A legume-specific duplication of the circadian clock gene ELF3 provides functional redundancy in pea and may help explain the importance of ELF3 genes in flowering time adaptation.
Abstract
Three pea (Pisum sativum) loci controlling photoperiod sensitivity, HIGH RESPONSE (HR), DIE NEUTRALIS (DNE), and STERILE NODES (SN), have recently been shown to correspond to orthologs of Arabidopsis (Arabidopsis thaliana) circadian clock genes EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO, respectively. A fourth pea locus, PHOTOPERIOD (PPD), also contributes to the photoperiod response in a similar manner to SN and DNE, and recessive ppd mutants on a spring-flowering hr mutant background show early, photoperiod-insensitive flowering. However, the molecular identity of PPD has so far remained elusive. Here, we show that the PPD locus also has a role in maintenance of diurnal and circadian gene expression rhythms and identify PPD as an ELF3 co-ortholog, termed ELF3b. Genetic interactions between pea ELF3 genes suggest that loss of PPD function does not affect flowering time in the presence of functional HR, whereas PPD can compensate only partially for the lack of HR. These results provide an illustration of how gene duplication and divergence can generate potential for the emergence of more subtle variations in phenotype that may be adaptively significant.
Plant responses to photoperiod are known to depend on interaction between light perception and the circadian clock. This interaction has been extensively explored in Arabidopsis (Arabidopsis thaliana), where the circadian clock consists of a network of genes that form several interlocking feedback loops and influences flowering time through control of several direct and indirect regulators of the florigen gene FLOWERING LOCUS T (FT; Andrés and Coupland, 2012; Song et al., 2012; Millar, 2016). While the overall architecture of the clock is complex and the details are still a matter of debate (Nagel and Kay, 2012; McClung, 2014), one group of genes has emerged as being particularly significant for clock entrainment and photoperiodism. The myb transcription factor gene LUX ARRHYTHMO (LUX; also known as PHYTOCLOCK1) and two other plant-specific genes EARLY FLOWERING3 (ELF3) and ELF4 have similar mutant phenotypes, exemplified by early, photoperiod-insensitive flowering, elongated hypocotyls, and loss of circadian rhythmicity under constant conditions (Hicks et al., 1996; Doyle et al., 2002; Hazen et al., 2005; Onai and Ishiura, 2005; Anwer et al., 2014). The proteins encoded by these genes form a complex termed the evening complex (EC), in which the ELF3 protein is suggested to act as a molecular scaffold and signaling hub, connecting ELF4 with LUX (Nusinow et al., 2011; Huang et al., 2016). ELF4 appears to have a role in determining the subcellular location of the complex (Herrero et al., 2012) and, thus, possibly in directing the activity of the LUX transcription factor. Loss of any one of these proteins compromises the EC function (Nusinow et al., 2011), an interaction that is likely to explain the generally similar phenotypes of elf3, elf4, and lux mutants.
The EC is integral to circadian clock function and gating of light input to the clock (Huang and Nusinow, 2016). EC proteins are involved in regulation of a number of clock-associated genes, including TIMING OF CAB EXPRESSION1 (TOC1), GIGANTEA (GI), LUX, PSEUDO RESPONSE REGULATOR7 (PRR7), and PRR9, and act to repress expression of these genes during the night (Huang and Nusinow, 2016). In some cases, this appears to reflect direct binding of LUX to LUX binding site elements in the promoters of these genes (Helfer et al., 2011; Nusinow et al., 2011; Chow et al., 2012; Herrero et al., 2012; Mizuno et al., 2014). In turn, the expression of EC genes and formation of the EC itself are also tightly regulated by the circadian clock through multiple regulatory mechanisms (Herrero et al., 2012; Choudhary et al., 2015). The ELF3, ELF4, and LUX promoters are bound by the key clock protein CCA1, a morning phased myb transcription factor that acts together with its paralog LATE ELONGATED HYPOCOTYL (LHY) to repress evening-phased genes during the day (Lu et al., 2012; Adams et al., 2015; Nagel et al., 2015; Kamioka et al., 2016). Autoregulation of the EC may also be achieved through binding of the LUX protein to its own promoter (Helfer et al., 2011). In addition to its regulation of and by other clock components, the EC is also regulated at multiple levels by light and temperature pathways, and this may be one important mechanism through which the clock can be entrained by environmental variation (Huang and Nusinow, 2016).
The EC proteins not only participate directly in clock function but also have a major role in several important clock-regulated outputs, although the molecular links between the EC genes and output pathways connecting EC activity to specific developmental processes have only been investigated in detail in a few cases. The most prominent example is the regulation of hypocotyl elongation in which the EC represses the expression of the growth-promoting PHYTOCHROME-INTERACTING FACTOR (PIF) transcription factors PIF4 and PIF5 during the night (Nusinow et al., 2011; Seaton et al., 2015). Although it is clear that the EC also plays a major role in control of photoperiodic flowering, the mechanism for this has not been fully elucidated. ELF3 has been shown to facilitate the control of GI protein stability by the CONSTITUTIVE PHOTOMORPHOGENIC1 ubiquitin ligase (Yu et al., 2008), and ELF4 controls GI localization within the nucleus and may prevent its access to target promoters (Kim et al., 2013). As GI is well known as an activator of FT (Suárez-López et al., 2001; Jung et al., 2007; Sawa and Kay, 2011), both of these could constitute mechanisms by which the EC could regulate FT and flowering time. In addition, although it is clear that participation in the EC is a key role for ELF3, ELF4, and LUX proteins, it also seems likely that the individual functions of ELF3 and ELF4 are not limited to this, as they have been reported to interact independently with other circadian clock, photoperiod pathway, and light signaling components (Liu et al., 2001; Kim et al., 2013; Kaiserli et al., 2015; Nieto et al., 2015).
The importance of EC genes for circadian rhythms and photoperiod responsiveness has been confirmed more recently in other plant groups, including legumes and cereals. Interestingly, in both crop groups, mutations in ELF3 genes have been shown to contribute to the expansion in range of species through alteration of photoperiod responsiveness and the associated impact on yield (Faure et al., 2012; Matsubara et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012; Alvarez et al., 2016; Lu et al., 2017). In crop species, a role for ELF4 function has been clearly established only in the legume species pea (Pisum sativum; Liew et al., 2009), but mutations in LUX orthologs have also been shown to cause early flowering and impaired photoperiod response in pea and in the cereals einkorn wheat (Triticum aestivum) and barley (Hordeum vulgare; Mizuno et al., 2012; Campoli et al., 2013; Liew et al., 2014), suggesting that the EC genes are likely to be intimately linked to the photoperiod response mechanism in both crop groups.
In the long-day legume species pea, the ELF3 ortholog HR has a central role in photoperiod adaptation, with a null mutation conferring partial loss of photoperiod responsiveness and earlier flowering under short photoperiods, and this appears to have been central to adaptation of the crop to spring sowing and expansion to higher latitudes (Weller et al., 2012). Naturally occurring and induced mutations in the LUX ortholog SN completely eliminate responsiveness to photoperiod (Liew et al., 2014), whereas a mutant for the ELF4 ortholog DNE enhances the effect of the hr mutation but has little effect when functional HR is present (Liew et al., 2009, 2014). In addition to these three EC homologs, mutations at two other loci confer early, photoperiod-insensitive flowering in pea: a dominant mutation in the PHYTOCHROME A (PHYA) gene (Weller et al., 2004) and recessive mutations at the PHOTOPERIOD (PPD) locus (Arumingtyas and Murfet, 1994; Taylor and Murfet, 1996; Murfet and Taylor, 1999), whose molecular identity is not yet known. In this study, we report that the ELF3 gene has undergone duplication in legumes and show that the second ELF3 gene, ELF3b, corresponds to the PPD locus. We also show that PPD is largely redundant with HR in control of flowering and confirm that unlike in Arabidopsis, none of the pea EC genes have a significant role in stem elongation.
RESULTS
PPD Contributes to Photoperiodic Flowering
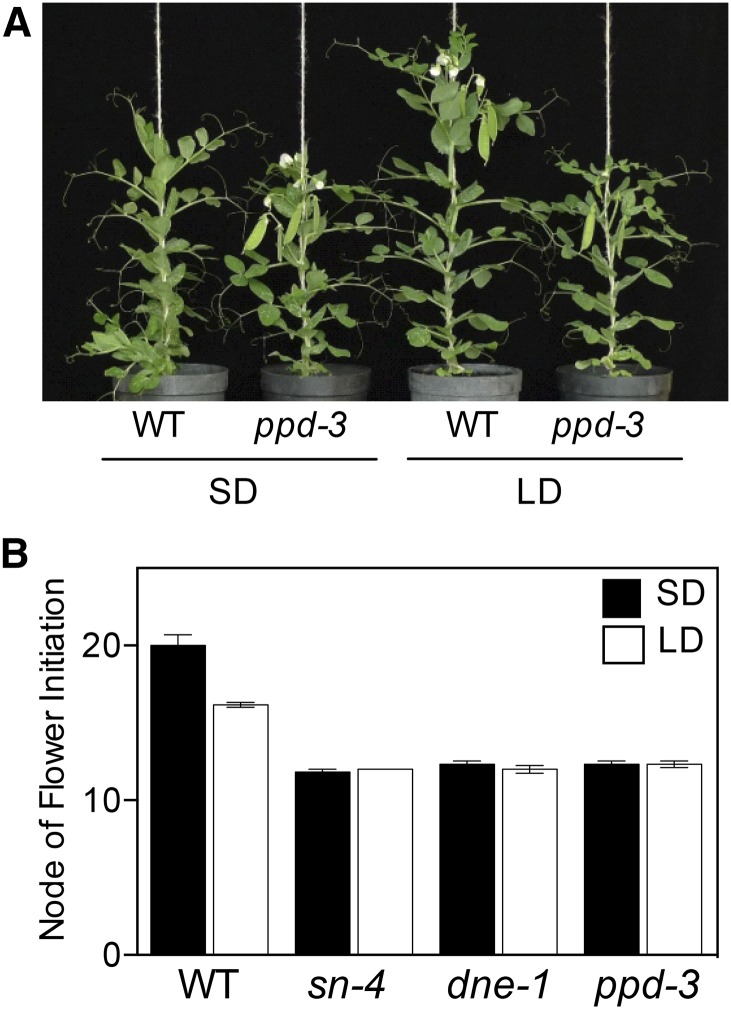
Two previously described recessive alleles, ppd-1 and ppd-2, were induced by gamma irradiation in pea cv Borek (Arumingtyas and Murfet, 1994; Taylor and Murfet, 1996). We subsequently identified a third recessive mutant allele, ppd-3, from EMS mutagenesis of the pea line NGB5839 (Hecht et al., 2007), which has been widely used as a reference wild-type line. Both of these parental lines carry the same hr mutation (Weller et al., 2012). Figure 1 and Supplemental Figure S1 confirm that in contrast to the photoperiod-responsive wild type (P < 0.001), the ppd-3 allele confers early flowering (P < 0.001) regardless of photoperiod, equivalent to ppd-1 and ppd-2 (Taylor and Murfet, 1996) and to sn and dne mutants (Murfet, 1971; King and Murfet, 1985; Liew et al., 2009, 2014). As all three ppd mutations appeared similar in their phenotypic effects, we used the ppd-3 mutant to explore the roles of PPD, as it was generated in the same genetic background as other relevant mutants, including sn and dne.
Figure 1.
Phenotypic comparison of photoperiod-insensitive early flowering pea mutants. A, Representative 7-week-old wild-type (WT) line NGB5839 and isogenic ppd-3 mutant plants. B, Comparison of the response to photoperiod in ppd-3 and similar mutants sn-4 and dne-1 in the NGB5839 genetic background. Data represent mean ± se for n = 5 to 6 plants. All plants were grown in the phytotron under SD (8L:16D) or LD (16L:8D) conditions using extended natural daylight. The progenitor line NGB5839 carries the hr mutation.
PPD Affects the Expression of FT Genes
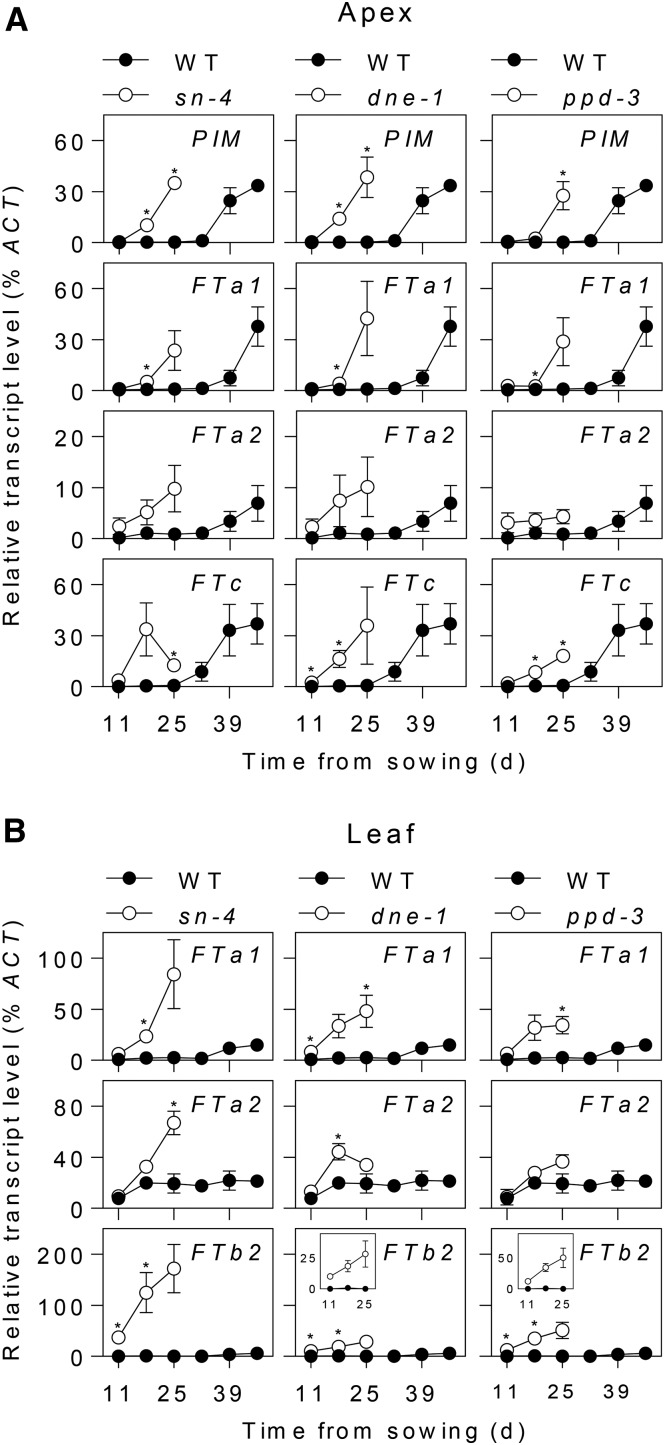
Previous studies have shown that the early flowering phenotypes of sn and dne mutants under short-day (SD) conditions are associated with elevated expression levels of several FT homologs, similar to the inductive effect of long days (LDs) observed for the wild type (Hecht et al., 2011; Liew et al., 2014). We used qRT-PCR to compare the expression of pea FT genes in ppd-3 and these other mutants over the course of development under SDs. Figure 2A shows that under SD conditions, the inflorescence identity marker PROLIFERATING INFLORESCENCE MERISTEM (PIM) is expressed 2 to 3 weeks earlier in apical tissue of sn-4, dne-1, and ppd-3 mutants than in the wild type, as expected in view of their early flowering phenotypes. The FTb2 gene appears to be the main target for photoperiod regulation of flowering in pea as it is strongly induced in leaf tissue during commitment to flowering under LDs but is not significantly expressed in SDs (Hecht et al., 2011). Figure 2B shows that in contrast to the wild type, all three mutants showed significant FTb2 expression in expanded leaf tissue under SDs, whereas FTb2 transcript was not detectable in equivalent wild-type tissue. Importantly, FTb2 transcript levels were detectable above background in all three mutants from the first time point at day 11, prior to the first detectable induction of PIM (at day 18 in sn-4 and dne-1 or day 25 in ppd-3).
Figure 2.
FT genes are misregulated in photoperiod-insensitive early flowering pea mutants. Developmental regulation of FT genes and the floral marker PIM in NGB5839 (WT) and the ppd-3 mutant are shown in comparison to sn-4 and dne-1 mutants. Plants were grown under an 8-h photoperiod in growth cabinets and samples harvested weekly until the appearance of visible flower buds. Transcript levels were determined in dissected shoot apex or uppermost fully expanded leaflet tissue and are shown relative to the ACTIN reference gene. Each sample consisted of pooled material from two plants, and each data point represents the mean ± se for n = 2 to 3. Asterisks indicate differences between wild-type and mutant values where P < 0.05.
A second pea FT gene, FTa1, has an important role in promotion of flowering, is expressed in expanded leaf tissue, and may contribute to a second mobile signal (Hecht et al., 2011). However, unlike FTb2, FTa1 does not appear to be directly involved in the response to photoperiod, as it shows basal expression under SDs, is relatively weakly induced by LD after floral commitment, and fta1 mutants, although late-flowering, are still capable of responding to photoperiod (Hecht et al., 2011). Nevertheless, expression of FTa1 was also elevated in leaf tissue of sn-4, dne-1, and ppd-3 mutants (Fig. 2B), and its induction in shoot apical tissue occurred 2 to 3 weeks earlier than the wild type, in parallel with PIM (Fig. 2A). As previously described, a third FT gene, FTc, is only expressed in shoot apex tissue and in wild-type plants is also induced in parallel with PIM under both SDs and LDs (Hecht et al., 2011; Fig. 2A). FTc induction also occurred 2 to 3 weeks earlier in all three mutants relative to the wild type. Although minor differences in the expression profiles and timing of PIM and FT induction were observed, these results overall show that early flowering in all three mutants is associated with a similar pattern of derepression of several FT genes.
PPD Affects the Maintenance of Circadian Rhythms
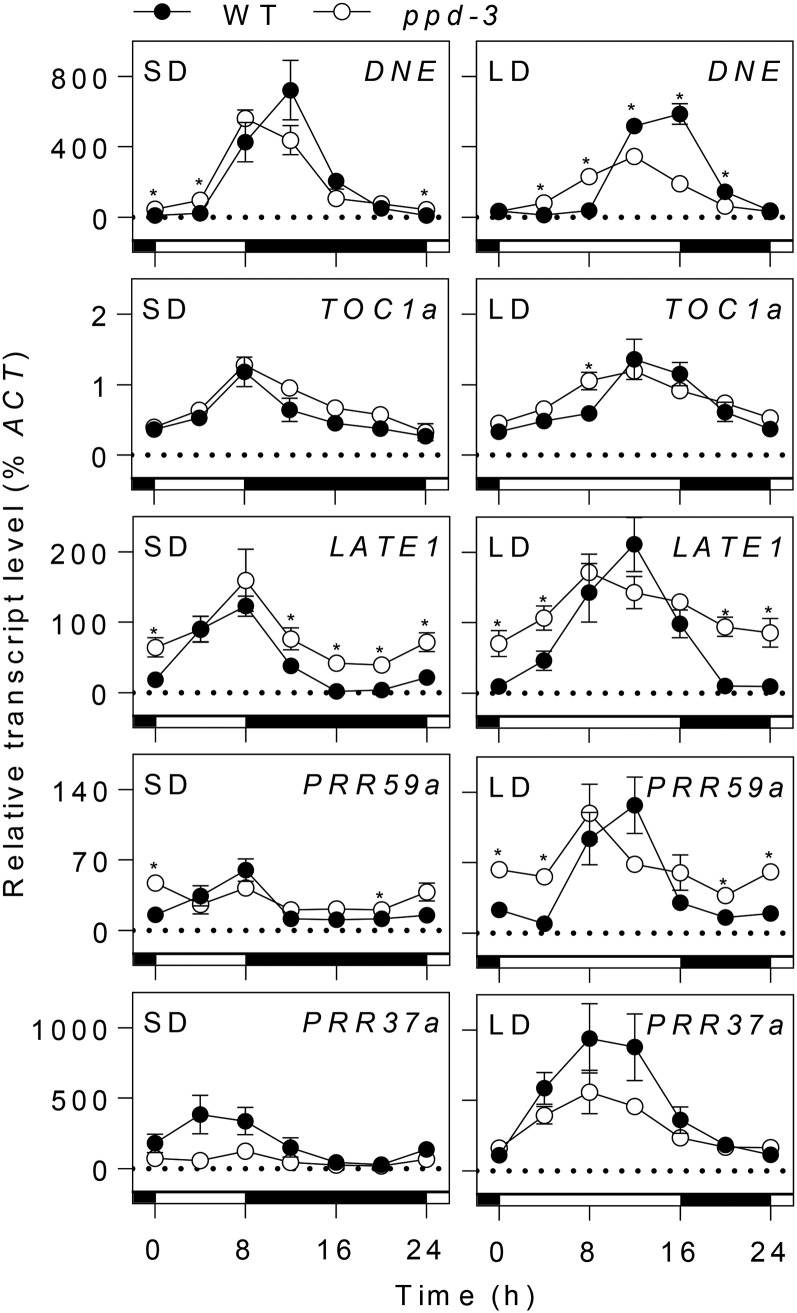
The similarity of the ppd mutant phenotype to those of the dne and sn mutants suggested that PPD, like SN and DNE, might also affect the circadian clock. To test this possibility, we compared diurnal and circadian expression patterns of key clock-related genes in the wild type and ppd-3. Figure 3 shows that the ppd-3 mutation influences the diurnal expression rhythms of several clock-associated genes under both conditions. Interestingly, the effects of ppd-3 were more pronounced under LD conditions where DNE, TOC1a, LATE1, and PRR59a expression rhythms showed a small but significant phase advance relative to the wild type (Supplemental Fig. S2). Trough expression levels for the evening genes LATE1 and PRR59a were also significantly higher in ppd-3 than in the wild type, a difference also apparent under SDs. In contrast, there was no clear evidence of any effect of ppd-3 on expression of PRR37a, consistent with earlier reports on sn and dne mutants (Liew et al., 2009, 2014).
Figure 3.
PPD affects diurnal expression rhythms of clock-related genes. Transcript levels were determined in the uppermost fully expanded leaf of 3-week-old NGB5839 (WT) and ppd-3 plants grown under SD (8L:16D) or LD (16L:8D) conditions at 20°C. Values are normalized to the transcript level of the ACTIN reference gene. Each sample consisted of pooled material from two plants, and each data point represents the mean ± se for n = 2 to 6. Light and dark periods are represented by white and black bars, respectively. Asterisks indicate differences between wild-type and mutant values where P < 0.05.
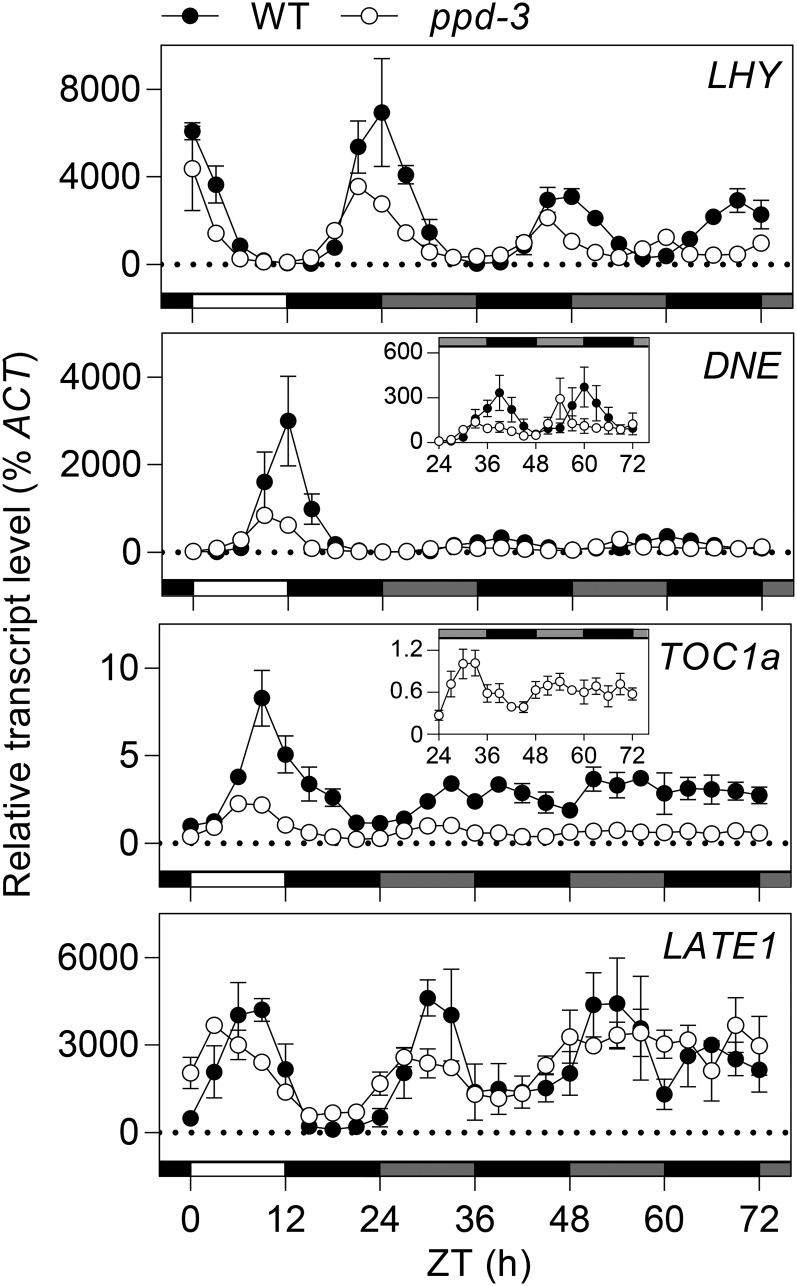
We also examined whether the ppd-3 mutation also affected expression rhythms after transfer of plants from entraining photoperiod cycles to constant darkness. Figure 4 shows that after transfer to constant darkness, rhythms of LHY, ELF4, and LATE1 were maintained for two cycles in both the wild type and ppd-3, but peaks occurred increasingly earlier in ppd-3, suggesting the mutant may affect the rhythmic period, and this was again confirmed through statistical analysis (Supplemental Fig. S2). Although clear rhythms of TOC1a expression in constant darkness were not apparent, the overall level of expression in ppd-3 remained lower than the wild type. Collectively these results are similar to those previously reported for the sn-4 (Liew et al., 2014) and dne-1 mutations (Liew et al., 2009), indicating a small but clear effect of the ppd-3 mutation on rhythmic expression of several key circadian clock components, which suggests that the primary role of PPD may also be related to clock function.
Figure 4.
PPD affects expression rhythms of clock-related genes in constant darkness. Transcript levels were determined in the uppermost fully expanded leaf of 3-week-old NGB5839 (WT) and ppd-3 plants entrained in SDs (12L:12D) at 20°C for 21 d before transfer to continuous darkness at Zeitgeber time (ZT) 24. Values are normalized to the transcript level of the ACTIN reference gene. Each sample consisted of pooled material from two plants, and each data point represents the mean ± se for n = 2 to 3. Light and dark are represented by white and black bars, respectively. The gray bars indicate the periods of subjective day during the period of continuous darkness. Zeitgeber time refers to the time since lights-on of the last full entraining cycle. Statistical analyses are presented in Supplemental Figure S2.
PPD Is the Second of Two Duplicate ELF3 Genes in Pea
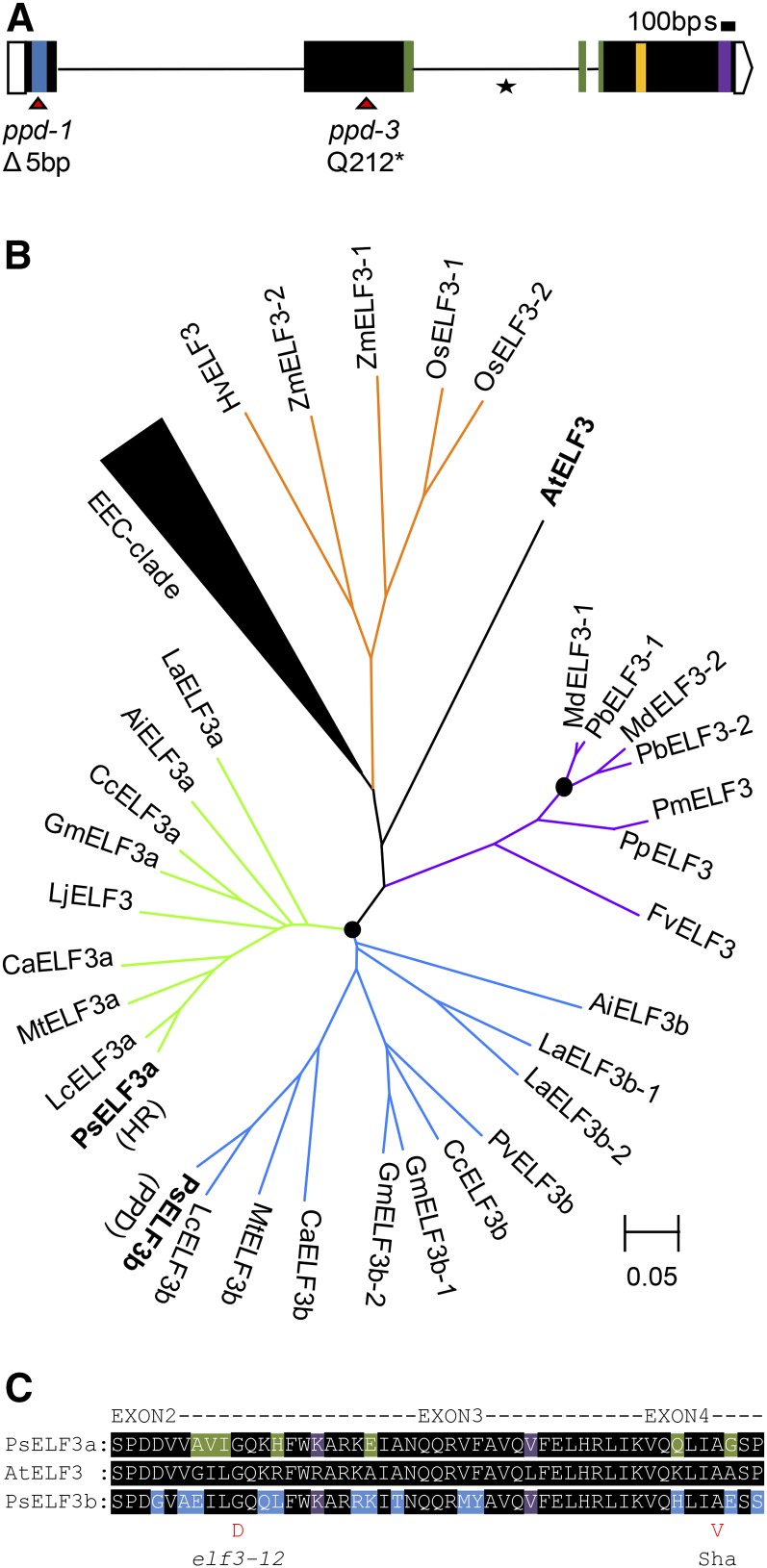
Initial studies reported that PPD was located toward the top of linkage group II near loci Aatp and Rms3 (Murfet and Taylor, 1999). We refined this position in the F2 of a cross between the ppd-3 mutant and cv Térèse, delimiting PPD to a region between markers PepTrans and ThiolP (Bordat et al., 2011). In the closely syntenic region of Medicago truncatula chromosome 1, these markers correspond to gene models Medtr1g009200 and Medtr1g018840 and define an interval of around 4.3 Mb containing ∼900 genes. Within this region we identified several genes potentially related to flowering time control, including two miR156 genes, the CONSTANS-like gene COLb (Wong et al., 2014), and a gene with strong similarity to the previously described HR/ELF3 gene (Weller et al., 2012), which we termed ELF3b. In view of the circadian clock-related role of PPD, we considered only the last two sequences as plausible candidates. We found that all three ppd alleles carried significant mutations in the ELF3b gene relative to their parental lines, whereas initial sequencing of COLb failed to identify any coding region polymorphisms and it was therefore dismissed as a candidate gene.
As shown in Figure 5A, the ppd-1 mutant carried a 5-bp deletion in exon 1 of ELF3b that results in a frame shift at codon 26 and a truncation of the protein after seven additional missense amino acids, while the ppd-3 mutant carried a nonsense mutation in exon 2 (C2383T), resulting in replacement of residue Q212 by a stop codon. Attempts to amplify ELF3b genomic and cDNA sequence from the ppd-2 mutant indicated the presence of a substantial rearrangement/insertion affecting the coding sequence, which we did not investigate further. The presence of deleterious mutations in each of three independent mutant alleles provides strong evidence that PPD is in fact equivalent to ELF3b.
Figure 5.
Multiple independent ppd mutants carry mutations in an ELF3-like gene. A, Diagram of the pea ELF3b gene showing details of mutations in the ppd-1 and ppd-3 mutants. Colored regions of the coding sequence represent conserved blocks as defined by Liu et al. (2001) and shown in Supplemental Figure S4. The asterisk indicates the site of a single polymorphism (a simple sequence repeat) between the two parental cultivars Borek and NGB5839. B, Phylogram of ELF3-like protein sequences from legumes and other selected species. The two clades of legume proteins are shown in green (ELF3a) and blue (ELF3b). ELF3 proteins from species in the family Rosaceae are shown in purple and those from grasses in orange. The black triangle represents a clade containing EEC-like sequences from the legume and Rosaceae species examined. Likely independent ELF3 duplications are indicated by black circles. All major clades have at least 75% bootstrap support (from 10,000 replications). Species abbreviations are as follows: Ai, Arachis ipaensis; Ca, Cicer arietinum; Cc, Cajanus cajan; Gm, Glycine max; La, Lupinus angustifolius; Lc, Lens culinaris; Lj, Lotus japonicus; Mt, Medicago truncatula; Ps, Pisum sativum; Pv, Phaseolus vulgaris; Fv, Fragaria vesca; Md, Malus domestica; Pb, Pyrus bretschneideri; Pm, Prunus mume; Pp, Prunus persica; At, Arabidopsis thaliana; Hv, Hordeum vulgare; Os, Oryza sativa; Zm, Zea mays. C, Sequence differences between PsELF3a (HR), AtELF3, and PsELF3b (PPD) in the conserved block II region. Location and nature of substitutions known to affect AtELF3 function are indicated. Sha, Location of substitution in ecotype Shakdara.
The existence of a second ELF3 homolog prompted us to examine the nature of ELF3-like genes in other legumes and related species. In Arabidopsis, ELF3 is a single-copy gene, and the next most closely related gene has been termed ESSENCE OF ELF3 CONSENSUS (EEC; Liu et al., 2001). Sequence searches in legumes and related Rosid taxa clearly identified homologs of ELF3 and of EEC. Figure 5B shows that in addition to an EEC homolog, most legumes contained two ELF3 homologs that represented two distinct clades: an ELF3a clade containing the previously described pea and lentil ELF3 genes (Weller et al., 2012) and a second clade containing the PPD/ELF3b gene. The two pea ELF3 proteins show ∼50% similarity to each other and 30% similarity to pea EEC. This duplication is present in both the major crop legume groups and in the more basal genistoid species narrow-leafed lupin (Lupinus angustifolius) and the dahlbergoid species wild peanut (Arachis ipaensis), but not in members of the Rosaceae, suggesting it is a relatively ancient, legume-specific feature. Evidence for ELF3 duplication was also present in the Rosaceae, but phylogenetic analysis clearly shows that this was independent from the legume event and much more recent (Fig. 5B). Within the legumes, certain species deviated from the expected standard complement of two genes, with Lotus japonicus and common bean (Phaseolus vulgaris) databases only providing evidence of one. However, the clear identity of these genes as ELF3a and ELF3b genes suggests that the second gene has either been lost from these species or is simply not represented in current genome builds. Soybean (Glycine max) also appears to have lost one of two ELF3a homoeologs, while narrow-leafed lupin has two copies of ELF3b (Fig. 5B).
ELF3 genes are defined by four short, highly conserved domains that have been designated blocks I to IV (Liu et al., 2001). The functional relevance of these domains remain largely unknown, but recent evidence suggests that block II is required for interaction with the ELF4 protein (Herrero et al., 2012; Saini et al., 2013), and mutations within block II have been shown to alter ELF3 function and cellular distribution (Anwer et al., 2014; Kolmos et al., 2011). We also assembled a more extensive collection of legume ELF3 sequences and examined the resulting alignment (Supplemental Fig. S4) for evidence of significant divergence in the region of block II. We found no differences perfectly distinguishing the ELF3a and ELF3b clades but found that ELF3b proteins from the temperate legume clade show a number of nonconservative amino acid substitutions relative to other legume ELF3 proteins and Arabidopsis ELF3 (Fig. 5C; Supplemental Fig. S4). Interestingly, these are clustered near highly conserved residues that are known to influence ELF3 function in Arabidopsis. Sequencing of the entire ELF3b coding sequence from selected diverse accessions of cultivated P. sativum var sativum and wild Pisum (Supplemental Table S1) showed no polymorphism in block II, suggesting that this region of ELF3b, although distinct from ELF3a, is highly conserved in pea germplasm.
PPD Only Affects Flowering in the Absence of Functional HR
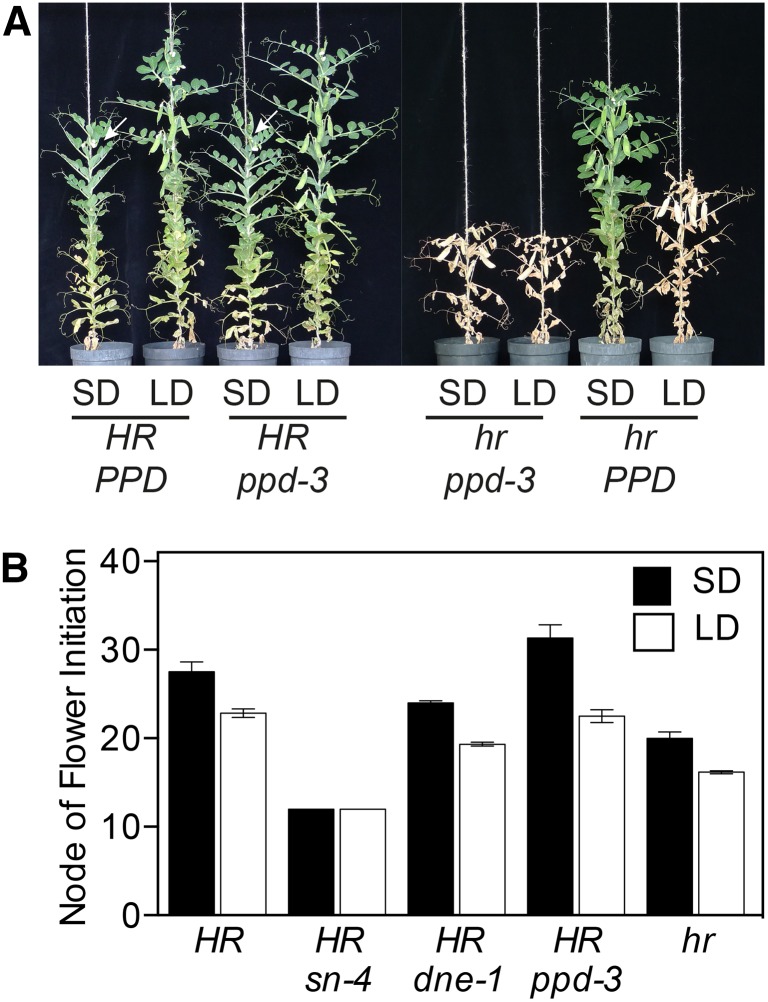
All three ppd mutant alleles were isolated in genetic backgrounds containing the hr mutation and therefore lack functional copies of both ELF3 paralogs. This indicates that loss of all ELF3-related activity also completely eliminates the ability of the plant to respond to photoperiod and shows that the partial photoperiod response of the hr mutant can be attributed to presence of the PPD gene. To examine the effect of the ppd mutation in isolation, we selected the HR ppd-3 genotype and compared it with an HR PPD isoline and also with near-isolines carrying single sn, dne, and hr mutations. Figure 6 and Supplemental Figure S5 show that in contrast to the clear promotion of flowering under SDs conferred by the ppd-3 mutation on an hr genetic background (Figs. 1 and 6A), the ppd-3 mutation alone has no effect in the presence of HR in either SDs or LDs (P > 0.05). This suggests that ELF3b plays a minor role that is subsidiary to ELF3a/HR, a difference that could conceivably be due to differences in protein structure and/or regulation. The ppd-3 mutation therefore shows a genetic interaction with HR similar to that of the dne-1 mutation, which also has minimal effect in the presence of the functional HR gene. These interactions contrast that of the sn-4 mutation, which is essentially epistatic to HR for flowering time (Liew et al., 2014; Fig. 6; Supplemental Fig. S4).
Figure 6.
Genetic interaction of PPD and HR. A, Representative 11-week-old PPD HR plants, single ppd and hr mutants, and double mutant ppd hr plants. Arrows represent node of flower initiation where pods are not yet clearly visible. B, Comparison of photoperiod response for initiation of flowering in single sn, dne, ppd, and hr mutants. Data represent mean ± se for n = 5 to 6 plants. All plants were grown in the phytotron under SD (8L:16D) or LD (16L:8D) conditions.
PPD and HR Have Similar Expression Patterns
In order to examine whether differences in ppd and hr mutant phenotypes might reflect differences in regulation of PPD and HR genes, we examined how transcript levels of both genes varied under different circumstances and conditions. Our previous results showed that ELF3a transcript is expressed at a relatively low level in leaf tissue and does not show a discernible diurnal rhythm (Liew et al., 2014). Supplemental Figure S6 shows that like ELF3a, ELF3b also appears to lack a clear diurnal rhythm in expression. Comparison of different tissue types and two different developmental time series indicated a higher expression level for ELF3b than ELF3a in leaf, apex, and stem tissues in certain experiments. but overall there was no major, consistent difference in expression level or pattern between the two ELF3 paralogs (Supplemental Fig. S6). ELF3a and ELF3b expression profiles from the pea gene expression atlas (Alves-Carvalho et al., 2015) also suggested no substantial differences. These observations indicate that the more dominant role of the HR gene relative to PPD does not simply reflect a higher expression level.
PPD Does Not Substantially Affect Stem Elongation
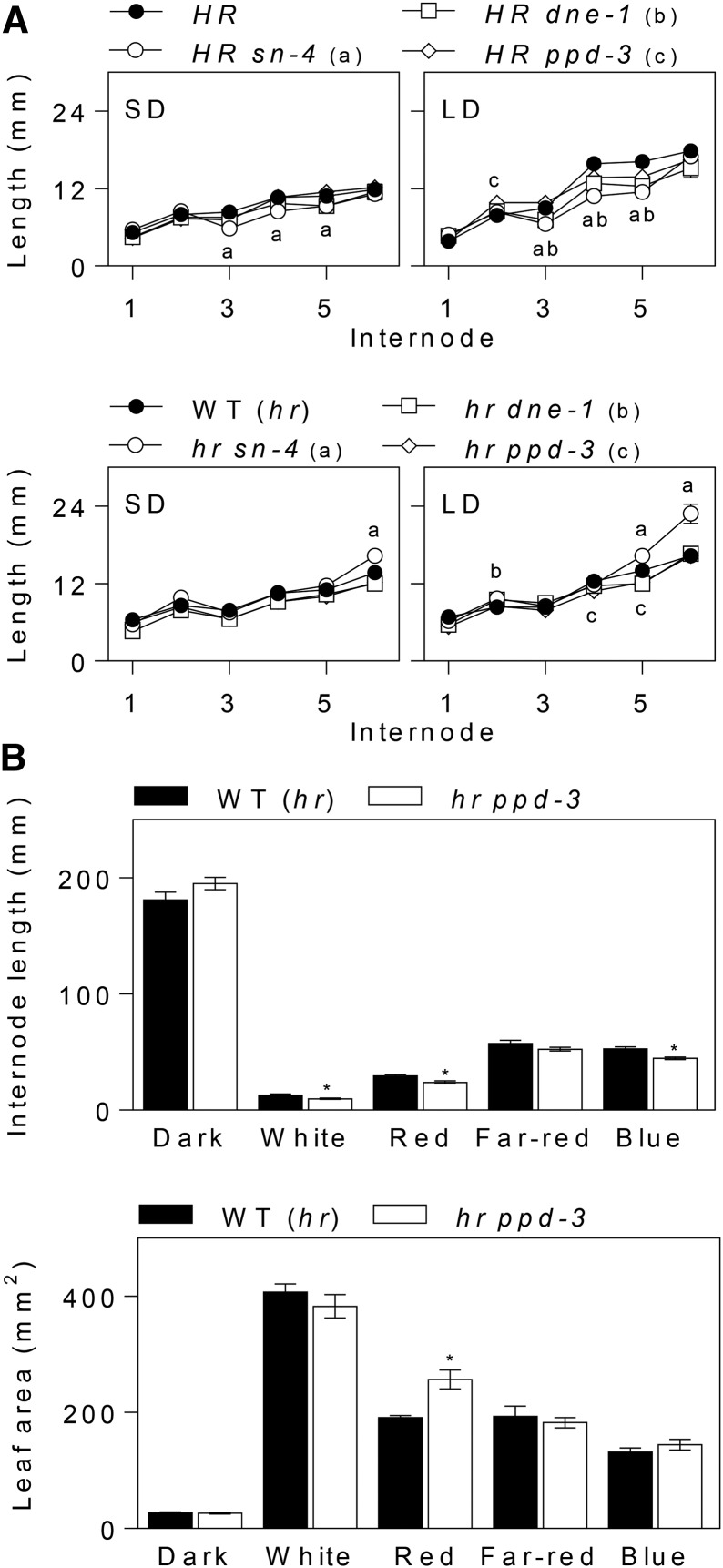
In Arabidopsis, the evening complex genes ELF3, ELF4, and LUX all affect seedling vegetative development in addition to photoperiodic flowering, with each of the single mutants showing a significant increase in hypocotyl elongation compared with the wild type under white light (Zagotta et al., 1996; Doyle et al., 2002; Hazen et al., 2005). Similar seedling phenotypes are also shown by elf3 mutants in barley, which have elongated coleoptiles and leaves and decreased chlorophyll content relative to ELF3 plants (Boden et al., 2014). We therefore examined whether the pea EC mutants displayed similar phenotypes. When grown under long- or short-day conditions, none of the four single mutants showed obvious elongation phenotypes at the seedling stage. Figure 7A shows that in plants grown under white light, length of the first two internodes was not affected in the sn and ppd mutants in a functional HR genetic background and only showed a small (12%) increase in ppd-3 under LDs. Several later internodes were actually slightly but significantly shorter relative to the wild type, in sn-4 under SDs and both sn-4 and dne-1 under LDs (Fig. 7A). We also examined their effect in the original (hr) genetic background, where all three mutants have clear effects on flowering time (Fig. 1), but again, no clear differences were observed (Fig. 7A) except for an increase in sn-4 over internodes 5 and 6 and dne-1 at internode 2 only in LDs. It is probable that this increase reflects the slightly earlier induction of FT genes of in sn-4 than other genotypes, as an increase in internode length after initiation of flowering is well documented in pea (Weller et al., 1997). We also examined the effect of the ppd-3 mutation on deetiolation phenotypes under monochromatic light to exclude the possibility that an effect on elongation under white light might be masked by its spectral complexity or the relatively high irradiance used. However, the results in Figure 7B show that under white light or lower irradiance monochromatic red, far-red, or blue light, certain early internodes of ppd-3 mutant plants were slightly shorter than the wild type, consistent with previous reports for sn-4 and dne-1 (Hecht et al., 2007; Liew et al., 2009). Regardless of these small differences, the overall similarity of the pea mutants shows that there is no substantial effect equivalent to the reduction in hypocotyl growth inhibition observed in Arabidopsis EC mutants.
Figure 7.
PPD and other EC genes do not affect stem elongation or photomorphogenesis. A, Comparison of genotypes differing at the SN, DNE, PPD, and HR loci for elongation of early stem internodes in plants grown under white light in SD (8L:16D) or LD (16L:8D). Data represent mean ± se for n = 5 to 6 plants. Values significantly different from the wild type (P < 0.05) are indicated by letters representing different genotypes. B, Effect of the ppd mutation on deetiolation phenotypes. NGB5839 (WT) and ppd-3 seedlings were grown for 14 d from sowing under continuous light or darkness. Internode length was measured as the length between nodes 1 and 3. Leaf area was estimated as the product of the length and width of a single leaflet from a leaf at node 3. Data represent mean ± se for n = 5 to 6 plants. Asterisks indicate differences between wild-type and mutant values where P < 0.05. All plants were grown in growth cabinets, in darkness, or under continuous white (100 µmol m−2 s−1), red, far-red, or blue light (15 µmol m−2 s−1).
DISCUSSION
It is becoming increasingly clear that the EC genes ELF3, ELF4, and LUX not only play an important role in circadian clock function but also provide an important component of adaptation in a number of crop species (Huang and Nusinow, 2016). In pea, an important model for temperate legume crops, a mutation in the pea ELF3 ortholog HR confers early flowering and a reduction in photoperiod response that is likely to have provided a key prehistoric adaptation to shorter growing seasons (Weller et al., 2012). A similar physiological function has been demonstrated for two other EC gene orthologs in pea, SN/LUX and DNE/ELF4, through analysis of naturally occurring and induced mutants (Liew et al., 2009, 2014). The sn and dne mutations effectively eliminate the residual response to photoperiod in an hr genetic background, and the same is true for mutations at a third locus, PPD (Arumingtyas and Murfet, 1994; Taylor and Murfet, 1996; Fig. 1). Our investigations show that, like sn and dne, ppd mutants do not affect photomorphogenesis but exhibit mild defects in rhythmic gene expression, suggesting a role specifically related to circadian clock function (Figs. 3, 4, and 7). Genetic mapping of PPD together with the analysis of multiple independent ppd mutant alleles (Fig. 5) has subsequently provided strong evidence that PPD is equivalent to ELF3b, a paralog of the HR gene ELF3a.
This observation gives a new perspective on the effect of the hr mutation. It has long been clear that the single known hr mutant allele reduces but does not eliminate the photoperiod response (Murfet, 1973), and the identification of hr as an apparent null mutation has implied a degree of redundancy must exist for ELF3 function (Weller et al., 2012; Liew et al., 2014). At the genomic level, this has been borne out by the identification of two groups of ELF3 genes in pea and other legumes, which are both distinct from a more distantly related clade containing orthologs of the Arabidopsis gene EEC (Fig. 5). Evidence for the ELF3 duplication is present in all of the crop legume genomes queried, indicating that it is relatively old, but it is not found in species in the closely related Rosaceae family, suggesting that it is most likely unique to the legume lineage. More specifically, the presence of the duplication in three distinct early branching Papilionoid legume groups (genistoid, dahlbergoid, and hologaleginoid clades) is consistent with an origin in the whole-genome duplication that occurred around 58 million years ago close to the base of this major legume clade (Pfeil et al., 2005; Lavin et al., 2005; Cannon et al., 2010; Young et al., 2011). A preliminary scan of other nonpapilionoid legume genomic resources suggests that this duplication is not present in the caesalpinoid legume Cercis (redbud) or the mimosoid Acacia, but a more comprehensive analysis will be needed to pinpoint its phylogenetic position more precisely.
Regardless of exactly how these two ELF3 genes first arose, the genetic interaction between the HR and PPD genes confirms their functional redundancy showing that the residual response in the hr mutant is eliminated by the ppd mutation (Fig. 6). In contrast, the effect of the ppd mutation is minimal when functional HR is present, implying that HR has the more substantial role (Fig. 6). Comparison of expression patterns revealed no major differences in tissue-specific, developmental, or diurnal expression between HR and PPD (Supplemental Fig. S6), suggesting that their difference in function could reflect an inherent difference in the activity of the two proteins. Alternatively, it is also possible that these broad expression patterns in whole leaves may not accurately reflect localized differences in expression in specific leaf tissues critical for their influence on flowering, as recently observed for a number of evening genes in the Arabidopsis clock (Endo et al., 2014).
Previous characterizations of pea EC mutants also identified differences in severity of apparent null sn and dne mutant phenotypes and raised the possibility that this might also reflect differences in redundancy (Liew et al., 2014). Interestingly, in both of these cases, the single mutant phenotypes are consistent with the structure of the gene family in question. For example, in Arabidopsis, LUX has a paralog NOX that can contribute to recruit the EC proteins to target promoters when LUX activity is absent (Dai et al., 2011; Helfer et al., 2011; Nusinow et al., 2011). In legumes, however, NOX genes are not present, and a null mutant for the single LUX ortholog in pea, SN, has a very strong early flowering phenotype and is completely insensitive to photoperiod (Liew et al., 2014), similar to the double ELF3 mutant hr ppd. Arabidopsis ELF4 is a member of a small gene family (Khanna et al., 2003) in which the closest paralog, ELF4-LIKE1 (EFL1), appears also to have ELF4 activity but three other ELF4-like genes do not (Kolmos et al., 2009). In legumes, ELF4 is also represented by small gene family (Liew et al., 2009), and the fact that in pea an apparent null mutant for the ELF4 ortholog DNE has only a minor effect on flowering when the other EC genes are intact (Liew et al., 2014) again implies a probable redundancy with one or more other members of the family.
While the question of redundancy may not have much importance for how the clock functions, it may be relevant to an understanding of the importance of EC genes in a natural setting. It is notable that ELF3a mutations are known to have provided adaptation in three different crop legumes (Weller et al., 2012; Lu et al., 2017) as well as in the ornamental species Lathyrus odoratus (V. Rajandran and J.L. Weller, unpublished data), and conserved quantitative trait loci positions suggest this could be true for several other legume species (Weller and Ortega, 2015). In contrast, ELF4 has not been implicated in natural variation for flowering time in any legume, and for LUX, natural mutants are only known in pea, have appeared only recently, and have a strong phenotype with limited value for large-scale production. It is reasonable to assume that the difference in prominence of EC mutants for adaptation mainly reflects the relative severity of the single mutant phenotypes. It also suggests that the simple duplication of the ELF3 gene may have provided a mechanism for achieving a partial loss of photoperiod responsiveness and an optimal compromise between generation of sufficient vegetative biomass to support yield and early completion of the life cycle in short season environments. However, it is also notable that ELF3 mutations provide adaptation in diploid cereals such as barley and einkorn wheat (Faure et al., 2012; Zakhrabekova et al., 2012; Alvarez et al., 2016) where ELF3 is single copy. This suggests that there could be another component to the adaptive advantage of elf3 mutations over other EC mutants, which could reflect unique roles of ELF3 that are conserved in these different crop groups. Broader physiological comparisons of EC mutants will in future be valuable in testing this idea.
Turning from questions of redundancy to the specific roles of EC genes, two output phenotypes are worth a brief consideration. In Arabidopsis, elongated hypocotyls under white light are a conspicuous feature of EC mutants and the EC has been shown to play an important role in photoperiod regulation of hypocotyl elongation, acting to repress transcription of growth-inhibiting PIF transcription factors in the evening (Nusinow et al., 2011). In contrast, none of the pea EC mutants or mutant combinations have substantial effect on elongation of early stem internodes under white or monochromatic light (Fig. 7). The fact that HR and DNE can complement the Arabidopsis elf3 and elf4 mutant elongation phenotypes, respectively (Liew et al., 2009; Weller et al., 2012), indicates that it is not due to an inherent difference in the structure of the pea and Arabidopsis proteins. It could instead result from a difference in the way that light signals regulating seedling stem elongation are gated in the two species and/or the activity of PIF genes in control of elongation. However, it is perhaps equally likely to reflect fundamental differences between Arabidopsis and pea in growth habit (rosette versus caulescent), germination mode (hypogeal versus epigeal), and seed size (small versus large), all of which might conceivably contribute to a reduced need for sensitive seedling elongation responses in pea versus Arabidopsis.
Early flowering under noninductive (SD) photoperiods remains the most striking overt phenotypic consequence of EC gene dysfunction in both Arabidopsis and pea, but the mechanisms through which EC genes influence flowering are surprisingly unclear. In Arabidopsis, photoperiod responsiveness depends significantly on the transcriptional rhythm of the CONSTANS (CO) gene and coincidence of CO expression with light under LD but not under SD conditions. Mutations in circadian clock genes such as TOC1 and LHY/CCA1 are considered to confer early flowering in SD through a shift in the phase of the CO expression rhythm to coincide with light and allow stabilization of the CO protein (Yanovsky and Kay, 2002; Mizoguchi et al., 2005). ELF3 is also reported to influence the daily transcriptional rhythm of CO in a similar manner, through protein-level regulation of the indirect CO activator GI (Yu et al., 2008). Regulation through GI could also potentially explain CO-independent effects of ELF3 (Lu et al., 2012). Whether similar mechanisms could also explain the early flowering of elf4 and lux mutants has not been addressed. However, ELF4 has been reported to influence the activity of GI by sequestering it to regions of the nucleus where it is unable to access the CO promoter (Kim et al., 2013). These results suggest that despite acting in some circumstances as part of a complex, EC components may have other roles and influence GI activity and flowering in distinct ways. Under SDs, the pea EC mutations confer early and elevated expression of several FT genes known to promote flowering, in a generally similar manner (Fig. 2). The lack of effect of ppd on expression on the CO ortholog COLa (Supplemental Fig. S7) is consistent with results from the dne mutant (Liew et al., 2009) and other recent reports indicating that COL genes are unlikely to participate in the photoperiod response mechanism in temperate legumes (Wong et al., 2014; Ridge et al., 2016). The GI ortholog LATE1 is also a key component of the photoperiod response pathway and is necessary for normal induction of FT genes (Hecht et al., 2007, 2011), and it is thus possible that EC genes may act to limit LATE1 function in some way. Some evidence for this is already apparent from analysis of gene expression rhythms, which have shown that GI transcript levels are significantly elevated during the nighttime trough phase by the dne, sn, and ppd mutations (Liew et al., 2009, 2014; Fig. 3). In future it will be interesting to learn how this elevated expression may be linked to elevated FT expression and whether other aspects of GI regulation are influenced by the SN, DNE, and HR/PPD genes.
MATERIALS AND METHODS
Plant Material
Origins of the ppd-1, ppd-2, sn-4, and dne-1 mutations have been described previously (King and Murfet, 1985; Arumingtyas and Murfet, 1994; Hecht et al., 2007). The ppd-3 mutant was obtained from EMS mutagenesis of line NGB5839 (Hecht et al., 2007), which also carries the hr mutation (Weller et al., 2012). Generation of HR, sn-4 HR, and dne-1 HR lines was described previously (Liew et al., 2014). The novel ppd-3 HR genotype was selected from a cross between ppd-3 and a near-isogenic line of NGB5839 carrying a functional HR allele (Weller et al., 2012), and its identity was verified in advanced generations by molecular genotyping using markers detailed in Supplemental Table S2.
General Growth Conditions
All plants were grown in a 1:1 mixture of dolerite chips and vermiculite topped with potting mix and received nutrient solution weekly. Plants for gene expression experiments (Figs. 2–4; Supplemental Figs. S6 and S7) were grown in growth cabinets at 20°C under 200 µmol m−2 s−1 white light from cool-white fluorescent tubes. Segregating progenies and plants for flowering and stem elongation experiments (Figs. 1, 6, and 7A) were grown in the phytotron and received an 8-h photoperiod of natural daylight either with (LD) or without (SD) an 8-h extension with white light (10 µmol m−2 s−1) from a mixture of fluorescent and incandescent sources (Hecht et al., 2007). Branches were periodically removed for the plants in the flowering experiment. Examination of deetiolation phenotypes (Fig. 7B) was conducted in growth chambers under light sources described previously (Hecht et al., 2007).
Mapping, Linkage, and Basic Genetic Analysis
Several markers used for linkage analysis were modified from gene-based markers described by Aubert et al. (2006) and Bordat et al. (2011). These markers were supplemented by newly designed markers targeted to introns of appropriate genes identified in the relevant interval of the Medicago truncatula genome (v4.0; www.jcvi.org/medicago/) and also present in pea sequence databases in GenBank (www.ncbi.nlm.nih.gov). Details of these markers and their method of detection are provided in Supplemental Table S2. The marker used to detect the hr allele has been described previously (Liew et al., 2009; Weller et al., 2012), and details of the marker used to follow the ppd-3 mutation are also provided in Supplemental Table S2.
Sequence and Expression Analysis
ELF3a and ELF3b genes in legumes were identified by BLAST searches of various sequence databases (bios.dijon.inra.fr/FATAL/cgi/pscam.cgi, www.jcvi.org/medicago/, www.phytozome.net, www.kazusa.or.jp/lotus, www.ncbi.nlm.nih.gov, legumeinfo.org/blast; knowpulse2.usask.ca/portal/blast) using the full-length amino acid sequence of Arabidopsis (Arabidopsis thaliana) ELF3 and M. truncatula ELF3a and ELF3b. All alignments were constructed with the MAFFT algorithm FFT-NS-ix 1000 in Geneious 8.1.8 using default settings. The neighbor-joining tree shown in Figure 5B was constructed from the alignment shown in Supplemental Figure S3 using ClustalX 2.1 (Thompson et al., 1997) with default settings and 10,000 bootstraps.
For all circadian and diurnal expression experiments (Figs. 2–4; Supplemental Figs. S5 and S6), plants were 3 weeks old at harvest, and harvested tissue consisted of both leaflets from the uppermost expanded leaf. Tissue harvests for the FT expression experiment presented in Figure 2 were performed as described by Hecht et al. (2011). RNA extraction, reverse transcription, and real-time PCR analysis were performed as described previously (Liew et al., 2009). Primers for expression analysis are presented in Supplemental Table S2.
Statistical Analysis
Significance of specified genotype effects were analyzed using a two-tailed Student’s t test, with the assumption of equal variance.
Estimates of rhythmic parameters in diurnal and circadian expression data for Figures 3 and 4 (presented in Supplemental Fig. S2) were obtained from the Biological Rhythms Analysis Software System (BRASS version 3.0; http://millar.bio.ed.ac.uk/PEBrown/BRASS/BrassPage.htm), which utilizes fast Fourier transform-nonlinear least squares analysis. Period and relative amplitude error analysis was conducted using default settings with the period range set from 10 to 35 h and a confidence interval of 75%. Phase analysis was conducted with FFT-NLL and the additional program Mfourfit using default settings with a spline curve option.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic comparison of photoperiod-insensitive early flowering pea mutants.
Supplemental Figure S2. Circadian gene rhythms analysis
Supplemental Figure S3. Alignment of selected ELF3-like protein sequences used to generate phylogenetic tree in Figure 5B.
Supplemental Figure S4. Alignment of ELF3-like protein sequences from 20 legume species with Arabidopsis ELF3.
Supplemental Figure S5. Phenotypic comparison of mutants for pea “evening complex” genes.
Supplemental Figure S6. Comparison of pea ELF3a and ELF3b gene expression.
Supplemental Figure S7. Diurnal regulation of group Ia CO-like genes in the ppd-3 mutant.
Supplemental Table S1. SNPs in PsELF3b coding sequence in selected P. sativum lines.
Supplemental Table S2. Primer and marker details.
Supplemental Table S3. Sequence details.
Supplementary Material
Acknowledgments
We thank Ian Cummings, Tracey Winterbottom, and Michelle Lang for help with plant husbandry.
Glossary
- EC
evening complex
- SD
short day
- LD
long day
Footnotes
This work was supported by the Australian Research Council through a Discovery Project grant to J.L.W.
Articles can be viewed without a subscription.
References
- Adams S, Manfield I, Stockley P, Carré IA (2015) Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS One 10: e0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J (2016) Genetic and physical mapping of the earliness per se locus Eps-A (m) 1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct Integr Genomics 16: 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Carvalho S, Aubert G, Carrère S, Cruaud C, Brochot AL, Jacquin F, Klein A, Martin C, Boucherot K, Kreplak J, et al. (2015) Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J 84: 1–19 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, Velikkakam James G, Nagy F, Davis SJ (2014) Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 3: 10.7554/eLife.02206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumingtyas E, Murfet IC (1994) Flowering in Pisum: a further gene controlling response to photoperiod. J Hered 85: 12–17 [Google Scholar]
- Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet 112: 1024–1041 [DOI] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM (2014) EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordat A, Savois V, Nicolas M, Salse J, Chauveau A, Bourgeois M, Potier J, Houtin H, Rond C, Murat F, et al. (2011) Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. G3 (Bethesda) 1: 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M (2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199: 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Ilut D, Farmer AD, Maki SL, May GD, Singer SR, Doyle JJ (2010) Polyploidy did not predate the evolution of nodulation in all legumes. PLoS One 5: e11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary MK, Nomura Y, Wang L, Nakagami H, Somers DE (2015) Quantitative circadian phosphoproteomic analysis of Arabidopsis reveals extensive clock control of key components in physiological, metabolic, and signaling pathways. Mol Cell Proteomics 14: 2243–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Helfer A, Nusinow DA, Kay SA (2012) ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 7: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN (2011) BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 23: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJM, Weller JL (2007) Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144: 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Laurie RE, Vander Schoor JK, Ridge S, Knowles CL, Liew LC, Sussmilch FC, Murfet IC, Macknight RC, Weller JL (2011) The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA (2016) Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics 15: 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Nusinow DA (2016) Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet 32: 674–686 [DOI] [PubMed] [Google Scholar]
- Jung J-H, Seo Y-H, Seo PJ, Reyes JL, Yun J, Chua N-H, Park C-M (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Páldi K, O'Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J (2015) Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev Cell 35: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kikis EA, Quail PH (2003) EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol 133: 1530–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lim J, Yeom M, Kim H, Kim J, Wang L, Kim WY, Somers DE, Nam HG (2013) ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Reports 3: 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King WM, Murfet IC (1985) Flowering in Pisum: a sixth locus, Dne. Ann Bot (Lond) 56: 835–846 [Google Scholar]
- Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ (2009) Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J 3: 350–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E, Herrero E, Bujdoso N, Millar AJ, Tóth R, Gyula P, Nagy F, Davis SJ (2011) A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell 23: 3230–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Liew LC, Hecht V, Laurie RE, Knowles CL, Vander Schoor JK, Macknight RC, Weller JL (2009) DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 21: 3198–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew LC, Hecht V, Sussmilch FC, Weller JL (2014) The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol 165: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zhao X, Hu Y, Liu S, Nan H, Li X, Fang C, Cao D, Kong L, Su T, et al. (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet (in press) [DOI] [PubMed] [Google Scholar]
- Lu SX, Webb CJ, Knowles SM, Kim SH, Wang Z, Tobin EM (2012) CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol 158: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M (2012) Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol 53: 709–716 [DOI] [PubMed] [Google Scholar]
- McClung CR. (2014) Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ. (2016) The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu Rev Plant Biol 67: 595–618 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Nitta M, Sato K, Nasuda S (2012) A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet Syst 87: 357–367 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55: 958–976 [DOI] [PubMed] [Google Scholar]
- Murfet IC. (1971) Flowering in Pisum: reciprocal grafts between known genotypes. Aust J Biol Sci 24: 1089–1102 [Google Scholar]
- Murfet IC. (1973) Flowering in Pisum. Hr, a gene for high response to photoperiod. Heredity 31: 157–164 [Google Scholar]
- Murfet IC, Taylor S (1999) Brief communication. Flowering gene Ppd in pea: map position and disturbed segregation of allele ppd-2. J Hered 90: 548–550 [Google Scholar]
- Nagel DH, Kay SA (2012) Complexity in the wiring and regulation of plant circadian networks. Curr Biol 22: R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: E4802–E4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K, Ishiura M (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972 [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54: 441–454 [DOI] [PubMed] [Google Scholar]
- Ridge S, Sussmilch FC, Hecht V, Vander Schoor JK, Lee R, Aubert G, Burstin J, Macknight RC, Weller JL (2016) Identification of LATE BLOOMER2 as a CYCLING DOF FACTOR homolog reveals conserved and divergent features of the flowering response to photoperiod in pea. Plant Cell 28: 2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini R, Szpotkowski K, Kozak M, Jaskolski M, Davis SJ (2013) Biochemical and structural studies of plant circadian clock proteins. BioTechnologia 94: 40–41 [Google Scholar]
- Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 11698–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton DD, Smith RW, Song YH, MacGregor DR, Stewart K, Steel G, Foreman J, Penfield S, Imaizumi T, Millar AJ, Halliday KJ (2015) Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Mol Syst Biol 11: 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Lee I, Lee SY, Imaizumi T, Hong JC (2012) CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J 69: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Taylor SA, Murfet IC (1996) Flowering in Pisum: Identification of a new ppd allele and its physiological action as revealed by grafting. Physiol Plant 97: 719–723 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Batge SL, Smith JJ, Kerckhoffs LHJ, Sineshchekov VA, Murfet IC, Reid JB (2004) A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol 135: 2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Liew LC, Hecht VF, Rajandran V, Laurie RE, Ridge S, Wenden B, Vander Schoor JK, Jaminon O, Blassiau C, et al. (2012) A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci USA 109: 21158–21163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Murfet IC, Reid JB (1997) Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol 114: 1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ortega R (2015) Genetic control of flowering time in legumes. Front Plant Sci 6: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Hecht VF, Picard K, Diwadkar P, Laurie RE, Wen J, Mysore K, Macknight RC, Weller JL (2014) Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front Plant Sci 5: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, Müller AH, Lundqvist J, Ahmann K, Dockter C, Matyszczak I, Kurowska M, Druka A, et al. (2012) Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA 109: 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.