HEMERA/pTAC12 accumulation in the nucleus requires localization to the plastids.
Abstract
HEMERA (HMR) is a nuclear and plastidial dual-targeted protein. While it functions in the nucleus as a transcriptional coactivator in phytochrome signaling to regulate a distinct set of light-responsive, growth-relevant genes, in plastids it is known as pTAC12, which associates with the plastid-encoded RNA polymerase, and is essential for inducing the plastomic photosynthetic genes and initiating chloroplast biogenesis. However, the mechanism of targeting HMR to the nucleus and plastids is still poorly understood. Here, we show that HMR can be directly imported into chloroplasts through a transit peptide residing in the N-terminal 50 amino acids. Upon cleavage of the transit peptide and additional proteolytic processing, mature HMR, which begins from Lys-58, retains its biochemical properties in phytochrome signaling. Unexpectedly, expression of mature HMR failed to rescue not only the plastidial but also the nuclear defects of the hmr mutant. This is because the predicted nuclear localization signals of HMR are nonfunctional, and therefore mature HMR is unable to accumulate in either plastids or the nucleus. Surprisingly, fusing the transit peptide of the small subunit of Rubisco with mature HMR rescues both its plastidial and nuclear localization and functions. These results, combined with the observation that the nuclear form of HMR has the same reduced molecular mass as plastidial HMR, support a retrograde protein translocation mechanism in which HMR is targeted first to plastids, processed to the mature form, and then relocated to the nucleus.
Deetiolation is a pivotal developmental transition when a seedling emerges from the ground and establishes a light-dependent photoautotrophic lifestyle or photomorphogenesis. In Arabidopsis (Arabidopsis thaliana), photomorphogenesis entails restriction of hypocotyl growth and promotion of leaf development and chloroplast biogenesis. These diverse morphological changes reflect massive transcriptional reprogramming in both the nuclear and plastidial genomes and sophisticated communications between these two genome-bearing subcellular compartments.
Light controls nuclear gene expression through a suite of photoreceptors (Chen et al., 2004; Kami et al., 2010), including the red (R) and far-red light-sensing phytochromes (phys) that are essential for establishing photomorphogenesis (Franklin and Quail, 2010). Phys are bilin-containing proteins that can be photoconverted between two relatively stable forms: an R light-absorbing inactive Pr form and a far-red light-absorbing active Pfr form (Rockwell et al., 2006; Nagatani, 2010; Burgie and Vierstra, 2014). Phys mediate almost all the reprogramming of the nuclear transcriptome in response to R light (Tepperman et al., 2006; Leivar et al., 2009; Hu et al., 2013). One of the first light responses at the cellular level is the translocation of photoactivated phys from the cytoplasm to the nucleus, where phys are further compartmentalized to subnuclear photosensory domains named photobodies (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999; Chen et al., 2003, 2005, 2010; Van Buskirk et al., 2012, 2014). A central mechanism by which phys control gene expression, possibly at photobodies, is by regulating the stability and activity of a group of nodal basic helix-loop-helix transcriptional regulators, the Phytochrome-Interacting Factors (PIFs; Shen et al., 2005; Al-Sady et al., 2006; Oh et al., 2006; Lorrain et al., 2008; Leivar and Quail, 2011; Park et al., 2012; Kaiserli et al., 2015; Qiu et al., 2015). Eight PIFs have been reported, including PIF1, PIF3-8, and PIL1 (PIF3-Like1), which perform overlapping and distinct antagonistic roles in photomorphogenesis (Ni et al., 1998; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004; Leivar et al., 2008a; Li et al., 2012; Luo et al., 2014). PIF1, PIF3, PIF4, PIF5, and PIF7 promote hypocotyl growth by activating growth-relevant genes, including genes in the biosynthesis and signaling of the plant growth hormone auxin (Huq and Quail, 2002; Huq et al., 2004; Al-Sady et al., 2008; Leivar et al., 2008a, 2008b; Lorrain et al., 2009; Shin et al., 2009; Li et al., 2012; Pfeiffer et al., 2014; Procko et al., 2016). PIF1, PIF3, and PIF5 inhibit chloroplast development by repressing nuclear-encoded photosynthetic genes (Huq et al., 2004; Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009; Kim et al., 2011). Most PIFs accumulate to high levels in dark-grown seedlings, and photoactivated phys mediate photomorphogenetic reprogramming by interacting directly with PIFs and repressing their stability and activity in the light (Shen et al., 2005; Al-Sady et al., 2006; Oh et al., 2006; Lorrain et al., 2008; Leivar and Quail, 2011; Park et al., 2012; Qiu et al., 2015).
Light also regulates the activity of plastid-encoded genes and, in turn, gene expression in plastids can influence nuclear genes through plastid-to-nucleus or retrograde signaling (de Souza et al., 2016; Woodson and Chory, 2008). The plastome in land plants carries over 120 genes encoding key components of the plastidial transcriptional, translational, and photosynthetic apparatuses (Sugiura, 1992; Wakasugi et al., 2001). Plastomic genes are transcribed by two types of plastid RNA polymerases: a phage-type nuclear-encoded plastid RNA polymerase (NEP) and a eubacterial-type plastid-encoded plastid RNA polymerase (PEP; Lerbs-Mache, 1993; Allison et al., 1996; Hricová et al., 2006; Liere et al., 2011). Although most plastid genes are transcribed by both the NEP and PEP (Legen et al., 2002; Demarsy et al., 2006, 2012), housekeeping genes are preferentially transcribed by the NEP, whereas photosynthetic genes are transcribed mainly by the PEP (Allison et al., 1996; Hajdukiewicz et al., 1997; Hübschmann and Börner, 1998). During deetiolation, phy signaling in the nucleus predominantly induces the plastid-encoded photosynthetic genes by the PEP (DuBell and Mullet, 1995; Chun et al., 2001; Thum et al., 2001). The PEP is comprised of eubacteria-like α, β, β’, and β” core subunits; a sigma factor; and a battery of plant-specific PEP-associated proteins (Pfalz and Pfannschmidt, 2013). Whereas the α, β, β’, and β” core subunits are encoded by the plastome, the sigma factors and PEP-associated proteins are encoded by nuclear genes (Hu and Bogorad, 1990; Pfannschmidt et al., 2000; Suzuki et al., 2004; Pfalz et al., 2006; Steiner et al., 2011; Melonek et al., 2012). Recent proteomic studies of purified soluble PEP-complex and membrane-bound transcriptionally active chromosome (TAC) from Arabidopsis and mustard (Sinapis alba) identified 12 PEP-associated proteins named PAPs and/or pTACs, which are essential for the activity of the PEP (Pfalz et al., 2006; Steiner et al., 2011; Pfalz and Pfannschmidt, 2013). The activity of the PEP can also influence phy-regulated nuclear gene expression through retrograde signaling (Woodson et al., 2013). Defects in chloroplast biogenesis, including deficiency in PEP activity, are able to repress the expression of the nuclear-encoded photosynthetic genes (Koussevitzky et al., 2007; Woodson et al., 2013; Martín et al., 2016). Despite the essential roles of PAPs/pTACs for the function of the PEP, their precise functions in PEP activity and roles in nuclear-plastidial communication are still not well understood.
Recent studies have uncovered that pTAC12, an essential PEP-associated protein (Pfalz et al., 2006; Steiner et al., 2011), is a key phy signaling molecule named HEMERA (HMR; Chen et al., 2010). Although the biochemical function of pTAC12/HMR in the PEP is still unclear, HMR in maize (Zea mays) was shown to bind single-stranded DNA and RNA and plays a role in the accumulation of the PEP (Pfalz et al., 2015). In addition, pTAC12/HMR interacts directly with two other PEP-associated proteins, pTAC14 and pTAC7 (Gao et al., 2011; Yu et al., 2013). Interestingly, both Arabidopsis and maize pTAC12/HMR are dual localized to the nucleus and plastids (Chen et al., 2010; Pfalz et al., 2015). The nuclear function of pTAC12/HMR was revealed through a forward genetic screen for mutants defective in the subnuclear localization of Arabidopsis phytochrome B (phyB) to photobodies (Chen et al., 2010). The hmr mutant represents the founding member of a new class of photomorphogenetic mutants that show a combination of long hypocotyl and albino phenotypes (Chen et al., 2010; Chen and Chory, 2011). HMR transduces light signals by interacting directly with photoactivated phys and all PIFs, and it acts as a transcriptional coactivator that regulates the degradation and activity of PIF1 and PIF3 (Galvão et al., 2012; Qiu et al., 2015). Knocking out PIF1, PIF3, PIF4, and PIF5 rescues hmr’s long hypocotyl phenotype but not its albino phenotype, indicating that HMR plays two separable roles: a PIF-dependent role in regulating hypocotyl elongation and a PIF-independent role in chloroplast biogenesis (Chen et al., 2010; Qiu et al., 2015). These two roles likely reflect the functions of HMR in the nucleus and plastids, respectively.
The discovery of HMR raises a new hypothesis that nuclear-plastidial communication could be coordinated by nuclear/plastidial dual-targeted proteins such as HMR. As an initial step to test this hypothesis, we characterized the subcellular localization signals of HMR and investigated the mechanisms that target HMR to the nucleus and plastids. We show that HMR can be directly imported into chloroplasts through a transit peptide, which is cleaved upon chloroplast import. However, the predicted nuclear localization signals of HMR are unexpectedly nonfunctional. We determined the mature form of HMR and showed that mature HMR retains the biochemical functions required for phytochrome signaling. Surprisingly, characterization of subcellular localization and function of mature HMR revealed an unexpected relationship between HMR’s plastidial localization and nuclear accumulation.
RESULTS
HMR Can Be Imported Directly into Chloroplasts by a Transit Peptide Residing in the N-Terminal 50 Amino Acids
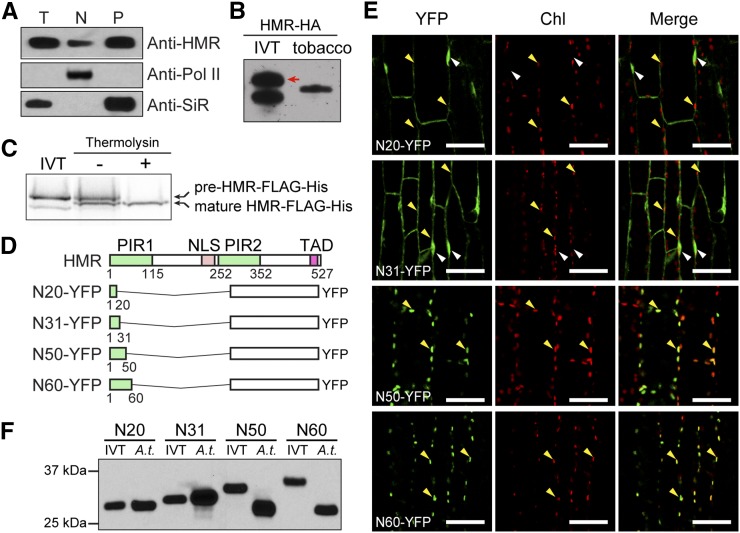
HMR isolated from the nucleus and chloroplasts show the same protein size (Fig. 1A; Chen et al., 2010; Pfalz et al., 2015). It was speculated that this mature form of HMR is smaller than the full-length HMR due to the loss of its transit peptide upon chloroplast import. However, it has not been demonstrated experimentally that HMR can be directly imported into chloroplasts. In fact, the ChloroP algorithm predicts that Arabidopsis HMR lacks a chloroplast-targeting sequence (Emanuelsson et al., 1999; Chen et al., 2010). Thus, we sought to determine whether HMR has an N-terminal transit peptide and, if so, whether this signal could be cleaved after chloroplast import. To address these questions, we compared the protein size of HA-tagged HMR (HMR-HA) transiently expressed in tobacco (Nicotiana benthamiana) with that of the full-length HMR-HA made by in vitro translation. The tobacco-expressed HMR-HA was clearly smaller than the full-length form, indicating that the mature HMR in vivo is a cleaved version (Fig. 1B). We then performed chloroplast import assays to confirm that HMR can be directly imported into chloroplasts. These experiments show that in vitro-translated HMR fused with FLAG and His tags (HMR-FLAG-His) could be imported into isolated Arabidopsis chloroplasts and resulted in smaller mature HMR-FLAG-His (Fig. 1C). To define HMR’s transit peptide, we fused various N-terminal fragments of HMR, ranging from 20 to 60 amino acids (N20-N60), to YFP (Fig. 1D) and generated transgenic lines in the Col-0 background expressing these recombinant YFP proteins. The ability of these HMR peptides in targeting YFP to the chloroplasts in the transgenic lines was examined by confocal microscopy. While N20-YFP and N31-YFP were localized in the nucleus and cytoplasm and failed to localize to the chloroplasts, both N50-YFP and N60-YFP were localized in the chloroplasts (Fig. 1E). In addition, N20-YFP and N31-YFP from the transgenic lines were the same sizes as their corresponding in vitro translated forms, whereas N50-YFP and N60-YFP from the transgenic lines were smaller than their respective in vitro translated forms (Fig. 1F). These results indicate that the N-terminal 50 amino acids of HMR are sufficient for chloroplast import. However, surprisingly, although N60-YFP is larger than N50-YFP, the mature forms of N50-YFP and N60-YFP had similar molecular mass (Fig. 1F), suggesting that N60-YFP was further proteolytically processed after the removal of the transit peptide. Together, these results demonstrate that Arabidopsis HMR can be directly targeted into the chloroplasts by a transit peptide, which resides in its N-terminal 50 amino acids. After cleavage of the transit peptide, the N terminus of HMR may be further proteolytically processed to yield mature HMR.
Figure 1.
HMR possesses an N-terminal transit peptide that is cleaved off in its mature form. A, Immunoblot analysis showing that HMR proteins from the nuclear and plastidial fractions have the same molecular mass. Total (T), nuclear (N), and plastidial (P) protein extracts were isolated from 4-d-old red light-grown Col-0 seedlings. HMR was detected by using anti-HMR antibodies. RNA polymerase II (Pol II) and SiR were used as controls for the nuclear and plastidial fractions, respectively. B, Immunoblot analysis showing that transiently expressed HMR-HA in tobacco migrated to a smaller-sized band compared with the in vitro translated (IVT) full-length HMR-HA. The red arrow indicates the full-length HMR-HA band. C, In vitro chloroplast import assays demonstrating that HMR can be imported directly into chloroplasts. HMR fused with FLAG and His tags, HMR-FLAG-His, was in vitro translated with 35S-Met and subjected to in vitro chloroplast import assays. After the import assay was completed, thermolysin was added to remove nonimported HMR-FLAG-His proteins. The protein extracts from the chloroplast import assays either with or without the thermolysin treatment were analyzed by SDS-PAGE and exposed to x-ray film. Whereas the sample without thermolysin treatment showed both full-length pre-HMR-FLAG-His outside of chloroplasts and mature HMR-FLAG-His in chloroplasts, the thermolysin-treated sample showed only the mature form with a reduced size. D, Schematic illustration of the domain structure of HMR and a series of N-terminal fragments of HMR fused with YFP. NLS, nuclear localization signal; PIR, phytochrome-A-interacting region; TAD, transcriptional activation domain. E, Confocal images showing the subcellular localization patterns of a series of N-terminal fragments of HMR fused with YFP in Arabidopsis transgenic lines. Chl, chlorophyll fluorescence. White arrows indicate nuclei, and yellow arrows indicate chloroplasts. F, Immunoblot analysis of the four recombinant proteins illustrated in D that were either prepared by in vitro translation or extracted from Arabidopsis transgenic lines (A.t.). The recombinant proteins were detected by using anti-GFP antibodies.
Mature HMR Lacks the N-Terminal 57 Amino Acids
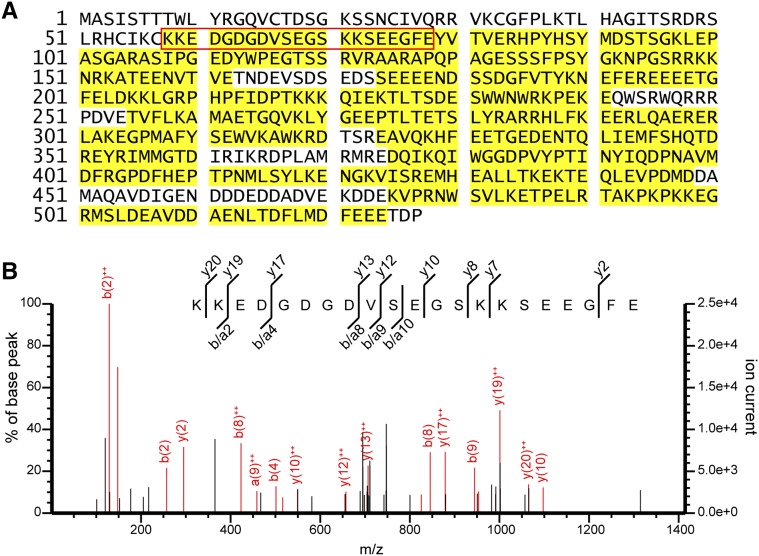
We then determined the mature form of HMR by mass spectrometry. We isolated HMR-HA from 4-d-old, red light-grown HMR-HA seedlings (Galvão et al., 2012; Qiu et al., 2015) by affinity purification with anti-HA antibody-conjugated agarose beads. HMR-HA proteins were separated by SDS-PAGE and stained with SimplyBlue SafeStain. The HMR band was excised from the gel and subjected to in-gel digestion with endoproteinase GluC, which cleaves peptide bonds C terminal to either Glu or Asp (Drapeau et al., 1972). The peptides digested from the excised band were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The LC-MS/MS analysis identified 94 unique HMR peptides (Supplemental Table S1), which spanned 76% of the full-length HMR sequence (Fig. 2A). The most N-terminal peptide identified is between amino acids 58 and 78, KKEDGDGDVSEGSKKSEEGFE (Fig. 2, A and B). Given the number of Glu and Asp residues within its sequence, this peptide likely resulted from incomplete digestion. Because the N-terminal residue preceding the peptide is a Cys, which cannot be cleaved by GluC, the cleavage between Cys-57 and Lys-58 was most likely catalyzed by an endogenous peptidase, suggesting that the mature form of HMR (HMRm) begins from Lys-58.
Figure 2.
Identification of HMRm by LC-MS/MS. A, LC-MS/MS analysis of HMR-HA. HMR-HA was immunoprecipitated from 4-d-old HMR-HA seedlings, digested with endoproteinase GluC, and analyzed by LC-MS/MS. Residues identified by MS analysis are highlighted in yellow. The red box indicates the N-terminal-most identified peptide. B, Annotated MS/MS spectrum (mascot ion score 26.7) of the peptide KKEDGDGDVSEGSKKSEEGFE on Q Exactive high-resolution mass spectrometer illustrating product ion coverage throughout sequence.
HMRm Retains Binding Activities with Phytochromes and PIFs
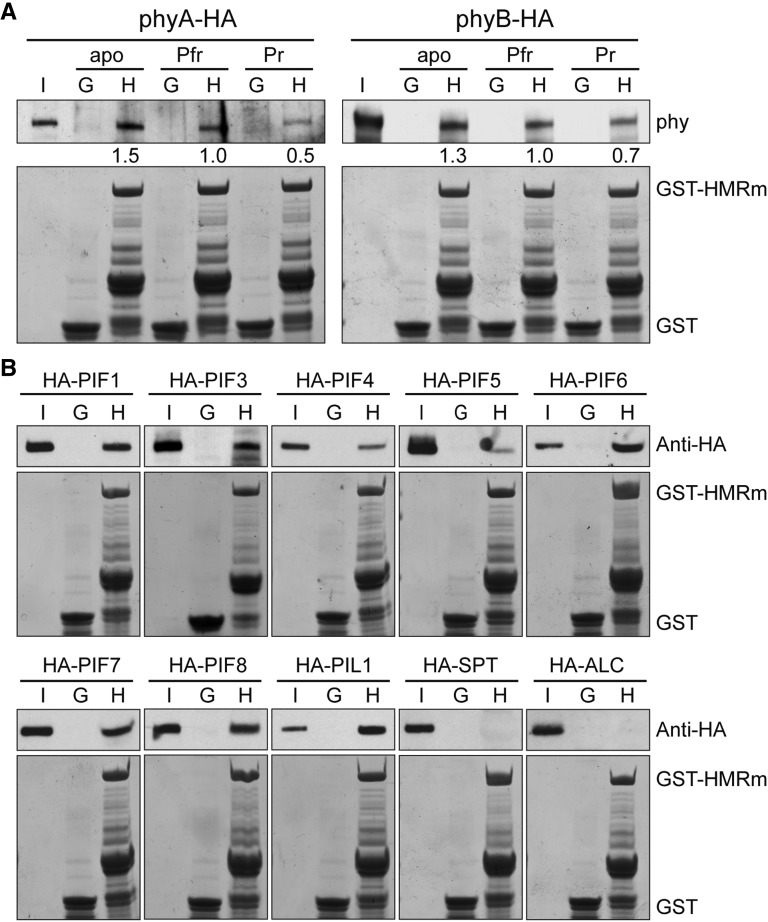
We showed previously that HMR acts as a phy signaling component in the nucleus via direct interactions with phys and PIFs. HMR interacts preferentially with the Pfr form of phyA and phyB (Galvão et al., 2012), and it binds to all seven PIFs and PIL1 (Qiu et al., 2015). Both the HMR-phy and HMR-PIF interactions are mediated by the N-terminal half of HMR, in particular the phyA-interacting region 1 (PIR1) within the N-terminal 115 amino acids. Therefore, it raised the question of whether HMRm retains the ability to interact directly with phys and PIFs. To address this question, we performed GST pull-down assays using Escherichia coli produced GST-HMRm as the bait to pull down in vitro translated HA-tagged phyA, phyB, PIFs, and PIL1. As shown in Figure 3A, GST-HMRm was able to pull down the apoprotein, Pr, and Pfr forms of phyA and phyB, and its interactions with the Pfr forms of phyA and phyB were stronger than those with the Pr forms. GST-HMRm could also pull down all PIFs and PIL1 but failed to pull-down SPATULA and ALCATRAZ, which lack PIFs’ conserved APB motif required for HMR interaction (Fig. 3B; Khanna et al., 2004; Qiu et al., 2015). These results indicate that HMRm retained the previously characterized biochemical properties in phytochrome signaling.
Figure 3.
HMRm interacts with phyA, phyB, and all PIFs in vitro. A, GST pull-down assays showing that HMRm interacts with phyA and phyB in vitro. The pull-down assays were performed with recombinant GST-HMRm or GST produced in E. coli to pull down in vitro-translated HA-tagged phyA and phyB (phyA-HA and phyB-HA) that were either apoproteins (apo) or holoproteins in the Pfr or Pr form. Input and bound phy fractions were detected by immunoblot using anti-phyA or -phyB antibodies (top). Immobilized GST-HMRm and GST are shown in the Coomassie blue-stained SDS-PAGE gels at the bottom. The relative amounts of the bound phy proteins are normalized to the amount of the corresponding GST-HMRm bait and are shown below the top. B, GST pull-down assays showing that HMRm interacts with all PIFs in vitro. E. coli-expressed recombinant GST-HMRm or GST were used to pull down in vitro-translated HA-tagged PIF1, PIF3-8, PIL1, SPATULA, and ALCATRAZ. Input and bound fractions of the prey proteins were detected by immunoblot using anti-HA antibodies (top). Immobilized GST-HMRm and GST are shown in the Coomassie blue-stained SDS-PAGE gels at the bottom. I, 10% input of the prey proteins; G, GST; H, GST-HMRm.
HMR’s Predicted Nuclear Localization Signals Are Nonfunctional
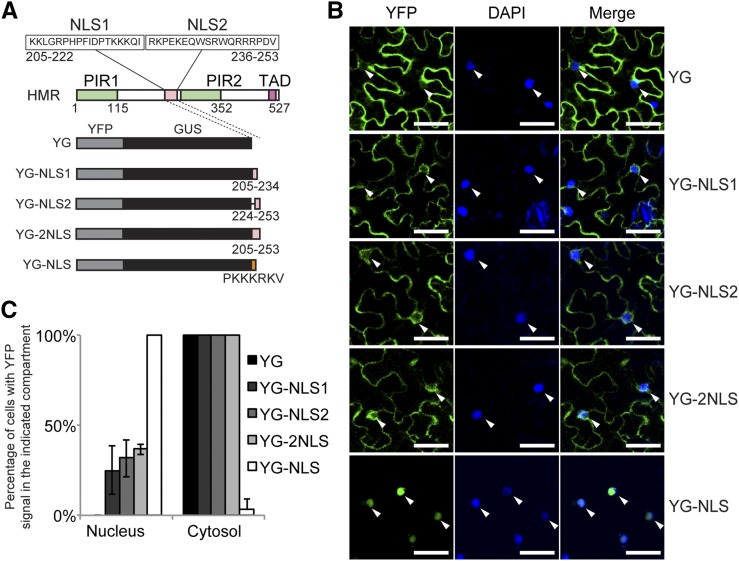
The observation that nuclear and plastidial HMR share the same Mr raises the possibility that HMR is targeted to the plastids first before translocating to the nucleus. If this hypothesis is correct, one might expect that deleting the transit peptide would block not only HMR’s chloroplast localization but also its nuclear accumulation. However, HMR does contain two predicted bipartite nuclear localization signals, NLS1 and NLS2 (Fig. 4A). We therefore tested whether these two nuclear localization signals are functional. To make a combined Mr greater than the nuclear pore’s passive diffusion limit of 60 kD (Wang and Brattain, 2007), we fused the two nuclear localization signals, either individually or combined, to YFP and GUS (YG; Fig. 4A). These recombinant proteins were transiently expressed in tobacco and their localization patterns were characterized by confocal microscopy. The results showed that neither NLS1, NLS2, nor combined NLS1/NLS2 (2NLS) was able to effectively import YG into the nucleus (Fig. 4, B and C). Although a small fraction of cells showed nuclear YFP signal, the majority of YG-NLS1, YG-NLS2, and YG-2NLS were localized to the cytoplasm, similar to the YG control (Fig. 4, B and C). These results indicate that the two predicted nuclear localization signals of HMR are nonfunctional.
Figure 4.
HMR’s predicted nuclear localization signals are nonfunctional. A, Schematic illustration of recombinant proteins of YG fused with either one or both of HMR’s predicted NLSs. YG with the SV40 NLS (PKKKRKV), YG-NLS, was used as a control for nuclear targeting. B, Confocal images of representative tobacco cells expressing the indicated recombinant proteins. Tissue was collected 3 d after agroinfiltration, stained with DAPI, and imaged by confocal microscopy. White arrowheads indicate nuclei. Scale bars = 50 μm. C, Quantification of subcellular localization data in B. The percentage of cells with YFP signal in the nucleus or cytoplasm for each of the recombinant proteins was determined by examining at least 60 DAPI- and YFP-positive cells. Error bars represent the sd of three independent experiments.
HMRm Could Rescue Neither the Albino nor Long Hypocotyl Phenotypes of hmr
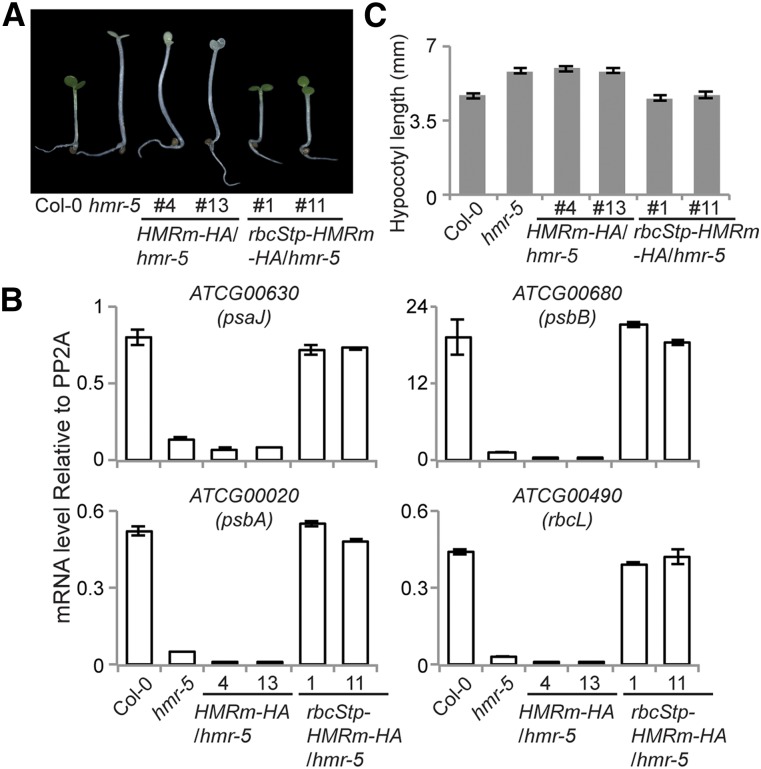
To examine the function of HMRm in vivo, we generated transgenic lines in the hmr-5 background expressing HA-tagged HMRm (HMRm-HA) under the 35S promoter, the same as the previously described HMR-HA line (Galvão et al., 2012). We analyzed two independent HMRm-HA transgenic lines. As expected, without the transit peptide, HMRm-HA could not rescue hmr-5’s albino phenotype (Fig. 5A) or its defects in the expression of PEP-dependent genes (Fig. 5B). However, HMRm also failed to rescue hmr-5’s long hypocotyl phenotype (Fig. 5, A and C), which presumably depends on the nuclear function of HMR (Qiu et al., 2015). These results suggest that HMRm alone is able to rescue neither the nuclear nor plastidial functions of HMR.
Figure 5.
HMRm requires a transit peptide to rescue both the tall and albino phenotypes of the hmr mutant. A, Images of 4-d-old seedlings grown under 10 μmol m−2 s−1 red light from Col-0, hmr-5, HMRm-HA/hmr-5, and rbcStp-HMRm-HA/hmr-5 lines. B, qRT-PCR data showing the transcript levels of representative PEP-dependent genes relative to PP2A in 4-d-old red light grown Col-0, hmr-5, HMRm-HA/hmr-5, and rbcStp-HMRm-HA/hmr-5 lines. Error bars represent the se of three replicates. C, Hypocotyl length measurements of seedlings in A. Error bars represent sd from at least 20 seedlings.
HMRm Failed to Rescue the Defects of HMR-Regulated Nuclear Genes in hmr
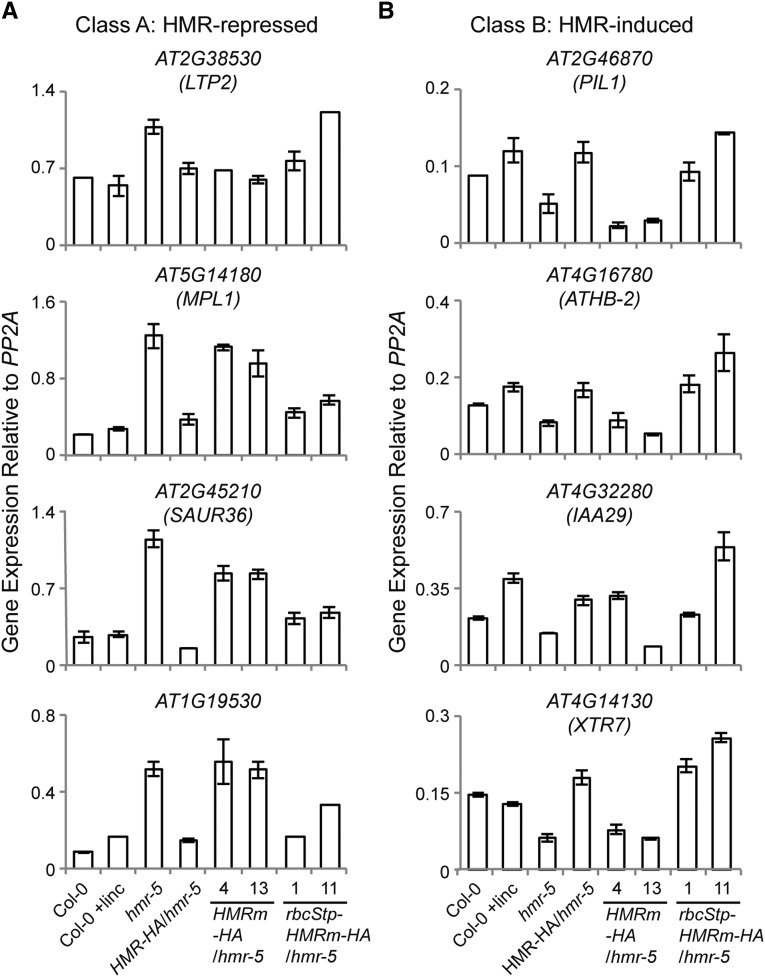
We showed previously that nuclear HMR is required for the regulation of a subset of growth-promoting PIF-induced direct target-genes: HMR-repressed Class A genes and HMR-induced Class B genes (Qiu et al., 2015). To demonstrate that HMRm-HA does not show nuclear function, we asked whether the HMRm-HA lines could rescue expression of the HMR-dependent nuclear genes. However, because the expression of light responsive genes in the nucleus is also controlled by retrograde signaling from plastids, deficiencies in chloroplast biogenesis could dramatically alter nuclear gene expression (Woodson and Chory, 2008). Therefore, one major challenge in characterizing the nuclear function of HMRm-HA in the albino HMRm-HA/hmr-5 lines was to distinguish whether alterations in nuclear gene expression were due to nuclear HMRm-HA or the concomitant defect in chloroplast biogenesis. To this end, we first tested whether the expression levels of the HMR-regulated genes were influenced by defects in chloroplast biogenesis through treatment with lincomycin, a potent inhibitor of chloroplast translation (Nott et al., 2006; Koussevitzky et al., 2007). Lincomycin inhibits the 70s ribosome, leading to the loss of the plastid-encoded core subunits of the PEP RNA polymerase. The resulting defects in expression of plastid-encoded photosynthetic genes subsequently lead to repression of nuclear-encoded photosynthesis-associated nuclear genes through retrograde signaling (Koussevitzky et al., 2007; Woodson et al., 2013). Interestingly, although the Class A and B genes are misregulated in the albino hmr-5 mutant, lincomycin treatment of Col-0 did not have a significant effect or the same effect on these HMR-regulated PIF-induced genes (Fig. 6). Therefore, these two groups of HMR-regulated PIF-induced genes can be used as markers for assessing the nuclear function of HMR. These results are consistent with a recent study by Martín et al., who showed that lincomycin, through retrograde signaling, controls PIF-repressed but not PIF-induced genes (Martín et al., 2016). We then examined the expression of the Class A and B genes in the HMRm-HA/hmr-5 lines. Compared with hmr-5, HMRm-HA/hmr-5 showed similar expression levels in three of the four Class A genes, At5g14180, At2g45210, and At1g19530, and in three of the four Class B genes, At2g46870, At4g16780, and At4g14130 (Fig. 6, A and B). These results support the conclusion that HMRm fails to rescue the nuclear defects of hmr-5.
Figure 6.
HMRm requires a transit peptide to regulate HMR-dependent genes in the nucleus. A, qRT-PCR analyses of the relative expression levels of representative HMR-repressed and PIF-induced Class A genes in 4-d-old Col-0, Col-0 with lincomycin (linc) treatment, hmr-5, HMR-HA/hmr-5, HMRm-HA/hmr-5, and rbcStp-HMRm-HA/hmr-5 lines grown in 10 μmol m−2 s−1 red light. B, qRT-PCR analyses of the relative expression levels of representative HMR-induced and PIF-induced Class B genes in 4-d-old Col-0, Col-0 with lincomycin treatment, hmr-5, HMR-HA/hmr-5, HMRm-HA/hmr-5, and rbcStp-HMRm-HA/hmr-5 lines grown in 10 μmol m−2 s−1 red light. Error bars represent the sd of three biological replicates.
HMRm Failed to Accumulate in the Nucleus
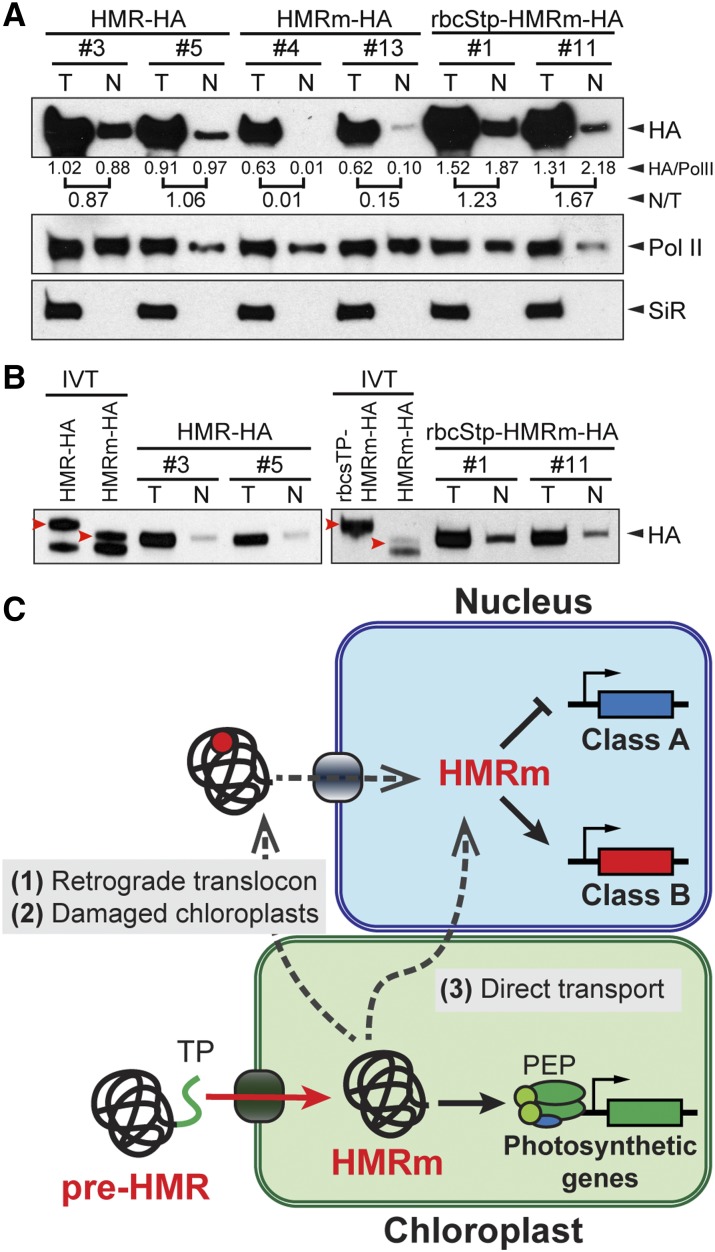
We then compared the relative amount of the nuclear fraction of HMRm-HA with that of HMR-HA. Strikingly, in the two HMRm-HA lines that we examined, the nuclear fractions of HMRm were more than 5-fold less than those of the HMR-HA lines (Fig. 7A). Consistent with the notion that HMR’s predicted nuclear localization signals are nonfunctional, these results demonstrate that HMRm-HA is unable to efficiently accumulate in the nucleus.
Figure 7.
HMR’s nuclear accumulation requires its localization to chloroplasts. A, Semiquantitative measurements of the nuclear fractions of HMR-HA, HMRm-HA, and rbcStp-HMRm-HA from transgenic lines. Total and nuclear HMR-HA protein fractions were isolated and analyzed with western blots using anti-HA antibodies. Pol II and SiR were used as nuclear and plastidial controls, respectively. Band intensities from the HA blots were normalized to their respective Pol II controls, and the nuclear to total ratio (N/T) was calculated for each line and is listed below the HA western blots. B, Immunoblots showing the endogenous HMR-HA proteins are the same size as or similar to the size of their corresponding in vitro translated mature forms. Left, The endogenous total and nuclear HMR-HA proteins are the same size as the in vitro translated (IVT) HMRm-HA and are clearly smaller than the in vitro translated full-length HMR-HA. Right, The endogenous rbcStp-HMRm-HA proteins are similar in size to the in vitro translated HMRm-HA and are smaller than the in vitro translated full-length rbcStp-HMRm-HA. Red arrowheads indicate the bands of full-length in vitro translated HMR proteins. C, model for the mechanism of nuclear and plastidial targeting of HMR. Upon translation in the cytoplasm, the HMR preprotein (pre-HMR) is imported into the chloroplasts by its transit peptide (TP, labeled in green). The TP and additional N-terminal residues are cleaved in the chloroplast to yield HMRm, which participates in the PEP-mediated transcriptional activation of plastid-encoded photosynthetic genes. HMRm can be then translocated to the nucleus by at least three possible ways: (1) with an unknown retrograde translocon, (2) due to leakage from damaged chloroplasts, and (3) through direct connections between chloroplasts and the nucleus, such as stromules. In the first two scenarios, HMRm could be posttranslationally modified (represented as the red circle) to facilitate its nuclear localization.
We had previously shown that when an YFP is fused to the N terminus of the full-length HMR to block the function of its transit peptide, YFP-HMR was localized to the nucleus and cytoplasm, and it fails to rescue the hmr mutant (Chen et al., 2010). To test whether the defect of YFP-HMR might also be due to the failure in nuclear accumulation, we reexamined YFP-HMR localization in the YFP-HMR lines. These results show that despite observable nuclear localization in 25% of the nuclei, YFP-HMR was mainly localized to the cytoplasm (Supplemental Fig. S1, A and B). As shown previously, YFP-HMR did not rescue the long hypocotyl phenotype of hmr-5 (Supplemental Fig. S1C); also, it did not rescue the defect in the expression of the HMR- and PIF-induced Class B genes (Supplemental Fig. S1D). These results support the notion that HMR does not contain a functional NLS and suggest that the reason why YFP-HMR did not rescue the nuclear defect of the hmr mutant is likely due to the lack of nuclear accumulation.
Fusing the Transit Peptide of rbcS to HMRm Rescues Its Defects in Nuclear Accumulation and Function
To further test the hypothesis that HMR’s nuclear accumulation is dependent on its plastidial localization, we asked whether fusing a heterologous transit peptide to HMRm would rescue its nuclear accumulation and function. To this end, we fused the transit peptide of the small subunit of Rubisco, rbcStp (Roesler et al., 1997), to HMRm-HA and generated transgenic lines in the hmr-5 background. We picked two independent rbcStp-HMRm-HA/hmr-5 lines for further characterization. Both transgenic lines rescued hmr-5’s albino phenotype (Fig. 5A) as well as its defects in the expression of PEP-dependent genes (Fig. 5B), indicating that HMRm is fully functional in plastids. Interestingly, rbcStp-HMRm-HA also rescued the long hypocotyl phenotype of hmr-5 (Fig. 5C). In addition, the expression of the Class A genes, AT5G14180, AT2G45210, and AT1G19530, as well as all of the Class B genes were rescued in the rbcStp-HMRm-HA/hmr-5 lines (Fig. 6). Therefore, rbcStp-HMRm-HA was able to rescue both the plastidial and nuclear defects of hmr-5. In line with these observations, the levels of nuclear HMR-HA in the rbcStp-HMRm-HA/hmr-5 lines were greatly enhanced compared with those in the HMRm-HA/hmr-5 lines and were comparable to those in the HMR-HA/hmr-5 lines (Fig. 7A). Also, similar to HMR-HA, the size of endogenous rbcStp-HMRm-HA was similar to, if not the same as, that of HMRm-HA (Fig. 7B), suggesting that the rbcS transit peptide was removed from rbcStp-HMRm-HA and that it was the mature form of rbcStp-HMRm-HA that had accumulated in the nuclei of the rbcStp-HMRm-HA/hmr-5 lines. Together, these results support the hypothesis that HMR’s nuclear accumulation depends on localization to the plastids.
DISCUSSION
HMR is the first identified phy signaling component dual targeted to the nucleus and plastids (Chen et al., 2010), which draws a direct link between the activity of the PEP RNA polymerase in the plastids and phy signaling in the nucleus (Pfalz et al., 2006, 2015; Chen et al., 2010; Galvão et al., 2012; Qiu et al., 2015). Molecular understanding of the mechanisms targeting HMR to the nucleus and plastids holds promise for gaining novel insight into the mechanisms of nuclear-plastidial communication (Krause and Krupinska, 2009; Krause et al., 2012). In this study, we have showed that HMR can be directly imported into chloroplasts, where it is processed to the mature form starting from Lys-58 (Figs. 1 and 2). HMRm retains the abilities to interact with phys and PIFs in phy signaling (Fig. 3; Galvão et al., 2012; Qiu et al., 2015). Unexpectedly, the predicted nuclear localization signals in HMR are nonfunctional (Fig. 4). We showed that HMR’s nuclear accumulation and function rely on its transit peptide (Figs. 5–7). Fusing the transit peptide of rbcS to HMRm rescues its defects in nuclear accumulation and function (Figs. 5–7). Together, these results, combined with the observation that nuclear HMR has the same molecular mass as plastidial HMR (Fig. 1; Chen et al., 2010; Pfalz et al., 2015), provide biochemical, cellular, and genetic evidence supporting a plastid-to-nucleus retrograde protein translocation mechanism, in which nuclear accumulation of HMR depends on localization to the plastids (Fig. 7C).
HMRm
HMR/pTAC12 was shown to be dual targeted to the nucleus and plastids in Arabidopsis and maize (Chen et al., 2010; Pfalz et al., 2015). It is intriguing that the nuclear and plastidial HMR proteins showed the same size (Fig. 1; Chen et al., 2010; Pfalz et al., 2015), which raised the possibility that HMRm is derived from the removal of its plastidial targeting transit peptide. However, it had not been experimentally shown that HMR could be directly imported into plastids, and HMRm had not been determined. In this study, we show that HMR can be directly imported into chloroplasts (Fig. 1C). Chloroplast localization leads to reduction in the size of HMR as well as recombinant proteins with N-terminal fragments of HMR in both tobacco and Arabidopsis (Fig. 1), demonstrating that full-length pre-HMR is proteolytically processed upon chloroplast localization. We determined that mature HMR begins from Lys-58. Several lines of evidence support this conclusion. First, when purified HMR-HA was subjected to Glu-C digestion and LC-MS/MS analysis, the most N-terminal fragment identified begins at Lys-58 (Fig. 2). Because Glu-C is not expected to cleave between Cys-57 and Lys-58, this peptide likely defines the N terminus of HMRm. Second, although the N-terminal 50 amino acids of HMR are sufficient to mediate chloroplast import, when the N-terminal 60 amino acids of HMR were fused to YFP, the mature protein was further processed to a residue between amino acids 50 and 60 (Fig. 1), which is consistent with the conclusion that HMRm begins at residue 58. Third, HMRm-HA from the HMR-HA/hmr-5 lines showed the same size as HMRm-HA from the HMRm-HA/hmr-5 lines (Fig. 7A). Fourth, HMRm retains the binding activities with phyA, phyB, PIFs, and PIL1 (Fig. 3). Lastly, fusing the transit peptide of rbcS to HMRm rescues both the nuclear and plastidial phenotypes of the hmr mutant, indicating that HMRm is functional in both the nucleus and plastids in vivo (Figs. 5–7). Based on these results, we conclude that HMRm in Arabidopsis begins from Lys-58.
Mechanism of Dual-Targeting HMR to Plastids and the Nucleus
It is well documented in yeast (Saccharomyces cerevisiae), animals, and plants that a single gene can produce proteins targeted to dual destinations (Karniely and Pines, 2005; Carrie et al., 2009; Krause et al., 2012). Dual distribution between subcellular compartments can be achieved by two general strategies. One strategy is that a single gene produces two protein variants that differ in subcellular targeting signals due to alternative transcription or translation initiation sites. For example, the Arabidopsis GSTF8 (glutathione S-transferase F8) gene utilizes multiple transcription start sites that produce two distinct protein forms: a large protein with an N-terminal transit peptide that localizes to the plastids and a small protein without the transit peptide that localizes to the cytoplasm (Thatcher et al., 2007). The alternative strategy is that a single protein can be targeted to two subcellular compartments. The dual-localization of HMR is likely due to the latter strategy for several reasons. First, HMR has not been found to produce alternative transcripts from distinct transcription start sites. In addition, because both the nuclear and plastidial HMR show the same size (Fig. 1; Chen et al., 2010; Pfalz et al., 2015), the two forms are unlikely to be produced by alternative transcripts like GSTF8. Second, although HMRm is smaller than full-length HMR, this is unlikely the result of alternative translation start sites. The second Met in HMR is at position 91, so if this Met were used as an alternative translational start site, it would produce a protein much smaller than the one observed. Furthermore, this second AUG is unlikely to be an initiator codon, because it does not contain the caA(A/C)aAUGGCg consensus sequence of plant translation initiator codons (Joshi et al., 1997). Taken together, the current evidence supports the idea that a single translational product of HMR is dual-targeted to plastids and the nucleus.
The best-recognized mechanism targeting a single protein to the nucleus and an organelle is that a single polypeptide harbors two targeting signals, a nuclear localization signal and an organellar targeting signal. In this case, the partitioning of the protein between these two compartments is determined by competition and accessibility of the two targeting signals (Karniely and Pines, 2005; Krause and Krupinska, 2009; Krause et al., 2012). For instance, yeast Pir1p contains both a nuclear localization signal and a mitochondrial targeting signal, and its partitioning to these two subcellular compartments is regulated by masking and unmasking of the nuclear localization signal by its binding partner Apn1p (Vongsamphanh et al., 2001). It was hypothesized that the dual targeting of HMR was mediated by its predicted nuclear localization signals and transit peptide (Pfalz et al., 2006; Chen et al., 2010); however, the functionality of these targeting signals had never been experimentally validated. In this study, we demonstrate that HMR possesses a functional transit peptide within its N-terminal 50 amino acids, which is required and sufficient for delivering a YFP reporter into the chloroplasts in Arabidopsis (Fig. 1). Surprisingly, neither of these two predicted nuclear localization signals in HMR is highly functional (Fig. 4). Consistent with this notion, HMRm fails to accumulate and exert functions in the nucleus (Figs 5–7). Together, these results argue against the original hypothesis of dual-targeting signals. On the contrary, these results, combined with the fact that both nuclear and plastidial HMR show the same size (Fig. 1; Chen et al., 2010; Pfalz et al., 2015), support the alternative model that HMR is first localized to and proteolytically processed in the plastids before being relocated to the nucleus, a mechanism that requires plastid-to-nucleus retrograde protein translocation (Fig. 7C). This plastid-to-nucleus retrograde protein translocation model has been proposed for the subcellular localization of another nuclear and plastidial localized protein, Whirly1 (Why1; Grabowski et al., 2008). Why1 is a single-stranded DNA and RNA binding protein (Desveaux et al., 2000, 2002; Prikryl et al., 2008). In the nucleus, Why1 binds to single-stranded telomeric DNA to modulate telomere length homeostasis (Yoo et al., 2007) and acts as a transcriptional regulator for pathogen responsive genes (Desveaux et al., 2000; Xiong et al., 2009). Plastidial Why1 is required for chloroplast biogenesis in maize (Prikryl et al., 2008) and plays a possible role in plastidial genome repair (Cappadocia et al., 2010, 2012). Interestingly, a fraction of Why1 is also associated with the pTAC complex (Pfalz et al., 2006; Melonek et al., 2010). Compared with the full-length protein, nuclear Why1 exhibits the same reduced molecular mass as the plastidial form (Grabowski et al., 2008). More importantly, plastomically expressed Why1 in tobacco could translocate to the nucleus, providing solid evidence that Why1, at least in tobacco, could move from plastids to the nucleus (Isemer et al., 2012). Here, we showed that nuclear accumulation of HMR is dependent on its localization to the plastids (Figs. 5–7). Although the localization data of plastomically expressed HMR are still missing, the results shown in this study strongly support the idea that HMR’s nuclear localization in Arabidopsis is mediated by a retrograde protein translocation pathway (Fig. 7C).
The mechanism of retrograde protein translocation from plastids to the nucleus is still largely unknown. There are at least three possible pathways (Fig. 7C). First, HMR can be retrieved from the plastids by retrograde movement through an unidentified translocon or secretory pathway. Second, HMR can be redistributed as a result of leakage from chloroplasts, either due to organellar damage or during autophagy (Woodson et al., 2015). Because HMRm cannot effectively accumulate in the nucleus, in these two scenarios, HMRm might be modified in plastids to enhance the activity of its own nuclear localization signals or to promote interactions with other proteins that facilitate HMR’s nuclear accumulation (Fig. 7C). Third, HMR can be transported from plastids to the nucleus through physical connections between these two compartments such as stromules (Kwok and Hanson, 2004). With the increasing number of described nuclear and plastidial dual-targeted proteins, it has become essential to understand the mechanisms of dual targeting to the nucleus and plastids and the significance of these proteins in both antero- and retrograde signaling between the two gene-bearing compartments (Krause and Krupinska, 2009; Krause et al., 2012). This study represents one of the initial attempts toward these goals.
MATERIALS AND METHODS
Plant Growth Conditions and Hypocotyl Measurements
Seeds were surface sterilized in 50% bleach with 0.01% Triton X-100 for 10 min and then washed four times with sterile double distilled water. Seeds were then plated on half-strength Murashige and Skoog with Gamborg’s vitamins (MSP0506; Caisson Laboratories), 0.5 mm MES (pH 5.7), and 0.8% (w/v) agar (A038; Caisson Laboratories). For lincomycin treatments, plates were supplemented with 0.5 mm lincomycin hydrochloride (L2774; Sigma-Aldrich). Seeds were stratified in the dark at 4°C for 4 d. For microscopy, hypocotyl measurements, nuclear fractionation, and RNA extraction, seedlings were grown at 21°C in an LED chamber (Percival Scientific), and light intensity was measured using an Apogee PS200 spectroradiometer (Apogee Instruments). For hypocotyl length measurements, an Epson Perfection V700 photo scanner was used to capture images of seedlings. At least 20 hypocotyls from each genotype and treatment were measured with NIH ImageJ software.
Plasmid Construction and Generation of Transgenic Lines
For producing HMR-HA by in vitro translation, HMR-HA was subcloned from pCHF3-HMR-HA (Qiu et al., 2015) and ligated into the XmaI and PstI sites of the pCMX-PL1 vector (Umesono et al., 1991).
The constructs for identifying HMR’s transit peptide were generated from pCHF3-YFP, which was generated by ligating YFP flanked by SalI and XhoI sites into the SalI site after the 35S promoter in pCHF3 (Fankhauser et al., 1999). The segments of HMR were amplified by PCR using the primers in Supplemental Table S2 and ligated into the BamHI and SalI sites of pCHF3-YFP. These constructs were then transformed into Col-0 plants with Agrobacterium tumefaciens strain GV3101 carrying the appropriate plasmids. Transformants were identified by screening on kanamycin plates.
Constructs producing recombinant HA-tagged prey proteins in vitro for GST pulldowns were previously described (Qiu et al., 2015). The bait construct encoding GST-tagged HMRm was cloned by amplifying HMRm by PCR with the primers in Supplemental Table S2 and ligating into the EcoRI and PstI sites of pET42b (Novagen).
For constructs for testing the predicted NLS of HMR, YFP without a stop codon was ligated into the KpnI and BamHI sites of pCHF3 to give pCHF3-YFP(N). Then GUS, lacking a stop codon and flanked by BglII and BamHI sites, was amplified by PCR with primers in Supplemental Table S2 and ligated into the BamHI site of pCHF3-YFP(N) to give pCHF3-YFP-GUS. The control constructs, YG-NLSsv40 and YG, were generated by ligating annealed oligonucleotides encoding the SV40 NLS or a stop codon (Supplemental Table S2) into the BamHI and SalI sites of pCHF3-YFP-GUS. Constructs YG-NLS1, YG-NLS2, and YG-2NLS were generated by amplifying segments of HMR by PCR with the primers in Supplemental Table S2 and then ligating them into the BamHI and SalI sites of pCHF3-YFP-GUS.
HMRm-HA was made by amplifying HMRm-HA by PCR with the primers described in Supplemental Table S2 using pCHF3-HMR-HA as a template. The PCR product was then inserted into the SalI site after the 35S promoter in pCHF3 by Gibson assembly (NEB). Similarly, rbcStp-HMRm-HA was cloned by amplifying the rbcStp from cDNA, and HMRm-HA from pCHF3-HMR-HA, by PCR with primers listed in Supplemental Table S2. Both fragments were then inserted simultaneously into the SalI site of pCHF3 by Gibson assembly (NEB). Transgenic lines were generated by transforming hmr-5 heterozygous plants with A. tumefaciens strain GV3101 carrying either pCHF3-HMRm-HA or pCHF3-rbcStp-HMRm-HA plasmids, and transgenic seedlings were identified by screening on kanamycin plates.
Chloroplast Import Assays
Import assays were performed as previously described (Li and Schnell, 2006). Substrate proteins for import were produced and labeled with [35S] Met using the TNT T7 Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions. Chloroplasts isolated from 12- to 14-d-old Col-0 plants were depleted of internal ATP by incubation in the dark for 30 min, followed by treatment with 400 nm nigericin to uncouple ATP generation. Energy-depleted chloroplasts corresponding to 30 μg of chlorophyll were incubated with [35S]-labeled proteins in the absence or presence of 1 mm ATP for 20 min at 26°C in import buffer (330 mm sorbitol, 50 mm HEPES-KOH, pH 7.5, 25 mm KOAc, and 5 mm MgOAc). Thermolysin treatment was then performed by diluting the import reactions with 3-fold excess ice-cold import buffer with or without 100 µg/mL thermolysin. The reactions were then incubated on ice for 30 min, and proteolysis was stopped with 1 mm phenylmethylsulfonyl fluoride, 0.05 mg/mL TLCK, and 10 mm EDTA. Chloroplasts were recovered by isolation over a 35% Percoll cushion and washed once with import buffer.
Protein Extraction and Immunoblots
Arabidopsis (Arabidopsis thaliana) seedlings or tobacco (Nicotiana benthamiana) leaves were harvested, immediately frozen in liquid N2, and stored at −80°C. For total protein extraction, tissue was ground in liquid nitrogen and then mixed with 2 volumes (mg/µL) extraction buffer containing 100 mm Tris-Cl, pH 7.5, 100 mm NaCl, 5 mm EDTA, 1% SDS, 5 mm dithiothreitol (DTT), 10 mm β-mercaptoethanol, 40 µm MG115 (Sigma-Aldrich), 40 µm MG132 (Sigma-Aldrich), 1× phosphatase inhibitor cocktail 3 (Sigma-Aldrich), and 1× EDTA-free protease inhibitor cocktail (Roche). Laemmli protein sample buffer was added to the crude extracts, which were then boiled for 10 min and centrifuged at 15,000g for 10 min. Then 20 to 30 μg of protein was separated by SDS-PAGE, transferred to nitrocellulose membranes, probed with the indicated primary antibodies, and then incubated with horseradish peroxidase-conjugated anti-goat, anti-mouse, or anti-rabbit secondary antibodies (Bio-Rad). Monoclonal anti-HA antibodies were used at 1:1,000 dilution (Roche). Polyclonal anti-HA antibodies were used for GST pulldown assays at 1:2,000 dilution (Abcam). Polyclonal anti-GFP antibodies were used at 1:2,000 dilution (Abcam). Monoclonal anti-phyB antibodies were used at 1:1,000 dilution. Polyclonal antibodies against chloroplast ferredoxin-sulfite reductase (SiR; Chi-Ham et al., 2002) were used at 1:1,000 dilution. Signals were detected by chemiluminescence using a SuperSignal kit (ThermoFisher Scientific).
Transient Expression in Tobacco
Proteins were transiently expressed in tobacco leaves as previously described (Roberts et al., 2011). A. tumefaciens strain GV3101 was transformed with each construct by electroporation. Cells were grown overnight, pelleted, and then resuspended in one-half the volume of the original culture of infiltration buffer, containing 10 mm MgCl2 and 10 mm MES. Cells were then induced with 200 μm acetosyringone (4′-hydroxy-3′,5′-dimethoxyacetophenone). Three hours after induction, cells were diluted to an optical density of 0.95 (600 nm). Experimental cells were mixed in equal volumes with A. tumefaciens expressing the P19 silencing suppressor before infiltration into the abaxial side of tobacco leaves. Samples were collected and stained with DAPI for microscopy three days after infiltration.
Affinity Purification and MS
To purify HMR-HA for transit peptide identification, HMR-HA lines were grown in a continuous white light incubator (Percival Scientific) for 96 h. Five grams of seedlings were collected and ground in liquid nitrogen with a mortar and pestle. The powder was then resuspended in coimmunoprecipitation buffer containing 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, and 1× EDTA-free protease inhibitor cocktail (Roche). The crude extract was centrifuged at 10,000g for 15 min, and the supernatant was then further cleared through a 0.45-µm filter. The clarified lysate was mixed with 50 µL of anti-HA conjugated agarose beads (ThermoFisher Scientific) and incubated at 4°C for 4 h. Washing and elution with 2× LDS loading buffer were done using an anti-HA spin column kit (ThermoFisher Scientific) according to the manufacturer’s directions.
The Duke Proteomics Core Facility received samples in LDS Loading buffer. Samples were reduced with 10 mm DTT at 70°C for 10 min prior to SDS-PAGE separation on a 4 to 12% Bis-Tris acrylamide gel (NuPAGE; ThermoFisher Scientific) with colloidal Coomassie blue staining. Bands were excised and subjected to standardized in-gel trypsin or protease V-8 (GluC) digestion. Extracted peptides were lyophilized to dryness and resuspended in 20 µL of 0.2% formic acid/2% acetonitrile.
Each sample was subjected to chromatographic separation on a Waters NanoAcquity UPLC equipped with a 1.7-µm BEH130 C18 75-µm i.d. × 250-mm reversed-phase column. The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. Following a 2-µL injection, peptides were trapped for 3 min on a 5-µm Symmetry C18 180-µm ID × 20-mm column at 5 µL/min in 99.9% A. The analytical column was then switched in-line and a linear elution gradient of 5% B to 40% B was performed over 60 min at 400 nL/min. The analytical column was connected to a fused silica PicoTip emitter (New Objective) with a 10-µm tip orifice and coupled to a Thermo Q Exactive Plus mass spectrometer through an electrospray interface operating in a data-dependent mode of acquisition. The instrument was set to acquire a precursor MS scan from m/z 375 to 1,600 with MS/MS spectra acquired for the 10 most abundant precursor ions. For all experiments, charge-dependent CID energy settings were employed and a 60-s dynamic exclusion was employed for previously fragmented precursor ions.
Raw LC-MS/MS data files were processed in Mascot distiller (Matrix Science) and then submitted to independent Mascot searches (Matrix Science) against a TAIR10 database containing both forward and reverse entries of each protein (32,790 forward entries). Search tolerances were 5 ppm for precursor ions and 0.02 D for product ions using semi-GluC specificity with up to seven missed cleavages. Carbamidomethylation (+57.0214 D on C) was set as a fixed modification, whereas oxidation (+15.9949 D on M) was considered a dynamic mass modification. All searched spectra were imported into Scaffold (v4.3; Proteome Software) and scoring thresholds were set to achieve a protein false discovery rate of 0.7% using the PeptideProphet algorithm.
Confocal Microscopy
Confocal microscopy was performed using a Zeiss LSM 510 inverted confocal microscope with either 10×/0.3, 20×/0.8, or 40×/0.95 Plan-Apochromat air objective. EYFP fluorescence was monitored using a 505- to 550-nm band-pass emission filter and 514-nm excitation from an argon laser. DAPI was monitored with a 420- to 480-nm band-pass emission filter and 405-nm diode laser excitation. Chlorophyll was observed with a long-pass 575-nm filter and 405-nm diode laser excitation.
Quantification of Nuclear Localization
For quantifying nuclear localization, YFP-GUS-tagged constructs were transiently expressed in tobacco, and confocal microscopy was used to take a Z-stack of images through the epidermal layer of each sample at 10× magnification. For each image, all DAPI-positive cells with YFP signal were marked in NIH ImageJ (https://imagej.nih.gov/nih-image/) and 20 cells were randomly chosen for analysis. All optical sections for each cell were examined for colocalization of DAPI and YFP. Localization was scored as “not nuclear” if there was no detectable YFP signal inside a given nucleus, or if in any optical section the YFP signal appeared as a ring around, but not within, a nucleus. Conversely, localization was scored as “nuclear” if YFP was clearly visible within the center of a nucleus in the optical section with maximum DAPI signal. Each cell was marked as having cytosolic localization if there was YFP signal visible around the cell’s border.
RNA Extraction and qRT-PCR
Seedlings for RNA extraction were collected, frozen in liquid nitrogen, and stored at −80°C before processing. Samples were ground in liquid nitrogen and RNA was extracted using the Quick-RNA MiniPrep kit with on-column DNase I digestion (Zymo Research). cDNA synthesis was performed with the Superscript II first-strand cDNA synthesis kit (Invitrogen), using oligo(dT) along with gene-specific primers for plastidial genes listed in Supplemental Table S3. For qRT-PCR, cDNA was mixed with FastStart Universal SYBR Green Master Mix (Roche) and primers. qRT-PCR reactions were performed in triplicate with a Roche LightCycler 96 thermal cycler. Primers for qRT-PCR of PEP-dependent genes are listed in Supplemental Table S3, while nuclear gene primers are listed in Supplemental Table S4.
GST Pull-Down Assays
Escherichia coli BL21(DE3) cells, expressing either GST or GST-HMRm from pET42b vectors (Novagen), were harvested by centrifugation at 5,000g for 10 min and then resuspended in E buffer containing 50 mm Tris-Cl (pH 7.5), 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% DMSO, 2 mm DTT, and bacterial protease inhibitor cocktail (Sigma-Aldrich). All subsequent purification and binding steps were carried out at 4°C. Cells were lysed by French press, and the lysate was centrifuged at 10,000g for 20 min. Proteins were then precipitated with 3.3 m ammonium sulfate and incubated for 4 h at 4°C. After centrifugation at 10,000g for 30 min, protein pellets were resuspended in E buffer. Insoluble protein was removed by centrifugation at 10,000g for 1 h, and the supernatant was dialyzed against E buffer overnight at 4°C.
To immobilize bait proteins, protein extracts were incubated with glutathione Sepharose beads (GE Healthcare) equilibrated in E buffer for 2 h, and then beads were washed four times in E buffer with 0.1% Nonidet P-40. HA-tagged prey proteins were produced in vitro using the TNT T7-coupled reticulocyte lysate system (Promega) according to the manufacturer’s protocol and with previously described plasmids (Qiu et al., 2015). TNT products were then diluted in E buffer with 0.1% Nonidet P-40 and incubated with bead-immobilized bait proteins at 4°C for 2 h. After binding, beads were washed four times in E buffer with 0.1% Nonidet P-40, and then protein was eluted by boiling in 1× Laemmli sample buffer. Bound proteins were separated by SDS-PAGE, and prey proteins were detected with immunoblots using either polyclonal goat anti-HA antibodies (Genescript) or monoclonal anti-phyB antibodies. Bait proteins were visualized by staining SDS-PAGE gels with Coomassie Brilliant Blue.
Nuclear Fractionation
Nuclei were isolated from Arabidopsis seedlings with some modifications to our previously described protocol (Galvão et al., 2012). Seedlings were ground in liquid nitrogen, and then 2 volumes (mg/µL) of nuclear extraction buffer containing 20 mm PIPES-KOH (pH 7.0), 10 mm MgCl2, 12% hexylene glycol, 0.25% Triton X-100, 5 mm β-mercaptoethanol, and 1× EDTA-free protease inhibitor cocktail (Roche) was added to the ground tissue. The crude lysate was filtered through a 40-µm cell strainer and incubated on ice for 10 min. The filtrate was then loaded on top of 2 mL of 30% Percoll (Sigma-Aldrich) and centrifuged at 700g for 5 min at 4°C. Filtration through Percoll gradients was repeated five times for each sample to ensure removal of all chloroplast debris. Nuclear pellets were resuspended in extraction buffer containing 100 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1% SDS, 5 mm EDTA, 5 mm DTT, 10 mm β-mercaptoethanol, and 1× EDTA-free protease inhibitor cocktail (Roche). Protein fractions were analyzed with western blots.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource or GenBank/EMBL data libraries under the following accession numbers: HEMERA/pTAC12 (At2g34640), PIF1 (At2g20180), PIF3 (At1g09530), PIF4 (At2g43010), PIF5 (At3g59060), PIF6 (At3g62090), PIF7 (At5g61270), PIF8 (At4g00050), PIL1 (At2g46970), ALCATRAZ (At5g67110), and SPATULA (At4g36930).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. YFP-HMR is localized mainly to the cytoplasm and fails to rescue the nuclear defect of the hmr mutant.
Supplemental Table S1. HMR peptides identified by LC-MS/MS analysis.
Supplemental Table S2. Oligonucleotides used for generating constructs used in this study.
Supplemental Table S3. Primers used for cDNA synthesis and qRT-PCR analysis of PEP-dependent plastidial genes.
Supplemental Table S4. Primers used for qRT-PCR analysis of nuclear-encoded genes.
Supplementary Material
Acknowledgments
We thank Dr. Akira Nagatani for phytochrome antibodies and Dr. Sabine Heinhorst for anti-SiR antibodies. We thank Dr. Erik Soderblom and the Duke University School of Medicine for the use of the Proteomics and Metabolomics Shared Resource, which provided the LC-MS/MS service. We thank Emily Yang and Keunhwa Kim for their critical comments and suggestions regarding the manuscript.
Glossary
- DTT
dithiothreitol
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- NEP
nuclear-encoded plastid RNA polymerase
- PEP
plastid-encoded plastid RNA polymerase
- R
red light
- SiR
sulfite reductase
Footnotes
This work was supported by a National Science Foundation grant (IOS-1051602 to M.C.), by National Institutes of Health grants R01GM087388 to M.C. and R01GM061893 to D.J.S., by an NIH fellowship (R01GM087388-02S1 to P.A.N.), and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through grant GBMF3405 to P.N.B.).
Articles can be viewed without a subscription.
References
- Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Kikis EA, Monte E, Quail PH (2008) Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Burgie ES, Vierstra RD (2014) Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26: 4568–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Maréchal A, Parent J-S, Lepage E, Sygusch J, Brisson N (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22: 1849–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Parent J-S, Zampini E, Lepage E, Sygusch J, Brisson N (2012) A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res 40: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J (2003) Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J (2005) Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Chi-Ham CL, Keaton MA, Cannon GC, Heinhorst S (2002) The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol Biol 49: 621–631 [DOI] [PubMed] [Google Scholar]
- Chun L, Kawakami A, Christopher DA (2001) Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol 125: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A, Wang JZ, Dehesh K (2016) Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol 68: 10.1146/annurev-arplant-042916-041007 [DOI] [PubMed] [Google Scholar]
- Demarsy E, Buhr F, Lambert E, Lerbs-Mache S (2012) Characterization of the plastid-specific germination and seedling establishment transcriptional programme. J Exp Bot 63: 925–939 [DOI] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol 142: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Allard J, Brisson N, Sygusch J (2002) A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Biol 9: 512–517 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N (2000) PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12: 1477–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau GR, Boily Y, Houmard J (1972) Purification and properties of an extracellular protease of Staphylococcus aureus. J Biol Chem 247: 6720–6726 [PubMed] [Google Scholar]
- DuBell AN, Mullet JE (1995) Differential transcription of pea chloroplast genes during light-induced leaf development (continuous far-red light activates chloroplast transcription). Plant Physiol 109: 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z-P, Yu Q-B, Zhao T-T, Ma Q, Chen G-X, Yang Z-N (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski E, Miao Y, Mulisch M, Krupinska K (2008) Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol 147: 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricová A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Bogorad L (1990) Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci USA 87: 1531–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC (2013) Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA 110: 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann T, Börner T (1998) Characterisation of transcript initiation sites in ribosome-deficient barley plastids. Plant Mol Biol 36: 493–496 [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemer R, Mulisch M, Schäfer A, Kirchner S, Koop H-U, Krupinska K (2012) Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett 586: 85–88 [DOI] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL (1997) Context sequences of translation initiation codon in plants. Plant Mol Biol 35: 993–1001 [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Páldi K, O’Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J (2015) Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev Cell 35: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Karniely S, Pines O (2005) Single translation--dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep 6: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Shin J, Lee SH, Kweon HS, Maloof JN, Choi G (2011) Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc Natl Acad Sci USA 108: 1729–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Krause K, Krupinska K (2009) Nuclear regulators with a second home in organelles. Trends Plant Sci 14: 194–199 [DOI] [PubMed] [Google Scholar]
- Krause K, Oetke S, Krupinska K (2012) Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int J Mol Sci 13: 11085–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok EY, Hanson MR (2004) Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep 23: 188–195 [DOI] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH (2008a) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008b) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (1993) The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc Natl Acad Sci USA 90: 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schnell DJ (2006) Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J Cell Biol 175: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S, Fankhauser C (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ (2014) COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, Matros A, Trösch M, Mock H-P, Krupinska K (2012) The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell 24: 3060–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, Mulisch M, Schmitz-Linneweber C, Grabowski E, Hensel G, Krupinska K (2010) Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232: 471–481 [DOI] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Nott A, Jung H-S, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G (2012) Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 72: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Holtzegel U, Barkan A, Weisheit W, Mittag M, Pfannschmidt T (2015) ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol 206: 1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18: 186–194 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G (2000) The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur J Biochem 267: 253–261 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Li M, Pasoreck EK, Long L, Shi Y, Galvão RM, Chou CL, Wang H, Sun AY, Zhang YC, et al. (2015) HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27: 1409–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, Zheng N, Hannink M, Genschik P, Liscum E (2011) Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3(NPH3). Plant Cell 23: 3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J (1997) Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol 113: 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A (1996) Nuclear localization activity of phytochrome B. Plant J 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. (1992) The chloroplast genome. Plant Mol Biol 19: 149–168 [DOI] [PubMed] [Google Scholar]
- Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P (2004) Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J 40: 164–172 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH (2006) phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Carrie C, Andersson CR, Sivasithamparam K, Whelan J, Singh KB (2007) Differential gene expression and subcellular targeting of Arabidopsis glutathione S-transferase F8 is achieved through alternative transcription start sites. J Biol Chem 282: 28915–28928 [DOI] [PubMed] [Google Scholar]
- Thum KE, Kim M, Christopher DA, Mullet JE (2001) Cryptochrome 1, cryptochrome 2, and phytochrome a co-activate the chloroplast psbD blue light-responsive promoter. Plant Cell 13: 2747–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65: 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk EK, Decker PV, Chen M (2012) Photobodies in light signaling. Plant Physiol 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk EK, Reddy AK, Nagatani A, Chen M (2014) Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol 165: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongsamphanh R, Fortier P-K, Ramotar D (2001) Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol Cell Biol 21: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, Tsudzuki T, Sugiura M (2001) The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth Res 70: 107–118 [DOI] [PubMed] [Google Scholar]
- Wang R, Brattain MG (2007) The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett 581: 3164–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salomé PA, Weigel D, Fitzpatrick JA, Chory J (2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J-Y, Lai C-X, Qu Z, Yang X-Y, Qin X-H, Liu G-Q (2009) Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression. Plant Mol Biol 71: 437–449 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HH, Kwon C, Lee MM, Chung IK (2007) Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J 49: 442–451 [DOI] [PubMed] [Google Scholar]
- Yu Q-B, Lu Y, Ma Q, Zhao T-T, Huang C, Zhao H-F, Zhang X-L, Lv R-H, Yang Z-N (2013) TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol Plant 148: 408–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.