Abstract
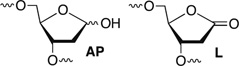
DNA is rapidly cleaved under mild alkaline conditions at apyrimidinic/apurinic sites, but the half-life is several weeks in phosphate buffer (pH 7.5). However, abasic sites are ~100-fold more reactive within nucleosome core particles (NCPs). Histone proteins catalyze the strand scission, and at superhelical location 1.5, the histone H4 tail is largely responsible for the accelerated cleavage. The rate constant for strand scission at an abasic site is enhanced further in a nucleosome core particle when it is part of a bistranded lesion containing a proximal strand break. Cleavage of this form results in a highly deleterious double-strand break. This acceleration is dependent upon the position of the abasic lesion in the NCP and its structure. The enhancement in cleavage rate at an apurinic/apyrimidinic site rapidly drops off as the distance between the strand break and abasic site increases and is negligible once the two forms of damage are separated by 7 bp. However, the enhancement of the rate of double-strand break formation increases when the size of the gap is increased from one to two nucleotides. In contrast, the cleavage rate enhancement at 2-deoxyribonolactone within bistranded lesions is more modest, and it is similar in free DNA and nucleosome core particles. We postulate that the enhanced rate of double-strand break formation at bistranded lesions containing apurinic/apyrimidinic sites within nucleosome core particles is a general phenomenon and is due to increased DNA flexibility.
Graphical abstract
DNA double-strand breaks are extremely deleterious to cells. Their repair is so essential to genomic stability and cell viability that two major pathways (NHEJ and homologous recombination) are present to repair them.1–3 These pathways are also desirable inhibition targets.4,5 Consequently, dsb formation is desirable when searching for cytotoxic molecules that damage DNA. Molecules that produce dsbs directly, particularly via a single chemical event, are rare and highly valued for their potential therapeutic benefits.6–8 Repair enzymes can convert bistranded lesions into dsbs, although the juxtaposition of the damaged sites greatly affects the efficiency of these processes.9–19 Abasic sites are chemically unstable and spontaneously yield strand breaks, which generate dsbs when the breaks are part of bistranded lesions.20–24 Although the half-lives for such processes in aqueous buffer are on the order of weeks, histone proteins within nucleosome core particles accelerate DNA cleavage at three forms of abasic site lesions, and excision of a fourth that is already incised.25–29 Strand scission is accelerated as much as 450-fold at oxidized abasic sites.27 Furthermore, dsbs are rapidly generated when two AP sites are present in NCPs within 3 bp of one another via intermediate bistranded lesions consisting of a ssb and one abasic site.25,30 Herein, the generality of this process was probed by examining the effects of the position of the bistranded lesion in the NCP, the structure of the DNA termini at the strand break, the juxtaposition of the strand break and abasic site, and the structure of the latter.
Bistranded lesions make up a subset of clustered lesions and are defined as two or more damaged sites on opposing strands that are within 1.5 helical turns.10,31–33 High local concentrations (“spurs”) of hydroxyl radical produced in the vicinity of DNA by the high energy of γ-radiolysis give rise to clustered lesions.34 In addition, a mechanism has been identified for forming bistranded lesions via interstrand radical transfer.35,36 However, there is no evidence reported of the involvement of such a process in cells. Extensive research has shown that bistranded clustered lesions inhibit base excision repair.32,37–39 The extent to which repair is impaired depends upon the structure of the lesions and their relative orientation within the duplex. Some small molecules that oxidatively damage DNA also produce bistranded lesions that are between 0 and 2 bp apart.7,40–42 For instance, the clustered lesions produced by the antitumor antibiotic neocarzinostatin contain the oxidized abasic site, 2-deoxyribonolactone, and a ssb on the opposite strand.43 The ssb is shifted 2 bp in the 3′-direction due to the molecule binding in the minor groove.
The half-lives of isolated L and AP measured within NCPs ranged from 3.3 to 30 h. However, the mechanisms for L and AP site cleavage in NCPs are different.26,29,30 The latter proceeds via Schiff base intermediates and resembles the lyase mechanism for incising AP sites utilized by BER enzymes; during cleavage at L, DNA–protein cross-links are generated as side products. In addition, the acceleration of strand scission at L within NCPs, ~20-fold faster than that of naked DNA, is more modest than that observed for AP. Prior to this investigation, no information was available regarding L reactivity within clustered lesions, such as those produced by neocarzinostatin.
EXPERIMENTAL PROCEDURES
Materials
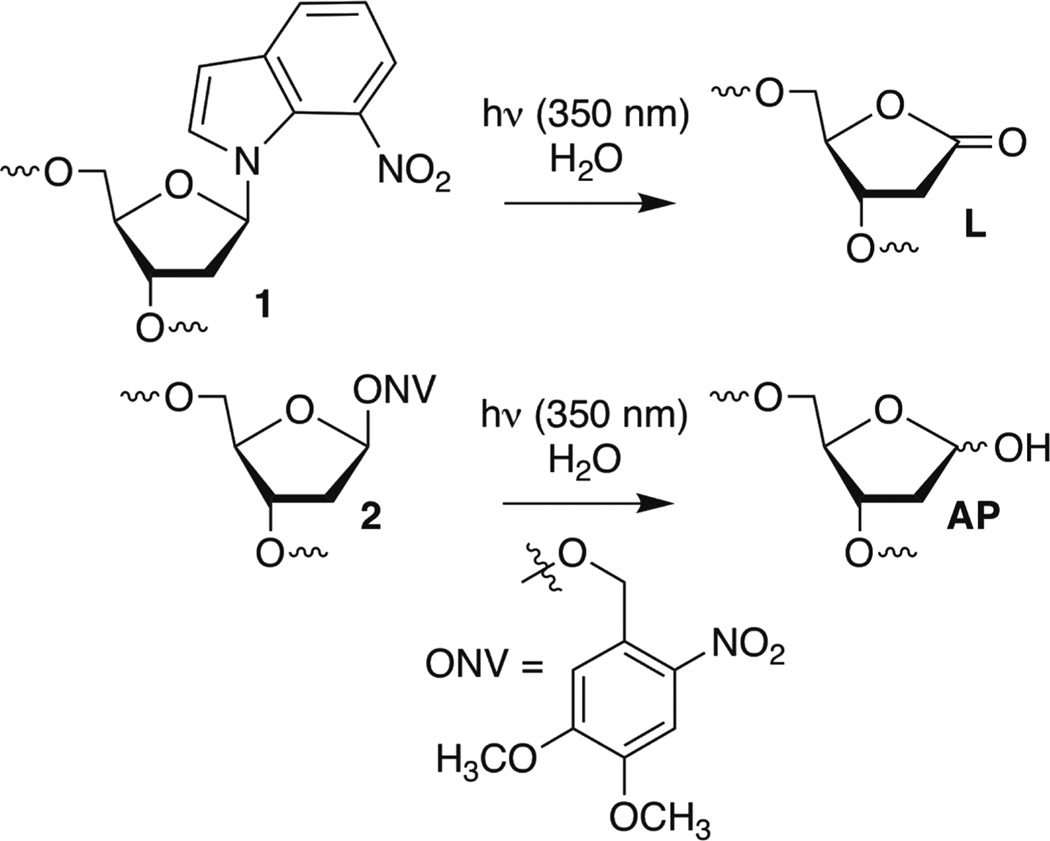
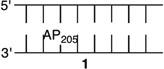
T4 polynucleotide kinase, T4 DNA ligase, and proteinase K were from New England Biolabs (Ipswich, MA). [γ-32P]ATP (specific activity of 4500 Ci mmol−1) was from MP Biomedicals (Santa Ana, CA). Oligonucleotides were synthesized using an Applied Biosystems Inc. 394 oligonucleotide synthesizer. DNA synthesis reagents were purchased from Glen Research (Sterling, VA). NaBH4 was purchased from Sigma-Aldrich. All experiments were conducted in clear siliconized tubes (Bio Plas Inc.). Quantification of radiolabeled oligonucleotides was conducted using a Molecular Dynamics Phosphorimager 840 equipped with ImageQuant TL software. Expression and purification of all the histone proteins, refolding of the histone octamer, and its purification were performed using reported procedures.44 DNA containing photolabile precursors 1 or 2 for L and AP, respectively (Scheme 1), were synthesized as previously described.25,26,30,45
Scheme 1.
Reconstitution of Nucleosome Core Particles (NCPs)
Salmon sperm DNA (10 µg) and 5′-32P-labeled 145 bp dsDNA (~1 pmol) were combined in a small siliconized tube to a final volume of 10 µL containing 2 M NaCl and 0.1 µg/µL BSA. The quantity of salmon sperm DNA is sufficient to form ~84 pmol of NCPs. Varying amounts of histone octamer (also in 2 M NaCl) were added to individual DNA samples (total volume around 11–12 µL) that were incubated at room temperature for 30 min before a series of dilutions using nucleosome buffer [10 mM HEPES and 1 mM EDTA (pH 7.5)] containing 0.1 µg/µL BSA at 30 °C in the heat block of a thermal cycler. The approximate octamer:DNA molar ratio was typically 0.8, 0.9, 1.0, and 1.1. Dilution data (volume of buffer added in microliters, incubation time in minutes): 1:12, 60; 2:6, 60; 3:6, 60; 4:10, 30; 5:10, 30; 6:20, 30; 7:50, 30; 8:100, 30. After the final dilution (total volume of ~225 µL), each sample was cooled to room temperature and any precipitate was pelleted via a brief (5 min) spin at 15000g. Each solution was then transferred to a fresh siliconized tube, and a small aliquot was analyzed by nucleoprotein gel electrophoresis [10 cm × 8 cm × 0.15 cm, 6%, acrylamide/bisacrylamide, 59:1, 0.2× TBE buffer, run at 4 °C using 0.2× TBE buffer; the gel was run under limiting voltage (200 V) until the bromophenol blue band migrated to the bottom] to determine the extent of reconstitution. Only samples that contained >90% reconstituted NCP were used in subsequent experiments. Reconstituted nucleosome core particles were stored at 4 °C, and under this condition, typically no dissociation was detected for up to 2 weeks.
General Procedure for Monitoring the Reactivity of AP and L in NCPs
NCPs prepared as described above were used directly in these experiments without adjustments to their concentrations following reconstitution [10 mM HEPES, 100 mM NaCl, 0.1 µg/µL BSA, and 1 mM EDTA (pH 7.5)]. NCPs containing AP or L precursor were photolyzed for 10 or 20 min, respectively, at room temperature in the cavity of a Rayonet photoreactor containing 16 lamps that emit maximally at 350 nm and immediately incubated at 37 °C for the duration of the time course experiment. Immediately after photolysis, four aliquots (4 µL) were removed from the reaction mixture. Two aliquots were treated with 0.1 N (final) NaOH at 37 °C for 30 min and then neutralized with 0.1 N HCl. All four aliquots were analyzed by denaturing PAGE as described below to determine the percent photoconversion (NaOH-treated aliquots) and background cleavage (untreated aliquots). When background cleavage in AP-containing NCPs was determined, the aliquots were treated with NaBH4 (0.1 M final concentration) at 4 °C for 1 h. NaBH4 treatment was omitted for experiments with L. All aliquots were treated with proteinase K (0.5 µL, 10 µg) for 15 min at room temperature prior to analysis by 8% denaturing PAGE (20 cm × 16 cm × 0.04 cm). The gel was run at 15 W (limiting conditions) until the bromophenol blue band migrated to the bottom. To monitor reactions in NCPs, aliquots were removed at appropriate times and stored at −78 °C until all aliquots were collected, at which time they were treated with NaBH4 as described above. The data collected for time course experiments were corrected to account for any remaining free DNA due to incomplete reconstitution. The amount of free DNA was determined using native PAGE. The total volume (as measured via phosphorimaging analysis) of all bands in any given lane (volume here is equivalent CPM, as determined by autoradiography) was multiplied by the fraction of remaining free DNA. This amount was then subtracted from the volume of intact DNA in the same lane. Reactions were similarly corrected for the fraction of unconverted photochemical precursor. The amount of product in a given aliquot based upon the fraction of background cleavage in aliquots removed immediately after photolysis was subtracted from the observed product. Data were fit linearly by plotting the ln(fraction of intact DNA) versus time using Origin 6.0.
General Procedure for Monitoring the Reactivity of AP and L in Free DNA
To study the reactivity of AP and L in free 145 dsDNA, 5′-32P-labeled 145 bp dsDNA (~1 pmol) was diluted to 225 µL in 10 mM HEPES (pH 7.5), 100 mM NaCl, and 1 mM EDTA. The solution was photolyzed (350 nm) for 10 min (for AP-containing DNA) or 20 min (for L-containing DNA) at room temperature and immediately incubated at 37 °C. Immediately after photolysis, two aliquots (4 µL) were removed from the reaction mixture and treated with 0.1 N (final) NaOH at 37 °C for 30 min. The samples were then neutralized with 0.1 N HCl and by denaturing PAGE to determine the percent photoconversion. Aliquots were removed at appropriate times and stored at −80 °C until all aliquots were collected. For AP-containing DNA, reactions were quenched with NaBH4 (0.1 M, final concentration) and analyzed directly by 8% denaturing PAGE (20 cm × 16 cm × 0.04 cm). NaBH4 treatment was not required for L-containing DNA. The gel was run under limiting power (15 W) until the bromophenol blue band migrated to the bottom. Reactions were corrected for the fraction of unconverted photochemical precursor. Data were fit using Origin.
RESULTS
Nucleosome Core Particle Construction
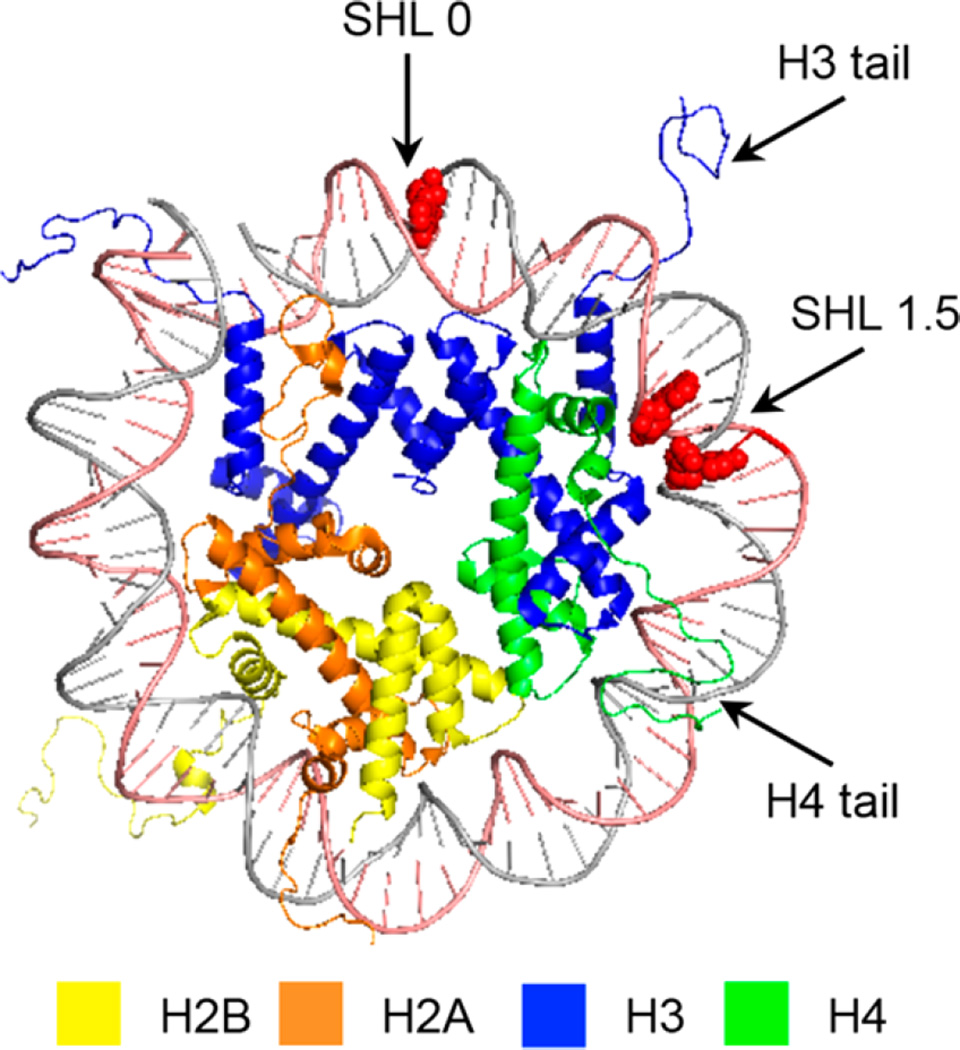
Nucleosome core particles were assembled via reconstitution of purified histone octamers with DNA in which the abasic site-containing strand was 5′-32P-labeled. The DNA sequences are based on the 601 strong positioning sequence discovered by Widom.46,47 AP and L were generated at positions 73, 89, or 205 of the 601 DNA from previously reported photochemical precursors (Figure 1). The requisite 145-nucleotide DNA containing 1 or 2 at a defined site was obtained by T4 DNA ligase-mediated ligation of chemically synthesized oligonucleotides. The full-length products were purified by denaturing PAGE. The latter two positions are on opposite strands in the SHL 1.5 region of the NCP, in which the DNA is kinked.47,48 This region is also a hot spot for DNA binding molecules.49 Position 73 is at the dyad axis of the NCP. The DNA is prepared by ligating chemically synthesized oligonucleotides, as previously described.25,26,30,45 The NCPs are reconstituted containing the lesions in protected form to guard against premature cleavage. The lesions were revealed via a brief photolysis at a λmax of 350 nm.
Figure 1.
General structural features of a nucleosome core particle showing one gyre of the DNA (Protein Data Bank entry 1kx5). Nucleotide positions at which abasic sites are incorporated are colored red; clockwise from the top are positions 73, 205, and 89, respectively.
Effect of Proximal Strand Break Termini on AP Reactivity in Bistranded Lesions
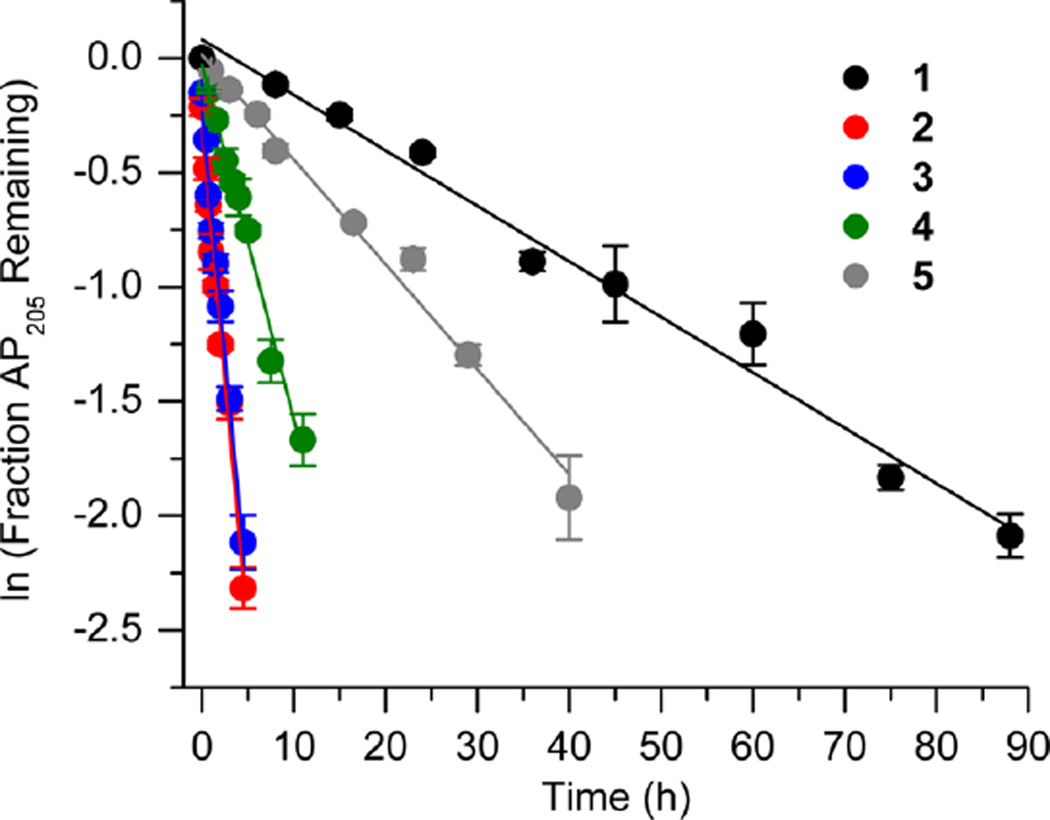
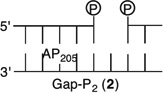
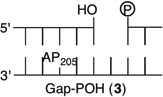
Initially, we investigated the reaction of an AP site within a bistranded lesion in a NCP that contained a ssb with a single-nucleotide gap. The 5′- and 3′-termini of the cleaved DNA surrounding the strand break contained phosphate groups [e.g., Gap-P2 (2) (Figure 2 and Table 1)].30 AP reactivity within the clustered lesion was accelerated ~20-fold compared to that of an isolated AP205 lesion (1). The accelerated reactivity of AP205 in the NCP was considerably greater than in free DNA, where the t1/2 was 1008 h when only the abasic site was present and 364 h when the single-nucleotide gap containing phosphate termini was present.30 Replacing one of the phosphate groups with a hydroxyl [Gap-POH (3)] had little to no effect on AP205 reactivity in the proximity of a single-nucleotide gap. The rate constants for AP205 disappearance in 2 and 3 are within experimental error of one another. It is notable that the structure of the single-nucleotide gap in 3 is that expected from AP excision by the BER enzymes Ape1 and Pol β. However, AP205 reacted more than 2-fold slower in Gap-OH2 (4), in which both termini at the gap contain hydroxyl groups, than in Gap-P2 (2). Nonetheless, AP205 still reacted more than 7 times faster in this bistranded lesion than when it was isolated within the NCP. Furthermore, the least reactive bistranded lesion in a gap was still far more reactive than AP205 when it was part of a bistranded lesion in which the opposite strand was nicked [nicked (5) (Table 1)]. AP205 reacts only twice as fast in the nicked substrate (5) as it does when present as an isolated lesion in a NCP. In free DNA, the nicked substrate is also less reactive (t1/2 = 400 h) than that containing a gap, but the difference is smaller than in the NCP.
Figure 2.
Representative plots of the effect of a nick or one-nucleotide gap (containing various termini) in the opposing strand on the rate of disappearance of AP205 in a NCP. The numbers 1–5 correspond to substrate numbers in Table 1.
Table 1.
AP205 Reactivity in NCPs as Part of an Isolated Lesion versus a Lesion Present in Bistranded Lesions
| kdis (10−6 s−1)a | t1/2 (h)a | Rel. Rate Gap : AP205(1)b |
|
|---|---|---|---|
 |
6.5 ± 1.1 | 29.6 ± 5.2 | - |
 |
128.3 ± 8.0 | 1.5 ± 0.1 | 19.7 |
 |
113.2 ± 6.3 | 1.7 ± 0.1 | 17.4 |
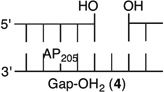 |
45.8 ± 5.7 | 4.2 ± 0.6 | 7.0 |
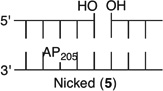 |
12.8 ± 1.4 | 15.0 ± 1.8 | 2.0 |
Values presented are averages ± the standard deviation of at least three experiments each consisting of three replicates.
The relative rate equals the average rate constant for AP205 disappearance in a NCP containing a bistranded lesion/1.
Effect of the Distance and Gap Size on AP on Reactivity
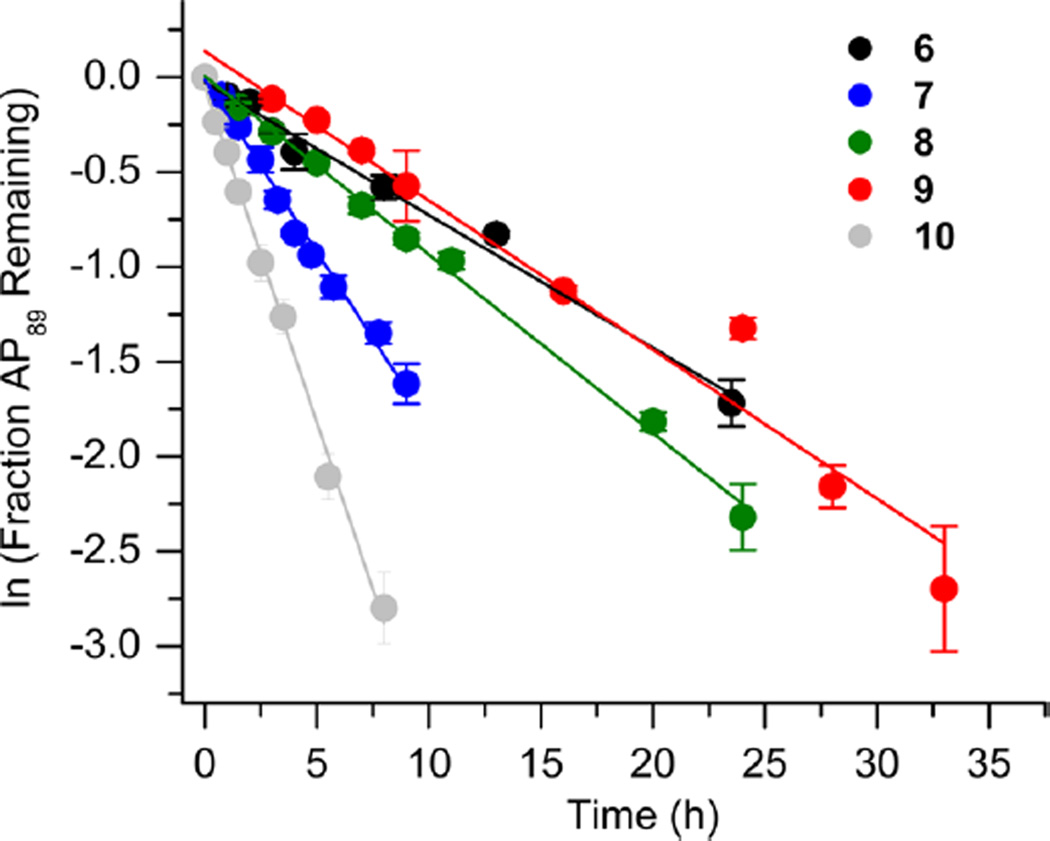
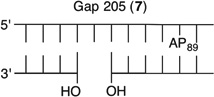
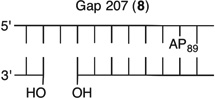
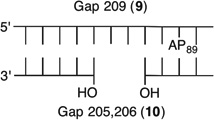
We reported that the reactivity at AP89 in a NCP (6) is accelerated 3-fold when a single-nucleotide gap containing phosphates at the 5′- and 3′-termini is introduced 3 bp away [gap position, 205; Gap 205 (7) (Figure 3 and Table 2)]. This effect is weaker than that observed in the complementary NCP in which AP205 is part of a bistranded lesion containing a gap centered at position 89 [4 (Table 1)]. The effect of the proximity of the single-nucleotide gap to AP89 on its reactivity was examined using three NCPs [7–9 (Figure 3 and Table 2)] in which the 5′- and 3′-termini at the gap contained hydroxyl groups. AP89 reacted 3 times faster when the single-nucleotide gap was centered 3 bp away [Gap 205 (7)] on the opposite strand. Increasing the size of the gap from one (7) to two (10) nucleotides at this site enhanced the rate constant for AP89 disappearance almost 2-fold. In contrast, the half-lives of AP89 in the corresponding free DNA containing a single nucleotide (t1/2 = 267 h) and two-nucleotide gap (t1/2 = 245 h) were essentially the same. However, the acceleration that can be attributed to the gap rapidly decreased when it was 5 bp [Gap 207 (8)] and 7 bp [Gap 209 (9)] removed from the nucleotide opposite AP89. When the gap was 7 bp from the abasic site, the rates of reaction of AP89 in the bistranded lesion containing NCP and that in which it was the only damage were within experimental error of one another.
Figure 3.
Representative plots of the effect of distance from a one-nucleotide or the presence of a two-nucleotide gap in the opposing strand on the rate of disappearance of AP89 in a NCP. The numbers 6–10 correspond to substrate numbers in Table 2.
Table 2.
Distance and Size Effects of a Gap in the Opposing Strand on AP89 Reactivity in NCPs
| kdis (10−5 s−1)a | t1/2 (h)a | Rel. Rate Gap : AP89b |
|
|---|---|---|---|
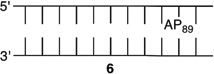 |
1.89 ± 0.12 | 10.2 ± 0.7 | - |
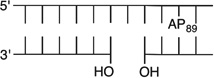 |
5.66 ± 1.05 | 3.4 ± 0.7 | 3.0 |
 |
2.50 ± 0.18 | 7.7 ± 0.6 | 1.3 |
 |
2.16 ± 0.13 | 8.9 ± 0.6 | 1.1 |
 |
10.13 ± 1.4 | 1.9 ± 0.3 | 5.4 |
Values presented are averages ± the standard deviation of at least three experiments each consisting of three replicates.
The relative rate equals the average rate constant for AP89 disappearance in a NCP containing a bistranded lesion/6.
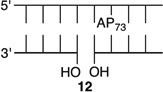
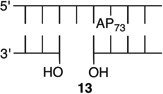
Effect of a Strand Break on AP Reactivity at the Dyad Axis in a NCP
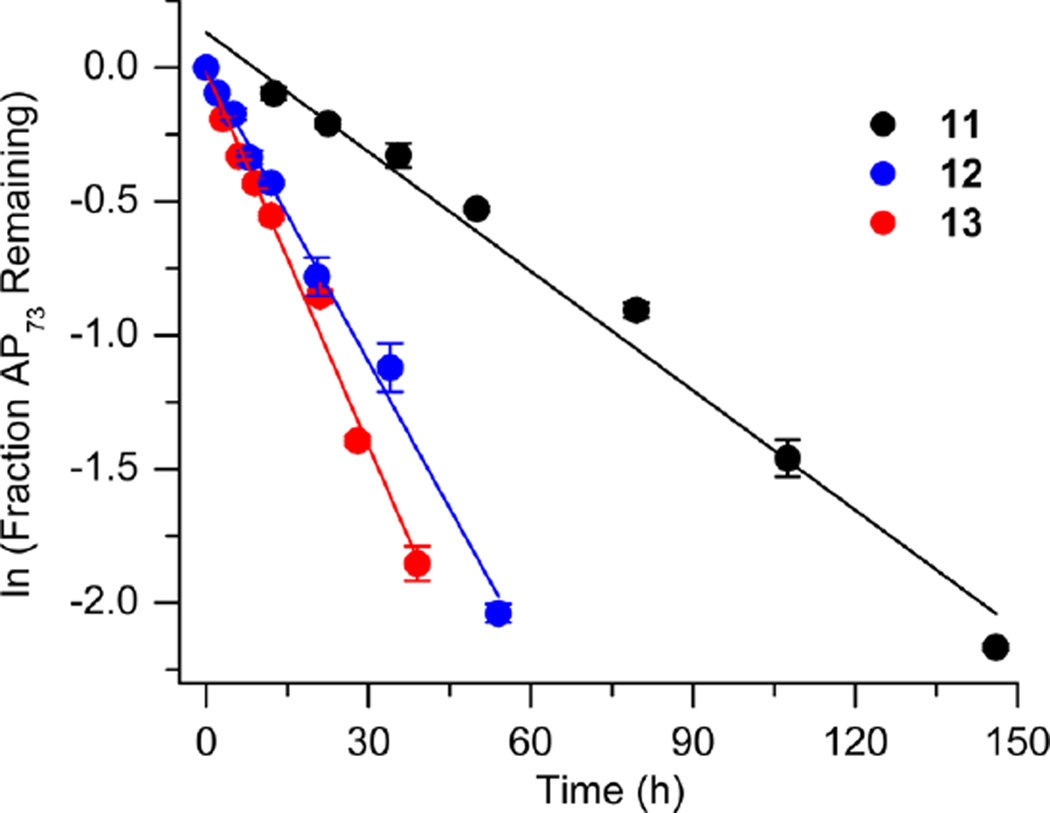
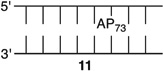
The amino-terminal tail of histone H3 is the most proximal flexible protein component to the dyad position (SHL 0). However, a lesion at this position [e.g., AP73 (11)] is farther from a protein tail (histone H4) than one at position 89. The bistranded lesions containing AP73 were constructed so that the strand break in the opposing strand is 1 bp closer to the abasic site than when the lesion was in the vicinity of SHL 1.5 (Figure 4 and Table 3). On the basis of the findings described above, we anticipated that this could enhance the effect of the bistranded lesion, but certainly not diminish it. Although the difference between the nick- and gap-containing substrates is smaller than when the abasic site is at SHL 1.5 [AP205 (Table 1)], the overall effect is qualitatively similar. AP73 reacts 3.2 times more rapidly in the gapped substrate (13) than when the lesion is isolated in the DNA (11). This is comparable to the acceleration observed at AP89 (Table 2) but smaller than in the gapped bistranded lesion containing AP205 (Table 1). Reaction at AP73 in the nicked substrate (12) is slightly slower than that in the gapped NCP (13).
Figure 4.
Representative plots of the effect of a nick or one-nucleotide gap in the opposing strand on the rate of disappearance of an abasic site at the dyad axis (AP73) in a NCP. The numbers 1–5 correspond to substrate numbers in Table 1. The numbers 11–13 correspond to substrate numbers in Table 3.
Table 3.
Reactivity of an Abasic Site at the Dyad Axis (AP73) in Various NCP Constructs
| kdis (10−6 s−1)a | t1/2 (h)a | Rel. Rate X : AP73 (11)b |
|
|---|---|---|---|
 |
3.80 ± 0.6 | 50.3 ± 8.7 | - |
 |
10.1 ± 0.3 | 19.0 ± 0.7 | 2.7 |
 |
12.7 ± 1.0 | 15.1 ± 1.2 | 3.4 |
Values presented are averages ± the standard deviation of at least three experiments each consisting of three replicates.
The relative rate equals the average rate constant for AP73 disappearance in a NCP containing a bistranded lesion/11.
Effect of a Strand Break on 2-Deoxyribonolactone Reactivity in a NCP
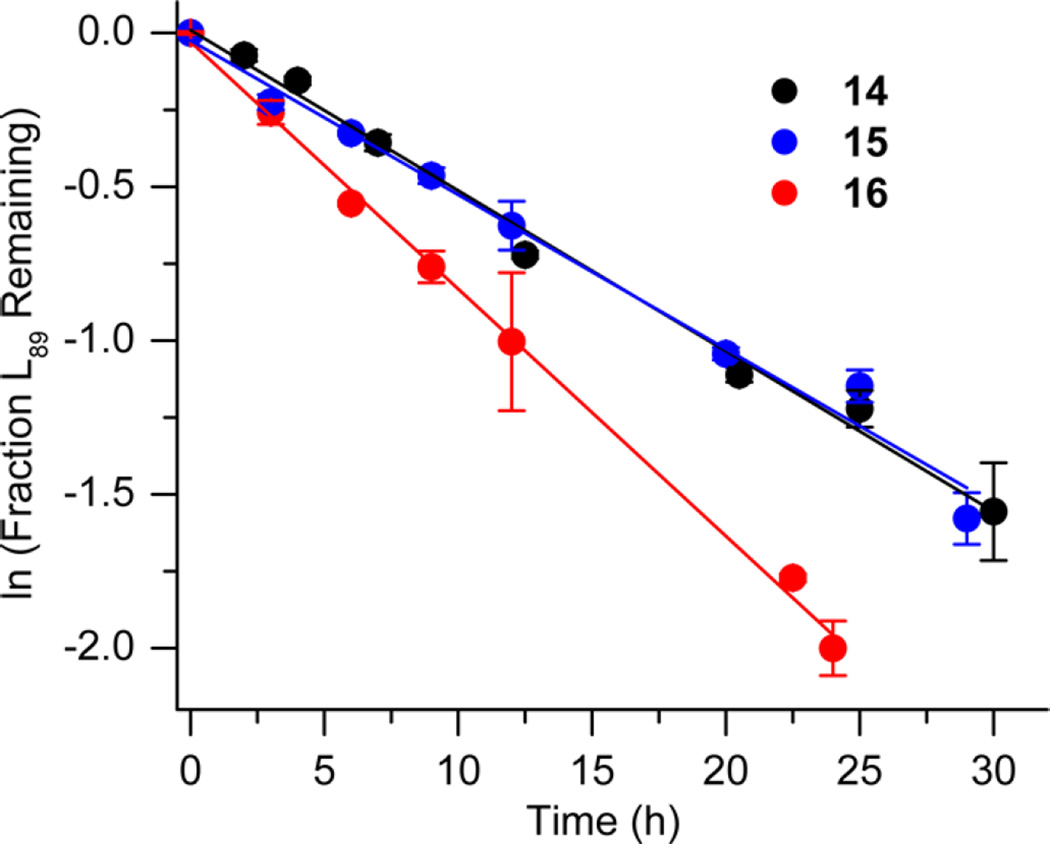
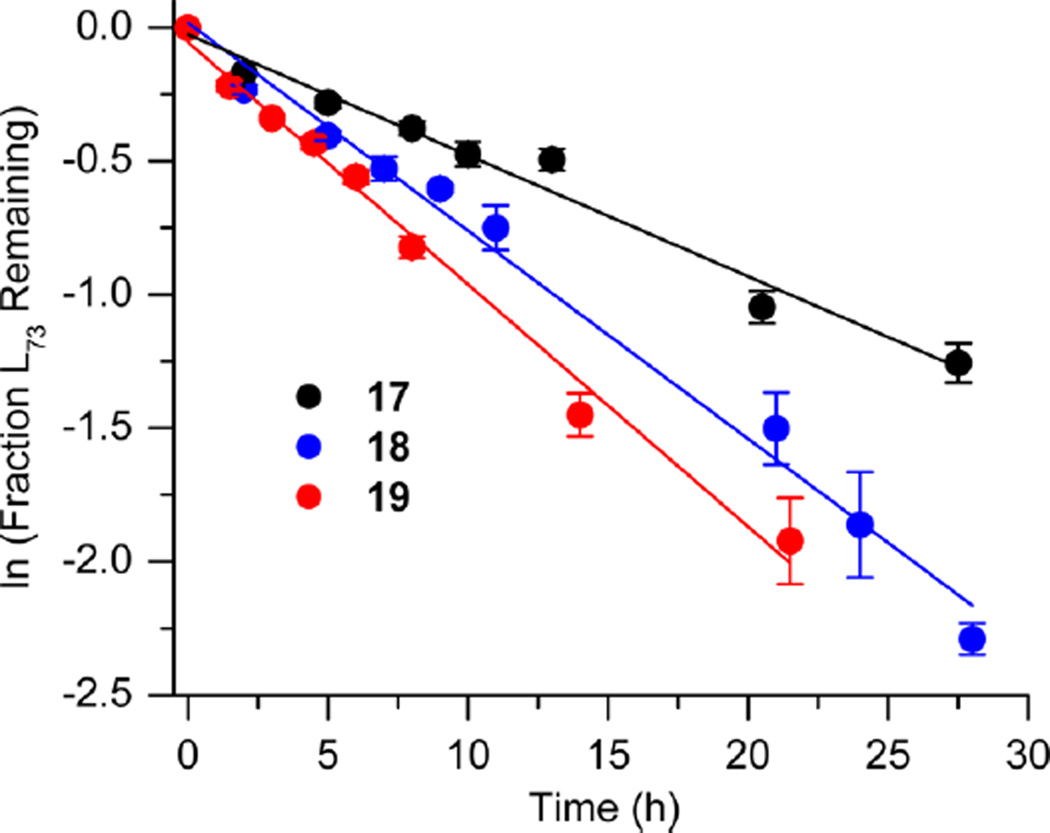
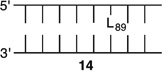
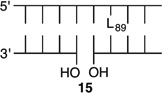
Bistranded lesions containing L were examined in which the nick (15 and 18) or a single-nucleotide gap (16 and 19) in the opposing strand was 2 bp removed. This separation, which is also oriented in the 3′-direction, was chosen to mimic the bistranded lesion produced by the antitumor antibiotic neocarzinostatin.7,43 The reactivities of isolated lactone in NCPs at SHL 1.5 [L89 (14) (Figure 5 and Table 4)] and the dyad axis [L73 (17) (Figure 6 and Table 5)] were very similar to one another and slightly slower than those previously reported.26 The effect of a strand break on L reactivity at either superhelical location was more modest than for strand cleavage at an AP site. The rate constant for strand scission at L89 in the presence of a nick (15) was within experimental error of that of a lesion when it was isolated in the NCP (14). 2-Deoxyribonolactone reactivity at the dyad axis (L73) was increased slightly in the presence of a nick [18 (Table 5)] but less so than was AP73 [12 (Table 3)]. Introducing a gap in the opposing strand (16 and 19) accelerated strand scission at L when the lesion was present at either SHL. However, the increase was <2-fold. In free DNA, the t1/2 of L89 in the absence of any damage in the opposing strand was 165 h. The lifetime of L89 decreased to 103 h in the presence of a gap at position 204, revealing that the increased reactivity at L89 in the presence of a single-nucleotide gap in the opposing strand within a NCP was comparable to that in free DNA.
Figure 5.
Representative plots of the effect of a nick or one-nucleotide gap in the opposing strand on the rate of disappearance of 2-deoxyribonolactone at SHL 1.5 (L89) in a NCP. The numbers 14–16 correspond to substrate numbers in Table 4.
Table 4.
Reactivity of a 2-Deoxyribonolactone Abasic Site at SHL 1.5 (L89) in Various NCP Constructs
| kdis (10−5 s−1)a | t1/2 (h)a | Rel. Rate X : L89 (14)b |
|
|---|---|---|---|
 |
1.58 ± 0.24 | 12.2 ± 2.1 | - |
 |
1.41 ± 0.07 | 13.6 ± 0.8 | 0.9 |
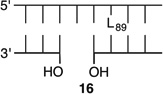 |
2.41 ± 0.27 | 8.0 ± 1.0 | 1.5 |
Values presented are averages ± the standard deviation of at least three experiments each consisting of three replicates.
The relative rate equals the average rate constant for L89 disappearance in a NCP containing a bistranded lesion/14.
Figure 6.
Representative plots of the effect of a nick or one-nucleotide gap in the opposing strand on the rate of disappearance of 2-deoxyribonolactone at SHL 1.5 (L73) in a NCP. The numbers 17–19 correspond to substrate numbers in Table 5.
Table 5.
Reactivity of a 2-Deoxyribonolactone Abasic Site at the Dyad Axis (L73) in Various NCP Constructs
| kdis (10−5 s−1)a | t1/2 (h)a | Rel. Rate X : L73 (17)b |
|
|---|---|---|---|
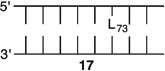 |
1.32 ± 0.09 | 14.6 ± 1.0 | - |
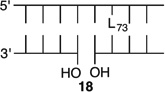 |
2.12 ± 0.14 | 9.1 ± 0.6 | 1.6 |
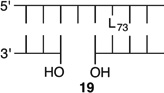 |
2.53 ± 0.24 | 7.6 ± 0.8 | 1.9 |
Values presented are averages ± the standard deviation of at least three experiments each consisting of three replicates.
The relative rate equals the average rate constant for L73 disappearance in a NCP containing a bistranded lesion/17.
DISCUSSION
Bistranded lesions, which are commonly produced by γ-radiolysis and some antitumor agents, are precursors to double-strand breaks, the most deleterious form of DNA damage. Two observations motivated this study. The first was that nucleosome core particles catalyze DNA cleavage at abasic sites as much as 450-fold.27 The second was that cleavage at an AP site in a NCP containing a strand break in the opposing strand was accelerated even further.30 The latter results in the formation of dsbs. The original report revealed that bistranded lesions in the region of the NCP where the DNA is kinked (SHL 1.5) result in significant additional acceleration of cleavage.25 The goal of this investigation was to examine the generality of this process and the source of the enhanced reactivity.
The experiments described herein illustrate that accelerated cleavage at an AP site is general but variable in magnitude. Bistranded lesions containing AP sites at three positions within the 601 strong positioning NCP all showed enhanced reactivity. However, the additional acceleration at an AP site within a bistranded lesion containing a single-nucleotide gap in which the termini contained hydroxyl groups ranged from 2.7 to 7.0. The number of sites probed within the NCP is small, and it is possible that the range may be even larger. The effect of end groups in bistranded lesions on the enhanced AP reactivity was examined for the site in which dsb was the greatest [AP205, gap at position 89 (2–4) (Table 1)]. The effects of the end groups on AP reactivity in gapped substrates were small compared to the overall acceleration, and all were considerably more reactive than when the opposing strand was “nicked” (5). The reactivity of AP in construct 3, which contains a 3′-hydroxyl terminus and 5′-phosphate, was within experimental error of the most reactive substrate (2) in which both termini were phosphorylated. The reactivity of AP205 in these bistranded lesions was inversely correlated with the anticipated stabilities of the duplexes.50,51 The nicked substrate (5), which lacks phosphate, should be the most stable duplex and least flexible. The gapped substrate containing phosphates at each terminus (2) should be the least stable. These data suggest that AP reactivity increases as the duplex stability decreases. Increased single strandedness also imparts greater flexibility on the DNA. The possibility that greater single strandedness and/or flexibility enhances AP reactivity is reinforced by the increased rate constant when the gap is increased from 1 bp (7) to 2 bp (10) (Table 2) and is consistent with recent nuclear magnetic resonance studies on abasic site-containing clustered lesions.52,53 In addition, the rapid decrease in the rate constant for AP89 reactivity as the distance between the single-nucleotide gap and the abasic site increases from 3 to 7 bp is also consistent with the single-stranded nature of the DNA contributing to reactivity. We hypothesize that the increased single strandedness and/or flexibility of the bistranded lesion allows the strand containing AP to adopt conformations that are conducive to reaction with the histone tail that is responsible for cleavage catalysis.25,30,54,55 This is also consistent with recent calculations concerning the association of DNA dynamics and bistranded lesion repair.10
2-Deoxyribonolactone exhibited a similar but smaller acceleration in reactivity (Tables 4 and 5) than AP within bistranded lesions at NCP locations. Duplexes containing bistranded lesions composed of L and another lesion are more flexible.31 The difference between the additional acceleration within the NCP and free DNA was insignificant. These observations are consistent with the differences in reactivity of L within NCPs from that of AP.26 The enhancement of 2-deoxyribonolactone reactivity in a NCP (~20-fold) is significantly smaller than that of AP (~100-fold). Hence, any additional acceleration in a bistranded lesion may also be smaller. Furthermore, the mechanism of L cleavage is different from that of AP. The latter requires Schiff base formation and may have more exacting requirements for protein accessibility that are accommodated in bistranded lesions.
Despite the lack of a significant acceleration of 2-deoxyribonolactone cleavage within NCPs that are part of bistranded lesions containing ssbs relative to free DNA, it is reasonable to expect that considerable levels of dsbs will form within cells. On the basis of the intracellular lifetime of bistranded lesions containing AP sites, the ~8–14 h half-life for such a process at two SHL locations is short enough that one could expect dsb formation to compete with repair.13 The same is also true for the more reactive bistranded lesions containing AP sites. The rate constants measured herein for strand scission at abasic sites that are proximal to single-nucleotide gaps are competitive with the known lifetimes of similar lesions within cells.13 Consequently, if bistranded lesions composed of a single-strand break and proximal abasic site are produced in cells, these data suggest that a significant fraction of them will be transformed into double-strand breaks via spontaneous reaction within chromatin. This would be a deleterious process, as double-strand breaks can result in the loss of an entire chromosome segment along with the gene, translocation of the broken DNA to the telomere of another chromosome, and ultimately cell death.
Supplementary Material
Acknowledgments
Funding
We are grateful for support of this research by the National Institute of General Medical Sciences (Grant GM-063028 to M.M.G.). M.D.P. was supported by the Program in Molecular Biophysics (T32 GM-008403).
We are grateful to Dr. Liwei Weng (University of Pennsylvania, Philadelphia, PA), Professor Chuanzheng Zhou (Nankai University, Tianjin, China), Professor Greg Bowman (Johns Hopkins University), Dr. Ilana Nodelman (Johns Hopkins University), and Professor Jonathan Sczepanski (Texas A&M, College Station, TX) for technical advice and helpful discussions.
ABBREVIATIONS
- AP
apurinic/apyrimidinic site
- L
2-deoxyribonolactone
- BER
base excision repair
- dsb
double-strand break
- ssb
single-strand break
- NHEJ
nonhomologous end joining
- SHL
superhelical location
- Ape1
apurinic endonuclease 1
- Pol β
DNA polymerase β
- BSA
bovine serum albumin
- NCP
nucleosome core particle
- PAGE
polyacrylamide gel electrophoresis
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Schematics describing the ligation of oligonucleotides to create the nucleosomal DNA and representative plots of previously unreported free DNA cleavage as a function of time (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Radhakrishnan SK, Jette N, Lees-Miller SP. Nonhomologous end joining: Emerging themes and unanswered questions. DNA Repair. 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 3.Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: Building up an integrated picture of the dynamic dsb repair super complex, one component and interaction at a time. DNA Repair. 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srivastava M, Raghavan SC. DNA double-strand break repair inhibitors as cancer therapeutics. Chem. Biol. 2015;22:17–29. doi: 10.1016/j.chembiol.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, Natarajan R, Kelkar M, De A, Choudhary B, Raghavan SC. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 6.Colis LC, Woo CM, Hegan DC, Li Z, Glazer PM, Herzon SB. The cytotoxicity of (−)-lomaiviticin a arises from induction of double-strand breaks in DNA. Nat. Chem. 2014;6:504–510. doi: 10.1038/nchem.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xi Z, Goldberg IH. DNA-damaging enediyne compounds. In: Kool ET, editor. Comprehensive natural products chemistry. Amsterdam: Elsevier; 1999. pp. 553–592. [Google Scholar]
- 8.Kennedy DR, Ju J, Shen B, Beerman TA. Designer enediynes generate DNA breaks, interstrand cross-links, or both, with concomitant changes in the regulation of DNA damage responses. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17632–17637. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sage E, Harrison L. Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bignon E, Gattuso H, Morell C, Dehez F, Georgakilas AG, Monari A, Dumont E. Correlation of bistranded clustered abasic DNA lesion processing with structural and dynamic DNA helix distortion. Nucleic Acids Res. 2016;44:8588–8599. doi: 10.1093/nar/gkw773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh V, Kumari B, Das P. Repair efficiency of clustered abasic sites by Ape1 in nucleosome core particles is sequence and position dependent. RSC Adv. 2015;5:23691–23698. [Google Scholar]
- 12.David-Cordonnier MH, Cunniffe SMT, Hickson ID, O’Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 13.Georgakilas AG, Bennett PV, Wilson DM, Sutherland BM. Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoietic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomax ME, Cunniffe S, O’Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 15.Georgakilas AG, Bennett PV, Sutherland BM. High efficiency detection of bi-stranded abasic clusters in g-irradiated DNA by putrescine. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh SK, Wang M, Staudt C, Iliakis G. Post-irradiation chemical processing of DNA damage generates double-strand breaks in cells already engaged in repair. Nucleic Acids Res. 2011;39:8416–8429. doi: 10.1093/nar/gkr463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asaithamby A, Hu B, Chen DJ. Unrepaired clustered DNA lesions induce chromosome breakage in human cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8293–8298. doi: 10.1073/pnas.1016045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiyama H, Fujiwara T, Ura A, Tashiro T, Yamamoto K, Kawanishi S, Saito I. Chemistry of thermal degradation of abasic sites in DNA. Mechanistic investigation on thermal DNA strand cleavage of alkylated DNA. Chem. Res. Toxicol. 1994;7:673–683. doi: 10.1021/tx00041a013. [DOI] [PubMed] [Google Scholar]
- 21.Roupioz Y, Lhomme J, Kotera M. Chemistry of the 2-deoxyribonolactone lesion in oligonucleotides: Cleavage kinetics and products analysis. J. Am. Chem. Soc. 2002;124:9129–9135. doi: 10.1021/ja025688p. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Sheppard TL. Half-life and DNA strand scission products of 2-deoxyribonolactone oxidative DNA damage lesions. Chem. Res. Toxicol. 2004;17:197–207. doi: 10.1021/tx034197v. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Stubbe J. Synthesis and characterization of oligonucleotides containing a 4′-keto abasic site. Biochemistry. 2004;43:5278–5286. doi: 10.1021/bi0495376. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Gil JM, Greenberg MM. Synthesis and characterization of oligonucleotides containing the C4′-oxidized abasic site produced by bleomycin and other DNA damaging agents. Angew. Chem., Int. Ed. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- 25.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou C, Greenberg MM. Histone-catalyzed cleavage of nucleosomal DNA containing 2-deoxyribonolactone. J. Am. Chem. Soc. 2012;134:8090–8093. doi: 10.1021/ja302993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou C, Sczepanski JT, Greenberg MM. Histone modification via rapid cleavage of C4′-oxidized abasic sites in nucleosome core particles. J. Am. Chem. Soc. 2013;135:5274–5277. doi: 10.1021/ja400915w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng L, Greenberg MM. Rapid histone-catalyzed DNA lesion excision and accompanying protein modification in nucleosomes and nucleosome core particles. J. Am. Chem. Soc. 2015;137:11022–11031. doi: 10.1021/jacs.5b05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett RAO, Swerdlow PS, Povirk LF. Spontaneous cleavage of bleomycin-induced abasic sites in chromatin and their mutagenicity in mammalian shuttle vectors. Biochemistry. 1993;32:3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- 30.Zhou C, Sczepanski JT, Greenberg MM. Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles. J. Am. Chem. Soc. 2012;134:16734–16741. doi: 10.1021/ja306858m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zálešák J, Constant J-F, Jourdan M. Nuclear magnetic resonance solution structure of DNA featuring clustered 2′-deoxyribonolactone and 8-oxoguanine lesions. Biochemistry. 2016;55:3899–3906. doi: 10.1021/acs.biochem.6b00396. [DOI] [PubMed] [Google Scholar]
- 32.Cunniffe S, O’Neill P, Greenberg MM, Lomax ME. Reduced repair capacity of a DNA clustered damage site comprised of 8-oxo-7,8-dihydro-2′-deoxyguanosine and 2-deoxyribonolactone results in an increased mutagenic potential of these lesions. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2014;762:32–39. doi: 10.1016/j.mrfmmm.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eccles LJ, Lomax ME, O’Neill P. Hierarchy of lesion processing governs the repair, double-strand break formation and mutability of three-lesion clustered DNA damage. Nucleic Acids Res. 2010;38:1123–1134. doi: 10.1093/nar/gkp1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milligan JR, Ng JYY, Wu CCL, Aguilera JA, Fahey RC, Ward JF. DNA repair by thiols in air shows two radicals make a double-strand break. Radiat. Res. 1995;143:273–280. [PubMed] [Google Scholar]
- 35.Taverna Porro ML, Greenberg MM. DNA double strand cleavage via interstrand hydrogen atom abstraction. J. Am. Chem. Soc. 2013;135:16368–16371. doi: 10.1021/ja409513q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taverna Porro ML, Greenberg MM. Double-strand breaks from a radical commonly produced by DNA-damaging agents. Chem. Res. Toxicol. 2015;28:810–816. doi: 10.1021/acs.chemrestox.5b00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mourgues S, Lomax ME, O’Neill P. Base excision repair processing of abasic site/single-strand break lesions within clustered damage sites associated with XRCC1 deficiency. Nucleic Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shikazono N, Pearson C, O’Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dizdaroglu M. Oxidatively induced DNA damage and its repair in cancer. Mutat. Res., Rev. Mutat. Res. 2015;763:212–245. doi: 10.1016/j.mrrev.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Povirk LF, Houlgrave CW. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: Specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry. 1988;27:3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- 41.Povirk LF, Han YH, Steighner RJ. Structure of bleomycin-induced DNA double-strand breaks: Predominance of blunt ends and single-base 5′ extensions. Biochemistry. 1989;28:5808–5814. doi: 10.1021/bi00440a016. [DOI] [PubMed] [Google Scholar]
- 42.Steighner RJ, Povirk LF. Bleomycin-induced DNA lesions at mutational hot spots: Implications for the mechanism of double-strand cleavage. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8350–8354. doi: 10.1073/pnas.87.21.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Povirk LF, Goldberg IH. Base substitution mutations induced in the Ci gene of lambda phage by neocarzinostatin chromophore: Correlation with depyrimidination hotspots at the sequence agc. Nucleic Acids Res. 1986;14:1417–1426. doi: 10.1093/nar/14.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2003;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 45.Kotera M, Roupioz Y, Defrancq E, Bourdat A-G, Garcia J, Coulombeau C, Lhomme J. The 7-nitroindole nucleoside as a photochemical precursor of 2′-deoxyribonolactone: Access to DNA fragments containing this oxidative abasic lesion. Chem. - Eur. J. 2000;6:4163–4169. doi: 10.1002/1521-3765(20001117)6:22<4163::aid-chem4163>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 46.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 47.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the ’601’ strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 48.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 49.Davey G, Wu B, Dong Y, Surana U, Davey CA. DNA stretching in the nucleosome facilitates alkylation by an intercalating antitumor agent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon K, Hobbs CA, Walter AE, Turner DH. Effect of a 5′-phosphate on the stability of triple helix. Nucleic Acids Res. 1993;21:601–606. doi: 10.1093/nar/21.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zálešák J, Lourdin M, Krejčí L, Constant JF, Jourdan M. Structure and dynamics of DNA duplexes containing acluster of mutagenic 8-oxoguanine and abasic site lesions. J. Mol. Biol. 2014;426:1524–1538. doi: 10.1016/j.jmb.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 53.Hazel RD, Tian K, de los Santos C. Nmr solution structures of bistranded abasic site lesions in DNA. Biochemistry. 2008;47:11909–11919. doi: 10.1021/bi800950t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weng L, Zhou C, Greenberg MM. Probing interactions between lysine residues in histone tails and nucleosomal DNA via product and kinetic analysis. ACS Chem. Biol. 2015;10:622–630. doi: 10.1021/cb500737y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sczepanski JT, Zhou C, Greenberg MM. Nucleosome core particle-catalyzed strand scission at abasic sites. Biochemistry. 2013;52:2157–2164. doi: 10.1021/bi3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.