Arabidopsis roots activate cytokinin signaling in response to shoot removal independently of reduced auxin signaling and induce chloroplast development and photosynthetic remodeling.
Abstract
The development of plant chloroplasts is regulated by various developmental, environmental, and hormonal cues. In Arabidopsis (Arabidopsis thaliana), chloroplast development is repressed in roots via auxin signaling. However, roots develop chloroplasts when they are detached from the shoot. In contrast to auxin, cytokinin positively affects chloroplast development in roots, but the role and signaling pathway of cytokinin in the root greening response remain unclear. To understand the regulatory pathways of chloroplast development in the plant stress response, we examined the mechanisms underlying the conditional greening of detached roots. In wild-type Arabidopsis roots, shoot removal activates type B ARABIDOPSIS RESPONSE REGULATOR (ARR)-mediated cytokinin signaling and induces chlorophyll accumulation and photosynthetic remodeling. ARR1 and ARR12 are essential for up-regulating nucleus- and plastid-encoded genes associated with chloroplast development in detached roots. In this process, WOUND INDUCED DEDIFFERENTIATION1 and class B GATA transcription factors (B-GATAs) act upstream and downstream of ARRs, respectively. Overexpression of B-GATAs promotes root greening, as does shoot removal, dependent on a light signaling transcription factor, LONG HYPOCOTYL5. Auxin represses the root greening response independent of ARR signaling. GNC-LIKE (GNL), a B-GATA, is strongly up-regulated in detached roots via ARR1 and ARR12 but is repressed by auxin, so GNL may function at the point of convergence of cytokinin and auxin signaling in the root greening response.
Like embryonic cells of higher plants that differentiate into various cell types, embryonic proplastids are converted into different types of plastids within cells (e.g. chloroplasts in leaves, amyloplasts in seeds and roots, and chromoplasts in flowers and fruits). Differentiated plastids have specific functions associated with the host cell types in addition to common roles in fundamental metabolism, such as the synthesis of amino acids and fatty acids (Jarvis and López-Juez, 2013). Because plastids are the site of various metabolic reactions and are involved in plant growth regulation, tight coordination between plastid and cell differentiation is essential for plant development.
Chloroplasts, which accumulate chlorophyll (Chl) and develop photosynthetic machinery in thylakoid membranes, convert solar energy into chemical energy and fix carbon dioxide into carbohydrates by photosynthesis. The biogenesis of chloroplasts is strictly regulated in response to the functional state of photosynthetic cells under various growth conditions to avoid photooxidative damage from unbalanced photosynthetic reactions. Chloroplast biogenesis requires a coordinated synthesis of photosynthetic proteins with Chl and other photosynthetic components. Photosynthesis-associated nuclear genes form tight coexpression networks with each other (Kobayashi et al., 2012b), which suggests that transcriptional regulation plays a pivotal role in regulating chloroplast biogenesis. Transcription factors known as positive regulators of chloroplast differentiation include LONG-HYPOCOTYL5 (HY5), GOLDEN2-LIKE (GLK), and class B GATA transcription factors (B-GATAs), including GATA, NITRATE-INDUCIBLE CARBON METABOLISM INVOLVED (GNC) and GNC-LIKE/CYTOKININ-RESPONSIVE GATA1 (GNL/CGA1). HY5 functions in photomorphogenesis downstream of light signaling pathways (Gangappa and Botto, 2016) and directly targets many of the light-inducible photosynthesis-associated genes (Lee et al., 2007; Toledo-Ortiz et al., 2014). Deficiency of HY5 causes down-regulation of the target photosynthetic genes in deetiolated seedlings and roots (McCormac and Terry, 2002; Kobayashi et al., 2012a, 2014). GLK transcription factors also directly up-regulate photosynthesis-associated genes, presumably by binding to their promoter regions (Waters et al., 2009).
Arabidopsis (Arabidopsis thaliana) has two GLK isoforms, GLK1 and GLK2, which function redundantly, and only the double mutant glk1 glk2 causes reduced expression of photosynthesis-associated genes with decreased Chl content in leaves (Fitter et al., 2002). Overexpression of Arabidopsis GLKs and their orthologs in rice (Oryza sativa) and tomato (Solanum lycopersicum) causes ectopic chloroplast development in nonphotosynthetic organs such as roots, fruits, and callus cells (Nakamura et al., 2009; Kobayashi et al., 2012a; Powell et al., 2012), so this family is one of the key factors in chloroplast differentiation.
Overexpression of GNC and GNL induces ectopic chloroplast development in roots, hypocotyls, and leaf epidermis of Arabidopsis (Chiang et al., 2012) and in leaf sheath epidermis of rice (Hudson et al., 2013). These factors can increase the number of chloroplasts in photosynthetic and nonphotosynthetic cells with their overexpression (Hudson et al., 2011; Chiang et al., 2012). The expression of GNC and GNL is induced by cytokinin via type B ARABIDOPSIS RESPONSE REGULATOR1 (ARR1) and ARR12 and is repressed by GA and auxin (Mara and Irish, 2008; Richter et al., 2010, 2013). Together with four other B-GATA paralogs, GNC and GNL are implicated in the regulation of chloroplast development in addition to various developmental processes related to light and several hormonal responses (Behringer and Schwechheimer, 2015). However, GNC and GNL up-regulate Chl biosynthesis genes indirectly and do not affect the expression of genes involved in chloroplast division (Hudson et al., 2011). How these factors participate in regulating chloroplast differentiation remains unclear.
Because chloroplasts are developmentally linked with photosynthetic tissues, various hormonal and environmental cues affect chloroplast differentiation. We reported that auxin transported from shoots represses chloroplast differentiation in Arabidopsis roots (Kobayashi et al., 2012a). When roots were detached from the shoot, they increased greening via reduced auxin signaling from the shoot. Indeed, roots deficient in auxin signaling showed an up-regulation of photosynthesis-associated genes along with increased GLK2 expression and HY5 protein level. In contrast to auxin, cytokinin positively affects chloroplast differentiation, because cytokinin deficiency down-regulates chloroplast-related genes in green tissues, whereas excess cytokinin up-regulates the genes in roots (Brenner and Schmülling, 2012). In fact, cytokinin treatment up-regulates photosynthesis-associated genes in Arabidopsis roots, whereas mutation in the cytokinin receptors ARABIDOPSIS HISTIDINE KINASE2 (AHK2) and AHK3 down-regulated such genes (Kobayashi et al., 2012a). The expression levels of key Chl biosynthesis genes were correlated with Chl content in roots, which suggests that transcriptional regulation plays a central role in controlling chloroplast development in roots. In addition, a recent study revealed that cytokinin up-regulates genes involved in Chl biosynthesis and light harvesting and enhances cotyledon greening during the etioplast-chloroplast transition in Arabidopsis (Cortleven et al., 2016). In this process, B-type ARRs (ARR1, ARR10, and ARR12) play an important role in up-regulating genes involved in Chl biosynthesis.
The conditional greening of detached roots indicates that plastids in roots can plastically differentiate into chloroplasts by light and hormone signaling as needed (Kobayashi et al., 2012a). This greening response may function to compensate for the loss of photosynthetic tissues. Indeed, we reported that photosynthesis in detached Arabidopsis roots contributed to carbon assimilation and the maintenance of biomass (Kobayashi et al., 2013). However, the underlying molecular mechanisms are largely unknown. To gain insight into regulatory mechanisms controlling chloroplast development in plants, we examined the signaling pathway involved in root greening after shoot removal in Arabidopsis. Our data suggest that shoot removal induces the quantitative and qualitative development of chloroplasts in roots, with cytokinin signaling via ARR1 and ARR12 functioning downstream of wounding signaling to up-regulate genes associated with chloroplast development, including chloroplast-related transcription factors and plastid-encoded genes.
RESULTS
Type B ARR-Mediated Cytokinin Signaling Promotes the Greening of Detached Roots Downstream of Wounding Signaling
To assess the involvement of cytokinin signaling in the greening of detached roots, first we examined whether cytokinin signaling is activated in roots in response to shoot removal. Cytokinin signaling was monitored in transgenic plants carrying a synthetic cytokinin reporter, TWO-COMPONENT-OUTPUT-SENSOR (TCS)::GFP (Müller and Sheen, 2008), and its improved version, TCSn::GFP (Zürcher et al., 2013), which harbored the concatemerized type B ARR-binding motifs. To induce root greening, shoots of 14-d-old seedlings were removed at the root-hypocotyl junction and detached roots were incubated for 7 d on agar medium under light. For the control, intact roots of 21-d-old seedlings continuously grown on agar medium were detached from shoots just before analysis.
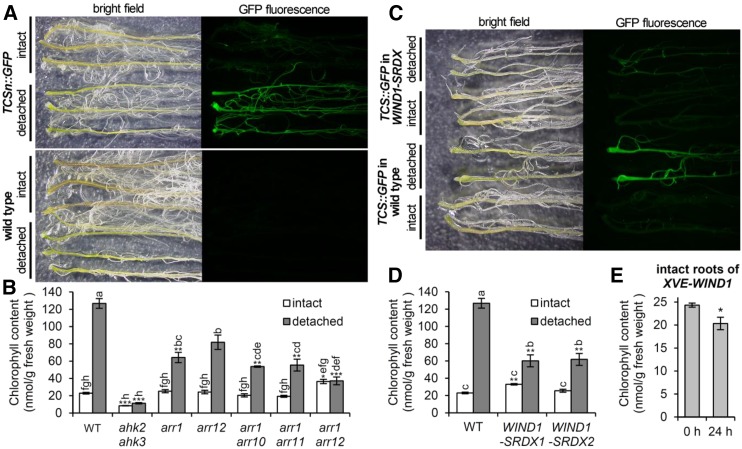
In TCSn::GFP transgenic plants, green fluorescence was brighter in detached roots than in intact roots (Fig. 1A). The same results were observed in the TCS::GFP line (Supplemental Fig. S1A). GFP fluorescence in detached roots was broadly observed, with that at the cutting site most prominent. Some detached roots produced callus at the cutting site, and in these cases, callus showed prominent TCS(n)::GFP fluorescence with relatively weak Chl fluorescence (Supplemental Fig. S1B). Under the same conditions, wild-type plants showed no green fluorescence in intact or detached roots (Fig. 1A), so the green fluorescence detected in detached TCSn::GFP roots was not autofluorescence from greenish cells but specifically reflected the type B ARR-mediated GFP expression.
Figure 1.
Cytokinin signaling functions in the greening of detached roots downstream of wounding signaling. A, GFP fluorescence in roots of the TCSn::GFP line and the wild-type control. B, Chl content in intact and detached roots of the wild type (WT) and mutants involved in cytokinin signaling. C, GFP fluorescence in roots of the TCS::GFP lines with the wild-type and WIND1-SRDX background. D, Chl content in intact and detached roots of the wild type and two independent lines of WIND1-SRDX plants. In A to D, for the detached root sample, roots were excised from 14-d-old seedlings and grown for 7 d, whereas for the intact root control, the shoots of 21-d-old seedlings were removed just before observation. E, Chl content in intact roots of the XVE-WIND1 line. Root Chl content is shown for 21-d-old seedlings treated with 17β-estradiol for 24 h compared with the untreated control (0 h). Student’s t test: *, P < 0.05. For B, D, and E, data are means ± se from three or more independent experiments. Different letters indicate significant differences by the Tukey-Kramer multiple comparison test (P < 0.05). Asterisks indicate significant differences from the wild type of each condition (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test after a Bonferroni correction for multiple comparisons).
Next, we evaluated the contribution of type B ARRs to the greening of detached roots by measuring Chl content in roots of several arr mutants (Fig. 1B). Consistent with a previous report (Kobayashi et al., 2012a), a double mutant for cytokinin receptors (ahk2 ahk3) showed a reduced Chl level in intact roots compared with the wild type. None of the arr mutants showed decreased Chl content in intact roots. Of note, Chl accumulation was slightly higher in the arr1 arr12 double mutant than in the wild type. As reported previously (Kobayashi et al., 2012a), Chl content in wild-type roots was increased substantially on shoot removal. By contrast, ahk2 ahk3 roots showed no noticeable increase in Chl content on shoot removal. Although Chl content was increased in detached roots of arr1, arr12, arr1 arr10, and arr1 arr11, the level was lower than in detached wild-type roots. Moreover, the arr1 arr12 double mutant showed no increase in root Chl content on shoot removal. Therefore, these type B ARRs are not essential for constitutive Chl accumulation in roots but are required for conditional root greening in response to shoot removal.
Type B ARR-mediated cytokinin signaling is activated in response to wounding stress via a transcription factor, WOUND INDUCED DEDIFFERENTIATION1 (WIND1; Iwase et al., 2011a). WIND1 cooperates with its close homologs (WIND2, WIND3, and WIND4) in a redundant manner to regulate wound-induced cellular reprogramming. Expression of WIND1-SUPERMAN REPRESSION DOMAIN (SRDX) chimeric proteins under the control of the WIND1 promoter dominantly repressed the function of WIND1 and presumably its homologs, which resulted in the strong impairment of reprogramming processes after wounding (Iwase et al., 2011a, 2015). Because WIND1-SRDX lines showed strongly suppressed activation of type B ARR-mediated cytokinin signaling at wounding sites (Iwase et al., 2011a), we tested whether WIND1 signaling is involved in the activation of type B ARR signaling in detached Arabidopsis roots. In contrast to the strong green fluorescence in detached roots of TCS::GFP plants in the wild-type background, green fluorescence was not remarkably enhanced by shoot removal in ProWIND1:WIND1-SRDX lines carrying the TCS::GFP gene. Moreover, we confirmed lower Chl accumulation in detached roots of two independent WIND1-SRDX lines than in detached wild-type roots (Fig. 1D), which suggests that WIND1 is involved in the root greening response.
To address whether WIND1 expression can solely activate Chl accumulation in roots, we tested β-estradiol-triggered overexpression of WIND1 in 21-d-old LexA-VP16-estrogen receptor (XVE)-WIND1 lines (Iwase et al., 2011a). A previous report showed that mature XVE-WIND1 seedlings treated with β-estradiol for long periods formed callus-like cell masses in shoots and roots, as did the stable WIND1 overexpression line (35S:WIND1; Iwase et al., 2011a). Moreover, induction of WIND1 in XVE-WIND1 seedlings for 1 d greatly enhanced a competence for shoot regeneration from roots (Iwase et al., 2015), which indicates that transient WIND1 induction strongly affects cellular differentiation states. In this study, WIND1 induction by β-estradiol for 24 h did not increase root Chl content and even decreased it slightly (Fig. 1E). These data are consistent with the report that ectopic WIND1 overexpression induced cell dedifferentiation and callus formation, thereby resulting in the degreening of green tissues (Iwase et al., 2011a). Therefore, although WIND1 is conditionally required for the greening of detached roots by activating type B ARR signaling in response to wounding, its ectopic overexpression would cause excessive reprograming effects to cells and degreening.
Type B ARR-Mediated Cytokinin Signaling Up-Regulates Genes Associated with Chloroplast Differentiation in Detached Roots
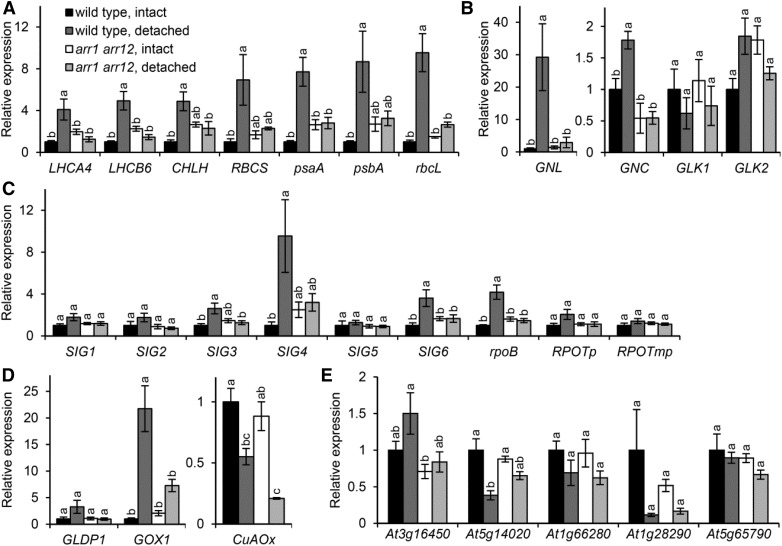
To assess whether shoot removal induces chloroplast-related gene expression in roots, we analyzed the expression of photosynthesis-associated nuclear genes in wild-type roots. LHCA4 and LHCB6 encode the light-harvesting complex subunits of PSI and PSII, respectively. CHLH encodes the H subunit of magnesium chelatase involved in Chl biosynthesis, and RBCS encodes the small subunit of Rubisco. All of these genes were strongly up-regulated in detached wild-type roots compared with intact roots (Fig. 2A). In addition, HEMA1, which encodes the major isoform of glutamyl-tRNA reductase involved in Chl biosynthesis, was up-regulated in roots on shoot removal, whereas the expression of HEMA2 encoding the minor isoform was not induced in detached roots (Supplemental Fig. S2). We also examined the expression of plastid-encoded photosynthetic genes. The genes psaA and psbA encode the core proteins of PSI and PSII, respectively, whereas rbcL encodes the Rubisco large subunit. In detached wild-type roots, the expression of psaA, psbA, and rbcL was increased 8- to 10-fold compared with intact roots (Fig. 2A). Therefore, shoot removal activates the formation of the photosynthetic machinery at the transcriptional level in roots.
Figure 2.
Expression of genes associated with chloroplast differentiation in intact and detached roots. Quantitative RT-PCR analysis of mRNA expression is shown for genes associated with photosynthesis (A), nuclear transcription (B), plastid transcription (C), mitochondrial and peroxisomal functions (D), and root tissues (E) in roots of the wild type and the arr1 arr12 double mutant. Data are means ± se fold difference from the untreated wild type after normalizing to ACTIN8 (n > 3). The sample preparation was as in Figure 1. Different letters indicate significant differences for each gene (P < 0.05, Tukey-Kramer multiple comparison test).
To elucidate the regulatory pathways involved in the greening of detached roots, we analyzed the expression of genes encoding nuclear transcription factors associated with chloroplast differentiation (GNC, GNL, GLK1, and GLK2; Fig. 2B). GNL was greatly up-regulated in wild-type roots on shoot removal, whereas the expression of GNC was increased only moderately. The expression of GLK1 and GLK2 was not increased significantly in detached roots.
Plastid-encoded photosynthetic genes such as psaA, psbA, and rbcL are transcribed mainly by plastid-encoded RNA polymerase (PEP). PEP genes, including rpoB, which encodes the β-chain of PEP, are transcribed by nucleus-encoded plastid RNA polymerase (NEP; De Santis-MacIossek et al., 1999). In PEP components, only sigma factors (SIGs), transcription initiation factors that are required for binding PEP to specific promoters of plastid genes, are encoded in the nuclear genome (Schweer et al., 2010). To address the mechanism involved in the up-regulation of plastid-encoded photosynthetic genes in detached roots, we investigated the expression of genes associated with plastid transcription (Fig. 2C). In addition to plastid-encoded rpoB, many SIG genes, particularly SIG4, were up-regulated in wild-type roots on shoot removal. These results were consistent with the increased expression of PEP-dependent photosynthetic genes (psaA, psbA, and rbcL) in detached roots. Meanwhile, the expression of the NEP genes RPOTp and RPOTmp was not increased significantly by shoot removal compared with intact roots.
Because peroxisomes and mitochondria transform their functions in concert with chloroplast differentiation during the development of photosynthetic tissues (Hayashi and Nishimura, 2006), we examined whether the expression of genes related to these organelles changes along with the expression of chloroplast-related genes during root greening (Fig. 2D). GOX1 encodes glycolate oxidase in mitochondria, and GLDP1 encodes the P protein of the Gly decarboxylase complex in peroxisomes. These genes are both involved in photorespiratory pathways and are expressed mainly in photosynthetic tissues (Kamada et al., 2003; Engel et al., 2007). CuAOx, which encodes putative copper amine oxidase localized in peroxisomes, is expressed specifically in roots (Kamada et al., 2003) and, thus, was used as a marker for nonphotosynthetic expression related to organelle functions. In wild-type detached roots, the expression of photorespiratory genes, particularly GOX1, was increased, whereas that of CuAOx was decreased (Fig. 2D).
To assess whether the shoot-like gene expression occurs globally in detached roots, we examined the expression of various root-specific genes (At3g16450, At5g14020, At1g66280, At1g28290, and At5g65790; Obayashi et al., 2004) that are not specifically responsive to cytokinin or wounding (Kilian et al., 2007; Winter et al., 2007). We found no general tendency in expression patterns of root-specific genes in response to shoot removal (Fig. 2E).
To ascertain whether type B ARR-mediated cytokinin signaling is involved in the transcriptional modification in detached roots, we examined gene expression in roots of the arr1 arr12 double mutant. In intact roots, the expression of some photosynthetic genes appeared to be increased in arr1 arr12 compared with the wild type (Fig. 2A), which is consistent with the slightly higher Chl accumulation in intact arr1 arr12 roots. However, in detached roots, the expression of these genes was substantially lower in arr1 arr12 than in the wild type. In addition, genes associated with plastid transcription were not up-regulated in mutant roots on shoot removal (Fig. 2C). In arr1 arr12, the expression of GLDP1, GOX1, GNC, and GNL was similar in detached and intact roots, whereas that of CuAOx was decreased by shoot removal as in the wild type (Fig. 2, B and D). Thus, ARR1 and ARR12 are required for the transcriptional induction of chloroplast differentiation in detached roots. Meanwhile, as in the wild type, arr1 arr12 showed no general tendency in expression patterns of root-specific genes on shoot removal (Fig. 2E).
Shoot Removal Changes the Quality of Photosynthetic Complexes in Root Chloroplasts
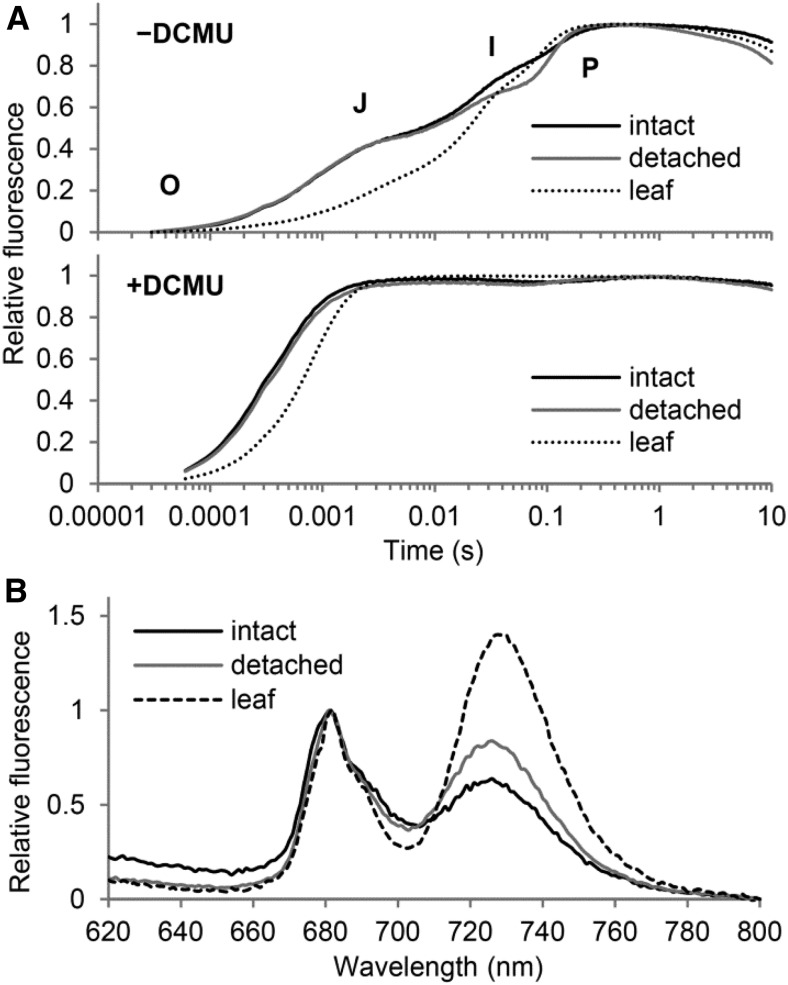
To evaluate the effect of shoot removal on the quality of the photosynthetic apparatus in root chloroplasts, we analyzed the transient kinetics of Chl fluorescence using a logarithmic timing series in the wild type (Fig. 3A). In a typical polyphasic fluorescence increase called the origin-inflection-intermediary peak-peak (OJIP) transient, the O-J phase and the J-I phase reflect the photochemical process in PSII and the reduction in the plastoquinone pool, respectively, whereas the I-P phase is related to the process in PSI (Schansker et al., 2005). The O-J transition occurred with higher fluorescence yield in both intact and detached roots than in leaves, which indicates an impaired electron transfer within PSII. However, unlike intact roots, detached roots showed enhanced inflection at the I transient, which resulted in lower Chl fluorescence at the I transient than in intact roots. Electron transport around PSI may somehow change in root chloroplasts on shoot removal, which improves electron transport in PSII. When the electron transport between the primary and secondary plastoquinones in PSII was inhibited with 3-(3,4-dichlorophenyl)-1,1-dimethylurea, both intact and detached roots showed a faster fluorescence increase than leaves. This result suggests that the size of the PSII antenna relative to the reaction center is larger in roots than in leaves, as described (Kobayashi et al., 2013), even after shoot removal.
Figure 3.
Comparison of photosystems in detached and intact roots and in leaves. A, Transient fluorescence induction kinetics of Chl in the absence (−) or presence (+) of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). Data are means from three independent experiments. Two inflections, J and I, are seen between the levels O (origin) and P (peak) only in the −DCMU samples. B, The 77K Chl fluorescence spectra in 2 μg mL−1 Chl fractions. Representative spectra from three independent experiments are shown. Data were normalized to the emission peak from PSII at 682 nm with background correction at 800 nm.
To address whether the state of Chl-protein complexes is changed in root chloroplasts in response to shoot removal, we measured Chl fluorescence spectra at 77K in membrane fractions containing 2 μg mL−1 Chl (Fig. 3B). Fluorescence emitted from PSI (FPSI), observed at 732 nm in the leaf sample, was slightly blue shifted in intact wild-type roots, as was found previously (Kobayashi et al., 2013). The same shift was observed in detached roots, so the coupling state between PSI and LHCI in roots was not changed by shoot removal. We observed maximal fluorescence emission from PSII (FPSII) at 682 nm in both leaf and root samples. The mean FPSI/FPSII ratio was lower in intact roots than in leaves (0.66 ± 0.03 versus 1.52 ± 0.20), which suggests less spillover of excitation energy from the PSII to the PSI antenna (Krause, 1991) or simply a lower abundance of PSI relative to PSII, as was suggested previously (Kobayashi et al., 2013). However, the FPSI/FPSII ratio was greater in detached roots than in intact roots (0.80 ± 0.04 versus 0.66 ± 0.03), which suggests that, with shoot removal, the energy distribution between PSII and PSI in root chloroplasts is partially improved to close to the leaf chloroplast status.
Cytokinin Signaling Improves Photosynthetic Efficiency in Detached Roots
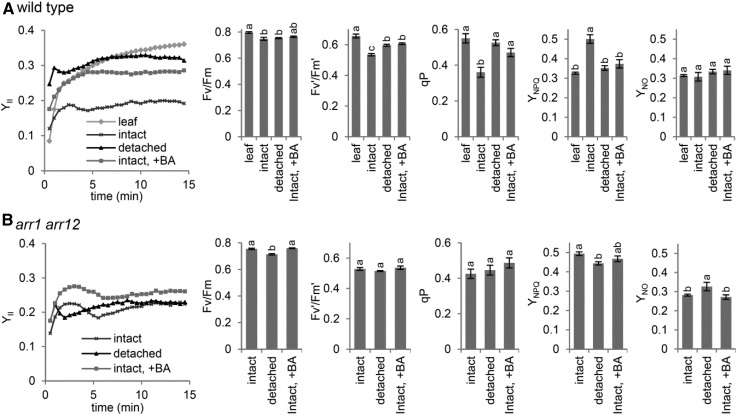
Because shoot removal affected photosynthetic components in roots (Fig. 3), we analyzed photosynthetic efficiency in detached roots by measuring Chl fluorescence with a fiber-optic pulse amplitude modulation (PAM) fluorometer. As reported previously (Kobayashi et al., 2013), the actual PSII efficiency (YII) under light was lower in intact roots than in leaves of the wild type. However, the YII value in roots was increased to close to the leaf level on shoot removal, although the induction profiles of YII differed between leaves and detached roots (Fig. 4A). Specifically, the detached roots showed a fast and transient increase in YII after actinic light illumination, which was not observed in leaves and intact roots, through an unknown mechanism. Meanwhile, the slightly lower Fv/Fm in intact roots than in leaves was not improved by shoot removal.
Figure 4.
Improvement of photosynthetic efficiency in detached roots via cytokinin signaling. Photosynthetic parameters are shown for 21-d-old detached roots and intact roots treated with 6-benzyladenine (BA) of the wild type (A) and the arr1 arr12 double mutant (B) compared with untreated intact roots. In A, wild-type leaves were used as a reference of the photosynthetically competent organ. Data are means ± se (n > 4). Fv/Fm, Maximum quantum yield of PSII; Fv′/Fm′, maximum quantum yield of PSII under light conditions; qP, coefficient of photochemical quenching; YNPQ, quantum yield of regulated energy dissipation; YNO, quantum yield of nonregulated energy dissipation. Slow induction kinetics under actinic light (intensity, 420 µmol photons m−2 s−1) are shown for YII, and other data were obtained after actinic light exposure for 15 min. Different letters indicate significant differences (P < 0.05, Tukey-Kramer multiple comparison test).
YII can be considered a product of two components: the qP, which represents the primary redox status and, thus, the openness of PSII, and Fv′/Fm′. Both of these values were lower in intact roots than in leaves, with qP more strongly decreased than Fv′/Fm′. However, these values in wild-type roots were increased to leaf levels after shoot removal, which suggests that electron transport downstream of PSII is ameliorated in detached roots. We also evaluated YNPQ and YNO, which represent the proportion of regulated and nonregulated dissipation of light energy in PSII, respectively (Kramer et al., 2004). As reported (Kobayashi et al., 2013), YNPQ was higher in intact roots than in leaves, whereas YNO was similar between roots and leaves. However, in detached roots, YNPQ decreased to a level similar to that in leaves. Thus, chloroplasts in detached roots may have photosynthetic characteristics close to those in leaves.
Next, we evaluated the involvement of cytokinin signaling in the regulation of photosynthetic activity in roots. Treatment of intact roots with a synthetic cytokinin, BA, induced changes in photosynthetic parameters similar to those in detached roots; that is, it increased YII, qP, and Fv′/Fm′ and decreased YNPQ compared with untreated roots (Fig. 4A). To further ascertain the role of cytokinin signaling, we analyzed photosynthetic activities in arr1 arr12 roots (Fig. 4B). Unlike the wild type, in arr1 arr12, neither shoot removal nor BA treatment greatly changed photosynthetic variables in roots. Thus, cytokinin signaling via ARR1 and ARR12 plays a central role in photosynthetic remodeling in detached roots.
Transcription Factors Involved in the Greening of Detached Roots
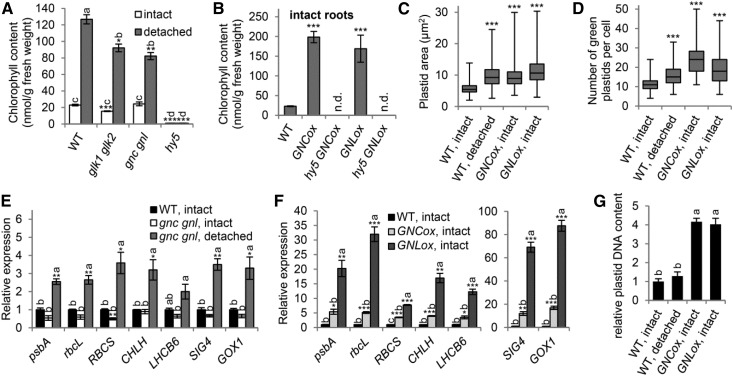
The removal of shoots induced a prominent expression of GNL in roots along with a slight increase in GNC expression via type B ARR signaling (Fig. 2B). To elucidate the role of these transcription factors in the greening of detached roots, we analyzed Chl content in roots of the gnc gnl mutant along with the glk1 glk2 mutant (Fig. 5A). Chl content was lower in intact roots of glk1 glk2 than in the wild type, as reported previously (Kobayashi et al., 2012a), whereas the content in intact gnc gnl roots was almost equivalent to the wild-type level. However, in detached roots, Chl content was lower in gnc gnl than in the wild type. Although Chl content increased 5.5-fold with shoot removal in wild-type roots, that in gnc gnl roots increased only 3.4-fold from the level in intact roots. Hence, GNC and GNL are conditionally required for Chl accumulation in roots in response to shoot removal. Chl content also was lower in detached glk1 glk2 roots than in wild-type roots. However, the magnitude of the increase in root Chl content on shoot removal was similar between glk1 glk2 and the wild type, which suggests that GLKs constitutively function in Chl accumulation in both intact and detached roots.
Figure 5.
Involvement of transcription factors in the root greening response after shoot removal. A and B, Chl content in intact and detached roots of each mutant line (A) and intact roots with GNC and GNL overexpression (GNCox and GNLox, respectively) in the wild type (WT) and hy5 mutant background (B). Data are means ± se from three or more independent experiments. n.d., Not determined. C and D, Size (C) and number (D) of green plastids in primary root cells. The horizontal line represents the median; the top and bottom of boxes represent the upper and lower quartiles, respectively; and whiskers represent the range (C, n > 900; D, n > 80). ***, P < 0.001 by Student’s t test. E and F, Expression of photosynthesis-associated genes in intact and detached roots of the gnc gnl double mutant (E) and intact roots of GNCox and GNLox (F). Data are means ± se fold difference from the intact wild-type root samples after normalizing to ACTIN8 (n = 3). G, Plastid DNA content in intact and detached root cells. The content of plastid DNA relative to nuclear DNA was determined by real-time PCR-based quantification of DNA content with rpoB-specific (plastid-encoded) and ACTIN8-specific (nucleus-encoded) primers. Data are means ± se fold difference from intact wild-type root samples (n = 3). Samples at 21 d old were used for all experiments. Different letters indicate significant differences by the Tukey-Kramer multiple comparison test (P < 0.05). Asterisks indicate significant differences from the wild type of each condition (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test after a Bonferroni correction for multiple comparisons).
We also investigated the effect of the overexpression of GNC (GNCox) and GNL (GNLox) on root greening. As reported previously (Chiang et al., 2012), overexpression of GNC and GNL greatly enhanced Chl accumulation in intact roots (Fig. 5B). Because enhanced Chl accumulation was already observed in roots of 10-d-old GNCox seedlings (Chiang et al., 2012), overexpression of these genes would increase the rate of Chl accumulation during root development. In these overexpression lines, the size and number of green plastids were increased in cells of the mature region of the primary root (Fig. 5, C and D; Supplemental Fig. S3A), which indicates the ectopic activation of chloroplast development in intact roots of these lines. In fact, Chiang et al. (2012) revealed that overexpression of GNC or GNL caused ectopic chloroplast development in the cortex in addition to increased chloroplast number in the pericycle cells in roots. Similarly, cells of detached wild-type roots showed increased size and number of green plastids.
We next examined whether GNC and GNL are involved in the transcriptional regulation of photosynthesis-associated genes in roots. Intact roots of the gnc gnl double mutant showed no or only slightly decreased expression of photosynthesis-associated genes compared with intact wild-type roots (Fig. 5E). After shoot removal, the gnc gnl mutant showed increased expression of these genes (Fig. 5E), but to a lesser extent than the wild type (Fig. 2). By contrast, overexpression of these GATA factors, particularly GNLox, substantially increased the expression of photosynthesis-associated genes in intact roots (Fig. 5F). As an exception, HEMA1 was not up-regulated in GNCox and GNLox roots, although its expression was induced by shoot removal via ARR1- and ARR12-mediated cytokinin signaling (Supplemental Fig. S2).
Because transcript levels of plastid-encoded genes may be linked with plastid DNA abundance in cells, we analyzed plastid DNA content in roots by quantitative PCR analysis. In the wild type, plastid DNA content did not differ largely in detached and intact roots (Fig. 5G; Supplemental Fig. S3B). These data suggest that the increased expression of plastid-encoded genes in detached wild-type roots is not attributed to the increased number of plastid genome copies but reflects a specific activation of plastid gene expression. Meanwhile, plastid DNA content was increased greatly in intact roots of GNCox and GNLox lines. The plastid DNA content was similar between these two lines (Fig. 5G), although GNLox roots showed much stronger expression of plastid-encoded genes than GNCox roots (Fig. 5F).
In contrast to GLKs and GATA factors, which are important but not essential for root greening, HY5 is indispensable for Chl accumulation in roots, as illustrated by a complete albino phenotype of hy5 mutant roots (Oyama et al., 1997; Usami et al., 2004; Kobayashi et al., 2012a). Moreover, we reported previously that overexpression of GLKs in the hy5 mutant background only slightly increased Chl accumulation in roots (Kobayashi et al., 2012a), which suggests that GLK factors require HY5 to maximize their function in the root. Thus, we evaluated the role of HY5 in root greening after shoot removal. The hy5 mutant did not accumulate Chl even in detached roots (Fig. 5A). Moreover, overexpression of GNC or GNL in the hy5 background (hy5 GNCox and hy5 GNLox) did not increase Chl content in roots (Fig. 5B). These data demonstrate that HY5 also is essential for the conditional root greening mediated by GATA factors.
B-GATAs Improve Photosynthesis in Roots
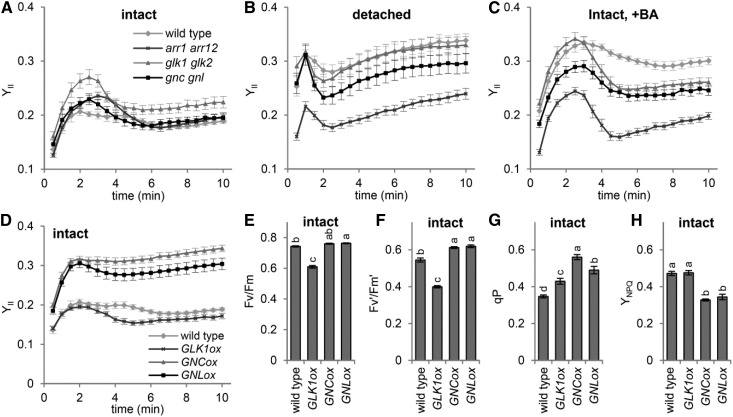
To examine the involvement of GLK and B-GATAs in regulating root photosynthesis, we analyzed the photosynthetic parameters of roots in glk1 glk2 and gnc gnl mutants along with the wild type and the arr1 arr12 mutant using an imaging PAM fluorometer. In this experiment, 28-d-old root samples were used to obtain sufficient Chl fluorescence signals for the PAM analysis. In intact roots, YII values were similar for all mutants and the wild type (Fig. 6A). In detached roots grown for 7 d without shoots, the wild type showed increased YII, whereas arr1 arr12 showed no remarkable increase in YII (Fig. 6B), which confirms the results in Figure 4. The glk1 glk2 double mutant showed a similar YII profile to the wild type in detached roots, so GLKs are not required for the remodeling of root photosynthesis on shoot removal. The gnc gnl roots also showed increased YII on shoot removal, although the values appeared slightly lower than that for the wild type. We also evaluated the effect of BA treatment on root photosynthesis. Consistent with the data in Figure 4, BA treatment increased the YII of roots in the wild type but not in arr1 arr12 (Fig. 6C). In both the glk1 glk2 and gnc gnl mutants, root YII was increased only moderately by BA treatment, so these factors are not essential but are partially important for the cytokinin-induced remodeling of root photosynthesis.
Figure 6.
Involvement of transcription factors in root photosynthesis in response to shoot removal. A to D, YII in intact (A and D), detached (B), and BA-treated intact (C) roots of each line. E to H, Photosynthetic parameters in intact roots of overexpression lines of each transcription factor compared with intact wild-type roots. Data are means ± se (n > 12). Slow induction kinetics under actinic light are shown for YII (A–D), and other data (E–H) were obtained after actinic light exposure (110 µmol photons m−2 s−1) for 10 min. Samples at 21 d old were used for all experiments. Different letters indicate significant differences (P < 0.05, Tukey-Kramer multiple comparison test).
We also analyzed photosynthetic activity in roots of GNCox and GNLox lines and the GLK1 overexpression line (GLK1ox). As reported previously (Kobayashi et al., 2013), in GLK1ox, Chl accumulation in roots was not accompanied by improved photosynthetic efficiency and even decreased Fv/Fm and Fv′/Fm′ (Fig. 6, D–F). By contrast, in intact roots, YII was higher for GNCox and GNLox than for the wild type (Fig. 6D). Higher YII in GNCox and GNLox roots was attributed mainly to the increased fraction of open PSII represented by higher qP and the decreased nonphotochemical energy dissipation represented by lower YNPQ (Fig. 6, G and H), as was observed in detached wild-type roots (Fig. 4A).
Auxin Signaling Regulates Root Greening Independently of Type B ARR Signaling
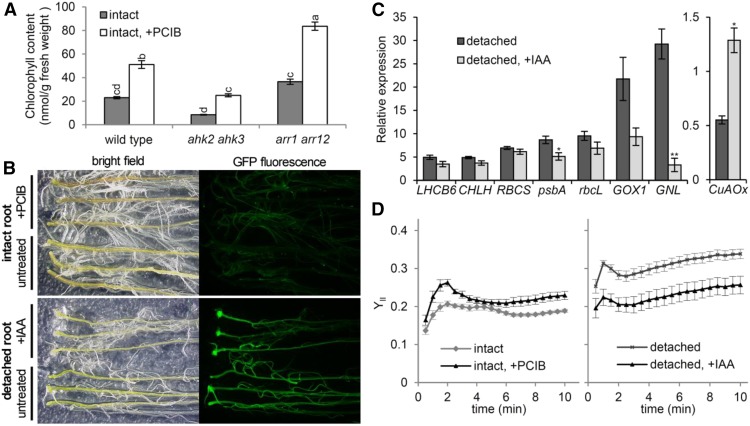
We reported previously that auxin is involved in the negative regulation of chloroplast development in roots (Kobayashi et al., 2012a). To examine the interaction of auxin and cytokinin signaling during the root greening response, we treated cytokinin mutants with an auxin signaling inhibitor, p-chlorophenoxyisobutyric acid (PCIB), and measured Chl content in roots. As reported previously (Kobayashi et al., 2012a), PCIB treatment increased Chl content in intact wild-type roots (Fig. 7A). Similarly, PCIB increased root Chl content in ahk2 ahk3 and arr1 arr12 mutants. Moreover, PCIB treatment did not increase type B ARR-mediated GFP fluorescence in intact roots of the TCSn::GFP line (Fig. 7B). These data indicate that the inhibition of auxin signaling by PCIB induces root greening independently of type B ARR-mediated cytokinin signaling. As well, the enhanced TCSn::GFP fluorescence in detached roots was not suppressed by treatment with an auxin, 1 μm IAA (Fig. 7B), which suggests no inhibitory effect of auxin on type B ARR activation during root greening. Thus, auxin signaling regulates Chl accumulation in roots independently of type B ARR activities.
Figure 7.
Auxin regulates root greening independent of cytokinin signaling. A, Chl content in intact roots of the wild type and cytokinin signaling mutants in the absence (intact) or presence of 10 μm PCIB (intact, +PCIB). Different letters indicate significant differences (P < 0.05, Tukey-Kramer multiple comparison test). B, GFP fluorescence in roots of the TCSn::GFP line. For the top images, roots of 21-d-old intact seedlings were grown in the absence (untreated) or presence of 10 μm PCIB for 7 d. Aerial parts were detached just before observation. For the bottom images, 21-d-old root samples were detached from 14-d-old seedlings and grown on agar medium containing no or 1 μm indole-3-acetic acid (IAA). C, Gene expression analysis in 21-d-old detached root samples treated with or without 1 μm IAA for 7 d. Data are means ± se fold difference from the 21-d-old intact wild-type root samples after normalizing to ACTIN8 (n = 3). Asterisks indicate significant differences from the untreated control (*, P < 0.05 and **, P < 0.01, Student’s t test). D, Slow induction kinetics of YII in intact roots of 28-d-old seedlings grown in the absence or presence of 10 μm PCIB for 7 d (left graph) and root samples detached from 21-d-old seedlings and grown on agar medium containing no or 1 μm IAA for 7 d (right graph). Data are means ± se (n > 12). Actinic light intensity was 110 µmol photons m−2 s−1.
Next, we examined whether exogenous auxin affects the expression of photosynthesis-associated genes during the greening of detached roots. IAA treatment suppressed the expression of psbA and GOX1, with a decreasing trend for LHCB6, CHLH, and rbcL expression in detached roots (Fig. 7C). Of note, IAA treatment inhibited the up-regulated GNL expression in detached wild-type roots. It also inhibited the suppressed CuAOx expression in roots with shoot removal. These data suggest that auxin signaling strongly affects the expression of some genes in detached roots.
We also evaluated photosynthetic activity in intact roots treated with PCIB and detached roots treated with IAA (Fig. 7D). PCIB treatment slightly increased YII in intact roots, but to a lesser extent than BA (Fig. 6C). Meanwhile, IAA treatment partially suppressed the increased YII in roots with shoot removal (Fig. 7D). These data suggest that auxin signaling functions to repress photosynthetic activation in roots in contrast to cytokinin signaling.
DISCUSSION
Root Greening as a Wounding Response via Cytokinin Signaling
We previously reported that the ahk2 ahk3 double mutant for cytokinin receptors showed reduced Chl accumulation in intact roots (Kobayashi et al., 2012a), which suggests the requirement for cytokinin signaling in chloroplast development in roots. By contrast, the type B arr mutants showed no reduction in Chl content in intact roots (Fig. 1B). Because the roles of the type B ARR family genes are highly redundant (Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008), the remaining ARR members may compensate for the loss of each ARR in mutants in terms of Chl accumulation in intact roots. Alternatively, cytokinin pathways may function in Chl accumulation in intact roots independently of type B ARR. Considering that the intact roots of the arr1 arr12 double mutant had higher Chl content (Fig. 1B) and higher expression of photosynthesis-associated genes than the wild type (Fig. 2A), complex mechanisms downstream of cytokinin reception would be involved in regulating chloroplast differentiation in intact roots. Cortleven et al. (2016) recently reported that the arr1 arr10 arr12 triple mutant showed an up-regulation of Chl biosynthesis genes and an increased greening rate of cotyledons during the etioplast-chloroplast transition, which is consistent with our data that loss of both ARR1 and ARR12 even increased Chl content in intact roots. An involvement of an as yet unknown compensatory mechanism in the greening phenotype of multiple arr mutants is assumed (Cortleven et al., 2016), and future investigations are required to elucidate the mechanism.
Meanwhile, the arr1 arr12 double mutant showed strongly inhibited conditional Chl accumulation in detached roots (Fig. 1B), so cytokinin signaling through ARR1 and ARR12 is specifically required for root greening after shoot removal. Because the dominant repression of WIND1 signaling by the WIND1-SRDX chimeric protein also partially impaired the greening of detached roots (Fig. 1D), WIND1-mediated wounding signaling may be involved in this regulation. WIND1 is an AP2/ERF transcription factor whose expression is strongly up-regulated within a few hours of wounding (Iwase et al., 2011a). WIND1, which plays a central role in regulating cell dedifferentiation after wounding, activates type B ARR-mediated cytokinin signaling at the wounding site, which is necessary for callus induction in response to wounding (Iwase et al., 2011a, 2011b). In fact, the dominant WIND1 repression in WIND1-SRDX inhibited type B ARR-mediated TCS::GFP expression in response to shoot removal (Fig. 1C). A similar result was observed in WIND1-SRDX hypocotyl explants, with repressed TCS::GFP expression and Chl accumulation (Iwase et al., 2011a). Thus, the WIND1-mediated wounding response would activate type B ARR signaling, thereby inducing Chl accumulation in detached roots (Fig. 8). However, forced overexpression of WIND1 causing degreening of green tissues reflects the ability of WIND1 to induce cell dedifferentiation and callus formation (Iwase et al., 2011a, 2011b).
Figure 8.
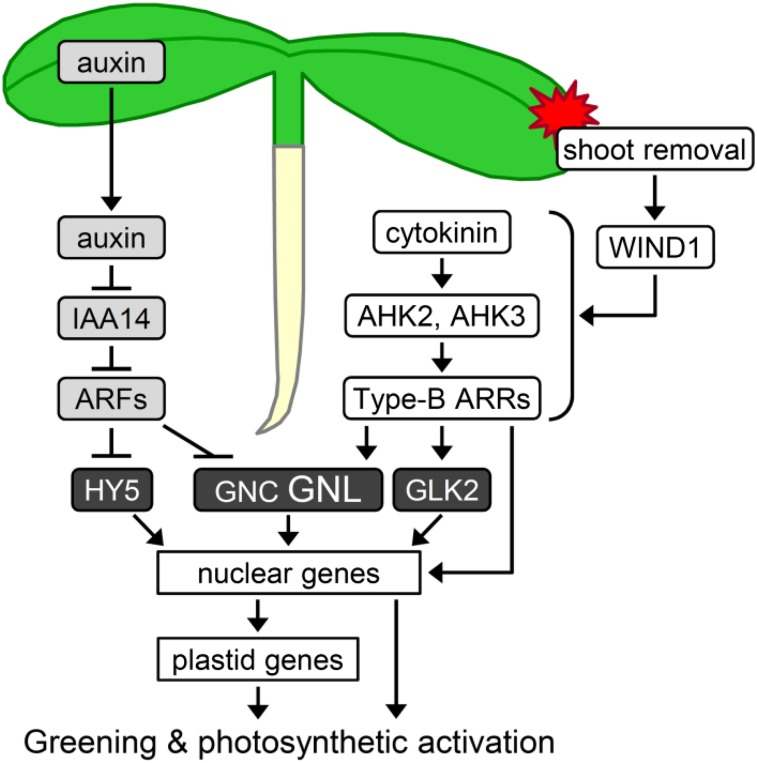
Model of the regulatory pathways of root greening after shoot removal. Chloroplast development in roots is positively regulated by cytokinin signaling via the His kinases AHK2 and AHK3 and negatively regulated by auxin signaling via INDOLE-3-ACETIC ACID INDUCIBLE14 (IAA14) and AUXIN RESPONSE FACTORs (ARFs). In intact roots, auxin transported from the shoot represses chloroplast development, but in detached roots, the negative auxin regulation is attenuated, and the wound-responsive transcription factor WIND1 is induced in response to shoot removal and activates the type B ARR-mediated cytokinin signaling pathway. Several transcription factors associated with chloroplast development (HY5, GNC, GNL, and GLK2) function downstream of auxin/cytokinin signaling to directly and indirectly up-regulate nucleus- and plastid-encoded genes associated with chloroplast development. Type B ARRs also directly regulate some photosynthesis-associated nuclear genes and induce Chl accumulation and photosynthetic activation in roots. Arrows and bars represent positive and negative regulation, respectively.
Consistently, artificial induction of WIND1 expression for 24 h even slightly decreased root Chl content (Fig. 1E). Moreover, despite strong type B ARR activities, calluses formed at the cutting site of some detached roots accumulated less Chl (Supplemental Fig. S1), as was observed in calluses formed in etiolated hypocotyls with strong WIND1 expression (Iwase et al., 2011a). Considering the fact that induction levels of WIND1 are strongly linked with dedifferentiation levels of tissues (Iwase et al., 2011a), there may be an optimum or threshold level of WIND1 in determining cell fate. Strict spatiotemporal regulation of WIND1 expression in response to shoot removal may be important for promoting callus formation and root greening at each specific site separately. We note that enhanced cytokinin signaling would not solely cause the degreening of dedifferentiating cells in callus or callus-like cell masses, because exogenous cytokinin treatment does not induce degreening but enhances Chl accumulation and photosynthesis in roots (Fig. 4; Kobayashi et al., 2012a). Some other factors induced by WIND1 at the callus-forming site may promote the degreening of dedifferentiating cells together with or independently of cytokinin signals.
Transcription Factors Functioning in Root Greening Downstream of Cytokinin Signaling
We showed that shoot removal substantially up-regulated GNL expression in wild-type roots (Fig. 2B). However, its up-regulation in detached roots was strongly inhibited in the arr1 arr12 double mutant, which suggests that shoot removal up-regulates GNL via type B ARR signaling. This result agrees with the finding that up-regulation of GNL by exogenous cytokinin treatment was impaired in the arr1 arr12 double mutant (Chiang et al., 2012). Moreover, overexpression of GNL or its paralog, GNC, caused ectopic chloroplast differentiation in the hypocotyl and roots (Chiang et al., 2012). In fact, we confirmed strong Chl accumulation, remarkable up-regulation of photosynthesis-associated genes, and increased number and size of green plastids in roots with GNL and GNC overexpression as in detached wild-type roots (Fig. 5). Therefore, activated type B ARR signaling by shoot removal likely up-regulates GNL expression, thereby inducing chloroplast development in detached roots (Fig. 8). However, the gnc gnl double mutant still showed increased Chl accumulation and photosynthetic gene expression in detached roots, despite lower levels than in the wild type (Fig. 5, A and E), so these GATA factors are important but not crucial for the greening of detached roots. In addition to GNC and GNL, four other similar B-GATA paralogs exist in Arabidopsis and function redundantly in part with GNC and GNL in the leaf greening process (Ranftl et al., 2016). Thus, the remaining B-GATAs in the gnc gnl mutant may compensate in part for the function of GNC and GNL in the root greening response.
In contrast to the conditional requirement of B-GATAs after shoot removal, GLK factors are required for Chl accumulation in both intact and detached roots (Fig. 5A). Moreover, detached roots of the glk1 glk2 double mutant showed no remarkable differences from the wild type in photosynthetic activity (Fig. 6B). These data suggest that GLKs are not specific for the root greening response after shoot removal. However, GLK2, which has a more important role in root Chl accumulation than GLK1 (Kobayashi et al., 2012a), was up-regulated in roots by cytokinin treatment (Kobayashi et al., 2012a). In addition, the glk1 glk2 mutant showed lower response to BA treatment in terms of root photosynthesis activity than the wild type (Fig. 6C). Thus, in addition to the GATA factors, GLK2 may contribute in part to the conditional root greening downstream of cytokinin signaling. In addition, ARR10 and ARR12 were recently shown to bind directly to the promoter regions of HEMA1 and LHCB6 (Cortleven et al., 2016). Of note, the HEMA1 expression in roots was induced by shoot removal via ARR1 and ARR12 but not by the overexpression of GNC and GNL (Supplemental Fig. S2). Thus, type B ARRs may directly up-regulate some photosynthesis-associated nuclear genes in roots in response to shoot removal.
We previously reported that complete loss of Chl in the hy5 mutant was not rescued by increased cytokinin signaling or inhibited auxin signaling (Kobayashi et al., 2012a). Moreover, overexpression of GLKs only slightly increased root Chl content in the hy5 background. Consistent with these results, shoot removal as well as overexpression of GNC and GNL did not increase root Chl content in the hy5 mutant (Fig. 5B). These data indicate that HY5 acts as a prerequisite for Chl accumulation in roots and that B-GATAs function thoroughly depending on HY5. Although Chl accumulation in the root is strictly regulated in a development- and region-dependent manner, HY5 protein globally accumulates in all root regions independently of developmental stage (Kobayashi et al., 2012a; Kobayashi and Masuda, 2013). Thus, HY5 widely accumulated in roots may support other factors, such as GLKs and B-GATAs, to regulate chloroplast development in specific areas and under particular conditions.
Up-Regulation of Photosynthesis-Associated Genes in Roots by Shoot Removal
In detached roots, all photosynthesis-associated nuclear genes investigated were strongly up-regulated in the wild type but not in the arr1 arr12 mutant (Fig. 2A), so ARR1 and ARR12 mediate the up-regulation of these genes after shoot removal. The expression of GOX1 and GLDP1, which are associated with the photorespiratory metabolism in mitochondria and peroxisomes, respectively, also was increased in detached roots in an ARR1- and ARR12-dependent manner (Fig. 2D), whereas the expression patterns of root-specific genes after shoot removal varied largely among genes (Fig. 2E). Thus, genes associated with photosynthesis, including the processes outside chloroplasts, may be the main targets of ARR1- and ARR12-mediated transcriptional remodeling during the root greening response.
In addition to the nuclear genes, plastid-encoded photosynthetic genes also were up-regulated via the ARR1- and ARR12-mediated signaling pathway (Fig. 2A). Our data are consistent with the finding that BA treatment of detached barley (Hordeum vulgare) leaves activates chloroplast transcription in the light (Zubo et al., 2008). Because ARR proteins are localized to the nucleus (Sakai et al., 2000; Hwang and Sheen, 2001), other factors acting downstream of cytokinin signaling would induce the expression of plastid-encoded genes. SIGs, which are encoded in the nucleus but function in plastids as transcription initiation factors of the PEP complex (Schweer et al., 2010), may be involved in the up-regulation of plastid-encoded photosynthetic genes. Indeed, many SIG genes, particularly SIG4, were up-regulated in roots on shoot removal via ARR1 and ARR12 (Fig. 2C). Consistent with these data, the expression of SIG4 was increased substantially in GNLox roots along with plastid-encoded psbA and rbcL (Fig. 5F), but induction of these genes in detached roots in the gnc gnl double mutant was weakened (Fig. 5E). These data suggest that GNL activates plastid gene expression downstream of ARR signaling during root greening. Of note, shoot removal increased the levels of plastid-encoded genes in roots without increasing plastid DNA content (Figs. 2A and 5G), which suggests that photosynthetic gene loci in plastid nucleoids are transcriptionally activated in detached root cells. In addition, the induction of plastid-encoded genes was stronger with GNLox than GNCox in roots (Fig. 5F), although these lines showed similar plastid DNA contents (Fig. 5G). Thus, unlike GNC, GNL can enhance the transcriptional activity of the photosynthetic gene loci in plastid nucleoids besides increasing plastid number and plastid genome copies in roots. B-GATAs may act on chloroplast-related genes in an indirect manner (Hudson et al., 2011), and the mode of function of B-GATAs in photosynthesis gene expression awaits future investigations.
Shoot Removal Improves Photosynthetic Electron Transport in Roots via Cytokinin Signaling
In addition to the increased Chl content and number and size of chloroplasts, photosynthetic efficiency was increased in wild-type roots on shoot removal (Fig. 4A), which indicates the quantitative and qualitative development of chloroplasts in detached roots. The mean FPSI/FPSII ratio was greater in detached roots than in intact roots (Fig. 3B), so light energy input to PSI is increased in root chloroplasts on shoot removal. This result is consistent with the data that electron transport around PSI was modified in root chloroplasts on shoot removal (Fig. 3A). Indeed, the PSII acceptor side limitation, represented by low qP, in root chloroplasts was improved to the leaf level in detached roots (Fig. 4A). In parallel to or as a result of the sufficient electron transport to PSI, thermal dissipation of light energy (YNPQ) decreased to the leaf level in detached roots. Thus, as shown by the change in FPSI/FPSII ratio, rearrangement of light energy input between PSII and PSI may improve the efficiency of electron transport downstream of PSII and decrease thermal energy dissipation within PSII in detached root chloroplasts. Because psaA, psbA, and rbcL were markedly up-regulated along with rpoB and SIGs in roots with shoot removal (Fig. 2), the global induction of plastid-encoded photosynthetic genes may increase photosynthetic efficiency in root chloroplasts. By contrast, the slow electron transport within PSII (Fig. 3A) as well as the low Fv/Fm (Fig. 4A) in root chloroplasts was not improved by shoot removal. Therefore, certain components in PSII may remain less efficient in roots even after shoot removal.
In wild-type roots, cytokinin treatment improved photosynthetic efficiency similar to that with shoot removal (Fig. 4A). By contrast, the photosynthetic modifications with shoot removal and cytokinin treatment were inhibited in arr1 arr12 roots (Fig. 4B). Moreover, the shoot removal-induced up-regulation of photosynthetic genes was strongly impaired in detached arr1 arr12 roots (Fig. 2). Therefore, cytokinin signaling via ARR1 and ARR12 plays a crucial role in the quality control of root chloroplasts and photosynthetic activities. In addition, GNC and GNL overexpression greatly improved photosynthetic efficiency in roots (Fig. 6). Both GNCox and GNLox increased the expression of plastid-encoded genes to similar or higher levels compared with nucleus-encoded photosynthetic genes (Fig. 5E), which may help to balance the amount of photosynthetic electron transport components. However, the effect of GNLox on photosynthetic gene expression was much stronger than that of GNCox, although Chl accumulation and photosynthetic activities in roots were similar between these two lines. Thus, the up-regulation of photosynthetic genes in GNCox may be sufficient to induce the remodeling of photosynthetic activity in roots. In contrast to GNCox and GNLox roots, GLK1ox roots showed lower PSII efficiency than wild-type roots, which may be attributed to the unbalanced photosynthetic machinery caused by the excessive expression of GLK1-targeted genes associated specifically with peripheral light-harvesting antennae (Waters et al., 2009; Kobayashi et al., 2013).
Involvement of Auxin Signaling in Root Greening after Shoot Removal
Our previous study suggested that auxin transported from the shoot, absent in detached roots, represses root greening (Kobayashi et al., 2012a). In fact, exogenous IAA treatment partially inhibited the up-regulation of photosynthesis-associated genes and photosynthetic remodeling in detached roots (Fig. 7, C and D). By contrast, inhibition of auxin signaling by PCIB slightly increased photosynthetic efficiency in intact roots (Fig. 7D). These data suggest that auxin signaling negatively regulates photosynthetic activity in roots. This regulation by auxin is independent of type B ARR activities (Fig. 7, A and B). However, IAA treatment strongly repressed the GNL expression in detached roots (Fig. 7C). GNC and GNL are negatively regulated by auxin signaling, and the expression of GNC and GNL is increased in some mutants defective in auxin signaling (Richter et al., 2013), including solitary root1, which has greenish roots (Kobayashi et al., 2012a). Thus, B-GATAs may function at the point of convergence of cytokinin and auxin signaling on chloroplast development in roots.
CONCLUSION
In combination with previous studies (Kobayashi et al., 2012a), we propose a model of regulatory pathways of root greening after shoot removal (Fig. 8). In intact seedlings, auxin transported from the shoot represses HY5 protein accumulation and GNL gene expression via IAA14 and its regulatory targets the ARFs in roots, which leads to the suppression of chloroplast-associated genes. After shoot removal, chloroplast-associated genes are released from the suppression by shoot-derived auxin. At the same time, shoot removal activates type B ARR-mediated cytokinin signaling in roots via the wound-responsive transcription factor WIND1. The cytokinin receptors AHK2 and AHK3 are essential for the root greening response, although the mechanism intertwining the wounding signaling by WIND1 and cytokinin signaling via AHKs remains elusive (Iwase et al., 2011a). Activated type B ARRs, particularly ARR1 and ARR12, strongly induce GNL and moderately induce GNC and GLK2 and also may directly increase the expression of chloroplast-associated nuclear genes (Cortleven et al., 2016). The up-regulation of nuclear genes associated with photosynthesis and plastid transcription is accompanied by an increased expression of plastid-encoded genes, which together result in Chl accumulation and photosynthetic activation in detached roots.
Land plants are at constant risk of serious damage or removal of photosynthetic tissues by physical attacks from herbivores and pathogens and by abiotic stresses such as heat and drought. We reported previously that root photosynthesis can contribute to carbon assimilation in Arabidopsis (Kobayashi et al., 2013). Thus, the root greening system regulated by cytokinin and auxin may function as a survival strategy of land plants upon the loss of main photosynthetic tissues.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia ecotype. ahk2-2 ahk3-3 (Higuchi et al., 2004), arr1-3, arr12-1, arr1-3 arr10-5, arr1-3 arr11-2, and arr1-3 arr12-1 (Mason et al., 2005; Argyros et al., 2008), TCS::GFP (Müller and Sheen, 2008), TCSn::GFP (Zürcher et al., 2013), WIND1-SRDX and XVE-WIND1 (Iwase et al., 2011a), glk1 glk2 (Fitter et al., 2002), GLK1ox (Waters et al., 2009), gnc gnl, GNCox, and GNLox (Chiang et al., 2012), and hy5-215 (Oyama et al., 1997) were described previously. The hy5 GNCox and hy5 GNLox lines were generated by crossing GNCox and GNLox with the hy5-215 mutant and segregating double homozygous lines. Plants were grown on Murashige and Skoog (MS) medium (adjusted to pH 5.7 with KOH) containing 1% (w/v) Suc solidified with 0.7% (w/v) Gelrite (Wako) on vertical plates at 23°C under continuous white light (40 µmol photons m−2 s−1). For analyses of detached roots, roots were excised from 14-d-old seedlings at the root-hypocotyl junction and further incubated for 7 d on MS medium under continuous white light. Intact roots of 21-d-old seedlings were used as the untreated control. For the imaging PAM analysis in Figures 6 and 7D, roots excised from 21-d-old seedlings were incubated for 7 d and used for analysis along with 28-d-old seedling controls. For the experiment in Figure 1E, 21-d-old XVE-WIND1 seedlings were transferred to MS medium containing 10 µm 17β-estradiol. For treatment with 1 µm BA, 1 µm IAA, or 10 µm PCIB, detached roots or intact seedlings of 14- or 21-d-old plants were transferred to MS medium containing each compound and grown for another 7 d. For experiments in Figures 3 and 4, leaves of 21-d-old Arabidopsis wild type were considered as photosynthetically competent tissues.
GFP Fluorescence Detection
The GFP fluorescence of TCS::GFP and TCSn::GFP roots was observed with a stereomicroscope (MZ 16FA; Leica) with a band-pass filter set (GFP Plants; Leica) and a color CCD camera (VB-7010; Keyence) in darkness. All images were obtained with the same microscope settings and CCD camera to compare fluorescence intensity fairly among samples. Background signals in blue and red channels were eliminated from images with the use of ZEN 2011 (Carl Zeiss), and brightness was linearly adjusted to the same extent for all images. We analyzed 21-d-old TCS::GFP and TCSn::GFP seedlings as the untreated control along with detached roots. In the untreated control, shoots were removed just before observation to clarify the fluorescence around the root-hypocotyl junction.
Real-Time PCR Analysis of Gene Expression
Total RNA was extracted by use of the RNeasy Plant Mini kit (Qiagen). Genomic DNA digestion and reverse transcription to obtain cDNA involved the use of the PrimeScript RT Reagent kit with gDNA Eraser (TaKaRa Bio). cDNA amplification involved the use of the Thunderbird PreMix kit (Toyobo) and 200 nm gene-specific primers listed in Supplemental Table S1. Signal detection and quantification involved the use of a MiniOpticon real-time PCR machine (Bio-Rad) as described (Kobayashi et al., 2013).
Real-Time PCR Analysis of Plastid DNA Content
Total DNA was extracted from root samples by use of the DNeasy Plant Mini kit (Qiagen). Plastid and nuclear DNA were amplified using rpoB-specific primers and ACTIN8-specific primers, respectively (Supplemental Table S1), by use of the Thunderbird PreMix kit (Toyobo) and the CFX Connect Real-Time System (Bio-Rad). Cycle threshold values of the amplified plastid DNA signals were normalized to those of nuclear DNA signals, and plastid DNA content in each root sample relative to that of intact wild-type roots was calculated by a PCR-based mathematical model (Pfaffl, 2001). The same result was obtained with the use of psaA-specific primers for plastid DNA and the use of CHLH-specific primers for nuclear DNA (Supplemental Fig. S3B). Three independent biological experiments were carried out for each sample.
Chl Determination and Photosynthetic Measurement
Spectroscopic Chl determination and kinetic analysis of transient Chl fluorescence induction were performed by measuring Chl fluorescence directly from root segments as described (Kobayashi et al., 2013). Fluorescence transients were normalized between minimal and maximal Chl fluorescence to range from 0 to 1.
To measure fluorescence emission spectra of Chl proteins at 77K (Fig. 4B), root samples were pulverized in liquid nitrogen and homogenized in a cold buffer (0.33 m sorbitol, 5 mm MgCl2, 5 mm EDTA, and 50 mm HEPES-KOH, pH 7.6). The homogenate was filtered through a single layer of Miracloth (Calbiochem) with gentle hand pressure. After centrifugation at 5,000g for 10 min at 4°C, the supernatant was discarded and the pellet was resuspended in a cold buffer to obtain 2 μg mL−1 Chl-containing membrane fractions. We should note the possibility that PSI-enriched small membrane fragments may remain in the supernatant under this mild centrifugation condition, which may slightly affect the FPSI/FPSII ratio. Chl fluorescence spectra of the membrane fractions from 620 to 800 nm were measured in liquid nitrogen by the use of an RF-5300PC spectrofluorometer (Shimadzu) under 435-nm excitation.
In Figure 4, photosynthetic parameters were analyzed by the use of a fiber-optic PAM fluorometer (Junior-PAM; Heinz Walz). The automated induction program provided by the WinControl-3 software (Heinz Walz) was used with default settings to measure light and saturation pulse. Single leaves or batches of 1-cm segments of primary roots from the root-hypocotyl junction were dark incubated for 15 min on the leaf clip. After measuring minimum Chl fluorescence (Fo) in the dark, maximal Chl fluorescence (Fm) was determined with a saturating pulse. Then, actinic light (420 µmol photons m−2 s−1) was supplied for 10 min with recording of stationary fluorescence (F), and saturating pulses were given every 20 s to determine maximum fluorescence (Fm′) under the actinic light. In Figures 6 and 7D, the IMAGING-PAM MAXI fluorometer and ImagingWin software (Heinz Walz) were used to analyze photosynthetic parameters under 110 µmol photons m−2 s−1 actinic light with saturating pulses given every 30 s. Photosynthetic parameters were calculated as follows (Maxwell and Johnson, 2000): Fo′ = Fo/(Fv/Fm + Fo/Fm′), Fv/Fm = (Fm − Fo)/Fm, Fv′/Fm′ = (Fm′ − Fo′)/Fm′, YII = (Fm′ − F)/Fm′, and qP = (Fm′ − F)/(Fm′ − Fo′). YII can be transformed into a product of qP and Fv′/Fm′: YII = (Fm′ − F)/Fm′ = qP × Fv′/Fm′. YNPQ and YNO were determined as described (Kramer et al., 2004).
Root Plastid Observation
Primary roots cut between 1 and 1.5 cm from the root-hypocotyl junction were vacuum infiltrated in an enzyme solution containing 2% (w/v) cellulase, 1% (w/v) macerozyme, 0.2% (w/v) pectolyase, 2 mm CaCl2, 10 mm KCl, 500 mm d-mannitol, and 5 mm MES-KOH (pH 5.8). Then, root samples were incubated in the same enzyme solution on a glass slide at room temperature for 3 h to digest cell walls. After fixing with 3.5% (w/v) glutaraldehyde, cells were loosened from tissues by tweezers and gently covered with a cover glass. The number of green plastids was counted in released or loosened cells with a light microscope (BX50; Olympus). Cells used for the measurement were selected randomly, but those obviously containing no plastids were eliminated from the data. Differential interference contrast images of the cells were taken with a digital camera (MC120 HD; Leica) on the BX50 microscope, and the size of green plastids was measured using ImageJ (https://imagej.nih.gov/ij/).
Accession Numbers
Sequence data from this article can be found in TAIR under the following accession numbers: CHLH (AT5G13630), CuAOx (AT1G31710), GLDP1 (AT4G33010), GLK1 (AT2G20570), GLK2 (AT5G44190), GNC (AT5G56860), GNL (AT4G26150), GOX1 (AT3G14420), HEMA1 (AT1G58290), HEMA2 (AT1G09940), LHCA4 (AT3G47470), LHCB6 (AT1G15820), psaA (ATCG00350), psbA (ATCG00020), rbcL (ATCG00490), RBCS (AT1G67090), rpoB (ATCG00190), RPOTp (AT2G24120), RPOTmp (AT5G15700), SIG1 (AT1G64860), SIG2 (AT1G08540), SIG3 (AT3G53920), SIG4 (AT5G13730), SIG5 (AT5G24120), and SIG6 (AT2G36990).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Fluorescence microscopy of roots from TCS::GFP and TCSn::GFP transgenic lines.
Supplemental Figure S2. Quantitative RT-PCR analysis of mRNA expression of HEMA1 and HEMA2 encoding glutamyl-tRNA reductase.
Supplemental Figure S3. Chloroplast development in detached wild-type roots and intact roots of GNCox and GNLox plants.
Supplemental Table S1. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank G. Eric Schaller (Department of Biological Sciences, Dartmouth College) for supplying the gnc gnl, GNCox, and GNLox lines, Krishna K. Niyogi (Department of Plant and Microbial Biology, University of California, Berkeley) for providing access to equipment for photosynthetic measurements, and Chika Ikeda (RIKEN Center for Sustainable Resource Science) for technical assistance.
Glossary
- Chl
chlorophyll
- PAM
pulse amplitude modulation
- qP
coefficient of photochemical quenching
- BA
6-benzyladenine
- PCIB
p-chlorophenoxyisobutyric acid
- IAA
indole-3-acetic acid
- MS
Murashige and Skoog
Footnotes
This work was supported by the Japan Society for the Promotion of Science (grant nos. JP24770055, JP26711016, JP24570042, and JP16K07393).
References
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer C, Schwechheimer C (2015) B-GATA transcription factors: insights into their structure, regulation, and role in plant development. Front Plant Sci 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Schmülling T (2012) Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A, Marg I, Yamburenko MV, Schlicke H, Hill K, Grimm B, Schaller GE, Schmulling T (2016) Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol 172: 464–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18: 477–489 [DOI] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu U, Morgenthal K, Weckwerth W, Pärnik T, Keerberg O, Bauwe H (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M (2006) Arabidopsis thaliana: a model organism to study plant peroxisomes. Biochim Biophys Acta 1763: 1382–1391 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi YM, Rothstein SJ (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Guevara DR, Hand AJ, Xu Z, Hao L, Chen X, Zhu T, Bi YM, Rothstein SJ (2013) Rice cytokinin GATA transcription factor1 regulates chloroplast development and plant architecture. Plant Physiol 162: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mita K, Nonaka S, Ikeuchi M, Koizuka C, Ohnuma M, Ezura H, Imamura J, Sugimoto K (2015) WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J Plant Res 128: 389–397 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011a) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Iwase A, Ohme-Takagi M, Sugimoto K (2011b) WIND1: a key molecular switch for plant cell dedifferentiation. Plant Signal Behav 6: 1943–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Kamada T, Nito K, Hayashi H, Mano S, Hayashi M, Nishimura M (2003) Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol 44: 1275–1289 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keränen M, Aro EM, Fukaki H, Ohta H, Sugimoto K, et al. (2012a) Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Fujii S, Sasaki D, Baba S, Ohta H, Masuda T, Wada H (2014) Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front Plant Sci 5: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T (2013) Spatial and temporal regulation of chloroplast development in Arabidopsis root. In Kuang T, Lu C, Zhang L, eds, Photosynthesis Research for Food, Fuel, and the Future. Springer, Berlin, pp 389–393 [Google Scholar]
- Kobayashi K, Obayashi T, Masuda T (2012b) Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal Behav 7: 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Sasaki D, Noguchi K, Fujinuma D, Komatsu H, Kobayashi M, Sato M, Toyooka K, Sugimoto K, Niyogi KK, et al. (2013) Photosynthesis of root chloroplasts developed in Arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors. Plant Cell Physiol 54: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE (2004) New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Krause GH. (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol 42: 313–349 [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara CD, Irish VF (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ (2002) Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J 32: 549–559 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Muramatsu M, Hakata M, Ueno O, Nagamura Y, Hirochika H, Takano M, Ichikawa H (2009) Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol 50: 1933–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Okegawa T, Sasaki-Sekimoto Y, Shimada H, Masuda T, Asamizu E, Nakamura Y, Shibata D, Tabata S, Takamiya K, et al. (2004) Distinctive features of plant organs characterized by global analysis of gene expression in Arabidopsis. DNA Res 11: 11–25 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Ranftl QL, Bastakis E, Klermund C, Schwechheimer C (2016) LLM-domain containing B-GATA factors control different aspects of cytokinin-regulated development in Arabidopsis thaliana. Plant Physiol 170: 2295–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Zourelidou M, Schwechheimer C (2013) Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci USA 110: 13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Schansker G, Tóth SZ, Strasser RJ (2005) Methylviologen and dibromothymoquinone treatments of pea leaves reveal the role of photosystem I in the Chl a fluorescence rise OJIP. Biochim Biophys Acta 1706: 250–261 [DOI] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G (2010) Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription: recent lessons from Arabidopsis thaliana. Eur J Cell Biol 89: 940–946 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A (2004) Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol 45: 1798–1808 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, et al. (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B (2013) A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.