Transgenic plants expressing H3K36M mutations have a dominant-negative effect on the endogenous histone methylation landscape, leading to pleiotropic developmental defects.
Abstract
Epigenetic modifications play critical roles in diverse biological processes. Histone Lys-to-Met (K-to-M) mutations act as gain-of-function mutations to inhibit a wide range of histone methyltransferases and are thought to promote tumorigenesis. However, it is largely unknown whether K-to-M mutations impact organismal development. Using Arabidopsis (Arabidopsis thaliana) as a model system, we discovered that a transgene exogenously expressing histone 3 Lys-36 to Met mutation (K36M) acts in a dominant-negative manner to cause global reduction of H3K36 methylation. Remarkably, this dominant repressive activity is dosage-dependent and causes strong developmental perturbations including extreme branching and early flowering by affecting the expression of genes involved in developmental and metabolic processes. Besides the established pathological roles of K-to-M mutations in tumor cells, we demonstrate a physiological outcome for K-to-M induced H3K36 hypomethylation. This study provides evidence for a conserved dominant-negative inhibitory role of histone K-to-M mutation across the plant and animal kingdoms. We also highlight the unique ability of K36M mutations to alter plant developmental processes leading to severe pleiotropic phenotypes. Finally, our data suggests K-to-M mutations may provide a useful strategy for altering epigenetic landscapes in organisms where histone methyltransferases are uncharacterized.
Genome compaction restricts, permits, and fine-tunes gene expression. Chromatin is composed of DNA coiled around nucleosomes and exists in relaxed or condensed forms (Luger et al., 1997). The Lys residues on histone H3 function not only as peptide building blocks, but also as a site of diverse posttranslational modifications (Strahl and Allis, 2000). Modifications also serve as a mark of the gene expression state and act as a platform for the active control of transcription. For example, methylated histone Lys residues are known to recruit histone-modifying enzymes, chromatin remodelers, and reader proteins to moderate chromatin state and transcription (He et al., 2011; Y. Xu et al., 2014).
Histone modification can be associated with either gene activation or repression, depending on its position and state. H3 Lys-36 trimethylation (H3K36me3) is a highly conserved histone mark and is primarily correlated with active gene expression in eukaryotes (Roudier et al., 2011; Wen et al., 2014). In Arabidopsis (Arabidopsis thaliana), H3K36me3 is enriched at the gene-body regions and plays an important role in regulating the expression of development-related genes involved in brassinosteroid signaling pathways, carbon and light responses, floral organ development, and flowering (Xu et al., 2008; Grini et al., 2009; He et al., 2011; Wang et al., 2014; Yang et al., 2014; Li et al., 2015). Deposition of H3K36 methylation is mediated by a collection of conserved SET domain-containing group (SDG) histone methyltransferases. Loss-of-function mutations in Arabidopsis SDG8 (also named “EFS/ASHH2”) result in a genome-wide reduction of both di- and tri-H3K36 methylation (H3K36me2/3) and defects in biological processes including proliferative branching (Dong et al., 2008; Cazzonelli et al., 2009), early flowering (Soppe et al., 1999; Zhao et al., 2005), reduced plant pathogen response (Berr et al., 2010), decreased carotenoid biosynthesis (Cazzonelli et al., 2009), and weakened innate immunity (Palma et al., 2010). SDG26 has also been shown to be required for H3K36me3 accumulation at individual loci (Berr et al., 2015), but does not appear to have an effect on global H3K36me3 levels (Liu et al., 2016). Notably, the reduced, but not completely, eliminated H3K36me2/3 in sdg8 mutants indicates the existence of yet-to-be-identified methyltransferase(s) that functions redundantly with SDG8. The increased H3K36me1 in sdg8 also suggests that distinct histone methyltransferase(s) may be responsible for H3K36me1 in Arabidopsis.
Previous studies have demonstrated that missense mutations of H3 Lys-27 to Met (K27M) exerted a dominant-negative effect on genome-wide endogenous H3K27me3 levels by inhibiting Polycomb Repressive Complex2 activity (Chan et al., 2013; Lewis et al., 2013). In humans, these gross changes in histone methylation levels were highly associated with pediatric glioma and high-grade astrocytoma formation (Chan et al., 2013; Lewis et al., 2013; Bechet et al., 2014). Similarly, exogenous expression of K27M and K9M was shown to reduce H3K27 and H3K9 methylation, respectively. The corresponding methylation losses lead to the derepression of several developmental genes for K27M and suppression of position-effect variegation for K9M in Drosophila melanogaster (Herz et al., 2014). Global reduction of endogenous H3K36 methylation by exogenously expressed K36M mutant protein was also observed in cultured cells (Lewis et al., 2013). Such K36M mutations were associated with 95% of chondroblastomas (Behjati et al., 2013). More recently, K36M mutation was shown to contribute to tumorigenesis by altering the expression of differentiation or cancer-related genes (Fang et al., 2016; Lu et al., 2016).
While K-to-M mutations promoting tumorigenesis have been investigated (Chan et al., 2013; Lewis et al., 2013; Fang et al., 2016; Lu et al., 2016), it is unknown whether K-to-M mutations influence organismal development. Using Arabidopsis as a model system, we report, to our knowledge, the first developmental phenotypic effects for the K36M mutation. We report a wide range of developmental defects for K36M mutant plants, including increased branching and early flowering. We further show that the K36M mutation reduces global H3K36 methylation and contributes to pleiotropic plant development, in part by altering expression of developmental- and metabolic-associated genes. Together, these data demonstrate that K36M acts as a driving mutation to alter plant development.
RESULTS
K36M Mutation Induces Pleiotropic Developmental Defects in Arabidopsis
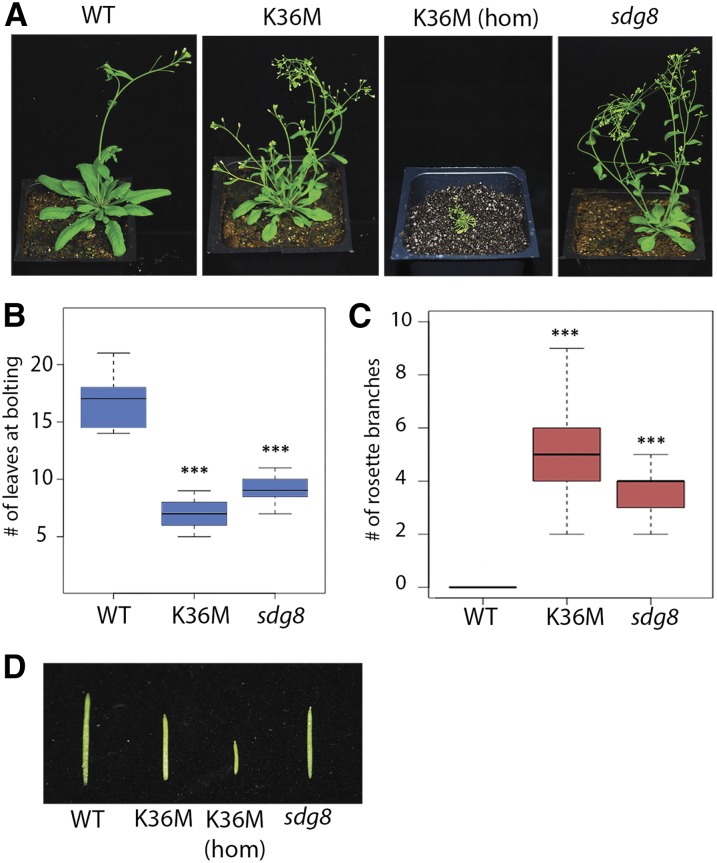
To investigate the biological consequences of K36M, we generated a transgene construct containing H3.3 K36M fused with a C-terminal 3xFLAG epitope tag under the control of its native promoter (Supplemental Fig. S1A). As a control, we produced a wild-type 3xFLAG-tagged H3.3 transgenic plant (referred to as the wild type), which had no phenotypic differences from the Columbia ecotype. As shown in Figure 1A, transgenic plants expressing heterozygous K36M (named K36M) exhibited a pleiotropic, highly stunted developmental phenotype compared to wild type. Interestingly, homozygous K36M transgenic plants (i.e., K36M hom) exhibit a gene-dosage-dependent phenotype with further stunting and infertility (Fig. 1A). Overall, we recovered a total of three independent transgenic lines, all of which displayed similar phenotypic defects (data not shown). Given that heterozygous K36M is sufficient to cause strong developmental defects and that homozygous K36M is nearly infertile, we focused our studies on heterozygous K36M. We noted an early flowering time (Fig. 1B) and a significant increase in rosette stem production in K36M (Fig. 1C). We also found that K36M mutants exhibited shorter siliques and a strong reduction in the number of fertilized ovules compared to the wild type (Fig. 1D; Supplemental Fig. S1, B and C). Interestingly, null mutants of the H3K36 histone methyltransferase SDG8 also phenocopy K36M heterozygous plants with increased shoot and stem branching, early flowering, and decreased fertility (Fig. 1; Soppe et al., 1999; Zhao et al., 2005; Dong et al., 2008; Cazzonelli et al., 2009; Grini et al., 2009). Although K36M plants showed phenotypic similarity to sdg8 null mutants, K36M (hom) exhibited stronger pleiotropic defects.
Figure 1.
K36M mutation induces pleiotropic developmental defects in Arabidopsis. A, Phenotypic analysis of transgenic plants expressing wild-type H3.3K36 3xFLAG (WT), heterozygous H3.3 K36M (K36M), homozygous H3.3 K36M (K36M hom), and sdg8 at 35 d after germination. B, K36M has a significantly lower number of leaves at bolting compared to the wild type (n > 30 plants). C, K36M shows significantly more rosette branches compared to the wild type (n = 30 plants). Rosette branches were measured from plants 7 d after main bolt production. D, K36M mutants show decreased silique length compared to the wild type. Student’s t test was used to calculate the P values (***P < 0.001).
K36M Alters the Expression of Development- and Metabolism-Associated Genes
To determine if the phenotypic defects in K36M were correlated with transcriptional alterations, we performed whole transcriptome analysis by mRNA sequencing (RNA-seq) in K36M and wild type at 21 d after germination (DAG; Supplemental Fig. S2A). We found a significant decrease in the expression of 920 genes and an increased expression of 295 genes in K36M compared to wild type (P < 0.05; Fig. 2A; Supplemental Tables S1 and S2). To examine the molecular pathways affected by the K36M mutation, we performed a gene ontology analysis on all differentially expressed genes. Consistent with phenotypic defects in K36M, we found significant enrichments of genes important for signal transduction (P = 7.2 × 10−13), hormone response (P = 1.2 × 10−3), oxidative stress response (P = 5.0 × 10−5), and flower development (P = 1.9 × 10−2; Fig. 2B). Specifically, K36M mutants showed altered expression of genes important for shoot branching and flowering (Fig. 2, C–E). We found that both the MAF1 and MAF4 floral repressor genes were downregulated in K36M and the inflorescence promoting genes (AG, AP1, FT, and PI) had increased expression (Fig. 2, C–E). Additionally, the expression of CRTISO (cis-to-trans carotenoid isomerase), a repressor of shoot branching (Cazzonelli et al., 2009), was downregulated in K36M (Fig. 2, D and E). SDG8, an H3K36me3 methytransferase known to be involved in flowering, showed no significant expression changes between K36M and wild type (Fig. 2, D and E). The transcriptional reductions of MAF1 and CRTISO were further confirmed to occur as early as 10 DAG (Supplemental Fig. S2B), correlating with the loss of H3K36me3 (Supplemental Fig. S2C). Thus, the pleiotropic developmental phenotypes in K36M are likely caused by misexpression of key development and metabolic-associated genes.
Figure 2.
K36M alters the expression of development- and metabolism-associated genes. A, Volcano plot displaying significantly upregulated and downregulated genes in K36M compared to the wild type (red dots, P < 0.05). The x axis represents the log2 value of K36M fragments per kilobase per million mapped reads/wild-type FPKM, and the y axis is the −log10 of the P value for the significance of differential expression. B, Gene Ontology analysis of significantly upregulated and downregulated genes presented as the −log10 of P value. C, Log2 FPKM of genes important for flowering in wild type and K36M. Asterisk marks each gene that is significantly differentially expressed in K36M. D, Semiquantitative gel-based RT-PCR showing expression of flowering genes at 21 d after germination. E, Quantitative RT-PCR showing the expression of genes involved in floral development and carotenoid synthesis. Each bar is the average of three biological replicates. Significance was determined via Student’s t test (*P < 0.05, **P < 0.01). FPKM, fragments per kilobase per million mapped reads.
K36M Mutation Exhibits a Dominant-Negative Effect on Endogenous H3K36 Methylation
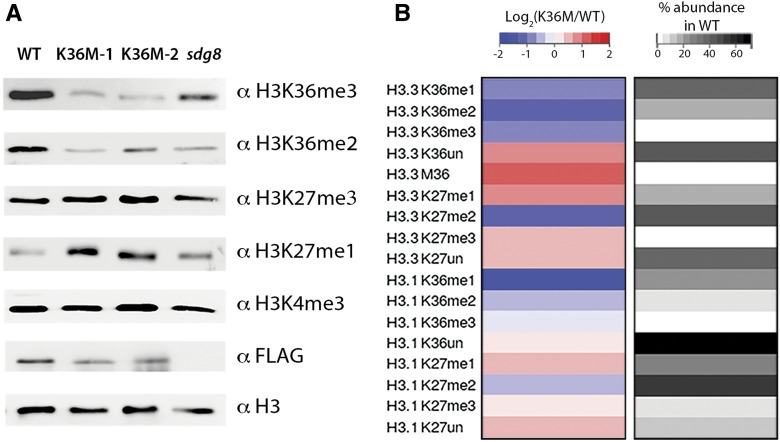
Given knowledge of K36M mutation inducing global reduction of H3K36 methylation in mammals (Lewis et al., 2013; Lu et al., 2016), we hypothesized that K36M may exert similar effects on the histone methylation landscape in plants. To test this hypothesis, we analyzed the accumulation level of various histone methylation marks in K36M by western blot. We included a null mutant of H3K36 methyltransferase sdg8 as a control, given the strong correlation of sdg8 mutant phenotype with the K36M phenotype (Fig. 1A). We found that H3K36me2 and H3K36me3 were substantially reduced in two independent K36M transgenic lines at 21 DAG (Fig. 3A). We further confirmed that the K36M reduction of H3K36me3 is independent of developmental stage (Supplemental Fig. S2C). The reduction of H3K36me3 in K36M appeared slightly stronger than that of sdg8. H3K4me3 showed no noticeable change in either K36M or sdg8. These observations were further confirmed by mass spectrometry analysis showing the reduction of H3K36 in all forms of methylation (me1, me2, and me3; Fig. 3B; Supplemental Fig. S3). This suggests that K36M mutant histones act as a dominant-negative inhibitor of endogenous H3K36 methylation. To further determine the potency of the K36M as an inhibitor, we searched for the peptide signature of M36, based on its mass difference from K36, using mass spectrometry. Consistent with the low amount of K36M in tumors and cultured cells (Schwartzentruber et al., 2012; Behjati et al., 2013; Fang et al., 2016; Lu et al., 2016), we found that K36M peptides were present at an abundance of 1.4% among total detected H3.3 peptides (Supplemental Fig. S3A). The low abundance of K36M peptides suggests that they act as potent inhibitors of endogenous H3K36 methylation.
Figure 3.
K36M mutation exhibits a dominant-negative effect on endogenous H3K36 methylation. A, Immunoblotting of histone methylation in the wild type (wild-type H3.3 3xFLAG transgene), two independent transgenic lines of K36M (K36M-1 and K36M-2), and sdg8. B, Heat map showing histone modifications on H3.3 K27-R40 and H3.1 K27-R40 peptides by mass spectrometry analysis. Each row is calculated as the log2 value of % abundance in K36M relative to % abundance in the wild type for a specific modification. The column on the right represents average % abundance of the same histone modifications detected in the wild type. Each modification is the average of triplicate technical replicates for K36M and the wild type.
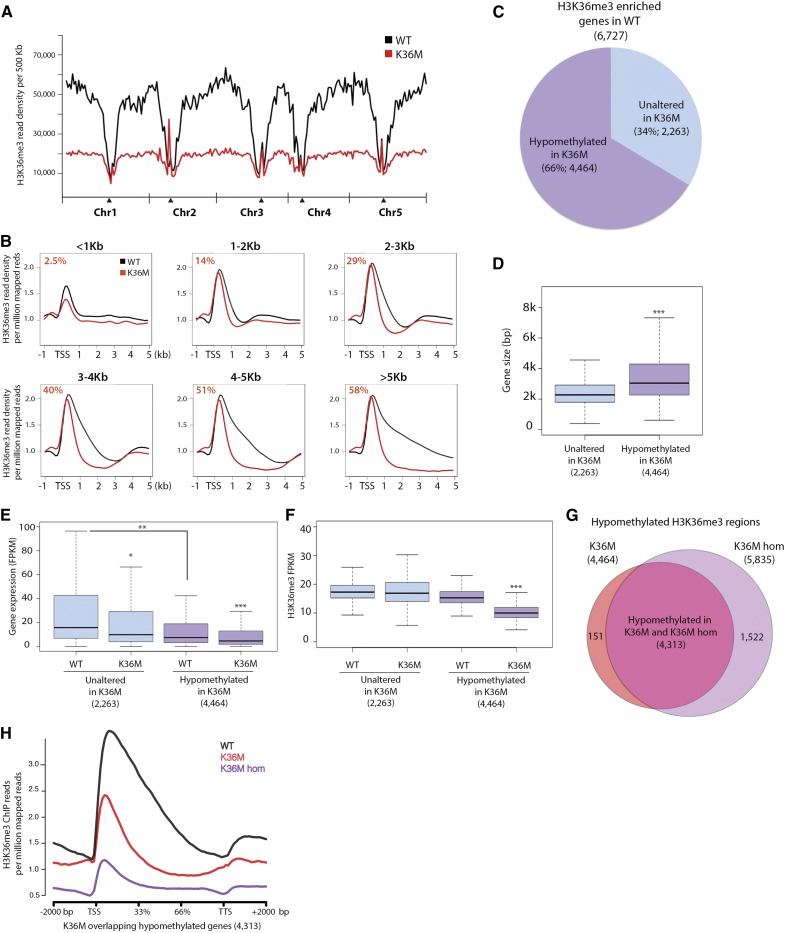
To determine whether K36M reduced H3K36 methylation at all or selected genomic loci, we profiled the genome-wide H3K36me3 distribution in K36M utilizing chromatin immunoprecipitation followed by sequencing (ChIP-seq). We observed a global reduction of H3K36me3 in the euchromatic arms of five Arabidopsis chromosomes after plotting library-normalized H3K36me3 density in 500-kb increments (Fig. 4A). To further determine where H3K36me3 loss was occurring (i.e. genes or intergenic regions), we first separated all genes in the genome into six groups by size and plotted library normalized H3K36me3 reads over them. We found a greater loss of H3K36me3 at the gene bodies of large genes than those of small genes (Fig. 4B; Supplemental Fig. S4A). Consistent with our RT-qPCR (Fig. 2E), we found the loss of H3K36me3 in genes (MAF1, CRTISO, and PIF5) involved in developmental and metabolic processes (Supplemental Fig. S4B). We also identified 6727 H3K36me3-enriched genes defined in the wild-type background (Supplemental Table S3) and found that 4464 were hypomethylated for H3K36me3 in K36M (Supplemental Table S4). This demonstrates that K36M affects many, but not all, H3K36me3-enriched genes (Fig. 4C). To identify the specific features of K36M target genes, we determined the size, expression, and H3K36me3 levels over two gene groups: 4,464 significantly hypomethylated genes in K36M and 2,263 genes with unaltered H3K36me3 levels in K36M (defined in Fig. 4C). We found that K36M-hypomethylated genes tended to be large with lower H3K36me3 levels and lower average expression than the unaltered genes (Fig. 4, D–F; Supplemental Fig. S4C). To determine if a gene dosage-dependent H3K36me3 loss could explain the stronger phenotype of homozygous K36M (K36M hom), we performed ChIP-seq of H3K36me3 in K36M hom plants. We noted that K36M and K36M hom hypomethylated genes (Supplemental Table S5) overlapped by 97%, with K36M hom displaying 1,522 additional hypomethylated genes (Fig. 4G). To determine if a quantitative difference in H3K36me3 level existed between K36M and K36M hom, we plotted library-normalized H3K36me3 ChIP-seq read density over the overlapping hypomethylated genes (4,313) and found that the overall H3K36me3 level was further reduced in K36M hom than in K36M (Fig. 4H). Together, K36M mutations exhibit a dosage-dependent dominant-negative effect on endogenous H3K36 methylation.
Figure 4.
K36M induces global reduction of H3K36me3. A, Chromosomal views show that H3K36me3 is enriched in euchromatic regions. The y axis represents H3K36me3 ChIP-seq read density over 500-kb increments normalized to total library reads. Chr1, Chr2, Chr3, Chr4, and Chr5 represent chromosomes 1 to 5, respectively. Black triangles indicate the location of centromeric regions. B, Metaplots of H3K36me3 ChIP-seq reads normalized to the total library reads over genes ranging from less than 1 kb (<1 kb), 1 to 2 kb, 2 to 3 kb, 3 to 4 kb, 4 to 5 kb, and larger than 5 kb (>5 kb). Value in left corner (red) designates the % of H3K36me3 hypomethylated genes in K36M compared to all genes of this size in the Arabidopsis genome. C, Pie chart demonstrates that the K36M mutation induces hypomethylation in many, but not all, H3K36me3-enriched genes. H3K36me3 hypomethylated genes are colored purple and genes that have unaltered H3K36me3 in K36M are blue. Box plots show the size (D), expression (E), and H3K36me3 levels (F) over the genes either H3K36me3 hypomethylated or unaltered in K36M defined in C. G, Overlap of H3K36me3 hypomethylated genes in heterozygous K36M and homozygous K36M (K36M hom). H, Metaplot of H3K36me3 normalized read count over hypomethylated overlapping genes defined in (G) in the wild type, K36M, and K36M hom. TTS: transcription termination site. Student’s t test was used to calculate the P values (*P < 0.05, **P < 0.01, and ***P < 0.001).
K36M Hypomethylated Genes Partially Overlap with SDG8 Targets
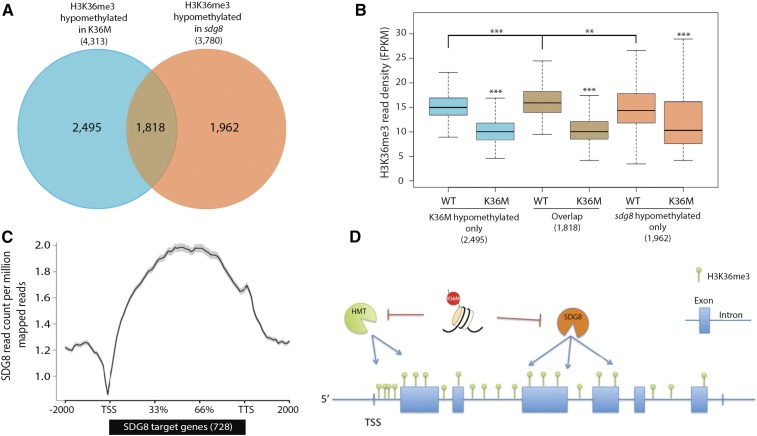
The known role of the SDG8 protein in H3K36me2/3 deposition and the similarity of developmental and molecular phenotypes between sdg8 and K36M plants prompted us to determine if effects of K36M mutations were dependent on the SDG8 pathway. To test this hypothesis and to minimize variability due to the growth condition, we determined the H3K36me3 profile in 21 DAG sdg8 plants grown side-by-side with K36M. We identified 3,780 genes with significant H3K36me3 reduction in sdg8 (Supplemental Table S6). Among them, 44% of the hypomethylated genes overlapped significantly with those reported previously (Li et al., 2015; hypergeometric test P value: 1.38 × 10−297; Supplemental Fig. S5). The different hypomethylated genes in the two datasets are likely due to plant developmental stage (21 DAG versus 14 DAG) and growth conditions (soil versus MS medium). When comparing the sdg8-hypomethylated genes (3,780) with the overlapped H3K36me3 hypomethylated genes in K36M and K36M hom (4,313 in Fig. 4G), we noted a significant number of genes are hypomethylated both in K36M and sdg8 (Fig. 5A; hypergeometric test P = 7.58 × 10−315). This suggests a potential SDG8-dependent mechanism of K36M action. Interestingly, K36M also caused a significant reduction of H3K36me3 in an additional 2,495 genes that did not overlap with sdg8-hypomethylated genes (Fig. 5A). These data suggest that the K36M mutation may also function through an SDG8-independent mechanism to inhibit endogenous histone methylation. Interestingly even at the SDG8 unique targets (1,962), there is significant reduction of H3K36me3 in K36M (Fig. 5B), suggesting that these genes are likely the weak K36M targets. Utilizing previously published data (Li et al., 2015), we also plotted the genomic location of SDG8 over its 728 direct target genes and found that SDG8 is specifically enriched at the gene body (Fig. 5C). This is consistent with our observations that the large amount of H3K36me3 is reduced within the gene body in K36M (Fig. 4B), supporting our hypothesis that K36M inhibits endogenous H3K36me3 partially via the SDG8 pathway (Fig. 5D). Given that K36M hom exhibits much stronger developmental defects than the sdg8 null mutant, our results suggest that K36M may also inhibit an unknown histone methyltransferase(s), which in turn leads to the reduction of H3K36me3 at the transcription start sites (TSS; Fig. 5D).
Figure 5.
K36M hypomethylated genes partially overlap with SDG8 targets. A, Overlap of H3K36me3 hypomethylated genes in K36M and sdg8 (hypergeometric P value = 7.58 × 10−315). B, Box plot of H3K36me3 reads over genes only hypomethylated in K36M, overlap, and only hypomethylated in sdg8. C, Metaplot shows that SDG8 primarily binds to the body of its 728 target genes. ChIP-seq of SDG8 is from Li et al. (2015). D, A working model for K36M action. K36M mutant histones likely act via a trans-mechanism to inhibit SDG8 activity and cause the reduction of H3K36me3 in the gene body regions, where SDG8 primarily functions. K36M may also inhibit an unknown histone methyltransferase), which leads to the reduction of H3K36me3 at the TSS. HMT, Methyltransferase; TTS, transcription terminal site. Student’s t test was used to calculate the P values (***P < 0.001, **P < 0.01).
DISCUSSION
In multicellular eukaryotes, gene expression is controlled through a hierarchy of mechanisms that function on the scale of individual genes to the entire genome. This study investigates the physiological consequences of disruption of one such regulatory mechanism, histone posttranslational modifications, using a histone K-to-M missense mutation. Our work revealed that a transgene exogenously expressing H3.3 K36M acts in a dominant-negative manner to induce global reduction of H3K36 methylation. Remarkably, this dominant repressive activity causes a strong developmental perturbation by affecting the expression of genes involved in developmental and metabolic processes. Besides its established pathological roles in tumor cells, we demonstrate a dominant-negative inhibitory function of K36M in plants that leads to severe developmental defects.
When comparing the phenotype of K36M with sdg8, we noted the transgenic plants expressing heterozygous K36M transgene show phenotypic similarity to sdg8 with increased branching, early flowering, and reduced fertility. Interestingly, homozygous K36M transgenic plants show greater expressivity with stunted growth and near infertility (Fig. 1A), suggesting that K36M acts in a gene-dosage-dependent manner to inhibit plant growth and development. This is consistent with the idea that the concentration of K-to-M oncohistone is important for its oncogenic potential (Jayaram et al., 2016). Although K36M appears phenotypically similar to sdg8, we also observed greater reduction of H3K36me3 in K36M mutants in our western-blot analysis (Fig. 3A). Our ChIP-seq further reveals that K36M preferentially targets genes with low levels of H3K36me3 when compared to all H3K36me3-enriched genes in the wild type. Unlike in cell culture models, we note a reduction, but not complete loss of H3K36me3 in K36M (Figs. 3 and 4D). The residual H3K36me3 may reflect the activity of other H3K36me3 methyltransferases that are unaffected by K36M inhibition. It may also be the result of utilizing whole plants as experimental material that contains a mixture of different cell types. This is consistent with previous observations of tissue-specific impairment of histone mutation in cancer cells (Schwartzentruber et al., 2012; Behjati et al., 2013; Wu et al., 2014).
Multiple lines of evidence suggest that the dominant-negative effect of the K36M mutant histone partially acts through SDG8 function. We found a large amount of K36M and SDG8 target genes overlap, and the reduction of H3K36me3 in K36M occurs most readily in the gene body where SDG8 is primarily localized (Figs. 4H and 5C). Furthermore, genes contributing to the stunted phenotype of K36M and sdg8 were misregulated in both mutants. Specifically, K36M mutants exhibited down-regulation of many flowering repressor genes (MAF1, MAF2, MAF4, and MAF5) and up-regulation of floral homeotic genes (AG, AP1, PI, FUL, and SEP3). Similar alterations in gene expression have been reported for the MAF floral repressors in sdg8 (Xu et al., 2008). These data provide compelling evidence that K36M mutants are developmentally primed for early flowering similarly to sdg8. Additionally, K36M leads to significant down-regulation of CRTISO, a carotenoid biosynthesis enzyme that controls branching inhibitor production. The down-regulation of CRTISO may reduce carotenoid precursors and strigolactone production, which could lead to increased axial branching patterns in K36M. Similar reduction of CRTISO expression was noted in sdg8 (Cazzonelli et al., 2009).
Studies of K36M mutation in cell culture models suggest that endogenous H3K36 methylation reduction occurred through the inhibition of H3K36me3-specific Lys methyltransferases (Fang et al., 2016; Lu et al., 2016). Similarly, mutations of other histone Lys (K9 and K27) to Met also cause specific reduction in corresponding methylation levels by inhibiting SET-domain-containing methyltransferases (Lewis et al., 2013; Brown et al., 2014; Jayaram et al., 2016). Thus, it is possible that K36M may act as a potent inhibitor of SDG8. K36M likely acts via a trans-mechanism to inhibit SDG8 function because H3.1 K36M, which has a distinct genomic localization from H3.3 K36M (Supplemental Fig. S6, A and B), also causes similar pleiotropic plant developmental defects (Supplemental Fig. S6C). This is in line with previous studies that the dosage of K36M peptide, but not the genomic location, is critical for its inhibitory function (Lu et al., 2016).
Besides the methyltransferase inhibition hypothesis, another possibility is that K36M may enhance the activity of an uncharacterized plant H3K36 demethylase(s) as suggested by a recent study (Herz et al., 2014). A third possibility is that K36M may inhibit K36 from becoming acetylated, which affects H3K36me3, gene expression, and development.
Although our data suggest that K36M likely acts through SDG8, it also appears to regulate endogenous H3K36me3 independent of SDG8. We found a large number of genes show reduced H3K36me3 levels in K36M but remain unchanged in sdg8 (Fig. 5A), suggesting that K36M may function as a potent inhibitor of other H3K36 methyltransferases. Besides SDG8, SDG4 and SDG26 have also been implicated in H3K36 methylation in Arabidopsis (Cartagena et al., 2008; Xu et al., 2008; Grini et al., 2009; Berr et al., 2015). However, the loss-of-function SDG26 mutant is phenotypically distinct from sdg8 and cannot induce global reduction of H3K36me3 (Xu et al., 2008). In contrast, sdg4 mutants have reduced fertility as a result of pollen tube growth suppression and exhibit global reduction of H3K4me2/3 and H3K36me2/3 by western blot (Cartagena et al., 2008). Although the precise genomic locations of H3K36me2/3 loss are unknown in sdg4, the similar phenotype of K36M and sdg4 suggests it is possible that K36M may affect SDG4 to cause the reduction of H3K36me3 at SDG8-independent genes.
While K36M induces global reduction of H3K36 methylation, we do not observe a significant difference of H3K27me3 in K36M compared to the wild type (Fig. 3), as has been noted in cell culture models (Lu et al., 2016). Instead, we noted a marked increase of H3K27me1 in K36M based on western blot and mass spectrometry (Fig. 3), suggesting a potential distinct mechanism between plants and animals. Interestingly, we found increased H3K27me1 in both replication-independent H3 variant H3.3 and replication-dependent H3.1 in K36M (Fig. 3B; Supplemental Fig. S3, A and B). The increase of H3K27me1 in K36M is unlikely due to an increased expression of H3K27me1 methyltransferase ATXR5 or ATXR6 because we do not observe a significant change of its expression based on our RNA-seq (Supplemental Fig. S7; Supplemental Tables S1 and S2). The precise mechanism for this increase is not known, but it appears possible that the conversion of H3K27me1 to H3K27me2 may be impaired in K36M as we noted a significant decrease of H3K27me2 accompanied with H3K27me1 increase in K36M (Fig. 3B). In Arabidopsis, H3K27me1 is strongly associated with heterochromatin (Jacob et al., 2009; Roudier et al., 2011), whereas H3K27me2/3 is more commonly found in euchromatic regions (Roudier et al., 2011). Another possibility is that severe reduction of H3K36 methylation may render the corresponding regions more like heterochromatin and less like euchromatin. This transformation of the local chromatin environment could make it more accessible for ATXR5/6 to deposit H3K27me1 at these regions. In line with this notion, a recent study proposes a redistribution model for the increased H3K27me3 in K36M in mammals (Lu et al., 2016). In the future, it will be interesting to uncover how K36M modulates the H3K27 methylation levels and how its activities function together with histone methyltransferases and/or other chromatin modifiers to modulate chromatin landscape.
Our study reveals that histone K-to-M mutations act as dominant-negative gain-of-function mutations in plants. K36M mutations lead to unique developmental aberrations including premature flowering, increased branching, and reduced fertility. Given its highly conserved inhibitory role across both plant and animal kingdoms, K-to-M mutation provides a potentially useful strategy to alter epigenetic landscapes in organisms where histone methyltransferases are uncharacterized or where genetic studies are challenging.
MATERIALS AND METHODS
Plant Growth Conditions
All plants were grown in long-day growth conditions (16 h light/8 h dark) at 21°C. Aerial tissues of 10-, 14-, 18-, and 21-d-old plants were harvested for experiments. sdg8 (efs-3) mutant plants were obtained as a gift from Dr. Richard Amasino (Kim et al., 2005).
Plasmid Construction and Transformation
Genomic DNA fragments containing full lengths of histone H3.3 (AT4G40040) and H3.1 (AT5G10390) with their 1-kb promoters were cloned into a pENTR/D-TOPO vector. The K36M mutation was created by site-directed mutagenesis using primers described in Supplemental Table S7. Constructs were recombined into a pEarleyGate302 binary vector (Earley et al., 2006; Chen et al., 2016) to create 3x FLAG fusions and transformed into AGL1 Agrobacterium tumefaciens and further into Arabidopsis (Arabidopsis thaliana) Columbia (Col-0) wild-type plants via the floral dip method (Clough and Bent, 1998). Transgenic plants were selected against using glufosinate (Basta) treatment.
Immunoblotting
Extracted histones or plant extracts were separated on SDS-PAGE gels, transferred to PVDF membrane, blocked in 3% BSA in TBS (50 mM Tris-Cl, 150 mm NaCl, and 0.1% Tween 20), incubated with primary antibody, and detected with horseradish peroxidase-conjugated α-mouse or α-rabbit secondary antibodies (PI31430 and PI31460; Thermo Fisher Scientific). Primary antibodies used included αH3K27me1 (07-448; Millipore), αH3K27me2 (07-452; Millipore), αH3K36me2 (ab9049; Abcam), αH3K36me3 (ab9050; Abcam), αH3 (ab1791; Abcam), and αFLAG (F3165; Sigma-Aldrich). Signal was detected through chemiluminescent imaging using an Imagequant LAS 4000 (GE Healthcare Life Sciences).
Chromatin Immunoprecipitation, Library Preparation, and Sequencing
Histone ChIP was performed as described by Lu et al. (2015). ChIP-seq libraries were prepared according to the Ovation Ultralow DR Multiplex System (cat. no. 0330; NuGEN). RNA-seq libraries were constructed using a TruSeq RNA Library Preparation Kit (no. RS-122-2002; Illumina). Libraries were sequenced on a HiSeq 2000 sequencing system (Illumina) in the UW-Madison Biotechnology Center.
Data Analysis
For ChIP-seq, reads were aligned to the Arabidopsis TAIR10 reference genome using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) with default parameters (Supplemental Table S8; Langmead and Salzberg, 2012). H3K36me3 peaks were called using SICER (Genomatix Software; S. Xu et al., 2014) and significantly hypomethylated regions were defined by using SICER-df-rb.sh with a window size of 200 bp and FDR cutoff < 0.01. Regions of hypomethylation were mapped to genes using custom scripts and the Bedtools intersect tool (http://bedtools.readthedocs.io/en/latest/content/tools/intersect.html; Quinlan, 2014) requiring 75% overlap of the hypomethylated region and the gene. Genome-wide density plots of ChIP signal were generated using custom Perl scripts that normalize each sample to its total aligned reads. Metaplots were generated using ngsplot (Shen et al., 2014) and box plots were produced using the R programming language (http://www.R-project.org). Data on SDG8-HA binding sites in sdg8-5 were obtained from Li et al. (2015). Overlap of K36M and sdg8 hypomethylated regions was performed by Venn-Diagram software (www.bioinformatics.lu/venn.php) after genome alignment, peak-calling, and gene mapping.
For RNA-seq, reads were aligned to the Arabidopsis TAIR10 genome using TopHat2 (https://ccb.jhu.edu/software/tophat/index.shtml) allowing two mismatches (Supplemental Table S8; Kim et al., 2013). Transcripts were assembled using Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) and a complete merged transcriptome was produced using Cuffmerge (http://software.broadinstitute.org/cancer/software/genepattern/modules/docs/Cuffmerge/3). Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/) arguments were used for differential expression analysis (Trapnell et al., 2010) and genes were called significant if they had a P < 0.05. Heat map analysis was conducted using the heatmap2 function of the R (https://cran.r-project.org/web/packages/gplots/index.html).
Accession Numbers
All RNA-seq and ChIP-seq data produced during this study were deposited into Gene Expression Omnibus under accession number GSE92449.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. K36M reduces fertility in Arabidopsis.
Supplemental Figure S2. Phenotypic and molecular analysis of K36M at different developmental stages.
Supplemental Figure S3. Mass spectrometry-based quantification of the abundance of histone modifications.
Supplemental Figure S4. Characterization of H3K36me3 hypomethylated genes in K36M.
Supplemental Figure S5. Venn diagram showing the overlap of H3K36me3 hypomethylated genes in sdg8.
Supplemental Figure S6. K36M mutant and wild type show similar localization patterns in the nucleus.
Supplemental Figure S7. Histone methyltransferase gene expression is unaffected in the K36M mutant.
Supplemental Table S1. Genes with increased expression in K36M heterozygous plants.
Supplemental Table S2. Genes with decreased expression in K36M heterozygous plants.
Supplemental Table S3. H3K36me3-enriched genes in wild type.
Supplemental Table S4. K36M heterozygous H3K36me3 hypomethylated genes.
Supplemental Table S5. K36M homozygous H3K36me3 hypomethylated genes.
Supplemental Table S6. sdg8 (efs-3) H3K36me3 hypomethylated genes.
Supplemental Table S7. Primers used in this study.
Supplemental Table S8. NGS alignment statistics.
Supplementary Material
Acknowledgments
We thank the UW-Madison Biotechnology Center DNA Sequencing Facility for high-throughput sequencing and Dr. Richard Amasino for sdg8 mutant seeds. We also thank Dr. Peter Lewis and members of the X. Zhong laboratory for discussions and critical comments on this manuscript.
Footnotes
1 Work in X.Z.’s laboratory was supported by the Alexander von Humboldt Foundation (as an Alfred Toepfer Faculty Fellow), a National Science Foundation CAREER award under NSF grant MCB-1552455, and a USDA and National Institute of Food and Agriculture grant under Hatch program no. 1002874. Work in J.M.D.’s laboratory was supported by the National Institutes of Health under NIH grant GM065386.
Articles can be viewed without a subscription.
References
- Bechet D, Gielen GG, Korshunov A, Pfister SM, Rousso C, Faury D, Fiset PO, Benlimane N, Lewis PW, Lu C, et al. (2014) Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol 128: 733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. (2013) Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Alioua A, Heintz D, Heitz T, Shen WH (2010) Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol 154: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Shafiq S, Pinon V, Dong A, Shen WH (2015) The trxG family histone methyltransferase SET DOMAIN GROUP26 promotes flowering via a distinctive genetic pathway. Plant J 81: 316–328 [DOI] [PubMed] [Google Scholar]
- Brown ZZ, Müller MM, Jain SU, Allis CD, Lewis PW, Muir TW (2014) Strategy for “detoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J Am Chem Soc 136: 13498–13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartagena JA, Matsunaga S, Seki M, Kurihara D, Yokoyama M, Shinozaki K, Fujimoto S, Azumi Y, Uchiyama S, Fukui K (2008) The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev Biol 315: 355–368 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lu L, Mayer KS, Scalf M, Qian S, Lomax A, Smith LM, Zhong X (2016) POWERDRESS interacts with HISTONE DEACETYLASE9 to promote aging in Arabidopsis. eLife 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dong G, Ma DP, Li J (2008) The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem Biophys Res Commun 373: 659–664 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fang D, Gan H, Lee JH, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J, et al. (2016) The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini PE, Thorstensen T, Alm V, Vizcay-Barrena G, Windju SS, Jørstad TS, Wilson ZA, Aalen RB (2009) The ASH1 HOMOLOG2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One 4: e7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Elling AA, Deng XW (2011) The epigenome and plant development. Annu Rev Plant Biol 62: 411–435 [DOI] [PubMed] [Google Scholar]
- Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A (2014) Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345: 1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD (2009) ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H, Hoelper D, Jain SU, Cantone N, Lundgren SM, Poy F, Allis CD, Cummings R, Bellon S, Lewis PW (2016) S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci USA 113: 6182–6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mukherjee I, Thum KE, Tanurdzic M, Katari MS, Obertello M, Edwards MB, McCombie WR, Martienssen RA, Coruzzi GM (2015) The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol 16: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Berr A, Chang C, Liu C, Shen WH, Ruan Y (2016) Interplay of the histone methyltransferases SDG8 and SDG26 in the regulation of transcription and plant flowering and development. Biochim Biophys Acta 1859: 581–590 [DOI] [PubMed] [Google Scholar]
- Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR, et al. (2016) Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352: 844–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Chen X, Sanders D, Qian S, Zhong X (2015) High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice. Epigenetics 10: 1044–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P, Hofius D, Petersen M, Mundy J (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 6: e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. (2014) BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. (2011) Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tönjes M, et al. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E (2014) ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Bentsink L, Koornneef M (1999) The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development 126: 4763–4770 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen J, Xie Z, Liu S, Nolan T, Ye H, Zhang M, Guo H, Schnable PS, Li Z, Yin Y (2014) Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol Plant 7: 1303–1315 [DOI] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. (2014) ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project (2014) The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46: 444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Grullon S, Ge K, Peng W (2014) Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 1150: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gan ES, Zhou J, Wee WY, Zhang X, Ito T (2014) Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res 42: 10960–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C (2014) Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24: 1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7: 1256–1260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.