Abstract
Vault nanoparticles represent promising vehicles for drug and probe delivery. Innately found within human cells, vaults are stable, biocompatible nanocapsules possessing an internal volume that can encapsulate hundreds to thousands of molecules. They can also be targeted. Unlike most nanoparticles, vaults are nonimmunogenic and monodispersed and can be rapidly produced in insect cells. Efforts to create vaults with modified properties have been, to date, almost entirely limited to recombinant bioengineering approaches. Here we report a systematic chemical study of covalent vault modifications, directed at tuning vault properties for research and clinical applications, such as imaging, targeted delivery, and enhanced cellular uptake. As supra-macromolecular structures, vaults contain thousands of derivatizable amino acid side chains. This study is focused on establishing the comparative selectivity and efficiency of chemically modifying vault lysine and cysteine residues, using Michael additions, nucleophilic substitutions, and disulfide exchange reactions. We also report a strategy that converts the more abundant vault lysine residues to readily functionalizable thiol terminated side chains through treatment with 2-iminothiolane (Traut’s reagent). These studies provide a method to doubly modify vaults with cell penetrating peptides and imaging agents, allowing for in vitro studies on their enhanced uptake into cells.
Keywords: vaults, nanoparticle, cell-penetrating transporter, imaging, chemical modification, protein cage
Graphical Abstract
First discovered in 1986, native vaults are the largest known ribonucleic protein complex (13 MDa) in eukaryotic organisms. Naturally occurring, vaults are ubiquitously found and highly conserved within eukaryotic organisms except for a few notable exceptions.1–4 Vaults are hollow, barrel-shaped structures composed of 78 copies of the major vault protein (MVP). These MVPs assemble into a protein cage of uniform size (67 nm × 40 nm by X-ray crystallography), with an internal cavity of approximately 3.87 × 107 Å3.5,6 In addition to the MVP (~100 kDa), native vaults also contain two other protein constituents: vault poly(ADPribose) polymerase (vPARP, ~193 kDa) and telomerase-associated protein 1 (TEP1, ~290 kDa), as well as several copies of small untranslated vault associated RNA (vRNA).7–12 However, the expression of MVP alone has been found to be sufficient for the formation of vault-like particles within insect cells, which lack endogenous vaults, when using a baculovirusinsect cell expression system.13 These recombinant MVP-only vaults retain native vault architecture and monodispersity, but lack vPARP, TEP1, and the vRNA. The function of native vaults has yet to be determined, although it has been hypothesized that they could play a role in intracellular transport, multidrug resistance, and/or cell signaling.4,14–16
Vaults show great potential for drug delivery and imaging due to their stability, biocompatibility (in general, mammalian cells already contain ~104–106 native vaults), and ability to be targeted.3,17,18 Vaults have been shown to encapsulate and protect a variety of cargos including fluorescent proteins, enzymes, gold nanoparticles, semiconducting polymers, and small molecule therapeutics.19–23 Previous work has demonstrated that vaults can be engineered to contain a lipid-rich core into which lipophilic compounds selectively associate. These bioengineered lipophilic vaults reversibly encapsulate therapeutic compounds including drugs currently in the clinic such as Bryostatin 1, a therapeutic lead for HIV/AIDS eradication and for the treatment of Alzheimer’s disease.24
Direct chemical derivatization of vaults can accommodate a wider range of modifications not accessible through molecular biological approaches. This method provides a complementary approach to creating modified vaults and offers a broader range of chemical bond forming opportunities, along with greater speed in accessing various diverse vault structures. However, few studies on chemical modifications of vault cysteine and lysine residues have been reported, and while noteworthy, these either do not provide information on the efficiency of vault modification25–27 or, in one case, report a low efficiency averaging one modification per MVP.28 The ability to selectively modify vault residues is critical to understanding the basic chemical reactivity patterns of vaults and thus key to the rational design of vaults exhibiting optimal physical properties and targeting abilities for clinical applications including imaging, diagnostics, and therapy.
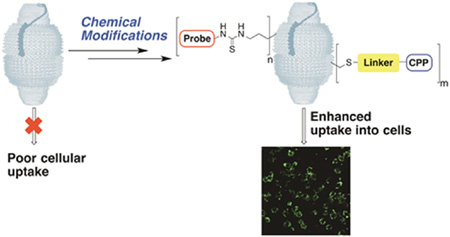
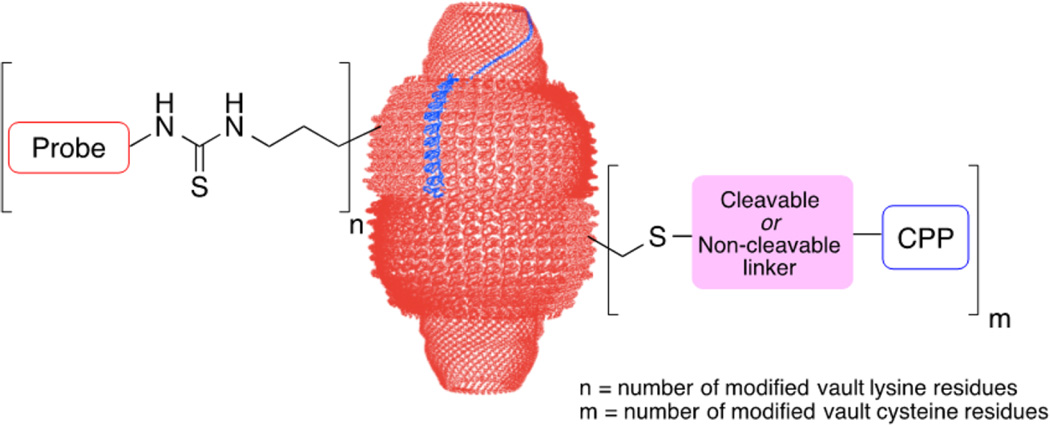
To address this opportunity, we have initiated studies on the covalent chemical modification of two types of recombinant vaults: human MVP-only vaults (hMVP vaults) and engineered human cysteine-rich vaults (CP-hMVP vaults), which contain an N-terminal cysteine-rich peptide (MAGCGCPCGCGA). The CP recombinant tag confers greater vault stability by way of introduction of four additional cysteine residues per MVP that are thought to associate, given their localized proximity, to one another within the vault luminal waist.29 With 78 MVP per vault, and each bearing a number of derivatizable amino acids, there are thousands of sites available for chemical modification using a variety of reagents.30 In this systematic chemical study, we have restricted our attention to derivatization of vault cysteine and lysine residues using three different reaction types: Michael additions, nucleophilic substitutions, and disulfide exchanges. In addition, we have investigated a strategy that allows for chemical conversion of the more abundant vault lysine residues into readily modifiable thiol functionalities using 2-iminothiolane (Traut’s reagent). Collectively these studies have enabled the selective synthesis of singly and doubly modified vaults, incorporating in the latter case both optical probes and cell penetrating transporters (Figure 1).
Figure 1.
Covalent attachment of cell penetrating peptides and fluorescent probes to vault nanoparticles.
Although vaults are found within eukaryotic cells, their cellular uptake is poor in many tested cell lines.12,28,31 To address this problem, we have drawn on our previous work on guanidinium-rich transporters as a method to enhance vault uptake.32 Inspired by the protein HIV-1 (TAT), which unlike many proteins readily enters cells, these guanidinium-rich molecular transporters have been used to deliver a diverse range of cargos, including small molecules, peptides, proteins, imaging agents, siRNA, plasmids, and liposomes across a variety of cellular and tissue barriers, including mammalian cell membranes, algal cell walls, human skin, and the blood brain barrier.32–38 Through an extensive reverse engineering study, we showed that this cellular uptake is not a function of the peptide backbone but rather the number and spatial array of guanidinium groups. This seminal finding led to the design cell-penetrating, guanidinuim-rich molecular transporters.39–41 Vaults have been previously bioengineered to incorporate a variant of this protein transduction domain, TAT48 (13 amino acid sequence derived from truncated version of the HIV-TAT protein: GRKKRRQRRRAHQ) encoded C terminus of rat MVP.32 These TAT48 CP-rMVP vaults (78 MVP chains per vault ×1 TAT peptide per MVP) demonstrated a qualitatively higher although unquantified cell uptake by confocal microscopy compared to vaults lacking the TAT48 tag. Seeking to circumvent the time and cost associated with bioengineered approaches and to accommodate a greater diversity of vault modifications with greater control over the number of modification sites than those allowed by biosynthesis, it led us to explore the direct chemical modification of vaults.
In this study, we report on the chemical modification of vaults and show that with the attachment of octaarginine cell penetrating peptides to the vault cysteine residues, the cellular uptake of vaults is enhanced, as visualized by optical imaging using vault lysine residues conjugated to fluorescent probes. This enhanced uptake offers promise for the use of vaults in drug encapsulation, protection, and delivery as well as in imaging and provides the foundation for future studies on attachment of targeting ligands. Here we describe a systematic study of vault modification, focused on the modification of vault lysine and cysteine residues, leading to the synthesis of vault nanoparticles modified with optical probes and cell penetrating transporters.
RESULTS AND DISCUSSION
Vault Side Chain Covalent Modification
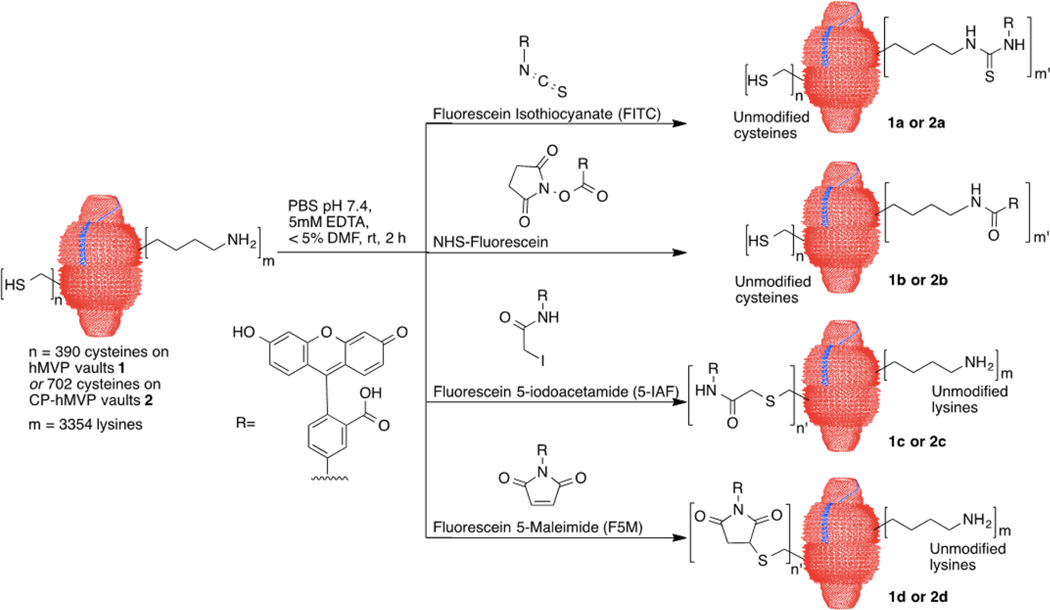
Covalent modifications of vaults were conducted systematically with (1) lysine amine groups, of which there are 43 per hMVP or CP-hMVP monomer, representing 3354 on a single vault; and (2) cysteine thiol groups, of which there are 5 per hMVP or 9 per CP-hMVP for a total of 390 or 702 thiol residues per vault, respectively. Fluorescein derivatives with different reactive functional groups were selected to determine vault reactivity based on an optical readout. Vault lysine residues were modified with fluorescein isothiocyanate (FITC) and fluorescein NHS-ester (NHS-Fluorescein). Vault cysteine thiols were reacted with either fluorescein 5-maleimide (F5M) or the haloacetyl derivative, fluorescein 5-iodoacetamide (5-IAF) to form thioethers (Scheme 1). Both hMVP vaults 1 and CP-hMVP vaults 2 were studied using the same strategy to obtain comparative information about the number of chemically modifiable sites on multiple types of recombinant vaults depending upon the reagent used.
Scheme 1.
Chemoselective Fluorescein Labeling of Vaults.
The number of conjugated fluorescein molecules per vault was quantified by UV–vis spectroscopy relative to the corresponding concentration of vault protein by a bicinchoninic acid assay (BCA) following chemical modification and removal of any unreacted fluorophore (Table 1, Supporting Information, Figure S1 for vault protein recovery). FITC labeled 187 lysines per hMVP vault (6% derivatization of all lysine residues on a single vault) and 143 lysines per CP-hMVP vault (4% derivatization of lysine residues). NHS fluorescein was more reactive, leading to the labeling of 471 sites per hMVP vault (14% derivatization) and 331 sites per CP-hMVP vault (10% derivatization). The thiol reactive probe 5-IAF modified about 19% of the cysteine residues regardless of the vault type, adding 73 probes per hMVP vault and 136 probes per CP-hMVP vault. Furthermore, the maleimide functionality proved more reactive than iodoacetamides. Up to 240 probes were added per hMVP vault (62% derivatization) via maleimide modification and up to 384 probes per CP-hMVP vault (55% derivatization). The increase in cysteine labeling of CP-hMVP vaults compared to hMVP vaults is consistent with labeling 1–2 of the four cysteine residues per cysteine-rich CP tag on the CP-hMVP vaults. This indicates that chemical labeling can occur internally within the vault lumen, which was anticipated given previous demonstration of small lipophilic drugs becoming internally bound within the previously engineered lipophilic-rich vault constructs.25 Lastly, the degree of labeling for all tested fluorescein derivatives equates to less than seven amino acid residues per each 100 kDa MVP monomer, thus representing minimal alteration to the MVP protein overall.
Table 1.
Vault Labeling Results Using Fluorescein Probes
| vault type | probe (reactive amino acid) |
mol excessa |
no. probes per vaultb (total available residues) |
percent derivatization (%) |
size (d·nm)c | |
|---|---|---|---|---|---|---|
| 1a | FITC (Lys) | 5:1 | 187 ± 16 (3354) | 6 ± 1 | 39.5 ± 10.4 | |
| 1b | NHS-Fluorescein (Lys) | 5:1 | 471 ± 78 (3354) | 14 ± 2 | 38.4 ± 10.7 | |
| hMVP vaults (1) | 1c | 5-IAF (Cys) | 20:1 | 73 ± 5 (390) | 19 ± 1 | 39.1 ± 10.4 |
| 1d | F5M (Cys) | 20:1 | 240 ± 19 (390) | 62 ± 5 | 37.7 ± 10.3 | |
| 2a | FITC (Lys) | 5:1 | 143 ± 78 (3354) | 4 ± 2 | 41.6 ± 9.5 | |
| 2b | NHS-Fluorescein (Lys) | 5:1 | 331 ± 39 (3354) | 10 ± 1 | 41.3 ± 9.3 | |
| CP-hMVP vaults (2) | 2c | 5-IAF (Cys) | 20:1 | 136 ± 47 (702) | 19 ± 7 | 42.0 ± 9.1 |
| 2d | F5M (Cys) | 20:1 | 384 ± 8 (702) | 55 ± 1 | 39.3 ± 9.9 |
Reaction ratio of fluorescein probe:vault amino acid.
The number of probes attached to the vaults was determined using UV–vis at 494 nm with 70,000 M−1 cm−1 as the extinction coefficient. All reactions were run in triplicate. The values are reported as the average of the measurements.
Size measurements determined by DLS.
In all labeling experiments, we used SDS-PAGE to confirm that the vault was covalently modified (Supporting Information, Figure S2 for results and procedure). By dynamic light scattering (DLS), the average vault diameter of unmodified hMVP and CP-hMVP vaults was determined to be 38 nm. After labeling with fluorescein, the particle size did not significantly change, remaining within 1–4 nm of the unmodified vaults, indicating that the vaults remained intact and monodispersed (Supporting Information, Figure S3 for DLS traces). Additionally, no precipitation of chemically modified vaults was observed indicating they remain stable and soluble. This is expected since even high-level chemical modifications should not out-weigh the inherent properties of the relatively larger vault nanoparticle relative to any attached cargos.
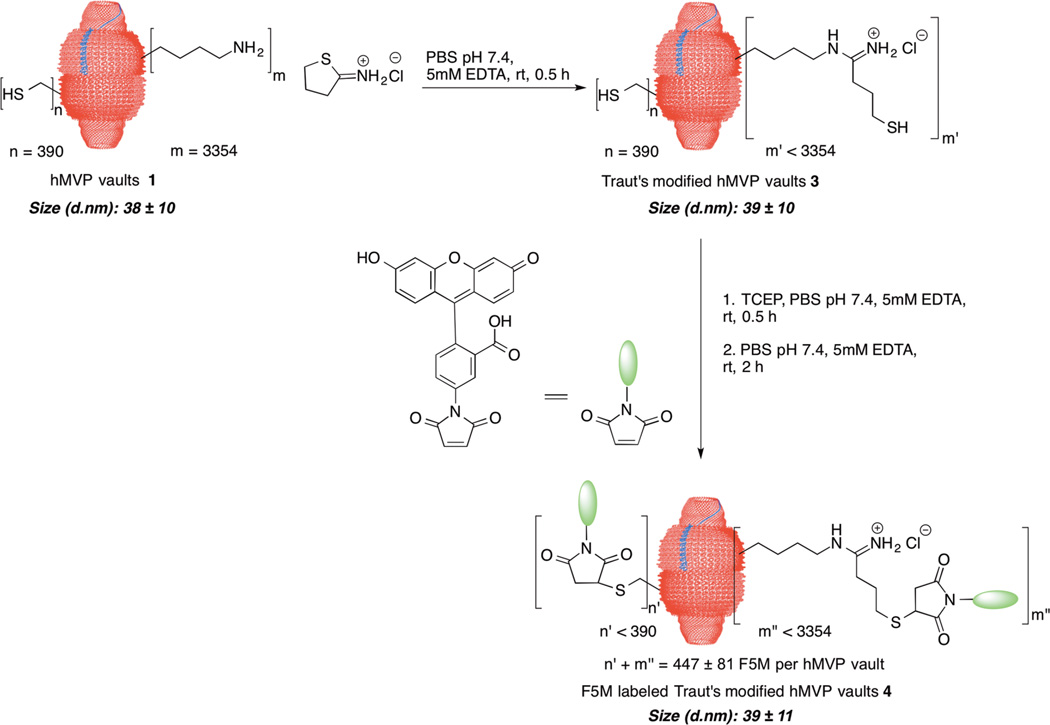
We next examined a strategy to convert the more abundant lysine residues into more nucleophilic thiol terminated groups. hMVP vaults were reacted with Traut’s reagent to convert the lysine amine residues into chain-extended thiols at a reaction ratio of 40:1 Traut’s reagent to vault lysine residues (Figure 2). The Traut’s modified hMVP vaults 3 were then reacted with tris(2-carboxyethyl)phosphine (TCEP) for 30 min to reduce any disulfides formed, followed by the thiol reactive probe, F5M. The F5M labeled Traut’s modified hMVP vaults 4 were found to incorporate 447 ± 81 probes per vault particle, almost double the number added for unmodified hMVP vaults. The size of the Traut’s modified hMVP vaults 3 and F5M labeled Traut’s modified hMVP vaults 4 was consistent with unmodified hMVP vaults 1 (Supporting Information, Figures S4–5 for DLS traces of Traut’s modified hMVP vaults as well as CP-hMVP vaults and electron microscopy images of Traut’s modified CP-hMVP vaults). Conversion of the more abundant lysine amines to more functionally useful thiols greatly adds to the chemistries available for vault modification. Additionally, this approach offers the potential for multiple labeling of the vault simply through initial modification of cysteine thiols followed by modification of Traut’s derived thiols.
Figure 2.
hMVP vault modification with Traut’s reagent followed by labeling with a fluorescent probe. Size measurements were determined by DLS. The number of probes attached to the vaults was determined using UV–vis at 494 nm with 70,000 M−1 cm−1 as the extinction coefficient. The labeling reaction was run in triplicate, and the reported values are of the average of the three trials.
Enhanced Cellular Uptake of Fluorescein Labeled Vaults
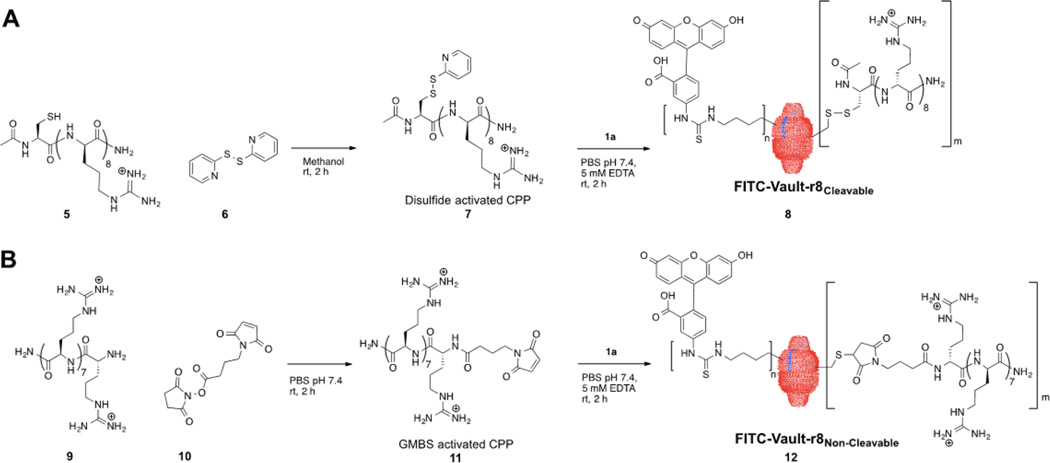
Having established the procedures for the chemical modification and optical tagging of vaults, we next set out to explore whether vaults could be modified with cell-penetrating peptides (CPPs) as a way to enhance their uptake into cells. We attached octaarginines onto the vault cysteine residues using either a redox cleavable linker or a noncleavable linker (Scheme 2). Both strategies provide a stable linkage between the protein carrier and the peptide cargo in the extracellular milieu. However, such disulfide linkages are readily cleaved after cellular entry due to the higher intracellular concentration of cytoplasmic glutathione.42
Scheme 2.
Synthesis of Doubly Modified hMVP Vaults with a Fluorescent Probe and CPPa
a(A) Activation of Ac-Cys-(dArg)8-NH2 using 2,2′-dipyridyl disulfide followed by attachment of the activated Cys-r8 CPP onto FITC vaults through a disulfide exchange with MVP cysteine residues. (B) Activation of CPP octaarginine using GMBS followed by the attachment of the activated octaarginine onto FITC vaults through a thiol-maleimide linkage.
For the cleavable method, an activated CPP 7 was prepared by reacting the Cys-r8 peptide 5 with 2, 2′-dipyridyl disulfide 6 (Scheme 2A). Before adding the activated CPP 7 to the vault protein, the vault lysine residues were labeled with FITC as described above to allow for subsequent imaging. This activated CPP 7 was then added onto the FITC vault cysteine thiols via a redox-sensitive disulfide-bond formation, providing a built-in strategy for intracellular release. FITC vaults were also used as a starting point for the noncleavable method (Scheme 2B). For this strategy, the octaarginine peptide 9 was activated by N-γ-maleimidobutyryl-oxysuccinimide ester (GMBS) 10 to add a maleimide group to form CPP 11 for subsequent attachment to the vault cysteines. For both the cleavable and noncleavable conjugation strategies, the activated peptides were reacted with the vault at a ratio of 2:1 activated peptide to vault cysteine residues. For sake of clarity, we have named these doubly chemically modified vaults FITC-Vault-r8Cleavable 8 and FITC-Vault-r8Noncleavable 12, respectively. We confirmed that vaults remained properly folded and intact after the CPP modifications by circular dichroism (CD), DLS, and SDS page gel (Supporting Information, Figures S6–8 for CD spectra, DLS traces, and SDS page gel of CPP modified vaults, respectively).
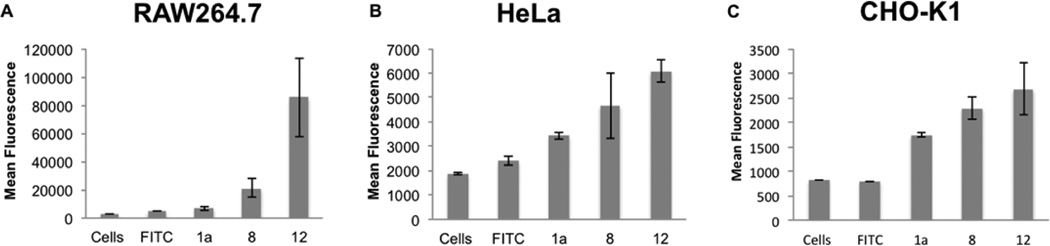
After purification of the modified vaults by ultracentrifugation, vault uptake was studied in RAW264.7, HeLa, and CHOK1 cells. Cells were treated with either FITC, FITC vaults, or CPP modified FITC vaults at a concentration of 10 µg vaults/500,000 cells at 37 °C for 16 h to allow for uptake. Significant increases of cellular uptake were observed by flow cytometry for CPP modified FITC vaults compared to FITC vaults without CPP conjugation (Figure 3). Importantly, the same batch of FITC vaults was used for the FITC vault sample and the CPP modified FITC-labeled vaults. FITC labeling was consistent with previous results, ~187 FITC/vault.
Figure 3.
Uptake of FITC, FITC vaults 1a, FITC-Vault-r8Cleavable 8, and FITC-Vault-r8Noncleavable 12 into (A) RAW264.7 cells, (B) HeLa cells, and (C) CHO-K1cells. Data represents mean ± SD, N ≥ 3 for all measurements.
The noncleavable vaults 12 (FITC-Vault-r8Noncleavable) exhibit the highest uptake in all cell lines (Figure 3). In the RAW264.7 cells, a macrophage cell line that internalizes unmodified vaults to some extent, a 3-fold increase in uptake was observed with the redox-sensitive vaults 8 (FITC-Vault-r8Cleavable) and a 10-fold increase with the noncleavable vaults 12 (FITC-Vaultr8Noncleavable) compared to FITC vaults 1a (Figure 2A). While determining the basis for the difference in cellular uptake between the FITC-Vault-r8-cleavable and -noncleavable systems is beyond the experimental scope of this study, it is possible that disulfide exchange and/or cleavage would occur, thus reducing the number of octaarginines on the cleavable system and thus its uptake into cells. In HeLa and CHO-K1 cells, there was a 3-fold (Figure 3B) and a 1.5-fold (Figure 3C) increase in uptake for the FITC-Vault-r8Noncleavable 12 compared to FITC vaults 1a, respectively. Neither of these cell lines showed significant uptake of the undecorated FITC vaults 1a, indicating the addition of cell penetrating peptides is necessary for improved uptake into these cell lines.
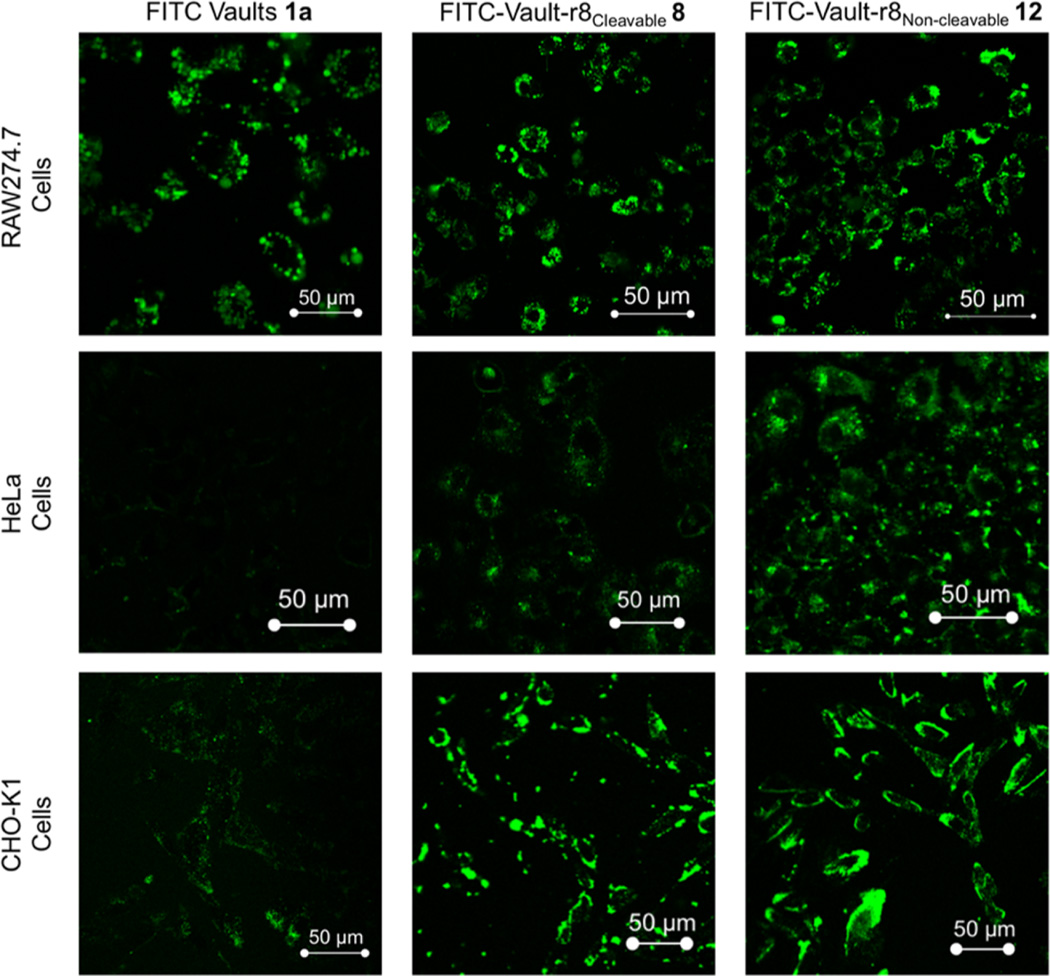
To analyze the above-described uptake results and to check for cellular internalization of chemically labeled fluorescent vaults with or without attached octaarginine CPPs, cells treated with modified vaults were visualized using confocal microscopy (Figure 4). RAW264.7, HeLa, and CHO-K1 cells were incubated with modified vaults (30 µg vaults/500,000 cells) for 16 h and then washed to remove any material not taken up by the cells. FITC-Vault-r8Cleavable 8 and FITC-Vaultr8Noncleavable 12 were efficiently internalized within all tested cell lines. By confocal microscopy, FITC vaults 1a clearly entered RAW264.7 cells but are poorly taken up by HeLa and CHO-K1 cells. This variation is attributed to the noted difference between the cell lines seen in the quantitative flow cytometry data (Figure 3). Under the tested confocal microscopy conditions, FITC vaults already saturate the imaging signal seen by RAW264.7 cells, thereby masking any signal improvement that occurs with the attachment of octaarginine CPP. However, since HeLa and CHO-K1 internalize FITC vaults less readily than RAW264.7 cells, a clear improvement in fluorescent signal is observed for the doubly labeled vaults. Thereby, demonstrating that the CPP labeling greatly improves uptake and internalization into these cells (Supporting Information, Figure S9 for overlay images).
Figure 4.
Confocal images of live RAW264.7 (top), HeLa (middle), and CHO-K1 (bottom) cells incubated with 30 µg of modified vaults/500,000 cells.
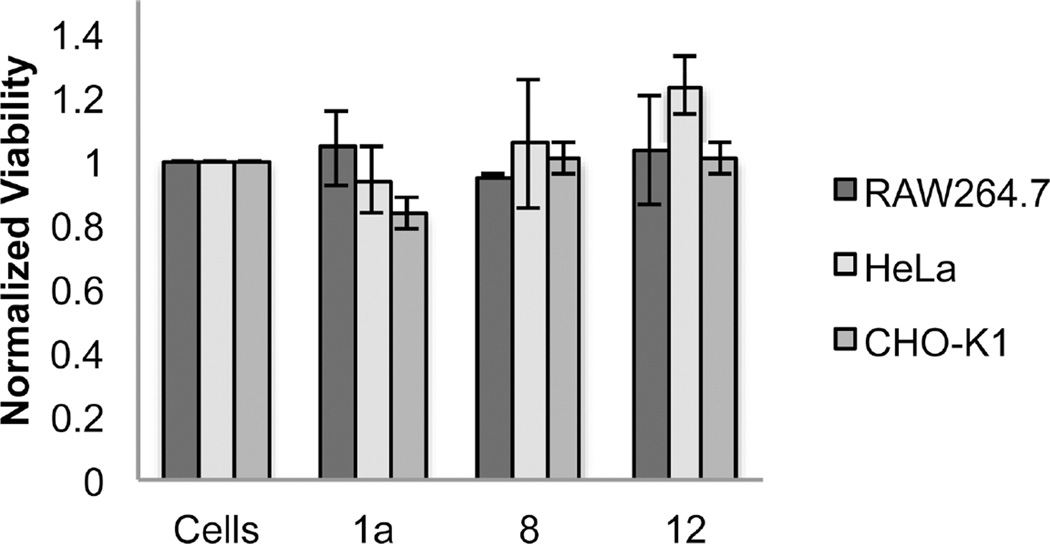
Additionally, the viability of RAW264.7, HeLa, and CHO-K1 cells following exposure to the chemically modified vaults was determined using an MTT assay. After incubation with FITC vaults 1a or either of the octaarginine decorated FITC vaults at various concentrations for 16 h, RAW264.7, HeLa, and CHOK1 cells maintained good cell viability (Figure 5) (Supporting Information, Figure S10 for MTT results at a series of concentrations).
Figure 5.
Normalized viability of cells treated with modified vaults at a concentration of 30 µg vaults/500,000 cells, as determined by MTT assay over 16 h. Data represents mean ± SD, N ≥ 3 for all measurements.
CONCLUSIONS
Given the formidable challenges and opportunities associated with drug delivery, considerable innovative effort in recent years has been directed at nanoparticles that can encapsulate and protect drugs and probes during administration and release the drug cargo either extracellularly after administration or after cell entry. Vaults provide a relatively under-explored space-enclosing nanocapsule for the protection and delivery of various cargos, including small molecules, peptides, proteins, siRNA, and mRNA. We previously reported the vault-mediated delivery of bryostatin-1, a therapeutic candidate now in the clinic for the treatment of Alzheimer’s disease and the eradication of HIV/AIDS.24 To address the ensuing questions associated with biodistribution and cellular uptake, we have reported here the chemistry needed to produce vaults that contain multimodal functionality, including imaging and enhanced cellular uptake, through the selective and differential labeling of various vault residues, which provides a fundamental foundation for future studies. Vault cysteine and lysine residues were selected in this initial study and were shown to be chemoselectively derivatized. Given the greater number of vault lysine residues and the versatile chemistry of thiols, we also demonstrate a simple, robust technique that can efficiently convert these more abundant residues into thiol terminated side chains. Using the chemistry we developed, we created vaults doubly modified with a fluorescein reporter probe and cell-penetrating octaarginine peptides attached via a redox-sensitive cleavable or noncleavable linker. Relative to unmodified vaults, the resultant modified vaults showed no adverse particle effects following chemical modification while clearly demonstrating increased optical properties and cellular uptake into cells of interest. These studies provide vaults with enhanced fluorescent labeling that are of immediate interest for imaging and biodistribution studies. Additionally, chemically labeled vaults containing guanidinium groups exhibit enhanced cellular uptake. More generally, this study provides a chemical foundation for predictable vault modification, as required for accessing a variety of vaults for use in imaging, therapeutic delivery, and basic biological research. It also offers a fast way to modify vaults for targeted delivery. These vault modifications and uptake studies along with our previous work on the loading of vaults collectively move this vault technology closer to therapeutic applications.
METHODS AND MATERIALS
Materials
Unless otherwise noted, all commercial reagents were purchased from Sigma-Aldrich, Thermo Fischer Scientific, VWR, or AnaSpec and used without additional purification. Octa-(d)-arginine was obtained from UCB bioproducts. hMVP and CP-hMVP were expressed using a baculovirus system in Sf9 insect cells, which do not contain endogenous vaults.13
Instrumental
NMR spectra were measured on a Varian 500 MHz magnetic resonance spectrometer. 1H chemical shifts are reported relative to the residual solvent peak (CD3OD = 3.31, ppm) as follows: chemical shift, multiplicity (s = singlet, bs = broad singlet, d = doublet, bd = broad doublet, t = triplet, q = quartet, p = pentet, m = multiplet), and integration. Matrix-assisted laser desorption mass spectra (MALDI) were done using an Applied Biosystems Voyager DE mass spectrometer. Mass spectra were obtained at the Vincent Coates Foundation mass spectrometry laboratory at Stanford University. Reverse-phase high-performance liquid chromatography (RP-HPLC) was performed with a Shimadzu LC-20AP using a semipreparative column (Restek Ultra II, C18, 250 × 10 mm). The products were eluted using a solvent gradient (solvent A = 0.1% TFA/H2O; solvent B = 0.1% TFA/CH3CN). UV–vis spectra were obtained on an Agilent Cary 6000i UV–vis-NIR with quartz cells or Nanodrop 1000. DLS was done using a Malvern Zetasizer Nano-S. CD was done on a Jasco J-815 CD spectrometer using a 1 mm quartz cuvette. Sodium dodecyl sulfate-poly(acrylamide) gel electrophoresis (SDS-PAGE) was accomplished on a Mini-Protean apparatus from Bio-Rad with 10–20% gradient poly(acrylamide) gels. Flow cytometry was performed on a FACScan instrument at the Stanford Shared FACS facility. Live cell imaging was performed using a Leica SP5 upright confocal microscope. Images were acquired from cells with a water immersion lens and analyzed using Las-af software. Scanning was done at an image resolution of 1024 × 1024 pixels and averaged 2–4 times using 25% 488 nm.
Vault Purification
Vaults (CP or hMVP) were purified from Sf9 insect cell pellets infected with baculoviruses encoding either hMVP or CP-hMVP following standard protocols.12 Purified vaults were analyzed by SDS-PAGE and electron microscopy to verify >95% particle purity. Vaults (0.5–1.0 mg aliquots) were lyophilized in PBS containing 10 mg/mL trehalose and stored at −20 °C prior to use.
Vault Stock Preparation
The vaults were resuspended at 4 °C in PBS 1× buffer (pH 7.4) with 5 mM EDTA for 30 min. At this time, the solution was spun down at 1000 × g for 2 min, and the supernatant was removed. The vault concentration of stock protein sample was determined by a bicinchoninic acid (BCA) assay (Pierce, Rockford, IL).
Fluorescein Labeling of Vault Nanoparticles
To a 418 µL solution of PBS 1× buffer (pH 7.4) with 5 mM EDTA was added recombinant vaults (6.5 pmol, 51.2 µg, 80 µL of a 0.64 mg/mL solution in pH 7.4 PBS 1× buffer with 5 mM EDTA). To this was added an amine or thiol reactive fluorescein probe (0.11 µmol or 0.055 µmol, respectively, as 50 mM stock solutions in DMF). Final reaction mixture was 4.4% DMF for lysine reactive probes and 2.2% for cysteine reactive probes. Attempted higher labeling ratios led to vault loss, presumably due to the higher percentage of DMF required to solubilize the fluorescent probes. Note for the CP vault reactions, additional thiol reactive fluorescein probes were added to account for the extra cysteine residues (0.1 µmol thiol reactive probe). The reaction mixture was incubated at room temperature for 2 h with rotation and protection from light. The mixtures were applied to Ultra-0.5 30K Centrifugal Filter Devices (Millipore) that were preequilibrated with 500 µL of PBS 1× buffer with 5 mM EDTA. Separation of unattached small molecules from modified vault proteins was achieved by centrifugal filtration for 9 min at 14,000 × g at room temperature, followed by four washes of 450 µL PBS 1× buffer with 5 mM EDTA for 8 min at 14,000 × g at room temperature. The purified vault was collected by inverse centrifugation for 2 min at 1000 × g at room temperature. Purified samples were analyzed for vault concentration using BCA protein assay (Pierce, Rockford, IL). The concentration of attached fluorescent probes was analyzed using an Agilent Cary 6000i UV–vis-NIR or Nanodrop 1000 at 494 nm, based on the extinction coefficient of 70,000 M−1 cm−1.
Traut’s Modification of Recombinant Vaults
To a 430 µL solution of PBS 1× buffer (pH 7.4) with 5 mM EDTA was added recombinant hMVP vault protein (51.0 pmol, 400 µg, 500 µL of a 0.8 mg/mL solution in pH 7.4 PBS 1× buffer with 5 mM EDTA). To this was added a 2-iminothiolane (6.9 µmol, 963 µg, 70 µL of a 100 mM in pH 7.4 PBS 1× buffer). The reaction mixture was incubated at room temperature for 0.5 h with rotation and protection from light. To reduce any potential disulfide linkages formed after introduction of the alkyl sulfhydryl groups by Traut’s reagent reactivity, tris(2-carboxyethyl)phosphine (TCEP) (5 µmol, 1.4 mg, 10 µL of 500 mM in pH 7.4 PBS 1× buffer) was added to the reaction mixture and allowed to react for 30 min. The mixtures were then applied to Ultra-0.5 30K Centrifugal Filter Devices (Millipore) that were preequilibrated with 500 µL of PBS 1× buffer with 5 mM EDTA. Separation of unreacted Traut’s reagent and TCEP from modified vault proteins was achieved by centrifugal filtration for 9 min at 14,000 × g at room temperature, followed by four washes of 450 µL PBS 1× buffer with 5 mM EDTA for 8 min at 14,000 × g at room temperature. The purified vault was collected by inverse centrifugation for 2 min at 1000 × g at room temperature. Purified samples were analyzed for vault concentration using BCA protein assay (Pierce, Rockford, IL). Same procedure was followed for modification of CP-hMVP vaults with Traut’s reagent.
Fluorescein Labeling of Traut’s Modified Recombinant Vaults
To a 440 µL solution of PBS 1× buffer (pH 7.4) with 5 mM EDTA was added Traut’s modified recombinant hMVP vault protein (4.2 pmol, 32.6 µg, 51 µL of a 0.64 mg/mL solution in pH 7.4 PBS 1× buffer with 5 mM EDTA). Importantly, Traut’s modified hMVP vaults were used immediately after the above-described TCEP reduction. To this were added thiol reactive fluorescein probe and fluorescein-5-maleimide (0.5 µmol, 214 µg, added via 10 µL of 50 mM solution in DMF). The reaction mixture was incubated at room temperature for 2 h with rotation and protection from light. The mixtures were applied to Ultra-0.5 30K Centrifugal Filter Devices (Millipore) that were pre-equilibrated with 500 µL wash of PBS 1× buffer with 5 mM EDTA. Separation of unattached small molecules (F5M) from modified vault proteins was achieved by centrifugal filtration for 9 min at 14,000 × g at room temperature, followed by four washes of 450 µL PBS 1× buffer with 5 mM EDTA for 8 min at 14,000 × g at room temperature. The purified vault was collected by inverse centrifugation for 2 min at 1000 × g at room temperature. Purified samples were analyzed for vault concentration using BCA protein assay. The concentration of attached fluorescent probes was analyzed using an Agilent Cary 6000i UV–vis-NIR or Nanodrop 1000 at 494 nm, based on the extinction coefficient of 70,000 M−1 cm−1.
Synthesis of CPP 7
To a solution of Ac-Cys-(dArg)8-NH2·8TFA 5 (5 mg, 2.2 µmol, 1 equiv) in 2:1 H2O/Methanol (1.5 mL, 1.5 mM) was added a solution of 2′2-dipyridyl disulfide 6 (1.5 mg, 6.8 µmol, 3 equiv). After reacting under argon for 2 h at room temperature, the peptide was purified by HPLC using a 1 to 30% solvent gradient of water/acetonitrile with 0.1% TFA. Appropriate fractions were isolated giving a yield of 74%. 1HNMR (500 MHz, CD3OD) δ = 8.45 (m, 1H), 8.08–8.26 (m, 3H), 7.83 (m, 2H), 7.26 (m, 1H), 4.16–4.46 (m, 10H), 3.14–3.25 (m, 16H), 2.04 (s, 4]3H), 1.57–2.00 (m, 32 H) ppm. MS (m/z): calculated for [C58H109N35O10S2] 1520.8; found (MALDI) 1521.1.
Synthesis of CPP 11
d-Octaarginine·8TFA 9 (5 mg, 2.3 µmol, 1 equiv) was dissolved in degassed PBS 1× (1.8 mL) in a 2 DR vial. To this was added N-γ-maleimidobutyryl-oxysuccinimide ester (GMBS) 10 (6.5 mg, 23 µmol, 10 equiv, in 0.2 mL DMF). After reacting under argon for 2 h at room temperature, the peptide was purified by HPLC using a 1 to 50% solvent gradient of water/acetonitrile with 0.1% TFA. Appropriate fractions were isolated giving a yield of 72%. 1HNMR (500 MHz, CD3OD) δ = 8.38 (m, 1H), 8.06–8.26 (m, 2H), 6.80 (s, 2H), 6.76 (s, 4H), 4.20–4.35 (m, 7H), 4.11 (m, 1H), 3.51 (m, 7H), 3.17 (m, 16H), 2.25 (m, 8H), 1.54–1.92 (m, 40H) ppm. MS (m/z): calculated for [C56H106N34O11]: 1431.69; found: 1433.0.
Bioconjugation of CPP 7 with FITC Labeled Vaults
To a 605 µL solution of PBS 1× buffer (pH 7.4) with 5 mM EDTA was added FITC labeled recombinant hMVP vault 1a (15.0 pmol, 123 µg, 350 µL of a 0.35 mg/mL solution in pH 7.4 PBS 1× buffer with 5 mM EDTA). To this was added a thiol reactive CPP 7 (12.2 nmol, 45 µL of 0.27 mM solution in PBS 1× buffer). The reaction mixture was incubated at room temperature for 2 h with rotation and protection from light before being purified by ultracentrifugation (100,000 × g at 4 °C for 2 h). After slow, gentle resuspension for 3 days 4 °C in 250 µL of PBS, the concentration of the FITC-vault-r8Cleavable 8 was determined to be 0.21 mg/mL.
Bioconjugation of CPP 11 with FITC Labeled Vaults
To a 474 µL solution of PBS 1× buffer (pH 7.4) with 5 mM EDTA was added FITC labeled recombinant hMVP vaults 1a (17.0 pmol, 130 µg, 500 µL of a 0.26 mg/mL solution in pH 7.4 PBS 1× buffer with 5 mM EDTA). To this was added a thiol reactive CPP 11 (13 nmol, 26 µL of 0.5 mM solution in PBS 1× buffer). The reaction mixture was incubated at room temperature for 2 h with rotation and protection from light before being purified by ultracentrifugation (100,000 × g at 4 °C for 2 h). After slow, gentle resuspension for 3 days 4 °C in 150 µL of PBS 1× buffer, the concentration of the FITC-vault-r8Noncleavable 12 was determined to be 0.25 mg/mL.
Cell Uptake Experiments by Flow Cytometry
Cells were grown and maintained in DMEM media (F12-K for CHO-K1 cells) with 10% fetal bovine serum (FBS) and Penn-strep. Prior to the addition of vaults, cells (50,000/well) were seeded in 12-well plates. The cells were treated at a concentration of 10 µg of vaults per 500,000 cells in serum free media and allowed to incubate for 16 h at 37 °C in 5% CO2. At this time, cells were washed with PBS 1× and then trypsin-EDTA and incubated at 37 °C for 10 min. Then serumcontaining media was added, and cells were pelleted out by centrifugation at 2000 rpm for 5 min. The supernatant was then removed, and the pelleted cells were resuspended in PBS 1× before fluorescence studies were performed. For the RAW264.7 cells, cell scrapers were used in replace of trypsin-EDTA. Fluorescence studies were done using a FACScan flow cytometer at 488 nm.
Confocal Microscopy for Imaging Cellular Uptake of Modified Vaults
Cells were plated onto 60 mm Petri dishes and incubated at 37 °C with 5% CO2 for 48 h in DMEM media (F12-K for CHO-K1 cells) with 10% FBS. Cells were washed three times with cold PBS before the addition of modified vaults (30 µg of vaults per 500,000 cells). After incubating at 37 °C with 5% CO2 in serum-free DMEM, cells were visualized under a confocal microscope. Live cell imaging was performed using a Leica sp5 upright confocal microscope. Images were acquired from cells with a water immersion lens and analyzed using Las-af software. Scanning was performed at an image resolution of 1024 × 1024 pixels and averaged 2–4 times using 25% 488 nm.
MTT Viability Assay
The cell viability of modified vaults in RAW264.7 cells, HeLa cells, and CHO-K1 cells were investigated by using 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The experiments were performed in 96-well tissue culture plates (Falcon) growing 5000 cells per well in DMEM antibiotic media (F-12K antibiotic media for CHO-K1 cells) supplemented with 10% FBS for 24 h prior to treatments. At this time, the cells were washed with PBS 1×, and the medium was changed to DMEM or F-12K without FBS. Vaults were then added to the wells at the indicated concentrations. Cells were incubated with the vaults for 16 h at 37 °C before performing the MTT assay. After the incubation period, cells were treated with MTT (10 µL, 5 mg/mL in DMEM medium per well) and further incubated for 2 h at 37 °C. At the end of the incubation time, 100 µL of solubilizing solution (10% Triton X-100, 90% 0.1 N HCl in isopropanol) was added to each well, and colorimetry data were obtained on a plate reader (570 nm). Normalized viability is obtained by comparing the absorbance at 570 nm produced by the treated cells with that of control cells (no vaults added).
DLS Size Determination of Labeled hMVP Vaults
A 100 µL solution of modified vaults with a concentration range of 0.06–0.15 mg/mL in pH 7.4 PBS 1× buffer was placed in a 70 µL disposable cuvette. The cuvette was then placed in a Malvern Zetasizer Nano-S instrument. The temperature was allowed to equilibrate at 25 °C for 120 s before each DLS measurement.
Supplementary Material
Acknowledgments
Funding
Support of this work through grants NIH R01 CA031841 (P.A.W.) and NIH R01 CA031845 (P.A.W.) from the National Institutes of Health is acknowledged. Additionally support was provided by the National Science Foundation through grant NSF CHE-1566423 (P.A.W.) and the Vault Nano Postdoctoral Research Fellowship (D.C.B.).
The authors acknowledge the use of instruments from the Cell Sciences Imaging Facility (CSIF) and the Stanford Shared FACS facility. We gratefully acknowledge L. Cegelski for tissue culture equipment and a plate reader; R. Zare for the use of the Malvern Zetasizer; and S. Boxer for the use of the Nanodrop 1000.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Results and procedures for SDS-PAGE gel, protein recovery, DLS traces, electron microscopy images of modified CP-hMVP vaults, and overlay of fluorescence and bright-field images from confocal microscopy (PDF)
The authors declare the following competing financial interest(s): L.H.R. and V.A.K. declare that they have financial interest in Vault Nano Inc. and that the Regents of the University of California have licensed intellectual property invented by L.H.R. and V.A.K. to Vault Nano Inc.
REFERENCES
- 1.Kedersha NL, Rome LH. Isolation and Characterization of a Novel Ribonucleoprotein Particle: Large Structures Contain a Single Species of Small RNA. J. Cell Biol. 1986;103:699–709. doi: 10.1083/jcb.103.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kedersha NL, Miquel MC, Bittner D, Rome LH. Vaults 2. Ribonucleoprotein Structures Are Highly Conserved among Higher and Lower Eukaryotes. J. Cell Biol. 1990;110:895–901. doi: 10.1083/jcb.110.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rome LH, Kickhoefer VA. Development of the Vault Particle as a Platform Technology. ACS Nano. 2013;7:889–902. doi: 10.1021/nn3052082. [DOI] [PubMed] [Google Scholar]
- 4.Rome L, Kedersha N, Chugani D. Unlocking Vaults: Organelles in Search of a Function. Trends Cell Biol. 1991;1:47–50. doi: 10.1016/0962-8924(91)90088-q. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka H, Tsukihara T. Structural Studies of Large Nucleoprotein Particles, Vaults. Proc. Jpn. Acad. Ser. B. 2012;88:416–433. doi: 10.2183/pjab.88.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T. The Structure of Rat Liver Vault at 3.5 Angstrom Resolution. Science. 2009;323:384–388. doi: 10.1126/science.1164975. [DOI] [PubMed] [Google Scholar]
- 7.Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-Kd Vault Protein, VPARP, is a Novel Poly(ADP-Ribose) Polymerase. J. Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kickhoefer VA, Stephen AG, Harrington L, Robinson MO, Rome LH. Vaults and Telomerase Share a Common Subunit, TEP1. J. Biol. Chem. 1999;274:32712–32717. doi: 10.1074/jbc.274.46.32712. [DOI] [PubMed] [Google Scholar]
- 9.Kickhoefer VA, Liu Y, Kong LB, Snow BE, Stewart PL, Harrington L, Rome LH. The Telomerase/Vault-Associated Protein TEP1 Is Required for Vault RNA Stability and Its Association with the Vault Particle. J. Cell Biol. 2001;152:157–164. doi: 10.1083/jcb.152.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kickhoefer VA, Searles RP, Kedersha NL, Garber ME, Johnson DL, Rome LH. Vault Ribonucleoprotein Particles from Rat and Bullfrog Contain a Related Small RNA That Is Transcribed by RNA Polymerase III. J. Biol. Chem. 1993;268:7868–7873. [PubMed] [Google Scholar]
- 11.Van Zon A, Mossink MH, Scheper RJ, Sonneveld P, Wiemer EA. The Vault Complex. Cell. Mol. Life. Sci. 2003;60:1828–1837. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the Major Vault Protein: Novel Roles in Signal Pathway Regulation and Immunity. Cell. Mol. Life. Sci. 2009;66:43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. Assembly of Vault-like Particles in Insect Cells Expressing Only the Major Vault Protein. J. Biol. Chem. 2001;276:23217–23220. doi: 10.1074/jbc.C100226200. [DOI] [PubMed] [Google Scholar]
- 14.Suprenant KA. Vault Ribonucleoprotein Particles: Sarcophagi, Gondolas, or Safety Deposit Boxes? Biochemistry. 2002;41:14447–14454. doi: 10.1021/bi026747e. [DOI] [PubMed] [Google Scholar]
- 15.Kickhoefer VA, Rajavel KS, Scheffer GL, Dalton WS, Scheper RJ, Rome LH. Vaults Are Up-Regulated in Multidrug-Resistant Cancer Cell Lines. J. Biol. Chem. 1998;273:8971–8974. doi: 10.1074/jbc.273.15.8971. [DOI] [PubMed] [Google Scholar]
- 16.Kickhoefer VA, Vasu SK, Rome LH. Vaults Are the Answer, What Is the Question? Trends Cell Biol. 1996;6:174–178. doi: 10.1016/0962-8924(96)10014-3. [DOI] [PubMed] [Google Scholar]
- 17.Esfandiary R, Kickhoefer VA, Rome LH, Joshi SB, Middaugh CR. Structural Stability of Vault Particles. J. Pharm. Sci. 2009;98:1376–1386. doi: 10.1002/jps.21508. [DOI] [PubMed] [Google Scholar]
- 18.Kickhoefer VA, Han M, Raval-Fernandes S, Poderycki MJ, Moniz RJ, Vaccari D, Silvestry M, Stewart PL, Kelly KA, Rome LH. Targeting Vault Nanoparticles to Specific Cell Surface Receptors. ACS Nano. 2009;3:27–36. doi: 10.1021/nn800638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, Rome LH. Engineering of Vault Nanocapsules with Enzymatic and Fluorescent Properties. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4348–4352. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Abad D, Kickhoefer VA, Rome LH, Mahendra S. Vault Nanoparticles Packaged with Enzymes as an Efficient Pollutant Biodegradation Technology. ACS Nano. 2015;9:10931–10940. doi: 10.1021/acsnano.5b04073. [DOI] [PubMed] [Google Scholar]
- 21.Goldsmith LE, Pupols M, Kickhoefer VA, Rome LH, Monbouquette HG. Utilization of A Protein ″Shuttle″ to Load Vault Nanocapsules with Gold Probes and Proteins. ACS Nano. 2009;3:3175–3183. doi: 10.1021/nn900555d. [DOI] [PubMed] [Google Scholar]
- 22.Ng BC, Yu M, Gopal A, Rome LH, Monbouquette HG, Tolbert SH. Encapsulation of Semiconducting Polymers in Vault Protein Cages. Nano Lett. 2008;8:3503–3509. doi: 10.1021/nl080537r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buehler DC, Toso DB, Kickhoefer VA, Zhou ZH, Rome LH. Vaults Engineered for Hydrophobic Drug Delivery. Small. 2011;7:1432–1439. doi: 10.1002/smll.201002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buehler DC, Marsden MD, Shen S, Toso DB, Wu X, Loo JA, Zhou ZH, Kickhoefer VA, Wender PA, Zack JA, Rome LH. Bioengineered Vaults: Self-Assembling Protein Shell–Lipophilic Core Nanoparticles for Drug Delivery. ACS Nano. 2014;8:7723–7732. doi: 10.1021/nn5002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu M, Ng BC, Rome LH, Tolbert SH, Monbouquette HG. Reversible pH Lability of Cross-Linked Vault Nanocapsules. Nano Lett. 2008;8:3510–3515. doi: 10.1021/nl080536z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, Jiang J, Said-Sadier N, Boxx G, Champion C, Tetlow A, Kickhoefer VA, Rome LH, Ojcius DM, Kelly KA. Activation of the NLRP3 Inflammasome by Vault Nanoparticles Expressing a Chlamydial Epitope. Vaccine. 2015;33:298–306. doi: 10.1016/j.vaccine.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto NM, Prabhakaran P, Rome LH, Maynard HD. Smart Vaults: Thermally-Responsive Protein Nanocapsules. ACS Nano. 2013;7:867–874. doi: 10.1021/nn3053457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai C-H, Wiethoff CM, Kickhoefer VA, Rome LH, Nemerow GR. Vault Nanoparticles Containing an Adenovirus-Derived Membrane Lytic Protein Facilitate Toxin and Gene Transfer. ACS Nano. 2009;3:691–699. doi: 10.1021/nn8008504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikyas Y, Makabi M, Raval-Fernandes S, Harrington L, Kickhoefer VA, Rome LH, Stewart PL. Cryoelectron Microscopy Imaging of Recombinant and Tissue Derived Vaults: Localization of the MVP N Termini and VPARP. J. Mol. Biol. 2004;344:91–105. doi: 10.1016/j.jmb.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 30.Boutureira O, Bernardes GJL. Advances in Chemical Protein Modification. Chem. Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Srinivasan A, Sun Y, Mrazek J, Shu Z, Kickhoefer VA, Rome LH. Vault Nanoparticles Engineered with the Protein Transduction Domain, TAT48, Enhances Cellular Uptake. Integr. Biol. (Cambridge) 2013;5:151–158. doi: 10.1039/c2ib20119d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanzl EG, Trantow BM, Vargas JR, Wender PA. Fifteen Years of Cell-Penetrating, Guanidinium-Rich Molecular Transporters: Basic Science, Research Tools, and Clinical Applications. Acc. Chem. Res. 2013;46:2944–2954. doi: 10.1021/ar4000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torchilin VP. Tat Peptide-Mediated Intracellular Delivery of Pharmaceutical Nanocarriers. Adv. Drug Delivery Rev. 2008;60:548–558. doi: 10.1016/j.addr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Wender PA, Huttner MA, Staveness D, Vargas JR, Xu AF. Guanidinium-Rich, Glycerol-Derived Oligocarbonates: A New Class of Cell-Penetrating Molecular Transporters That Complex, Deliver, and Release siRNA. Mol. Pharmaceutics. 2015;12:742–750. doi: 10.1021/mp500581r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyman JM, Geihe EI, Trantow BM, Parvin B, Wender PA. A Molecular Method for the Delivery of Small Molecules and Proteins across the Cell Wall of Algae Using Molecular Transporters. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13225–13230. doi: 10.1073/pnas.1202509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seward GK, Wei Q, Dmochowski IJ. Peptide-Mediated Cellular Uptake of Cryptophane. Bioconjugate Chem. 2008;19:2129–2135. doi: 10.1021/bc8002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green M, Loewenstein PM. Autonomous Functional Domains of Chemically Synthesized Human Immunodeficiency Virus Tat Trans-Activator Protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 38.Frankel AD, Pabo CO. Cellular Uptake of the Tat Protein from Human Immunodeficiency Virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 39.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The Design, Synthesis, and Evaluation of Molecules that Enable or Enhance Cellular Uptake: Peptoid Molecular Transporters. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vives L, Brodin P, Lebleu BA. A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine Enters Cells More Efficiently than Other Polycationic Homopolymers. J. Pept. Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 42.Jones LR, Goun EA, Shinde R, Rothbard JB, Contag CH, Wender PA. Releasable Luciferin-Transporter Conjugates: Tools for the Real-Time Analysis of Cellular Uptake and Release. J. Am. Chem. Soc. 2006;128:6526–6527. doi: 10.1021/ja0586283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.