Tomato antiviral resistance protein Tm-22 functions on the plasma membrane independent of the plasmodesmata localization of its avirulence protein.
Abstract
The tomato Tobacco mosaic virus resistance-22 (Tm-22) gene encodes a coiled-coil-nucleotide binding site-Leu-rich repeat protein lacking a conventional plasma membrane (PM) localization motif. Tm-22 confers plant extreme resistance against tobamoviruses including Tobacco mosaic virus (TMV) by recognizing the avirulence (Avr) viral movement protein (MP). However, the subcellular compartment where Tm-22 functions is unclear. Here, we demonstrate that Tm-22 interacts with TMV MP to form a protein complex at the PM. We show that both inactive and active Tm-22 proteins are localized to the PM. When restricted to PM by fusing Tm-22 to the S-acylated PM association motif, the Tm-22 fusion protein can still induce a hypersensitive response cell death, consistent with its activation at the PM. Through analyses of viral MP mutants, we find that the plasmodesmata (PD) localization of the Avr protein MP is not required for Tm-22 function. These results suggest that Tm-22-mediated resistance takes place on PM without requirement of its Avr protein to be located to PD.
Plants have evolved an efficient R (Resistance)-gene mediated innate immune system to prevent pathogen invasion (Jones and Dangl, 2006; Dodds and Rathjen, 2010). The R gene products, mostly R proteins, directly or indirectly recognize an Avirulence (Avr) protein from pathogens to activate a resistance signal and to trigger a robust immune response. This process frequently leads to local programmed cell death, referred to the hypersensitive response (HR; Collier and Moffett, 2009; Cui et al., 2015).
Most R proteins belong to the NBS-LRR protein family, containing a central nucleotide binding site (NBS) domain and a C-terminal Leu-rich repeat (LRR) domain (Takken and Goverse, 2012; Qi and Innes, 2013). Based on the type of the N-terminal domains, NBS-LRR proteins are mainly classified into two subclasses, i.e. the CC-NBS-LRR (CC: coiled-coil domain) and TIR-NBS-LRR (TIR: Toll/IL-1 receptor domain) proteins. The N-terminal domains play important roles in initiating signal transduction, interacting with cofactors and recognizing Avr proteins (Burch-Smith et al., 2007; Sacco et al., 2007; Tameling and Baulcombe, 2007; Maekawa et al., 2011; G.F. Wang et al., 2015). The NBS domain, also called the NB-ARC domain, is further divided into three subdomains: NB, ARC1, and ARC2 (Rairdan and Moffett, 2006; Rairdan et al., 2008). The NBS domain is shared by R proteins and metazoan apoptosis factors Apaf-1 and CED-4, and appears to act as a molecular switch that regulates the activity of NBS-LRR proteins through binding and hydrolyzing nucleotides (Tameling et al., 2006). In general, the LRR domain is required for host cells to recognize specific pathogens (Dodds et al., 2006; Ravensdale et al., 2012) and to keep R proteins from self-activation through the interaction with NBS (Rairdan and Moffett, 2006).
Different plant NBS-LRR proteins have different subcellular localization patterns that are important for their function (Qi and Innes, 2013). Several R proteins have a nucleocytoplasmic distribution and are relocated to the nucleus to regulate defense gene expression upon infection by pathogens (Wirthmueller et al., 2007; Caplan et al., 2008). In the presence of incompatible pathogens, barley (Hordeum vulgare) MLA protein in the nucleus triggers resistance response (Shen et al., 2007), while the cytoplasmic MLA induces production of cell death signals (Bai et al., 2012). However, the potato (Solanum tuberosum) Rx1 protein recognizes viral coat protein and elicits resistance in the cytoplasm, but the nuclear Rx1 balances this activity in different conditions (Slootweg et al., 2010; Tameling et al., 2010). By contrast, some R proteins are persistently localized to the endomembrane through their N-terminal motifs (Takemoto et al., 2012), although others are relocated from the cytoplasm to the endosomal compartments, for example, the potato R3a upon perception of the recognized effector AVR3aKI (Engelhardt et al., 2012). A subset of NBS-LRR proteins, such as Arabidopsis (Arabidopsis thaliana) RPM1 and RPS5, are localized to the plasma membrane (PM). The activated RPM1 resides at the PM (Boyes et al., 1998; Gao et al., 2011) together with its cofactor RIN4 and effectors AvrB and AvrRPM1 (Nimchuk et al., 2000; Mackey et al., 2002). The N-terminal acylation of the RPS5 CC domain and the RPS5′s guardee PBS1 (which also contains an N-terminal S-acylation signal) are required for their PM localization (Ade et al., 2007; Qi et al., 2012, 2014).
The tomato (Solanum lycopersicum) R gene Tm-22 encodes a CC-NBS-LRR protein, conferring durable and extreme resistance to tobamoviruses including Tobacco mosaic virus (TMV) and Tomato mosaic virus (ToMV; Lanfermeijer et al., 2003). The Tm-22-mediated extreme resistance manifests no visible lesions after viral infection (Zhang et al., 2013). To achieve this, Tm-22 perceives its Avr protein, i.e. the viral movement protein (MP; Meshi et al., 1989; Weber and Pfitzner, 1998), and its LRR domain is involved in this recognition (Lanfermeijer et al., 2005; Kobayashi et al., 2011). Rubisco small subunit and Type I J-Domain NbMIP1 proteins are involved in Tm-22-mediated extreme resistance and viral movement (Du et al., 2013; Zhao et al., 2013).
Plant viruses encode MPs to facilitate cell-to-cell movement via plasmodesmata (PD). The PD channels span across cell wall, and both the PM and the endoplasmic reticulum (ER) are continuous through the channels. TMV MP is localized to PD, ER, and PM and also binds to the cytoskeleton (Ding et al., 1992; Moore et al., 1992; McLean et al., 1995; Heinlein et al., 1998; Peiró et al., 2014). MPs accumulate in PD to increase the PD size exclusion limit; they are associated with the ER membrane at viral replication sites and are involved in targeting and transferring the ER-associated viral replication complex to PD (Wolf et al., 1989; Citovsky et al., 1990, 1992; Waigmann et al., 1994). Intriguingly the Tm-22-mediated resistance is not expressed in protoplasts that do not have PD (Motoyoshi and Oshima, 1975). Due to this, it has long been hypothesized that Tm-22 functions in PD (Meshi et al., 1989).
In this study, we demonstrate that Tm-22 functions at the PM, but its function is independent of PD localization of its cognate Avr protein MP.
RESULTS
Tm-22 Is Associated with Its Avr Protein TMV MP in Vivo
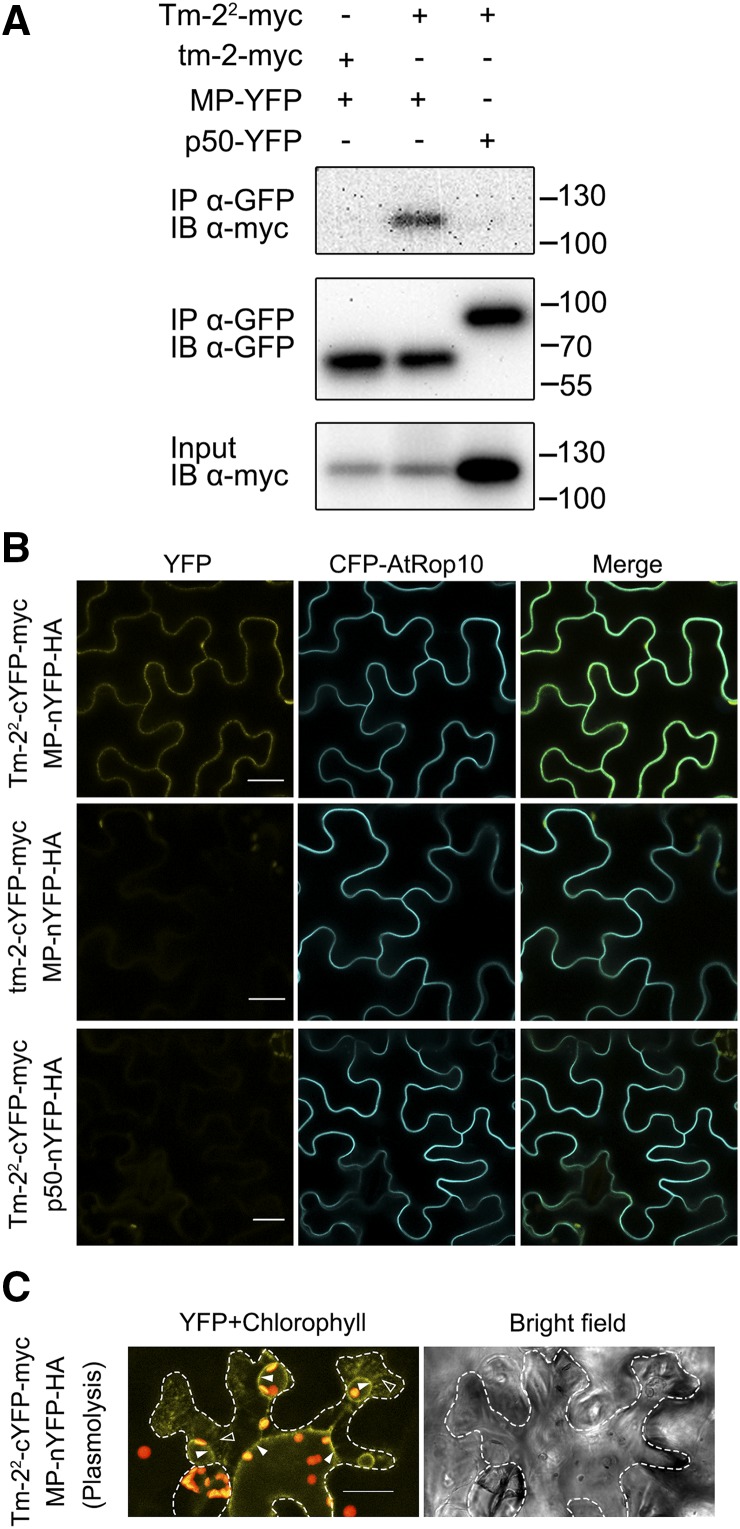
R proteins recognize their Avr proteins through either direct or indirect interaction, and R-Avr protein pairs usually form a complex and are colocalized to a similar cellular compartment in plant cells (Axtell and Staskawicz, 2003; Burch-Smith et al., 2007; Cesari et al., 2013; Le Roux et al., 2015; Sarris et al., 2015). We tested whether this is also the case for Tm-22 and MP. Because it is notably difficult to detect the expression of Tm-22 and MP due to the rapid HR, we used LaCl3 treatment to inhibit Tm-22-MP-mediated cell death. LaCl3 blocks the PM Ca2+ channel and Ca2+ influx, which is an essential process before cell death (Grant et al., 2000). We generated myc-tagged Tm-22 (Tm-22-myc) and YFP-tagged TMV MP (MP-YFP) expression cassettes, both of which were driven by the CaMV 35S promoter. Tm-22-myc and MP-YFP were transiently coexpressed in Nicotiana benthamiana leaves followed by infiltration of 2 mm LaCl3 or water at 16 h postinoculation (hpi). As seen in Supplemental Figure S1, LaCl3 compromised the HR mediated by coexpression of Tm-22 and MP. Next, we performed a coimmunoprecipitation (co-IP) assay of Tm-22 and MP using susceptible protein tm-2 from tomato (Lanfermeijer et al., 2003, 2005) as a negative control (there are only 38 different amino acids between Tm-22 and tm-2). TMV p50 (the helicase domain of the TMV replicase proteins) is localized to the cytoplasm and the nucleus (Padmanabhan et al., 2013) and is not recognized by Tm-22. We also included viral protein p50 as a negative control of MP. Although LaCl3 inhibited HR cell death, the expression of Tm-22-myc was reduced in the presence of MP (Fig. 1A), which may be due to defense-mediated translation suppression. Nevertheless, the co-IP assay reveals that Tm-22-myc can form a complex with MP-YFP, but not with p50-YFP, while susceptible tm-2 is not associated with MP (Fig. 1A). These results also suggest that Tm-22 and TMV MP can be localized to the same cellular compartment in plant cells.
Figure 1.
Tm-22 forms a complex with TMV MP in N. benthamiana. A, Tm-22 coimmunoprecipitates with MP. Tm-22-myc was transiently expressed with MP-YFP or p50-YFP in N. benthamiana leaves, and tm-2-myc was also coexpressed with MP-YFP. Agroinfiltration is followed by LaCl3 treatment at 16 hpi. Protein extracts from the infiltrated leaves at 36 hpi were subjected to anti-GFP immunoprecipitation (IP) followed by immunoblotting (IB) with the indicated antibodies. The size of protein molecular weight markers (kD) is on the right. B, The BiFC assays show the interaction of Tm-22 with MP. Tm-22-cYFP-myc was coexpressed with MP-nYFP-HA or p50-nYFP-HA, and tm-2-cYFP-myc was coexpressed with MP-nYFP-HA. All the combinations were coexpressed with the PM fluorescent marker CFP-AtROP10. C, YFP fluorescence from the interaction of Tm-22-cYFP-myc with MP-nYFP-HA was observed after cell plasmolysis. Cell plasmolysis was performed by treatment of 5% NaCl for 5 min. Red color indicates the chloroplast autofluorescence. Hechtian strands, typical connections of PM-cell wall, are indicated by outlined triangles, and the retracted PM is indicated by filled triangles. The cell wall is highlighted by dotted lines. Bars = 20 μm.
These findings were further confirmed by bimolecular fluorescence complementation (BiFC). To perform BiFC, Tm-22 and tm-2 were fused with cYFP-myc (C-terminal domain of YFP and myc tag) to generate Tm-22-cYFP-myc and tm-2-cYFP-myc, and viral MP and p50 were fused with nYFP-HA (N-terminal domain of YFP and HA tag) to generate MP-nYFP-HA and p50-nYFP-HA. These fusion proteins were detected by western blots (Supplemental Fig. S2). The positive interaction indicated by yellow fluorescence was only detected when Tm-22-cYFP-myc was coexpressed with MP-nYFP-HA, but not in other combinations (Fig. 1B). Interestingly, the yellow fluorescence appeared as a thin line circumventing the cells and coincided with the PM labeled by the CFP-tagged PM marker CFP-AtROP10 (Lavy and Yalovsky, 2006). After cell plasmolysis, YFP fluorescence resulted from the Tm-22-MP interaction labeled the plasma membrane extensions (called Hechtian strands; Oparka, 1994) connecting the plasma membrane to the cell wall (Fig. 1C), also indicating the interaction occurred on the PM.
Taken together, these data suggest that Tm-22 forms a complex with MP on the PM where Tm-22 functions to trigger defense.
Tm-22 Is a PM-Localized Protein
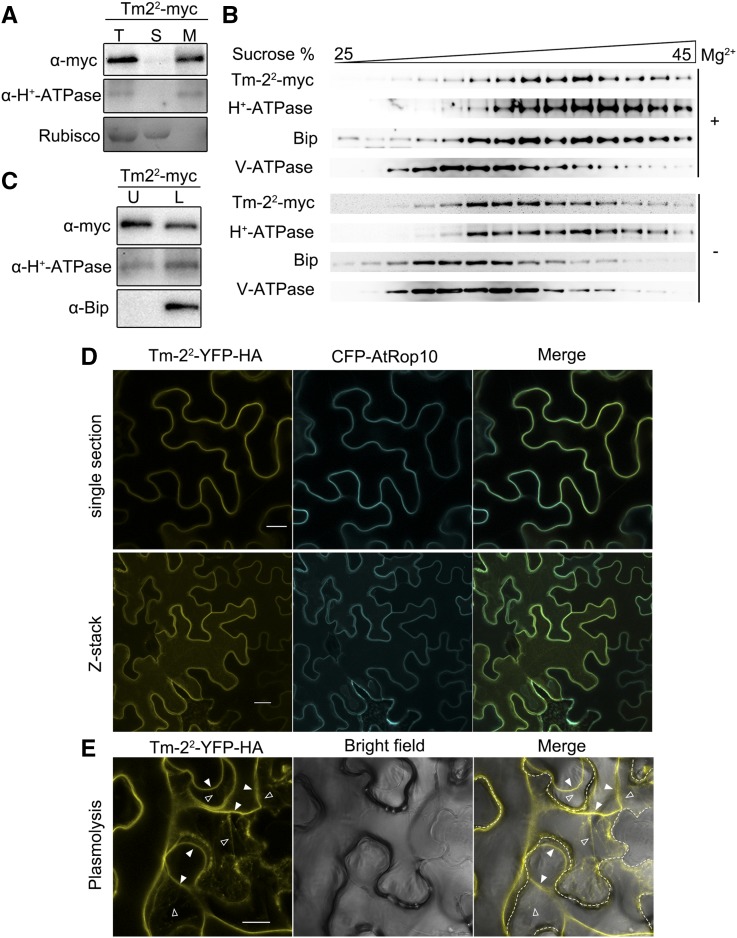
We examined the PM localization of Tm-22 and tm-2 using a cell fractionation method. Total protein extract from the leaves expressing Tm-22-myc or tm-2-myc was separated into soluble and microsomal membrane fractions by ultracentrifugation. We found that Tm-22-myc was mostly detected in microsomal membrane fraction (Fig. 2A), while tm-2-myc was detected in both soluble and microsomal membrane fractions (Supplemental Fig. S3).
Figure 2.
Tm-22 is localized to the PM. A, Tm-22-myc is a membrane-associated protein. The total protein (T) extracted from leaves expressing Tm-22-myc was fractionated into soluble (S) and membrane (M) fractions by ultracentrifugation at 100,000g. B, Tm-22 cofractionated with the PM marker through Suc gradients centrifugation. C, Tm-22-myc is a PM-located protein. Upper phase (U) and lower phase (L) were obtained by aqueous two-phase partitioning. Fractions in A to C were analyzed by western blot using antibodies against myc epitope, H+-ATPase (PM marker), BiP (ER marker), and V-ATPase (tonoplast marker). Rubisco (soluble protein marker) was stained by Ponceau S. D, Confocal images illustrated that Tm-22-YFP-HA colocalized with CFP-AtRop10 at the PM. Upper, single image intersecting the epidermal cells; lower, projection from Z-stack images. E, Tm-22-YFP-HA was also detected in the Hechtian strands after plasmolysis. Hechtian strands are indicated by outlined triangles, and the retracted PM is indicated by filled triangles. The cell wall is highlighted by dotted lines. Bars = 20 μm.
Furthermore, we performed Suc-density gradient centrifugation using N. benthamiana microsomes containing Tm-22-myc. The centrifugation was performed in the presence (+Mg2+) or absence (−Mg2+) of magnesium ions. Removal of Mg2+ ions results in destabilization of some membrane proteins especially for ribosomes and redistribution of membrane proteins in Suc gradients. This treatment particularly affects the distribution of ER and Golgi membrane proteins (Chen et al., 2002). Fractions from the Suc gradient were subsequently analyzed by western blot using antibodies against Tm-22-myc or marker proteins specific for PM, ER, and tonoplast. ER marker BiP diagnostically shifted from high density to low density when Mg2+ removed, but tonoplast marker V-ATPase was always abundant in fractions of lower Suc density (Fig. 2B). However, Tm-22 proteins were cofractionated with PM marker H+-ATPase in either the presence or the absence of Mg2+ (Fig. 2B). Cofractionation of Tm-22 with PM marker H+-ATPase was further confirmed using the aqueous two-phase partitioning approach to isolate the PM from microsomal membranes. Microsomal membrane fraction containing Tm-22-myc was further subjected to a two-phase solution. The PM marker H+-ATPase and ER marker BiP were both detected in the lower phase, while only the PM marker H+-ATPase and Tm-22-myc were detected in the upper phase (Fig. 2C). Tm-22-myc copartitioned with H+-ATPase, indicating that Tm-22 was associated with the PM.
We also identified the PM localization of Tm-22 by confocal analysis. For this purpose, we generated N-terminal YFP-tagged Tm-22 (YFP-Tm-22) and C-terminal YFP-4×HA-tagged Tm-22 (Tm-22-YFP-HA). These constructs were driven by the 35S promoter and agroinfiltrated into N. benthamiana leaves to test whether YFP-Tm-22 or Tm-22-YFP-HA could confer resistance against TMV. Tm-22-YFP-HA, but not YFP-Tm-22, conferred resistance against TMV-GFP and also induced HR in the presence of MP (Supplemental Fig. S4), suggesting that Tm-22-YFP-HA is functional. Furthermore, Tm-22-YFP-HA or tm-2-YFP-HA was coexpressed with CFP-AtRop10 in N. benthamiana leaves for confocal analysis. In a single section or z-stack of multiple sections, Tm-22-YFP-HA and tm-2-YFP-HA were detected at the cell periphery and colocalized with the PM marker CFP-AtRop10 (Fig. 2D; Supplemental Fig. S3). In addition, tm-2-YFP-HA was also localized in the cytoplasm (Supplemental Fig. S3). Following plasmolysis of cell, the fluorescence of Tm-22-YFP-HA or tm-2-YFP-HA was detected in Hechtian strands (Fig. 2E; Supplemental Fig. S3). Collectively, these results clearly suggest that Tm-22 is a PM-localized protein, while tm-2 localized to both PM and cytoplasm.
Tm-22 Is a Peripheral Membrane Protein
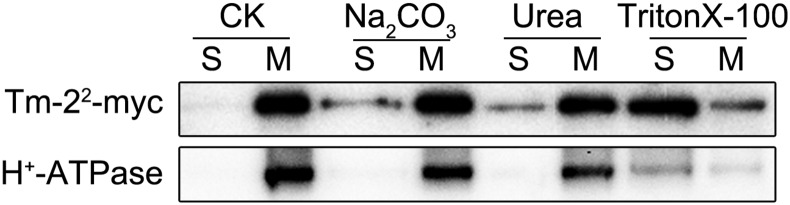
To investigate whether Tm-22 is an integral or peripheral membrane protein, we extracted the Tm-22-myc-containing microsomal membrane fraction using extraction buffer (as a control), Na2CO3 (pH = 11), 2 m urea, or 1% Triton X-100 (Boyes et al., 1998). The mild detergent Triton X-100 releases both peripheral and integral proteins, while the treatment of Na2CO3 or urea only releases peripheral proteins. As expected, Triton X-100 treatment released both the integral membrane protein PM H+-ATPase and Tm-22 into the soluble fractions; the treatment of Na2CO3 or urea did not release H+-ATPase into the soluble fractions. However, the treatment of Na2CO3 or urea was able to release Tm-22 into the soluble fractions (Fig. 3). Thus, we conclude that Tm-22 is peripherally associated with the PM.
Figure 3.
Tm-22 is a peripheral membrane protein. Microsomal membranes purified from leaves expressing Tm-22-myc were treated to release peripheral membrane proteins as indicated. The remaining membranes (M) and the newly soluble proteins (S) were analyzed by immunoblot with indicated antibodies. CK, The extraction buffer.
The Activated Tm-22 Is Also Localized to the PM
We tested the subcellular localization of the activated Tm-22 under two different conditions. First, we used TMV MP to activate Tm-22. For this purpose, we coexpressed Tm-22-myc with MP-YFP in N. benthamiana. Second, we employed a Tm-22 autoactive mutation in the conserved MHD motif. The conserved MHD motif (IHD in Tm-22) in ARC2 subdomain is required for coordinating nucleotide and controlling subdomain interaction, and mutations in this conserved motif generally lead to activation of NBS-LRR proteins independent of Avr proteins (Bendahmane et al., 2002; Howles et al., 2005; van Ooijen et al., 2008; Engelhardt et al., 2012). We generated a myc-tagged MHD mutant D481V. Similar to coexpression of Tm-22 and MP, the autoactive mutant D481V alone induced cell death (Fig. 4A), and the D481V mutant-mediated cell death was blocked by LaCl3 (Supplemental Fig. S1).
Figure 4.
The activated Tm-22 resides at the PM. A, HR cell death is induced by Tm-22 in the presence of TMV MP and autoactive MHD mutant D481V, but not by wild-type Tm-22 alone in N. benthamiana. Cell death was visualized by trypan blue staining (lower panel) at 48 hpi. Solid line circles indicate cell death; dashed line circles indicate no obvious cell death. B, Cell fractionation assays show that activated Tm-22 is associated with the membrane. Soluble (S) and microsomal membrane (M) fractions were separated by ultracentrifugation. C, Aqueous two-phase partitioning assays show that the activated Tm-22 is partitioned in the PM phase. Upper phase (U) and lower phase (L) were obtained by aqueous two-phase partitioning of microsomal membrane fractions from B.
Next, we used cell fractionation and aqueous two-phase partitioning to analyze the subcellular localization of the activated Tm-22. Cell fractionation assays showed that either the activated Tm-22 by coexpression with MP or D481V mutant remained in the microsomal membrane fraction, the same as the inactive Tm-22 (Fig. 4B). Microsomal membrane was further purified by aqueous two-phase partitioning. The activated Tm-22 appeared in upper phase and copartitioned with the PM marker H+-ATPase (Fig. 4C).
Through Suc density gradient centrifugation, we also found that the activated Tm-22 by coexpression with MP was still cofractionated with the PM marker protein (Supplemental Fig. S5). Similarly, we examined the localization of MP when coexpressed with Tm-22. MP-YFP was detected in the PM fraction and cofractionated with the PM marker protein H+-ATPase (Fig. 4C; Supplemental Fig. S5). To further investigate the colocalization of Tm-22 and MP, we constructed CFP-tagged MP (MP-CFP) and RFP-tagged AtRop10 as the PM marker. Confocal microscopy assays further showed the presence of MP-CFP in the PM besides PD when coexpressed with either Tm-22-YFP-HA or tm-2-YFP-HA (Supplemental Fig. S6), indicating that Tm-22 did not affect MP localization. In addition, Tm-22-YFP-HA and MP-CFP colocalized at the PM (Supplemental Fig. S6), consistent with the previous observations that MP is able to be localized at the PM (Moore et al., 1992; Heinlein et al., 1998; Kahn et al., 1998; Lewis and Lazarowitz, 2010; Amari et al., 2014). All these results indicate that the activated Tm-22 is located at the PM.
Targeting Tm-22 to the PM Retains Its Function to Induce HR Cell Death
To rule out the possibility that Tm-22 may act at other non-PM cellular sites, we used the S-acylated PM association domain to restrict Tm-22 to PM. The C-terminal domain of AtRop10 is sufficient for association of proteins with PM due to the S-acylation (Lavy and Yalovsky, 2006). This domain has only 25 amino acid residues consisting of polybasic region and GC-CG box, while the substitution of the five nonpolar residues in GC-CG box by charged REDER residues can block the PM association (Fig. 5A). To confirm the function of this domain, the C terminus of YFP was tagged with this motif sequence (Rop tag) or its REDER mutant (mRop tag, as control) to generate YFP-Rop and YFP-mRop, which were then transiently expressed in N. benthamiana. As expected, YFP-Rop was observed to surround cells as a thin line, while YFP-mRop was scattered in cytoplasm and nucleus (Fig. 5B). After plasmolysis of cells, YFP-Rop, but not YFP-mRop, was detected in Hechtian strands (Fig. 5B), indicating that Rop, the C-terminal domain of AtRop10, can be used to confine YFP to the PM.
Figure 5.
Plasma membrane-tethered Tm-22 retains effector-mediated HR function. A, Schematic representations of Rop tag and mRop tag. B, Confocal images show the localization of YFP-Rop or YFP-mRop in normal condition or after plasmolysis. C, Confocal images show the localization of Tm-22-YFP-Rop or Tm-22-YFP-mRop in normal condition or after plasmolysis. Hechtian strands are indicated by outlined triangles, and the retracted PM is indicated by filled triangles. The cell wall is highlighted by dotted lines. D, Both Tm-22-YFP-Rop and Tm-22-YFP-mRop induced cell death when coexpressed with MP. Cell death was visualized by trypan blue staining (right).
Furthermore, Tm-22-YFP was fused with Rop or mRop tag to generate Tm-22-YFP-Rop or Tm-22-YFP-mRop. Both Tm-22-YFP-Rop and Tm-22-YFP-mRop were detected in the PM through confocal imaging and plasmolysis (Fig. 5C). When coexpressed with MP, Tm-22-YFP-Rop and Tm-22-YFP-mRop induced a similar extent of HR cell death (Fig. 5D). In addition, we also fused Tm-22-myc with Rop or mRop tag. Both Tm-22-myc-Rop and Tm-22-myc-mRop stayed in the membrane fraction and triggered MP-dependent cell death (Supplemental Fig. S7). These results suggest that restricting Tm-22 to PM does not affect its function, further confirming that Tm-22 functions on the PM.
Tm-22 Requires All Domains for Its Membrane Association
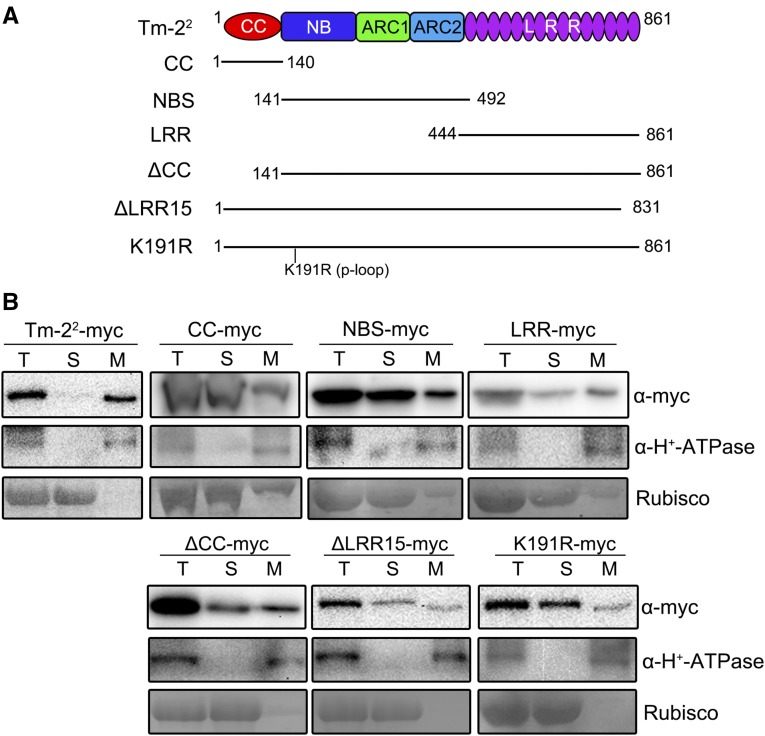
The PM localization of some CC-NBS-LRR proteins including Arabidopsis RPS2, RPS5, and rice (Oryza sativa) Pit is reported to depend on acylation of CC domain (Qi et al., 2012; Kawano et al., 2014). However, no conventional transmembrane domains or acylation sites were predicted for Tm-22 protein. To determine motifs required for PM localization of Tm-22, we separately expressed the CC, NBS, and LRR domains of Tm-22 with a C-terminal myc tag in N. benthamiana leaves (Fig. 6A). Expression of these single domains did not induce any obvious cell death (Supplemental Fig. S8). We then investigated the subcellular localization of the CC, NBS, and LRR domains. Surprisingly, none of them was found completely in the microsomal membrane fraction (Fig. 6B, upper panel). Instead, the CC, NBS, and LRR domains were all detected partially in soluble fraction.
Figure 6.
Tm-22 requires CC, NBS, and LRR domains for the PM localization. A, Schematic diagram of Tm-22 mutants used for cell fractionation. B, Cell fractionation analysis of Tm-22 or its mutants. The total proteins extracted from N. benthamiana leaves expressing myc-tagged Tm-22 mutants were fractionated by ultracentrifugation at 100,000g. The fractions were detected by immunoblotting with anti-myc and anti-H+-ATPase antibodies. Rubisco was stained by Ponceau S.
We further examined the subcellular localization of two truncated Tm-22 proteins. To achieve this, we generated Tm-22 with a deletion of the CC domain (ΔCC) or of the last Leu-rich repeat motif (ΔLRR15; Fig. 6A). Expression of these two deletion mutants did not cause cell death. Those two mutant proteins were detected in both soluble and membrane fractions (Fig. 6B). Thus, both CC and LRR domains affect the Tm-22 PM localization. In addition, the deletion mutants also affect MP-mediated cell death and the autoactivity of D481V mutant (Supplemental Fig. S8).
We also checked the effect of NBS domain on the PM localization of Tm-22. We focused on the P-loop motif in the NBS domain, which is required for ATP binding and NBS-LRR protein function (Dinesh-Kumar et al., 2000; Bendahmane et al., 2002; Tameling et al., 2006). Indeed, P-loop motif is important for Tm-22-mediated resistance (Supplemental Fig. S8). Moreover, the cell fractionation assay revealed that the K191R mutant was partially soluble (Fig. 6B). These results indicate mutations in the NBS domain can also affect Tm-22 membrane association.
Taken together, these results suggest that Tm-22 requires all domains for its proper PM localization.
The Function of Tm-22 Is Independent of the PD Localization of TMV MP
In plant cells, MP is not only localized to membranes, but also accumulates in PD. PD localization of MP is essential for TMV movement (Kahn et al., 1998; Boyko et al., 2000; Liu and Nelson, 2013). Moreover, Tm-22-mediated virus resistance is observed in tissues and whole plants, but not in protoplasts in which no PD exists (Motoyoshi and Oshima, 1975). It has been hypothesized that viral MP accumulation in PD is required for Tm-22-mediated virus resistance (Meshi et al., 1989).
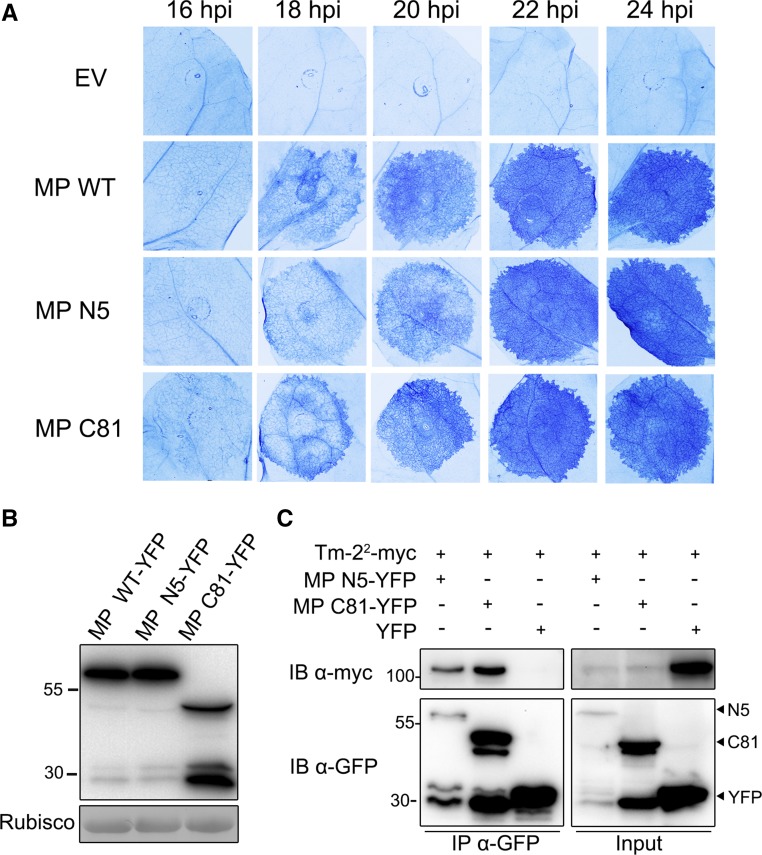
To test this hypothesis, we used MP dysfunctional mutants N5 and C81, which failed to target to PD and impaired viral movement (Boyko et al., 2000; Kotlizky et al., 2001). N5 mutant was generated by deleting the N-terminal three to approximately five amino acids of MP, and C81 was generated by deleting the C-terminal 81 amino acids. We analyzed the localization and function of C-terminal YFP-tagged MP or its two dysfunctional mutants. Consistent with the previous reports (Boyko et al., 2000; Kotlizky et al., 2001), N5 and C81 cannot target to PD (Supplemental Fig. S9).
We then tested the speed and robustness of HR induced by MP and its dysfunctional mutants in Tm-22 transgenic line TM#1 (Zhang et al., 2013). All the constructs containing wild-type MP, N5, or C81 mutants were expressed in the same leaves. We monitored HR at the interval of 2 h from 16 to 24 hpi and then stained the leaves with trypan blue to facilitate observation. We found that wild-type and mutant MP started to induce HR cell death at about 16 to ∼18 hpi. Finally, all MP variants exhibited a similar intensity of HR cell death at 24 hpi (Fig. 7A). Expression of MP and its mutants was confirmed at protein level by immunoblotting (Fig. 7B). These results revealed that MP mutants without PD accumulation did not affect Tm-22-mediated cell death. Furthermore, we observed that Tm-22 can be coimmunoprecipitated by those MP mutants, although the expression of Tm-22 was reduced during the activated resistance response (Fig. 7C). Thus, Tm-22 interacts with MP and functions to induce HR and cell death independent of PD accumulation of MP.
Figure 7.
TMV MP mutants defective in targeting plasmodesmata still trigger Tm-22-mediated cell death. A, The extent of HR cell death induced by MP and its mutants was analyzed in transgenic Tm-22 plants. MP (MP WT) and its PD-targeting defective mutants (MP N5 and MP C81) tagged with C-terminal YFP were agroinfiltrated in Tm-22 transgenic N. benthamiana, and leaves from 16 to 24 hpi were stained by trypan blue. B, Total protein from wild-type N. benthamiana leaves expressing MP or its mutants at 24 hpi was extracted and detected with anti-GFP antibody. C, Tm-22-myc can coimmunoprecipitate with MP mutant N5 or C81. YFP was employed as a negative control. Proteins were immunoprecipitated with anti-GFP beads, and the immunoblotting was performed with indicated antibodies. The sizes of protein molecular weight markers (kD) are indicated.
DISCUSSION
Where and how an R protein recognizes its Avr protein to initiate defense response are two critical questions in the research of plant-pathogen interaction. R proteins are localized to diverse subcellular compartments and use a different strategy to detect their Avr proteins. Here, we found that Tm-22 interacts with MP in planta, is localized to the PM, and performs its antiviral function independent of the accumulation of viral MP in PD.
Tm-22 Is a Peripheral Membrane Protein Associated with the PM
In plants, an R protein usually forms a complex and is colocalized with its Avr counterpart for recognition and conferring resistance (Dodds et al., 2006; Krasileva et al., 2010). In this study, we detected the specific interaction between wild-type Tm-22 and TMV MP in vivo using both co-IP and BiFC assays (Fig. 1). Tm-22 and MP coexist in the same complex situated in the same subcellular compartment.
It has been well documented that viral MP is associated with ER and PM, accumulates in PD for cell-to-cell movement of plant viruses, and also binds to cytoskeleton (Ding et al., 1992; Moore et al., 1992; McLean et al., 1995; Heinlein et al., 1998; Peiró et al., 2014). Using confocal microscopy and biochemical approaches, we demonstrate that Tm-22 is located to the PM in inactive and autoactive forms (Figs. 2 and 4). Considering that Tm-22 can be disassociated from membrane by mild treatments that are only able to release peripheral membrane proteins (Fig. 3), we conclude that Tm-22 is a peripheral PM-associated protein. These findings are consistent with previous reports that several PM-localized R proteins, such as Arabidopsis RPM1 and HRT, are shown to be peripherally associated with the PM (Boyes et al., 1998; Jeong et al., 2010), and RPS2 is an integral membrane protein (Axtell and Staskawicz, 2003).
To investigate whether the active Tm-22 is restricted to the PM, we tested the localization of Tm-22 under two different activating conditions. We found that Tm-22 in the presence of MP does not release from the PM, and both Tm-22 and MP are colocalized to the PM (Fig. 4; Supplemental Fig. S6), consistent with the BiFC data that Tm-22 interacts with viral MP at the PM (Fig. 1). Furthermore, we generated an autoactive Tm-22 mutation in its MHD motif that induced cell death independent of the Avr protein (Fig. 4A). This autoactive mutant also resides at the PM (Fig. 4, B and C). These data suggest that the active form of Tm-22 persistently remains at the PM without cellular relocalization to induce HR cell death.
Furthermore, we tethered Tm-22 to the PM by fusing Tm-22 with the PM association domain from AtRop10. This domain efficiently targets soluble YFP to the PM (Fig. 5B). Expression of Tm-22 fused with a functional or dysfunctional PM domain showed no difference in inducing cell death when coexpressed with MP (Fig. 5D). The additional PM association domain does not affect Tm-22-mediated HR, suggesting that the action site for Tm-22 to activate defense signaling pathway is PM rather than other subcellular localizations. Similarly, RPM1 can also be tethered to the PM membrane by a CBL (calcineurin B-like protein) tag that contains dual-lipid modification sites, and the RPM1 fusion protein can still retain its HR function (Gao et al., 2011).
In this study, we consistently observed accumulation of Tm-22-YFP-HA in PM, but not in the interspersed spots as reported previously for Tm-22-YFP (Du et al., 2013). However, a through confocal microscopy assay indicated that Tm-22-YFP also accumulated in PM in addition to some interspersed dots along with cell membranes (Supplemental Fig. S10). This discrepancy may be caused by the protein level, time after infiltration, extra tag sequence, or some other unknown technical differences. Nonetheless, both Tm-22-YFP-HA and Tm-22-YFP are sufficient to confer TMV resistance and induce HR in the presence of MP (this study; Du et al., 2013). These data suggest that the accumulation in PM, but not in the interspersed dots, is responsible for Tm-22 function.
Like Tm-22, all PM-localized R proteins reported so far belong to the CC-NBS-LRR subfamily. Compared to non-PM-localized R proteins, which seem to have translocation ability, PM-localized R proteins tend to transduce defense signaling at the PM. Indeed, it has been demonstrated that Arabidopsis NBS-LRR proteins RPM1, RPS2, and RPS5 and rice Pit activate production of downstream signals at the PM (Axtell and Staskawicz, 2003; Kawano et al., 2010; Gao et al., 2011; Qi et al., 2012). It is possible that Tm-22 may be attached to the PM by interacting with some integral PM proteins to activate downstream defense signaling at the PM.
PM seems to be a scaffold to assemble some R proteins and downstream components for signal transduction. In rice, a small GTPase OsRac1 interacts with R protein Pit at the PM and transduces signal through a defensome with various downstream proteins (Kawano et al., 2010). NDR1 may be one potential component of the signaling pathway that is localized to the PM and involved in the resistance conferred by many CC-NBS-LRR proteins (Aarts et al., 1998; Day et al., 2006). Nevertheless, it remains to be elucidated how PM-associated R proteins initiate the cascade of the signaling transduction pathway in plant defense.
Tm-22 Requires CC, NBS, and LRR Domains for Its Proper PM Localization
N-terminal motifs of several R proteins play essential roles in localization, especially in membrane localization (Takemoto et al., 2012). Arabidopsis RPS5 and RPS2 are found to localize at the PM through myristoylation and palmitoylation in its N-terminal CC domain (Qi et al., 2012). In the case of rice resistance protein Pit, substitution of palmitoylated amino acids in its CC domain results in lack of the PM localization and loss of function (Kawano et al., 2014). However, the PM-localized mechanism of other R proteins without acylation is still unknown. No acylation sites in CC domain or other domains of Tm-22 were found. Surprisingly, no single domain was found to be completely responsible for Tm-22 PM localization. In addition, CC or LRR domain-truncated Tm-22 only partially lost their PM localization. Interestingly, a single amino acid mutation in P-loop of NBS domain also partially abrogated Tm-22 PM localization, but completely failed to induce HR cell death, similar to that of RPM1 P-loop mutant (Gao et al., 2011). These results suggest that all domains may be required for Tm-22 PM localization. However, the precise contribution of the CC, NBS, or LRR domain to target Tm-22 to the PM and by which mechanism these domains make such contribution need to be further investigated.
Tm-22 Functions Independent of PD Localization of Viral MP
TMV MP is not only associated with cell membranes, but also specifically targets PD to increase its size exclusion limit in order to translocate the ribonucleocomplex through the PD (Wolf et al., 1989; Citovsky et al., 1992; Ding et al., 1992). An early study has demonstrated that Tm-22 resistance is not expressed in protoplasts that lack cell walls and PD (Motoyoshi and Oshima, 1975). It has been proposed that MP accumulation in PD is required for Tm-22 resistance and Tm-22 blocks cell-to-cell movement of viruses (Meshi et al., 1989). In addition, the N gene-mediated resistance against TMV is also not expressed in protoplasts (Otsuki et al., 1972), but both N and its Avr protein p50 localize in the cytoplasm and nucleus, and p50 does not participate in cell-to-cell movement. In this study, we found that two MP mutants N5 (ΔN3-5 amino acids) and C81 (1–187 amino acids) were still able to induce HR cell death in transgenic Tm-22 N. benthamiana plants (Fig. 7B) and form a protein complex with Tm-22 (Fig. 7D). However, the two MP mutants N5 and C81 are unable to target PD, suggesting that PD accumulation of MP is not necessary for Tm-22 recognition.
MPs of TMV and ToMV have 77% amino acid sequence identity. They are conserved in the N-terminal region (1–211 amino acids) but have high polymorphisms in their C-terminal regions. Both MPs induce Tm-22-mediated HR cell death. We found that N-terminal 187 amino acids of TMV MP were sufficient to trigger Tm-22-mediated HR cell death. Consistent with this finding, the ToMV MP deletion mutant (1–188 amino acids) can induce HR cell death in tomato containing Tm-22 (Weber et al., 2004). These findings suggest that the N-terminal but not C-terminal region of MP is responsible for Tm-22-mediated resistance. However, it has been reported that deletion of C-terminal 30 amino acids in ToMV MP (1–234 amino acids) breaks the Tm-22 resistance to ToMV infection (Weber and Pfitzner, 1998). It is possible that the C-terminal domain of MP affects exposure of protein structures that are recognized by Tm-22.
MATERIALS AND METHODS
Plant Materials and Plasmids
Transgenic Nicotiana benthamiana line TM#1 contains Tm-22 gene with native promoter and terminator, and confers an extreme resistance against ToMV and TMV (Zhang et al., 2013). Wild-type and transgenic N. benthamiana plants were grown in growth rooms at 25°C under a 16-h-light/8-h-dark cycle.
For generating T-DNA expression vectors, a ligation-independent cloning (LIC) cassette containing ccdB gene and a chloramphenicol-resistance gene flanking LIC adaptors with ApaI site was PCR amplified using pYL436 as a template and then inserted into pCAMBIA-nLUC or pCAMBIA-cLUC, respectively (Chen et al., 2008), to generate pLIC-nLUC or pcLUC-LIC. pLIC-myc, pLIC-HA, pLIC-YFP-HA, or pLIC-YFP was generated by replacing nLUC sequence of pLIC-nLUC with 4×myc, 3×HA, YFP-4×HA, or YFP sequence, respectively. pYFP-LIC was generated by replacing cLUC sequence of pcLUC-LIC with YFP sequence. Before LIC cloning, LIC vectors were digested with ApaI and treated with T4 DNA polymerase in the presence of dTTP. Tm-22-cYFP, tm-2-cYFP, MP-nYFP, p50-nYFP, Tm-22 MHD (D481V) mutant, and P-loop (K191R) mutant were generated by overlapping PCR. Tm-22 deletion mutants and TMV MP dysfunctional mutants were generated by specific primers. All PCR products treated with T4 DNA polymerase in the presence of dATP were cloned into the treated LIC vectors as described (Zhao et al., 2016). Rop or mRop tag was amplified by PCR, digested and ligated into the related plasmids. Primers used for plasmid construction in this study are listed in Supplemental Table S1.
Agrobacterium tumefaciens-Mediated Transient Expression and LaCl3 Treatment
A. tumefaciens-mediated transient expression was performed by agroinfiltration approach (Y. Wang et al., 2015). GV3101 strains containing the relevant expression vector were grown overnight, collected by centrifugation, and resuspended to an optical density of OD600 = 1.0 in infiltration buffer (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone, pH 5.6). Agrobacteria suspensions were infiltrated into 4- to 5-week-old N. benthamiana leaves with needleless syringes. To inhibit cell death, 2 mm LaCl3 in distilled water was infiltrated into leaves at 16 h postagroinfiltration.
Membrane Fractionation
N. benthamiana leaves were homogenized in a mortar on ice with extract buffer (0.33 m Suc, 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 5 mm DTT, and 1× protease inhibitor cocktail). The lysate was filtered with one layer of Miracloth and was centrifuged at 10,000g for 10 min at 4°C. The supernatant was ultracentrifuged at 100,000g for 1 h at 4°C to obtain soluble and microsomal membrane fractions.
Aqueous two-phase partitioning to purify the PM was performed as described (Liu et al., 2009). Briefly, microsomal membrane was suspended in partitioning buffer (0.33 m Suc, 5 mm potassium phosphate, pH 7.8, 1 mm DTT, and 0.1 mm EDTA) and loaded into polymer solution with a DexT500/PEG4000 concentration of 6.2% (w/w) for partitioning.
The 20 to 50% (w/w) Suc gradients in the presence or absence of Mg2+ were modified from Michael Weaver et al. (2006). The gradient without Mg2+ contained 10 mm Tris, pH 7.5, 1 mm DTT, and 4 mm EDTA, while for the gradient containing 7 mm Mg2+, the concentration of EDTA was reduced to 2 mm.
For solubility test, the membrane fraction was resuspended in either extraction buffer as a control, 2 m urea buffer (extraction buffer plus 2 m urea), alkaline buffer (100 mm Na2CO3, pH 11, 0.33 m Suc, 5 mm EDTA, 5 mm DTT, and 1× protease inhibitor cocktail), or Triton X-100 buffer (extraction buffer plus 1% Triton X-100) for 1 h at 4°C and then ultracentrifuged at 100,000g for 1 h at 4°C to separate new supernatant and pellet fractions. The control proteins were detected by the antibodies anti-BiP (Santa Cruz), anti-H+-ATPase (Agrisera), and anti-V-ATPase (Agrisera).
Protein Analyses and Co-IP
For protein analysis, total proteins from N. benthamiana leaves were extracted with a ratio of 1:2 of Laemmli buffer and then separated by SDS-PAGE for western blot using the indicated antibodies (Du et al., 2013). For co-IP assays, total proteins of 2 g leaf tissues were extracted using prechilled 2.5× IP buffer (10% [v/v] glycerol, 25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 10 mm DTT, 1× protease inhibitor cocktail, and 0.2% [v/v] NP-40). Protein extracts were incubated with 30 µL GFP-trap_A (ChromoTek) beads for 4 h at 4°C. The beads were washed four times with ice-cold IP buffer at 4°C and then boiled in 50 µL 2× Laemmli buffer. IP samples were analyzed by SDS-PAGE, immunoblotted using anti-myc (Abmart) or anti-GFP (ChromoTek) antibodies, and detected using an ECL western blotting substrate or SuperSignal West Femto Maximum Sensitivity Substrate (Pierce).
Confocal Microscopy
We used an agroinfiltration approach to transiently express proteins in N. benthamiana for confocal imaging. The leaves were detached at 36 or 48 hpi, and confocal imaging was performed using an inverted Zeiss LSM 710 laser scanning microscope. For z-stack projection, a series of z-stack images were collected and then projected and processed by using ImageJ.
Trypan Blue Staining
N. benthamiana leaves were boiled for 10 min in a 2:1 mixture of ethanol and staining stock solution (mix 10 g phenol, 10 mL glycerol, 10 mL lactic acid, 10 mL water, and 20 mg trypan blue together) for staining. The leaves were then washed with destaining solution (2.5 g/mL chloral hydrate in water).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Tm-22 (AAQ10736.1); tm-2 (AAQ10734.1); TMV MP (BAF93925.1); AtRop10 (AT3G48040).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used in this study.
Supplemental Figure S1. LaCl3 treatment inhibited Tm-22-mediated cell death.
Supplemental Figure S2. Expression of fusion proteins for BiFC.
Supplemental Figure S3. tm-2 is localized in the cytoplasm and at the PM.
Supplemental Figure S4. N-terminal tag affects Tm-22 function.
Supplemental Figure S5. Suc gradient analysis of Tm-22 in the presence of MP.
Supplemental Figure S6. MP and Tm-22 or tm-2 are colocalized at the PM.
Supplemental Figure S7. Rop tag does not affect Tm-22 function.
Supplemental Figure S8. Tm-22 mutants failed to induce cell death in the presence or absence of MP.
Supplemental Figure S9. Subcellular localization of TMV MP and mutants.
Supplemental Figure S10. Confocal images of Tm-22-YFP.
Supplementary Material
Glossary
- HR
hypersensitive response
- PM
plasma membrane
- TMV
Tobacco mosaic virus
- ToMV
Tomato mosaic virus
- MP
movement protein
- PD
plasmodesmata
- ER
endoplasmic reticulum
- hpi
hours postinoculation
- co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
Footnotes
This work was supported by the National Natural Science Foundation of China (31530059, 31421001, 31470254, 31300134, and 31370180), the National Basic Research Program of China (2014CB138400), and the National Transgenic Program of China (2016ZX08009-003-001 and 2016ZX08009001-004).
Articles can be viewed without a subscription.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari K, Di Donato M, Dolja VV, Heinlein M (2014) Myosins VIII and XI play distinct roles in reproduction and transport of tobacco mosaic virus. PLoS Pathog 10: e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FL, Schulze-Lefert P, Shen QH (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V, van der Laak J, Ferralli J, Suslova E, Kwon MO, Heinlein M (2000) Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein. J Virol 74: 11339–11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J, Padmanabhan M, Dinesh-Kumar SP (2008) Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135 [DOI] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Knorr D, Schuster G, Zambryski P (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Wong ML, Shaw AL, Prasad BV, Zambryski P (1992) Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 4: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Staskawicz BJ (2006) NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18: 2782–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Tham WH, Baker BJ (2000) Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc Natl Acad Sci USA 97: 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Haudenshield JS, Hull RJ, Wolf S, Beachy RN, Lucas WJ (1992) Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Du Y, Zhao J, Chen T, Liu Q, Zhang H, Wang Y, Hong Y, Xiao F, Zhang L, Shen Q, Liu Y (2013) Type I J-domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLoS Pathog 9: e1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt S, Boevink PC, Armstrong MR, Ramos MB, Hein I, Birch PR (2012) Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell 24: 5142–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chung EH, Eitas TK, Dangl JL (2011) Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA 108: 7619–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howles P, Lawrence G, Finnegan J, McFadden H, Ayliffe M, Dodds P, Ellis J (2005) Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact 18: 570–582 [DOI] [PubMed] [Google Scholar]
- Jeong RD, Chandra-Shekara AC, Barman SR, Navarre D, Klessig DF, Kachroo A, Kachroo P (2010) Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc Natl Acad Sci USA 107: 13538–13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kahn TW, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy RN (1998) Domains of the TMV movement protein involved in subcellular localization. Plant J 15: 15–25 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, Kawasaki T, Shimamoto K (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Fujiwara T, Yao A, Housen Y, Hayashi K, Shimamoto K (2014) Palmitoylation-dependent membrane localization of the rice resistance protein pit is critical for the activation of the small GTPase OsRac1. J Biol Chem 289: 19079–19088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto-Katou A, Katou S, Hirai K, Meshi T, Ohashi Y, Mitsuhara I (2011) Identification of an amino acid residue required for differential recognition of a viral movement protein by the Tomato mosaic virus resistance gene Tm-2(2). J Plant Physiol 168: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Kotlizky G, Katz A, van der Laak J, Boyko V, Lapidot M, Beachy RN, Heinlein M, Epel BL (2001) A dysfunctional movement protein of tobacco mosaic virus interferes with targeting of wild-type movement protein to microtubules. Mol Plant Microbe Interact 14: 895–904 [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfermeijer FC, Dijkhuis J, Sturre MJ, de Haan P, Hille J (2003) Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol Biol 52: 1037–1049 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Warmink J, Hille J (2005) The products of the broken Tm-2 and the durable Tm-2(2) resistance genes from tomato differ in four amino acids. J Exp Bot 56: 2925–2933 [DOI] [PubMed] [Google Scholar]
- Lavy M, Yalovsky S (2006) Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J 46: 934–947 [DOI] [PubMed] [Google Scholar]
- Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci USA 107: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Nelson RS (2013) The cell biology of Tobacco mosaic virus replication and movement. Front Plant Sci 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- McLean BG, Zupan J, Zambryski PC (1995) Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T, Motoyoshi F, Maeda T, Yoshiwoka S, Watanabe H, Okada Y (1989) Mutations in the tobacco mosaic virus 30-kD protein gene overcome Tm-2 resistance in tomato. Plant Cell 1: 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael Weaver L, Swiderski MR, Li Y, Jones JD (2006) The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J 47: 829–840 [DOI] [PubMed] [Google Scholar]
- Moore PJ, Fenczik CA, Deom CM, Beachy RN (1992) Developmental changes in plasmodesmata in transgenic tobacco expressing the movement protein of tobacco mosaic virus. Protoplasma 170: 115–127 [Google Scholar]
- Motoyoshi F, Oshima N (1975) Infection with tobacco mosaic virus of leaf mesophyll protoplasts from susceptible and resistant lines of tomato. J Gen Virol 29: 81–91 [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363 [DOI] [PubMed] [Google Scholar]
- Oparka KJ. (1994) Plasmolysis: new insights into an old process. New Phytol 126: 571–591 [Google Scholar]
- Otsuki Y, Shimomura T, Takebe I (1972) Tobacco mosaic virus multiplication and expression of the N gene in necrotic responding tobacco varieties. Virology 50: 45–50 [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh-Kumar SP (2013) Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog 9: e1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiró A, Martínez-Gil L, Tamborero S, Pallás V, Sánchez-Navarro JA, Mingarro I (2014) The Tobacco mosaic virus movement protein associates with but does not integrate into biological membranes. J Virol 88: 3016–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Dubiella U, Kim SH, Sloss DI, Dowen RH, Dixon JE, Innes RW (2014) Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on s-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol 164: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Innes RW (2013) Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol 4: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P (2008) The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale M, Bernoux M, Ve T, Kobe B, Thrall PH, Ellis JG, Dodds PN (2012) Intramolecular interaction influences binding of the Flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog 8: e1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco MA, Mansoor S, Moffett P (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Rafiqi M, Hurley U, Lawrence GJ, Bernoux M, Hardham AR, Ellis JG, Dodds PN, Jones DA (2012) N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol Plant Microbe Interact 25: 379–392 [DOI] [PubMed] [Google Scholar]
- Takken FL, Goverse A (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol 15: 375–384 [DOI] [PubMed] [Google Scholar]
- Tameling WI, Baulcombe DC (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59: 1383–1397 [DOI] [PubMed] [Google Scholar]
- Waigmann E, Lucas WJ, Citovsky V, Zambryski P (1994) Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA 91: 1433–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Ji J, El-Kasmi F, Dangl JL, Johal G, Balint-Kurti PJ (2015) Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog 11: e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zheng X, Yu B, Han S, Guo J, Tang H, Yu AY, Deng H, Hong Y, Liu Y (2015) Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy 11: 2259–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Ohnesorge S, Silber MV, Pfitzner AJ (2004) The Tomato mosaic virus 30 kDa movement protein interacts differentially with the resistance genes Tm-2 and Tm-2(2). Arch Virol 149: 1499–1514 [DOI] [PubMed] [Google Scholar]
- Weber H, Pfitzner AJ (1998) Tm-2(2) resistance in tomato requires recognition of the carboxy terminus of the movement protein of tomato mosaic virus. Mol Plant Microbe Interact 11: 498–503 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Zhang Y, Jones JD, Parker JE (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Wolf S, Deom CM, Beachy RN, Lucas WJ (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao J, Liu S, Zhang DP, Liu Y (2013) Tm-22 confers different resistance responses against tobacco mosaic virus dependent on its expression level. Mol Plant 6: 971–974 [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu Q, Hu P, Jia Q, Liu N, Yin K, Cheng Y, Yan F, Chen J, Liu Y (2016) An efficient Potato virus X-based microRNA silencing in Nicotiana benthamiana. Sci Rep 6: 20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Liu Q, Zhang H, Jia Q, Hong Y, Liu Y (2013) The rubisco small subunit is involved in tobamovirus movement and Tm-22-mediated extreme resistance. Plant Physiol 161: 374–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.