RING-type E3 ligase controls clade A protein phosphatase 2C, a core ABA component indirectly or directly at the transcriptional and posttranslational levels in ABA and drought signaling.
Abstract
Ubiquitin-mediated protein modification occurs at multiple steps of abscisic acid (ABA) signaling. Here, we sought proteins responsible for degradation of the pepper (Capsicum annuum) type 2C protein phosphatase CaADIP1 via the 26S proteasome system. We showed that the RING-type E3 ligase CaAIRF1 (Capsicum annuum ADIP1 Interacting RING Finger Protein 1) interacts with and ubiquitinates CaADIP1. CaADIP1 degradation was slower in crude proteins from CaAIRF1-silenced peppers than in those from control plants. CaAIRF1-silenced pepper plants displayed reduced ABA sensitivity and decreased drought tolerance characterized by delayed stomatal closure and suppressed induction of ABA- and drought-responsive marker genes. In contrast, CaAIRF1-overexpressing Arabidopsis (Arabidopsis thaliana) plants exhibited ABA-hypersensitive and drought-tolerant phenotypes. Moreover, in these plants, CaADIP1-induced ABA hyposensitivity was strongly suppressed by CaAIRF1 overexpression. Our findings highlight a potential new route for fine-tune regulation of ABA signaling in pepper via CaAIRF1 and CaADIP1.
Global warming has resulted in climate change and extreme temperatures, leading to environmental stresses that interfere with normal plant growth and development. Plants have evolved elaborate defense mechanisms to enable survival under unfavorable environmental conditions, such as cold, high salinity, and drought. Drought, caused by water-deficit conditions, severely influences normal plant growth, leading to decreased crop yield. To adapt to drought stress, plants activate various defense mechanisms, including stomatal closure, water content reduction, and abscisic acid (ABA) synthesis and accumulation (Lee and Luan, 2012; Golldack et al., 2014). ABA, an important regulator of plant responses to abiotic stresses, especially drought stress, plays a key role in plant growth and development (Osakabe et al., 2014). ABA signaling regulates the expression levels of various stress-related genes, including transcription factors and ubiquitin ligase (E3) involved in osmotic adjustment and modification of root hydraulic conductivity (Sirichandra et al., 2009); however, the ABA signal transduction pathway has not yet been fully elucidated. Previous studies have investigated the sequence of ABA signal transduction from ABA perception to response. Initiation of ABA signaling in plant cells is dependent on the presence of ABA receptors and ABA signal delivery to downstream components. In this process, pyrabactin resistance/pyrabactin resistance-like/regulatory component of ABA receptor (PYR/PYL/RCAR) proteins function as ABA receptors (Ma et al., 2009; Park et al., 2009). The downstream events of ABA perception are protein phosphorylation and dephosphorylation, associated with Suc nonfermenting 1-related subfamily 2 (SnRK2) protein kinases and type 2C protein phosphatases (PP2Cs), respectively. SnRK2 protein kinases, including SnRK2.2, SnRK2.3, and SnRK2.6 (OST1), positively regulate ABA signaling. Conversely, group A PP2Cs negatively regulate ABA signaling (Lee and Luan, 2012). PYR/PYL/RCAR proteins interact with and inhibit group A PP2Cs (Szostkiewicz et al., 2010). Increased ABA levels are perceived by PYR/PYL/RCAR proteins, which in turn inhibit protein phosphatase-kinase interactions, leading to the release of SnRK2 protein kinases; these SnRK2-type kinases promote specific gene expression and ion channel activation via phosphorylation of target proteins, including transcription factors and SLAC1 anion channels, thereby enabling plants to adapt to adverse environments (Lee et al., 2009; Lee and Luan, 2012; Lim et al., 2015a).
Protein degradation via the ubiquitination pathway is an important mechanism involved in regulating adaptation and defense responses to abiotic stresses. Ubiquitination is a key posttranslational regulatory mechanism involving the sequential action of three enzymes: ubiquitin (Ub)-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub ligase (E3; Ciechanover and Schwartz, 1998). Initially, Ub is activated by E1; the activated Ub is then transferred to E2; finally, Ub is attached to the target protein by E3 ligase. In these processes, E3 ligase determines and recruits target proteins. Ubiquitination is an intrinsic mechanism involving hundreds or thousands of distinct E3 ligases with diverse target proteins in eukaryotic cells (Vierstra, 2009; Sadanandom et al., 2012). E3 ligases can be classified into two types based on their subunit compositions. The really interesting new gene (RING), homology to E6-AP C terminus, and plant U-box (PUB) E3 ligases consist of a single subunit, whereas the S-phase kinase-associated protein/cullin/F-box and CULLIN4-damaged-specific DNA binding protein1 ligases consist of a multisubunit (Stone et al., 2005; Pazhouhandeh et al., 2011; Irigoyen et al., 2014; Seo et al., 2014). To date, more than 1,400 E3 ligases have been identified in Arabidopsis (Arabidopsis thaliana; Vierstra, 2009). The Arabidopsis genome encodes 242 RING E3 ligases, which have frequently been shown to function in adaptation to biotic and abiotic stresses. RING E3 ligases positively or negatively regulate abiotic stress in diverse monocot and dicot plants, including rice (Oryza sativa), maize (Zea mays), hot pepper (Capsicum annuum), and Arabidopsis; hence, they are widely conserved in plants (Lyzenga and Stone, 2012; Li et al., 2013; Zhao et al., 2014; Park et al., 2015). Recently, RING-type E3 ligase RSL1 was reported to be involved in ubiquitination and degradation of ABA receptors, including PYR1 and PYL4, at the plasma membrane (Bueso et al., 2014). Additionally, CRL4-type E3 ligase complex was found to degrade nuclear PYL8 ABA receptor (Irigoyen et al., 2014). Moreover, as an interacting partner of several PYR/PYL/RCAR proteins, including PYR1 and PYL8, clade A PP2C ABI1 interacted with and was ubiquitinated by the U-box E3 ligases PUB12 and PUB13 only in the presence of ABA and PYR1 (Nishimura et al., 2010; Kong et al., 2015). Similarly, ABA promotes interaction of PP2CA, HAB2, and ABI2 with the E3 ligase RGLG1 and RGLG5, but in vitro ubiquitination of these PP2Cs by RGLG1 and RGLG5 occurs in the absence of ABA receptors (Wu et al., 2016). The role of E3 ligases in response to drought stress via the ABA-signaling pathway has been extensively studied in various plants, but their precise function remains unclear.
We previously reported that the pepper protein phosphatase CaADIP1 negatively regulates ABA. We further demonstrated that CaADIP1 is associated with drought sensitivity via alteration of ABA-responsive gene expression and regulation of the stomatal aperture (Lim and Lee, 2016). Here, we isolated the RING-type E3 ligase CaAIRF1 (Capsicum annuum ADIP1 Interacting RING Finger Protein 1), which interacts with CaADIP1 and is involved in CADIP1 degradation. Our findings indicate that CaAIRF1 positively regulates ABA signaling via its E3 ligase activity by influencing CaADIP1 stability.
RESULTS
ABA-Promoted CaADIP1 Degradation via the 26S Proteasome
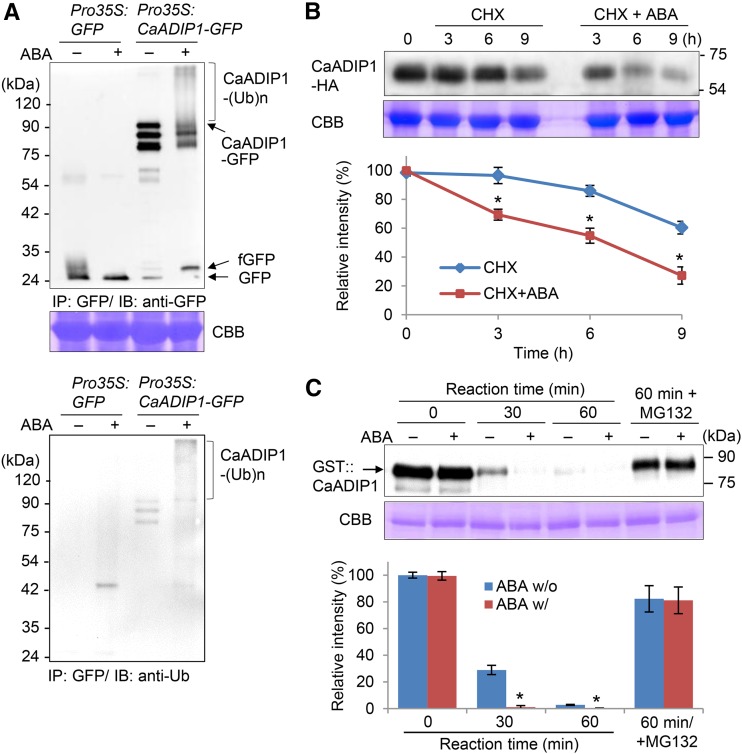
We investigated the mechanism whereby the pepper type 2C protein phosphatase CaADIP1 is regulated in response to ABA treatment. First, we performed an in vivo ubiquitination assay using the leaves of tobacco (Nicotiana benthamiana) plants transiently expressing Pro-35S:CaADIP1-GFP and Pro-35S:GFP as a negative control. To prevent proteasomal degradation of the fusion proteins, we applied MG132, which is an inhibitor of 26S proteasome-mediated proteolysis. ABA treatment resulted in polyubiquitination of the CaADIP1 protein (Fig. 1A). To investigate the mechanism whereby ABA influences CaADIP1 protein stability, we performed an in vivo degradation assay. The leaves of tobacco plants transiently expressing Pro-35S:CaADIP1-HA were infiltrated with cycloheximide (CHX) to inhibit subsequent protein synthesis. ABA treatment led to a marked increase in CaADIP1 protein degradation (>70% vs. 40%; Fig. 1B). Next, we conducted a cell-free degradation assay using ABA-treated pepper leaves. Immunoblotting analysis revealed gradual degradation of the CaADIP1-GST recombinant protein. After incubation for 30 and 60 min with crude protein extracts prepared from nontreated leaves, the CaADIP1-GST levels were >30% and <5%, respectively, of the initial level (Fig. 1C). In comparison, the CaADIP1-GST levels were markedly reduced by ABA treatment at all time points; after 30 min, most of the protein was degraded. This CaADIP1 degradation was mostly attenuated by application of MG132 to the incubation mixtures. After 60 min of incubation with MG132, <20% of the CaADIP1 protein was degraded. These data indicate that ABA promotes CaADIP1 degradation via the 26S proteasome pathway.
Figure 1.
ABA-promoted degradation of the CaADIP1 protein in vitro. A, Polyubiquitination of the CaADIP1 protein. Leaves of tobacco plants harboring Pro-35S:CaADIP1-GFP were harvested 6 h after treatment with 100 μm ABA. At 12 h before sample harvesting, 50 μm MG132 was infiltrated into the leaves. Protein extracts were immunoprecipitated using a GFP-trap, followed by immunoblot analysis. Polyubiquitinated CaADIP1 protein was detected using anti-GFP (top) and anti-Ub (bottom). Coomassie blue staining (CBB) staining indicates equal loading of protein extract. fGFP, fragmented GFP. B, ABA-promoted in vivo degradation of CaADIP1. Leaves of tobacco plants harboring Pro-35S:CaADIP1-HA were infiltrated with 50 μm CHX 2 d after agroinfiltration and harvested at the indicated time points after treatment with 100 μm ABA. Immunoblot analysis was performed using anti-HA antibody, and the relative intensities of the CaADIP1-HA fusion proteins were measured with Image J 1.46r (http://imagej.nih.gov/ij) software. CBB staining indicates equal loading of total protein. C, Cell-free degradation of the CaADIP1 protein. The GST-tagged CaADIP1 (500 ng) protein was incubated for the indicated periods with crude extracts prepared from the leaves of ABA-treated 4-week-old pepper plants. Immunoblot analysis was performed using anti-GST antibody (top). CBB staining indicates equal loading of crude extract (bottom). The relative intensities of the GST-CaADIP1 fusion proteins were measured. All data represent the mean ± sd of three independent experiments; asterisks indicate significant differences (Student’s t test; P < 0.05).
Interaction of CaADIP1 with RING-Type E3 Ligase CaAIRF1
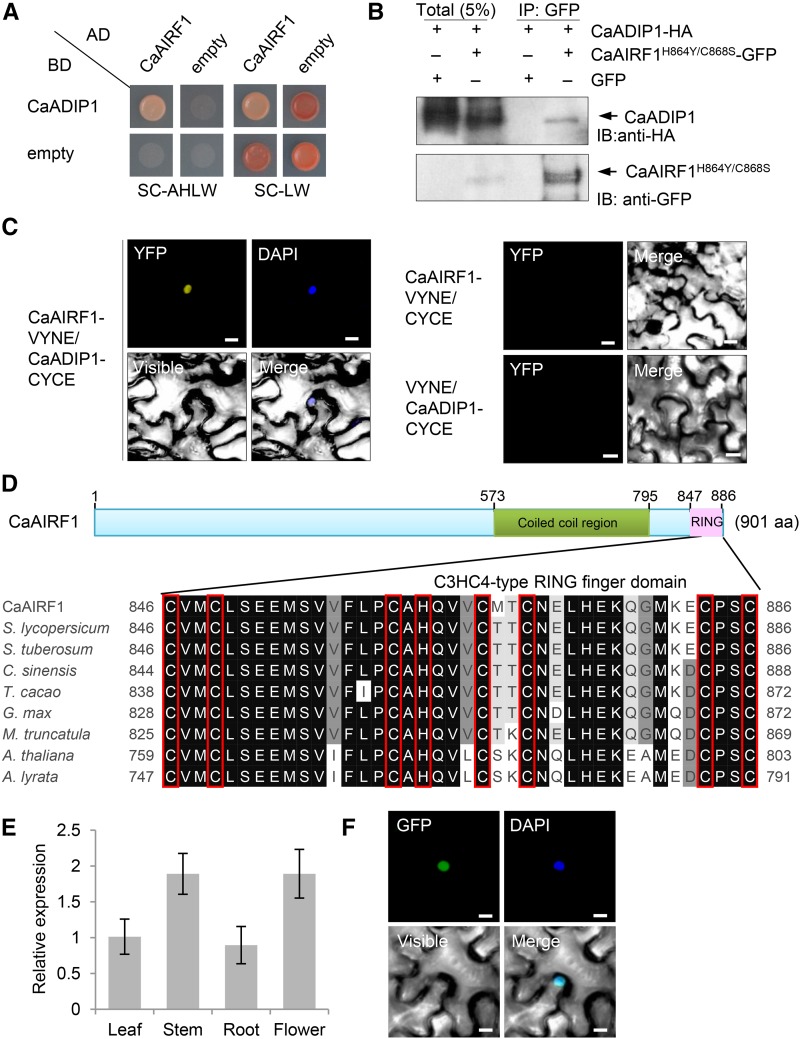
Next, we performed a yeast two-hybrid (Y2H) screen assay to isolate candidate proteins responsible for the 26S proteasome-mediated CaADIP1 degradation. We used a cDNA library constructed from the leaves of pepper plants treated with ABA as prey and CaADIP1 as bait (Lim and Lee, 2016). We selected CaAIRF1 from the CaADIP1-interacting proteins and reconfirmed the CaADIP1-CaAIRF1 interaction using a one-by-one Y2H assay (Fig. 2A), coimmunoprecipitation (co-IP) assay (Fig. 2B), and bimolecular fluorescence complementation (BiFC) analysis (Fig. 2C). In particular, coexpression of CaADIP1-CYCE with CaAIRF1-VYNE resulted in yellow fluorescence predominantly in the nuclei. For co-IP assay, we used the CaAIRF1H864Y/C868S protein, which shows loss of E3 ligase activity (Fig. 2B). When GFP-tagged intact CaAIRF1 was expressed, we observed no signal of interaction between CaAIRF1 and CaADIP1.
Figure 2.
Interaction of CaADIP1 with CaAIRF1. A, Yeast two-hybrid assay of interactions between CaADIP1 and CaAIRF1. Interaction was indicated by growth on selection medium (SC-adenine-His-Leu-Trp; left); growth on SC-LW was used as a control (right). B, Co-IP of CaADIP1-HA and CaAIRF1H864Y/C868S-GFP. For efficient interaction with CaADIP1, CaAIRF1H864Y/C868S protein showing loss of E3 ligase activity was used instead of intact CaAIRF1. CaADIP1-HA was coexpressed with CaAIRF1H864Y/C868S-GFP or GFP alone (negative control) in the leaves of tobacco. Immunoblot analysis was performed using anti-HA and anti-GFP antibodies. C, BiFC assay of interactions between CaADIP1 and CaAIRF1. CaADIP1-VYNE was coexpressed with CaAIRF1-CYCE in the leaves of tobacco. For the negative control, CaADIP1-VYNE and CaAIRF1-CYCE were coexpressed with CYCE and VYNE, respectively. Scale bar = 10 μm. D, Domain organization of deduced amino acids in the CaAIRF1 protein. The conserved domain was analyzed using the SMART Web site (http://smart.embl-heidelberg.de) and framed in the green (coiled-coil region) and pink (RING domain) boxes (top). Multiple alignment analysis of the RING domains of CaAIRF1 and its homologous proteins was performed using ClustalW2 (bottom). Amino acid residues of RING domains are shaded according to the percentage identity in ClustalW2; key residues are boxed in red (see also Supplemental Fig. S1). E, Organ-specific expression of CaAIRF1 in pepper plants. The pepper Actin1 (CaACT1) gene was used as an internal control. F, Subcellular localization of CaAIRF1 using transient expression of the GFP fusion protein in tobacco. White bar = 10 μm.
CaAIRF1 encodes a 901-amino acid protein and contains a C3HC4-type RING finger domain at the C-terminal end; this domain is highly conserved in various species of flowering plants (Fig. 2D; Supplemental Fig. S1). CaAIRF1 was expressed in all the examined tissues (Fig. 2E); moreover, the expression levels were markedly high in the stems and flowers. To investigate the subcellular localization of CaAIRF1, we transiently expressed GFP-tagged CaAIRF1 in tobacco leaves. The GFP fluorescent signal was detected in the nucleus and overlapped with the blue DAPI signal (Fig. 2F), indicating targeting of CaAIRF1 to the nucleus.
Enhanced Expression of CaAIRF1 in Response to ABA, H2O2, Drought, and High Salinity
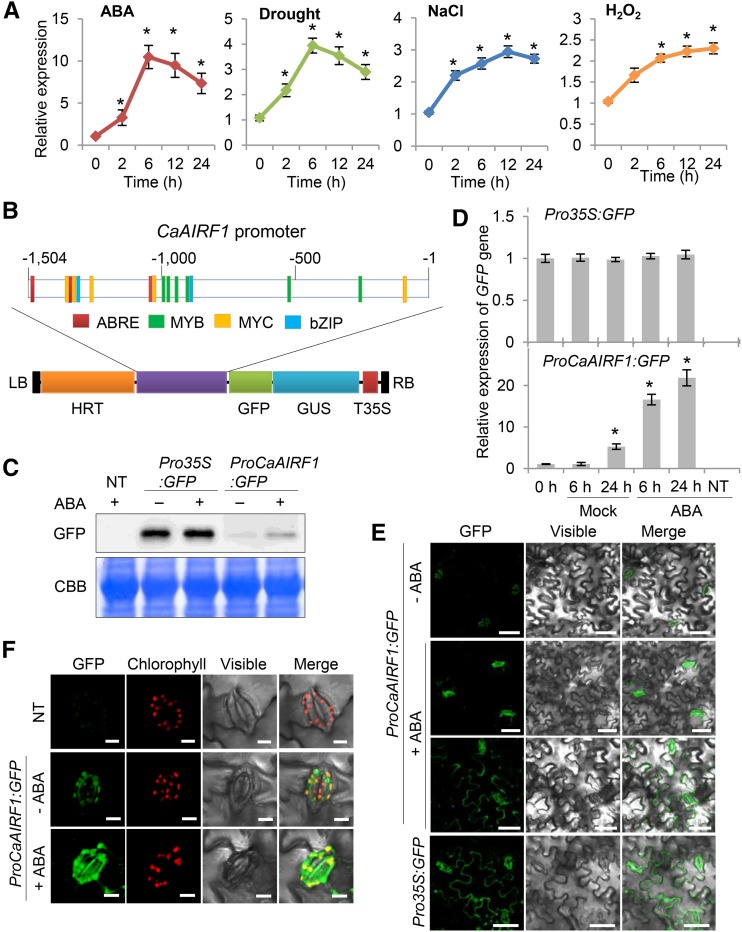
CaADIP1 is strongly induced by ABA, drought, and NaCl; moreover, it modulates ABA sensitivity and drought tolerance (Lim and Lee, 2016). Hence, we investigated whether CaAIRF1 is associated with ABA signaling and abiotic stress responses. First, we performed quantitative RT-PCR (qRT-PCR) analysis to examine the CaAIRF1 expression patterns in the leaves of pepper plants treated with ABA, H2O2, drought, or high salinity. After exposure to ABA and drought, CaAIRF1 transcripts were strongly expressed and reached maximum levels after 6 h. On the other hand, after exposure to H2O2 and NaCl, the CaAIRF1 expression levels increased gradually for up to 24 h (Fig. 3A).
Figure 3.
ABA-promoted expression of the pCaAIRF1::EGFP fusion gene. A, RT-PCR analysis of CaAIRF1 expression. The expression pattern of CaAIRF1 was analyzed in the leaves of pepper plants treated with ABA (100 μm), H2O2 (100 μm), drought, or NaCl (200 mm). The pepper Actin1 (CaACT1) gene was used as an internal control. B, Schematic representation of the transfer DNA (T-DNA) region of the pHGWFS7 vector containing a 1,504-bp upstream region of CaAIRF1. The colored boxes indicate cis-regulatory elements, associated with ABA signaling, in the CaAIRF1 promoter sequence. HRT, hygromycin phosphotransferase; LB, left border; RB, right border. C and D, Induction of GFP expression in response to ABA. Leaves were harvested from nontransgenic (NT) and transgenic tobacco plants 24 h after treatment with 50 μm ABA. Immunoblot (C) and qRT-PCR (D) analyses were performed using an tobacco leaf harboring the ProCaAIRF1::EGFP fusion gene. The polyclonal anti-GFP antibody was used. For qRT-PCR, the expression level of the GFP gene was normalized with that of CaACT1. Data represent the mean ± sd of three independent experiments; asterisks indicate significant differences (Student’s t test; P < 0.05). E and F, GFP signals from the leaves of tobacco (E) and pepper (F) plants. After agroinfiltration, leaves were treated with 50 μm ABA for 24 h and subjected to microscopic analysis. White bar = 50 μm (E) and 10 μm (F).
Next, we analyzed known cis-regulatory elements within the 1.5-kb promoter sequence upstream of the CaAIRF1 coding region, retrieved from the Sol Genomics Network (https://solgenomics.net). In silico analysis revealed that this region contained three ABA-responsive element-like sequences (CCACGTGG), six MYB binding sites (CA/TAACCA), four MYC binding sites (CACATG), and two bZIP binding sequences (ACACNNG; Fig. 3B). We subsequently isolated and fused the 1,504-bp promoter sequence with the GFP reporter gene to monitor the expression pattern of CaAIRF1 in plant cells after ABA treatment. Western-blot analysis revealed that ProCaAIRF1:GFP was expressed in tobacco leaf epidermal cells; moreover, the expression level was enhanced by ABA (Fig. 3C). Consistently, CaAIRF1 promoter-driven GFP gene expression was significantly induced after ABA treatment (Fig. 3D). Microscopic examination revealed that the GFP signal was detected predominantly in guard cells and partially in whole cells (<20%) prepared from the leaves of tobacco (Fig. 3E) and pepper plants (Fig. 3F). Consistent with the transcriptional and translational changes, the GFP intensities in whole cells and guard cells prepared from ABA-treated leaves were higher than in those prepared from nontreated leaves (whole cells, 3.0- to 3.8-fold increase; guard cells, 2.8- to 3.5-fold increase).
Identification of CaAIRF1 as an E3 Ligase
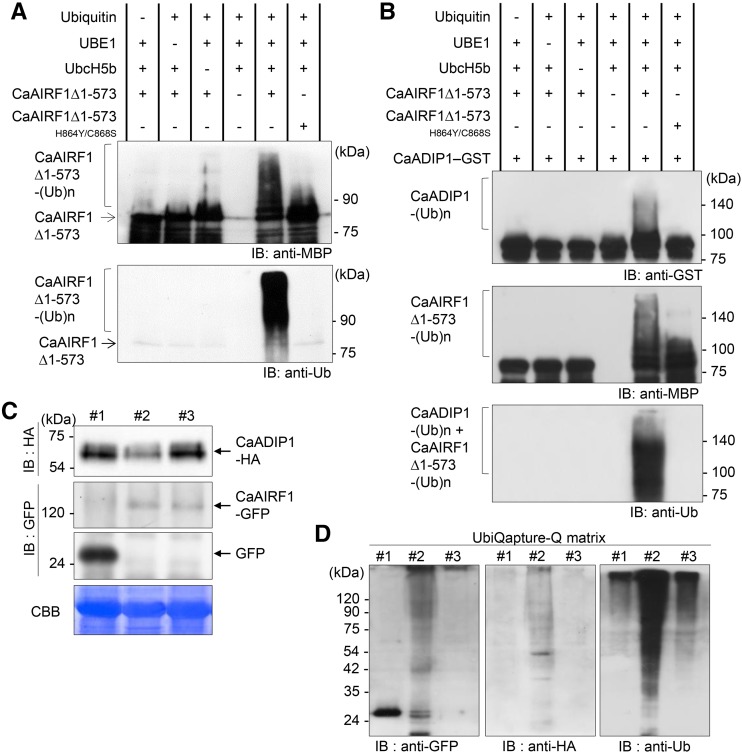
Many RING domain-containing proteins function as E3 ligases; hence, we examined whether CaAIRF1 has E3 ligase activity. When maltose-binding protein (MBP)-tagged CaAIRF1 was expressed in Escherichia coli, the fusion proteins were enriched in the insoluble fraction, and the elution level was very low. Enhancement of the protein solubility led to loss of MBP-CaAIRF1 activity. To circumvent this problem and clarify the functional role of CaAIRF1, we performed deletion analysis of CaAIRF1 using the Y2H system. Similar to the full-length CaAIRF1 protein, the truncated protein CaAIRF1∆1-573 interacted with CaADIP1 (Supplemental Fig. S2A); we subsequently used this protein in an in vitro self-ubiquitination assay. Immunoblot analysis using anti-MBP and anti-Ub antibodies revealed a high-Mr smear only when CaAIRF1∆1-573 was present with all ubiquitination components, including Ub, ATP, E1 (Arabidopsis UBE1), and E2 (Arabidopsis UBCH5b; Fig. 4A). To disrupt E3 ligase activity, we substituted His-864 and Cys-868 for Tyr-864 and Ser-868, respectively, in the RING domain. This mutation led to loss of E3 ligase activity in CaAIRF1 (Fig. 4A), but did not influence the subcellular localization and interaction with CaADIP1 (Supplemental Fig. S3). Our data indicate that CaAIRF1 has E3 ligase activity and that this activity is dependent on the presence of an intact RING domain.
Figure 4.
CaAIRF1-mediated degradation of CaADIP1. A, In vitro E3 Ub ligase activity of the CaAIRF1∆1-573 protein. MBP-tagged recombinant CaAIRF1∆1-573 and double-amino acid substitution mutant CaAIRF1∆1-573H864Y/C868S proteins were assayed for E3 activity in the presence or absence of Arabidopsis E1, E2, Ub, and ATP. Immunoblot analyses were performed using anti-MBP (top) and anti-Ub (bottom) antibodies. B, In vitro ubiquitination of CaADIP1 by CaAIRF1. MBP-CaAIRF1∆1-573 and MBP-CaAIRF1∆1-573H864Y/C868S were incubated for 2 h with GST-CaADIP1 in the presence or absence of E1, E2, and Ub. Immunoblot analyses were performed using anti-GST antibody. C and D, CaAIRF1-mediated polyubiquitination of CaADIP1 in vivo. In the tobacco leaves, Pro-35S:CaADIP1-HA was coexpressed with Pro-35S:GFP (#1), Pro-35S:CaAIRF1-GFP (#2), or Pro-35S:CaAIRF1H864Y/C868S-GFP (#3). At 12 h before sample harvesting, 50 μm MG132 was infiltrated into the leaves. C, Total protein extracts were prepared 3 d after agroinfiltration and subjected to immunoblot analysis using anti-HA and anti-GFP antibodies. D, Using the UbiQapture-Q matrix, all ubiquitinated proteins were isolated from the same protein samples. Immunoblot analysis was performed using anti-GFP (right), anti-HA (middle), and anti-Ub (right) antibodies.
Role of CaAIRF1 in the Degradation of CaADIP1 via the Ub-26S Proteasome System
Based on the Y2H and in vitro self-ubiquitination assays, we postulated that CaAIRF1 is responsible for degradation of CaADIP1. To validate this hypothesis, we performed an in vitro ubiquitination assay using CaADIP1 as a substrate for CaAIRF1. When the CaADIP1-GST protein was incubated with CaAIRF1 in the presence of E1, E2, and Ub, we observed a high-Mr smear (Fig. 4B, top). To confirm whether this ubiquitination is mediated by CaAIRF1, western-blot analysis was performed using anti-MBP antibody. CaAIRF1∆1-573 protein had ubiquitination activity only in presence of all ubiquitination components (Fig. 4B, middle), indicating that CaAIRF1 can ubiquitinate CaADIP1. Western blot with anti-Ub antibody showed all the ubiquitinated proteins of CaAIRF1∆1-573 as well as CaADIP1 (Fig. 4B, bottom). In contrast, CaAIRF1∆1-573H864Y/C868S, which lacks E3 ligase activity but strongly interacts with CaADIP1 (Supplemental Fig. S2B), did not ubiquitinate CaADIP1 (Fig. 4B). We also examined CaAIRF1-mediated polyubiquitination of CaADIP1 in vivo. Pro-35S:CaADIP1-HA was coexpressed with Pro-35S:GFP, Pro-35S:CaAIRF1-GFP, and Pro-35S:CaAIRF1H864Y/C868S-GFP in tobacco leaves. Immunoblot analysis showed a relatively low level of CaADIP1 protein in the presence of intact CaAIRF1 (Fig. 4C). To isolate ubiquitinated proteins, the same protein samples were precipitated with the UbiQapture-Q matrix. As predicted, the CaADIP1 protein was polyubiquitinated only by intact CaAIRF1 (Fig. 4D).
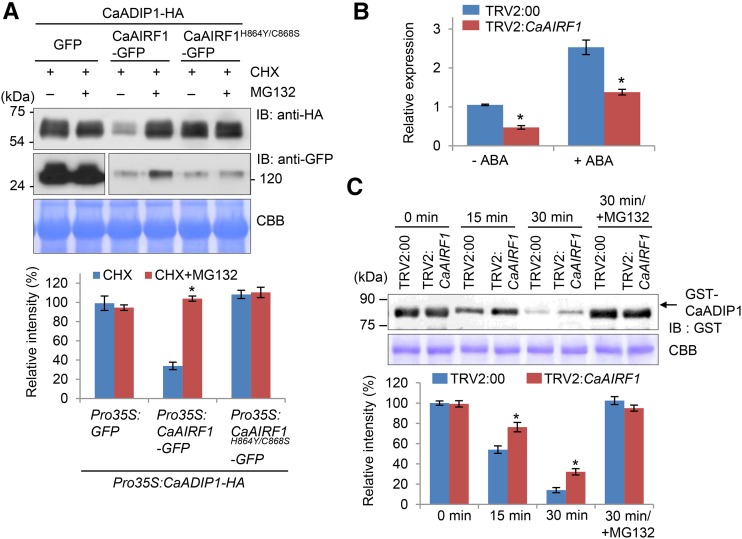
Next, we performed a cell-free degradation assay to investigate the mechanism whereby CaAIRF1 influences CaADIP1 protein stability in the presence of CHX (an inhibitor of protein synthesis) and/or MG132 (an inhibitor of 26S proteasome-mediated proteolysis). When co-overexpressed with CaAIRF1, >60% of the CaADIP1 protein was degraded in tobacco leaves; however, when coexpressed with CaAIRF1H864Y/C868S, CaADIP1 stability was not affected (Fig. 5A). Consistent with our data for the ABA-promoted degradation of CaADIP1 (Fig. 1, B and C), CaAIRF1-mediated CaADIP1 degradation was completely blocked by MG132 application. To examine the role of CaAIRF1 in the degradation of CaADIP1 in pepper cells, we used the tobacco rattle virus (TRV)-based virus-induced gene silencing system. CaAIRF1 transcript accumulation was approximately 50% lower in CaAIRF1 gene-silenced pepper plants (TRV2:CaAIRF1) than in TRV2:00 control plants, even after treatment with ABA (Fig. 5B). To investigate the mechanism whereby the decrease in expression levels of CaAIRF1 influences CaADIP1 stability, we performed a cell-free degradation assay by incubating crude protein extracts prepared from the leaves of TRV2:00 and TRV2:CaAIRF1 plants with CaADIP1-GST. Immunoblot analysis revealed that CaADIP1 degradation was partially suppressed by weak expression of CaAIRF1; after incubation for 30 min, the CaADIP1 levels were 26% to 33% and 41% to 50% of the initial levels in the leaves of TRV2:00 and TRV2:CaAIRF1 plants, respectively (Fig. 5C). Additionally, CaADIP1 protein degradation was completely blocked by MG132 application. Our data indicate that CaAIRF1 mediates CaADIP1 degradation via the Ub-26S proteasome system.
Figure 5.
CaAIRF1-mediated degradation of CaADIP1 in a 26S proteasome-dependent manner. A, In vivo assay of CaAIRF1-mediated degradation of CaADIP1. Through agroinfiltration, Pro-35S:CaADIP1-HA was coexpressed with Pro-35S:GFP, Pro-35S:CaAIRF1-GFP, or Pro-35S:CaAIRF1H864Y/C868S-GFP in the leaves of tobacco plants. At 6 h before sample harvesting, 50 μm CHX and 50 μm MG132 were infiltrated into the leaves. Protein extracts were subjected to immunoblot analysis. The relative intensities of the CaADIP1-HA fusion proteins were measured using Image J 1.46r (http://imagej.nih.gov/ij) software (bottom). CBB staining indicates equal loading of crude extract. B, Expression level of CaAIRF1 in the leaves of CaAIRF1-silenced pepper plants. TRV2:CaAIRF1 and TRV2:00 control pepper plants were treated with 10 μm ABA; after 24 h, leaves of each line were harvested. The relative expression level of CaAIRF1 was examined using qRT-PCR analysis and normalized to that of CaACT1 as an internal control gene. Data represent the mean ± sem of three independent experiments. C, Cell-free degradation assay for CaADIP1. The GST-CaADIP1 (500 ng) protein was incubated for 30 min with crude extracts prepared from the leaves of 4-week-old TRV:CaAIRF1 pepper plants. Immunoblot analysis was performed using anti-GST antibody (top). CBB staining indicates equal loading of crude extract. The relative intensities of the GST-CaADIP1 fusion proteins were measured using Image J 1.46r (http://imagej.nih.gov/ij) software (bottom). Data represent the mean ± sd of three independent experiments; asterisks indicate significant differences (Student’s t test; P < 0.05).
Reduced ABA Sensitivity in the Leaves of CaAIRF1-Silenced Pepper Plants
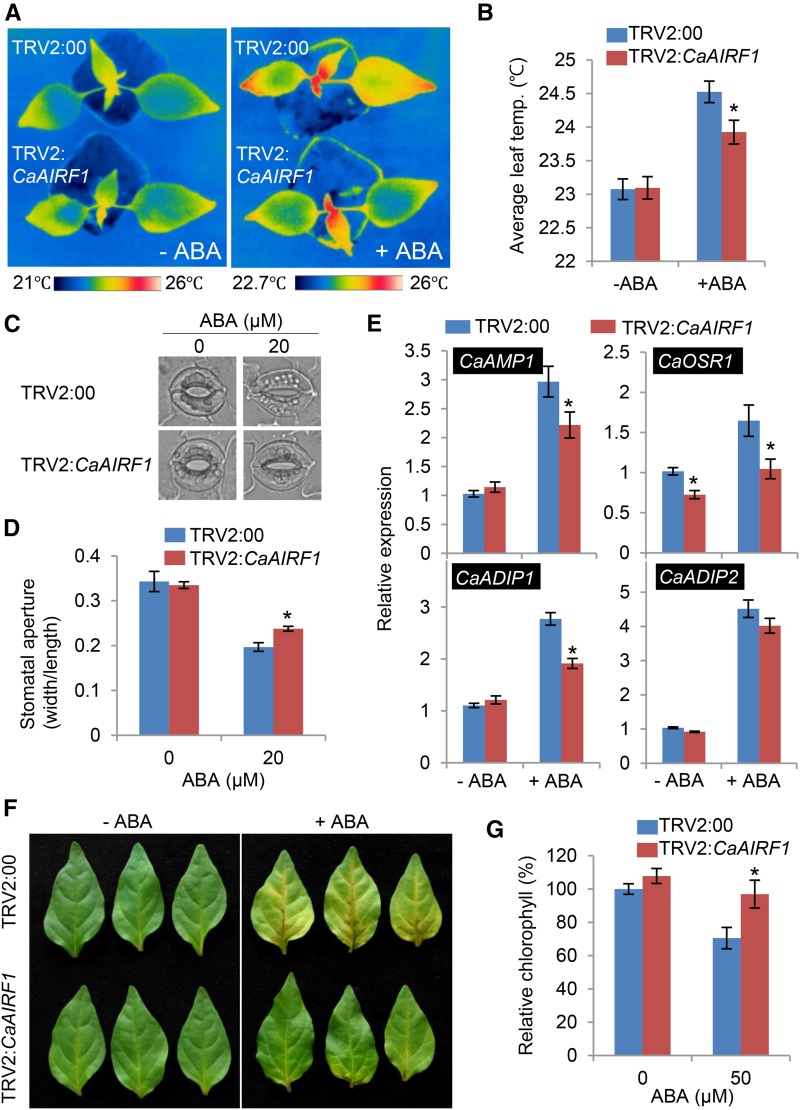
Microscopic examination revealed that ABA induced CaAIRF1 expression predominantly in guard cells and partially in whole leaf cells (Fig. 3). To examine the involvement of CaAIRF1 in ABA-mediated stomatal regulation, we measured the leaf temperature as an indirect indication of stomatal aperture. We used control and CaAIRF1-silenced pepper plants having fully expanded first and second leaves. In the absence of ABA, the average leaf temperature did not differ significantly between control and CaAIRF1-silenced plants (Fig. 6, A and B). Treatment with ABA led to an increase in average leaf temperatures; however, the leaf temperatures were significantly lower in TRV2:CaAIRF1 plants than in TRV2:00 plants. These differences were also observed in older plants having four fully expanded leaves (Supplemental Fig. S4). Stomatal closure leads to a decrease in evaporative cooling and consequently elevated leaf temperatures (Schroeder et al., 2001); hence, we postulated that after ABA treatment, CaAIRF1-silenced pepper plants exhibit delayed stomatal closure. To validate this hypothesis, we incubated leaf peels of TRV2:00 and TRV2:CaAIRF1 plants with ABA and measured the stomatal apertures. In the absence of ABA, the stomatal apertures did not differ significantly between TRV2:00 and TRV2:CaAIRF1 plants. However, after ABA treatment, the stomatal apertures of TRV2:CaAIRF1 plants were larger than those of TRV2:00 plants (Fig. 6C); in comparison with nontreated plants, the average stomatal apertures of TRV2:00 and TRV2:CaAIRF1 plants were decreased by 42% and 28%, respectively (Fig. 6D).
Figure 6.
Suppression of ABA-mediated stomatal closing and ABA-responsive gene expression in CaAIRF1-silenced pepper leaves. A and B, Representative thermographic images of CaAIRF1-silenced pepper plants 6 h after treatment with 50 μm ABA (A); the mean leaf temperature was measured using 10 plants of each line (B). Data represent the mean ± sd of three independent experiments. C and D, Stomatal apertures in control and CaAIRF1-silenced pepper plants treated with ABA. Leaf peels harvested from 4-week-old plants of each line were incubated for 2 h in SOS buffer containing 20 μm ABA. Representative images were taken (C) and the stomatal apertures were measured under the microscope (D). Data represent the mean ± se of three independent experiments. E, RT-PCR analysis of ABA-responsive gene expression in the leaves of TRV2:00 and TRV2:CaAIRF1 pepper plants. Four-week-old plants of each line were treated with 10 μm ABA; after 24 h, leaves of each line were harvested. The relative expression level (∆∆CT) of each gene was normalized to that of CaACT1 as an internal control gene. Data represent the mean ± se of three independent experiments. F and G, Floating leaf assay of TRV2:00 and TRV2:CaAIRF1 pepper plants. The first and second fully expanded leaves of each plant line (4 weeks old) were floated in SOS buffer containing 50 μm ABA. After 6 d, representative images were taken (F) and the relative chlorophyll content of each leaf was measured (G). Data represent the mean ± sd of three independent experiments. Asterisks indicate significant differences (Student’s t test; P < 0.05). For all experiments, the first and second leaves of pepper plants were used.
To investigate the mechanism whereby decreased ABA sensitivity in CaAIRF1-silenced pepper plants influences the expression of ABA- and drought-responsive genes, we conducted qRT-PCR analysis (Fig. 5B). Previously, we showed that CaAMP1 and AtRD29B homolog CaOSR1 were significantly induced by ABA and drought stress (Lee and Hwang, 2009; Lim et al., 2015c). Hence, we measured the expression levels of these genes as genetic markers. ABA treatment triggered CaAMP1 and CaOSR1 transcript accumulation; however, the expression levels were lower in TRV2:CaAIRF1 plants than in TRV2:00 plants (Fig. 6E). Moreover, in the absence of ABA, we determined low expression levels of CaOSR1. Next, we measured the expression levels of CaADIP1 and its homologous gene CaADIP2 (Ca05g16320), which are induced by ABA and drought stress (Lim and Lee, 2016). Each of these genes showed a similar expression pattern to that of CaAMP1 andCaOSR1; further, the expression level of CaADIP2 was slightly lower in TRV2:CaAIRF1 plants than in TRV:00 plants (Fig. 6E). Finally, we floated fully expanded leaves detached from TRV2:00 and TRV2:CaAIRF1 plants in stomatal opening solution (SOS) buffer containing 50 μm ABA. The leaves turned yellow only in the presence of ABA (Fig. 6F). In comparison with the leaves of TRV2:00 plants, the leaves of TRV2:CaAIRF1 plants appeared bright green and not yellow. Consistently, the chlorophyll content of these leaves was higher than that of control leaves (Fig. 6G). Our data indicate that CaAIRF1 functions as a positive regulator of ABA-mediated responses, including stomatal closure, induction of stress-responsive genes, and leaf senescence.
Decreased Drought Tolerance of CaAIRF1-Silenced Pepper Plants
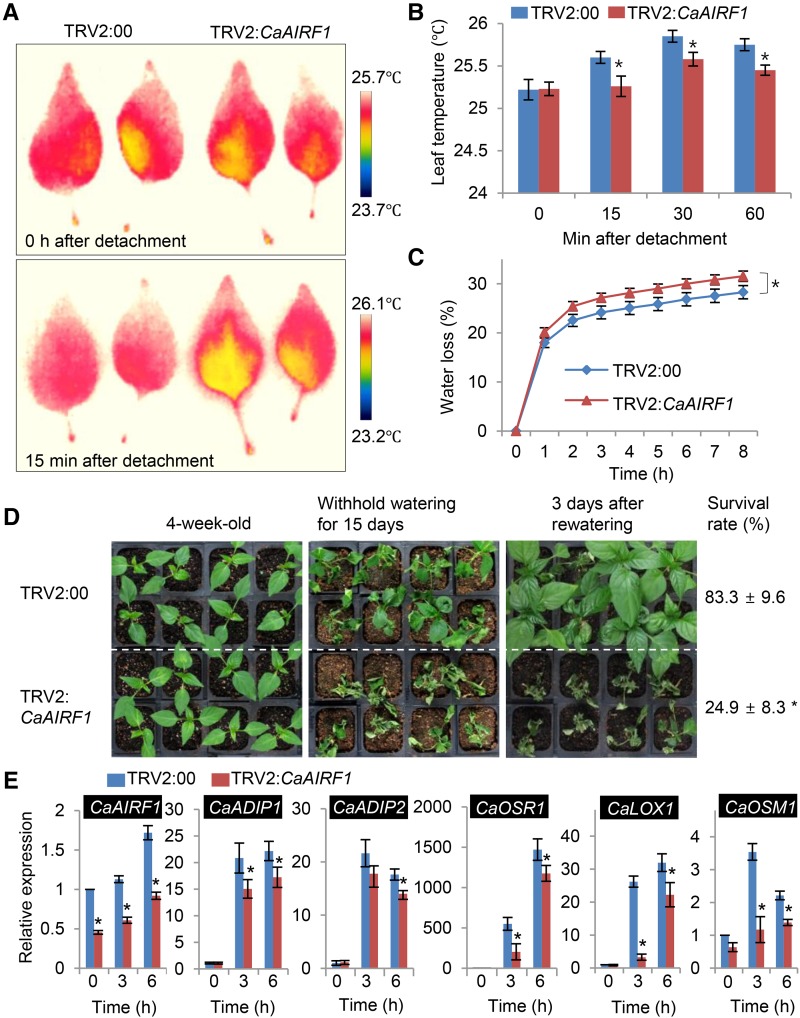
We analyzed the physiological and molecular responses of CaAIRF1-silenced pepper plants to drought stress. First, we detached leaves from TRV2:00 and TRV2:CaAIRF1 plants grown under irrigated conditions and monitored the leaf temperatures of these plants. The leaf temperatures increased rapidly; however, at all time points, the leaf temperatures of TRV2:CaAIRF1 plants were lower than those of TRV2:00 plants (Fig. 7, A and B). Concomitantly, we monitored transpirational water loss by measuring the fresh weight loss of detached leaves. Consistent with the leaf temperatures, 8 h after detachment, leaf fresh weight losses were higher in TRV2:CaAIRF1 plants (31.5%) than in TRV2:00 plants (28.2%; Fig. 7C).
Figure 7.
Increased susceptibility of CaAIRF1-silenced pepper plants to dehydration stress. A and B, Representative thermographic images of detached leaves from TRV2:00 and TRV2:CaAIRF1 pepper plants (A); the mean leaf temperature was measured in the first and second leaves of each line (n = 10) (B). Data represent the mean ± sd of three independent experiments. C, Water loss from the leaves of TRV2:00 and TRV2:CaAIRF1 pepper plants at various time points after detachment of leaves. The leaf fresh weights of each line were measured 8 h after detachment of leaves. Data represent the mean ± se of three independent experiments, each evaluating 16 plants. D, Dehydration sensitivity of CaAIRF1-silenced pepper plants. Four-week-old TRV2:CaAIRF1 and TRV2:00 pepper plants were subjected to dehydration stress by withholding watering for 15 d. Representative images were taken before (left) and after (middle) dehydration stress and 3 d after rewatering (right). The survival rate was measured 3 d after rewatering. Data represent the mean ± sd of three independent experiments. E, RT-PCR analysis of ABA-responsive gene expression in dehydrated leaves of TRV2:00 and TRV2:CaAIRF1 pepper plants. Four-week-old plants of each line were subjected to dehydration stress by removal of their roots. The relative expression level (∆∆CT) of each gene was normalized to that of CaACT1 as an internal control gene. Data represent the mean ± se of three independent experiments. Asterisks indicate significant differences (Student’s t test; P < 0.05).
To investigate drought sensitivity, we subjected 4-week-old TRV2:00 and TRV2:CaAIRF1 plants to drought stress by withholding watering for 15 d (Fig. 7D). Under well-watered conditions, we observed no phenotypic differences between TRV2:00 and TRV2:CaAIRF1 plants. However, after drought stress, TRV2:CaAIRF1 plants displayed a more wilted phenotype than control plants. Moreover, upon rewatering, growth was resumed by >83.3% of TRV2:00 plants but by only 24.9% of TRV2:CaAIRF1 plants. To examine whether decreased drought tolerance in TRV2:CaAIRF1 plants is accompanied by altered expression of ABA- and/or drought-responsive genes, we conducted qRT-PCR analysis of these genes, including CaADIP1, CaADIP2, CaOSR1, CaLOX1, and CaOSM1 (Fig. 7E). Under drought conditions, CaAIRF1 transcription was gradually increased; however, the expression level was significantly lower in TRV2:CaAIRF1 plants than in TRV2:00 plants. Concomitantly, the expression levels of ABA- and/or drought-responsive genes were lower in TRV2:CaAIRF1 plants than in TRV2:00 plants. Taken together, our results indicate that silencing of CaAIRF1 increases drought sensitivity by enhancing transpirational water loss from leaves and suppressing induction of ABA and/or drought-responsive genes.
Enhanced ABA Sensitivity and Increased Drought Tolerance of CaAIRF1-Overexpressing Arabidopsis Plants
To further investigate the involvement of CaAIRF1 in the plant response to ABA, we generated transgenic Arabidopsis plants overexpressing CaAIRF1 under the control of the strong constitutive Pro-35S promoter. We selected two independent T3 homozygous transgenic progenies (Pro-35S:CaAIRF1) for phenotypic analyses (Supplemental Fig. S5). Under normal growth conditions, we observed no significant difference in seed germination or seedling development between wild-type plants and transgenic lines (Supplemental Fig. S7). However, in the presence of ABA, seed germination rates, seedling establishment, and root lengths of Pro-35S:CaAIRF1 plants were lower than those of wild-type plants (Supplemental Fig. S7).
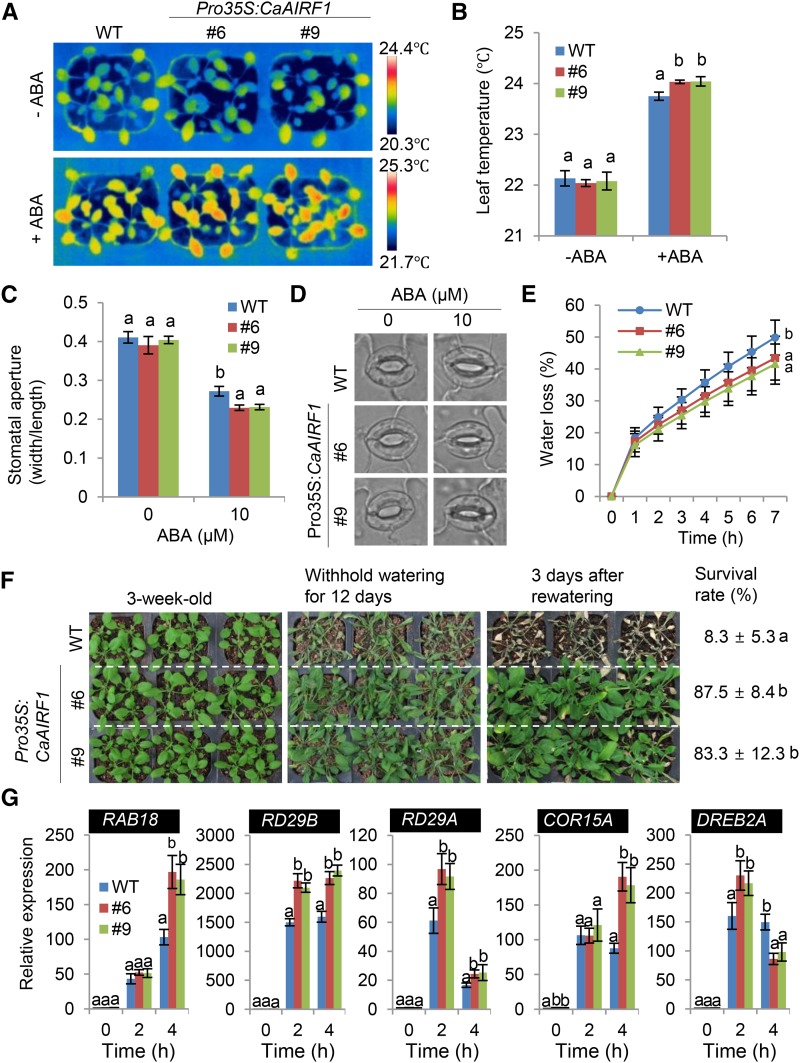
To examine whether CaAIRF1 overexpression influences ABA-mediated stomatal closure, we measured changes in leaf temperature and stomatal aperture after treatment with ABA. Before ABA treatment, leaf temperatures did not differ significantly between Pro-35S:CaAIRF1 and wild-type plants. However, after 6 h of ABA treatment, the leaf temperatures of Pro-35S:CaAIRF1 plants were higher than those of wild-type plants (Fig. 8, A and B). To monitor ABA-induced stomatal closure, we incubated leaf peels of 3-week-old Pro-35S:CaAIRF1 and wild-type plants in SOS buffer supplemented with 10 μm ABA. In comparison with nontreated plants, the stomatal apertures of Pro-35S:CaAIRF1 and wild-type plants were decreased by 41.2% to 42.8% and 33.7%, respectively (Fig. 8, C and D), indicating that CaAIRF1 overexpression increases stomatal closure in response to ABA. We subsequently measured the fresh weight loss of leaves from 3-week-old Pro-35S:CaAIRF1 and wild-type plants 7 h after detachment. In comparison with nontreated plants, the leaf fresh weight losses in Pro-35S:CaAIRF1 and wild-type plants were 44.6% to 46.5% and 53.4%, respectively (Fig. 8E), indicating that increased stomatal closure in Pro-35S:CaAIRF1 plants leads to reduced transpirational water loss.
Figure 8.
Enhanced tolerance of Pro-35S:CaAIRF1 transgenic Arabidopsis plants to dehydration stress. A and B, Representative thermographic images of Pro-35S:CaAIRF1 transgenic Arabidopsis lines and wild-type (WT) plants 6 h after treatment with 50 μm ABA (A); the mean leaf temperatures of the three largest leaves were measured using 10 plants of each line (B). Data represent the mean ± sd of three independent experiments. C and D, Stomatal apertures in Pro-35S:CaAIRF1 transgenic Arabidopsis lines and wild-type plants treated with ABA. Leaf peels harvested from 3-week-old plants of each line were incubated for 2 h in SOS buffer containing 10 μm ABA. The stomatal apertures were measured under the microscope (C) and representative images were taken (D). Data represent the mean ± se of three independent experiments. E, Transpirational water loss from the leaves of Pro-35S:CaAIRF1 transgenic lines and wild-type plants. The fresh weights of each line were measured 7 h after detachment of leaves. Data represent the mean ± se of three independent experiments, each evaluating 16 plants. F, Dehydration sensitivity of Pro-35S:CaAIRF1 transgenic plants. Three-week-old wild-type and transgenic plants were subjected to dehydration stress by withholding watering for 12 d. Representative images were taken, and the percentages of plants that survived were measured after rehydration for 3 d. Data represent the mean ± se of three independent experiments. G, Expression analysis of dehydration-responsive genes in the leaves of Pro-35S:CaAIRF1 transgenic lines and wild-type plants. The relative expression level (∆∆CT) of each gene was normalized to that of Actin8 as an internal control gene. Data represent the mean ± se of three independent experiments. Different letters indicate significant differences (ANOVA; P < 0.05).
To investigate the influence of CaAIRF1 overexpression on drought tolerance, we randomly grew Pro-35S:CaAIRF1 and wild-type plants in soil for 3 weeks under well-watered conditions. We observed no significant phenotypic differences between Pro-35S:CaAIRF1 plants and wild-type plants (Fig. 8F, left). However, when we subjected plants to drought stress by withholding watering for 12 d, Pro-35S:CaAIRF1 plants displayed a less wilted phenotype than wild-type plants (Fig. 8F, middle); moreover, upon rewatering, Pro-35S:CaAIRF1 plants resumed growth faster than wild-type plants (Fig. 8F, right). Three days after rewatering, the survival rates of Pro-35S:CaAIRF1 and wild-type plants were 83.3% to 87.5% and 8.3%, respectively (Fig. 8F). This increased drought tolerance of Pro-35S:CaAIRF1 plants was accompanied by up-regulation of drought-responsive marker genes. qRT-PCR analysis revealed higher expression levels of RAB18, RD29B, RD29A, COR15A, and DREB2A in dehydrated leaves of Pro-35S:CaAIRF1 plants than in those of wild-type plants (Fig. 8G). Our data indicate that CaAIRF1 positively regulates drought tolerance in Arabidopsis and pepper plants by modulating the expression of drought-responsive marker genes and ABA-mediated stomatal closure.
CaAIRF1 Suppression of CaADIP1-Induced ABA Hyposensitivity
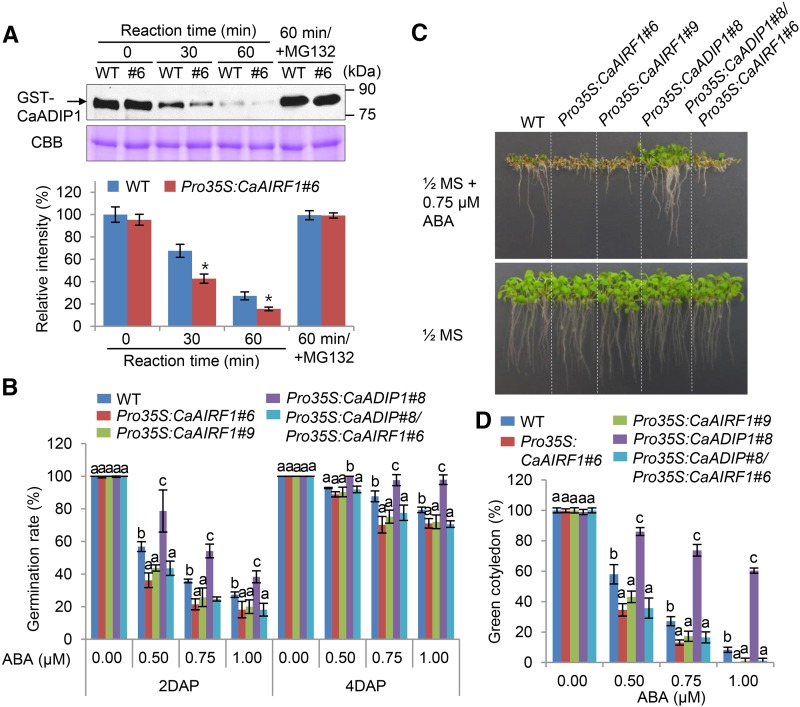
To more fully elucidate the functional relationship between CaAIRF1 and CaADIP1, we performed a cell-free degradation assay using Pro-35S:CaAIRF1 plants. Purified CaADIP1-GST protein was incubated with crude protein extracts prepared from the leaves of Pro-35S:CaAIRF1 #6 and wild-type plants. Consistent with our data for the ABA-promoted degradation of CaADIP1 (Fig. 1, B and C), western blotting revealed gradual degradation of the CaADIP1-GST protein. After 60 min of incubation, the CaADIP1-GST protein was degraded by 85% (Fig. 9A). This pattern was accelerated by overexpression of CaAIRF1; after incubation for 60 min, the levels of CaADIP1-GST protein in wild-type and Pro-35S:CaAIRF1 #6 plants were reduced by 73% and 85%, respectively. However, after treatment with MG132, the level of CaADIP1 remained unchanged. We observed a similar pattern of degradation in the leaves of Pro-35S:CaAIRF1 #9 plants (Supplemental Fig. S7). Our data indicate that CaAIRF1 mediates degradation of CaADIP1 via the 26S proteasome system. Moreover, they imply that CaAIRF1 regulates CaADIP1-induced ABA hyposensitivity (Lim and Lee, 2016). To validate this hypothesis, we generated Pro-35S:CaAIRF1/Pro-35S:CaADIP1 double transgenic plants by crossing the two independent lines Pro-35S:CaADIP1 (line #8) and Pro-35S:CaAIRF1 (line #6). RT-PCR analysis revealed that double transgenic plants showed high transcript levels of CaAIRF1 and CaADIP1 (Supplemental Fig. S5B). We germinated wild-type, Pro-35S:CaAIRF1, Pro-35S:CaADIP1, and Pro-35S:CaAIRF1/Pro-35S:CaADIP1 seeds on Murashige and Skoog (MS) medium supplemented with various concentrations of ABA. In the absence of ABA, we determined no significant differences in germination rates (Fig. 9B). However, in the presence of ABA, Pro-35S:CaAIRF1 and Pro-35S:CaADIP1 plants exhibited hypersensitive and hyposensitive phenotypes, respectively. As predicted, the germination rate of Pro-35S:CaAIRF1/Pro-35S:CaADIP1 plants did not differ significantly from that of the two Pro-35S:CaAIRF1 plant lines; this pattern was also observed at the seedling stage. Seven days after plating, the root length and rate of cotyledon greening in Pro-35S:CaAIRF1/Pro-35S:CaADIP1 plants did not differ significantly from that of the two Pro-35S:CaAIRF1 plant lines (Fig. 9, C and D). Our data indicate that CaAIRF1 inhibits CaADIP1-induced ABA hyposensitivity during the seed germination and seedling stages.
Figure 9.
CaAIRF1-mediated inhibition of CaADIP1 function during germination and seedling development. A, Cell-free degradation assay for CaADIP1. The GST-CaADIP1 protein (500 ng) was incubated for the indicated periods with crude extracts prepared from the leaves of 4-week-old plants of the Pro-35S:CaAIRF1 transgenic line #6 in the presence or absence of MG132. Immunoblot analysis was performed using anti-GST antibody (top). CBB staining indicates equal loading of crude extract. The relative intensities of the GST-CaADIP1 fusion proteins were measured using Image J 1.46r (http://imagej.nih.gov/ij) software (bottom). Data represent the mean ± sd of three independent experiments; asterisks indicate significant differences (Student’s t test; P < 0.05). B, Germination rates of transgenic lines and wild-type (WT) plants on 0.5× MS medium supplemented with various concentrations of ABA. The numbers of seeds with emerged radicles were counted 2 d and 4 d after plating (DAP). Data represent the mean ± sd of three independent experiments, each evaluating 50 seeds. C, Seedling growth of transgenic lines and wild-type plants exposed to ABA. The seedlings were grown vertically in 0.5× MS containing 0.75 μm ABA. After 7 d, representative images were taken. D, Rate of cotyledon greening of transgenic lines and wild-type plants exposed to ABA. The numbers of seedlings in each line with expanded cotyledons were counted 7 d after plating. Data represent the mean ± sd of three independent experiments, each evaluating 45 seeds. Different letters indicate significant differences (ANOVA; P < 0.05).
DISCUSSION
An increasing number of studies have shown that the ubiquitin-proteasome system is involved in regulation of the ABA-signaling pathway at multiple steps. Several ABA signaling-related E3 ligases have been isolated from Arabidopsis; the identified targets of these ligases are mainly restricted to the final stage of ABA signaling, and they include transcription factors such as ABF2 (Kim et al., 2004), ABI3 (Zhang et al., 2005), and ABI5 (Seo et al., 2014). Recent studies have identified different types of E3 ligases that modulate the stability of core ABA-signaling components involved in the initial stage of ABA perception. The single subunit E3 ligase RSL1 and the E3 ligase complex CRL4 (Cullin4-RING E3 ligase) mediate ubiquitination and protein degradation in some ABA receptor PYR/PYL/RCAR proteins, including PYR1, PYL4, PYL8, and PYL9 in Arabidopsis (Bueso et al., 2014; Irigoyen et al., 2014). The U-box E3 ligases PUB12 and PUB13 interact with and ubiquitinate clade A PP2C ABI1 only in the presence of ABA and PYR1 (Kong et al., 2015). RING-type E3 ligase RGLG1 and RGLG5 interact with PP2CA, HAB2, and ABI2, which is promoted by exogenous ABA, and ubiquitinate these PP2Cs in the absence of ABA receptor (Wu et al., 2016). Here, we have demonstrated that the pepper RING-type E3 ligase CaAIRF1 interacts with and ubiquitinates CaADIP1, which is homologous to Arabidopsis HAB1.
A core ABA-signaling pathway is well established in Arabidopsis; in this pathway, at least six clade A PP2Cs—ABI1, ABI2, HAB1, HAB2, AHG1, and PP2CA/AHG3—negatively regulate ABA (Lee and Luan, 2012). The negative regulatory role of PP2Cs has been reported in several plant species, including Arabidopsis and rice, indicating evolutionary conservation of PP2Cs (González-García et al., 2003; Singh et al., 2015). Previously, we identified the ABA-induced protein phosphatase CaADIP1 in pepper plants. CaADIP1 plays a negative role in the ABA-signaling and drought stress responses; moreover, CaADIP1-induced ABA hyposensitivity is inhibited by the RCAR3/PYL8 homolog CaRLP1 (Lim and Lee, 2016). In the current study, we used a cell-free degradation assay to show that the CaADIP1 protein is degraded in pepper plants, mainly via the 26S proteasome system. Our data provide a valuable new insight into modulation of CaADIP1 in plant cells.
Based on a Y2H screening assay with CaADIP1 as bait, we selected CaAIRF1 as a candidate protein responsible for CaADIP1 degradation. In particular, CaAIRF1 is clustered in the same clade as E3 ligase proteins from the family Solanaceae (Supplemental Fig. S8); moreover, CaAIRF1 shares the highest identity (89%) and similarity (93%) with putative E3 ubiquitin-protein ligase containing RING domain RF298 from tomato (Solanum lycopersicum) and potato (Solanum tuberosum; Supplemental Fig. S1). CaAIRF1 is localized in the nucleus, where it interacts with CaADIP1 (Fig. 2, C and F). Our in vivo and in vitro ubiquitination assays revealed that CaAIRF1 is self-ubiquitinated and ubiquitinates CaADIP1 (Fig. 4). Hence, we postulated that the RING-type E3 ligase CaAIRF1 regulates CaADIP1 stability in the nucleus. We examined CaAIRF1-mediated CaADIP1 degradation using CaAIRF1-silenced pepper plants and Pro-35S:CaAIRF1 transgenic Arabidopsis plants. Our data suggest that the level of CaADIP1 is determined by the expression level of CaAIRF1 in pepper and Arabidopsis leaves (Figs. 5 and 9). Intriguingly, 60% of the CaADIP1 protein was degraded after 30 min of incubation with crude protein extracts prepared from the leaves of wild-type Arabidopsis plants (Fig. 9A). This finding may be explained by the existence of CaAIRF1 homologs in Arabidopsis. Our sequence similarity analysis revealed that CaAIRF1 shares 38.2% identity and 53.2% similarity with Arabidopsis E3-Ub ligase (At4g03000; Supplemental Fig. S1).
In contrast to CaADIP1, CaAIRF1 functions as a positive regulator of the ABA-signaling and drought stress responses, including stomatal closure and induction of stress-responsive genes. CaAIRF1-silenced pepper plants exhibited reduced ABA sensitivity and decreased drought tolerance. However, 35S promoter-driven expression of CaAIRF1 led to a contrasting phenotype (Figs. 8 and 9). The ABA-hypersensitive phenotype exhibited by Pro-35S:CaAIRF1 plants is likely derived from CaAIRF1-mediated modulation of AtPP2CA. CaAIRF1 interacted with only one of nine clade A PP2Cs from Arabidopsis, namely, AtPP2CA, as prey (Supplemental Fig. S9A). In our cell-free degradation assay, AtPP2CA-GST was more completely degraded by crude proteins extracted from Pro-35S:CaAIRF1 plants than by those extracted from wild-type plants (Supplemental Fig. S9B). Consistently, our in vivo ubiquitination assay revealed that CaAIRF1 can ubiquitinate AtPP2CA (Supplemental Fig. S9C), indicating that CaAIRF1 modulates PP2C stability in Arabidopsis and pepper plants. The observed phenotypic differences in response to ABA and drought stress facilitated elucidation of the genetic relationship between CaAIRF1 and CaADIP1. In a previous study, we observed no phenotypic differences between control and CaADIP1-silenced pepper plants in response to drought stress, because of the complexity and functional redundancy of pepper PP2Cs (Lim and Lee, 2016). Additionally, we were unable to induce simultaneous silencing of CaAIRF1 and CaADIP1 in pepper plants. Owing to the molecular and technical restrictions involved in genetic investigation of pepper plants, we generated transgenic Arabidopsis plants overexpressing CaAIRF1 and CaADIP1. CaAIRF1 is associated with degradation of CaADIP1, and therefore we postulated that co-overexpression of CaAIRF1 and CaADIP1 confers enhanced ABA sensitivity, similar to that of the Pro-35S:CaAIRF1 transgenic lines. As predicted, CaADIP1-induced ABA hyposensitivity was strongly inhibited by CaAIRF1 overexpression in Pro-35S:CaADIP1/Pro-35S:CaAIRF1 double mutants (Fig. 9, B–D). Our results indicate that CaAIRF1 acts upstream of CaADIP1 in ABA signaling, thereby supporting our hypothesis that CaAIRF1 mediates negative modulation of CaADIP1 stability.
In response to ABA and drought stress, CaAIRF1 altered the expression levels of ABA- and drought stress-responsive genes (Figs. 6E, 7E, and 8G; Supplemental Fig. S10). Several studies have demonstrated that expression levels of stress-responsive genes are closely related to stress tolerance in Arabidopsis. However, data regarding this relationship in pepper plants are lacking. Hence, we selected six pepper genes—CaAMP1 (Lee and Hwang, 2009), CaOSM1 (Hong et al., 2004), CaOSR1 (Lim et al., 2015c), CaLOX1 (Lim et al., 2015b), CaADIP1, and CaADIP2 (Lim and Lee, 2016)—for which the expression levels were previously shown to be associated with ABA sensitivity and drought tolerance. CaAIRF1-silenced pepper plants showed consistent expression patterns (Fig. 7E). Increased ABA sensitivity in Pro-35S:CaAIRF1 plants was accompanied by enhanced expression at 2 h after treatment of the ABA-responsive genes RAB18 and RD29B (Supplemental Fig. S10A). However, we observed no significant difference in expression of the ABA biosynthesis-related gene NCED3 between wild-type and Pro-35S:CaAIRF1 plants, either in the presence or absence of ABA (Supplemental Fig. S10B). Thus, the increased ABA sensitivity shown by Pro-35S:CaAIRF1 plants is unlikely to be derived from altered ABA accumulation. In ABA-mediated drought signaling, CaAIRF1 influences the expression levels of stress-responsive genes directly or indirectly; however, the mechanisms underlying this regulatory process remain unclear.
Interestingly, CaADIP1 was regulated by CaAIRF1 at the transcriptional level in pepper plants, whereas the expression level of CaAIRF1 was not determined by CaADIP1 (Fig. 6E; Supplemental Fig. S11). After ABA treatment, the expression levels of CaAIRF1 in CaADIP1-silenced pepper plants did not differ from those of control plants, indicating that CaAIRF1 acts upstream of CaADIP1 at the transcriptional and posttranslational levels. CaADIP1 and CaAIRF1 are strongly induced in pepper leaves by ABA (Lim and Lee, 2016). Hence, our findings indicate that ABA can regulate the CaADIP1 level positively or negatively according to the expression stage of CaADIP1. Similar to CaADIP1, many clade A PP2Cs in Arabidopsis are induced by ABA (Bhaskara et al., 2012). Thus, ABA triggers strong induction of PP2C genes to inhibit ABA signaling, thereby reinstating normal plant physiological conditions. In this regard, low expression levels of CaADIP1 may be manifested as reduced ABA sensitivity in CaAIRF1-silenced pepper plants. In contrast, ABA also promotes degradation of CaADIP1 via CaAIRF1-mediated ubiquitination. This different pattern between the transcriptional and protein levels has also been demonstrated for ABI1 (Kong et al., 2015). As a negative regulator, ABA-induced degradation of PP2C proteins such as CaADIP1 and ABI1 may be essential for ABA signal transduction.
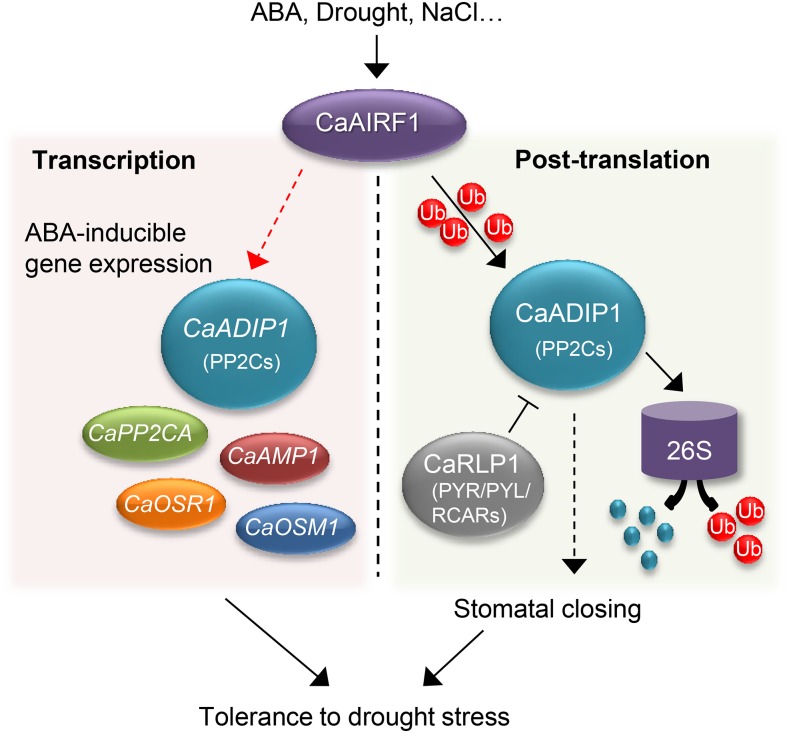
In summary, our findings highlight a potential new route to fine-tune regulation of the ABA-signaling pathway and drought stress response in pepper plants via CaAIRF1 and CaADIP1. We propose that the RING-type E3 ligase CaAIRF1 interacts with and ubiquitinates the clade A PP2C CaADIP1 (Fig. 10). As the upstream partner, CaAIRF1 regulates CaADIP1 at the transcriptional and posttranslational levels. ABA and stress conditions induce CaAIRF1 gene expression and promote CaADIP1 transcript accumulation; whether this occurs directly or indirectly is not clear from the current study. Additionally, CaAIRF1 promotes CaADIP1 degradation via the Ub-26S proteasome system, leading to enhanced ABA sensitivity and increased drought tolerance. Together with the PYR/PYL/RCAR proteins, ubiquitin–proteasome system-mediated regulation of PP2Cs is a key regulatory mechanism in ABA signaling. Identification of additional E3 ligases and target PP2Cs of CaAIRF1 will help to clarify the route to fine-tune regulation at the early stage of the ABA-signaling pathway.
Figure 10.
Schematic representation of the functional role of CaAIRF1 in the ABA-signaling pathway that mediates drought stress tolerance in pepper plants. We proposed that CaAIRF1 plays a positive role in ABA signaling via regulation of CaADIP1 protein stability and CaADIP1 transcript accumulation. This leads to alteration of downstream responses, including ABA-inducible gene expression and stomatal closure, and consequently controls plant tolerance to drought stress. Dashed lines indicate direct or indirect actions of CaAIRF1 and CaADIP1. Arrows indicate promotion actions; lines with end bar indicate inhibitory actions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of pepper (Capsicum annuum cv Nockwang) and tobacco (Nicotiana benthamiana) were sown in a steam-sterilized compost soil mix (peat moss, perlite, and vermiculite, 5:3:2, v/v/v), sand, and loam soil (1:1:1, v/v/v). The pepper plants were raised in a growth room at 27 ± 1°C under white fluorescent light (130 µmol photons m−2 s−1; 16 h/d). The tobacco plants were raised in a growth chamber at 25 ± 1°C and under a 16-h-light/8-h-dark cycle and white fluorescent light (130 µmol photons m−2 s−1). Arabidopsis (Arabidopsis thaliana; ecotype Col-0) seeds were germinated on MS salt (Duchefa Biochemie) supplemented with 1% Suc and Microagar (Duchefa Biochemie) in a growth chamber at 24°C and under a 16-h light/8-h dark cycle. Arabidopsis seedlings were grown in a 9:1:1 ratio of peat moss, perlite, and vermiculite under controlled environmental conditions (130 μmol photons m−2 s−1; 24°C; 60% relative humidity; 16-h-light/8-h-dark cycle). All seeds were vernalized at 4°C for 2 d to synchronize germination.
Virus-Induced Gene Silencing
CaAIRF1-silenced pepper plants were generated using the tobacco rattle virus (TRV)-based virus-induced gene silencing system as described previously (Lim and Lee, 2014). A 321-bp fragment of the CaAIRF1 cDNA was inserted into the pTRV2 vector and introduced into Agrobacterium tumefaciens strain GV3101 via electroporation. A. tumefaciens strain GV3101 containing pTRV1, pTRV2:00, and pTRV2:CaAIRF1 was coinfiltrated into the fully expanded cotyledons of pepper plants (OD600 = 0.2 for each construct). Plants were placed in a growth room at 24°C and maintained under a 16-h-light/8-h-dark cycle for growth and spread of the virus.
Generation of Transgenic Arabidopsis Plants
A. tumefaciens-mediated transformation of Arabidopsis was performed using the floral dip method (Clough and Bent, 1998); all mutants generated in this study were in the Col-0 background. For Pro-35S:CaAIRF1, a 2,709-bp full-length cDNA sequence of CaAIRF1 (CA1g23600, Pepper Genome Database version 1.55) was integrated into the pK2GW7 binary vector. To isolate Pro-35S:CaAIRF1 transgenic lines, seeds harvested from the putative transformed plants were sown on MS agar plates containing 50 μg mL−1 of kanamycin. T3 homozygous lines were used for further phenotypic analysis. For inducing co-overexpression of CaAIRF1 and CaADIP1, Pro-35S:CaAIRF1 mutant line #6 (male) was crossed with Pro-35S:CaADIP1 line #8 (female; Lim and Lee, 2016). Transgenic mutants were initially selected on MS agar plates containing 50 μg mL−1 of kanamycin and 25 μg mL−1 of phosphinothricin. Based on PCR analysis, F2 plants showing coamplification of CaAIRF1 and CaADIP1 were selected and grown to obtain homozygous F3 seeds for further analysis.
ABA, H2O2, Drought, and NaCl Treatments and Phenotypic Analyses
To investigate CaAIRF1 expression in pepper plants, six-leaf stage pepper plants were treated with ABA (100 μm), H2O2 (100 μm), drought, or NaCl (200 mm) as described previously (Lim et al., 2015c). Pepper leaves were harvested at 0, 2, 6, 12, and 24 h after each treatment. For qRT-PCR analysis, four-leaf stage CaAIRF1-silenced pepper plants and 4-week-old Pro-35S:CaAIRF1 transgenic Arabidopsis mutants were treated with 10 and 50 μm ABA, or were carefully removed from the soil before being subjected to dehydration stress and harvested at the indicated time points after treatment.
For the seedling growth test, 100 seeds of wild-type and Pro-35S:CaAIRF1 transgenic Arabidopsis lines were sown on plates containing MS agar medium supplemented with various concentrations of ABA; seedlings with green cotyledons were counted 7 d later. Concomitantly, seedlings from each line were vertically grown on MS plates for 7 d and their root lengths were measured. The seedlings were grown at 24°C under a light intensity of 130 μmol photons m−2 s−1 and a 16-h-light/8-h-dark cycle.
Dehydration tolerance assays were conducted as described previously (Lim et al., 2015c). Briefly, four-leaf stage CaAIRF1-silenced pepper plants and 2-week-old Pro-35S:CaAIRF1 Arabidopsis seedlings were subjected to dehydration stress by withholding watering for 15 or 12 d, respectively. The survival rates of the plants were calculated 3 d after rewatering. To measure transpirational water loss, leaves were detached from four-leaf stage pepper plants and 3-week-old Arabidopsis plants, and the loss of fresh weight was determined at the indicated time points. All the experiments were performed at least in triplicate.
Thermal Imaging
Thermal imaging analysis was performed as described previously (Lim et al., 2015c). Four-week-old pepper plants having fully expanded first and second leaves and 3-week-old Arabidopsis plants were treated with 50 μm ABA or subjected to dehydration stress by withholding watering for 15 or 12 d, respectively. Thermal images were obtained using an infrared camera (FLIR systems; T420), and leaf temperatures were measured with FLIR Tools+ version 5.2 software.
Stomatal Aperture Bioassay
The stomatal aperture bioassay was performed as described previously (Lim and Lee, 2014). Briefly, epidermal peels were stripped from rosette leaves of 3-week-old Arabidopsis plants and the first and second leaves of pepper plants 10 d after agroinfiltration; the peels were then floated in SOS (50 mm KCl, 10 mm MES-KOH, pH 6.15, and 10 µm CaCl2) in the light. After incubation for 3 h, the buffer was replaced with fresh SOS containing 10 or 20 μm ABA. After an additional 2.5 h of incubation, 100 stomata in each individual sample were randomly observed under a Nikon Eclipse 80i microscope, and the width and length of stomata were measured using Image J 1.46r (http://imagej.nih.gov/ij). Each experiment was performed in triplicate.
RNA Isolation and qRT-PCR
Total RNA isolation and RT-PCR analyses were performed as described previously, using the Arabidopsis and pepper leaf tissues that had been treated with ABA or subjected to drought stress (Lim et al., 2015c). cDNA was synthesized using a Transcript First Strand cDNA Synthesis kit (Roche) with 1 µg of total RNA according to the manufacturer’s instructions. For qRT-PCR analysis, the synthesized cDNA was amplified in a CFX96 Touch Real-Time PCR detection system (Bio-Rad) with iQSYBR Green Supermix and specific primers (Supplemental Table S1). All reactions were performed in triplicate. The relative expression level of each gene was calculated using the ∆∆Ct method, as described previously (Livak and Schmittgen, 2001). The Arabidopsis actin8 (AtACT8) and pepper actin 1 (CaACT1) genes were used for normalization.
Floating Leaf Assay and Measurement of Chlorophyll Content
The first and second fully expanded leaves of CaAIRF1-silenced plants and control pepper plants (4 weeks old) were floated in SOS buffer containing 50 μm ABA and incubated at 24°C under a light intensity of 130 μmol photons m−2 s−1 and a 16-h-light/8-h-dark cycle. After 6 d, the chlorophyll content was determined spectrophotometrically according to the following formula: Chla + b = (5.24 × A664) + (22.24 × A648) as described previously (Lim et al., 2015b).
Subcellular Localization Analysis
The coding regions of CaAIRF1 without the stop codon were inserted into the GFP-fused binary vector p326GFP. A. tumefaciens strain GV3101 carrying each construct was combined with strain p19 (1:1 ratio; OD600 = 0.5) and coinfiltrated into fully expanded leaves of 5-week-old tobacco plants. At 3 d after infiltration, leaf discs were cut and the lower epidermal cells were examined under a confocal microscope (510 UV/Vis Meta; Zeiss) equipped with LSM Image Browser software.
Y2H Assay
Y2H assays were conducted as described previously (Lee et al., 2007). The cDNA fragments of CaADIP1 or CaAIRF1 were subcloned into pGBKT7 or pGADT7 vectors, respectively, and introduced into the yeast strain AH109 using the lithium acetate-mediated transformation method (Ito et al., 1983). To evaluate the interaction between bait and prey proteins, transformant candidates were selected on SC-Leu-Trp and SC-adenine-His-Leu-Trp media. Next, 10-fold serial dilutions were prepared from each yeast cell culture (OD600 = 0.5), and 5 μL of each sample was spotted onto SC-Leu-Trp medium or SC-adenine-His-Leu-Trp medium.
Bimolecular Fluorescence Complementation Assay
The bimolecular fluorescence complementation assay was performed as described previously (Lim and Lee, 2016). To generate the BiFC constructs, full-length cDNAs of CaAIRF1 and CaADIP1 without stop codons were subcloned into Pro-35S:VYNE and Pro-35S:CYCE vectors using the LR reaction (Waadt et al., 2008). For transient expression, A. tumefaciens strain GV3101 harboring each construct was syringe-infiltrated into the leaves of 5-week-old tobacco plants (OD600 = 0.5). At 3 d after infiltration, microscopic analysis was performed as described above.
Coimmunoprecipitation Assay
The co-IP assay was performed as described previously (Lim and Lee, 2016). Pro-35S:CaADIP1-HA and Pro-35S:CaAIRF1∆1-573H864Y/C868S-GFP or Pro-35S:GFP as an empty vector control were coexpressed in the leaves of tobacco plants using agroinfiltration as described above. After 3 d, leaves were harvested. Proteins were extracted using buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, 0.1% Nonidet P-40, and 13 Complete protease inhibitor (Roche). The supernatants were collected after centrifugation at 16,000g at 4°C and used in the co-IP experiment. Protein extracts were incubated with prewashed GFP-trap magnetic beads (Chromotek) for 1 h at 4°C with rotation. After washing three times with 500 μL of extraction buffer, beads were resuspended in 100 μL of 2× SDS sample buffer and subjected to SDS-PAGE. Immunodetection was performed using anti-HA (1:1,000; Santa Cruz Biotechnology) and anti-GFP (1:1,000; Santa Cruz Biotechnology) antibodies.
Purification of Recombinant Protein
Expression of the MBP-CaAIRF1∆1-573 and MBP-CaAIRF1∆1-573H864Y/C868S recombinant proteins in bacterial cells was performed as described previously (Park et al., 2015). The full-length CaAIRF1 cDNA sequence was inserted into the pMAL-c2X vector (New England Biolabs) and introduced into Escherichia coli strain c+cell. The MBP-CaAIRF1 fusion protein was induced and purified according to the manufacturer’s instructions (New England Biolabs). For CaADIP1, the coding sequence of CaADIP1 was introduced into the pGEX4T-3 vector (GE Healthcare Bio-Sciences) and was then transformed into E. coli strain c+cell. Recombinant GST-CaADIP1 fusion proteins were affinity purified from bacterial lysates, using Glutathione-Sepharose 4 Fast Flow (GE Healthcare Bio-Science) according to the manufacturer’s instructions. The purified protein concentration was measured using a Pierce BCA protein assay kit (Thermo Scientific), and the purified proteins were stored at −20°C until use in the in vitro ubiquitination assay.
In Vitro and in Vivo Ubiquitination Assays
The in vitro self-ubiquitination assay was performed as described previously (Park et al., 2015). The purified MBP-CaAIRF1∆1-573 or MBP-CaAIRF1∆1-573H864Y/C868S (500 ng) was incubated with recombinant human UBE1 (Boston Biochemicals), recombinant his-tagging human UbcH5b (Enzo Life Science), and bovine ubiquitin (Sigma-Aldrich) in reaction buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.05 mm ZnCl2, 1 mm Mg-ATP, 0.2 mm dithiothreitol [DTT], 10 mm phosphocreatine, and 0.1 units of creatine kinase [Sigma-Aldrich]) at 30°C for 3 h. To examine whether CaAIRF1 mediates CaADIP1 ubiquitination, 50 ng of the GST-CaADIP1 fusion protein were added to the ubiquitination mixture and the mixture was incubated for 2 h. The reacted proteins were separated using SDS-PAGE and analyzed using immunoblotting with anti-Ub (Santa Cruz Biotechnology), anti-MBP (New England Biolabs), and anti-GST (Santa Cruz Biotechnology) antibodies.
For the in vivo ubiquitination assay, Pro-35S:CaADIP1-HA and Pro-35S:CaAIRF1-GFP or Pro-35S:GFP as an empty vector control were coexpressed in the leaves of tobacco plants as described above. To prevent protein degradation, 50 μm MG132 was infiltrated 12 h before sampling. To exclude any possible effects of further protein synthesis on CaADIP1 stability, 50 μm CHX was infiltrated (with MG132) 16 h before sampling. Each leaf sample was extracted with native extraction buffer [50 mm Tris-MES (pH 8.0), 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, 5 mm DTT, and Complete protease inhibitor (Roche)] (Liu et al., 2010). Following immunoprecipitation with GFP-trap magnetic beads (Chromotek), purified proteins were subjected to SDS-PAGE and immunoblot analysis with anti-Ub (Santa Cruz Biotechnology) and anti-GFP (Santa Cruz Biotechnology) antibodies. Ubiquitinated proteins were purified from the extracts using the UbiQapture-Q matrix (Enzo Life Science) according to the manufacturer’s instructions.
Cell-Free Degradation Assay
Crude protein extracts were prepared from the leaves of 4-week-old pepper plants that had been treated with 100 µm ABA for 12 h and from seedlings of Pro-35S:CaAIRF1 transgenic Arabidopsis line #6, using extraction buffer (25 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm DTT, 0.1% Triton X-100, 10 mm ATP, and 10 mm NaC) The GST-CaADIP1 fusion proteins (500 ng) were incubated with crude protein extracts (50 µg of total protein) for 0.5 h and 1 h and additionally for 1 h in the presence of 50 µm MG132. To evaluate protein degradation, immunoblotting was performed using anti-GST. All assays were independently repeated three times with two replicates per trial.
Generation of the ProCaAIRF1:GFP Fusion Construct
The sequences of the upstream region of CaAIRF1 were obtained from the pepper genome database (https://solgenomics.net/), and three specific primers were designed (Supplemental Table S1). In the first-round PCR with forward and reverse1 primers, the 1,874-bp DNA sequences, including the 370-bp coding sequence at the 5′ end of CaAIRF1, were amplified from genomic DNA prepared from pepper leaves and used as a template for the second nested amplification. The 1,504-bp CaAIRF1 promoter was specifically isolated using forward and reverse2 primers (Supplemental Table S1) and integrated into the pHGWFS7 binary vector to generate the ProCaAIRF1:GFP construct for agroinfiltration.
Statistical Analyses
To determine significant differences between genotypes, statistical analyses were performed using Student’s t test or one-way ANOVA, followed by Fisher’s lsd test. Results were considered significant at P < 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, GenBank/EMBL databases, or the Pepper Genome database under the following accession numbers: pepper CaAIRF1 (CA01g23600; KU748268), CaACT1 (CA12g08730; GQ339766), CaADIP1 (CA03g35550; KM403424), CaADIP2 (CA05g16320; KU748269), CaOSR1 (CA03g17780; KT693385), CaAMP1 (CA03g19350; AY548741), CaOSM1 (CA00g28760; AY262059), and CaLOX1 (CA00g46890; FJ377708); Arabidopsis Actin8 (At1g49240), RAB18 (At5g66400), RD29B (At5g52300), RD29A (At5g52310), COR15A (At2g42540), and DREB2A (At5g05410); Solanum lycopersicum RF298-like (XP_004229474), Solanum tuberosum RF298-like (XP_006365281), Citrus sinensis RF298-like isoform X1 (XP_006489214), Glycine max RF298-like (XP_003542905), Medicago truncatula RF298-like (AES64310), Theobroma cacao RING/U-box (XP_007035383), Arabidopsis thaliana RF298 (NP_192209; AT4G03000), and Arabidopsis lyrata protein-binding protein (XP_002874883).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Multiple alignments of the CaAIRF1 protein.
Supplemental Figure S2. Interaction of CaAIRF1∆1-573 with CaADIP1.
Supplemental Figure S3. Interaction of CaAIRF1H864Y/C868S with CaADIP1 in the nucleus.
Supplemental Figure S4. Thermal imaging analysis of CaAIRF1-silenced pepper plants.
Supplemental Figure S5. Expression of CaAIRF1 and CaADIP1 genes in Arabidopsis transgenic lines.
Supplemental Figure S6. Enhanced ABA sensitivity of Pro-35S:CaAIRF1 transgenic Arabidopsis plants during germination and seedling development.
Supplemental Figure S7. Cell-free degradation assay for CaADIP1.
Supplemental Figure S8. Phylogenetic tree analysis of CaAIRF1 and its homologous proteins in higher plants.
Supplemental Figure S9. Interaction of CaAIRF1∆1-573 with Arabidopsis PP2CA protein.
Supplemental Figure S10. Relative expression levels of ABA biosynthesis and ABA-responsive genes in the leaves of Pro-35S:CaAIRF1 transgenic Arabidopsis plants.
Supplemental Figure S11. Relative expression levels of the CaAIRF1 gene in the leaves of CaADIP1-silenced pepper plants.
Supplemental Table S1. Sequences of primers used in this study.
Supplemental Data Set S1. Sequence alignment in fasta format.
Supplementary Material
Glossary
- ABA
abscisic acid
- BiFC
bimolecular fluorescence complementation
- CHX
cycloheximide
- co-IP
coimmunoprecipitation
- DTT
dithiothreitol
- MS
Murashige and Skoog
- qRT-PCR
quantitative RT-PCR
- SOS
stomatal opening solution
- Ub
ubiquitin
- Y2H
yeast two-hybrid
Footnotes
This work was supported by grants from “the Next-Generation BioGreen 21 Program for Agriculture and Technology Development (project no. PJ01101001),” by the Rural Development Administration, and by the Research Foundation of Korea funded by the government of the Republic of Korea (NRF-2015R1A2A2A01002674).
References
- Bhaskara GB, Nguyen TT, Verslues PE (2012) Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 160: 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Rodriguez L, Lorenzo-Orts L, Gonzalez-Guzman M, Sayas E, Muñoz-Bertomeu J, Ibañez C, Serrano R, Rodriguez PL (2014) The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J 80: 1057–1071 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL (1998) The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci USA 95: 2727–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Rodríguez D, Nicolás C, Rodríguez PL, Nicolás G, Lorenzo O (2003) Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol 133: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Jung HW, Lee BK, Lee SC, Lee YK, Hwang BK (2004) An osmotin-like protein gene, CAOSM1, from pepper: differential expression and in situ localization of its mRNA during pathogen infection and abiotic stress. Physiol Mol Plant Pathol 64: 301–310 [Google Scholar]
- Irigoyen ML, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, et al. (2014) Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, et al. (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Hwang BK (2009) Functional roles of the pepper antimicrobial protein gene, CaAMP1, in abscisic acid signaling, and salt and drought tolerance in Arabidopsis. Planta 229: 383–391 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Luan S (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35: 53–60 [DOI] [PubMed] [Google Scholar]
- Li Z, Hu Q, Zhou M, Vandenbrink J, Li D, Menchyk N, Reighard S, Norris A, Liu H, Sun D, et al. (2013) Heterologous expression of OsSIZ1, a rice SUMO E3 ligase, enhances broad abiotic stress tolerance in transgenic creeping bentgrass. Plant Biotechnol J 11: 432–445 [DOI] [PubMed] [Google Scholar]
- Lim CW, Baek W, Jung J, Kim JH, Lee SC (2015a) Function of ABA in stomatal defense against biotic and drought stresses. Int J Mol Sci 16: 15251–15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CW, Han SW, Hwang IS, Kim DS, Hwang BK, Lee SC (2015b) The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol 56: 930–942 [DOI] [PubMed] [Google Scholar]
- Lim CW, Hwang BK, Lee SC (2015c) Functional roles of the pepper RING finger protein gene, CaRING1, in abscisic acid signaling and dehydration tolerance. Plant Mol Biol 89: 143–156 [DOI] [PubMed] [Google Scholar]
- Lim CW, Lee SC (2014) Functional roles of the pepper MLO protein gene, CaMLO2, in abscisic acid signaling and drought sensitivity. Plant Mol Biol 85: 1–10 [DOI] [PubMed] [Google Scholar]
- Lim CW, Lee SC (2016) Pepper protein phosphatase type 2C, CaADIP1 and its interacting partner CaRLP1 antagonistically regulate ABA signalling and drought response. Plant Cell Environ 39: 1559–1575 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63: 599–616 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Osakabe K, Shinozaki K, Tran LS (2014) Response of plants to water stress. Front Plant Sci 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Lim CW, Baek W, Lee SC (2015) RING type E3 ligase CaAIR1 in pepper acts in the regulation of ABA signaling and drought stress response. Plant Cell Physiol 56: 1808–1819 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M, Molinier J, Berr A, Genschik P (2011) MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S (2012) The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Seo KI, Lee JH, Nezames CD, Zhong S, Song E, Byun MO, Deng XW (2014) ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 26: 695–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Jha SK, Bagri J, Pandey GK (2015) ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One 10: e0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J (2009) The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot 60: 1439–1463 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang X, Peirats-Llobet M, Belda-Palazon B, Wang X, Cui S, Yu X, Rodriguez PL, An C (2016) Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 28: 2178–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang H, Cui P, Ding F, Wang G, Li R, Jenks MA, Lü S, Xiong L (2014) The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in Arabidopsis. Plant Physiol 165: 1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.