Abstract
The epicardium promotes neovascularization and cardiomyocyte regeneration by generating vascular smooth muscle cells (SMCs) and producing regenerative factors after adult heart infarction. It is therefore a potential cell resource for repair of the injured heart. However, the epicardium also participates in fibrosis and scarring of the injured heart, complicating its use in regenerative medicine. In this study, we report coexpression of TBX18 and WT1 in the majority of epicardial cells during mouse embryonic epicardial development. Furthermore, we describe a convenient chemically defined, immunogen-free, small molecule-based method for generating TBX18+/WT1+ epicardial-like cell populations with 80% homogeneity from human pluripotent stem cells by modulation of the WNT and retinoic acid signaling pathways. These epicardial-like cells exhibited characteristic epicardial cell morphology following passaging and differentiation into functional SMCs or cardiac fibroblast-like cells. Our findings add to existing understanding of human epicardial development and provide an efficient and stable method for generating both human epicardial-like cells and SMCs.
Keywords: : human pluripotent stem cell, epicardial cell, WNT, RA, smooth muscle cell
Introduction
Epicardial cells are required for embryonic heart formation. The proepicardium, a transient embryonic structure composed of epicardial progenitor cells, develops at the inflow of the heart tube and localizes posterior to the sinus venosus (SV) and atria [1]. Although the precise origin of the proepicardium is unclear, several genetic lineage-tracing analyses have suggested that the proepicardium and myocardium develop from a common cardiogenic precursor pool [2]. At the looping stage of heart development, proepicardial cells migrate onto the heart tube and cover the heart surface, thereby forming the epicardium [3].
A subset of epicardial cells undergo epithelial-to-mesenchymal transition (EMT) and invade the subepicardium and myocardial layer as epicardium-derived cells (EPDCs) to generate cardiac fibroblasts (CFs), smooth muscle cells (SMCs), and coronary endothelial cells that contribute to coronary vessel formation [4–7]. In addition, embryonic epicardial cells promote cardiomyocyte proliferation and maturation by producing paracrine factors, including retinoic acid (RA), fibroblast growth factor (FGF)-9, and insulin-like growth factor-2 [8–10].
In the adult mammalian heart, the proliferative ability of cardiomyocytes is not retained and the vast majority of epicardial cells have lost expression of the embryonic epicardial marker genes TBX18 and WT1. Myocardial infarction (MI) and thymosin β4 injection can reactivate expression of these two genes in adult epicardium and subepicardium, leading to neovascularization in infarcted ventricular tissue [11–14]. Furthermore, transplantation of adult human EPDCs into a mouse infarction model has been shown to improve heart function and vascularization [15,16]. Thus, epicardial cells and EPDCs, particularly TBX18- and WT1-expressing epicardial cells, are potential cell sources for repair of injured heart tissue using cell-based transplantation.
Recently, two groups reported WT1+ epicardial cell differentiation methods that used the cytokine basic FGF (bFGF), bone morphogenetic protein (BMP) 4, activin A, vascular endothelial growth factor, and WNT3A in an animal product-containing medium [17,18]. Our differentiation method is based on an understanding of signals involved in epicardial development.
Early studies in chicken embryo showed expression of retinaldehyde dehydrogenase 2 (RALDH2), the rate-limiting enzyme for RA synthesis, in the posterior part of the lateral mesoderm, which includes proepicardial precursors, the atria, and SV progenitors [19,20]. At this developmental stage, WNT-8c and the competitive WNT inhibitor, Crescent, are distributed at opposite ends of the embryo. The posterior part of the heart field, which develops into the atria, is located within the Crescent-expressing region, while proepicardial precursors are located within the anterior edge of the WNT-expressing zone [21]. Therefore, we hypothesized that RA is required for both atrium and proepicardium development, with WNT signals inducing the fate separation of atrial myocytes and epicardial cells.
In this study, we showed that WNT activated WT1 expression in human pluripotent stem cell (hPSC)-derived cardiac progenitor cells (CPCs), and RA promoted TBX18 expression in the WT1+ cell population. Manipulating the WNT and RA signaling pathways with small molecular compounds in a chemically defined albumin-free medium led to ∼80% of hPSCs differentiating into TBX18+/WT1+ epicardial-like cells. These cells possessed epicardial cell characteristics, exhibited a cobblestone-like morphology following passaging, and differentiated into functional SMCs or CF-like cells when induced with transforming growth factor β1 (TGFβ1) and bFGF. This method could potentially be used to efficiently and stably generate minimally pathogenic epicardial cells and coronary SMCs for clinical cardiovascular tissue engineering and heart regeneration in the future.
Materials and Methods
Animals and tissue sections
Institute for Cancer Research mice were obtained from the Center for Animal Research, Institute of Biophysics, Chinese Academy of Sciences. All experiments with mice were performed according to protocols approved by the Institute of Biophysics. Pregnant females were sacrificed and embryonic hearts (E10.5, E12.5, and E14.5) were harvested. Isolated hearts were embedded in the Jung Tissue Freezing Medium (No. 020108926; Leica, Nussloch, Germany), sectioned at 8 μm, and mounted on microscope slides (No. FRC-05; Matsunami, Osaka, Japan).
hPSC culture and passage
H7 and H9 human embryonic stem cells (hESCs; WiCell Research Institute, Madison, WI), and xeno-free and virus-free (XVF) human-induced pluripotent stem cells (hiPSCs; human foreskin fibroblast [HFF]-derived XVF1 and XVF2, generated within our laboratory), were maintained in an E8 medium [22] containing 2 ng/mL TGFβ1 (No. 100-21; PeproTech, Rocky Hill, NJ) and 100 ng/mL bFGF (No. 100-18B; PeproTech). hPSCs were passaged onto vitronectin-coated plates at a 1:6 ratio every 6 days using collagenase IV (No. 17104-019; Gibco, Grand Island, NY) digestion.
hPSC differentiation
Two days before differentiation, hPSCs were digested to single cells by 0.48 mM ethylene diamine tetraacetic acid (EDTA), then plated as a monolayer on vitronectin-coated 24-well plates at a density of 2.5 × 105 cells/well. When the cells reached full confluence, differentiation procedures were initiated. A chemically defined S12 differentiation medium was developed within our laboratory (unpublished results). On day (D) 0, the culture medium was changed from E8 to S12 medium (without insulin) supplemented with 6 μM CHIR99021 (CHIR; No. 4423; Tocris, Bristol, UK) to induce mesoderm formation. CHIR was withdrawn after 24 h to avoid excessive WNT activation. On D3, the medium was changed to S12 (without insulin) and supplemented with 5 μM IWR1 (No. I0161; Sigma, St. Louis, MO). During D5–8, cells were cultured with 1 μM RA (No. R2625; Sigma) to induce atrial progenitor cell formation. After D8, CPCs were cultured in S12 to promote cardiomyocyte differentiation. For epicardial differentiation, media were supplemented with an additional 5 μM CHIR for D5–8. On D8, the cells were passaged once at a ratio of 2:5.
Monolayer passage and differentiation of SMCs and CFs
For generation of epithelial-like cells (EPLCs), D14 cultures were trypsinized and plated onto vitronectin-coated plates at a density of 2 × 104 cells/cm2. After an initial 24-h recovery period, the S12 medium was changed every 2 days until analysis. As indicated in Figures 4A and 6A, supplementation of TGFβ1 (5 ng/mL) and bFGF (10 ng/mL) in the culture medium was used for SMC and fibroblast differentiation. For the maturation of SMCs, cells were exposed to 5 ng/mL TGFβ1 for 6 days. To generate CFs, further culturing of bFGF-induced cells with bFGF in 10% fetal calf serum (Gibco)-containing S12 medium was performed for 6 days.
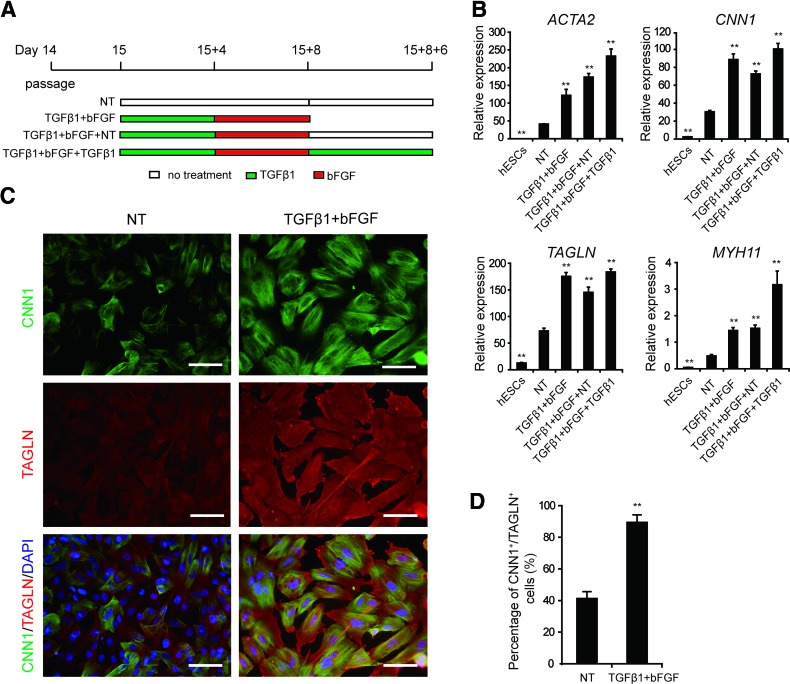
FIG. 4.
EPLCs have the potential to differentiate into SMC-like cells. (A) Schematic of the protocol used for SMC induction. (B) qRT-PCR analysis of expression of the SMC markers ACTA2, CNN1, TAGLN, and MYH11 in cells treated with the indicated factors. Gene expression was normalized to TBP. Error bars represent SEM; n = 3; **p < 0.01 versus untreated cultures, analyzed by the Student's t-test. (C) Immunofluorescence staining of CNN1 and TAGLN in D15 + 8 NT and TGFβ1+bFGF-induced EPLC cultures. Scale bars, 100 μm. (D) Flow cytometry analysis of the proportion of CNN1+/TAGLN+ cells in D15 + 8 NT and TGFβ1+bFGF-induced EPLC cultures. Error bars represent SEM; n = 3; **p < 0.01. bFGF, basic fibroblast growth factor; NT, no treated; SMC, smooth muscle cell; TGFβ1, transforming growth factor β1. Color images available online at www.liebertpub.com/scd
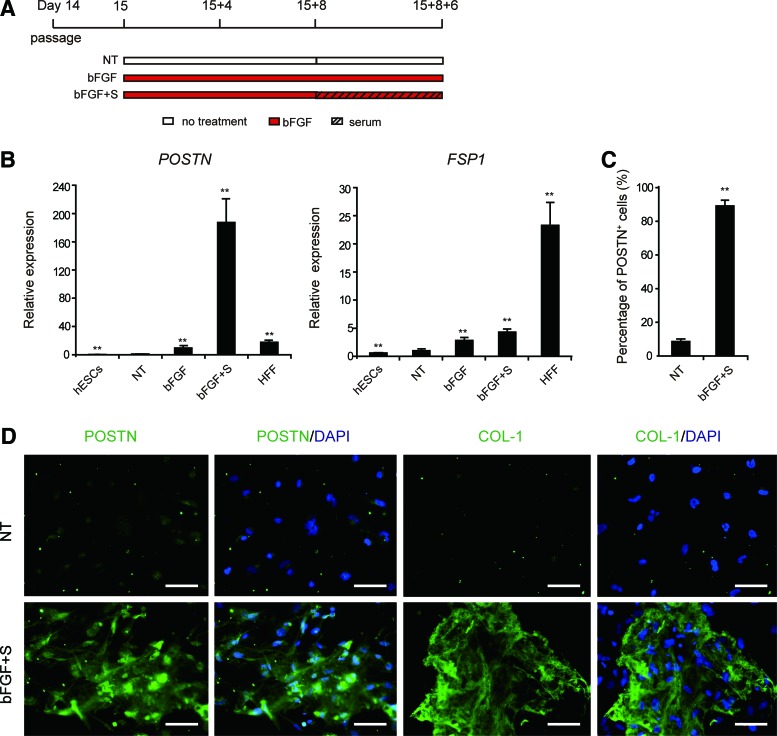
FIG. 6.
EPLCs have the potential to differentiate into CF-like cells. (A) Schematic of the protocol used for CF-like cell induction. (B) qRT-PCR analysis of expression of the fibroblast markers POSTN and FSP1 in hESCs, HFFs, and EPL-derived cells. Gene expression was normalized to TBP. Error bars represent SEM; n = 3; **p < 0.01 versus untreated cultures, analyzed by the Student's t-test. (C) Flow cytometry analysis of the proportion of POSTN+ cells in untreated and bFGF+S-induced cultures. Error bars represent SEM; n = 3; **p < 0.01. (D) Immunofluorescence staining of POSTN and COL-1 in untreated and bFGF+S-induced cultures. Scale bars, 100 μm. CF, cardiac fibroblast; HFFs, human foreskin fibroblasts. Color images available online at www.liebertpub.com/scd
Quantitative real-time polymerase chain reaction
Total RNA was extracted with TRIzol® reagent (No. 15596; Life Technologies, Carlsbad, CA) and 1 μg RNA was reverse transcribed into complementary DNA in a 20-μL volume of PrimeScript RT reagent with gDNA Eraser (No. RR047A; Takara, Shiga, Japan). A QuantiFast SYBR® Green PCR Kit (No. 204057; Qiagen, Hilden, Germany) was used for qPCR in a Rotor-Gene Q 2plex Real-Time PCR Machine (No. 9001620; Qiagen). Relative gene expression was calculated by normalizing values to the housekeeping gene TATA binding protein. Primer sequences are listed in Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd.
Flow cytometry
Differentiated cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.4% Triton™ X-100 (No. T8787; Sigma). After blocking in 5% donkey or goat serum, cells were stained with primary antibodies against ISL1 (diluted 1:1,000; sc23590; Santa Cruz, Dallas, TX), cTnT (0.5 μg/mL; MAB1874; R&D, Minneapolis, MN), WT1 (diluted 1:1,000; ab89901; Abcam, Cambridge, UK), CNN1 (diluted 1:10,000; C2687; Sigma), TAGLN (diluted 1:1,000; ab14106; Abcam), POSTN (diluted 1:1,000; ab14041; Abcam), or KDR-PE (10 μL/106 cells; FAB357P; R&D).
Alexa Fluor® 488 donkey anti-goat IgG (705-545-147; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), Alexa Fluor 488 goat anti-rabbit IgG (111-545-003; Jackson), Alexa Fluor 488 goat anti-mouse IgG (115-545-003; Jackson), and PE goat anti-rabbit IgG (GR200G-09C; Sungene Biotech, Tianjin, China) were used as secondary antibodies. Goat IgG (sc3887; Santa Cruz), Rabbit IgG (ab199376; Abcam), mouse IgG1-PE (IC002P; R&D), and mouse IgG1 (M5284; Sigma) were used as isotype controls. Samples were assessed using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) and data were analyzed using FlowJo (Treestar).
Immunofluorescence and microscopy
Slides containing cells or heart sections were fixed in 4% PFA for 15 min and then permeabilized with 0.4% Triton X-100 for 15 min. Slides were then blocked with 5% goat or donkey serum in phosphate-buffered saline for 1 h and then incubated with primary antibodies against WT1 (diluted 1:1,000; ab89901; Abcam), TBX18 (diluted 1:100; sc17869; Santa Cruz), cTnT (0.5 μg/mL; MAB1874; R&D), ZO1 (diluted 1:100; 339100; Life Science), CNN1 (diluted 1:10,000; C2687; Sigma), TAGLN (diluted 1:1,000; ab14106; Abcam), POSTN (diluted 1:1,000; ab14041; Abcam), or COL-1 (diluted 1:200; ab90395; Abcam) overnight at 4°C.
Slides were then incubated with the relevant secondary antibody: Alexa Fluor 594 goat anti-rabbit IgG (111-585-003; Jackson), Alexa Fluor 488 goat anti-mouse IgG (115-545-003; Jackson), Alexa Fluor 488 donkey anti-goat IgG (705-545-147; Jackson), Alexa Fluor 594 donkey anti-rabbit IgG (711-585-152; Jackson), or Alexa Fluor 488-goat anti-rabbit IgG (111-545-003; Jackson) for 1 h at room temperature. Nuclei were counterstained by incubation with DAPI (0.5 μg/mL; D3571; Life Technologies) for 1–3 min. Immunofluorescence images were visualized and captured using an Olympus DP71 camera (Tokyo, Japan). Proportional analysis of TBX18+/WT1+ cells was performed using an ImageXpress® Micro Widefield High-content Screening System (Molecular Devices, Sunnyvale, CA). Bright field images were visualized and captured using a Zeiss AX10 microscope.
Calcium assay
Cells were preloaded with the calcium-sensitive molecular probe Fluo-4 AM (2.5 μM; F14201; Life Technologies) in Tyrode's solution consisting of 140 mM NaCl, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose (pH 7.4) at 37°C for 30 min. Cells were then trypsinized and washed with Tyrode's solution. Calcium was measured by recording changes in mean fluorescent intensity before and after the addition of carbachol (100 μM; 51-83-2; Sigma) using a FACSCalibur instrument. Relative mean fluorescent intensity was normalized to the value obtained at 0 min (before carbachol addition).
Calcium imaging
Primary human coronary artery SMCs (HCASMCs, ATCC) and EPL-SMCs were preloaded with 2.5 μM Fluo-4 AM in Tyrode's solution at 37°C for 30 min. Cells were then washed thrice with Tyrode's solution. Calcium imaging was performed using a Leica TCS SP5 confocal microscope with 488-nm excitation. Images with 1,024 × 1,024 pixel resolution were continuously acquired using a 40× objective and combined into a continuous sequence representing a 260-s period. All calcium transient recordings were performed using the same excitation and acquisition settings. Each recording consisted of a 100-s baseline, and a 160-s recording period following the addition of 15 μM phenylephrine (No. S2569; Selleck, Houston, TX). Three independent experiments were performed for TGFβ1+bFGF+TGFβ1-induced cultures and no treated (NT) cultures.
Fluorescence intensity (F) of EPL-SMCs exhibiting calcium transients was measured and then normalized to the average intensity during the last 52 s before agonist addition (baseline, F0). The proportion of functional EPL-SMCs was determined by counting cells exhibiting calcium transients and dividing by the total number of cells in the field of view. Kinetic parameters of the Ca2+ signal (tmax: time to peak, t1/2on: time to half-peak, t1/2off: time from the peak to half recovery) were determined using Microsoft Excel software [23]. Twelve EPL-SMCs were randomly selected from fields of three independent experiments for assessment of change in fluorescent cell surface, as assessed by ImageJ software (National Institutes of Health, Bethesda, MD).
Results
Coexpression of TBX18 and WT1 is a reliable marker for early embryonic epicardial cells
The proepicardium/epicardium consists of heterogeneous cell populations that differentially express several marker genes, including TBX18, WT1, Sema3D, SCX, TCF21, and NFATC1, either alone or in various combinations [7,24,25]. Among these, TBX18 and WT1 are two well-established epicardium marker genes used separately in lineage-tracing studies to identify cells descended from the proepicardium/epicardium [26–30]. However, whether these two genes label the same epicardial cell population has not been confirmed. In addition to expression in the epicardium, TBX18 is also expressed in the SV myocardium and neighboring mesenchyme [31], and WT1 is expressed in the endocardium and cardiac endothelial cells [29,32]. Therefore, expression of TBX18 or WT1 alone is not a restrictive criterion for the identification of hPSC-differentiated epicardial cells in vitro.
To address this question, we examined expression patterns of TBX18 and WT1 in mouse E10.5–E14.5 embryonic hearts with double immunofluorescence staining. Our results indicate that the majority of epicardial cells at the outer myocardial layer were double positive for TBX18 and WT1, with few WT1 single positive cells in the myocardial layer of the E14.5 heart (Supplementary Fig. S1). This finding confirms that TBX18 and WT1 label the vast majority of embryonic epicardial cells, supporting their coexpression as a stringent criterion for the identification of hPSC-differentiated epicardial cells in vitro.
WNT signaling switches CPCs from atrial myocytes to WT1+ noncardiomyocyte cells
To test our hypothesis that RA and WNT signaling are required for proepicardial differentiation, we activated WNT signaling during differentiation of hESCs into atrial myocytes. hESCs were differentiated into atrial myocytes using a small molecule-based cardiac differentiation method developed by Lian et al., with RA addition on day (D) 5 [33,34]. Briefly, hESCs were treated with the WNT signaling pathway agonist CHIR (6 μM) to induce mesodermal differentiation over the first 24 h. On D3, cells were exposed to the WNT signaling pathway antagonist IWR1 (5 μM) for 48 h to generate CPCs [33]. Cells were then treated with 1 μM RA from D5–8 to induce differentiation into atrial myocytes. hESCs were differentiated into epicardial cells using the atrial myocyte differentiation protocol, except that cells were treated with RA and CHIR from D5 to 8 (Fig. 1A).
FIG. 1.
WNT signaling determines the WT1+ noncardiomyocyte fate of CPCs. (A) Schematic of the protocol for epicardial differentiation of hPSCs using CHIR99021 (CHIR), IWR1, and RA. (B) qRT-PCR analysis of expression of M markers (T and MESP1) and CPC markers (ISL1 and NKX2.5) on day (D) 2 and D5 cultures. (C) Flow cytometric analysis of the proportions of ISL1+ and KDR+ cells in D5 cultures. Error bars represent standard error of mean (SEM); n = 3. (D) qRT-PCR analysis of expression of the epicardial markers TBX18 and WT1 in D14 cultures subjected to the indicated treatments. RA, 1 μM; IWR1, 5 μM; CHIR, 0–12 μM; –, DMSO was used as a vehicle control. (E) Flow cytometric analysis of the proportion of WT1+ cells in D14 cultures. Error bars represent SEM; n = 3; *p < 0.05, **p < 0.01 versus RA-treated cultures. (F) qRT-PCR analysis of cTnT expression in D14 cultures treated with RA or RA/CHIR. (G) Flow cytometric analysis of the proportion of cTnT+ cells in D14 cultures treated with RA or RA/CHIR. Error bars represent SEM; n = 3; **p < 0.01 versus RA-treated cultures. (H) Immunofluorescence staining of cTnT and WT1 in D14 cultures treated with RA or RA/CHIR. Scale bars, 100 μm. Quantitative real-time polymerase chain reaction (qRT-PCR) values shown are relative to the housekeeping gene TBP. Error bars represent SEM; n = 3; *p < 0.05, **p < 0.01 versus hESC or RA-treated cultures, analyzed by the Student's t-test. CPCs, cardiac progenitor cells; hESC, human embryonic stem cell; hPSCs, human pluripotent stem cells; M, mesoderm; RA, retinoic acid; TBP, TATA binding protein. Color images available online at www.liebertpub.com/scd
Quantitative real-time polymerase chain reaction (qRT-PCR) revealed a substantial increase in the expression of mesodermal markers (T and MESP1) on D2. After 5 days of differentiation, expression levels of ISL1 and NKX2.5, two markers of CPCs [35,36], were significantly increased (Fig. 1B). Flow cytometry analysis of cardiac progenitor marker expression indicated that 94.2% of cells were Isl1 positive, and 62.3% of cells expressed KDR, a cardiovascular progenitor marker [37] (Fig. 1C). qRT-PCR results for D14-differentiated cultures indicated that, in the presence of RA, expression levels of both TBX18 and WT1 were significantly upregulated following treatment with CHIR on D5. Indeed, cultures treated with 1 μM RA and 5 μM CHIR (RA/CHIR) exhibited a 24-fold increase of TBX18 and an 88-fold increase of WT1 expression compared with cultures treated with RA alone (Fig. 1D).
Meanwhile, expression of the cardiomyocyte marker cTnT was significantly reduced in RA/CHIR-treated cultures (Fig. 1F). Flow cytometry analysis showed that 90.6% of cells were WT1 positive and less than 3% of cells were cTnT positive in RA/CHIR-treated cultures (Fig. 1E, G). Double immunofluorescence staining also showed that the majority of cells in RA/CHIR-treated cultures were WT1+/cTnT− noncardiomyocytes, while the majority of cells in RA-treated cultures were WT1−/cTnT+ cardiomyocytes (Fig. 1H). As previously reported [34,38], single-cell patch clamp analysis for action potentials indicated that more than 90% of RA-differentiated cardiomyocytes produced atrial-like action potentials (unpublished results). Taken together, these results demonstrate that WNT signaling activates WT1 gene expression in CPCs, whereby switching cell fate from atrial myocyte to WT1+ noncardiomyocyte.
WNT and RA act synergistically to promote epicardial cell fate specification
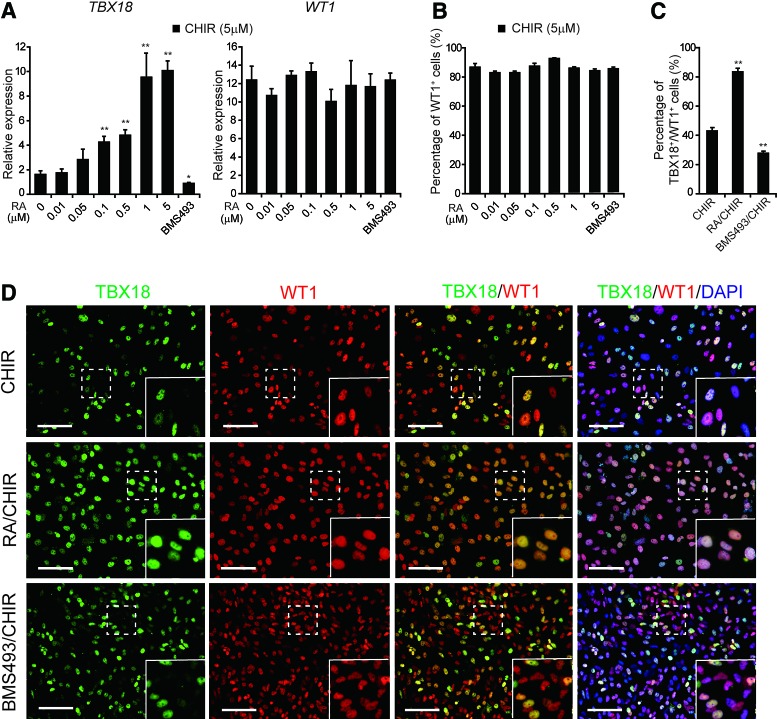
We next investigated the roles of RA in regulating WT1 and TBX18 expression. In the presence of 5 μM CHIR, addition of RA to cultures had no significant effect on WT1 expression or differentiation of WT1+ cells (Fig. 2A, B). Interestingly, addition of RA to CHIR-treated cultures upregulated TBX18 expression in a dose-dependent manner, with a fivefold increase observed with 1 μM RA (Fig. 2A). Using high-content imaging assays (no TBX18 antibodies tested were suitable for flow cytometry), we found that 27.8% of cells were double positive for TBX18 and WT1 in cultures treated with CHIR and the RA inhibitor BMS493 versus 83.5% in RA/CHIR-treated cultures (Fig. 2C, D).
FIG. 2.
WNT and RA act synergistically to specify TBX18+/WT1+ cell fate. (A) qRT-PCR analysis of TBX18 and WT1 expression in D14 cultures following treatment with 5 μM CHIR and the indicated concentrations of RA. BMS493, 5 μM. Gene expression was normalized to TBP. Error bars represent SEM; n = 3; *p < 0.05, **p < 0.01 versus CHIR (5 μM)-treated cultures, analyzed by the Student's t-test. (B) Flow cytometry analysis of the proportion of WT1+ cells in D14 cultures. Error bars represent SEM; n = 3. (C) High-content imaging assays of the proportion of TBX18+/WT1+ cells in D14 cultures treated with CHIR, RA/CHIR, or BMS493/CHIR. Error bars represent SEM; n = 3; **p < 0.01 versus CHIR (5 μM)-treated cultures. (D) Immunofluorescence staining of TBX18 and WT1 in D14 cultures treated with CHIR, RA/CHIR, or BMS493/CHIR. Yellow in the inset panels indicates TBX18 and WT1 coexpression. Scale bars, 100 μm. Color images available online at www.liebertpub.com/scd
These results indicate that CHIR activates WT1 expression, while RA promotes TBX18 expression in the WT1+ cell population, suggesting that simultaneous activation of WNT and RA signaling pathways efficiently drives CPC differentiation into TBX18+/WT1+ cells, the major epicardial cell population of the embryonic heart. We designated these D14 TBX18+/WT1+ cells derived from RA/CHIR treatment as proepicardium-like cells (pEPLCs).
Morphological and molecular characterization of hPSC-derived epicardial-like cells
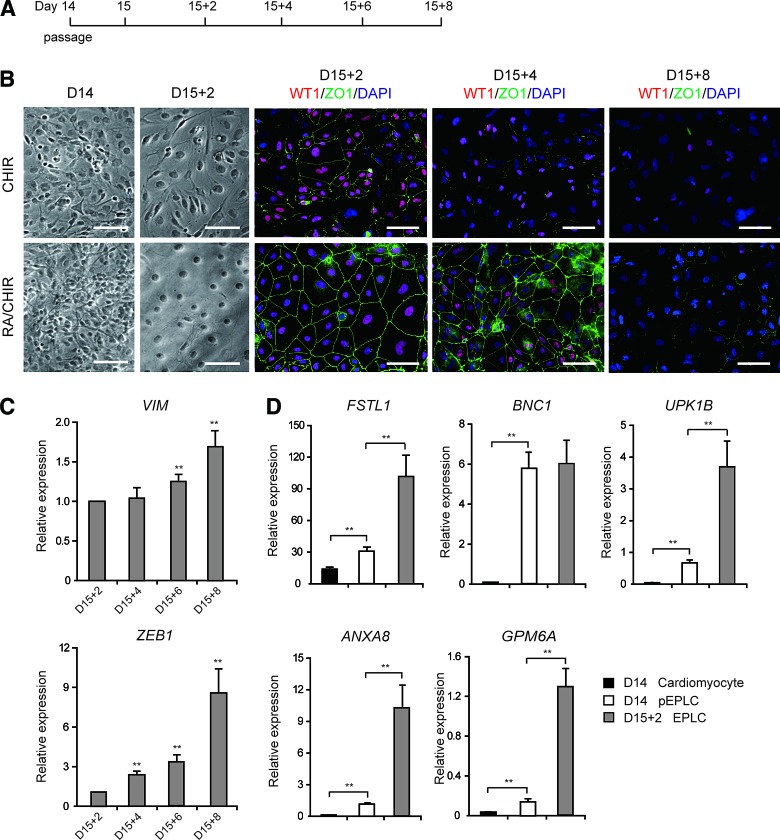
During embryonic heart development, the epicardium forms an epithelial-like sheet expressing ZO1, a marker of epithelial tight junctions [39]. D14 pEPLCs initially lack epithelial-like morphology; however, following passage at low density (2.5 × 104 cells/cm2) (Fig. 3A), these cells formed an epithelial monolayer with cobblestone morphology and expressed ZO1 along cell borders (Fig. 3B, D15 + 2). However, expression of WT1 and ZO1 quickly declined after D15 + 4 (Fig. 3B), indicating that these cells may undergo EMT spontaneously [40,41]. Consistent with this hypothesis, expression levels of the mesenchymal markers VIM and ZEB1 increased in passaged pEPLCs after D15 + 2 (Fig. 3C).
FIG. 3.
Characteristics of EPLCs. (A) Schematic of protocol for EPLC generation. (B) Bright field and immunofluorescence staining micrographs of CHIR- and RA/CHIR-treated cultures. RA/CHIR-treated cultures exhibited cobblestone-like morphology and high expression of the epithelial marker ZO1 at D15 + 2 after passage. Scale bars, 100 μm. (C) qRT-PCR analysis of expression of the EMT markers VIM and ZEB1 at various time points. Error bars represent SEM; n = 3; **p < 0.01 versus D15 + 2 cells, analyzed by the Student's t-test. (D) qRT-PCR analysis of expression of the epicardial genes BNC1, UPK1B, ANXA8, and GPM6A in D15 + 2 EPLCs, D14 pEPLCs, and cardiomyocytes. Gene expression was normalized to TBP. Error bars represent SEM; n = 3; **p < 0.01 versus relative controls, analyzed by the Student's t-test. pEPLCs, proepicardium-like cells. Color images available online at www.liebertpub.com/scd
In contrast, cells treated with CHIR alone and plated at the same density as passaged pEPLCs did not develop typical epithelial morphology, and displayed impaired ZO1 expression (Fig. 3B). qRT-PCR analysis of expression of the epicardium-produced cardiomyocyte regeneration factor FSTL1 [42] and the epicardium-specific genes BNC1, UPK1B, ANXA8, and GPM6A [43] showed that passaged D15 + 2 pEPLCs exhibited higher expression levels of epicardial genes than D14 pEPLCs and D14 cardiomyocytes (Fig. 3D). Collectively, these results indicate that RA/CHIR-induced TBX18+/WT1+ pEPLCs can recapitulate in vivo epicardium development and express epicardial-specific markers following passage. We henceforth refer to these post-D15 passaged pEPLCs as EPLCs.
Epicardial-like cells are capable of generating functional SMCs
During cardiac development, epicardial cells have the potential to differentiate into SMCs and CFs [4,5]. TGFβ1 and bFGF regulate epicardial EMT and induce SMC differentiation [17]. To further differentiate hESC-derived EPLCs into SMCs, D15 EPLCs were treated with TGFβ1+bFGF for 8 days as previously reported [17] (Fig. 4A). qRT-PCR analysis indicated that TGFβ1+bFGF treatment increased expression of the SMC marker genes ACTA2, CNN1, TAGLN, and MYH11 by ∼2.5-fold (Fig. 4B). Immunofluorescence staining further confirmed increased levels of CNN1 and TAGLN protein expression in D15 + 8 TGFβ1+bFGF-induced EPLCs compared with D15 + 8-untreated EPLCs (NT) (Fig. 4C). Flow cytometry also showed that 89.4% of D15 + 8 TGFβ1+bFGF-induced EPLCs were CNN1+/TAGLN+, while only 41.3% of D15 + 8 NT cells were CNN1+/TAGLN+ (Fig. 4D).
To verify the identity of SMC-like cells, we used the calcium-sensitive dye Fluo-4 AM and carbachol stimulation to functionally characterize EPLC-derived cells by examining Ca2+ flux, which regulates smooth muscle contraction [18]. In contrast with HeLa cells (used as a negative control), in which no significant response was elicited following 30 s of exposure to carbachol, D15 + 8 NT cells showed a weak response and TGFβ1+bFGF-induced cultures showed a marked response (Fig. 5A). TGFβ1 promotes maturation of SMCs in vitro [44]. As expected, additional TGFβ1 treatment for 6 days increased expression of MYH11, a marker of mature contractile SMCs, by twofold in D15 + 8 + 6 TGFβ1+bFGF+TGFβ1-induced EPLCs (Fig. 4A, B). Ca2+ signaling analysis also showed that TGFβ1+bFGF+TGFβ1-induced EPLCs exhibited a higher level of calcium activity than TGFβ1+bFGF-induced EPLCs (Fig. 5A), indicating that TGFβ1 and bFGF promote differentiation of EPLCs into SMCs, and additional TGFβ1 treatment promotes maturation of TGFβ1+bFGF-induced SMCs.
FIG. 5.
Functional assay of EPLC-derived SMCs (EPL-SMCs). (A) Time-course of change in relative mean fluorescence intensity of Fluo-4-preloaded EPLC-derived cells, determined by flow cytometry after carbachol addition. Error bars represent SEM; n = 3/group; *p < 0.05, **p < 0.01 versus untreated cultures, analyzed by the Student's t-test. (B) Total proportion of functional EPLC-SMCs displaying calcium transients after phenylephrine addition in D15 + 8 + 6 NT and TGFβ1+bFGF+TGFβ1-induced cultures. Error bars represent SEM; n = 3/group; **p < 0.01 versus untreated cultures, analyzed by the Student's t-test. (C) Calcium imaging examining the functional response of EPLC-SMCs and HCASMCs to phenylephrine. Average fluorescence intensity normalized to baseline (F/Fo) of five random cells exhibiting calcium transients was measured. Peaks correspond to active calcium cycles in response to 15 μM phenylephrine added at 100 s, eliciting different responses. (D) Amplitude of calcium transients after phenylephrine addition. Error bars represent SEM; n = 5 cells; **p < 0.01 versus HCASMCs, analyzed by the Student's t-test. (E) Kinetic parameters of calcium transients after phenylephrine addition. Error bars represent SEM; n = 5 cells; **p < 0.01 versus HCASMCs, analyzed by the Student's t-test. (F) Fluo-4 AM-loaded EPLC-SMCs and HCASMCs displayed a change in cell surface area following phenylephrine stimulation. Scale bars, 100 μm. (G) Percentage change in cell surface area. Error bars represent SEM; n = 12 cells; **p < 0.01 versus D15 + 8 + 6 cells that did not exhibit calcium transients, analyzed by the Student's t-test. HCASMCs, human coronary artery smooth muscle cells; NS, no significant difference. Color images available online at www.liebertpub.com/scd
To visually and accurately test the calcium-handling activity of EPLC-SMCs, we measured calcium transients following stimulation with phenylephrine in D15 + 8 + 6 cultures. HCASMCs were used as a positive control (Fig. 5C; Supplementary Videos S1 and S2). More cells displayed calcium transients in TGFβ1+bFGF+TGFβ1-induced cultures (>80%) than in NT cultures (∼40%), indicating that TGFβ1+bFGF+TGFβ1 promotes EPLC differentiation into functional SMCs (Fig. 5B).
Next, we analyzed the amplitude and dynamics of phenylephrine-induced calcium transients in NT cultures, TGFβ1+bFGF+TGFβ1-induced cultures, and HCASMCs using a method as described previously [23]. We found no significant difference in peak amplitudes of relative fluorescence intensity (F/F0) between TGFβ1+bFGF+TGFβ1-induced cultures and HCASMCs (Fig. 5D), but the kinetic parameter values [23] of Ca2+ transients (tmax, t1/2on, and t1/2off) in TGFβ1+bFGF+TGFβ1-induced cultures were higher than those in HCASMCs (Fig. 5E), suggesting that EPLC-SMCS had a weaker calcium-handling activity than primary HCASMCs.
Contraction was assessed with ImageJ software by measuring the difference in cell surface area before and after phenylephrine treatment. As shown in Figure 5F and G, after application of phenylephrine, similar cell contractions occurred in both TGFβ1+bFGF+TGFβ1-induced cultures (29.5%) and HCASMCs (33.8%), while only a 9.1% reduction in cell surface area was observed in NT cultures. Taken together, these findings demonstrate that EPLCs have the potential to differentiate into functional SMCs and TGFβ1+bFGF+TGFβ1 promotes the SMC fate determination of EPLCs.
Epicardial-like cells are capable of generating CF-like cells
To investigate their potential for differentiating into CF-like cells, hESC-derived EPLCs were treated with bFGF for 14 days to induce fibroblast differentiation [17] (Fig. 6A). qRT-PCR analysis indicated that bFGF treatment increased expression of POSTN, a specific marker of embryonic CFs [45,46] (Fig. 6B). Furthermore, culturing EPLCs with bFGF in a serum (S)-containing medium (bFGF+S) for an additional 6 days after D15 + 8 dramatically enhanced expression of POSTN, such that levels were 17 times higher than those in HFFs. Conversely, expression of FSP1, a fibroblast marker gene absent in CFs [47], was much lower in bFGF+S-induced cells than in HFFs (Fig. 6B). Flow cytometry indicated that 89.2% of bFGF+S-induced EPLCs were POSTN+ (Fig. 6C). Furthermore, bFGF+S-induced EPLCs not only expressed POSTN but also secreted COL-1, a matrix protein secreted by CFs in the heart [48] (Fig. 6D). Collectively, these findings demonstrate that EPLCs possess the potential to differentiate into CF-like cells.
To demonstrate the reproducibility of this protocol, we differentiated pEPLCs and EPLCs from another hESC line (H9) and two hiPSC lines (XVF1 and XVF2) with the same approach. Double immunofluorescence staining showed that both of these lines generated a high percentage of TBX18+/WT1+ cells (∼80%, assessed by high-content imaging assay; Supplementary Fig. S2A, B) and these cells could give rise to hPSC-EPLCs following passage (Supplementary Fig. S2C). The hPSC-EPLCs could be differentiated into CNN1+/TAGLN+ SMCs or POSTN+ CFs by treating with TGFβ1+bFGF or bFGF+S (Supplementary Fig. S2D, E). Flow cytometric analysis further revealed that the induction efficiency for production of SMCs (>80%) and CFs (∼85%) from H9-EPLCs and hiPSC-EPLCs was similar to that for H7-EPLCs using this protocol (Supplementary Fig. S2F, G).
Discussion
WT1 and TBX18 are two key genes that regulate epicardium development. In a WT1−/− mouse model, the epicardium was missing over much of the heart, with no EDPCs or vessels observed [49]. Tbx18−/− mouse epicardium had a rough epithelial appearance and impaired coronary vasculogenesis [50]. Indeed, coronary vasculature formation is responsive to Tbx18-dependent gene targets in the epicardium [50]. Our results demonstrate that a major subpopulation of epicardial cells homogeneously express both TBX18 and WT1. These findings indicate that TBX18+/WT1+ is a more authentic marker for epicardial cells than WT1+ alone and expression of TBX18 in epicardial cells is necessary for their coronary vasculature fate, which supports neovascularization of damaged heart tissue. Furthermore, TBX18+/WT1+ epicardial cell-derived SMCs have great potential for drug testing, building vascular disease models from patient hiPSCs, and possibly generation of artificial coronary vessels.
Results reported by Iyer et al. indicated that RA treatment plays a major role in generating epicardium, which consists of a heterogeneous mix of WT1+ and TCF21+ cells [18]. In contrast, Witty et al. reported a method for generating high percentages of WT1+ cells without the addition of exogenous RA [17]. In our system, highly homogenous TBX18+/WT1+ epicardial cells (∼80% of total cell population) were induced by treatment with RA and WNT signaling agonists.
Our results suggest a collaborative relationship between WNT and RA signaling in epicardial cell fate determination. The WNT signaling agonist switches the CPC from RA-induced atrial myocyte to epicardium. In the presence of WNT signaling, RA treatment of CPCs did not affect WT1 expression, but rather increased the proportion of TBX18+ cells within the WT1+ cell population. This finding implies that WNT, not RA, is the dominant factor for epicardial determination. Consistent with this hypothesis, double-null Dkk1−/−/Dkk2−/− mouse mutant models exhibited embryonic heart tissue that preferentially formed epicardium rather than myocardium [51], while RALDH2−/− mice have visually normal proepicardium [52].
It is difficult to ascertain the role of RA in epicardium determination by comparing our findings with those of Witty et al. [17], as they did not quantify the proportion of TBX18+ cells. In Iyer et al.’s study, RA played a more important role than WNT in inducing WT1 expression. To some extent, this result was inconsistent with findings in mutant models [51,52]. Considering that the concentration and time-course of RA treatment in Iyer et al.’s study was very high and prolonged, it is possible that there is more than one way to induce epicardium differentiation, as occurred in vivo [30,31,53].
Whether BMP signaling is involved in proepicardium development in vivo remains unclear. In Witty et al.’s study, WT1+ cells could be generated by activating BMP or WNT signaling pathways in mesoderm. However, activation or inhibition of the BMP signaling pathway did not change the cardiomyocyte fate of differentiating cells in our system (unpublished results). This difference may be caused by the generation of distinct mesoderm before epicardium induction. In Witty et al.’s system, WNT signaling was not inhibited, which led to KDR+/PDGFRα+ lateral plate mesoderm generation. In contrast, WNT signaling was inhibited in our system to generate ISL1+ cardiac mesoderm. It can be speculated that selective combinations of different factors, which may give rise to the different parts of mesoderm, appear equally capable of generating epicardial-like cells. This speculation is consistent with findings that there may be multiple origins of epicardial progenitors [30,53,54]. Moreover, Witty et al.’s data also suggested that BMP and WNT signaling pathways act independently in epicardium determination. Whether WT1+ cells generated by each method in Witty et al.’s study were identical is currently unknown. More in-depth study of signaling pathways activated by these various treatments will aid greater understanding of epicardial origin and heterogeneity.
Epicardial cells have been demonstrated as a potential cell source for heart injury therapy. TBX18 and WT1 regulate formation of both epicardium and coronary vessels in mouse embryonic heart [49,50], and their reactivation in adult heart is involved in the reprogramming of adult epicardial cells into embryonic epicardial cells, facilitating heart regeneration after MI [55–57]. Previous studies have shown that transplantation of adult human EPDCs into mouse MI models improved heart function and vascularization [15,16]. These findings raise the hope of a potential application for TBX18+/WT1+ epicardial cells in cell-based therapies for MI. Furthermore, a recent report showed that epicardial FSTL1 delivery was critical for cardiomyocyte regeneration in adult mouse and swine MI models [42]. Thus, our FSTL1-expressing TBX18+/WT1+ epicardial cells may represent an alternative cell resource or additional cell type that could be used in the generation of cardiomyocyte patches for cell-based repair of MI. Nevertheless, although cardiomyocytes are very sparse in our differentiated cultures, arrhythmogenic foci would likely be generated after transplantation therapy. Furthermore, epicardial cell purification with an epicardium-specific surface marker, such as CD44 [40], may improve availability.
This study developed a novel culture method combining small molecules and a chemically defined medium for reliable and efficient generation of epicardial cells from hPSCs. Methods developed by both Witty et al. and Iyer et al. used cytokines, including bFGF, BMP4, activin A, vascular endothelial growth factor, and WNT3A, for epicardium differentiation in albumin-containing media [17,18]. We speculate that using small molecular compounds instead of cytokines makes our differentiation system more stable and economical. Furthermore, maintenance and differentiation of stem cells were carried out in chemically defined, xeno-free, albumin-free media, thereby reducing the risks of pathogenic and immunogenic contamination, which may make these cells more suitable for future clinical application. Taken together, our findings add to existing understanding of epicardial development and provide a simplified method for differentiation of epicardial cells.
Supplementary Material
Acknowledgments
The authors thank Dr. Guangju Ji and Dr. Zhihai Qin of the Institute of Biophysics for support with instrument use, and Dr. Ting Xie of the University of Kansas School of Medicine for assistance with the article. This work was supported by the Hi-Tech Research and Development Program of China (863 Program; Y286021001), the National Program on Key Basic Research Project (973 Program; Y197061001, 2010CB945204), the National Natural Science Foundation of China (Y4JM201001, Y4JY281001, Y5JY181001), the Strategic Priority Research Program of the Chinese Academy of Sciences (Y1CF062001), and the Beijing Municipal Science and Technology Project (Y4DG021001).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Manner J, Perez-Pomares JM, Macias D. and Munoz-Chapuli R. (2001). The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs 169:89–103 [DOI] [PubMed] [Google Scholar]
- 2.van Wijk B. and van den Hoff M. (2010). Epicardium and myocardium originate from a common cardiogenic precursor pool. Trends Cardiovasc Med 20:1–7 [DOI] [PubMed] [Google Scholar]
- 3.Schlueter J. and Brand T. (2011). Origin and fates of the proepicardium. Aswan Heart Cent Sci Pract Ser 2011:11 doi: http://dx.doi.org/10.5339/ahcsps.2011.11
- 4.Dettman RW, Denetclaw W, Jr, Ordahl CP. and Bristow J. (1998). Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol 193:169–181 [DOI] [PubMed] [Google Scholar]
- 5.Guadix JA, Carmona R, Munoz-Chapuli R. and Perez-Pomares JM. (2006). In vivo and in vitro analysis of the vasculogenic potential of avian proepicardial and epicardial cells. Dev Dyn 235:1014–1026 [DOI] [PubMed] [Google Scholar]
- 6.Cano E, Carmona R, Ruiz-Villalba A, Rojas A, Chau YY, Wagner KD, Wagner N, Hastie ND, Munoz-Chapuli R. and Perez-Pomares JM. (2016). Extracardiac septum transversum/proepicardial endothelial cells pattern embryonic coronary arterio-venous connections. Proc Natl Acad Sci U S A 113:656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA. and Tabin CJ. (2012). Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell 22:639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J. and Ornitz DM. (2005). Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell 8:85–95 [DOI] [PubMed] [Google Scholar]
- 9.Li P, Cavallero S, Gu Y, Chen TH, Hughes J, Hassan AB, Bruning JC, Pashmforoush M. and Sucov HM. (2011). IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 138:1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weeke-Klimp A, Bax NA, Bellu AR, Winter EM, Vrolijk J, Plantinga J, Maas S, Brinker M, Mahtab EA, et al. (2010). Epicardium-derived cells enhance proliferation, cellular maturation and alignment of cardiomyocytes. J Mol Cell Cardiol 49:606–616 [DOI] [PubMed] [Google Scholar]
- 11.Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, et al. (2011). Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest 121:1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR. and Riley PR. (2007). Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 445:177–182 [DOI] [PubMed] [Google Scholar]
- 13.Bock-Marquette I, Shrivastava S, Pipes GCT, Thatcher JE, Blystone A, Shelton JM, Galindo CL, Melegh B, Srivastava D, Olson EN. and DiMaio JM. (2009). Thymosin β4 mediated PKC activation is essential to initiate the embryonic coronary developmental program and epicardial progenitor cell activation in adult mice in vivo. J Mol Cell Cardiol 46:728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smart N, Risebro CA, Clark JE, Ehler E, Miquerol L, Rossdeutsch A, Marber MS. and Riley PR. (2010). Thymosin beta4 facilitates epicardial neovascularization of the injured adult heart. Ann N Y Acad Sci 1194:97–104 [DOI] [PubMed] [Google Scholar]
- 15.Winter EM, Grauss RW, Hogers B, van Tuyn J, van der Geest R, Lie-Venema H, Steijn RV, Maas S, DeRuiter MC, et al. (2007). Preservation of left ventricular function and attenuation of remodeling after transplantation of human epicardium-derived cells into the infarcted mouse heart. Circulation 116:917–927 [DOI] [PubMed] [Google Scholar]
- 16.Winter EM, van Oorschot AA, Hogers B, van der Graaf LM, Doevendans PA, Poelmann RE, Atsma DE, Gittenberger-de Groot AC. and Goumans MJ. (2009). A new direction for cardiac regeneration therapy: application of synergistically acting epicardium-derived cells and cardiomyocyte progenitor cells. Circ Heart Fail 2:643–653 [DOI] [PubMed] [Google Scholar]
- 17.Witty AD, Mihic A, Tam RY, Fisher SA, Mikryukov A, Shoichet MS, Li RK, Kattman SJ. and Keller G. (2014). Generation of the epicardial lineage from human pluripotent stem cells. Nat Biotechnol 32:1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer D, Gambardella L, Bernard WG, Serrano F, Mascetti VL, Pedersen RA, Talasila A. and Sinha S. (2015). Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development 142:1528–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, Rosenthal N. and Xavier-Neto J. (2003). A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development 130:5363–5374 [DOI] [PubMed] [Google Scholar]
- 20.Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA. and Buckingham ME. (2008). Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development 135:1157–1167 [DOI] [PubMed] [Google Scholar]
- 21.Marvin MJ, Di Rocco G, Gardiner A, Bush SM. and Lassar AB. (2001). Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev 15:316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, et al. (2011). Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai K, Chang Y, Wei B, Liu Q, Leblais V, Fischmeister R. and Ji G. (2014). Phosphodiesterase types 3 and 4 regulate the phasic contraction of neonatal rat bladder smooth myocytes via distinct mechanisms. Cell Signal 26:1001–1010 [DOI] [PubMed] [Google Scholar]
- 24.Combs MD, Braitsch CM, Lange AW, James JF. and Yutzey KE. (2011). NFATC1 promotes epicardium-derived cell invasion into myocardium. Development 138:1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braitsch CM, Combs MD, Quaggin SE. and Yutzey KE. (2012). Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol 368:345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C. and Kispert A. (2009). Tbx18 and the fate of epicardial progenitors. Nature 458:E8–E9; discuss E9–E10 [DOI] [PubMed] [Google Scholar]
- 28.Zhou B. and Pu WT. (2012). Genetic Cre-loxP assessment of epicardial cell fate using Wt1-driven Cre alleles. Circ Res 111:e276–e280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudat C. and Kispert A. (2012). Wt1 and epicardial fate mapping. Circ Res 111:165–169 [DOI] [PubMed] [Google Scholar]
- 30.Zhou B, von Gise A, Ma Q, Rivera-Feliciano J. and Pu WT. (2008). Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem Biophys Res Commun 375:450–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF. and Christoffels VM. (2010). The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res 87:92–101 [DOI] [PubMed] [Google Scholar]
- 32.Duim SN, Kurakula K, Goumans MJ. and Kruithof BP. (2015). Cardiac endothelial cells express Wilms’ tumor-1: Wt1 expression in the developing, adult and infarcted heart. J Mol Cell Cardiol 81:127–135 [DOI] [PubMed] [Google Scholar]
- 33.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ. and Palecek SP. (2013). Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc 8:162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, Xu Y, Cao H, Meng Q, et al. (2011). Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res 21:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ. and Chien KR. (2009). Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460:113–117 [DOI] [PubMed] [Google Scholar]
- 36.Lints TJ, Parsons LM, Hartley L, Lyons I. and Harvey RP. (1993). Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119:419–431 [DOI] [PubMed] [Google Scholar]
- 37.Ema M, Takahashi S. and Rossant J. (2006). Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 107:111–117 [DOI] [PubMed] [Google Scholar]
- 38.Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, Gkatzis K, Elliott DA, Chuva de Sousa Lopes SM, et al. (2015). Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med 7:394–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeichi M, Nimura K, Mori M, Nakagami H. and Kaneda Y. (2013). The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PLoS One 8(2): e57829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bax NA, van Oorschot AA, Maas S, Braun J, van Tuyn J, de Vries AA, Groot AC. and Goumans MJ. (2011). In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFbeta-signaling and WT1. Basic Res Cardiol 106:829–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olivey HE, Mundell NA, Austin AF. and Barnett JV. (2006). Transforming growth factor-beta stimulates epithelial-mesenchymal transformation in the proepicardium. Dev Dyn 235:50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei K, Serpooshan V, Hurtado C, Diez-Cunado M, Zhao M, Maruyama S, Zhu W, Fajardo G, Noseda M, et al. (2015). Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 525:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochmann L, Sarathchandra P, Mori F, Lara-Pezzi E, Lazzaro D. and Rosenthal N. (2010). Revealing new mouse epicardial cell markers through transcriptomics. PLoS One 5(6): e11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross JJ, Hong Z, Willenbring B, Zeng L, Isenberg B, Lee EH, Reyes M, Keirstead SA, Weir EK, Tranquillo RT. and Verfaillie CM. (2006). Cytokine-induced differentiation of multipotent adult progenitor cells into functional smooth muscle cells. J Clin Invest 116:3139–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snider P, Standley KN, Wang J, Azhar M, Doetschman T. and Conway SJ. (2009). Origin of cardiac fibroblasts and the role of periostin. Circ Res 105:934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lajiness JD. and Conway SJ. (2014). Origin, development, and differentiation of cardiac fibroblasts. J Mol Cell Cardiol 70:2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong P, Christia P, Saxena A, Su Y. and Frangogiannis NG. (2013). Lack of specificity of fibroblast-specific protein 1 in cardiac remodeling and fibrosis. Am J Physiol Heart Circ Physiol 305:H1363–H1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majkut S, Dingal PC. and Discher DE. (2014). Stress sensitivity and mechanotransduction during heart development. Curr Biol: CB 24:R495–R501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore AW, McInnes L, Kreidberg J, Hastie ND. and Schedl A. (1999). YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 126:1845–1857 [DOI] [PubMed] [Google Scholar]
- 50.Wu SP, Dong XR, Regan JN, Su C. and Majesky MW. (2013). Tbx18 regulates development of the epicardium and coronary vessels. Dev Biol 383:307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips MD, Mukhopadhyay M, Poscablo C. and Westphal H. (2011). Dkk1 and Dkk2 regulate epicardial specification during mouse heart development. Int J Cardiol 150:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin SC, Dolle P, Ryckebusch L, Noseda M, Zaffran S, Schneider MD. and Niederreither K. (2010). Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc Natl Acad Sci U S A 107:9234–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlueter J. and Brand T. (2013). Subpopulation of proepicardial cells is derived from the somatic mesoderm in the chick embryo. Circ Res 113:1128–1137 [DOI] [PubMed] [Google Scholar]
- 54.Bressan M, Liu G. and Mikawa T. (2013). Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science 340:744–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD. and Yutzey KE. (2013). Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol 65:108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Limana F, Bertolami C, Mangoni A, Di Carlo A, Avitabile D, Mocini D, Iannelli P, De Mori R, Marchetti C, et al. (2010). Myocardial infarction induces embryonic reprogramming of epicardial c-kit+ cells: Role of the pericardial fluid. J Mol Cell Cardiol 48:609–618 [DOI] [PubMed] [Google Scholar]
- 57.Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, et al. (2011). De novo cardiomyocytes from within the activated adult heart after injury. Nature 474:640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.