Domestication-selective GmZF351 encoding tandem CCCH zinc finger protein promotes seed oil accumulation in soybean.
Abstract
Seed oil is a momentous agronomical trait of soybean (Glycine max) targeted by domestication in breeding. Although multiple oil-related genes have been uncovered, knowledge of the regulatory mechanism of seed oil biosynthesis is currently limited. We demonstrate that the seed-preferred gene GmZF351, encoding a tandem CCCH zinc finger protein, is selected during domestication. Further analysis shows that GmZF351 facilitates oil accumulation by directly activating WRINKLED1, BIOTIN CARBOXYL CARRIER PROTEIN2, 3-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, DIACYLGLYCEROL O-ACYLTRANSFERASE1, and OLEOSIN2 in transgenic Arabidopsis (Arabidopsis thaliana) seeds. Overexpression of GmZF351 in transgenic soybean also activates lipid biosynthesis genes, thereby accelerating seed oil accumulation. The ZF351 haplotype from the cultivated soybean group and the wild soybean (Glycine soja) subgroup III correlates well with high gene expression level, seed oil contents and promoter activity, suggesting that selection of GmZF351 expression leads to increased seed oil content in cultivated soybean. Our study provides novel insights into the regulatory mechanism for seed oil accumulation, and the manipulation of GmZF351 may have great potential in the improvement of oil production in soybean and other related crops.
Soybean (Glycine max), a major crop for oil and protein resources, accounts for 56% of total oilseed production in the world (Wilson, 2008). Given that soybean seed contains about 190 to 230 g kg−1 oil on a dry weight basis (Bellaloui et al., 2015), it is of great importance to adopt traditional breeding and genetic engineering approaches to breed high-oil-content varieties to satisfy the increasing demand for soybean oil.
In plants, triacylglycerols (TAGs) represent a relatively high proportion compared with other compounds in seed oil. TAG accumulation is initiated from de novo synthesis of fatty acids (FAs) in plastid, and many key enzymes are involved in this process, including acetyl-CoA carboxylase and 3-ketoacyl-acyl carrier protein synthase (KAS). FAs then are transferred into the endoplasmic reticulum and esterified on a glycerol backbone to form TAGs via the Kennedy pathway, which are catalyzed by glycerol 3-phosphate acyltransferase, lysophosphatidic acid acyltransferase, phosphatidic acid phosphohydrolase, and diacylglycerol acyltransferase (Baud and Lepiniec, 2010).
Besides regulation by key enzyme activity, seed oil accumulation is precisely controlled by transcription factors that determine the expression profiles of the biosynthetic genes. Among them, four regulators, LEAFY COTYLEDON1 (LEC1), LEC2, FUSCA3, and ABSCISIC ACID INSENSITIVE3, globally harness seed maturation and the accumulation of seed reserves in Arabidopsis (Arabidopsis thaliana; Baud and Lepiniec, 2009). Mutations of these regulators lead to severe defects in seed oil (Mendoza et al., 2005; Mu et al., 2013; Roscoe et al., 2015). The carbon flux from glycolysis, which is promoted by WRINKLED1 (WRI1), exerts a positive effect on storage oil level (Cernac and Benning, 2004; Baud et al., 2009). WRI1 encodes an AP2/EREBP transcription factor and enhances oil accumulation through up-regulation of the expression of genes involved in both glycolysis and FA biosynthesis, such as BIOTIN CARBOXYL CARRIER PROTEIN2 (BCCP2), PKpα, and PKpβ1 (Baud et al., 2009; To et al., 2012). WRI1 functions downstream of LEC1 and LEC2 to specify their roles in the regulatory pathway (Baud et al., 2007; Mu et al., 2008). In addition to these, other transcription factors have been uncovered to manipulate oil content in Arabidopsis, including GLABRA2, TRANSPARENT TESTA1 (TT1), TT2, and bZIP67 (R. Chen et al., 2012; Mendes et al., 2013; Liu et al., 2014; Chen et al., 2015).
Soybean seed oil content is a key agronomical trait targeted by artificial selection and may be controlled by plenty of selective regions in the soybean genome. A set of selection sweeps related to oil content, embodied FA synthesis genes, were unveiled (Zhou et al., 2015). Previously, we identified several soybean transcriptional regulators promoting seed oil synthesis. Overexpression of GmDOF4 and GmDOF11 increased lipid content in seeds of transgenic Arabidopsis plants via the direct activation of lipid biosynthesis genes and the repression of storage protein genes (Wang et al., 2007). Overexpression of GmDOF4 in Chlorella ellipsoidea also enhanced lipid contents (Zhang et al., 2014). GmMYB73 functioned as a repressor for GL2, a negative regulator of oil accumulation (Shen et al., 2006), and relieved GL2-inhibited expression of PLDα1 to accelerate the conversion of phosphatidylcholine to TAG (Liu et al., 2014). Another transcription factor, GmbZIP123, also elevated lipid contents in seeds of transgenic Arabidopsis plants by activating Suc transporter genes and cell wall invertase genes for sugar translocation and sugar breakdown, respectively (Song et al., 2013). Recently, through comparative RNA sequencing (RNA-seq) analysis in developing seeds from cultivated soybean and wild soybean (Glycine soja), we identified two gene coexpression networks in cultivated soybean for seed trait regulation and found that GmNFYA in one network increases seed oil content in transgenic Arabidopsis plants (Lu et al., 2016).
From our previous RNA-seq analysis for seed-preferred genes (Song et al., 2013) and the gene coexpression networks established (Lu et al., 2016), we identified an additional gene, GmZF351, encoding tandem CCCH zinc finger proteins. GmZF351 enhanced the oil contents in seeds of transgenic Arabidopsis plants and transgenic soybean plants through the activation of lipid biosynthesis-related genes. Domestication appeared to target the GmZF351 locus and thus brought about a boost to seed oil content in cultivated soybean. Our findings shed light on the regulatory mechanisms of oil accumulation during soybean domestication and may be useful for improving soybean seed quality.
RESULTS
GmZF351 Is a Domestication Gene Associated with Seed Oil Accumulation
Soybean seed oil content was targeted by artificial selection, as evidenced by the fact that cultivated soybean seeds have higher oil content than wild soybean (Zhou et al., 2015). In our previous study, we established two cultivar-specific gene coexpression networks based on the analysis of transcriptomes from developing seeds of cultivated and wild soybeans (Lu et al., 2016). Glyma06g44440 was found in one network, and it was also a seed-preferred gene whose abundance was at least 10-fold higher in seeds than those in roots, leaves, or stems (Song et al., 2013). The gene at the Glyma06g44440 locus encoded a protein of the tandem CCCH zinc finger (TZF) family and was named GmZF351 (where 351 indicates the total amino acids in the protein). Moreover, GmZF351 was located at the known quantitative trait locus, namely SEED OIL PLUS PROTEIN1-2, by searching the soybean quantitative trait locus map (http://soybase.org/). Thus, GmZF351 function was then investigated in detail.
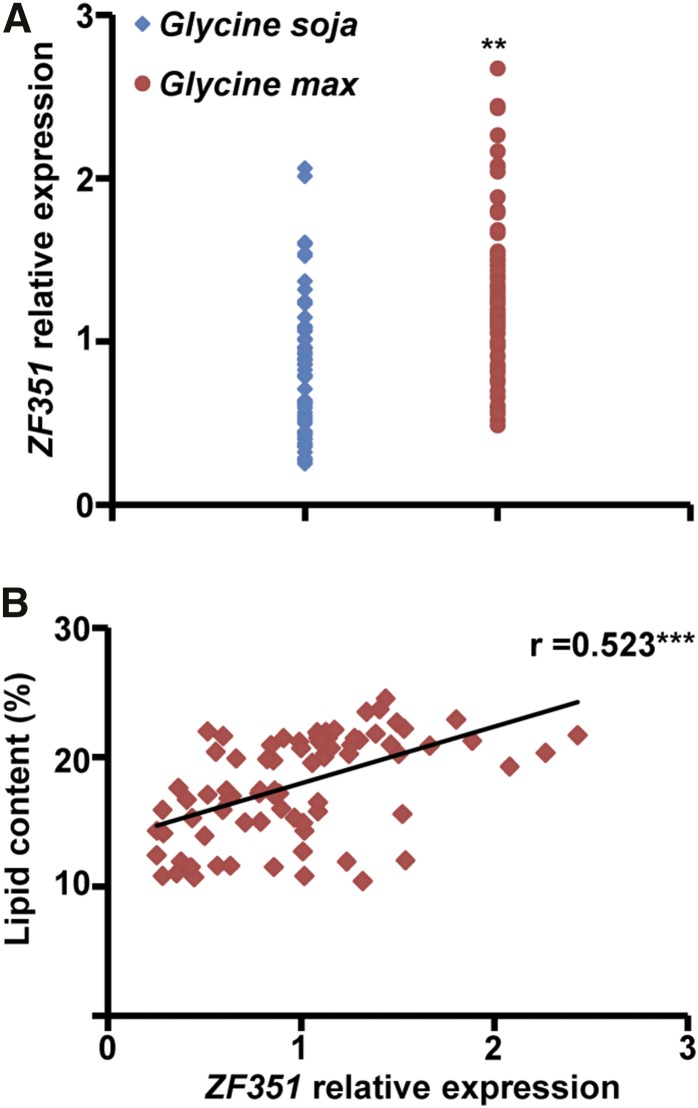
Because GmZF351 was identified in the cultivar-specific gene coexpression network, we compared the gene expression levels of ZF351 at the H5 stage of developing seeds between wild soybean and cultivated soybean accessions. ZF351 expression in the cultivated soybean was significantly higher on average than that in the wild soybean (Fig. 1A). We further examined the relationship of gene expression with seed lipid contents in all accessions and found that ZF351 expression positively correlated with seed lipid contents (Fig. 1B). Additionally, ZF351 expression contributed to the seed lipid contents by 27.4%. These results suggest that ZF351 may play a major role in seed oil accumulation during the domestication process.
Figure 1.
ZF351 expression and correlation with seed oil content in wild and cultivated soybeans. A, ZF351 transcript levels in H5 seeds of wild soybean accessions (n = 51) and cultivated soybean accessions (n = 48). B, Pearson correlation analysis (r) between seed oil content of soybeans and ZF351 expression level of H5 seeds. Asterisks indicate significant differences (**, P < 0.01 and ***, P < 0.001).
Since ZF351 showed higher expression in cultivated soybean than in wild soybean, it was possible that the difference in expression levels was relevant to the variations of promoter regions. We then sequenced ∼4-kb genomic regions of ZF351 in the 51 cultivated soybean and 48 wild soybean accessions (Supplemental Table S1) and calculated the nucleotide diversity of the promoters and coding sequences (CDSs). The promoter region of cultivated soybean (nucleotide diversity [π] = 0.000488) harbored about 7% of the variation found in wild soybean (π = 0.007322), while the CDS region of cultivated soybean (π = 0.000638) had about 11% of the variation found in wild soybean (π = 0.005831). Significantly negative Tajima’s D value (−2.588) for the promoter of ZF351 in cultivated soybean indicated strong selection at the ZF351 locus during domestication from wild to cultivated soybeans. In addition, the CDS region evolved neutrally during the domestication process (Table I). These results indicate that the ZF351 promoter region appears to be selected during soybean domestication. Since ZF351 expression is positively related to seed oil content, it is most likely that GmZF351 is associated with an increase of seed oil content during domestication.
Table I. Summary of the nucleotide diversity of ZF351 in cultivated soybean and wild soybean.
m indicates single-nucleotide polymorphism (SNP). θ indicates Watterson’s estimator of θ per site, which is calculated based on the total number of segregating sites, and values are θ × 103. π indicates average pairwise differences per site between sequences, and values are π × 103. πGm/πGs indicates the relative genetic diversity between cultivated soybean (Gm) and wild soybean (Gs), and values are percentages. D indicates Tajima’s D, which is calculated based on m and π.
| Gene Locus | Sequence Loci | Cultivated Soybean |
Wild Soybean |
πGm/πGs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| m | θ | π | D | m | θ | π | D | |||
| ZF351 | 5′ promoter | 20 | 2.520 | 0.488 | −2.588 | 90 | 6.967 | 7.322 | 0.180 | 7% |
| CDS | 5 | 1.067 | 0.638 | −0.986 | 24 | 5.066 | 5.831 | 0.490 | 11% | |
Molecular Characterization of GmZF351
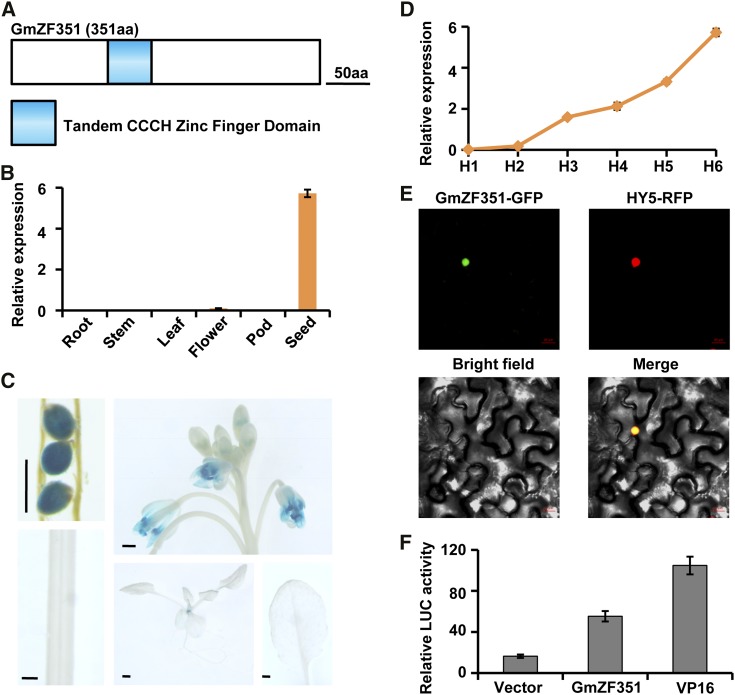
GmZF351 possessed one plant-specific TZF motif and, thus, belonged to the tandem CCCH zinc finger protein subfamily (Fig. 2A; Supplemental Fig. S1). Phylogenetic analysis revealed that it was clustered with SOMNUS (Supplemental Fig. S1), which was expressed specifically in seeds of Arabidopsis (Kim et al., 2008). GmZF351 also was expressed abundantly in soybean seeds and weakly in flower (Fig. 2B). The tissue-specific expression pattern of GmZF351 was examined using transgenic plants containing the GUS reporter gene driven by the GmZF351 promoter. In agreement with reverse transcription (RT)-quantitative PCR (qPCR) data, GUS activity was detected mainly in seeds and flowers but only slightly in seedlings. GUS activity was not detected in leaves or roots (Fig. 2C). In addition, the GmZF351 transcript accumulated steadily at stage H1 to H6 during seed development, implying its possible role in seed oil accumulation (Fig. 2D).
Figure 2.
Molecular characterization of GmZF351. A, Schematic representation of GmZF351 structure. aa, Amino acids. B, GmZF351 expression in various organs of soybean plants. Error bars indicate sd (n = 3). C, Histochemical analysis of GmZF351 promoter activity. The GUS reporter is driven by the GmZF351 promoter in transgenic Arabidopsis plants. Three GUS-expressing lines were investigated, and all exhibit a similar pattern. Bars = 0.5 mm. D, Expression pattern of GmZF351 at six seed developmental stages. Error bars indicate sd (n = 3). E, Subcellular localization of the GmZF351 protein revealed by transient expression assay in tobacco leaves. Bars = 20 μm. F, Transcriptional activation activity of GmZF351 in the transient Arabidopsis protoplast assay. Error bars indicate sd (n = 4).
To determine the subcellular location of GmZF351, its CDS was fused to GFP. Then, the fusion gene under the control of the 35S promoter was transformed into tobacco (Nicotiana benthamiana) leaves using a transient expression system. GmZF351-GFP was colocalized with nuclear transcription factor HY5 (red fluorescent protein [RFP]; Zhang et al., 2015), suggesting that GmZF351 is targeted to the nucleus of cells (Fig. 2E).
TZF proteins in Arabidopsis were found to possess transcription activation activity in a yeast system (Chai et al., 2015). To examine whether GmZF351 possessed transcription activation activity, a dual-luciferase reporter assay was performed using Arabidopsis protoplasts with Renilla luciferase (LUC) as the internal control. Compared with the negative control pRT-BD vector, the effector plasmid 35S:GAL4BD-GmZF351 enhanced the reporter LUC activity by approximately 2-fold, suggesting that GmZF351 has transactivation capacity to promote the expression of reporter genes in protoplasts (Fig. 2F).
GmZF351 Enhances Seed Oil Content in Transgenic Arabidopsis Plants
To investigate whether GmZF351 could affect seed oil content, we generated GmZF351-overexpressing transgenic Arabidopsis lines. The FLAG tag was fused to the protein for later analysis. Gene expression revealed by RT-qPCR with At4g12590 encoding an endoplasmic reticulum membrane protein complex subunit-like protein and AtACTIN2 as internal controls (Maeo et al., 2009; Dekkers et al., 2012) was largely consistent between different transgenic lines (Supplemental Fig. S2). Three homozygous lines with relatively high expression of GmZF351 (OE-3, OE-19, and OE-39) were selected for further analysis (Supplemental Fig. S2). Three-week-old GmZF351 transgenic plants showed a mild dwarf phenotype, whereas 5-week-old transgenic plants showed no obvious phenotypic change in comparison with Columbia-0 during rosette growth (Supplemental Fig, S3).
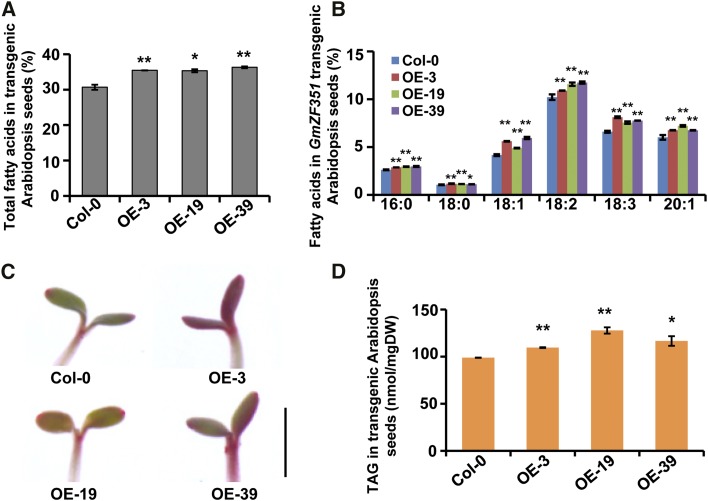
The transgenic seeds were subjected to FA determination. Overall, significant increases were observed in total FA contents in all GmZF351-overexpressing lines (Fig. 3A; Supplemental Fig. S2). In comparison with Columbia-0 (Col-0), the 35S:GmZF351 lines OE-3, OE-19, and OE-39 showed 15%, 15%, and 18% increases in FA content, respectively. Each FA composition also was compared, and most of the compositions exhibited significantly higher contents in transgenic lines than in the Col-0 plant (Fig. 3B). Furthermore, when 7-d-old seedlings were stained with Fat Red 7B, which is used for lipid and fat staining, we observed that the red color intensity was strengthened in young cotyledons and hypocotyls of overexpressing plants, suggesting a higher level of lipid in these plants compared with the control (Fig. 3C). TAG represents the major component of seed oil, and its level also was measured. The accumulation of TAG was enhanced significantly in mature seeds of GmZF351 transgenic plants. The TAG content was increased by up to 29% in seeds of the 35S:GmZF351 lines compared with Col-0 (Fig. 3D). These results indicate that the overexpression of GmZF351 leads to the accumulation of lipids in seeds and young vegetative tissues.
Figure 3.
Overexpression of GmZF351 increases seed oil content in transgenic Arabidopsis plants. A, Total FA contents in seeds of GmZF351 transgenic plants. Error bars indicate sd (n = 4). B, FA composition in seeds of GmZF351 transgenic plants. Error bars indicate sd (n = 4). C, Fat Red 7B staining of GmZF351 transgenic plants. Bar = 2 mm. D, TAG contents in seeds of GmZF351 transgenic plants. Error bars indicate sd (n = 3). DW, Dry weight. Asterisks indicate significant differences compared with Col-0 (*, P < 0.05 and **, P < 0.01).
Lipid Biosynthesis Genes Are Regulated by GmZF351
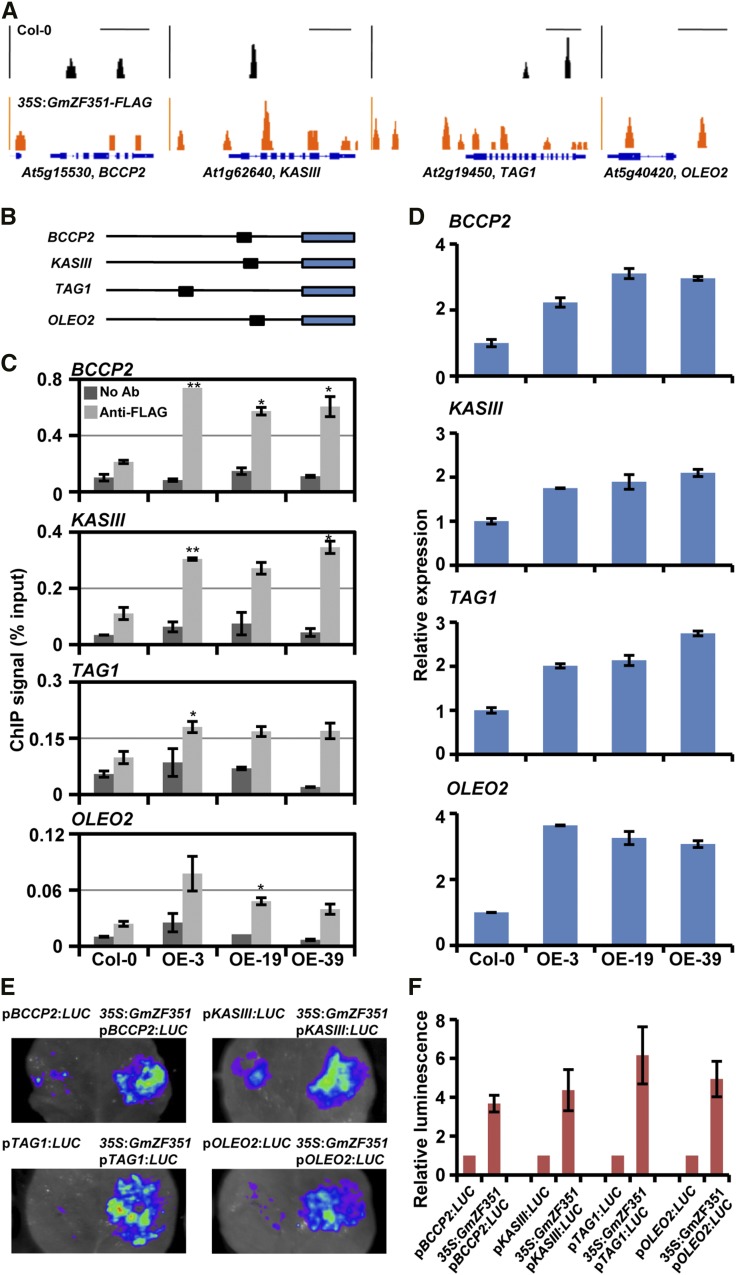
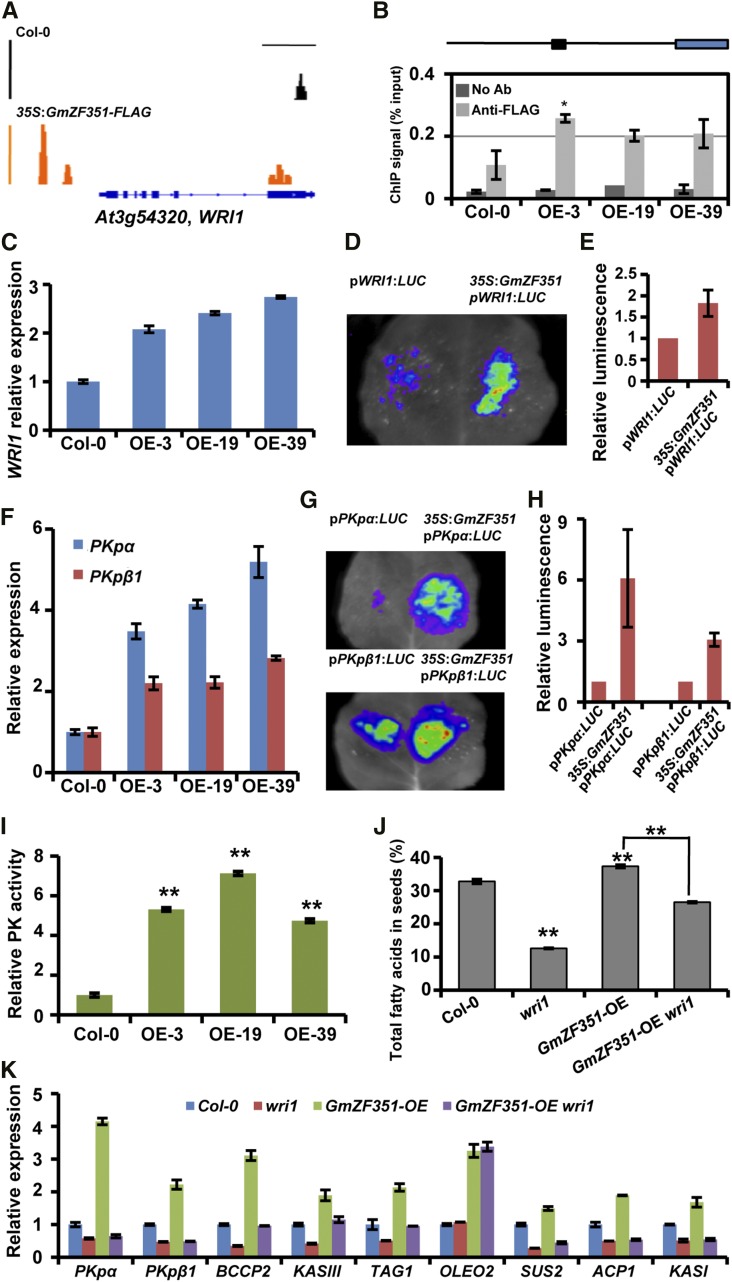
Because GmZF351 not only had strong transcription activation activity but also was located in nuclei, we hypothesized that the accumulation of lipid in GmZF351-overexpressing plants might result from the direct activation of downstream genes. We then performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis to explore the potential target genes of GmZF351 using transgenic lines of 35S:GmZF351-FLAG (OE-19 and OE-39). Multitudes of loci with a 2-fold increase in read number were identified in all biological replicates in comparison with Col-0 (Supplemental Table S2). Four candidate genes that encoded key enzymes/proteins in lipogenesis and lipid storage, BIOTIN CARBOXYL CARRIER PROTEIN2 (BCCP2), KASIII, DIACYLGLYCEROL O-ACYLTRANSFERASE1 (TAG1), and OLEOSIN2 (OLEO2), were identified from thousands of loci (Fig. 4A). We further designed specific primers for each gene according to the positions of peaks in their promoter regions (Fig. 4B) and validated gene enrichment by chromatin immunoprecipitation (ChIP)-qPCR in young siliques of transgenic Arabidopsis and Col-0 plants. The ChIP signals were enriched significantly at the promoter region of these genes in the GmZF351 transgenic plants, indicating that GmZF351 can bind to the promoters of the four lipid biosynthesis genes (Fig. 4C). However, the FaTA gene, which was identified in the ChIP-seq analysis and encoded an enzyme in the termination of FA synthesis by hydrolyzing the acyl carrier protein (ACP) complexes, was not enriched in ChIP-qPCR validation at the peak in the FaTA promoter region in GmZF351 transgenic plants compared with that in Col-0 (Supplemental Fig. S4). The At3g22960 (PKpα) promoter fragments (F1–F5) were used as negative controls, which were neither identified in ChIP-seq analysis nor enriched in ChIP-qPCR validation (Supplemental Fig. S4). All these results indicate that GmZF351 specifically targets promoters of several lipid biosynthesis/storage genes.
Figure 4.
GmZF351 activates the expression of BCCP2, KASIII, TAG1, and OLEO2 by binding to the promoter regions. A, ChIP-seq genome browser views of GmZF351 occupancy at four selected loci in Col-0 and 35S:GmZF351-FLAG transgenic plants. Orange peaks represent read numbers in transgenic line OE-19, and black peaks represent read numbers in Col-0. Bars indicate 1-kb regions in the genome. B, Positions of the promoter sequences in GmZF351-binding genes identified by ChIP-seq. The black lines and blue rectangles indicate 2-kb promoter regions and CDS regions, respectively. The black rectangles indicate promoter fragments used for ChIP-qPCR analysis. C, ChIP-qPCR analysis of the GmZF351-binding genes. Chromatin immunoprecipitated without antibody was used as a negative control, while isolated chromatin before immunoprecipitation was used as an input control. The ChIP signal (% input) was quantified as a percentage of input DNA by qPCR. Asterisks indicate significant differences of anti-FLAG ChIP signal in GmZF351-OE plants compared with those in Col-0 (*, P < 0.05 and **, P < 0.01). Error bars indicate sd (n = 3). D, Expression of BCCP2, KASIII, TAG1, and OLEO2 in GmZF351 transgenic Arabidopsis plants. RNA samples were prepared from 5-d-old seedlings. The mRNA level (relative to At4g12590) of each gene in Col-0 was set to 1. Error bars indicate sd (n = 3). E, GmZF351 activates the promoter activity of BCCP2, KASIII, TAG1, and OLEO2 genes by transient expression assay in tobacco leaves. The promoters of downstream genes are fused with LUC as reporters. The A. tumefaciens harboring reporter vector was mixed with A. tumefaciens harboring 35S:GmZF351-FLAG and infiltrated into tobacco leaves. A. tumefaciens harboring pGWB412 was used as a negative control. LUC images were taken 2 d after infiltration. F, Quantitative analysis of the luminescence intensity in E. Luminescence intensity is analyzed with IndiGo software. Error bars indicate sd (n = 5).
The expression of BCCP2, KASIII, TAG1, and OLEO2 as further examined by RT-qPCR, and these genes showed obvious elevation of expression in transgenic plants in comparison with Col-0 (Fig. 4D). These results indicate that GmZF351 can enhance the expression of downstream genes.
Due to the transcriptional activation ability of GmZF351, we then tested whether GmZF351 could activate the expression of the four genes, BCCP2, KASIII, TAG1, and OLEO2, in an in vivo transient expression system. The 1.5-kb promoter sequences of BCCP2, KASIII, TAG1, and OLEO2 were fused to the LUC gene, and the agrobacteria harboring the resulting reporter plasmids were infiltrated into N. benthamiana leaves together with the agrobacteria harboring the effector plasmid 35S:GmZF351-FLAG or the empty vector control. Infiltration of agrobacteria harboring the effector 35S:GmZF351-FLAG led to a significant increase in reporter LUC activity (Fig. 4, E and F), supporting that GmZF351 could trigger the activation of BCCP2, KASIII, TAG1, and OLEO2 in vivo. All these results indicate that GmZF351 directly activates the expression of the above lipid biosynthesis genes to regulate the accumulation of seed oil.
GmZF351 Positively Regulates Lipid Biosynthesis by Enhancing WRI1 Activity
In addition to genes in the lipid biosynthesis pathway, we also identified the transcription factor gene WRI1 as a potential direct target of GmZF351 by ChIP-seq (Fig. 5A). WRI1 has been found to act upstream of BCCP2 and KASIII to orchestrate lipid biosynthesis (Santos-Mendoza et al., 2008). We then examined the WRI1 ChIP signal and its gene expression in GmZF351-overexpressing transgenic plants. The ChIP signal intensity and expression of the WRI1 gene were higher in the transgenic plants than in Col-0 (Fig. 5, B and C). The GmZF351 effector plasmid also enhanced pWRI1:LUC reporter activity when agrobacteria harboring each plasmid were cotransfected into tobacco leaves, indicating that GmZF351 activates the promoter activity of the WRI1 gene (Fig. 5, D and E).
Figure 5.
GmZF351 directly activates WRI1 and WRI1-regulated gene expression. A, ChIP-seq genome browser views of GmZF351 occupancy at the WRI1 locus in Col-0 and 35S:GmZF351-FLAG transgenic plants. B, The WRI1 promoter region is enriched by ChIP-qPCR in GmZF351 transgenic plants. The schematic diagram represents the location of the promoter fragment (black rectangle) used for ChIP-qPCR analysis in the 2-kb promoter region of WRI1 (black line). The blue rectangle indicates the CDS region of WRI1. No Ab samples were immunoprecipitated without antibody and served as a negative control, while isolated chromatin before immunoprecipitation was used as an input control. The ChIP signal (% input) was quantified as a percentage of input DNA by qPCR. Error bars indicate sd (n = 3). The asterisk indicates a significant difference of anti-FLAG ChIP signal in GmZF351-OE plants compared with those in Col-0 (*, P < 0.05). C, RT-qPCR analyses of WRI1 expression. RNA samples were prepared from 5-d-old seedlings. The WRI1 mRNA level relative to At4g12590 in Col-0 was set to 1. Error bars indicate sd (n = 3). D, GmZF351 activates WRI1 promoter activity by transient expression assay in tobacco leaves. A. tumefaciens harboring pWRI1:LUC was mixed with A. tumefaciens harboring 35S:GmZF351-FLAG and infiltrated into tobacco leaves. A. tumefaciens harboring pGWB412 was used as a negative control. The LUC image was taken 2 d after infiltration. E, Quantitative analysis of the luminescence intensity in D. Luminescence intensity was analyzed with IndiGo software. Error bars indicate sd (n = 5). F, RT-qPCR analyses of PKpα and PKpβ1 expression in Col-0 and GmZF351 transgenic Arabidopsis lines. Others are as in C. G, GmZF351 activates PKpα and PKpβ1 promoter activity by transient expression assay in tobacco leaves. A. tumefaciens harboring pPKpα:LUC or pPKpβ1:LUC was mixed with A. tumefaciens harboring 35S:GmZF351-FLAG and infiltrated into tobacco leaves. Others are as in D. H, Quantitative analysis of the luminescence intensity in G. Others are as in E. I, Pyruvate kinase (PK) activity of Col-0 and GmZF351 transgenic Arabidopsis plants. Pyruvate kinase activity in Col-0 was set to 1. Error bars indicate sd (n = 4). Asterisks indicate significant differences from Col-0 (**, P < 0.01). J, Total FA contents in seeds of Col-0, wri1, GmZF351-OE, and GmZF351-OE wri1 plants. Error bars indicate sd (n = 4). The values from GmZF351-OE wri1 were only compared with GmZF351-OE. Asterisks indicate significant differences compared with Col-0 or between the compared pairs (**, P < 0.01). K, Expression levels of PKpα, PKpβ1, BCCP2, KASIII, TAG1, OLEO2, SUS2, ACP1, and KASI in 5-d-old seedlings of Col-0, wri1, GmZF351-OE (OE-19), and GmZF351-OE wri1 plants. The mRNA levels (relative to At4g12590) of each gene in Col-0 were set to 1. Error bars indicate sd (n = 3).
Since GmZF351 activated WRI1 expression and WRI1 could promote the expression of PKpα and PKpβ1, which encoded subunits of pyruvate kinase to regulate glycolysis for the production of acetyl-CoA, which is the starting material for FA biosynthesis (Andre et al., 2007), we were interested to investigate whether PKpα and PKpβ1 gene expression was actually increased in the GmZF351-overexpressing plants. Figure 5 showed that both PKpα and PKpβ1 expression was elevated in overexpressing plants. A transient expression assay further demonstrated that GmZF351 enhanced the promoter activity of PKpα and PKpβ1 genes (Fig. 5, G and H). Enzyme activities of pyruvate kinase in developing seeds of Col-0 and overexpressing lines also were measured, and the activities increased significantly in all GmZF351 transgenic plants (Fig. 5I).
To further examine whether WRI1 functioned downstream of GmZF351, we generated a GmZF351-OE wri1 plant and analyzed the FA content in seeds. The total FA level was significantly lower in GmZF351-OE wri1 plants than in GmZF351-OE plants, indicating that GmZF351 function in oil accumulation is partially dependent on WRI1 (Fig. 5J). Consistently, the expression of WRI1-regulated PKpα, PKpβ1, SUS2, ACP1, and KASI was elevated in GmZF351-OE and was nearly unaltered in GmZF351-OE wri1 compared with wri1, suggesting that the promotion of expression of PKpα, PKpβ1, SUS2, ACP1, and KASI by GmZF351 fully/largely depends on WRI1 function (Fig. 5K; Supplemental Fig. S5; Baud et al., 2009; Maeo et al., 2009). BCCP2, KASIII, and TAG1 were not only targets of GmZF351 but also downstream genes of WRI1 (Maeo et al., 2009). Transcripts of BCCP2, KASIII, and TAG1 were accumulated in GmZF351-OE wri1 in comparison with wri1, suggesting that the promotion of their expression by GmZF351 is only partially dependent on WRI1 (Fig. 5K; Supplemental Fig. S5). In addition, the induction of OLEO2 by GmZF351 did not require WRI1 function (Fig. 5K; Supplemental Fig. S5). Examination of the above gene expression with AtACTIN2 as an internal control produced similar results in two GmZF351-OE lines (Supplemental Fig. S5). All these results indicate that GmZF351 promotes oil accumulation in seeds through the activation of WRI1-independent and WRI1-dependent gene expression in the lipid biosynthesis pathway.
GmZF351 Elevates Seed Oil Content in Seeds of Transgenic Soybean Plants
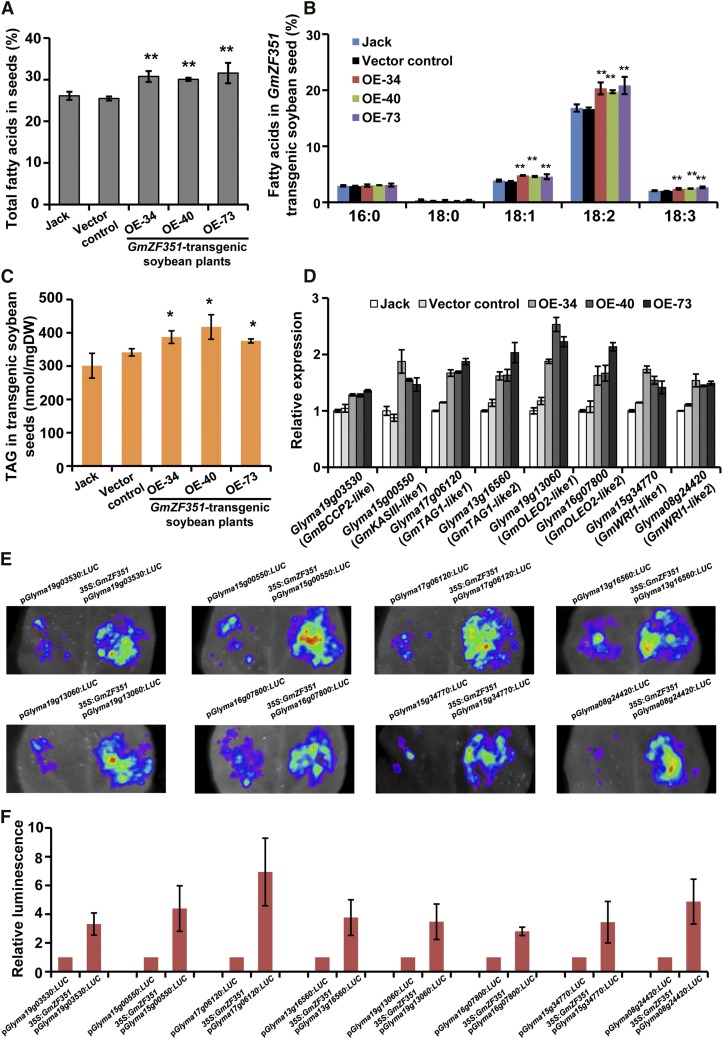
To further study GmZF351 function in soybean, GmZF351, under the control of the 35S promoter, was transformed into soybean, and single-copy transgenic soybean plants were generated (Supplemental Fig. S6). Three higher expressors (OE-34, OE-40, and OE-73) were used for further analysis. The three individual transgenic lines exhibited up to 20% increased total FA content in seeds compared with the original cv Jack or vector control plants (Fig. 6A). The major FA compositions also were elevated in these lines (Fig. 6B). As to TAG content, it was enhanced significantly in seeds of GmZF351 transgenic soybean plants. The TAG content was increased by 28%, 38%, and 24% in seeds of the 35S:GmZF351 lines OE-34, OE-40, and OE-73, respectively, compared with cv Jack (Fig. 6C). Transgenic soybean plants showed a mild dwarf phenotype at the V1 stage but they did not exhibit aberrant morphology at the R3 stage compared with cv Jack and vector control plants (Supplemental Fig. S7; Fehr and Caviness, 1977).
Figure 6.
Overexpression of GmZF351 increases seed oil content in transgenic soybean plants. A, Total FA contents in seeds of various plants. The cv Jack, vector control, and GmZF351 transgenic soybean plants (OE-34, OE-40, and OE-73) were used. Error bars indicate sd (n = 4). Asterisks indicate significant differences compared with cv Jack (**, P < 0.01). B, FA composition in seeds of various plants. Others are as in A. C, TAG contents in seeds of GmZF351 transgenic plants. Error bars indicate sd (n = 3). Asterisks indicate significant differences compared with cv Jack (*, P < 0.05). DW, Dry weight. D, Enhanced expression of lipid biosynthesis genes and regulatory genes in GmZF351 transgenic soybean. Transcript levels of BCCP2 homolog (Glyma19g03530), KASIII homolog (Glyma15g00550), TAG1 homologs (Glyma13g16560 and Glyma17g06120), OLEO2 homologs (Glyma19g13060 and Glyma16g07800), and WRI1 homologs (Glyma15g34770 and Glyma08g24420) were detected in H5 stage seeds from various plants. The cv Jack, vector control, and GmZF351 transgenic soybean plants (OE-34, OE-40, and OE-73) were used. The mRNA level (relative to GmTUBULIN) of each gene in cv Jack was set to 1. Error bars indicate sd (n = 3). E, GmZF351 activates the promoter activities of BCCP2 homolog (Glyma19g03530), KASIII homolog (Glyma15g00550), TAG1 homologs (Glyma13g16560 and Glyma17g06120), OLEO2 homologs (Glyma19g13060 and Glyma16g07800), and WRI1 homologs (Glyma15g34770 and Glyma08g24420) by transient expression assay in tobacco leaves. The promoters of downstream genes were fused with LUC as reporters. A. tumefaciens harboring a reporter vector was mixed with A. tumefaciens harboring 35S:GmZF351-FLAG and infiltrated into tobacco leaves. A. tumefaciens harboring pGWB412 was used as a negative control. LUC images were taken 2 d after infiltration. F, Quantitative analysis of the luminescence intensity in E. Luminescence intensity was analyzed with IndiGo software. Error bars indicate sd (n = 5).
Since WRI1, BCCP2, KASIII, TAG1, and OLEO2 were regulated by GmZF351 in transgenic Arabidopsis, we further examined whether the homologous genes were similarly regulated by GmZF351 in soybean. A set of soybean genes including BCCP2 homolog (Glyma19g03530), KASIII homologs (Glyma18g44350, Glyma09g41380, and Glyma15g00550), TAG1 homologs (Glyma13g16560 and Glyma17g06120), OLEO2 homologs (Glyma19g13060 and Glyma16g07800), and WRI1 homologs (Glyma15g34770 and Glyma08g24420) were selected for expression analysis based on the high homology and functional prediction from the SFGD database (http://bioinformatics.cau.edu.cn/SFGD/). Except for the KASIII homologs (Glyma18g44350 and Glyma09g41380), the transcript levels of all the other genes increased during the development of soybean seeds (Supplemental Fig. S8), and the expression patterns were very similar to that of GmZF351 (Fig. 2D), indicating that these genes may be regulated by GmZF351. The expression of these genes was then examined in GmZF351 transgenic soybean plants, and transcripts of the WRI1 homologs (Glyma15g34770 and Glyma08g24420), BCCP2 homolog (Glyma19g03530), KASIII homolog (Glyma15g00550), TAG1 homologs (Glyma13g16560 and Glyma17g06120), and OLEO2 homologs (Glyma19g13060 and Glyma16g07800) were all increased markedly in GmZF351 transgenic soybean plants (Fig. 6D), whereas the expression of another two KASIII homologs (Glyma18g44350 and Glyma09g41380) was not altered (Supplemental Fig. S9). A transient expression assay was performed to test whether GmZF351 could activate these downstream genes. As shown in Figure 6, in contrast to the effector vector control, the LUC activities driven by the promoters of all eight genes are apparently augmented by at least 3-fold with the inclusion of the 35S:GmZF351 effector. As predicted, pGlyma18g44350:LUC and pGlyma09g41380:LUC could not be activated by the 35S:GmZF351 effector (Supplemental Fig. S9). These results indicate that GmZF351 can exert a great boost on seed oil accumulation by activating the expression of WRI1 homologs and other lipid biosynthesis genes of soybean.
Haplotype Variation in the ZF351 Promoter Associates with Differences in Gene Expression and Seed Oil Content
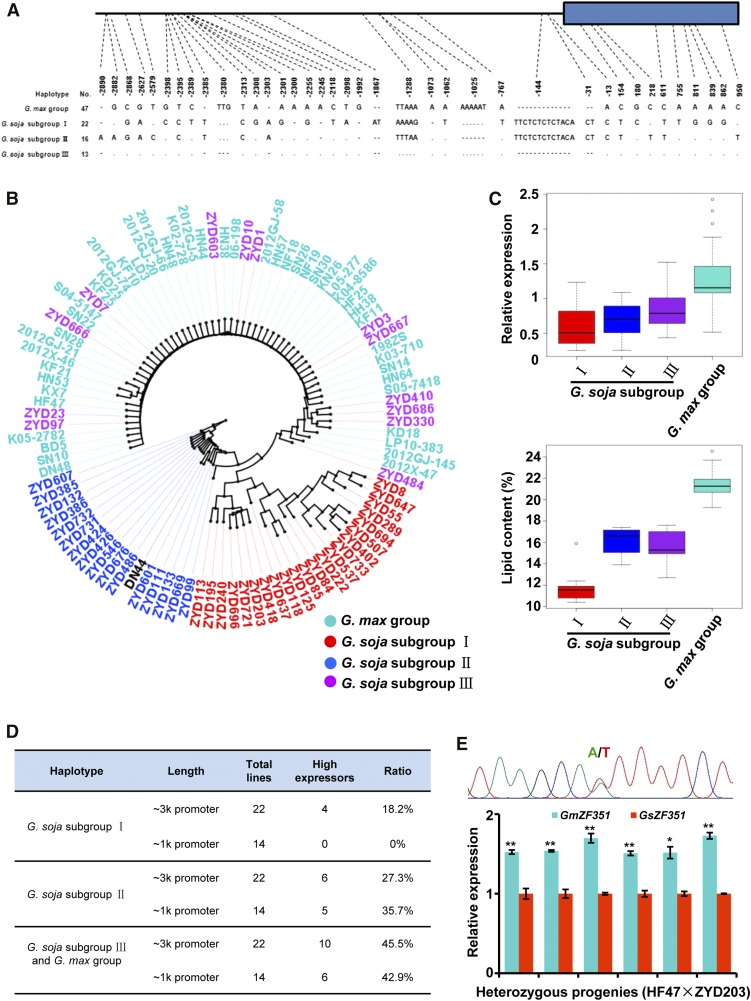
Considering the higher ZF351 expression in cultivated soybean than in wild soybean (Fig. 1A), the good correlation between ZF351 expression and seed oil content (Fig. 1B), and ZF351 function in lipid accumulation in transgenic soybean seeds (Fig. 6), it was most likely that the higher expression of GmZF351 would lead to higher oil contents in soybean. We then investigated the potential genetic basis of the artificial selection that led the cultivated soybean to generally have higher ZF351 expression than the wild soybean by examining the sequence variations of the ZF351 locus. The main SNPs and insertions/deletions, which jointly existed in the ZF351 sequences within one haplotype, were identified and compared. Almost all the GmZF351 genes from cultivated soybean, except cv DN44, belonged to the same group (Fig. 7, A and B). From the GsZF351 genes in wild soybean, three subgroups (I, II, and III) were classified (Fig. 7A). In comparison with the cultivated soybean group, wild soybean subgroup I showed the greatest variation in haplotype. Interestingly, wild soybean subgroup III shared almost the same sequence with the cultivated soybean group, suggesting that the wild soybean subgroup III haplotype may be the target of artificial selection and may have been fixed in a single domestication event (Guo et al., 2010). The haplotype of wild soybean subgroup II was the same as that of cv DN44 (Fig. 7, A and B).
Figure 7.
ZF351 haplotypes and their correlation with promoter activity, gene expression, and oil content. A, Haplotype variation of the cultivated soybean group and three wild soybean subgroups in the ∼4-kb ZF351 genomic region. No. represents the number of soybean accessions. The black line and the blue rectangle indicate the promoter region and the CDS region, respectively. B, Phylogenetic tree of ZF351 promoter sequences from 48 cultivated soybeans and 51 wild soybeans. The red accessions indicate the wild soybean subgroup I, the blue accessions indicate the wild soybean subgroup II, the purple accessions indicate the wild soybean subgroup III, and the turquoise accessions indicate the cultivated soybean group. C, ZF351 transcript levels and seed oil contents of cultivated soybean and wild soybean in relation to ZF351 haplotypes. D, Promoter activities of different haplotypes revealed by the percentage of high expressors among all the promoter-GUS transgenic lines. High expressors indicate transgenic Arabidopsis lines with GUS expression (relative to AtACTIN2) in excess of 0.009 as determined by RT-qPCR. RNA samples were prepared from 5-d-old seedlings. The effects of different promoter lengths also were compared. E, TaqMan RT-qPCR analysis of allele-specific expression. Transcript levels of GmZF351 (from HF47) and GsZF351 (from ZYD203) in six seeds of heterozygous progeny are shown. The diagram at top indicates the confirmation of a heterozygous genotype by sequencing, marked by an SNP (A/T). The expression of the GsZF351 allele in each seed was set to 1. GmTUBULIN was used as an internal control. Error bars indicate sd (n = 3). Asterisks indicate significant differences between two alleles in each seed (*, P < 0.05 and **, P < 0.01).
Phylogenetic analysis was further performed to reveal the relationship of the soybean accessions using the neighbor-joining method based on an ∼-3-kb promoter region. Except for cv DN44, other cultivated soybean cultivars were clustered in the same branch with inclusion of 13 wild soybeans from the wild soybean subgroup III (Fig. 7B). Wild soybean subgroup I and II accessions formed the corresponding clusters, with the exception that cv DN44 was included in wild soybean subgroup II (Fig. 7B). These results indicate that the grouping based on sequence haplotypes is consistent with the distribution of accessions in phylogenetic analysis.
We further analyzed the correlation of ZF351 haplotypes with gene expression level and lipid content. The gene expression values and lipid contents for each haplotype were pooled and compared. As shown in Figure 7, the ZF351 transcript level was highest in the cultivated soybean group. In the case of the wild soybean subgroups, the ZF351 expression value decreased gradually from subgroup III to subgroup I. Regarding lipid content, the cultivated soybean group had the highest lipid content, whereas the wild soybean subgroup I had the lowest content, with wild soybean subgroups II and III being in between (Fig. 7C). These results suggest that the ZF351 haplotype of the cultivated soybean group is associated with higher gene expression and higher oil contents compared with the wild soybean subgroups, whereas the wild soybean subgroup I haplotype is relevant to lower gene expression and lower oil contents.
To determine whether the ZF351 expression difference was due to the variation in promoter activity, we constructed pZF351:GUS using ∼3-kb promoter regions of different haplotype groups and transformed them into Arabidopsis to detect GUS expression by RT-qPCR. We determined the transgenic line with GUS expression in excess of 0.009 as a high expressor. The percentage of high expressors in transgenic lines was compared for each haplotype group (Fig. 7D). The transgenic lines harboring the ZF351 promoter of the cultivated soybean group and the wild soybean subgroup III showed the highest percentage of high expressors, whereas the transgenic lines harboring the ZF351 promoter of the wild soybean subgroup I had the lowest percentage (Fig. 7D). We also examined the ∼1-kb promoter activity by generating the corresponding pZF351:GUS transgenic lines, and a similar distribution was observed for high expressors (Fig. 7D), suggesting that an ∼1-kb sequence is sufficient to maintain the difference of the ZF351 promoter activity among haplotype groups. These results indicate that the ZF351 expression difference in wild and cultivated soybeans is most likely due to the difference in promoter activity derived from sequence variations.
Since differences in promoter activity led to different ZF351 expression in wild and cultivated soybeans, we were interested to compare the expression of different ZF351 alleles in F1 or F2 segregated heterozygous seeds derived from a cross between wild and cultivated soybeans. We used segregated F2 heterozygous seeds from a cross between HF47 (cultivated soybean group) and ZYD203 (wild soybean subgroup I) for an analysis of the allele-specific expression of ZF351. The F1 population was generated in our previous effort. We amplified and sequenced an ∼500-bp promoter region of ZF351 from F2 seeds to identify the heterozygotes. Heterozygous seeds were determined by the A/T SNP at 767 bp upstream of the start codon (Fig. 7E, top). In order to quantify mRNA levels of the GmZF351 allele (HF47) and the GsZF351 allele (ZYD203), we performed an accurate and specific transcript quantification assay based on TaqMan RT-qPCR in six F2 heterozygous seeds. In agreement with the promoter activity, the expression of the GmZF351 allele with A at −767 bp is significantly higher than that of the GsZF351 allele with T at −767 bp (Fig. 7E, bottom). TaqMan probes have been tested in the two parents for ZF351 allele specificity (Supplemental Fig. S10). All these results indicate that the ZF351 allele from cultivated soybean has higher expression than the ZF351 allele from wild soybean in the same genetic background.
The function of GsZF351 from the wild soybean was studied further. In spite of several SNPs in the coding region, overexpression of GsZF351 improved the FA content in transgenic Arabidopsis seeds as well as GmZF351, so we could rule out the possibility that the ZF351 coding regions caused the difference of seed oil contents between cultivated and wild soybeans (Supplemental Fig. S11).
Taken together, the presence of the GmZF351 promoter leads to an improvement of seed oil content in cultivated soybean and may have been inherited from ancient alleles reserved in the wild soybean during domestication.
DISCUSSION
Agronomic traits of soybean have experienced strong artificial selection, as exemplified by the great boost in seed oil content from wild soybean to cultivated soybean. Here, we analyzed a cultivar-specific gene coexpression network and seed-preferred genes from RNA-seq data to identify potential regulators participating in seed oil accumulation. We discovered that a plant-specific TZF protein, GmZF351, increases seed oil content by directly activating WRI1 and lipid biosynthesis genes. The GmZF351 locus may have been selected during soybean domestication and contributes to increased lipid content in cultivated soybean seeds.
The improvement of seed oil in cultivated soybean is attributed in large part to the fact that domestication imposes selective pressure on a multitude of genes controlling oil biosynthesis. Nevertheless, a large number of them encode enzymes participating in lipogenesis (Zhou et al., 2015). Our genetic and functional analyses reveal that GmZF351 is a domestication-selective regulator conferring improvement of seed oil content in cultivated soybean. The following evidence supports this conclusion. First, ZF351 expression is relatively higher in seeds of cultivated soybean accessions than in wild soybean accessions and is positively correlated with seed oil content (Fig. 1). Second, the parameters of genetic diversity and Tajima’s D test indicate that ZF351 has been subjected to selection (Table I). Third, the ZF351 haplotype of the cultivated soybean group and wild soybean subgroup III correlates with high gene expression, lipid content, and promoter activity (Fig. 7, C and D). Fourth, GmZF351 enhances seed oil accumulation in transgenic Arabidopsis and transgenic soybean through the activation of WRI1, BCCP2, KASIII, TAG1, and OLEO2 expression (Figs. 3–6). Considering that wild soybean may be difficult to transform, we choose to ectopically express GmZF351 in cultivated soybean. Although the overexpression of GmZF351 in soybean may not be a strict complementation test for the validation of domestication, GmZF351 could be predicted to consistently improve seed oil content in wild soybean as well as in cultivated soybean. In addition, the weak lines of 35S:GmZF351 transgenic soybean plants (OE-64, OE-98, and OE-101) show no significant difference in seed oil content compared with cv Jack and a vector control (Supplemental Fig. S6). This may demonstrate, at least in part, that the expression of ZF351 is causative for the improvement of seed oil content in cultivated soybean.
Recently, through RNA-seq analysis and gene coexpression network analysis, we proposed a mechanism of selection by expression for seed traits during soybean domestication. From the networks, two genes, GmGA20OX and GmNFYA, which enhance seed weight and seed oil content, respectively, show higher expression in cultivated soybean than in wild soybean (Lu et al., 2016). In this study, GmZF351 also followed the mechanism of selection by expression. Actually, we found that GUS expression driven by a promoter from the cultivated soybean group and wild soybean subgroup III is obviously higher than that of wild soybean subgroup I. Furthermore, the GmZF351 allele exhibits higher expression than the GsZF351 allele (wild soybean subgroup I haplotype) in the same genetic background of soybean (Fig. 7E). The fixation of the wild soybean subgroup III haplotype happens in almost all cultivated soybeans. Given that the causal mutations within an ∼1-kb promoter sequence of GmZF351 adequately result in changes in expression level, it is possible that an SNP at 767 bp upstream from the start codon is involved. Mutation from T (wild soybean subgroup I) to A generates a CAAT box within the cultivated soybean group and the wild soybean subgroup III haplotype that may signal the binding site not only for RNA polymerase to influence the frequency of transcriptional initiation but also for NFYs to regulate transcriptional activity (Kusnetsov et al., 1999). Actually, GmNFYA is located in the network harboring GmZF351. It is possible, therefore, that GmNFYA may activate GmZF351 and, hence, other genes in the lipid biosynthesis pathway for oil accumulation. This speculation should be investigated further. It should be noted that, although the promoter sequences are almost the same between the cultivated soybean group and the wild soybean subgroup III haplotype, the expression of ZF351 and seed oil content are both higher in the former group than in the latter group. This discrepancy may be due to the fact that there are other specific regulators promoting ZF351 expression in cultivated soybean.
Therefore, we were interested to test if GmNFYA and GmZF351 can be mutually regulated at the transcription level in cultivated soybean. The selection by expression mechanism also may be applied to other genes driving agronomic trait evolution. For pod shattering resistance, a NAC-type transcription factor gene, SHAT1-5, is selected for its much higher expression in cultivated soybean than in wild soybean due to the disruption of a repressor. This higher expression promotes the thickening of fiber cap cells in cultivated soybean to avoid shattering (Dong et al., 2014). For stem growth habit, Dt1 and Dt2 also may have been subjected to selection by expression during soybean domestication (Tian et al., 2010; Ping et al., 2014).
TZF proteins play pivotal roles in many developmental processes, such as AtTZF4/AtTZF5/AtTZF6 in seed germination (Bogamuwa and Jang, 2013), AtTZF1/AtTZF10/AtTZF11 in abiotic stress tolerance (Sun et al., 2007; Lin et al., 2011), PEI1 in embryo formation (Li and Thomas, 1998), and AtC3H14/AtC3H15 in male fertility and secondary wall thickening (Chai et al., 2015). Our study reveals that the TZF protein GmZF351 in soybean is involved in regulating lipogenesis. However, the fact that lipogenesis-related genes account for only a small portion of the GmZF351 targets implies that GmZF351 may play many other roles in plant development and/or stress responses. Sequence variations in regions other than the conserved TZF domain may lead to diverse functions within the same family (Supplemental Fig. S1A). Recently, based on comparative genome, transcriptome, and differential expression analyses between four high-oil dicots and three low-oil grasses, GmZF351 was predicted to be a candidate regulator in seed oil accumulation, which is in agreement with our results here (Zhang et al., 2016). As a homolog of GmZF351, SOMNUS (TZF4) shows very similar molecular characteristics in terms of subcellular location and tissue specificity in Arabidopsis (Kim et al., 2008). Furthermore, TZF4 also promotes seed oil accumulation (Supplemental Fig. S12), indicating that the function of TZF4 in oil biosynthesis also is conserved in soybean. Nevertheless, ectopic expression of SOMNUS in Arabidopsis strongly suppresses plant growth (Bogamuwa and Jang, 2013), whereas GmZF351-overexpressing soybean plants only exhibit a mild dwarf phenotype in V1 stage and no obvious difference in R3 stage compared with cv Jack or a vector control (Supplemental Fig. S7), suggesting that GmZF351 function is partially distinct from that of TZF4. Similarly, GmNFYA-overexpressing Arabidopsis plants exhibit higher oil content in seeds compared with Col-0 (Lu et al., 2016); however, overexpression of its homolog AtNFYA9 causes defective seeds (Mu et al., 2013). Polyploidy in soybean that leads to the incremental diversity of transcription factors may be a force driving the conversion of gene function (Schmutz et al., 2010; Salman-Minkov et al., 2016).
Our study identifies GmZF351 for seed oil accumulation in soybean, which may represent a master regulator in soybean compared with our previously identified oil-promoting genes GmDOF4, GmDOF11, GmbZIP123, and GmMYB73. GmDOF4 and GmDOF11 only regulate a few genes related to oil biosynthesis, GmbZIP123 promotes Suc translocation, while GmMYB73 indirectly enhances PLDα1 expression for TAG conversion. Ectopically expressing GmZF351 not only magnifies the supply of acyl-CoA via WRI1 but also directly promotes lipid synthesis and storage through BCCP2, KASIII, TAG1, and OLEO2. In addition, overexpression of GmZF351 also induces the lipid biosynthesis gene ACP1 and KASI in a WRI1-dependent manner. It is worth noting that, as a downstream gene of WRI1, SUS2 is still mildly induced in GmZF351-OE wri1 compared with wri1 (Fig. 5K), suggesting that GmZF351 may actually up-regulate SUS2 expression to modulate sucrolysis and the Suc-hexose ratio through both WRI1-independent and -dependent means (Maeo et al., 2009; Angeles-Nunez and Tiessen, 2010). Considering that alteration of sugar levels by the GmbZIP123 transcription factor also affects seed oil content (Song et al., 2013), it is possible that GmZF351 may regulate oil content through SUS2-mediated sugar metabolism.
From our analyses here, GmZF351 fine-tunes the whole lipid biosynthesis pathway, which is similar to the function of GmNFYA (Lu et al., 2016). So far, the functions of WRI1, BCCP2, KASIII, TAG1, and OLEO2 in lipid biosynthesis are clear in Arabidopsis. As to the downstream genes in soybean, Glyma15g34770 encodes a WRI1 homolog of AP2/EREBP and coexpresses with plenty of lipid biosynthesis-related genes (Yu et al., 2014). WRI1 homologs in many plant species, such as ZmWRI1 and BnWRI1, are proved to participate in lipogenesis (Shen et al., 2010; Li et al., 2015). Therefore, we speculate that the up-regulation of Glyma15g34770 (GmWRI1-like1) and Glyma08g24420 (GmWRI1-like2) may consistently enhance seed oil accumulation. The domestication-selective KASIII homolog Glyma15g00550 may play a vital role in the increase of seed oil content from wild soybean to cultivated soybean (Zhou et al., 2015). Diacylglycerol acyltransferase is the rate-limiting enzyme for oil production of soybean, as supported by a previous study (Settlage et al., 1998). Although the roles of the BCCP2, KASIII, and TAG1 homologs have not been established, their enzymatic functions indicate that they are well suited to increase seed oil content. Overexpression of the oleosin genes Glyma19g13060 and Glyma16g07800 increases seed lipid content in transgenic rice (Oryza sativa; Liu et al., 2013). In addition, based on SoyBase data (http://soybase.org/), Glyma19g03530 (GmBCCP2-like) and Glyma13g16560 (GmTAG1-like2) loci are located in the known oil-related quantitative trait loci (Supplemental Table S3).
In contrast to the specific regulation of the KASIII homolog (Glyma15g00550) by GmZF351, two KASIII homologs, Glyma18g44350 and Glyma09g41380, with largely invariant expression patterns in developing seeds, exhibit unchanged transcript levels in GmZF351 transgenic soybean plants compared with cv Jack and vector control plants, suggesting specific but not comprehensive regulation of GmZF351 on soybean polyploidy genes. Since a high GmZF351 level may promote lipogenesis by activating the biosynthesis genes BCCP2 homolog Glyma19g03530, KASIII homolog Glyma15g00550, TAG1 homologs Glyma13g16560 and Glyma17g06120, and OLEO2 homologs Glyma19g13060 and Glyma16g07800, as well as the regulatory genes WRI1 homologs Glyma08g24420 and Glyma15g34770, GmZF351 may be a significant pivot controlling oil biosynthesis in soybean. It should be noted that, while GmZF351 significantly enhances seed oil content, the expression of lipid biosynthesis genes is not strongly induced by GmZF351 overexpression. Therefore, other mechanisms of downstream genes/components may contribute to the oil accumulation. Recent studies demonstrate that the extension of WRI1 expression in the mid phase of seed development and increased WRI1 stability through C-terminal manipulation or interaction with 14-3-3 proteins play more critical roles in seed oil accumulation than the strength of WRI1 expression and lead to significant enhancement of seed oil content (Chen et al., 2013; Ma et al., 2015, 2016; Kanai et al., 2016). Considering that the function of GmZF351 partially depends on WRI1, it is possible that GmZF351 may fine-tune the temporal expression of WRI1 and lipid biosynthesis genes and/or may facilitate the stabilization of the WRI1 protein as well as the promotion of oil content. These possibilities should be investigated further. Moreover, unlike tt2 (M. Chen et al., 2012), GmZF351 improves seed oil accumulation without decreasing seed size in both transgenic Arabidopsis and transgenic soybean (Supplemental Fig. S13). GmZF351 increases TAG content by up to 38% in soybean seeds and by up to 29% in Arabidopsis seeds, indicating its great potential application for improving seed oil in dicotyledonous plants by genetic manipulation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Soybean plants were grown in the experimental station in Beijing. F2 heterozygous soybean (Glycine spp.) seeds were harvested from the F1 population, which had been generated by a cross between cultivated soybean HF47 (Glycine max group) and wild soybean ZYD203 (Glycine soja subgroup I) in 2015. Roots, stems, leaves, flowers, pods, and seeds were collected and stored at −80°C for RNA/DNA isolation.
The GmZF351-OE wri1 mutant plant was obtained by crossing of the GmZF351 transgenic Arabidopsis (Arabidopsis thaliana) line OE-19 with wri1-3. Arabidopsis seeds were sterilized with 1% NaClO for 12 min and then plated on one-half-strength Murashige and Skoog medium for germination. The seedlings were grown in a green chamber at 22°C with a photoperiod of 16 h of light/8 h of dark. Arabidopsis plants were harvested and stored at −80°C for RNA/DNA isolation.
Population Genetic Analysis
We amplified PCR products from wild soybean and cultivated soybean genomic DNA using KOD FX polymerase (TOYOBO). The alignments of nucleotide sequences were performed using ClustalX software (Thompson et al., 1997). Watterson’s estimator and π were calculated by MEGA5 using ∼4-kb GmZF351 genomic sequences (Tamura et al., 2011). Tajima’s D value was estimated to examine the sequence departure of ZF351 from neutral evolution, also in MEGA5. Phylogenetic trees were generated using the ∼3-kb promoter sequence of ZF351 based on the neighbor-joining method with PHYLIP (Retief, 2000). Primers for PCR are listed in Supplemental Table S4.
Plant Transformation
The full-length open reading frame (ORF) of GmZF351 (GenBank accession no. XM_003526219) without stop codon was amplified by RT-PCR from cv HN44 using KOD FX polymerase (TOYOBO). The PCR fragment was cloned into the pCR8/GW/TOPO entry vector (Invitrogen) and subsequently recombined into pGWB412 plasmid to generate the 35S:GmZF351-FLAG construct. For the promoter-GUS analysis, the ∼1-kb promoter region of GmZF351 and ∼3-kb promoter region of GmZF351 were amplified and cloned into pCR8/GW/TOPO entry vector (Invitrogen), following by recombining into pGWB433 (Invitrogen) to drive GUS reporter expression. The ∼1-kb and ∼3-kb promoters of ZF351 representing three sequence haplotypes were cloned from cv HN44, ZYD486, and ZYD203. Primers for PCR are listed in Supplemental Table S4. Plasmids were transformed into Agrobacterium tumefaciens GV3101, and then Arabidopsis plant transformation was performed as described (Wei et al., 2015). For soybean transformation, the amplified sequence of the GmZF351 ORF was digested by SalI and SpeI and then ligated with the corresponding digested pCAMBIA1301 vector (CAMBIA; http://www.cambia.org). The final construct was transferred into the soybean cv Jack using the cotyledonary node-A. tumefaciens-mediated transformation method (Zeng et al., 2004).
ChIP-Seq and ChIP-qPCR Analysis
The ChIP experiment was performed as described (R. Chen et al., 2012) with minor modifications. Col-0 and two individual lines of 35S:GmZF351-FLAG siliques were cross-linked in 1% formaldehyde. The protein-DNA complex was immunoprecipitated with anti-FLAG antibody (EarthOx), and the precipitated DNA was used for subsequent sequencing and qPCR. ChIP-seq libraries were prepared using a ChIP-seq DNA sample prep kit (Illumina) according to the manufacturer’s protocol. Sequencing was performed using Genome Analyzer IIx (Illumina). ChIP-seq reads were aligned to the Arabidopsis genome build TAIR10 by Bowtie 2, allowing up to two mismatches (Langmead and Salzberg, 2012). Peak identification was performed using HOMER version 4.5, a software suite for ChIP-seq analysis (Heinz et al., 2010). We used a cutoff of false discovery rate < 0.05 for ChIP-seq analysis. The enriched peaks were visualized using the IGV program (Robinson et al., 2011). For ChIP-qPCR, the ChIP signal was normalized to input. Primers were designed according to binding DNA fragment discovery by sequencing and are listed in Supplemental Table S4. Experiments were done with three biological replicates.
RT-qPCR Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. First-strand cDNA was prepared with 5 µg of total RNA using oligo(dT)20 primer and SuperScript III reverse transcriptase (Invitrogen). To detect the expression of ZF351 and downstream genes, qPCR was performed using SYBR qPCR Mix (TOYOBO) on the LightCycler480 System (Roche). Gene expression in Arabidopsis was normalized to two reference genes, At4g12590 (GenBank accession no. NM_117329.3) and AtACTIN2 (GenBank accession no. NM_112764.3). Gene expression in soybean was normalized to GmTUBULIN (GenBank accession no. XM_003520891). Experiments were done with three biological replicates. For the detection of allele-specific expression in populations from the cross (HF47 × ZYD203), TaqMan RT-qPCR was performed using Probe qPCR Mix (TOYOBO) on the LightCycler480 System (Roche) according to the manufacturer’s protocol. cDNA products from triplicate RT reactions of single F2 seeds were used for TaqMan analysis. The TaqMan probes and corresponding primer set of ZF351 were designed using Primer Express software (Applied Biosystems) based on a C/T SNP at 154 bp downstream of the start codon in the coding region. The TaqMan probe and corresponding primer set of reference gene GmTUBULIN were designed from the CDS without differences between HF47 and ZYD203. Reporter (5′ end) and quencher (3′ end) dyes for the TaqMan probes were FAM and TAMRA (6-carboxytetramethyl rhodamine), respectively. The relative expression of GmZF351 from HF47 and GsZF351 from ZYD203 was calculated by 2ΔCt [ΔCt = Ct(GmTUBULIN) − Ct(ZF351)]. Primers and probes for TaqMan RT-qPCR are listed in Supplemental Table S4. The primer sets for At4g12590, SUS2, ACP1, and KASI were described previously (Maeo et al., 2009; Dekkers et al., 2012).
Subcellular Localization of GmZF351
The ORF of GmZF351 in the pCR8/GW/TOPO entry vector was recombined into pGWB405 (Invitrogen) to generate 35S:GmZF351-GFP constructs. 35S:HY5-RFP was used as a nucleus marker. These constructs were transformed into A. tumefaciens strain GV3101 and introduced into tobacco (Nicotiana tabacum) leaves by infiltration as described (Song et al., 2013). The tobacco plants were cultured for 3 d and then observed with a Zeiss LSM 710 confocal microscope.
Transcriptional Activation Assay
The ORF of GmZF351 was cloned into pRT-BD to fuse with the GAL4 DNA-binding domain (BD) under the control of the 35S promoter. 35S:GAL4BD-VP16 and 35S:GAL4BD were used as positive and negative controls, respectively. The reporter plasmid contained a firefly LUC gene driven by the minimal 35S promoter and five GAL4 binding elements. Plasmid pTRL harboring the Renilla LUC gene was selected as an internal control. The effectors, reporter, and internal control were cotransfected into Arabidopsis protoplasts, and relative LUC activity was determined according to a previously described protocol (Wei et al., 2015). Experiments were done with four biological replicates.
Histochemical GUS and Fat Red 7B Staining
Plant tissues were collected from transgenic plants containing pGmZF351:GUS. GUS staining was performed as described (Wang et al., 2015). The tissues immersed in GUS staining solution were incubated at 37°C overnight and then decolorized by 75% ethanol to remove the chlorophyll. Fat Red staining was performed by incubating seedlings in 0.1% (w/v) Fat Red 7B (Sigma-Aldrich) solution for 3 h at room temperature. Samples were then rinsed with 70% ethanol. The strained samples were observed with a stereomicroscope (Leica).
FA and TAG Content Analysis
Arabidopsis seeds (10 mg) or fine powder of soybean seeds (10 mg) were prepared for FA isolation. The FA was extracted with 1 mL of extraction buffer (2.5% [v/v] H2SO4 in CH3OH) at 85°C for 1 h. The supernatant (500 µL) was mixed with 300 µL of hexane and 600 µL of 0.9% (w/v) NaCl. After gentle rotation and centrifugation, the organic phase was removed to a clean tube and dried by natural volatilization. FA methyl esters were redissolved in 200 µL of ethyl acetate and analyzed immediately with a gas chromatography system (GC-2014; Shimadzu). The temperature was initiated at 170°C and maintained for 5 min, then increased by 2°C min−1 to 210°C. Peaks corresponding to each FA species were identified by FA methyl ester analytical standard (catalog no. 18920-1AMP; Supelco). Concentrations of FA species were normalized against the internal control heptadecanoic acid (Sigma-Aldrich). Experiments were done with four biological replicates. TAG content analysis was performed as described (Liu et al., 2014). Experiments were done with three biological replicates.
Transient Expression Assays in Nicotiana benthamiana Leaves
The 1.5-kb promoters of WRI1, BCCP2, KASIII, TAG1, and OLEO2 were cloned into the pCR8/GW/TOPO entry vector (Invitrogen). Then, the promoters were fused with the luciferase reporter gene LUC through LR reactions into the plant binary vector pGWB435. The effector construct was the above-described 35S:GmZF351-FLAG. For assays of soybean genes, the 3-kb promoters of Glyma19g03530, Glyma15g00550, Glyma13g16560, Glyma17g06120, Glyma19g13060, Glyma16g07800, Glyma08g24420, Glyma15g34770, Glyma18g44350, and Glyma09g41380 were cloned into the pCAMBIA2300 vector with fusion of LUC by recombination using the Seamless Assembly Cloning Kit (Clone Smarter). The effector construct was 35S:GmZF351-FLAG. The transient expression assays were performed as described (Wang et al., 2014). Agrobacteria harboring empty effector vector were coinfiltrated with reporters as a negative control. The PCR primers are listed in Supplemental Table S4.
Pyruvate Kinase Enzyme Assay
Eleven-day-old siliques from Col-0 and three individual lines of 35S:GmZF351-FLAG were harvested and frozen in liquid nitrogen. Pyruvate kinase activity was measured with the Pyruvate Kinase Activity Assay Kit (Sigma-Aldrich) following the manufacturer’s instructions. Experiments were done with four biological replicates.
Statistical Analysis
The data were analyzed with Student’s t test using SPSS 15.0 (SPSS).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (www.arabidopsis.org) and the Plant Genome Database (www.plantgdb.org) under the following accession numbers: GmZF351 (Glyma06g44440), BCCP2 (AT5G15530), KASIII (AT1G62640), TAG1 (AT2G19450), OLEO2 (AT5G40420), WRI1 (AT3G54320), PKpα (At3g22960), PKpβ1 (AT5G52920), SUS2 (AT5G49190), ACP1 (AT3G05020), KASI (AT5G46290), FaTA (AT3G25110), AtACTIN2 (AT3G18780), GmTUBULIN (Glyma03g15020), and SOMNUS (AT1G03790).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence features and phylogenetic analysis of GmZF351.
Supplemental Figure S2. Expression level of GmZF351 and total FA contents of seeds in GmZF351 transgenic Arabidopsis plants.
Supplemental Figure S3. Morphology of GmZF351 transgenic Arabidopsis.
Supplemental Figure S4. GmZF351 ChIP analysis of the At3g22960 promoter and FaTA promoter.
Supplemental Figure S5. Expression levels of GmZF351 downstream genes in various Arabidopsis plants.
Supplemental Figure S6. Expression levels of GmZF351 and total FA contents of seeds in GmZF351 transgenic soybean plants.
Supplemental Figure S7. Morphology of GmZF351 transgenic soybean plants.
Supplemental Figure S8. Expression pattern of potential downstream genes in six developmental stages of soybean seeds.
Supplemental Figure S9. Overexpression of GmZF351 does not influence the expression of Glyma18g44350 and Glyma09g41380.
Supplemental Figure S10. Expression of GmZF351 and GsZF351 in seeds of HF47 and ZYD203 as determined by TaqMan RT-qPCR.
Supplemental Figure S11. Function of GsZF351 from ZYD203 in seed oil accumulation.
Supplemental Figure S12. Overexpression of SOMNUS/TZF4 improves seed oil accumulation in transgenic Arabidopsis seeds.
Supplemental Figure S13. Morphology of GmZF351 transgenic Arabidopsis seeds and GmZF351 transgenic soybean seeds.
Supplemental Table S1. Accessions of cultivated soybean and wild soybean used in this study.
Supplemental Table S2. Nearest gene regions identified in the ChIP-seq experiment.
Supplemental Table S3. Quantitative trait loci for seed oil overlapped with downstream genes of GmZF351 in soybean.
Supplemental Table S4. Primers used in this work.
Supplementary Material
Acknowledgments
We thank Dr. Sebastien Baud (Jean-Pierre Bourgin Institute, University of Paris-Saclay) and Dr. Jyan-Chyun Jang (Center for Applied Plant Sciences, Ohio State University) for wri1-3 mutant and TZF4-OE seeds, respectively.
Glossary
- TAG
triacylglycerol
- FA
fatty acid
- RNA-seq
RNA sequencing
- CDS
coding sequence
- RT
reverse transcription
- qPCR
quantitative PCR
- Col-0
Columbia-0
- ChIP-seq
sequencing chromatin immunoprecipitation
- ChIP
chromatin immunoprecipitation
- SNP
single-nucleotide polymorphism
- ORF
open reading frame
Footnotes
This work was supported by the Chinese Academy of Sciences Leading Project (grant no. XDA08020106), the National Transgenic Research Projects (grant nos. 2016ZX08009003-004, 2016ZX08004003-005, and 2014ZX0800926B), the Key R&D Project (grant nos. 2016YFD0100304 and 2016YFD0100504), the 973 Projects (grant nos. 2013CB835205 and 2015CB755702), the National Natural Science Foundation of China (grant no. 31671258), and the State Key Laboratory of Plant Genomics.
Articles can be viewed without a subscription.
References
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles-Nunez JG, Tiessen A (2010) Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 232: 701–718 [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L (2010) Physiological and developmental regulation of seed oil production. Prog Lipid Res 49: 235–249 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, To A, Rochat C, Lepiniec L (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J 60: 933–947 [DOI] [PubMed] [Google Scholar]
- Bellaloui N, Bruns HA, Abbas HK, Mengistu A, Fisher DK, Reddy KN (2015) Agricultural practices altered soybean seed protein, oil, fatty acids, sugars, and minerals in the midsouth USA. Front Plant Sci 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa S, Jang JC (2013) The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ 36: 1507–1519 [DOI] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chai G, Kong Y, Zhu M, Yu L, Qi G, Tang X, Wang Z, Cao Y, Yu C, Zhou G (2015) Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J Exp Bot 66: 2595–2609 [DOI] [PubMed] [Google Scholar]
- Chen LY, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H (2013) Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang Z, Zhu Y, Li Z, Hussain N, Xuan L, Guo W, Zhang G, Jiang L (2012) The effect of TRANSPARENT TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol 160: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang B, Li C, Kulaveerasingam H, Chew FT, Yu H (2015) TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol 169: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53: 28–37 [DOI] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5: 3352. [DOI] [PubMed] [Google Scholar]
- Fehr WR, Caviness CE (1977) Stages of Soybean Development. Agricultural and Home Economics Experiment Station, Iowa State University, Ames, IA [Google Scholar]
- Guo J, Wang Y, Song C, Zhou J, Qiu L, Huang H, Wang Y (2010) A single origin and moderate bottleneck during domestication of soybean (Glycine max): implications from microsatellites and nucleotide sequences. Ann Bot (Lond) 106: 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Mano S, Kondo M, Hayashi M, Nishimura M (2016) Extension of oil biosynthesis during the mid-phase of seed development enhances oil content in Arabidopsis seeds. Plant Biotechnol J 14: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnetsov V, Landsberger M, Meurer J, Oelmuller R (1999) The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J Biol Chem 274: 36009–36014 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Shao JH, Tang SH, Shen QW, Wang TH, Chen WL, Hong YY (2015) Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front Plant Sci 6: 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10: 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J 65: 253–268 [DOI] [PubMed] [Google Scholar]
- Liu WX, Liu HL, Qu LQ (2013) Embryo-specific expression of soybean oleosin altered oil body morphogenesis and increased lipid content in transgenic rice seeds. Theor Appl Genet 126: 2289–2297 [DOI] [PubMed] [Google Scholar]
- Liu YF, Li QT, Lu X, Song QX, Lam SM, Zhang WK, Ma B, Lin Q, Man WQ, Du WG, et al. (2014) Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol 14: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li QT, Xiong Q, Li W, Bi YD, Lai YC, Liu XL, Man WQ, Zhang WK, Ma B, et al. (2016) The transcriptomic signature of developing soybean seeds reveals genetic basis of seed trait adaptation during domestication. Plant J 86: 530–544 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Grix M, Mantyla JJ, Yang Y, Benning C, Ohlrogge JB (2015) Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J 83: 864–874 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Mantyla JJ, Yang Y, Ohlrogge JB, Benning C (2016) 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J 88: 228–235 [DOI] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MS, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Mu JY, Tan HL, Hong SL, Liang Y, Zuo JR (2013) Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant 6: 188–201 [DOI] [PubMed] [Google Scholar]
- Mu JY, Tan HL, Zheng Q, Fu FY, Liang Y, Zhang JA, Yang XH, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping JQ, Liu YF, Sun LJ, Zhao MX, Li YH, She MY, Sui Y, Lin F, Liu XD, Tang ZX, et al. (2014) Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26: 2831–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retief JD. (2000) Phylogenetic analysis using PHYLIP. Methods Mol Biol 132: 243–258 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56: 1215–1228 [DOI] [PubMed] [Google Scholar]
- Salman-Minkov A, Sabath N, Mayrose I (2016) Whole-genome duplication as a key factor in crop domestication. Nat Plants 2: 16115. [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Settlage SB, Kwanyuen P, Wilson RF (1998) Relation between diacylglycerol acyltransferase activity and oil concentration in soybean. J Am Oil Chem Soc 75: 775–781 [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60: 377–387 [DOI] [PubMed] [Google Scholar]
- Song QX, Li QT, Liu YF, Zhang FX, Ma B, Zhang WK, Man WQ, Du WG, Wang GD, Chen SY, et al. (2013) Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J Exp Bot 64: 4329–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci USA 107: 8563–8568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Joubes J, Barthole G, Lecureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY (2007) The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52: 716–729 [DOI] [PubMed] [Google Scholar]
- Wang L, Li Q, Lei Q, Feng C, Gao Y, Zheng X, Zhao Y, Wang Z, Kong J (2015) MzPIP2;1: an aquaporin involved in radial water movement in both water uptake and transportation, altered the drought and salt tolerance of transgenic Arabidopsis. PLoS ONE 10: e0142446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Li QT, Chen HW, Zhang WK, Ma B, Chen SY, Zhang JS (2014) Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol 14: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Zhang YQ, Tao JJ, Chen HW, Li QT, Zhang WK, Ma B, Lin Q, Zhang JS, Chen SY (2015) The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J 81: 871–883 [DOI] [PubMed] [Google Scholar]
- Wilson RF. (2008) Soybean: market-driven research needs in genetics and genomics. In G Stacey, ed, Genetics and Genomics of Soybean. Springer, New York, pp 3–15 [Google Scholar]
- Yu J, Zhang Z, Wei J, Ling Y, Xu W, Su Z (2014) SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genomics 15: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P, Vadnais DA, Zhang Z, Polacco JC (2004) Refined glufosinate selection in Agrobacterium-mediated transformation of soybean Glycine max (L.) Merrill. Plant Cell Rep 22: 478–482 [DOI] [PubMed] [Google Scholar]
- Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hao Q, Bai L, Xu J, Yin W, Song L, Xu L, Guo X, Fan C, Chen Y, et al. (2014) Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol Biofuels 7: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang SB, Li QG, Song J, Hao YQ, Zhou L, Zheng HQ, Dunwell JM, Zhang YM (2016) An integrated bioinformatics analysis reveals divergent evolutionary pattern of oil biosynthesis in high- and low-oil plants. PLoS ONE 11: e0154882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, et al. (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33: 408–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.