The abundance of drought-regulated miRNAs is affected by grafting in grapevine.
Abstract
Grapevine (Vitis vinifera) is routinely grafted, and rootstocks inducing drought tolerance represent a source for adapting vineyards to climate change in temperate areas. Our goal was to investigate drought stress effects on microRNA (miRNA) abundance in a drought-resistant grapevine rootstock, M4 (Vitis vinifera × Vitis berlandieri), compared with a commercial cultivar, Cabernet Sauvignon, using their autografts and reciprocal grafts. RNA extracted from roots and leaves of droughted and irrigated plants of different graft combinations was used to prepare cDNA libraries for small RNA sequencing and to analyze miRNAs by quantitative real-time polymerase chain reaction (RT-qPCR). Measurements of leaf water potential, leaf gas exchange, and root hydraulic conductance attested that, under irrigation, M4 reduced water loss in comparison with cultivar Cabernet Sauvignon mostly through nonhydraulic, root-specific mechanisms. Under drought, stomatal conductance decreased at similar levels in the two genotypes. Small RNA sequencing allowed the identification of 70 conserved miRNAs and the prediction of 28 novel miRNAs. Different accumulation trends of miRNAs, observed upon drought and in different genotypes and organs, were confirmed by RT-qPCR. Corresponding target transcripts, predicted in silico and validated by RT-qPCR, often showed opposite expression profiles than the related miRNAs. Drought effects on miRNA abundance differed between the two genotypes. Furthermore, the concentration of drought-responsive miRNAs in each genotype was affected by reciprocal grafting, suggesting either the movement of signals inducing miRNA expression in the graft partner or, possibly, miRNA transport between scion and rootstock. These results open new perspectives in the selection of rootstocks for improving grapevine adaptation to drought.
The silencing phenomena mediated by small noncoding RNAs finely tune physiological and biochemical processes that tightly regulate plant development (Li and Zhang, 2016) and adaptation to the surrounding environment (Kruszka et al., 2012; Sunkar et al., 2012). Among small noncoding RNAs, microRNAs (miRNAs) constitute the most studied group of endogenous posttranscriptional silencing effectors, which serve as guides for either the cleavage or translational inhibition of complementary target transcripts (Voinnet, 2009; Rogers and Chen, 2013). Plant miRNAs are single-stranded RNA molecules of approximately 21 nucleotides in length generated by DICER-LIKE1 (DCL1) enzymes from stem-loop precursors encoded by endogenous MIR genes (Nozawa et al., 2012). In recent years, increasing experimental evidence has suggested a role for plant miRNAs as crucial mediators in the regulation of molecular signaling cascades upon exposure to abiotic stresses, particularly drought and salinity (for review, see Ding et al., 2013; Ferdous et al., 2015; Zhang, 2015). Indeed, in many herbaceous plants, such as Arabidopsis (Arabidopsis thaliana; Liu et al., 2008; Song et al., 2013; Xu et al., 2014), tobacco (Nicotiana tabacum; Frazier et al., 2011), tomato (Lycopersicon esculentum; Candar-Cakir et al., 2016), Medicago truncatula (Trindade et al., 2010; Wang et al., 2011), cotton (Gossypium hirsutum; Wang et al., 2013), soybean (Glycine max; Kulcheski et al., 2011), and various cereals (for review, see Budak et al., 2015), as well as in woody species, such as poplar (Populus spp.; Ren et al., 2012; Li et al., 2013a; Shuai et al., 2013) and fruit trees (Solofoharivelo et al., 2014), conserved and novel miRNA families have been shown to dynamically respond to abiotic stress. Although these findings contributed greatly to identify a large source of stress-related miRNAs, further efforts are needed to advance current understanding of the interrelated molecular pathways controlling miRNA biogenesis and action (Borges and Martienssen, 2015; Reis et al., 2015). Progress in this direction was made recently by the identification of stress-responsive regulatory sequences located within the MIR gene promoters (Devi et al., 2013) and by the discovery of salt stress-responsive miR-SSR markers (Mondal and Ganie, 2014).
Grapevine (Vitis vinifera) is a major fruit crop worldwide, and it features a relatively high tolerance to drought, determined by a still poorly explored array of morphological, biochemical, and molecular mechanisms (Cramer et al., 2007, 2013; Perrone et al., 2012; Ferrandino and Lovisolo, 2014; Corso et al., 2015). Previous studies identified conserved and novel miRNAs in grapevine (Carra et al., 2009; Mica et al., 2010; Pantaleo et al., 2010, 2016; Wang et al., 2012), and a grapevine miRNA expression atlas was published recently (Belli Kullan et al., 2015). However, no studies specifically addressing the identification of drought stress (DS)-responsive miRNAs have been reported in Vitis spp. Further investigation of such miRNAs could indeed offer important clues to their specific function within the context of regulatory networks promoting grapevine adaptation to stressful environments.
Grafting is an ancient agronomical technique widely used in horticulture for diverse purposes, such as vegetative propagation, reduction of juvenility, and tolerance to soil pathogens. European grapevines are routinely grafted on interspecific hybrid rootstocks in order to control infestation by phylloxera (Daktulosphaira vitifoliae; Mudge et al., 2009). In parallel, rootstocks affect scion growth vigor and resistance to abiotic stresses (Berdeja et al., 2015; Lovisolo et al., 2016; Lavoie-Lamoureux et al., 2017). Specific rootstocks induce tolerance to DS (Carbonneau, 1985; Soar et al., 2006; Koundouras et al., 2008; Tramontini et al., 2013), and it was shown that particular genetic regions in their genome are linked to the induction of stress tolerance (Marguerit et al., 2012). Thus, rootstock management is considered a promising tool to enhance the resilience of grapevine to water scarcity (Berdeja et al., 2015). A recent transcriptomic study demonstrated that grafting can determine stock-specific transcript concentration changes in the grapevine scion (Corso et al., 2015), and this could depend at least in part on modifications in the concentration of miRNAs, as shown recently in grafts of cucumber (Cucumis sativus) and pumpkin (Cucurbita moschata; Li et al., 2014).
In this study, we provide new insights on the regulatory effect exerted by DS on the abundance of grapevine miRNAs. In particular, we identified, by the use of high-throughput next-generation sequencing technology and in silico analysis, novel putative miRNAs in roots and leaves of two grapevine genotypes, cv Cabernet Sauvignon (CS), a widely known commercial variety, and the recently selected, water stress-tolerant Vitis hybrid M4. We further quantified miRNA expression levels under DS by applying a well-established stem-looped quantitative real-time PCR (RT-qPCR) approach and focusing on a group of selected conserved and novel miRNAs. These analyses were performed in autografted as well as in heterografted plants in order to unravel changes in miRNA accumulation induced by grafting. We show that several conserved and novel miRNAs, potentially influencing biological processes that affect the whole-plant capacity to cope with DS, are modulated by the imposed treatment. Moreover, we report that heterografting alters miRNA abundance in the graft partners, possibly due to the movement of stress signals between them.
RESULTS
Water Status and Gas Exchange in Autografted and Heterografted Grapevines
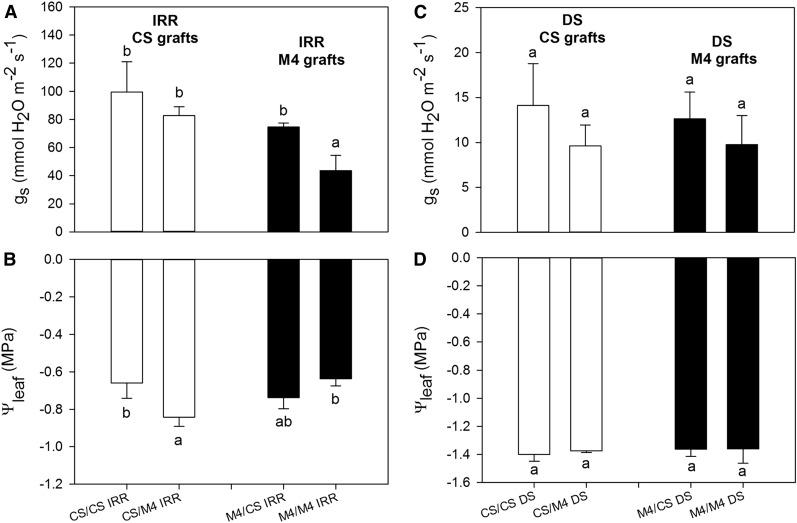
In order to outline the physiological behavior of autografted CS and M4 plants under DS, we withdrew irrigation and followed dynamic changes in leaf water potential (Ψleaf) and stomatal conductance (gs; Fig. 1). Upon irrigation, gs rates were significantly lower in M4/M4 vines than in CS/CS vines (Fig. 1A). Nevertheless, when severe DS occurred (about 10 d after the treatment imposition), gs decreased strongly up to 1 order of magnitude in both the autografts (Fig. 1C). Accordingly, Ψleaf was significantly lower in droughted plants than in irrigated plants (Fig. 1, B and D).
Figure 1.
Measurements of gs (A and C) and Ψleaf (B and D) in autografted and heterografted plants of CS and M4 genotypes in irrigated conditions (IRR; A and B) and after 10 d of DS (C and D). Lowercase letters above and below the bars denote significant differences (P < 0.05) attested using Tukey’s honestly significant difference (HSD) test. Error bars represent se (n = 10).
Physiological responses to drought also were investigated on the reciprocal heterografts of the two genotypes (CS/M4 and M4/CS), with the final goal to obtain more information on the relative role of either root or shoot in reducing gs of M4 autografted plants. Irrigated M4/CS and CS/M4 vines showed comparable gs values, slightly lower than CS/CS vines but significantly higher than those measured in M4/M4 plants (Fig. 1A). Under irrigation, Ψleaf was significantly more negative in heterografts than in autografts, and in particular, the lowest values were observed for the CS/M4 combination (Fig. 1B). In heterografted plants, drought treatment strongly reduced gs (Fig. 1C) together with an evident decrease in Ψleaf values, which were close to −1.4 MPa on average (Fig. 1D). The observed trends in gs were well mirrored by leaf transpiration rates measured on the tested plants (data not shown).
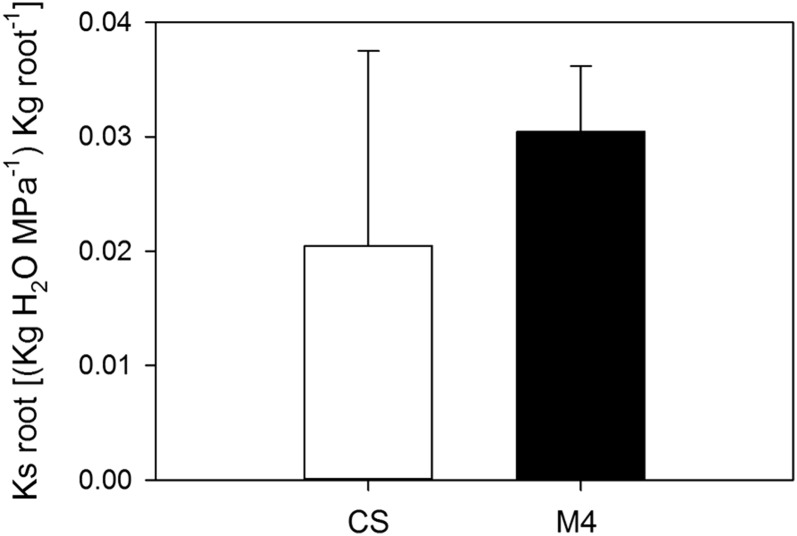
As the results showed that autografted M4 plants lost less water than CS autografts under irrigated conditions, we compared the specific root hydraulic conductance (root ks) of M4 with that of CS upon well-watered conditions. The results indicated that root ks of M4 was higher, although not significantly, than in CS (Fig. 2), suggesting that factors other than hydraulic signals should contribute to control gs, and hence transpiration, of this genotype.
Figure 2.
Specific hydraulic conductance (i.e. root kh), with reference to root dry weight (Ks), in M4 and CS roots under irrigated control conditions. Error bars represent se (n = 10). No significant differences were observed in the pairwise comparison when performing Student’s t test (P < 0.05).
miRNA Sequencing and Identification
In order to identify novel candidate miRNAs affected by DS, we prepared eight small RNA libraries from pooled low molecular weight (LMW) RNA samples extracted from roots and leaves of CS and M4 autografted plants, either irrigated or exposed to DS treatment. When subjected to SOLiD deep sequencing, the libraries yielded more than 180 million raw reads in total. After removing low-quality sequences, adapters, small sequences (fewer than 17 nucleotides), noncoding RNAs, and other contaminants, an average of more than 7 million nonredundant reads, with length ranging from 17 to 25 nucleotides, was obtained for each library and aligned to the Vitis genome (Vitis 12X database version V1 [12X V1]; http://genomes.cribi.unipd.it/grape/). The sequences perfectly matching at least one genome locus were then analyzed to either retrieve conserved/known miRNAs, by further mapping these sequences on miRBase (version 21; http://www.mirbase.org/), or identify novel candidate miRNAs using two dedicated computational softwares (miRCat and miRDeep-P; for details, see “Materials and Methods”). A summary of small RNA sequencing analysis is reported for each library in Table I.
Table I. Statistics of small RNA sequencing data analysis.
Small RNA libraries were obtained from leaf (L) and root (R) samples of CS and M4 autografted vines upon DS or irrigated (IRR) conditions.
| Library Name | Total Raw Reads | Adapter Trimminga | Contaminant Filteringa | Noncoding RNA Filteringb | Unique Reads Matching the Grape Genomec |
|---|---|---|---|---|---|
| CS IRR L | 20,579,159 | 16,922,667 | 16,053,719 | 9,680,892 | 1,365,464 |
| CS DS L | 23,614,667 | 15,125,262 | 13,881,849 | 6,290,048 | 679,650 |
| CS IRR R | 16,015,312 | 13,289,525 | 11,884,302 | 6,197,784 | 247,795 |
| CS DS R | 13,677,360 | 8,040,667 | 7,188,734 | 2,985,063 | 67,588 |
| M4 IRR L | 27,596,925 | 21,264,151 | 19,223,470 | 7,904,996 | 759,658 |
| M4 DS L | 26,706,483 | 22,914,129 | 20,900,171 | 8,748,394 | 179,474 |
| M4 IRR R | 27,028,350 | 21,354,251 | 17,907,958 | 9,592,832 | 682,325 |
| M4 DS R | 29,588,911 | 21,875,311 | 18,629,316 | 11,112,533 | 1,518,832 |
Sequences obtained after quality, adapter, and size (read lengths of less than 17 nucleotides) filtering. bSequences obtained after filtering noncoding RNAs (e.g. tRNA, rRNA, snoRNA, and snRNA). cTotal of unique sequences from conserved and novel miRNAs matching the grape genome (12X V1 GGDB CRIBI) and/or miRBase (version 21).
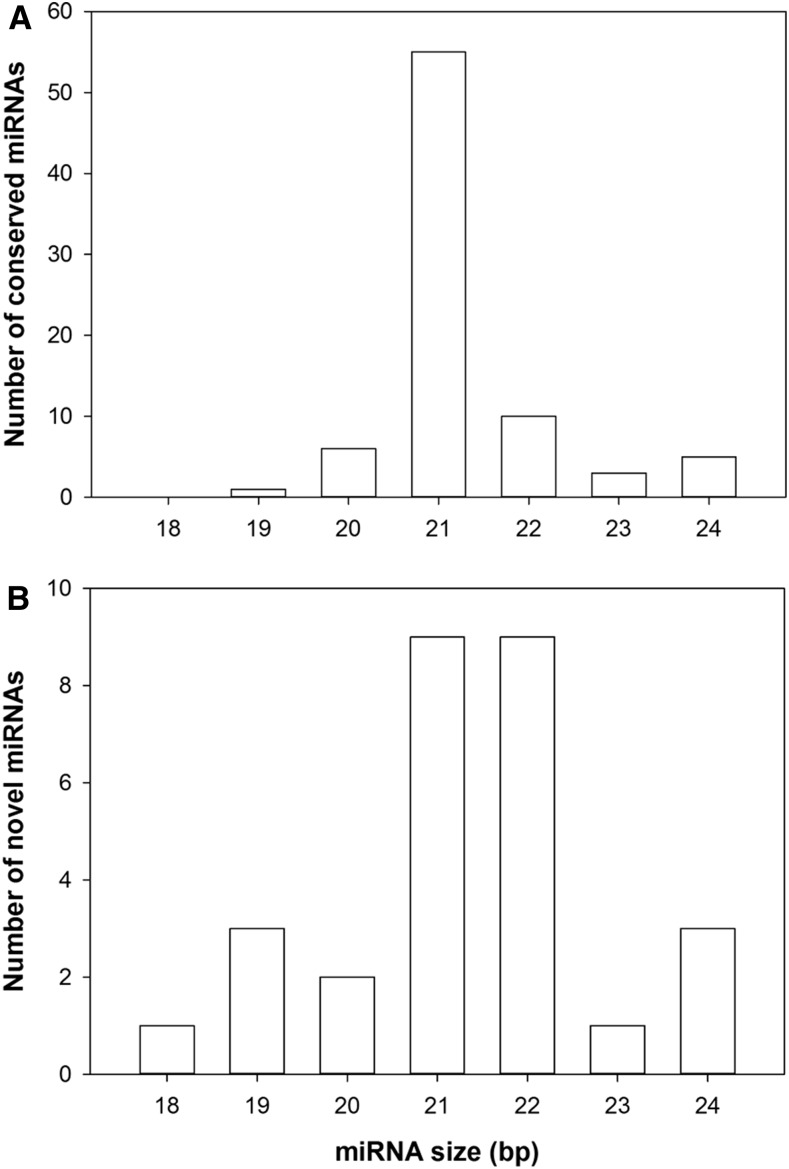
Sequencing elaboration allowed the identification of 635 miRNAs, and filtering above a threshold value (100 or more unique normalized reads) reduced the number to 98. Among these, 70 were classified as conserved/known miRNAs belonging to 42 families (miRBase annotation version 21; Supplemental Table S3), whereas 28 were predicted as putative novel miRNAs (Supplemental Table S4). The majority (70%) of conserved miRNA sequences fell within the 21-nucleotide category (Fig. 3A), one of the most abundant size classes for known grapevine DCL1-derived products (Pantaleo et al., 2010; Wang et al., 2012; Belli Kullan et al., 2015). Among novel candidate miRNAs, two major (each above 30%) size categories were observed at 21 and 22 nucleotides (Fig. 3B).
Figure 3.
Size distribution of conserved (A) and novel (B) miRNAs identified by SOLiD sequencing analysis.
Drought-Induced Changes of Conserved miRNA Accumulation in Autografted Plants
Based on results from the sequencing experiment (Supplemental Table S3), the concentration of a group of conserved or grape-specific known miRNAs was analyzed by RT-qPCR in autografted plants. The conserved miR156, miR159, miR393, and miR396 and the grape-specific miR3624 (Carra et al., 2009) were chosen because they were highly expressed in our sequencing data set (miR159, miR396, and miR3624; Supplemental Table S3) and/or were reported previously to play a key role in the regulation of abiotic stress responses in plants (miR156, miR159, miR393, and miR396; Covarrubias and Reyes, 2010; Ferdous et al., 2015, and refs. therein; Zhang, 2015).
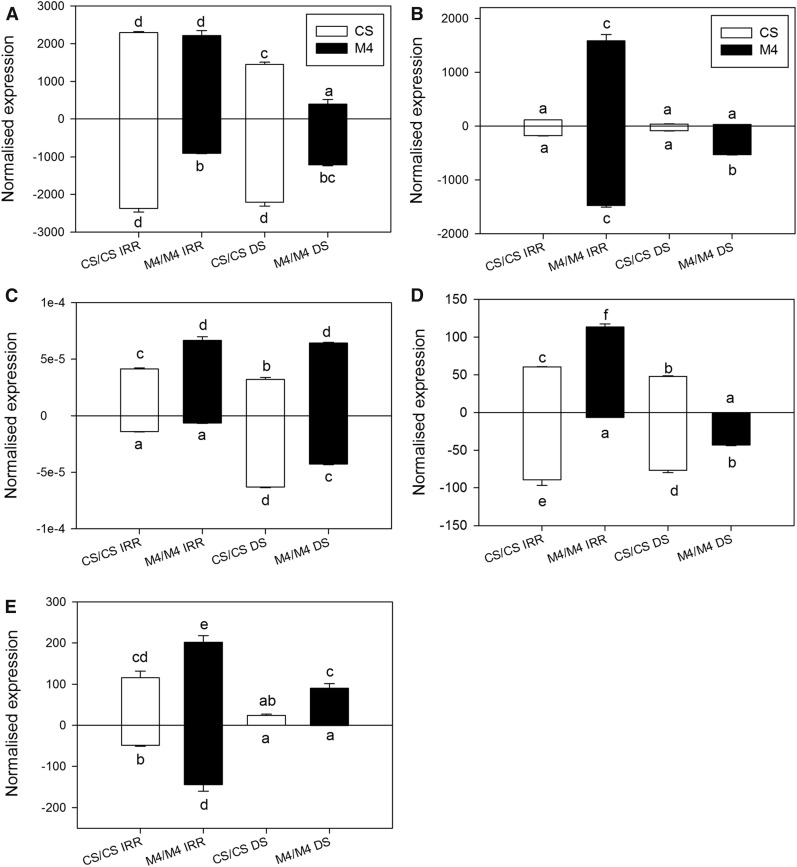
In control plants, these miRNAs were detected at comparable concentrations in both genotypes, with the exception of miR159, which was expressed at a lower level in CS compared with M4, independently of tissue type (Fig. 4B). DS induced significant expression changes in these miRNAs, and in some cases, accumulation trends were common for the two genotypes (Fig. 3). For instance, miR156 abundance was lowered significantly by drought in the leaves of both genotypes, whereas in roots, it was not affected significantly (Fig. 4A). Conversely, miR393 increased in DS root samples of both CS and M4, while in DS leaves, it was slightly less abundant (Fig. 4C). A negative effect of DS also was evident for miR3624, particularly in roots, where this miRNA was either much less accumulated (M4) or almost absent (CS; Fig. 4E).
Figure 4.
Expression changes occurring in a group of conserved miRNAs. RT-qPCR analyses are shown for miR156 (A), miR159 (B), miR393 (C), miR396 (D), and miR3624 (E) on leaf (top bars) and root (bottom bars) samples obtained from CS and M4 autografted vines upon irrigated (IRR) and DS conditions. The small nuclear RNA U6 (VvsnRU6) was used as the endogenous control gene for the normalization procedure. Lowercase letters above and below the bars denote significant differences (P < 0.05) attested using Tukey’s HSD test. Error bars represent se (n = 3).
On the contrary, two miRNAs showed distinct expression patterns in CS and M4 genotypes. miR159 abundance did not change in CS samples independently of the treatment and tissue type, whereas in M4 plants, this miRNA was much less abundant in DS tissues (Fig. 4B). miR396 was accumulated upon drought in M4 roots, while in M4 leaves and in CS vines, the treatment significantly lowered its levels (Fig. 4D).
Quantification of Target Transcripts of Conserved miRNAs
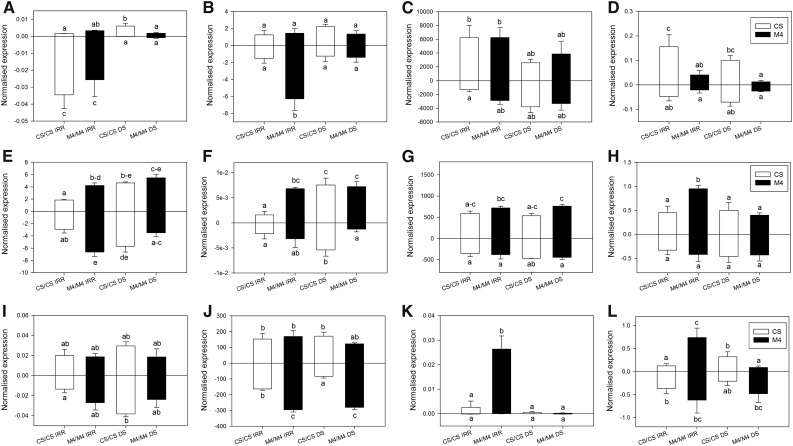
In order to characterize the downstream effects of transcriptional changes occurring in conserved miRNA accumulation upon stress, we quantified selected targets by RT-qPCR (for details, see Supplemental Table S6). In many cases, as expected, the expression of the miRNA and its target gene followed opposite trends, although this was not always the rule. In all conditions, the abundance of miR156 was inversely correlated to the zinc finger protein CONSTANS5-like target mRNA (VIT_04s0008g07340; Supplemental Fig. S1A). In the case of miR159, accumulation of the target transcripts VIT_19s0090g00590, encoding the putative grape ortholog of Arabidopsis MYB101, VIT_09s0002g08370, encoding TOPLESS-RELATED PROTEIN4-like, and VIT_00s0287g00040, encoding GATA TRANSCRIPTION FACTOR26-like, also followed divergent patterns from the miRNA (Supplemental Fig. S1, B–D). The miR393 target mRNA, encoding the auxin-responsive factor TIR1-like (VIT_14s0068g01330), exhibited a divergent pattern of expression in leaves (Supplemental Fig. S1E), while the analysis of miR396 target transcript, encoding a growth-regulating factor (VIT_00s0494g00010), displayed an expression profile opposite to that of miR396 in M4 but not in CS (Supplemental Fig. S1F). Besides the genes targeted by these four conserved miRNAs, the target transcript of the grape-specific miR3624 (Carra et al., 2009) was analyzed. The expression profile of this gene, encoding a metal ion-binding protein (VIT_00s0194g00340), followed an opposite trend when compared with the cognate miRNA (Supplemental Fig. S1G).
Changes of Novel miRNA Abundance in Autografted Plants
We selected a group of novel candidate miRNAs showing important changes in the number of sequencing reads (Supplemental Table S4) upon water stress application (n_22, n_47, n_55, n_191, n_231, n_312, n_327, n_337, n_346, n_479, n_520, and n_622), and we analyzed their expression in autografted plants by RT-qPCR. The relative abundance of these putative miRNAs was variable: n_55, n_327, and n_479 were the most abundant (more than 102 times higher than the small nuclear RNA U6), followed by n_47, n_231, n_337, and n_622, while n_22, n_191, n_312, n_346, and n_520 were the least concentrated (Fig. 5).
Figure 5.
Expression changes occurring in a group of selected novel candidate miRNAs. RT-qPCR analyses are shown for n_22 (A), n_47 (B), n_55 (C), n_191 (D), n_231 (E), n_312 (F), n_327 (G), n_337 (H), n_346 (I), n_479 (J), n_520 (K), and n_622 (L) on leaf (top bars) and root (bottom bars) samples collected from CS and M4 autografted vines upon irrigated (IRR) and DS conditions. VvsnRU6 was used as the endogenous control gene for the normalization procedure. Lowercase letters above and below the bars denote significant differences (P < 0.05) attested using Tukey’s HSD test. Error bars represent se (n = 3).
In control conditions, the relative concentration of the analyzed miRNAs was modulated by genotype and tissue type. Expression of n_191 was significantly greater in CS than in the corresponding M4 tissues (Fig. 5D). Tissue specificity independent of genotype was observed in n_22, which was significantly more abundant in roots (Fig. 5A). A group of putative miRNAs showed tissue specificity in one genotype only. In CS, expression in roots was lower for n_55 and n_191 (Fig. 5, C and D) and higher for n_622 (Fig. 4I). In M4, n_337 and n_520 were less abundant in roots (Fig. 5, G, H, and K), whereas n_47, n_231, and n_479 were more concentrated (Fig. 5, B, E, and L).
In a few cases, water stress activated the expression of putative miRNAs: n_231 was positively induced by drought, with the exception of M4 root, where it was significantly repressed (Fig. 5E). Upon stress, three miRNAs underwent significant overexpression in CS/CS plants: n_312 in leaves and roots (Fig. 5F), n_622 in leaves (Fig. 5L), and n_346 in roots (Fig. 5I).
The abundance of a larger group of putative miRNAs decreased upon DS. The concentration of the root-specific n_22 candidate miRNA decreased to very low levels upon drought in CS/CS and M4/M4 (Fig. 5A). On the contrary, n_622 showed a genotype-dependent stress response in leaves, and it increased significantly in CS, whereas it was much less accumulated in M4 (Fig. 5L). The stress treatment significantly decreased the abundance of n_479 in CS roots (Fig. 5J), while the abundance of n_337 and n_520 was significantly lower upon stress in M4 leaves (Fig. 5, H and K).
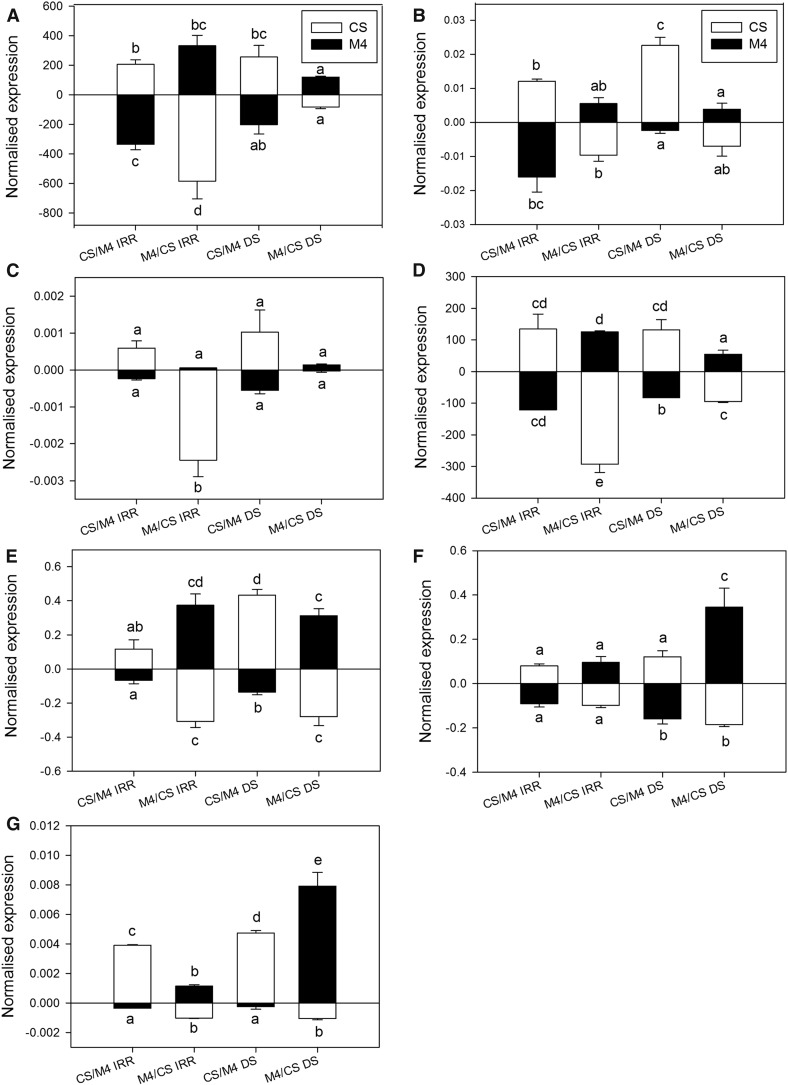
Changes of Novel miRNA Abundance in Heterografted Plants
The expression profiles of seven candidate miRNAs (n_55, n_191, n_312, n_327, n_337, n_346, and n_520) were investigated further in the reciprocal scion/stock combinations CS/M4 and M4/CS (Fig. 6).
Figure 6.
Expression changes of novel candidate miRNAs in heterografted plants. Expression profiles are shown for n_55 (A), n_191 (B), n_312 (C), n_327 (D), n_337 (E), n_346 (F), and n_520 (G) in leaf (top bars) and root (bottom bars) samples collected from CS and M4 heterografted vines (CS/M4 and M4/CS) upon irrigated (IRR) and DS conditions. VvsnRU6 was used as the endogenous control gene for the normalization procedure. Lowercase letters above and below the bars denote significant differences (P < 0.05) attested using Tukey’s HSD test. Error bars represent se (n = 3).
In control conditions, the CS-specific (in autografted plants) n_191 was either equally distributed (M4/CS) or more abundant in M4 tissues (M4/CS; Fig. 6B). Also, the tissue specificity observed for n_55, n_191, n_337, and n_520 in autografted plants was lost in both heterografted combinations (Fig. 6, A, B, E, and G). On the contrary n_312, equally present in both genotypes and tissues in the autografted combinations, was nearly absent in M4 tissues in the reciprocal grafts (Fig. 6C).
In a few cases, the effects of drought on miRNA concentration observed in autografts were detected in the corresponding genotypes and tissues of heterografts, as in the case of the up-regulation of n_346 in CS roots in both autografts and heterografts (Fig. 6F). In two cases (n_312 in CS roots and n_520 in M4 leaves), the effects of drought observed in autografted plants were reversed in the same tissues of heterografts (Fig. 6, C and G). Nevertheless, in most cases, significant up- or down-regulation trends in heterografted plants were observed for putative miRNAs whose expression was not affected by drought in autografted plants. As an example, n_337 was induced by drought treatment in leaves and roots of CS/M4 heterografts (Fig. 6E), an effect not observed previously in autografts (Fig. 5H). Another interesting case was n_346, for which a significant stress-induced up-regulation was observed in almost all tissues collected from heterografts (Fig. 6F), different from what was observed for autografted vines (Fig. 5K). Conversely, drought significantly decreased the expression of n_55 in CS roots and in M4 roots and leaves collected from heterografts (Fig. 6A), but not in the corresponding tissues of autografts (Fig. 5C). Significant effects of drought also were noticed for n_191 in both leaves and roots of CS/M4 grafts (Fig. 6B) and for n_312 exclusively in the root (CS) of the opposite graft combination (Fig. 6C), but not in the corresponding tissues of autografted vines (Fig. 5, D and F). Unlike what was described in autografted plants (Fig. 5G), drought negatively affected the expression of n_327 in heterografts, with particular evidence in the roots of both graft combinations (Fig. 6D).
In Silico Prediction of Novel miRNA Target Genes and Quantification of Related Expression Changes
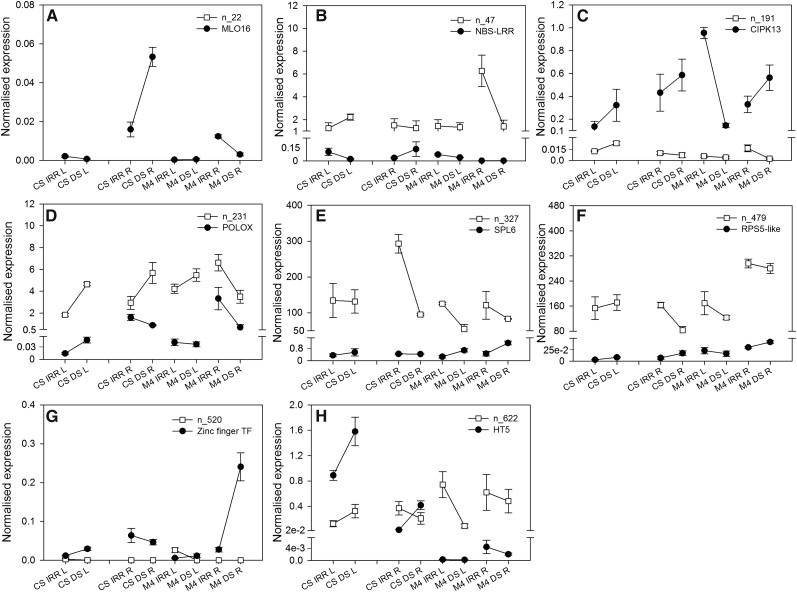
Target gene sequences of the putative novel miRNAs analyzed in this study were predicted in silico using two computational algorithms in parallel, psRNATarget and Target-align. Some of the miRNAs targeted more than one transcript (i.e. n_622, n_479, and n_327). For n_312 and n_337, no targets could be predicted. The Gene Ontology annotation showed that target transcripts encoded proteins, enzymes, or transcription factors involved in a broad range of biological processes, among which stress defense and stress signaling response mechanisms were the most common hits (Supplemental Table S5).
The transcriptional changes of six of these predicted target genes, chosen among the most interesting target genes in terms of possible role in the context of stress response regulation (n_22, n_47, n_191, n_231, n_312, n_479, n_520, and n_622), were assessed by RT-qPCR analysis (Fig. 7). Overall, target transcripts showed opposite expression trends when compared with those of the related novel miRNAs, supporting negative posttranscriptional regulation driven by the miRNA. This was the case in CS samples: for VIT_ 12s0035g0026 (Fig. 7A), encoding a calmodulin-binding protein (MLO16), target of n_22; for VIT_09s0096g00830, target of n_47, encoding a NBS-LRR defense protein (Fig. 7B); for transcripts of a polyphenol oxidase (VIT_00s0480g00100), target gene of n_231 (Fig. 7D); and for a squamosa promoter binding protein-like transcription factor gene (SPL6; VIT_01s0011g00130), targeted by n_327 (Fig. 7E). The same pattern was observed in M4 samples for the CBL interacting binding protein kinase gene CIPK13 (VIT_13s0067g0248), target of n_191 (Fig. 7C); or in both genotypes, as in the case of a zinc finger transcription factor gene (VIT_06s0004g06440), targeted by n_520 (Fig. 7G). Transcriptional regulation of target genes occurred only in specific tissues. This happened for VIT_09s0002g04950 (Fig. 7F), target gene of n_479, which encodes an RPS5-like defense protein belonging to the NBS-LRR superfamily: in leaves, its expression trend followed that of its putative cognate miRNA, whereas in the root of both genotypes, expression changes were opposite (Fig. 7F). A similar result was observed in CS root upon both control and stress conditions for transcripts of the hexose transporter HT5 (VIT_05s0020g03140), targeted by n_622 (Fig. 7H).
Figure 7.
Expression patterns of novel miRNA target transcripts. RT-qPCR profiles are shown for n_22 (A), n_47 (B), n_231 (C), n_327 (D), n_191 (E), n_520 (F), n_479 (G), and n_622 (H) target genes in correlation with each related novel miRNA, measured in leaf (L) and root (R) samples collected from either CS and M4 autografted (A–C and F–H) or heterografted (D and E) vines upon irrigated (IRR) and DS conditions. The ubiquitin (VvUBI) and actin (VvACT1) genes were used as endogenous controls for the normalization procedure of target transcripts, while VvsnRU6 was the endogenous control for miRNA expression analysis. Error bars represent se (n = 3).
DISCUSSION
Conserved and Novel Grapevine miRNAs Respond to DS in the Two Genotypes
In order to separate the shoot-controlled physiological processes from those regulated at the root level, we investigated the effects of stress on autografts and heterografts of two grapevine genotypes, CS and M4. Under irrigated conditions, gs was significantly lower in M4 than in CS, and both genotypes significantly decreased gs with DS. Heterografted plants also showed significant decreases in gs with DS, and there was no significant difference in the response with rootstock.
The root ks in M4 was higher but not significantly different from that in CS, suggesting that the lower gs induced by M4 may not rely on hydraulic signals but rather on other factors, such as signaling molecules. We reported previously that grapevine rootstocks control gs when water potential decreases (Tramontini et al., 2013) and that root-originated abscisic acid (ABA) limits gs during water stress and upon rehydration after drought events (Lovisolo et al., 2008; Tramontini et al., 2014).
During the last few years, an emerging number of drought-related miRNAs have been reported in several crop plants, such as barley (Hordeum vulgare; Hackenberg et al., 2015), rice (Oryza sativa; Zhou et al., 2010; Barrera-Figueroa et al., 2012), cotton (Xie et al., 2015), potato (Solanum tuberosum; Zhang et al., 2014), maize (Zea mays; Li et al., 2013b), tomato (Candar-Cakir et al., 2016), cowpea (Vigna unguiculata; Barrera-Figueroa et al., 2011), peach (Prunus persica; Eldem et al., 2012), and poplar (Li et al., 2011; Ren et al., 2012). Recently, Sun et al. (2015) described changes in grape miRNA abundance induced by cold stress and Pantaleo et al. (2016) characterized novel miRNAs in virus-infected grapevine contemporaneously subjected to water stress. However, to the best of our knowledge, this is the first time that the effects of drought alone were analyzed on grape miRNA populations.
As the accumulation of miRNAs can vary significantly among diverse organs or tissues (Ferdous et al., 2015), we investigated miRNA expression changes in roots and leaves of two genotypes. Our analysis of conserved miRNAs confirms their response to osmotic stress and variation among tissue types. The grape-specific miR3624 was inhibited by drought, while its target gene, encoding a metal ion-binding protein, increased greatly, in accordance with the known up-regulation of metallothionein-induced abiotic stress (Kim et al., 2014). miR393 positively responded to drought exclusively in the roots of the tested genotypes. This is consistent with previous work showing the root-specific up-regulation of this miRNA (Gao et al., 2016) and revealing its essential role in the adaptation of lateral root growth to DS (Chen et al., 2012). Additional evidence supporting the stress-mediated regulation of miR393 is the presence of specific drought-induced cis-elements in the promoter region of the related MIR gene (Devi et al., 2013). In Arabidopsis, miR393 is a key component of the auxin signaling cascade and tunes the expression of TIR1/AFB2 genes (Si-Ammour et al., 2011); this function also was confirmed in other plant species (Ferreira et al., 2012; Xia et al., 2012). In our study, however, the expression of the homologous grape target transcript, encoding the auxin-responsive factor TIR1-like, was inversely correlated to the abundance of miR393 only in leaves, implying that, in root, other stress-induced regulatory mechanisms could interfere with the miRNA inhibitory activity (Rogers and Chen, 2013).
miR156, miR159, and miR396 are abundant in Arabidopsis plants exposed to osmotic stress (Covarrubias and Reyes, 2010), while in our experiments, they are mostly down-regulated by drought. Data on the regulation of conserved miRNAs divergent from that observed in Arabidopsis have been reported before: upon water stress, a lower accumulation of miR156 and miR393 was observed in cotton (Xie et al., 2015) and in cowpea (Barrera-Figueroa et al., 2011), while the expression of miR396, another drought-induced miRNA in Arabidopsis (Liu et al., 2008), was inhibited in rice (Zhou et al., 2010). miR156 is one of the most highly conserved miRNAs in plants and is involved primarily in the control of floral transition phases by targeting members of the SPL transcription factor family, as demonstrated experimentally in Arabidopsis (Wu and Poethig, 2006) and tomato (Ferreira e Silva et al., 2014). Besides SPL genes, other species-specific targets were predicted in grapevine by degradome analysis (Pantaleo et al., 2010). Here, we showed that one of these predicted genes, encoding a zinc finger CONSTANS5-like protein, is induced by stress while miR156 decreases, supporting its status as a target of this miRNA. In our analysis, miR396 levels increased upon drought exclusively in M4 root, while GRL1-like1, a member of the growth-regulating factor family, already shown to be targeted by miR396 in Arabidopsis (Liu et al., 2009) and grapevine (Pantaleo et al., 2016), followed an opposite trend in the same genotype. The diverse expression profiles observed for miR396 and its target in M4 and CS tissues are in accordance with other reports on genotypes with different sensitivity to DS (Kulcheski et al., 2011; Candar-Cakir et al., 2016). Grape targets of miR159 (Pantaleo et al., 2010) include the GAMYB transcription factors MYB33, MYB65, and MYB101 (Achard et al., 2004), which take part in the signaling cascades induced by ABA accumulation in the presence of stress (Reyes and Chua, 2007). In our study, three putative miR159 targets, encoding a TOPLESS4-like protein, a GATA transcription factor, and the putative Vitis ortholog of AtMYB101, showed an opposite expression profile than the cognate miRNA, supporting the hypothesis that, in grapevine, miR159 may control additional molecular pathways than in Arabidopsis.
Thus, conserved miRNAs affected by stress in Arabidopsis may be involved in stress defense mechanisms in grapevine, but their modes of response and action seem to be different from those in the model plant. Our findings also confirm that members of a miRNA family can oppositely respond to stress in different plants and that their abundance can vary greatly in different organs or tissues (e.g. miR393) under water deficit (Zhang, 2015). Nevertheless, it should be mentioned that these variations among species could depend on experimental conditions, such as the timing and severity of DS (Frazier et al., 2011; Budak et al., 2015).
Besides conserved miRNAs, we identified 12 novel putative miRNAs whose abundance was affected significantly by DS. These novel miRNAs add to the emerging number of miRNAs identified in Vitis spp. (Carra et al., 2009; Mica et al., 2010; Pantaleo et al., 2010, 2016; Wang et al., 2012). The relative concentrations of these novel putative miRNAs were mostly lower than those of conserved miRNAs measured in the same samples. A similar situation was observed previously for novel miRNAs isolated in this and other plant species (Mica et al., 2010; Li et al., 2011; Ren et al., 2012; Ge et al., 2013). Moreover, since miRNAs belonging to low-abundance categories have emerging big roles in plant developmental processes (Wang and Guo, 2015), we hypothesize that the analyzed novel miRNAs that are less abundant than conserved miRNAs could function to modulate the cross talk between plant development and the abiotic stress response. For instance, the novel candidate miRNA n_22 was predicted to target an MLO calmodulin-binding protein (MLO16)-encoding transcript belonging to a plant gene family involved in pathogen defense and abiotic stress response processes (Piffanelli et al., 2002). Upon drought, MLO16 transcriptional rates increased in CS root, while n_22 decreased accordingly. Grape-specific miRNAs have already been shown to target members of the NBS-LRR superfamily (Carra et al., 2009). Since miRNA-regulated NBS-LRR genes are involved in immune responses (Li et al., 2012), a role for these proteins in the response to abiotic stress also is conceivable. Here, NBS-LRR transcripts were predicted to be targets of n_47 and n_479 and showed transcriptional trends opposite to the expression profiles of these miRNAs. NBS-LRR gene biological functions often overlap (Meyers et al., 2003), and interestingly, it was reported that miRNAs can effectively control members of this protein family by triggering the biogenesis of phased secondary small interfering RNAs that, in turn, target NBS-LRR genes (Zhai et al., 2011). This strengthens the power of miRNAs as mediators of the integrated network of stress-induced molecular signals (Atkinson and Urwin, 2012).
The novel candidate miRNA n_231 was predicted to target a chloroplastic polyphenoloxidase (PPO)-encoding gene, repressed upon drought, whereas the related miRNA was strongly activated. PPO enzymes play a role in the control of reactive oxygen species (Sharma et al., 2012), and inhibition of the PPO gene, putatively mediated by n_231, could counteract drought-induced oxidative stress damage. The candidate miRNA n_622 has three putative targets, including the hexose transporter HT5, which was functionally characterized in grapevine as a homolog of the Arabidopsis sugar transporter STP13 (Hayes et al., 2007, 2010). This could support an induction of sugar signaling pathways, which, in woody species, can facilitate adaptation and/or recovery in response to dehydration (Secchi et al., 2016).
A gene encoding CIPK13 and a transcription factor, SPL6, belonging to the squamosa promoter-binding protein-like superfamily were predicted and analyzed as target sequences of n_191 and n_327, respectively. In higher plants, CIPKs form an interacting network with CBL calcium sensors, which control calcium-dependent molecular cascades involved in environmental stress adaptation (Weinl and Kudla, 2009) and were reported as target transcripts of species-specific miRNAs tied to abiotic stress tolerance (Jeong and Green, 2013). Accordingly, in our experiments, the CIPK13 gene was, in most cases, activated significantly upon stress and showed an expression profile opposite to that of n_191. Overall, expression levels of SPL6 increased in drought-stressed plants and were divergent from those of n_327. The predicted target gene of n_520 encodes a zinc-finger domain protein, which belongs to a wide group of transcription factors directing unique biological processes in plants, such as flower development, light-regulated morphogenesis, and pathogen responses (Takatsuji, 1998). The mentioned target exhibits an increasing expression in M4 roots and leaves upon drought occurrence, where contemporaneously low levels of n_520 were observed.
The occurrence of opposite RT-qPCR trends between target transcripts and related miRNAs suggests their interaction in planta, but for some targets (RPS5-like, HT5, and CIPK13), this was not observed, especially upon stress conditions. However, the quantification of target mRNAs does not provide information on the possible effects of miRNAs on protein synthesis through translational repression, a widespread mechanism in Arabidopsis (Brodersen et al., 2008) and recently reported in woody species (Li et al., 2013a), which could be effective in grape as well. Furthermore, it must be taken into account that stress could affect the expression of target genes through regulation mechanisms independent of miRNA-mediated silencing (Hirayama and Shinozaki, 2010).
Most of the predicted target genes, especially those of conserved miRNAs, encoded proteins involved in embryogenesis and cell development (e.g. SPL genes), apparently not linked to stress defense. These data are not surprising, since plants coping with a changing environment must regulate the transcription of genes tied to cell proliferation, development, and control of phenological phases (Sunkar et al., 2012). This is supported both by recent work on Brachypodium distachyon (Bertolini et al., 2013), where the role of miRNAs upon drought was mostly correlated to the regulation of leaf cell growth reprogramming and development, and by more specific studies unraveling the induction of stress tolerance strategies through the miRNA-mediated regulation of key transcription factors tied to plant development (Stief et al., 2014).
The Accumulation of Some Novel miRNAs Is Differently Modulated in Heterografted Plants
Grafting is performed routinely in grapevine management to obtain resistance against phylloxera, but grafting on specific rootstocks also can optimize plant growth rate, fruit quality, and tolerance to abiotic stresses. In pumpkin and cucumber, it has been shown that reciprocal grafting modifies the abundance of several miRNAs and predicted target genes (Li et al., 2014). As grapevine miRNAs are responsive to DS and are differently expressed in specific genotypes, reciprocal grafting of these genotypes could modify the scion or stock concentration of these miRNAs. To test this hypothesis, we assessed the concentrations of a group of novel putative miRNAs in reciprocal grafts of the stress-tolerant M4 with the stress-sensitive CS.
Grafting affected miRNA abundance, particularly for some of the analyzed novel candidates. This is the case for n_191, which in autografted vines was more abundant in CS, while in heterografted plants, significant differences in the accumulation of this miRNA between stock and scion were evident upon stress when M4 was used as stock. Another interesting case is n_520, which, in autografted vines, was present nearly exclusively in the leaves of M4 and only under irrigated conditions, whereas in heterografted plants, a relatively high abundance of this miRNA was present in CS root and leaves. However, the down-regulation of this putative miRNA observed upon drought in M4 was not noticed in CS tissues of heterografted plants. Thus, upon grafting, n_520 was present in heterografted CS, but it was not affected by drought.
Some degree of immune response to grafting is experienced even in these compatible genotypes, and a recent report demonstrated the up-regulation of stress responses in heterografting with nonself rootstocks (Cookson et al., 2014). This general response also could be the basis of the changes in expression that we observed for some candidate miRNAs. The abundance of n_55, n_191, and n_312 was 1 order of magnitude lower in heterografted plants than in autografted plants. Thus, a nonself response could affect the accumulation of specific miRNAs. The movement of hormones between stock and scion has been demonstrated previously (Foo et al., 2007), and many signals, such as ABA, commonly move across the graft junction in grapevine (Ferrandino and Lovisolo, 2014). This implies that altered signal fluxes induced by grafting could affect the regulation of miRNA expression in either one or both of the graft partners. Accordingly, several research studies have elucidated that miRNAs are key players in hormone (e.g. ABA and auxin) signaling cascades driving the response to DS (Ding et al., 2013; Zhang, 2015).
Finally, results obtained from the analysis of phloem sap have supported the hypothesis that, under stress, miRNAs can accumulate in phloem vessels, thus potentially serving as long-distance signaling molecules (Pant et al., 2008; Marín-González and Suárez-López, 2012). Testing of this possibility is not straightforward in grapevine and most other plants where the application of genetic approaches, such as the generation of specific mutant lines used as rootstock in grafting experiments, is still limited due to the absence of effective transformation protocols and long reproductive cycles (Gambino and Gribaudo, 2012). It is worth noting that, until now, small RNAs have been detected in the phloem and not in the xylem sap (Pant et al., 2008); thus, direct movement should explain the presence of scion-specific miRNAs in stocks but not the reverse. In this context, an interesting result concerns miRNA n_312, whose abundance was similar in CS and M4 autografted plants, but it accumulated to different extents in heterografts, with higher concentration in M4 (either leaf or root) heterografted plants, particularly upon stress.
Taken together, our findings highlight changes in the accumulation patterns of conserved and novel miRNAs dependent on DS, tissue type, and grapevine genotype. Moreover, the analyses of diverse grafting combinations reveal the existence of even more complex genotype-dependent molecular networks that tune stress-related miRNA abundance and mode of action. These effects could be triggered in a grafted genotype through the interaction of signaling molecules (e.g. ABA) produced by the other graft component.
MATERIALS AND METHODS
Plant Material and Experimental Setup
Two-year-old autografted grapevines of Vitis vinifera ‘Cabernet Sauvignon’, M4 [a new hybrid genotype selected at the University of Milan by crossing (V. vinifera × Vitis berlandieri) × V. berlandieri ‘Resseguier n.1’ (Meggio et al., 2014)], and reciprocal grafting combinations (heterografts: CS shoot/M4 root and M4 shoot/CS root) were grown in a greenhouse under partially controlled climate conditions (20 plants per graft combination). The temperature in the greenhouse was maintained in the 26°C to 35°C range, and natural light/dark cycles were followed. Maximum photosynthetic photon flux density ranged between 1,330 and 1,580 µmol m−2 s−1. Grapevine genotypes often show a capacity to explore diverse volumes of soils, and for this reason, we used a limited substrate volume in all the grafting conditions tested. Each plant grew in a 5-L pot filled with a substrate composed of a sandy loam soil (pH 7; available phosphorus, 7.9 mg kg−1; organic matter, 1.37%; cation-exchange capacity, 4.58 mE 100 g−1):expanded clay:peat mixture (2:1:1, w/w). From bud break (February 15) to the beginning of the experimental period (July 24), plants were irrigated twice per week to maintain water container capacity. A DS treatment was applied during a period of high atmospheric evaporative water demand (vapor pressure deficit averaging 25 mbar bar−1). Half of the plants of each graft combination were maintained at container capacity (Lovisolo and Schubert, 1998) and used as irrigated controls, whereas the other 40 plants were subjected to a DS treatment by withholding irrigation for a period of about 10 d. During the treatment, Ψleaf and gs were measured daily. Droughted plants were stressed until the measured Ψleaf and gs values were lower than −1.2 MPa and 0.05 mmol water m−2 s−1, respectively, which can be considered levels of severe water stress according to grapevine adaptations to water stress (Lovisolo et al., 2010).
Leaf Gas-Exchange and Ψleaf Measurements
The gs was measured on adult, nonsenescing leaves well exposed to direct sunlight (photosynthetic photon flux density of 400–700 nm) using a portable gas-exchange fluorescence system (GFS-3000; Heinz Walz). The measurements were performed using an 8-cm2 leaf chamber with artificial irradiation (1,200 µmol photons m−2 s−1), setting the chamber temperature at 26°C to avoid overheating of the leaf. CO2 and relative humidity were maintained at greenhouse conditions (420 µL L−1 and 50%, respectively). Measurements were taken on two leaves per plant between 9 am and 12 noon on each experimental day. After measuring gs, the same leaves were excised and used to determine Ψleaf using a Scholander-type pressure chamber (Soil Moisture Equipment).
Measurements of Root kh
Root kh was measured on well-watered rootstocks of CS/M4 and M4/CS plants under laboratory conditions by means of an HPFM-XP Hydraulic Conductance Flowmeter (Dynamax; http//www.dynamax.com/) following the protocol described by Lovisolo and Tramontini (2010). Five transient measurements were run by increasing the pressure up to 0.4 MPa at a rate of ∼5 KPa s−1 and by measuring the instantaneous flow. The root kh corresponds to the slope of the linear regression obtained by plotting flow data versus pressure data. Root ks was calculated by normalizing the root kh on the root dry weight.
Small RNA Library Preparation and Sequencing
Leaf and root samples were collected from DS and irrigated autografted plants of both genotypes (CS/CS and M4/M4), weighed (about 1 g each), immediately frozen in liquid nitrogen, and stored at −80°C. In detail, leaf samples were always taken from adult, nonsenescing leaves, whereas roots were sampled by collecting the whole root system. Root and leaf samples obtained from all the plants (n = 10) of each condition were pooled together, and LMW RNA was extracted following the protocol by Carra et al. (2007). The integrity and quantity of LMW RNA were checked with a 2100 Bioanalyzer (Agilent). Only LMW RNA samples obtained from autografted plants, representing eight tested conditions (2 genotypes × 2 organs × 2 treatments), were processed further for library preparation; only one replicate per condition was considered for next-generation sequencing analysis. LMW RNA samples were processed according to the protocol for the SOLiD Total RNA-Seq Kit (Applied Biosystems) with minor modifications: the first step was the ligation of adaptors that ensure the directionality of sequencing; after the reverse transcription step, a size selection was done on a 10% acrylamide denaturing gel. At the end of the process, the obtained cDNA libraries were amplified by emulsion PCR, barcoded, and sequenced using the SOLiD platform (Applied Biosystems). At the end of the sequencing process, raw sequence data were trimmed to remove adaptor sequences, filtered for quality, and aligned against the Rfam 11 database (http://rfam.xfam.org/; Gardner et al., 2011) to exclude from the genomic mapping contaminants and small noncoding RNAs not belonging to the miRNA clade (i.e. tRNAs, rRNAs, scRNAs, snoRNAs, snRNAs, and RDPs). The unique number of reads was obtained by mapping the filtered sequences on the grapevine reference genome (12X V1; http://genomes.cribi.unipd.it/grape/) and, in parallel, on miRBase (http://www.mirbase.org/; version 21) to allow the detection of already annotated miRNA sequences among the predicted novel ones. Finally, to compare the number of reads obtained for each predicted miRNA among the different libraries, a normalization step was performed, and the relative number of miRNA reads was calculated for each miRNA, as follows:
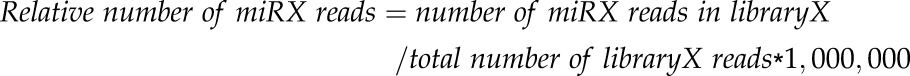 |
in which miRX is a predicted known or novel miRNA, libraryX is one of the sequenced library samples, and 1,000,000 is a constant value.
Novel miRNAs and their precursors were identified on the reference grapevine genome by applying two dedicated prediction softwares, mirCat (Moxon et al., 2008) and miRDeep-P (Yang and Li, 2011). Precursor sequences of the predicted novel miRNAs were further processed using MFOLD version 2.3 (http://unafold.rna.albany.edu/?q=mfold; Zuker, 2003) to analyze the folding of secondary hairpin structures. Conserved and known miRNAs were identified through alignment on miRBase (Kozomara and Griffiths-Jones, 2014) by setting the PASS algorithm in order to consider only the alignments showing no mismatches with the miRBase entries and with a minimum coverage of 17 bp, because in miRBase the size range of annotated sequences is between 17 and 26 nucleotides.
Prediction of Novel miRNA Target Genes
Putative target genes of candidate miRNAs were predicted in silico by applying two computational approaches: psRNATarget (http://plantgrn.noble.org/psRNATarget/; Dai and Zhao, 2011) and Target-align (http://www.leonxie.com/targetAlign.php; Xie and Zhang, 2010).
psRNATarget is a recent evolution of a previous software, miRU (Zhang, 2005), which calculates the score of complementarity between the small RNA and its target gene and the target site accessibility, which is assessed by calculating the energy required to unpair the secondary structure around a small RNA target site on the mRNA. Target gene sequences were retrieved by alignment with 12X V1 using the default setting parameters of psRNATarget, with the only exception that of the maximum expectation value, for which a cutoff threshold of 2 was applied. The Target-align algorithm, which was also developed as an implementation of the Smith-Waterman algorithm, as well as psRNAtarget, scores for a specific miRNA and its potential targets considering all potential factors that can affect the identification of miRNA targets (number of consecutive mismatches, base site restrictions, number of G:U wobbles, and number of gaps), according to the indications reported by Jones-Rhoades and Bartel (2004).
Predicted target sequences were then aligned to the M4 resequenced genome (http://genomes.cribi.unipd.it/grape/serres/) in order to check for nucleotide mismatches in the region of miRNA complementarity, M4 being a hybrid Vitis genotype.
Target genes were also annotated functionally using the Blast2GO software (version 2.7.1; http://blast2go.com/b2ghome) with default parameters.
Real-Time PCR Analysis
Expression changes of conserved and novel miRNAs and of target transcripts were determined on leaf and root samples collected from both autografts and heterografts by stem-looped RT-qPCR assay.
Total RNA was extracted in triplicate from autografted and heterografted plants of the two genotypes, following the protocol by Chang et al. (1993) with only slight modifications. RNA samples were quantified and qualified with the 2100 Bioanalyser and treated with DNase I, RNase-free (50 units μL−1; Fermentas) to avoid any risk of DNA contamination. First-strand cDNA was synthesized starting from 10 µg of total RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) following the manufacturer’s instructions. Only in the case of miRNA-specific cDNA was the retrotranscription protocol modified using a stem-loop primer (rather than random primers) specific for each of the target miRNAs. The use of stem-loop primers in RT-qPCR experiments is an effective and well-developed strategy for the detection of miRNAs, first reported by Chen et al. (2005) and Varkonyi-Gasic et al. (2007) and applied successfully in many research experiments addressing the study of abiotic stress-responsive miRNAs (Shen et al., 2010; Eldem et al., 2012; Xie et al., 2014; Candar-Cakir et al., 2016; Gao et al., 2016; Yadav et al., 2016). A list of the stem-loop primers used in this study is given in Supplemental Table S1.
Real-time PCR was carried out in the StepOnePlus RT-qPCR System (Applied Biosystems) using the SYBR Green (Applied Biosystems) method for quantifying amplification results. Thermal cycling conditions were as follows: an initial denaturation phase at 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min (only in the case of miRNA expression analysis, a step at 58°C for 15 s was added to the cycling stage). Specific annealing of primers was checked on dissociation kinetics performed at the end of each RT-qPCR run.
Expression levels of miRNAs and of the related target transcripts were quantified after normalization to either VvsnRU6 or VvUBI and VvACT1 genes used as internal controls, respectively. Gene-specific primers used in real-time PCR experiments are listed in Supplemental Table S2. Three biological replicates were run for each RT-qPCR experiment.
In the case of novel miRNA expression analysis, RT-qPCR products were purified, cloned into the pGEM-T vector (Promega), and sequenced using T7 and SP6 forward and reverse primers in order to further ascertain the accuracy of the amplification results.
Statistical Analyses
Significant differences among treatments were statistically analyzed by applying a one-way ANOVA test, using Tukey’s HSD posthoc test to separate means when ANOVA results were significant (P < 0.05). Significant differences of pairwise comparisons were assessed by Student’s t test.
The SPSS statistical software package (SPSS; version 22) and Sigma Plot software (Systat Software) were used to run the statistical analyses reported above and to elaborate figure charts, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression patterns of conserved miRNA target transcripts.
Supplemental Table S1. Stem-loop primers used in this study for miRNA-specific RT-qPCR assay.
Supplemental Table S2. Oligonucleotides used in this study for RT-qPCR analysis of miRNAs and related target transcripts.
Supplemental Table S3. Conserved and known miRNAs identified by high-throughput SOLiD sequencing in CS and M4 root and leaf samples upon irrigated control and DS conditions.
Supplemental Table S4. Putative novel miRNAs identified by high-throughput SOLiD sequencing in CS and M4 root and leaf samples upon irrigated control and DS conditions.
Supplemental Table S5. Target transcripts of the novel candidate miRNAs analyzed in this study.
Supplemental Table S6. Target transcripts of the conserved/known miRNAs analyzed in this study.
Supplementary Material
Acknowledgments
We thank Giorgio Gambino (IPSP-CNR) for helpful discussion and support during miRNA molecular analyses, Tiziano Strano (University of Torino) for grapevine maintenance, and Luca Espen and Giovanbattista Simone Di Lorenzo (University of Milano) for providing grafted plants.
Glossary
- miRNA
microRNA
- CS
Cabernet Sauvignon
- RT-qPCR
quantitative real-time PCR
- DS
drought stress
- LMW
low molecular weight
- ABA
abscisic acid
- HSD
honestly significant difference
Footnotes
This work was supported by the AGER foundation (SERRES Project, grant no. 2010-2105).
Articles can be viewed without a subscription.
References
- Achard P, Herr A, Baulcombe D, Harberd N (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3544 [DOI] [PubMed] [Google Scholar]
- Barrera-Figueroa BE, Gao L, Dio NN, Wu Z, Ehlers JD, Roberts PA, Close TJ, Zhu JK, Liu R (2011) Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol 11: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Figueroa BE, Gao L, Wu Z, Zhou X, Zhu J, Jin H, Liu R, Zhu JK (2012) High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol 12: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli Kullan J, Pinto D, Bertolini E, Fasoli M, Zenoni S, Tornielli G, Pezzotti M, Meyers B, Farina L, Pe M, et al. (2015) miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics 16: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdeja M, Nicolas P, Kappel C, Dai ZW, Hilbert G, Peccoux A, Lafontaine M, Ollat N, Gomes E, Delrot S (2015) Water limitation and rootstock genotype interact to alter grape berry metabolism through transcriptome reprogramming. Hortic Res 2: 15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini E, Verelst W, Horner DS, Gianfranceschi L, Piccolo V, Inzé D, Pè ME, Mica E (2013) Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol Plant 6: 423–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Mol Cell Biol 16: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Budak H, Kantar M, Bulut R, Akpinar BA (2015) Stress responsive miRNAs and isomiRs in cereals. Plant Sci 235: 1–13 [DOI] [PubMed] [Google Scholar]
- Candar-Cakir B, Arican E, Zhang B (2016) Small RNA and degradome deep sequencing reveals drought- and tissue-specific microRNAs and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J 14: 1727–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonneau A. (1985) The early selection of grapevine rootstocks for resistance to drought conditions. Am J Enol Vitic 36: 195–198 [Google Scholar]
- Carra A, Gambino G, Schubert A (2007) A cetyltrimethylammonium bromide-based method to extract low-molecular-weight RNA from polysaccharide-rich plant tissues. Anal Biochem 360: 318–320 [DOI] [PubMed] [Google Scholar]
- Carra A, Mica E, Gambino G, Pindo M, Moser C, Pe ME, Schubert A (2009) Cloning and characterization of small non-coding RNAs from grape. Plant J 59: 750–763 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chen C, Ridzon D, Broomer A, Zhou Z, Lee D, Nguyen J, Barbisin M, Xu N, Mahuvakar V, Andersen M, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhuofu L, Xiong L (2012) A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett 586: 1742–1747 [DOI] [PubMed] [Google Scholar]
- Cookson S, Moreno M, Hevin C, Mendome L, Delrot S, Magnin N, Trossat-Magnin C, Ollat N (2014) Heterografting with nonself rootstocks induces genes involved in stress responses at the graft interface when compared with autografted controls. J Exp Bot 65: 2473–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corso M, Vannozzi A, Maza E, Vitulo N, Meggio F, Pitacco A, Telatin A, D’Angelo M, Feltrin E, Negri A, et al. (2015) Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. J Exp Bot 66: 5739–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias AA, Reyes JL (2010) Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ 33: 481–489 [DOI] [PubMed] [Google Scholar]
- Cramer GR, Ergul A, Grimplet J, Tillett RL, Tattersall EAR, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, et al. (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7: 111–134 [DOI] [PubMed] [Google Scholar]
- Cramer GR, Van Sluyter SC, Hopper DW, Pascovici D, Keighley T, Haynes PA (2013) Proteomic analysis indicates massive changes in metabolism prior to the inhibition of growth and photosynthesis of grapevine (Vitis vinifera L.) in response to water deficit. BMC Plant Biol 13: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao P (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi SJ, Madhav MS, Kumar GR, Goel AK, Umakanth B, Jahnavi B, Viraktamath BC (2013) Identification of abiotic stress miRNA transcription factor binding motifs (TFBMs) in rice. Gene 531: 15–22 [DOI] [PubMed] [Google Scholar]
- Ding Y, Tao Y, Zhu C (2013) Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot 64: 3077–3086 [DOI] [PubMed] [Google Scholar]
- Eldem V, Celikkol Akcay U, Ozhuner E, Bakir Y, Uranbey S, Unver T (2012) Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS ONE 7: e50298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous J, Hussain SS, Shi BJ (2015) Role of microRNAs in plant drought tolerance. Plant Biotechnol J 13: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandino A, Lovisolo C (2014) Abiotic stress effects on grapevine (Vitis vinifera L.): focus on abscisic acid-mediated consequences on secondary metabolism and berry quality. Environ Exp Bot 103: 138–147 [Google Scholar]
- Ferreira TH, Gentile A, Vilela RD, Lacerda Costa GG, Dias LI, Endres L, Menossi M (2012) MicroRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 7: e46703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira e Silva GF, Silva EM, Azevedo Mda S, Guivin MA, Ramiro DA, Figueiredo CR, Carrer H, Peres LE, Nogueira FT (2014) microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J 78: 604–618 [DOI] [PubMed] [Google Scholar]
- Foo E, Morris S, Parmenter K, Young N, Wang H, Jones A, Rameau C, Turnbull C, Beveridge C (2007) Feedback regulation of xylem cytokinin content is conserved in pea and Arabidopsis. Plant Physiol 143: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TP, Sun G, Burklew CE, Zhang B (2011) Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol 49: 159–165 [DOI] [PubMed] [Google Scholar]
- Gambino G, Gribaudo I (2012) Genetic transformation of fruit trees: current status and remaining challenges. Transgenic Res 21: 1163–1181 [DOI] [PubMed] [Google Scholar]
- Gao F, Wang N, Li H, Liu J, Fu C, Xiao Z, Wei C, Lu X, Feng J, Zhou Y (2016) Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci Rep 6: 34601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. (2011) Rfam: Wikipedia, clans and the ‘decimal’ release. Nucleic Acids Res 39: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge A, Shangguan L, Zhang X, Dong Q, Han J, Liu H, Wang X, Fang J (2013) Deep sequencing discovery of novel and conserved microRNAs in strawberry (Fragaria × ananassa). Physiol Plant 148: 387–396 [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Gustafson P, Langridge P, Shi BJ (2015) Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol J 13: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MA, Davies C, Dry IB (2007) Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. J Exp Bot 58: 1985–1997 [DOI] [PubMed] [Google Scholar]
- Hayes MA, Feechan A, Dry IB (2010) Involvement of abscisic acid in the coordinated regulation of a stress-inducible hexose transporter (VvHT5) and a cell wall invertase in grapevine in response to biotrophic fungal infection. Plant Physiol 153: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Jeong DH, Green PJ (2013) The role of rice microRNAs in abiotic stress responses. J Plant Biol 56: 187–197 [Google Scholar]
- Jones-Rhoades M, Bartel D (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kim SH, Jeong JC, Ahn YO, Lee HS, Kwak SS (2014) Differential responses of three sweetpotato metallothionein genes to abiotic stress and heavy metals. Mol Biol Rep 41: 6957–6966 [DOI] [PubMed] [Google Scholar]
- Koundouras S, Tsialtas I, Zioziou E, Nikolaou N (2008) Rootstock effects on the adaptive strategies of grapevine (Vitis vinifera L. cv. Cabernet-Sauvignon) under contrasting water status: leaf physiological and structural responses. Agric Ecosyst Environ 128: 86–96 [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka K, Pieczynski M, Windels D, Jarmolowski A, Kulinsk SZ, Vazquez F (2012) Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol 16: 1664–1672 [DOI] [PubMed] [Google Scholar]
- Kulcheski FR, de Oliveira LF, Molina LG, Almerao MP, Rodrigues FA, Marcolino J, Barbosa JF, Stolf-Moreira R, Nepomuceno AL, Marcelino-Guimarães FC, et al. (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie-Lamoureux A, Sacco D, Risse PA, Lovisolo C (2017) Factors influencing stomatal conductance in response to water availability in grapevine: a meta-analysis. Physiol Plant http://dx.doi.org/10.1111/ppl.12530 [DOI] [PubMed] [Google Scholar]
- Li B, Duan H, Li H, Deng XW, Yin W, Xia X (2013a) Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol Biol 81: 525–539 [DOI] [PubMed] [Google Scholar]
- Li B, Qin Y, Duan H, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62: 3765–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li Y, Bai L, Zhang T, He C, Yan Y, Yu X (2014) Grafting-responsive miRNAs in cucumber and pumpkin seedlings identified by high-throughput sequencing at whole genome level. Physiol Plant 151: 406–422 [DOI] [PubMed] [Google Scholar]
- Li C, Zhang B (2016) MicroRNAs in control of plant development. J Cell Physiol 231: 303–313 [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard J, Cohn M, Tung J, Sun H, Kumar P, Baker B (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Fu F, An M, Zhou S, She Y, Li W (2013b) Differential expression of microRNAs in response to drought stress in maize. J Integr Agric 12: 1414–1422 [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D (2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Liu H, Tian X, Li Y, Wu C, Zheng C (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovisolo C, Lavoie-Lamoureux A, Tramontini S, Ferrandino A (2016) Grapevine adaptations to water stress: new perspectives about soil/plant interactions. Theor Exp Plant Physiol 28: 53–66 [Google Scholar]
- Lovisolo C, Perrone I, Carra A, Ferrandino A, Flexas J, Medrano H, Schubert A (2010) Drought-induced changes in development and function of grapevine (Vitis spp.) organs and in their hydraulic and non-hydraulic interactions at the whole-plant level: a physiological and molecular update. Funct Plant Biol 37: 98–116 [Google Scholar]
- Lovisolo C, Perrone I, Hartung W, Schubert A (2008) An abscisic acid-related reduced transpiration promotes gradual embolism repair when grapevines are rehydrated after drought. New Phytol 180: 642–651 [DOI] [PubMed] [Google Scholar]
- Lovisolo C, Schubert A (1998) Effects of water stress on vessel size and xylem hydraulic conductivity in Vitis vinifera L. J Exp Bot 49: 693–700 [Google Scholar]
- Lovisolo C, Tramontini S (2010) Methods for assessment of hydraulic conductance and embolism extent in grapevine organs. In Delrot S, Medrano H, Or E, Bavaresco L, Grando S, eds, Methodologies and Results in Grapevine Research. Springer, Dordrecht, The Netherlands, pp 71–85 [Google Scholar]
- Marguerit E, Brendel O, Lebon E, Van Leeuwen C, Ollat N (2012) Rootstock control of scion transpiration and its acclimation to water deficit are controlled by different genes. New Phytol 194: 416–429 [DOI] [PubMed] [Google Scholar]
- Marín-González E, Suárez-López P (2012) “And yet it moves”: cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci 196: 18–30 [DOI] [PubMed] [Google Scholar]
- Meggio F, Prinsi B, Negri A, Di Lorenzo G, Lucchini G, Pitacco A, Failla O, Scienza A, Cocucci M, Espen L (2014) Biochemical and physiological responses of two grapevine rootstock genotypes to drought and salt treatments. Aust J Grape Wine Res 20: 310–323 [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mica E, Piccolo V, Delledonne M, Ferrarini A, Pezzotti M, Casati C, Del Fabbro C, Valle G, Policriti A, Morgante M, et al. (2010) High throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifera. BMC Genomics 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal TK, Ganie SA (2014) Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene 535: 204–209 [DOI] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V (2008) A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- Mudge K, Janick J, Scofield S, Goldschimdt EE (2009) A history of grafting. Hortic Rev (Am Soc Hortic Sci) 35: 437–493 [Google Scholar]
- Nozawa M, Miura S, Nei M (2012) Origins and evolution of microRNA genes in plant species. Genome Biol Evol 4: 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B, Buhtz A, Kehr J, Scheible W (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J 62: 960–976 [DOI] [PubMed] [Google Scholar]
- Pantaleo V, Vitali M, Boccacci P, Miozzi L, Cuozzo D, Chitarra W, Mannini F, Lovisolo C, Gambino G (2016) Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci Rep 6: 20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone I, Pagliarani C, Lovisolo C, Chitarra W, Roman F, Schubert A (2012) Recovery from water stress affects grape leaf petiole transcriptome. Planta 235: 1383–1396 [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins N, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RS, Eamens AL, Waterhouse PM (2015) Missing pieces in the puzzle of plant microRNAs. Trends Plant Sci 20: 721–728 [DOI] [PubMed] [Google Scholar]
- Ren Y, Chen L, Zhang Y, Kang X, Zhang Z, Wang Y (2012) Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct Integr Genomics 12: 327–339 [DOI] [PubMed] [Google Scholar]
- Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchi F, Pagliarani C, Zwieniecki MA (2016) The functional role of xylem parenchyma cells and aquaporins during recovery from severe water stress. Plant Cell Environ http://dx.doi.org/10.1111/pce.12831 [DOI] [PubMed] [Google Scholar]
- Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygene species, oxidative damage, and antioxidative defence mechanism in plants under stressful conditions. J Bot 2012: 217037 [Google Scholar]
- Shen J, Xie K, Xiong L (2010) Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics 284: 477–488 [DOI] [PubMed] [Google Scholar]
- Shuai P, Liang D, Zhang Z, Yin W, Xia X (2013) Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics 14: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F Jr, Vazquez F (2011) Mir393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol 157: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soar CJ, Dry PR, Loveys BR (2006) Scion photosynthesis and leaf gas exchange in Vitis vinifera L. cv. Shiraz: mediation of rootstock effects via xylem sap ABA. Aust J Grape Wine Res 12: 82–96 [Google Scholar]
- Solofoharivelo MC, van der Walt AP, Stephan D, Burger JT, Murray SL (2014) MicroRNAs in fruit trees: discovery, diversity and future research directions. Plant Biol 16: 856–865 [DOI] [PubMed] [Google Scholar]
- Song JB, Gao S, Sun D, Li H, Shu XX, Yang ZM (2013) miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol 13: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Baurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Fan G, Su L, Wang W, Liang Z, Li S, Xin H (2015) Identification of cold-inducible microRNAs in grapevine. Front Plant Sci 6: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Takatsuji H. (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontini S, Döring J, Vitali M, Ferrandino A, Stoll M, Lovisolo C (2014) Soil water-holding capacity mediates hydraulic and hormonal signals of near-isohydric and near-anisohydric Vitis cultivars in potted grapevines. Funct Plant Biol 41: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Tramontini S, Vitali M, Centioni L, Schubert A, Lovisolo C (2013) Rootstock control of scion response to water stress in grapevine. Environ Exp Bot 93: 20–26 [Google Scholar]
- Trindade I, Capitão C, Dalmay T, Fevereiro MP, Santos DM (2010) miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231: 705–716 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton E, Hellens R (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenenesis and activity of plant miRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang C, Han J, Liu C, Kibet KN, Kayesh E, Shangguan L, Li X, Fang J (2012) Identification of microRNAs from Amur grape (Vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genomics 13: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JJ, Guo HS (2015) Cleavage of INDOLE-3-ACETIC ACID INDUCIBLE28 miRNA by microRNA847 upregulates auxin signalling to modulate cell proliferation and lateral organ growth in Arabidopsis. Plant Cell 27: 574–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang Q, Zhang B (2013) Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530: 26–32 [DOI] [PubMed] [Google Scholar]
- Wang TZ, Chen L, Zhao MG, Tian QY, Zhang WH (2011) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, Wang Y, Zhang M (2012) OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 7: e30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Stewart CN Jr, Taki FA, He Q, Liu H, Zhang B (2014) High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol J 12: 354–366 [DOI] [PubMed] [Google Scholar]
- Xie F, Wang Q, Sun R, Zhang B (2015) Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J Exp Bot 66: 789–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Zhang B (2010) Target-align: a tool for plant microRNA target identification. Bioinformatics 26: 3002–3003 [DOI] [PubMed] [Google Scholar]
- Xu MY, Zhang L, Li WW, Hu XL, Wang MB, Fan YL, Zhang CY, Wang L (2014) Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J Exp Bot 65: 89–101 [DOI] [PubMed] [Google Scholar]
- Yadav A, Khan Y, Prasad M (2016) Dehydration-responsive miRNAs in foxtail millet: genome-wide identification, characterization and expression profiling. Planta 243: 749–766 [DOI] [PubMed] [Google Scholar]
- Yang X, Li L (2011) miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 27: 2614–2615 [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot 66: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yang J, Wang Z, Wen Y, Wang J, He W, Liu B, Si H, Wang D (2014) Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS ONE 9: e95489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33: W701–W704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61: 4157–4168 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.