 intersubtype recombinants in Central Myanmar: Geographical hot spot of extensive recombination. AIDS 2003;17:2077–2087 [DOI] [PubMed] [Google Scholar]
- 6.Pang W, Zhang C, Duo L, et al. : Extensive and complex HIV-1 recombination between B', C and CRF01_AE among IDUs in south-east Asia. AIDS 2012;26:1121–1129 [DOI] [PubMed] [Google Scholar]
- 7.Han X, An M, Zhao B, et al. : High prevalence of HIV-1 intersubtype B'/C recombinants among injecting drug users in Dehong, China. PLoS One 2013;8:e65337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Yang L, Ma Y, et al. : Emerging variability in HIV-1 genetics among recently infected individuals in Yunnan, China. PloS One 2013;8:e60101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Ye M, Duo L, et al. : First description of two new HIV-1 recombinant forms CRF82_cpx and CRF83_cpx among drug users in Northern Myanmar. Virulence 2016:0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson DL, Anderson JP, Bradac JA, et al. : HIV-1 nomenclature proposal. Science 2000;288:55–56 [DOI] [PubMed] [Google Scholar]
- 11.Zhou YH, Liang YB, Pang W, et al. : Diverse forms of HIV-1 among Burmese long-distance truck drivers imply their contribution to HIV-1 cross-border transmission. BMC Infect Dis 2014;14:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragonnet-Cronin M, Hodcroft E, Hue S, et al. : Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013;14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lole KS, Bollinger RC, Paranjape RS, et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou YH, Chen X, Liang YB, et al. : Near Full-Length Identification of a Novel HIV-1 CRF01_AE/B/C Recombinant in Northern Myanmar. AIDS Res Hum Retroviruses 2015;31:845–850 [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Jia Y, Xu Q, Zheng YT, Zhang C: Phylodynamics of HIV-1 unique recombinant forms in China-Myanmar border: Implication for HIV-1 transmission to Myanmar from Dehong, China. Infect Genet Evol 2012;12:1944–1948 [DOI] [PubMed] [Google Scholar]
- 16.Beyrer C, Razak MH, Lisam K, Chen J, Lui W, Yu XF: Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. AIDS 2000;14:75–83 [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Wei H, Hsi J, et al. : Identification of a novel HIV Type 1 circulating recombinant form (CRF65_cpx) composed of CRF01_AE and subtypes B and C in Western Yunnan, China. AIDS Res Hum Retroviruses 2014;30:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Feng Y, Miao Z, et al. : Near-Full-Length Genome Sequences of a Novel HIV-1 Circulating Recombinant Form, CRF01_AE/B'/C (CRF78_cpx), in Yunnan, China. AIDS Res Hum Retroviruses 2016;32:601–606 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.