Preprotein-binding sites are mapped to the dimer interface and the switch II region of the Toc159 GTPase domain.
Abstract
Most chloroplast proteins are synthesized in the cytosol as higher molecular weight preproteins and imported via the translocons in the outer (TOC) and inner (TIC) envelope membranes of chloroplasts. Toc159 functions as a primary receptor and directly binds preproteins through its dimeric GTPase domain. As a first step toward a molecular understanding of how Toc159 mediates preprotein import, we mapped the preprotein-binding regions on the Toc159 GTPase domain (Toc159G) of pea (Pisum sativum) using cleavage by bound preproteins conjugated with the artificial protease FeBABE and cysteine-cysteine cross-linking. Our results show that residues at the dimer interface and the switch II region of Toc159G are in close proximity to preproteins. The mature portion of preproteins was observed preferentially at the dimer interface, whereas the transit peptide was found at both regions equally. Chloroplasts from transgenic plants expressing engineered Toc159 with a cysteine placed at the dimer interface showed increased cross-linking to bound preproteins. Our data suggest that, during preprotein import, the Toc159G dimer disengages and the dimer interface contacts translocating preproteins, which is consistent with a model in which conformational changes induced by dimer-monomer conversion in Toc159 play a direct role in facilitating preprotein import.
Over 95% of chloroplast proteins are encoded by the nuclear genome, translated in the cytosol as a higher molecular mass preprotein with an N-terminal transit peptide, and then posttranslationally imported into chloroplasts. The translocon on the chloroplast envelope specifically recognizes the transit peptides and translocates the preproteins into chloroplasts. Translocon components located in the outer and inner envelope membranes are called TOC and TIC (for translocon at the outer and inner envelope membrane of chloroplasts) proteins, respectively (Schnell et al., 1997). The TOC complex includes two homologous GTPases, Toc34 and Toc159, and a β-barrel-type channel, Toc75 (Kessler et al., 1994; Schnell et al., 1994). Toc34 contains a cytosolically exposed GTPase domain and a C-terminal membrane-anchoring segment (Kessler et al., 1994). Toc159 has a similar topology, with an N-terminal acidic domain (A domain), followed by a GTPase domain (G domain) in the cytosol, and a C-terminal membrane domain (M domain; Hirsch et al., 1994; Chen et al., 2000). The import process can be divided roughly into three stages. First, preproteins associate with the chloroplast surface in an energy-independent low-affinity manner. Then, this association is followed by stable binding of preproteins to the translocon and translocation of transit peptides across the outer membrane, stimulated by the presence of ATP in the micromolar range. Finally, when a millimolar concentration of ATP is present in the stroma, preproteins are further translocated across the envelope into the stroma and are processed into their mature size (for review, see Li and Chiu, 2010; Shi and Theg, 2013; Paila et al., 2015).
Multiple lines of evidence support that Toc159 functions as an initial receptor for preproteins during the import process. It has been shown that the preprotein of the small subunit of ribulose 1,5-bisphosphate carboxylase (prRBCS), with a Cys in its transit peptide for cross-linker conjugation, was cross-linked to Toc159 both in the absence of ATP and in the presence of 75 µm ATP (Perry and Keegstra, 1994; Ma et al., 1996), indicating that transit peptides are in contact with Toc159 at the first two stages of the import process. The preprotein of ferredoxin (prFd), which only has Cys at the N terminus of its mature region, also was cross-linked to Toc159 in the presence of 75 µm ATP, indicating that the mature region of preproteins remains in contact with Toc159 during stable preprotein binding (Ma et al., 1996). The Toc159 G domain (Toc159G) was further shown to be the domain that binds transit peptides (Smith et al., 2004). Genetic evidence also supports that Toc159 is essential for chloroplast biogenesis. The ppi2 mutants, with knockout mutations in the Arabidopsis (Arabidopsis thaliana) Toc159 gene (AtToc159), are albino and cannot survive on soil (Bauer et al., 2000). However, despite the importance of Toc159, how Toc159 binds preproteins (i.e. the location of the preprotein-binding site within the Toc159G domain) is still unknown.
The functions of both Toc159 and Toc34 seem to be regulated by dimerization and GTP binding, but the exact mechanism is unclear, although more in vitro information is available for Toc34. The crystal structure of the pea (Pisum sativum) Toc34 G domain (Toc34G) shows typical features of a small GTPase, including the G1 motif (P loop) and the switch I and switch II regions, which in other GTPases are the main parts that move upon activation by GTP. Pea Toc34G has been crystalized as a dimer, with GDP associated with each monomer being stabilized by residues from both monomers (Sun et al., 2002). A conserved Arg residue of each monomer inserts into the active site and contacts the β-phosphate of GDP bound by the reciprocal monomer (Sun et al., 2002). Therefore, this Arg resembles the catalytic Arg finger of many GTPase-activating proteins (GAP; Ahmadian et al., 1997), so each Toc34 monomer was proposed to function as the GAP for the reciprocal monomer in the dimeric complex (Sun et al., 2002). However, dimerization only increases the GTPase activity of Toc34G by 2- to 6-fold in vitro (Yeh et al., 2007), so a requirement for other activation factors, such as co-GAP, also has been proposed (Koenig et al., 2008b). In vivo, mutations that specifically disrupt dimerization but not the basal GTPase activity of AtToc33, the Arabidopsis ortholog of Toc34, reduce the rate of preprotein import into chloroplasts. Chloroplasts from these monomeric AtToc33 mutant plants actually have an increased affinity for preproteins but have a reduced efficiency of membrane translocation of preproteins (Lee et al., 2009). Therefore, it has been suggested that the conformational change arising from AtToc33 dimer formation may produce the necessary signal for the dissociation of bound preproteins, which results in preprotein translocation into the Toc75 channel (Lee et al., 2009).
Residues involved in Toc34 dimerization are conserved in Toc159G (Sun et al., 2002); indeed, Toc159G also forms dimers in solution (Yeh et al., 2007). However, information on the physiological significance of the Toc159G dimer is not yet available. It has been shown that a mutant AtToc159 protein with slightly increased GTP-binding affinity, but a 90% reduction in GTP hydrolysis, can still fully complement the growth defect of the ppi2 mutants (Wang et al., 2008). Chloroplasts from these AtToc159-mutant transgenic plants actually show an increased preprotein import rate relative to wild-type chloroplasts due to the ability of the mutant AtToc159 protein to stabilize the formation of a GTP-dependent preprotein-binding intermediate (Wang et al., 2008).
To understand how dimerization and GTP binding in Toc159 regulate preprotein binding, it is crucial to determine the location of the preprotein-binding site on Toc159G. In this study, we mapped the preprotein-binding sites on the Arabidopsis Toc159G (AtToc159G). We used the artificial protease 1-(p-bromoacetamidobenzyl) EDTA, iron (III) chelate (FeBABE), conjugated to purified preproteins, to cleave AtToc159G after preprotein binding. We identified two sites on AtToc159G that are in close proximity to preproteins. Using the Toc34 crystal structure as a template to predict the structure of AtToc159G, one of the contact sites was shown to reside at the dimer interface and the other at the switch II region of AtToc159G. These contact sites were confirmed by Cys-Cys cross-linking analyses in vitro, and the contact site at the dimer interface was further confirmed in vivo using transgenic plants expressing AtToc159 with Cys engineered into designated positions.
RESULTS
Recombinant prRBCS-GST Specifically Binds AtToc159G
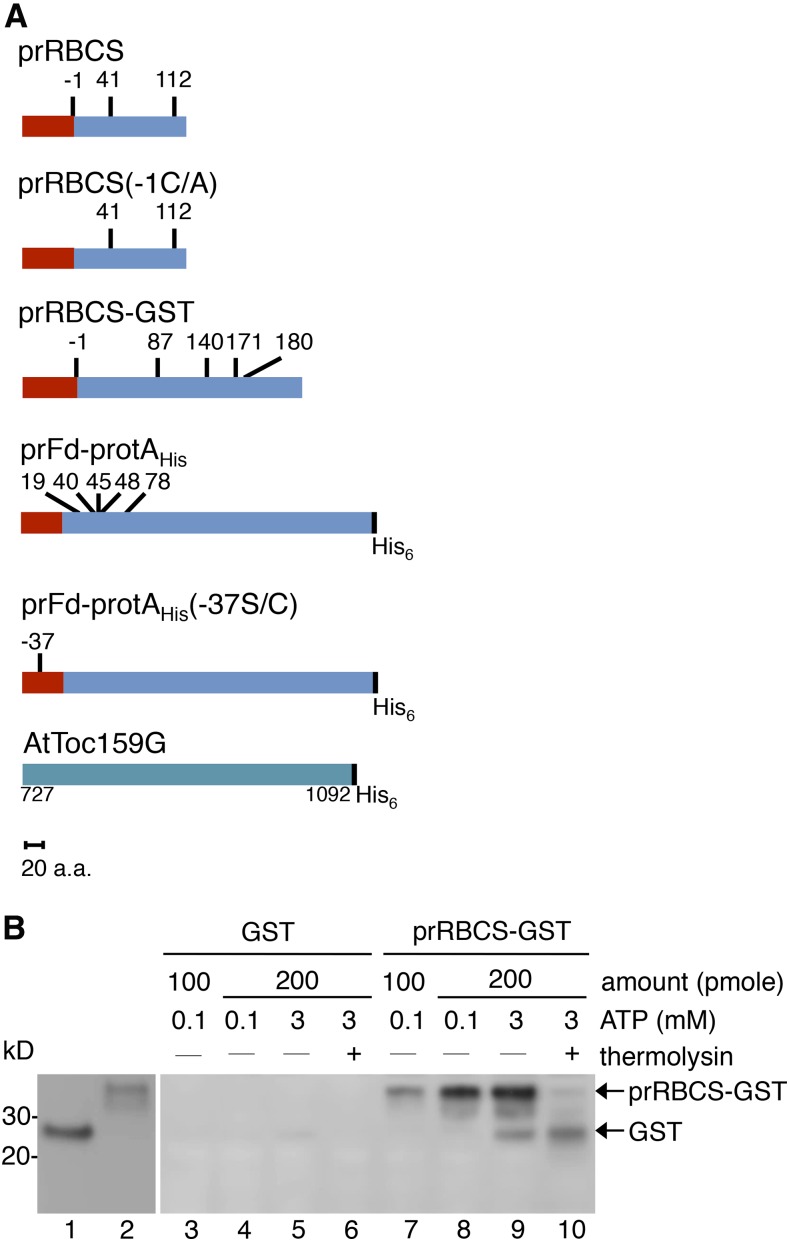
To map the binding site of preproteins on Toc159, we constructed the chimeric preprotein prRBCS-GST (Fig. 1A), which is composed of the prRBCS transit peptide fused to the N terminus of GST. The prRBCS transit peptide has been shown to bind Toc159 (Perry and Keegstra, 1994; Kouranov and Schnell, 1997; Smith et al., 2004). We first verified the binding and import competency of prRBCS-GST expressed and purified from Escherichia coli. GST without the transit peptide was used as a negative control. Both proteins were purified from the soluble fraction of E. coli cell lysates. The purified proteins were incubated with isolated pea chloroplasts in the presence of 0.1 or 3 mm ATP to assay binding or import into chloroplasts, respectively. Intact chloroplasts were reisolated after the reactions and analyzed by SDS-PAGE and immunoblotting. Under conditions of 0.1 mm ATP, prRBCS-GST bound to chloroplasts in a dosage-dependent manner (Fig. 1B, lanes 7 and 8). Under conditions of 3 mm ATP, prRBCS-GST was imported into chloroplasts and processed into a protein equivalent in size to GST, and this protein was thermolysin resistant, indicating that it was located within chloroplasts (Fig. 1B, lanes 9 and 10). GST did not bind to nor was imported into chloroplasts (Fig. 1B, lanes 3–6). These results show that the prRBCS-GST recombinant protein can specifically bind to and be imported into chloroplasts.
Figure 1.
The prRBCS transit peptide directs the specific binding and import of prRBCS-GST into chloroplasts. A, Schematic representation of constructs used in this study. The red rectangles represent the transit peptides, and the blue rectangles represent the mature protein regions or the passenger proteins. The numbers above each construct indicate the positions of Cys residues, with the first residue of the mature protein designated as +1 and residues in the transit peptide designated with negative numbers. The AtToc159G recombinant protein construct, composed of AtToc159 residues 727 to 1,092 with a C-terminal His6 tag, also is depicted. All constructs are drawn to scale. a.a., Amino acids. B, Binding and import of prRBCS-GST into isolated chloroplasts. GST (lane 1) and prRBCS-GST (lane 2) were purified from the soluble fraction of E. coli. Two different amounts were incubated with ATP-depleted chloroplasts in the presence of 0.1 and 3 mm ATP for the binding and import reactions, respectively. Samples from the import reaction were further treated with 0.2 mg mL−1 thermolysin. Samples were analyzed by SDS-PAGE and immunoblotting with anti-GST antibody.
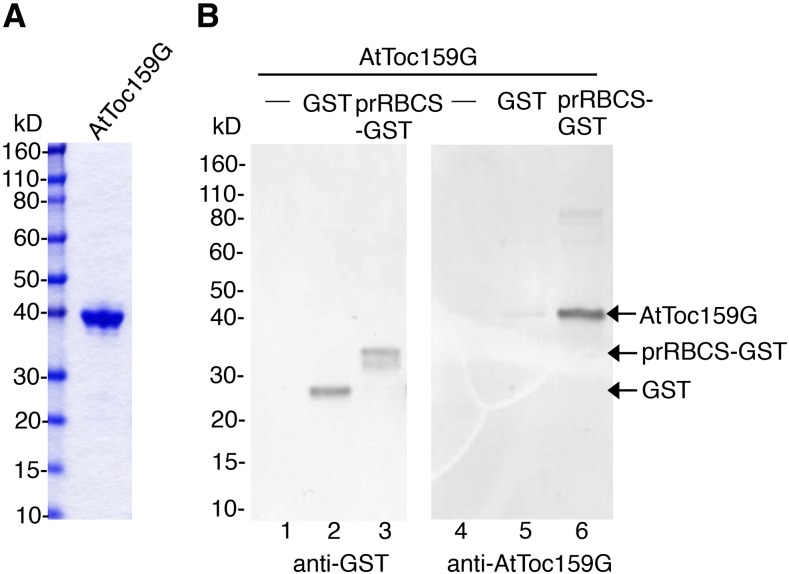
It has been shown that the GTPase domain of Toc159 is sufficient to specifically recognize and bind transit peptides (Smith et al., 2004). To examine whether prRBCS-GST can be specifically recognized and bound by purified AtToc159G, we expressed AtToc159G with a C-terminal His6 tag (Fig. 1A) and purified the protein from the soluble fraction of E. coli (Fig. 2A). GST or prRBCS-GST was mixed with AtToc159G, repurified with glutathione-conjugated Sepharose (GSH resin), and analyzed by SDS-PAGE and immunoblotting. As shown in Figure 2B, AtToc159G was specifically pulled down by prRBCS-GST.
Figure 2.
Recombinant prRBCS-GST binds to AtToc159G. A, AtToc159G was expressed and purified from E. coli. Protein integrity was analyzed by SDS-PAGE, and the gel was stained with Coomassie Blue. B, Purified AtToc159G was incubated with GST or prRBCS-GST. Proteins were pulled down by GSH resin and analyzed by SDS-PAGE. The bound proteins were visualized by immunoblotting using the anti-GST or anti-AtToc159G antibody as indicated. The anti-AtToc159G antibody was raised in rats against the purified AtToc159G protein as shown in A.
There Are Two Preprotein-Binding Sites on AtToc159G
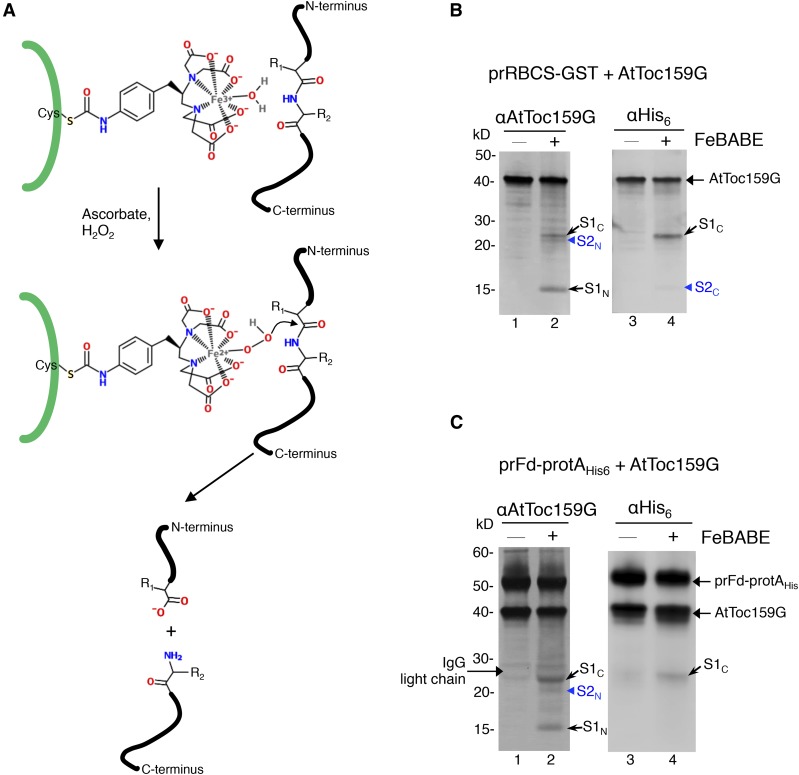
For initial mapping of the preprotein-binding site on AtToc159G, we used the heterobifunctional chelating agent FeBABE. At one end, this molecule has a sulfhydryl-reactive group that can be conjugated to a Cys, and at the other end, it has an EDTA-chelated iron that can cleave a peptide bond within a distance of 12 Å when activated by ascorbate and hydrogen peroxide (Fig. 3A). Thus, FeBABE cleavage analysis can be used to map the approximate contact region between two polypeptides (Chen and Hahn, 2003).
Figure 3.
Cleavage of AtToc159G by FeBABE-conjugated preproteins. A, Schematic representation of the FeBABE cleavage mechanism. FeBABE was conjugated onto the thiol group of a Cys residue in the preprotein represented by the thick green line. In the presence of ascorbate and hydrogen peroxide (H2O2), the hydroxyl group of FeBABE becomes nucleophilic and attacks a peptide bond within a distance of 12 Å. B, prRBCS-GST with or without FeBABE conjugation was incubated with AtToc159G. Protein complexes were purified by GSH resin, and FeBABE cleavage was activated. The reactions were analyzed by SDS-PAGE and immunoblotting using the anti-AtToc159G or anti-His6 antibody. S1C and S2C, AtToc159G C-terminal fragments generated by FeBABE cleavage; S1N and S2N, AtToc159G N-terminal fragments generated by FeBABE cleavage. C, The experiment was performed as described in B, except that prFd-protAHis or FeBABE-conjugated prFd-protAHis was used and IgG Sepharose was used to pull down prFd-protAHis and bound AtToc159G.
We conjugated FeBABE to the Cys residues of prRBCS-GST. AtToc159G was incubated with FeBABE-prRBCS-GST and GSH resin. After incubation, FeBABE-prRBCS-GST and AtToc159G bound to FeBABE-prRBCS-GST were repurified by pelleting the GSH resin. After washing and resuspending of the resin, the FeBABE cleavage reaction was activated by adding ascorbate and hydrogen peroxide. The reactions were terminated by the addition of SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting and hybridized with antibodies against AtToc159G and the His6 tag to visualize the cleaved fragments. Three AtToc159G fragments of 24, 21, and 15 kD were observed (Fig. 3B, lane 1), and these fragments were not observed when prRBCS-GST without FeBABE was used to bind AtToc159G (Fig. 3B, lane 2).
The 24-kD fragment (Fig. 3B, lane 2, S1C for site 1 C-terminal fragment) also was recognized by the anti-His6 antibody (Fig. 3B, lane 4), indicating that it is a C-terminal fragment. The 15-kD fragment (Fig. 3B, lane 2, S1N for site 1 N-terminal fragment) was not recognized by the anti-His6 antibody, indicating that it is an N-terminal fragment. Together, these two fragments constituted approximately the entire length of AtToc159G, indicating that AtToc159G had been cut by FeBABE-prRBCS-GST at about 15 kD from the N terminus. The signal for the 21-kD fragment (Fig. 3B, lane 2, arrowhead, S2N) was weaker than those for the other two fragments, and this fragment was not recognized by the anti-His6 antibody, indicating that it is an N-terminal fragment. A very faint signal, slightly larger than 15 kD, was detected by the anti-His6 antibody (Fig. 3B, lane 4, arrowhead, S2C). S2C migrated only slightly above S1N, and its signal might have been covered by that of S1N when the immunoblots were hybridized with the anti-Toc159G antibody. S2N and S2C together were slightly smaller than the full length of AtToc159G. It is possible that some AtToc159G molecules were cut at 21 kD from the N terminus. The resulting C-terminal fragment may have been cut further, and the final C-terminal fragment was only about 15 kD. We then calculated the positions of cleavage: 15 kD from the N terminus would be close to residue 864 of AtToc159G, and 21 kD from the N terminus would be close to residue 924.
We further verified these results with another preprotein, prFd-protAHis, which is a chimeric preprotein with the transit peptide of prFd fused to Staphylococcus protein A (Fig. 1A). This chimeric preprotein has been used to show preprotein cross-linking to Toc159 and Toc75 during import (Ma et al., 1996) and was shown to bind specifically to AtToc159G in vitro (Smith et al., 2004). We incubated AtToc159G with FeBABE-conjugated prFd-protAHis and repurified FeBABE-prFd-protAHis and bound AtToc159G by IgG Sepharose before activating the cleavage reaction. When analyzed by immunoblotting with the antibody against AtToc159G, three fragments similar in size to those from the FeBABE-prRBCS-GST cleavage reaction were observed (Fig. 3C, lane 2). As described for FeBABE-prRBCS-GST, the largest fragment of approximately 24 kD also was recognized by the anti-His6 antibody (Fig. 3C, lanes 2 and 4, S1C), whereas the 15-kD fragment was not (Fig. 3C, lane 2, S1N), suggesting that FeBABE-conjugated prFd-protAHis also cleaved AtToc159G at ∼15 kD from the N terminus. The weaker 21-kD fragment also was observed (Fig. 3C, lane 2, arrowhead, S2N), but the amount of its corresponding C-terminal fragment was most likely too low to be detected by the anti-His6 antibody (Fig. 3C, lane 4). These results indicate that there is one major region (15 kD from the N terminus) and one minor region (21 kD from the N terminus) on AtToc159G that are in close proximity to preproteins during preprotein binding. Despite the varied locations of Cys residues on prRBCS-GST and prFd-protAHis (Fig. 1A), these two preproteins cleaved at similar regions on AtToc159G.
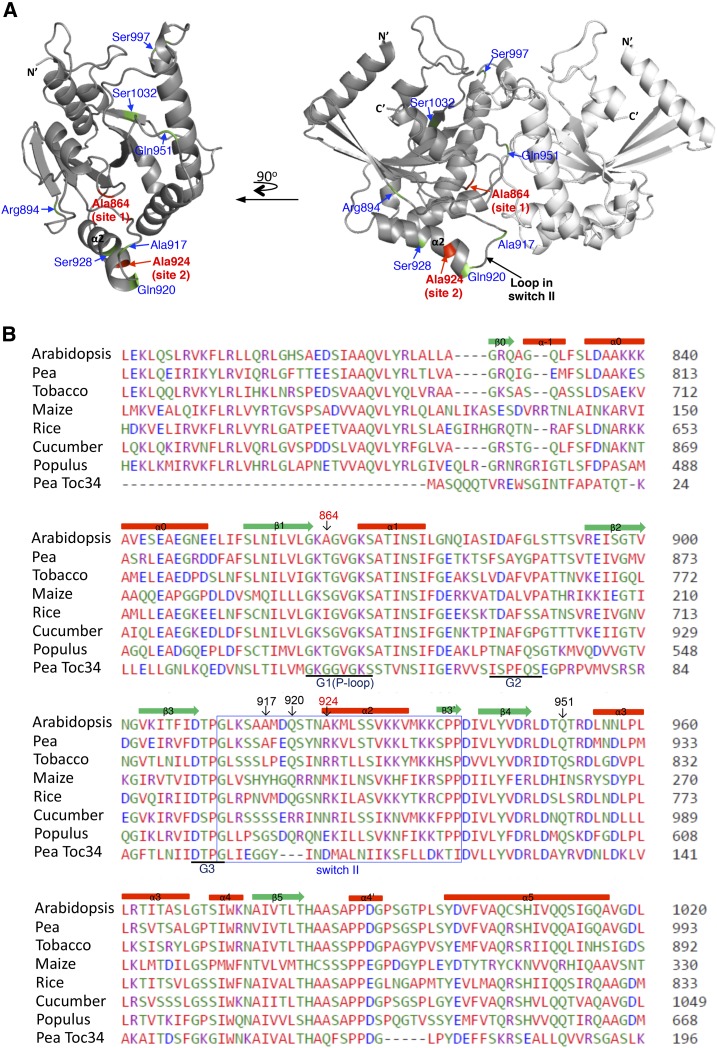
To visualize the localizations of the two cleavage sites on AtToc159G, we used the crystal structure of pea To34G (Sun et al., 2002) as a template to generate a structural model for AtToc159G (Fig. 4A). Residue 864 is on the dimer interface of AtToc159G (Fig. 4, highlighted in red and labeled as site 1) and is a nonconserved residue in the G1 motif. Residue 924 is on α-helix 2, close to the central loop in switch II (Fig. 4, highlighted in red and labeled as site 2). In the structure of Toc34G, the corresponding residue is located right at the junction between α-helix 2 and the central loop in switch II. In the predicted structure of AtToc159G, residue 924 is positioned at the second turn of α-helix 2 (Fig. 4A) because Toc159 has an insertion of three amino acids compared with Toc34 in this region (Fig. 4B), so α-helix 2 extends one more turn.
Figure 4.
AtToc159G structure model and sequence alignment. A, The two deduced sites cleaved by FeBABE-conjugated preproteins are colored in red. Additional residues mutated to Cys are indicated and colored in green. The N- and C-terminal ends, α-helix 2, and the central loop in switch II also are indicated. At left, one of the monomers is rotated 90° horizontally to show the dimer interface. B, Sequence alignment of the G domain region of Toc159 homologs from representative higher plant species and the pea Toc34 G domain. Secondary structure elements are marked above the sequence, and G1 to G3 motifs are marked below the sequence, all of which are labeled according to the pea Toc34 crystal structure (Protein Data Bank no. 1h65). The predicted switch II is boxed. Deduced residues cleaved by FeBABE and residues that generated significant cross-linking to preproteins when mutated to Cys are indicated with red and black numbers, respectively. Accession numbers for the sequences shown are as follows: At4g02510 (Arabidopsis Toc159), AAF75761 (pea Toc159), XP_009769991 (tobacco [Nicotiana tabacum] Toc159), NP_001147969 (maize [Zea mays] Toc159), XP_015639221 (rice [Oryza sativa] Toc159), XP_004152365 (cucumber [Cucumis sativus] Toc159), XP_002312975 (Populus spp. Toc159), and Q41009 (pea Toc34).
Preprotein Binding to AtToc159G Reduces the Dimerization of AtToc159G
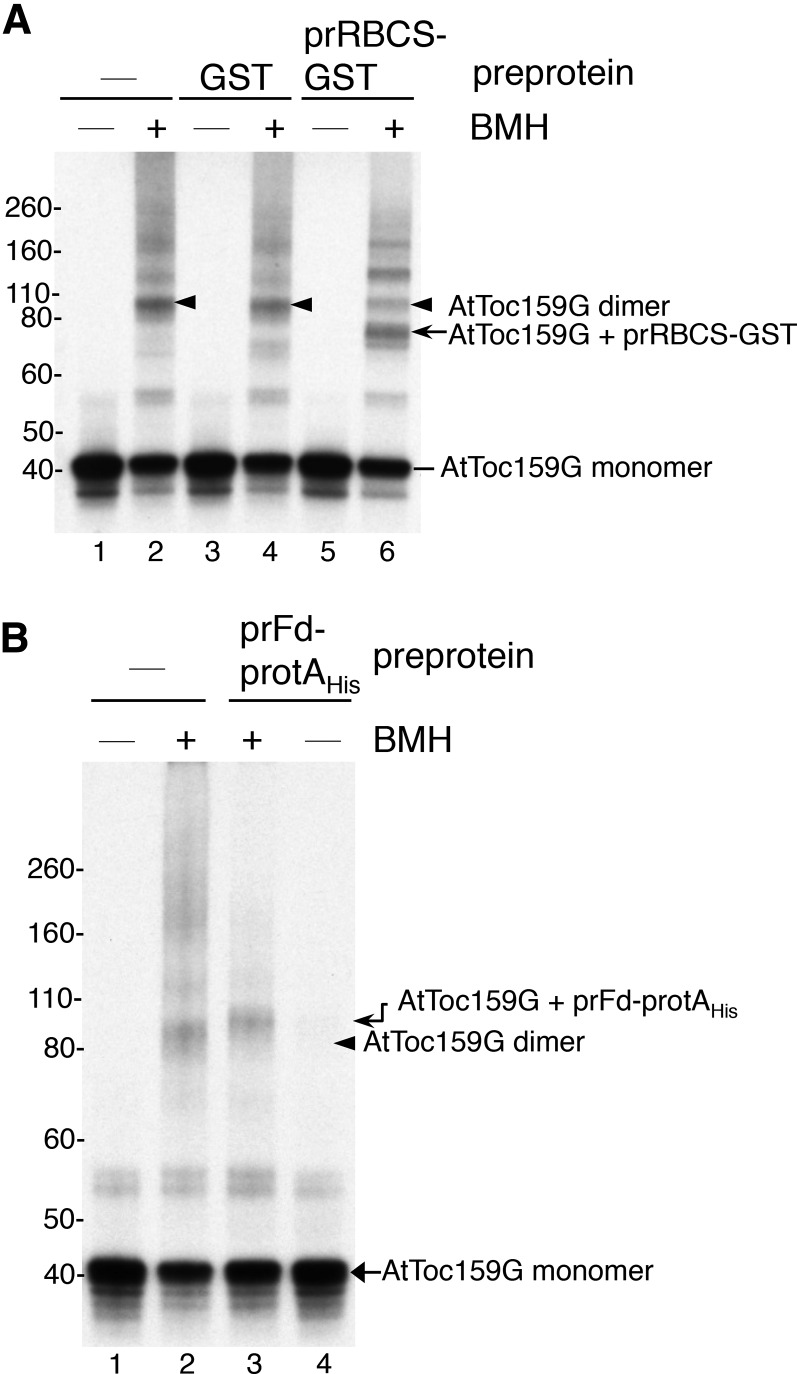
It has been shown that pea Toc159G forms homodimers in vitro (Yeh et al., 2007). One of the deduced preprotein-binding sites from our FeBABE cleavage experiments is on the dimer interface of AtToc159G (Fig. 4, residue 864). If our results are correct, preprotein binding would be expected to interrupt AtToc159G dimerization. To test this inference, we performed in vitro cross-linking analyses with the homobifunctional Cys-Cys cross-linker bismaleimidohexane (BMH). We incubated purified prRBCS-GST with in vitro-translated [35S]Met-labeled AtToc159G and then added BMH. The cross-linking reaction was analyzed by SDS-PAGE and fluorography. The molecular mass of AtToc159G is 40.8 kD and that of prRBCS-GST is 34.2 kD (for their mobility on SDS-PAGE, see Fig. 2). In the absence of prRBCS-GST, some AtToc159G molecules were cross-linked to form a product of ∼90 kD that most likely represented AtToc159G dimers (Fig. 5A, lanes 2, 4, and 6, arrowheads). The amount of dimer was not affected when GST was added (Fig. 5A, lane 4), but when prRBCS-GST was added, the amount of AtToc159G dimer was reduced significantly (Fig. 5A, lane 6). A cross-linked product corresponding to approximately the size of AtToc159G plus prRBCS-GST was then observed (Fig. 5A, lane 6, arrow). The identity of this cross-linked product was confirmed by affinity purification of the cross-linking reactions using GSH resin (Supplemental Fig. S3A). This AtToc159G-prRBCS-GST adduct, but not the AtToc159G dimer, was pulled down by GSH resin (Supplemental Fig. S3A, right, lane 4). The two higher molecular mass products (Supplemental Fig. S3A, right, lane 4, asterisks) most likely represented additional prRBCS-GST cross-linked to the AtToc159G-prRBCS-GST adduct, since higher molecular mass prRBCS-GST multimers were revealed when the gels were stained with Coomassie Blue (Supplemental Fig. S3A, left, lane 4). Similar results were obtained when prFd-protAHis was used (Fig. 5B), with AtToc159G dimer signal almost completely disappearing and being replaced by the AtToc159G-prFd-protAHis adduct with a slightly higher molecular mass (Fig. 5B, compare lanes 2 and 3). The molecular mass of prFd-protAHis is 46.6 kD, so an adduct of AtToc159G-prFd-protAHis is expected to be slightly bigger than the AtToc159G dimer. This adduct was specifically pulled down by IgG Sepharose, confirming that it indeed contained prFd-protAHis (Supplemental Fig. S3B, right, lane 4). The reduction of AtToc159G dimer levels by preprotein binding is in agreement with our FeBABE cleavage results showing that one of the preprotein-binding sites is located at the dimer interface of AtToc159G.
Figure 5.
Binding of preproteins reduces AtToc159G dimerization. A, Purified GST or prRBCS-GST was incubated with in vitro-translated [35S]Met-labeled AtToc159G. BMH was added to cross-link protein complexes. Reactions were analyzed by SDS-PAGE and fluorography. The cross-linked AtToc159G dimer is indicated by arrowheads. The cross-linked adduct of AtToc159G-prRBCS-GST is indicated by the arrow. B, Binding and cross-linking experiments were performed as in A, except that purified prFd-protAHis was used.
The Two Preprotein-Binding Sites Are at the Dimer Interface and the Switch II Region of AtToc159G
We next performed more Cys-Cys cross-linking assays to further verify the preprotein-binding sites. The four endogenous Cys residues of AtToc159G were first substituted with Ala. Exposed residues from different sides of AtToc159G were then chosen and mutated to Cys (Fig. 4A, the locations of the residues replaced by Cys are highlighted in green) to create AtToc159G mutants containing a single Cys at designated positions. In addition, to verify the FeBABE cleavage results, residue 864 of site 1 at the dimer interface and residue 924 of site 2 on α-helix 2 were mutated to Cys. For site 2, we also replaced Gln-920 and Ser-928 with Cys. Gln-920 is one of the residues in the three-amino acid insertion unique to AtToc159 in this region (Fig. 4B) and is closer to the central loop of switch II. Ser-928 is a bit farther away from the central loop. In some small GTPases, the conformational change upon GTP activation in this part of α-helix 2 is so small that it is not considered part of switch II (Wittinghofer and Vetter, 2011). All the single-Cys AtToc159G mutants were first expressed in E. coli and purified to perform GTPase assays to verify that the mutations did not cause aberrant conformational changes. All mutants, except the Cys-864 mutant, had a similar GTPase activity to that of wild-type AtToc159G (Supplemental Fig. S1). The Cys-864 mutant had about a 50% reduction in its GTPase activity (Supplemental Fig. S1), similar to previously reported results (Wang et al., 2008). An Ala-to-Arg substitution at residue 864 results in normal or increased GTP binding but a reduction in GTP hydrolysis (Smith et al., 2002). However, this mutant fully complemented the AtToc159 knockout mutation (Wang et al., 2008), indicating that a mutation at residue 864 does not affect the function of AtToc159. We also verified whether the Ala-864-to-Cys mutation is likely to affect AtToc159G dimer conformation. We used the PISA server (Krissinel and Henrick, 2007) to measure the dimer interface surface area of the two protein models. The surface area buried is 1,252.4 Å2 for the wild-type AtToc159G dimer and 1,253.0 Å2 for the Cys-864 mutant, suggesting that the Ala-864-to-Cys mutation is unlikely to change the accessibility of the dimer interface.
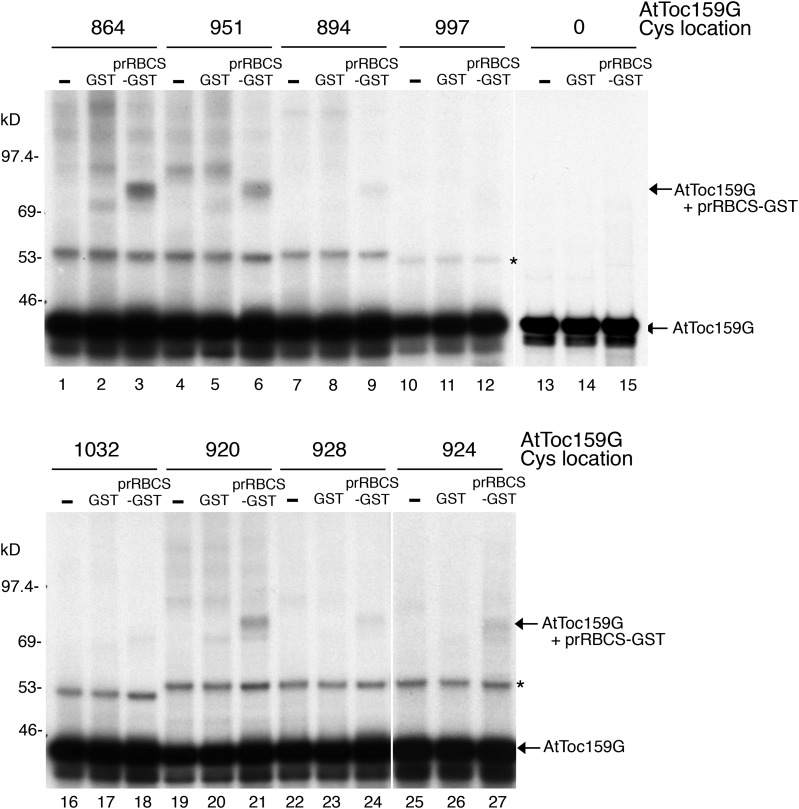
To determine which side of AtToc159G is in the vicinity of bound preproteins, [35S]Met-labeled single-Cys mutants of AtToc159G were incubated with prRBCS-GST and then BMH was added. Incubation with GST was used as a negative control. The cross-linking reactions were then analyzed by SDS-PAGE. Among the single-Cys mutants, those containing a Cys at the dimer interface (residues 864 and 951) showed significant cross-linking to prRBCS-GST (Fig. 6, lanes 3 and 6), whereas those containing a Cys on the other sides (residues 894, 997, and 1,032) showed very little cross-linking (Fig. 6, lanes 9, 12, and 18). For the three Cys residues on α-helix 2, residue 920 showed the highest amount of cross-linking, whereas residue 928 showed the lowest amount of cross-linking to AtToc159G, suggesting that residues closer to the central loop in switch II might be nearer to preproteins. An AtToc159G mutant with no Cys residue (Fig. 6, lanes 13–15, designated as 0) generated no cross-linking product, confirming the specificity of the BMH cross-linking reactions.
Figure 6.
prRBCS-GST was cross-linked to Cys residues at the AtToc159G dimer interface and switch II. prRBCS-GST or GST was incubated with [35S]Met-labeled single-Cys AtToc159G. After BMH cross-linking, reactions were analyzed by SDS-PAGE and fluorography. The locations of the single Cys residues are indicated above the lanes. A nonspecific cross-linked product from a protein in the reticulocyte lysate is indicated by the asterisk.
The Mature Portion Preferentially Makes Contact with the Dimer Interface, and the Transit Peptide Interacts with Both Sites Similarly
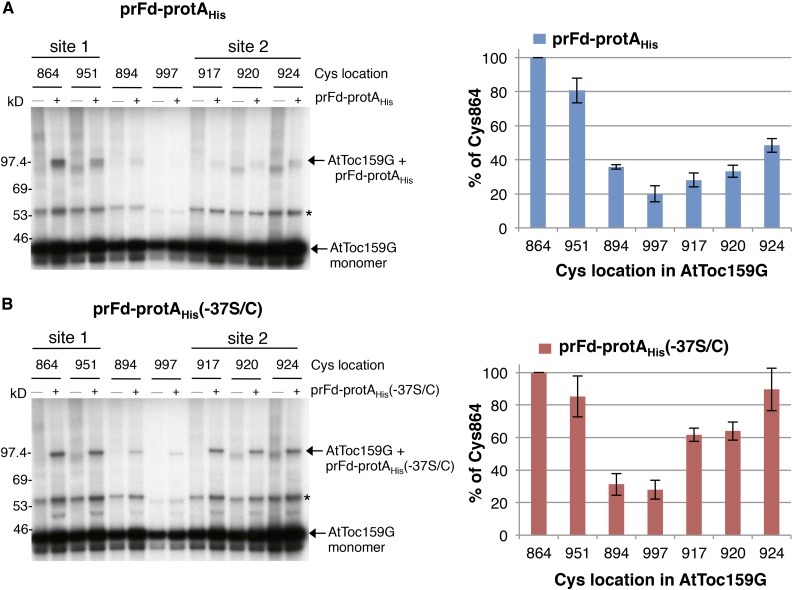
The prRBCS-GST construct we used has one Cys in the transit peptide and four Cys residues in the mature region, whereas the prFd-protAHis construct has a cluster of five Cys residues at the N terminus of the mature region close to the transit peptide (Fig. 1A). Therefore, the two binding sites mapped by FeBABE cleavage and BMH cross-linking could have resulted from AtToc159G contacting only the Cys in the mature region of the two preproteins used. We next verified whether transit peptides bind to the two sites. Due to difficulties in purifying prRBCS-GST with no Cys in the mature region, we generated a prFd-protAHis mutant, prFd-protAHis(-37S/C), in which all Cys residues in the mature region were replaced by Ala and the Ser at residue −37 in the transit peptide was replaced by Cys (Fig. 1A; the first residue of the mature protein is designated as +1, residues in the transit peptide are designated with negative numbers, and residue −37 is the 11th residue from the initiation Met of the preprotein). We purified prFd-protAHis and prFd-protAHis(-37S/C) from E. coli and incubated them with the [35S]Met-labeled AtToc159G single-Cys mutants. An additional AtToc159G single-Cys mutant, Cys-917, having a single Cys in the central loop of switch II, also was created and tested. After BMH cross-linking, samples were analyzed by SDS-PAGE and fluorography. As shown in Figure 7A, wild-type prFd-protAHis showed more cross-linking to AtToc159G mutants having Cys around site 1 (i.e. at the dimer interface; residues 864 and 951) than to AtToc159G mutants having Cys close to site 2 at switch II (residues 917, 920, and 924). In contrast, prFd-protAHis(-37S/C) cross-linked similarly to AtToc159G mutants having Cys in both regions (Fig. 7B). These results suggest that the mature portion of a preprotein preferentially makes contact with the dimer interface, whereas the transit peptide interacts similarly to the dimer interface and switch II.
Figure 7.
The mature region of prFd-protAHis preferentially contacts the dimer interface of AtToc159G. A, prFd-protAHis was incubated with various [35S]Met-labeled single-Cys AtToc159G mutants. After BMH cross-linking, reactions were analyzed by SDS-PAGE and fluorography. The locations of the single Cys residues are indicated above the lanes. Cys-864 and Cys-951 are located around FeBABE cleavage site 1 on the dimer interface, whereas Cys-917, Cys-920, and Cys-924 are located around cleavage site 2 in the switch II region. The cross-linked adduct of AtToc159G-prFd-protAHis is indicated by an arrow. A nonspecific cross-linked product from a protein in the reticulocyte lysate is indicated by the asterisk. B, Binding and cross-linking experiments were performed as described in A, except that prFd-protAHis (-37S/C) was used. Quantification of the cross-linked products is shown at right for each preprotein. The value for each single-Cys AtToc159G preprotein cross-linked product was normalized to the amount of cross-linking observed for AtToc159G Cys-864 to the same preprotein. Values plotted are means ± sd of three independent experiments.
Preproteins Make Contact with the Dimer Interface of AtToc159G When Bound to Chloroplasts
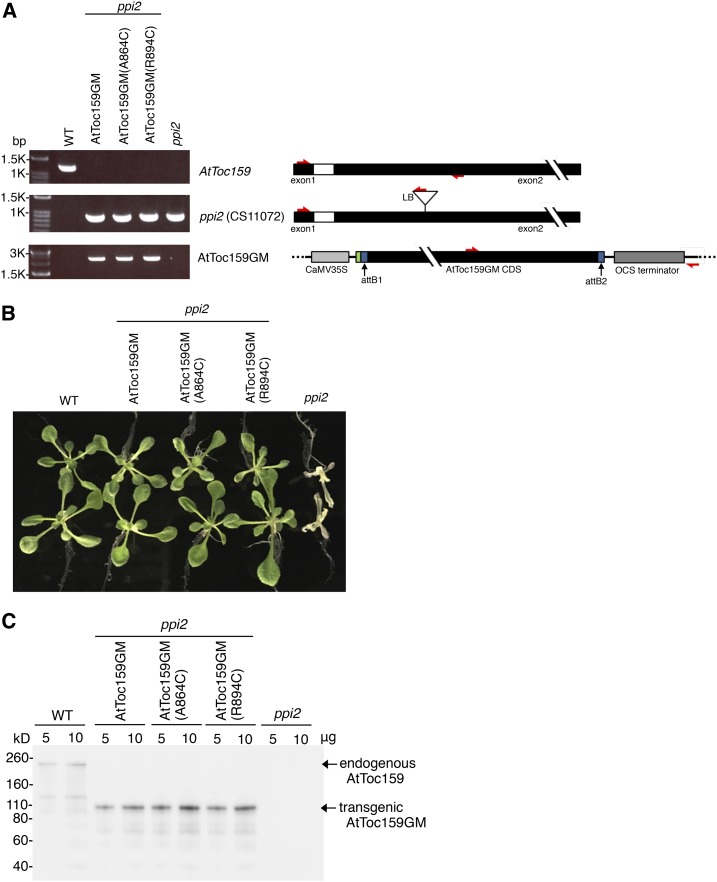
We next tried to verify the in vitro mapping results in chloroplasts. To achieve this, we generated Arabidopsis transgenic plants expressing, in the ppi2 mutant, AtToc159 variants carrying a Cys replacement at designated positions. Because both the transit peptide and the mature regions contact the AtToc159G dimer interface in vitro, we first focused on verifying preprotein binding to the dimer interface. It has been shown that the G and M domains of AtToc159 (AtToc159GM) are sufficient to complement the ppi2 mutant (Agne et al., 2009). We engineered a transgene construct encoding AtToc159GM (AtToc159 residues 727–1,503) driven by the CaMV 35S promoter or the same construct carrying the A864C mutation, designated as AtToc159GM(A864C). In addition, our in vitro BMH cross-linking results showed that Cys placed at residue 894 showed little cross-linking to preproteins (Fig. 6, lane 9), so the same construct carrying the R894C mutation, designated as AtToc159GM(R894C), was used as a negative control. These constructs were introduced into the ppi2 heterozygous plants using Agrobacterium tumefaciens-mediated transformation. Stable transgenic lines homozygous for the ppi2 mutation and containing the introduced AtToc159GM constructs were selected through antibiotic screening and genotyping using wild-type- and ppi2-specific primer pairs as well as a primer pair specific for the transgene (Fig. 8A). All three AtToc159GM constructs fully rescued the ppi2 mutant, and the transgenic plants were indistinguishable from the wild-type plants (Fig. 8B), indicating that all three AtToc159GM proteins were as functional as the wild-type full-length AtToc159. Immunoblotting using the anti-Toc159 antibody showed the absence of endogenous AtToc159 protein in the untransformed ppi2 plants, the presence of full-length AtToc159 in wild-type plants, and the 86-kD AtToc159GM protein in the three transgenic lines (Fig. 8C).
Figure 8.
Genotypic and phenotypic analyses of transgenic plants. A, Total genomic DNA was isolated from the ppi2 mutant, the corresponding wild-type plant (WT), and three transgenic lines containing the transgene encoding the different AtToc159GM variants transformed into the ppi2 mutant. DNA was amplified by PCR using primer pairs specific for the endogenous AtToc159 gene, the ppi2 T-DNA insertion, or the transgene encoding the introduced AtToc159GM. Primer positions are depicted at right with red arrows. The attB1 and attB2 sites resulting from the LR recombination reaction are shown in blue, and the FLAG tag from the pEarleyGate 202 vector is shown in green. B, Phenotypes of 12-d-old wild-type, AtToc159GM transgenic, and ppi2 mutant plants grown on Murashige and Skoog (MS) medium. C, AtToc159 protein expression. Protein extracts from 12-d-old plants as shown in B were analyzed by SDS-PAGE and immunoblotting using anti-AtToc159 antibody.
We then used [35S]Met-labeled prRBCS to perform BMH cross-linking experiments with isolated chloroplasts. We first tested chloroplasts isolated from wild-type untransformed plants and from the AtToc159GM-transformed ppi2 plants. Isolated chloroplasts were incubated with prRBCS in the dark at room temperature for 15 min. Intact chloroplasts were reisolated, and BMH was added. The cross-linking reactions were quenched by adding 4× NuPAGE sample buffer, and the samples were analyzed by SDS-PAGE and fluorography. Although Toc159 has been shown to cross-link to preproteins both in the absence of ATP and in the presence of micromolar concentrations of ATP (Perry and Keegstra, 1994; Ma et al., 1996), the amount of prRBCS bound to chloroplasts in the absence of ATP was very low, making reliable quantification difficult (data not shown). Therefore, prRBCS was bound to chloroplasts under 100 µm ATP. Two strong cross-linked signals in the 80- to 100-kD region were observed (Fig. 9A, lanes 1 and 2). Chloroplasts from the AtToc159GM transgenic plants were then solubilized with 1% LDS. The solubilized chloroplasts were diluted to reduce the LDS concentration to 0.1% and then immunoprecipitated using the anti-Toc75 or anti-Toc159 antibody. Both signals were precipitated by the anti-Toc75 antibody but not by the anti-Toc159 antibody, indicating that they were products of prRBCS cross-linked to Toc75 (Fig. 9A, lanes 3 and 4). Since AtToc159GM is about 86 kD, an AtToc159GM-prRBCS cross-linked product would be expected to be slightly larger than the Toc75-prRBCS cross-linked products. Indeed, a very faint signal located above the two Toc75-prRBCS signals at about 110 kD was observed (Fig. 9A, lane 2, blue circle). This 110-kD signal was absent in the wild-type plants, in which a faint 260-kD signal was observed instead (Fig. 9A, lane 1, blue circle), suggesting that the 110-kD signal may represent AtToc159GM cross-linked to prRBCS and the 260-kD signal may be full-length AtToc159 cross-linked to prRBCS. However, the signal was too weak to be reliably quantified.
Figure 9.
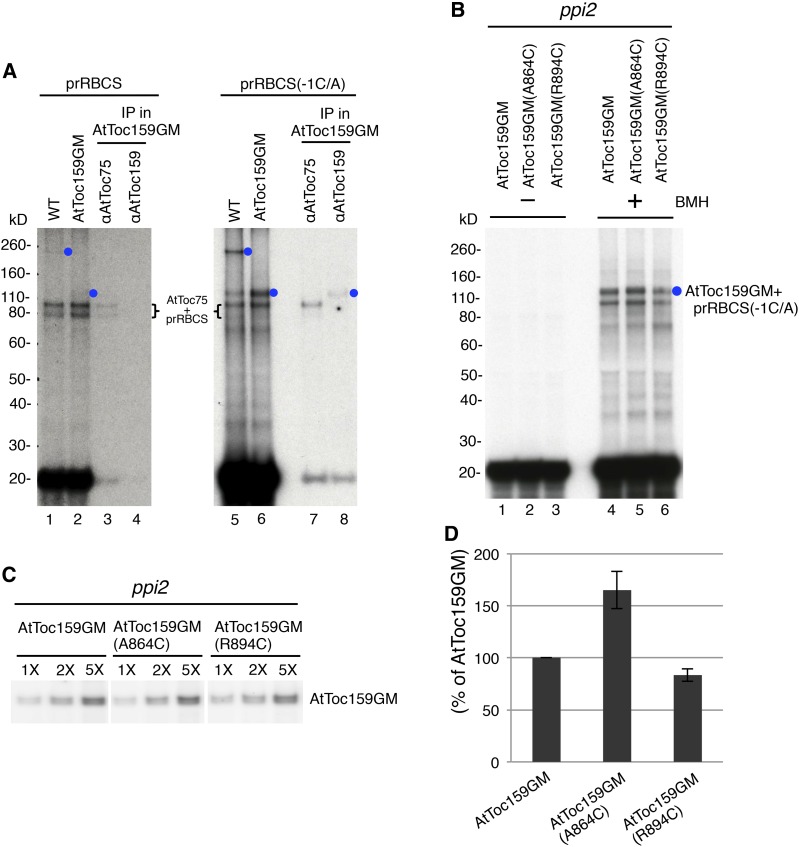
Cys placed at the dimer interface results in increased cross-linking of AtToc159 to preproteins bound on chloroplasts. A, prRBCS(-1C/A) exhibited more cross-linking to AtToc159 than prRBCS did. Chloroplasts were isolated from wild-type plants (WT) or AtToc159GM transgenic plants. [35S]Met-labeled prRBCS or prRBCS(-1C/A) was bound to the chloroplasts under 100 µm ATP and cross-linked to the chloroplasts with BMH. Samples were analyzed by SDS-PAGE (lanes 1, 2, 5, and 6). Part of the samples from the AtToc159GM transgenic plant chloroplasts was solubilized with 1% lithium dodecyl sulfate (LDS) and subjected to immunoprecipitation (IP) using anti-AtToc75 or anti-AtToc159 antibody. The immunoprecipitates were analyzed by SDS-PAGE (lanes 3, 4, 7, and 8). B, AtToc159GM(A864C) showed more cross-linking to preproteins. Chloroplasts were isolated from AtToc159GM, AtToc159GM(A864C), and AtToc159GM(R894C) transgenic plants. [35S]Met-labeled prRBCS(-1C/A) was bound to the chloroplasts under 100 µm ATP. Intact chloroplasts were reisolated. Part of the samples was analyzed directly to obtain the values of total prRBCS(-1C/A) bound on chloroplasts (lanes 1–3). Another part of the samples was used to perform BMH cross-linking assays (lanes 4–6). Samples were analyzed by SDS-PAGE and fluorography. C, Levels of AtToc159GM in transgenic plant chloroplasts. Chloroplasts from binding and cross-linking experiments as shown in B were serially diluted and analyzed by SDS-PAGE and immunoblotting using anti-AtToc159 antibody. The samples were analyzed on the same blotting membrane and developed for the same amount of time with intervening lanes removed. D, Quantification of the amount of cross-linking between AtToc159GM and prRBCS(-1C/A). Binding and cross-linking experiments as shown in B were performed, and the amount of AtToc159GM-prRBCS(-1C/A) cross-linked adduct (blue circle in B) was quantified by a phosphorimager. The value obtained was first normalized to the amount of prRBCS(-1C/A) bound to the chloroplasts of each transgenic line without BMH added (lanes 1–3 in B) and then further normalized to the amount of AtToc159GM in each transgenic plant as shown in C. Values plotted are means ± sd of three independent experiments.
Previously, we noticed that when the Cys at the −1 position (which is the only Cys in the prRBCS transit peptide) was replaced with Ala, a strong cross-linked signal was observed at about 110 kD (i.e. above the prRBCS-Toc75 adduct; Huang et al., 2016; the prRBCS variant was named prRBCS[-1C/A]). We suspect that this 110-kD cross-linked product may represent cross-linking of prRBCS(-1C/A) to the major pea Toc159 degradation product, which is about 86 kD and essentially composed of Toc159GM. Therefore, we tried performing BMH cross-linking experiments using prRBCS(-1C/A) with isolated Arabidopsis chloroplasts under 100 µm ATP. As shown in Figure 9A, the amount of 260-kD cross-linked product in the wild-type chloroplasts, and the amount of 110-kD cross-linked product in the AtToc159GM transgenic plant chloroplasts, were both increased significantly (lanes 5 and 6, blue circles). Chloroplasts from the AtToc159GM transgenic plants containing cross-linked prRBCS(-1C/A) were further solubilized with LDS and then immunoprecipitated using the anti-Toc75 or anti-Toc159 antibody. The two bands located between 80 and 100 kD were again precipitated by the anti-Toc75 antibody. The 110-kD band was precipitated by the anti-Toc159 antibody (Fig. 9A, lane 8, blue circle), indicating that this band represented the prRBCS(-1C/A)-AtToc159GM cross-linked adduct. Due to the better cross-linking efficiency of prRBCS(-1C/A) to Toc159, we used this preprotein to compare cross-linking to chloroplasts isolated from the three different transgenic plants. We also tested several other preproteins, but none offered better binding and cross-linking efficiency than prRBCS(-1C/A). It is not clear why prRBCS(-1C/A) shows more cross-linking to AtToc159GM than prRBCS does. It is possible that, with 100 µm ATP, the transit peptides of most bound prRBCS molecules are already in the Toc75 channel so their Cys at the −1 position would be cross-linked to Toc75 and their Cys in the mature region would be cross-linked to AtToc159GM. Cross-linked products containing three protein species (i.e. Toc75, AtToc159GM, and prRBCS) may be more difficult to solubilize or may have difficulties migrating into the separating gels.
To test whether the AtToc159G dimer interface makes contact with preproteins, chloroplasts were isolated from the AtToc159GM, AtToc159GM(A864C), and AtToc159GM(R894C) transgenic plants and incubated with prRBCS(-1C/A) under 100 µm ATP, and BMH cross-linking experiments were performed. The 110-kD cross-linked adduct of prRBCS(-1C/A)-AtToc159GM was observed in chloroplasts from all three transgenic lines (Fig. 9B, lanes 4–6, blue circle). To compare the amount of cross-linking in each line, the amount of the 110-kD prRBCS(-1C/A)-AtToc159GM cross-linked product was quantified and normalized to total prRBCS(-1C/A) bound to the chloroplasts before BMH was added (Fig. 9B, lanes 1–3) and the amount of AtToc159GM in each sample (Fig. 9C). Cross-linking of AtToc59GM to prRBCS(-1C/A) indicated that at least one of the endogenous Cys residues in AtToc159GM was in close proximity to some of the bound prRBCS(-1C/A). However, in chloroplasts from the AtToc159GM(A864C) transgenic line, in which Ala-864 at the dimer interface was replaced with Cys, we observed a 60% increase of BMH-mediated cross-linking to prRBCS(-1C/A) (Fig. 9, B, lane 5, and D). The increase was not observed in chloroplasts from the AtToc159GM(R894C) transgenic line. These results showed that residue 864 at the dimer interface was in close proximity to the mature region of prRBCS(-1C/A) when prRBCS(-1C/A) bound to chloroplasts.
DISCUSSION
Toc159 functions as the primary receptor for preprotein import into chloroplasts. As a first step in understanding how Toc159 mediates preprotein import, we mapped the preprotein-binding sites on Toc159. Using FeBABE cleavage and Cys-Cys cross-linking, we show that there are two sites on AtToc159G that are in close proximity to preproteins: one is on the dimer interface, and the other is in the switch II region. Preprotein binding to the dimer interface is further supported by data showing that preprotein binding to AtToc159G reduces the population of the AtToc159G dimer. Furthermore, in stable transgenic plants in which a Cys residue was placed at the AtToc159G dimer interface, BMH-mediated Cys-Cys cross-linking of AtToc159GM to preproteins bound to chloroplasts was increased significantly.
We further found that the transit peptide and the mature region of a preprotein were detected differentially at the two sites; the mature region was detected preferentially at the dimer interface, whereas the transit peptide binds similarly to both sites. It is possible that the transit peptide first binds to the dimer interface and then moves downward to bind to switch II, while the mature region then occupies the vacated dimer interface. In vivo, the transit peptide may subsequently contact Toc75 and then be translocated across the outer membrane through Toc75 when micromolar ATP is available.
It has been shown that transit peptides also reduce the dimerization of Toc34 (Oreb et al., 2011; Lumme et al., 2014; Holbrook et al., 2016), suggesting that the two GTPases may interact with preproteins in a similar manner. Furthermore, by comparing the monomeric conformation of Arabidopsis AtToc33G with the dimeric conformation of pea Toc34G, it was noted that significant conformational differences exist between the monomeric and dimeric forms in switch II (Koenig et al., 2008a). Therefore, if Toc159 behaves similarly to Toc34, monomer-dimer conversion would affect conformations in both the dimer interface and switch II (i.e. the two regions that we show to be in close proximity to preproteins).
The functional mechanism of the Toc159 and Toc34 receptor GTPases in mediating preprotein import is complicated and not fully understood. Results from different research groups have generated differing models. In vitro analyses suggest that preprotein binding induces the dissociation of Toc34 dimers and promotes the exchange of bound GDP for GTP (Oreb et al., 2011; Lumme et al., 2014; Holbrook et al., 2016), suggesting that preproteins may initially bind to the GDP-bound Toc34 dimer and then act like a GTP/GDP exchange factor (GEF) for Toc34 by inducing dimer dissociation (Li et al., 2007; Oreb et al., 2011). Interestingly, many GEFs interact extensively with the switch II region of their target GTPase (Cherfils and Chardin, 1999; Cherfils and Zeghouf, 2013), and our results here also showed preproteins contacting switch II of AtToc159G. However, other groups have proposed that transit peptides function as the GAP, not GEF, for Toc34 (Jelic et al., 2002; Reddick et al., 2007). In vivo, chloroplasts from transgenic plants expressing an AtToc33 mutant that could not dimerize have a higher affinity for preproteins but have reduced efficiency in subsequent preprotein translocation into the Toc75 channel (Lee et al., 2009), suggesting that the conformational change arising from AtToc33 dimer formation is related to preprotein translocation into the Toc75 channel (Lee et al., 2009). For Toc159, mutants that bind but do not hydrolyze GTP actually have an increased preprotein-binding and import rate, suggesting that the GTP-bound form of AtToc159 is the active component in stable preprotein binding to the TOC translocon (Wang et al., 2008).
One thing that most models share is that conformational changes brought about by the monomer-dimer conversion of Toc159G should affect the import of preproteins. Therefore, our data showing the binding of preproteins to the dimer interface and switch II provide a critical piece of evidence that is required for these models. Preproteins may recognize the Toc159G monomers directly or interact with the dimers and then induce dimer opening. Binding of preproteins to the dimer interface may then facilitate either GDP-to-GTP exchange or GTPase activation. If preprotein binding induces GDP-to-GTP exchange, as has been suggested (Li et al., 2007; Oreb et al., 2011), and the preprotein temporarily remains on the dimer interface after the dimer disengages, as we showed here, then a Toc159 mutant with a higher affinity for GTP but reduced hydrolysis would indeed have increased GTP-stimulated preprotein binding, as shown previously (Wang et al., 2008). Conformational changes in the dimer interface and switch II brought about by redimerization would result in the dissociation of bound preproteins and subsequent preprotein translocation into Toc75. Therefore, a mutant that could not dimerize would have a higher affinity for bound preproteins but reduced translocation efficiency into Toc75, as shown previously (Lee et al., 2009).
Our work offers the initial localization information for preprotein on Toc159. Residues in AtToc159G that show good cross-linking to preproteins are surface exposed but do not seem to be strictly conserved (Fig. 4B). More residues in AtToc159G and more preproteins with Cys in different positions need to be tested to produce a better picture for the path of a preprotein on AtToc159 during import. More work is also required to understand the nature of the Toc159G preprotein-binding sites that allows AtToc159 to bind to diverse transit peptide sequences.
MATERIALS AND METHODS
Growth of Pea and Arabidopsis and Chloroplast Isolation
Pea (Pisum sativum ‘Green Arrow’) seeds were imbibed overnight and grown on vermiculite for 7 to 10 d under a 12-h photoperiod at 20°C with a light intensity of ∼150 μmol m−2 s−1. Sterilized Arabidopsis (Arabidopsis thaliana) seeds were plated on 0.3% Gelrite-solidified Murashige and Skoog medium containing Gamborg’s B5 vitamin and 2% Suc as described previously (Su and Li, 2010). Pea and Arabidopsis seedlings were harvested and chloroplasts were isolated as described previously (Perry et al., 1991; Chu and Li, 2011).
Purification of Recombinant Proteins and Antibody Production
Plasmids were transformed into the Escherichia coli BL21(DE3)pLysS strain, and protein expression was induced using 1 mm IPTG. After incubation at 37°C for 3 h, the bacteria were harvested and lysed using an M-110P Microfluidizer (Microfluidics). The bacterial lysate was centrifuged at 12,000g for 30 min, and soluble recombinant proteins were purified by passing the supernatant fraction through affinity purification columns. All the recombinant proteins used in this study are in the soluble fractions. For the purification of GST and prRBCS-GST, GSH resin (GE Healthcare) was used. For the purification of AtToc159G (AtToc159 residues 727–1,092), prFd-protAHis, and prFd-protAHis(-37S/C), Talon affinity resin (Clontech) was used. The purified recombinant proteins were dialyzed against 50 mm HEPES-NaOH, pH 7.5, and protein concentration was measured with the BCA Kit (Thermo).
The same AtToc159G construct described above was used to produce antigen to generate the rat anti-AtToc159G antibody used in Figures 2 and 3. The plasmid encoding AtToc159G was transformed into the E. coli BL21(DE3)pLysS strain, and AtToc159G was overexpressed and purified using Talon resin as described above and shown in Figure 2A. The rat anti-AtToc159G antibody was used at a 1:2,000 dilution. This antibody specifically recognizes AtToc159 (Supplemental Fig. S2). To detect various forms of AtToc159 in Arabidopsis plants in Figures 8 and 9, a previously described anti-AtToc159 antibody (Tu et al., 2004) generated against the M domain of AtToc159 was used at a 1:2,000 dilution.
Plasmid Construction, in Vitro Translation of Preproteins, and Antibody Preparation
The coding region for residues 727 to 1,092 of AtToc159 (At4g02510), corresponding to the GTPase domain, was amplified by PCR and cloned into the NcoI/XhoI site of pET21d, resulting in translational fusion of the His6 tag to the C terminus of AtToc159G. For prRBCS-GST, the coding region for the pea prRBCS transit peptide was amplified by PCR and cloned into the unique EcoNI site at the N terminus of the GST coding region in plasmid pGEX-5X-1. All the sequences were confirmed by sequencing. Plasmids encoding prRBCS (Lubben and Keegstra, 1986) and prFd-protAHis (Smith et al., 2004) have been described.
Preprotein Binding and Import into Chloroplasts and Postimport Analyses
Isolated chloroplasts were incubated with 100 or 200 pmol of GST or prRBCS-GST, as indicated, in the presence of 0.1 mm ATP in import buffer (330 mm sorbitol in 50 mm HEPES-KOH, pH 8) on ice for 10 min to assay protein binding to chloroplasts. Intact chloroplasts were reisolated by loading the reactions onto a 40% Percoll cushion and centrifuged at 2,900g for 6 min at 4°C. For import assays, isolated chloroplasts were incubated with 200 pmol of GST or prRBCS-GST in the presence of 3 mm ATP at room temperature for 10 min, and intact chloroplasts were reisolated as described above. Thermolysin treatments of chloroplasts after import reactions were performed as described (Perry et al., 1991), and intact chloroplasts were reisolated as described above. Samples were analyzed by SDS-PAGE (NuPAGE 4-12% Bis-Tris Gel; Invitrogen), and the bound GST and prRBCS-GST were visualized by immunoblotting using an anti-GST antibody (Invitrogen).
In Vitro Pull-Down Assays Using GSH Resin
To bind GST or prRBCS-GST onto GSH resin, 75 pmol of purified GST or prRBCS-GST was incubated with 5 μL of washed GSH resin in 200 μL of binding buffer (50 mm HEPES-NaOH, pH 8) containing 5 mm MgCl2 at room temperature for 30 min. Seventy-five picomoles of purified wild-type AtToc159G was then added and incubated for another 30 min. The GSH resin was pelleted and washed with 1 mL of wash buffer (50 mm HEPES-NaOH, pH 8, 5 mm MgCl2, and 150 mm NaCl) three times. The bound proteins were eluted from the GSH resin using 30 μL of 1× NuPAGE sample buffer. The samples were analyzed by SDS-PAGE, and the bound proteins were visualized by immunoblotting using the anti-GST antibody or anti-AtToc159G antibody.
FeBABE Conjugation
FeBABE (Dojino) conjugation was performed as described previously (Datwyler and Meares, 2000). In brief, purified recombinant preproteins were desalted by gel filtration as described (Su and Li, 2010) to remove any possible reducing agents. One hundred sixty picomoles of desalted preprotein was then incubated with 800 pmol of FeBABE in the FeBABE conjugation buffer (20 mm MOPS-KOH, pH 8, and 10% [v/v] glycerol) at room temperature for 30 min. Free FeBABE was then removed by centrifugation at 3,900g for 6 min using an Amicon Ultra-0.5 concentrator (Merck Millipore). FeBABE-conjugated preproteins in the concentrator were resuspended in conjugation buffer and stored at −70°C. Protein concentrations of the FeBABE-conjugated preproteins were measured with the BCA Kit (Thermo).
In Vitro Pull-Down Assays for FeBABE Cleavage
For prRBCS-GST, 75 pmol of prRBCS-GST or FeBABE-conjugated prRBCS-GST was incubated with 5 μL of GSH resin in 200 μL of binding buffer at room temperature for 30 min. Seventy-five picomoles of purified wild-type AtToc159G was then added and further incubated for another 30 min. The GSH resin was pelleted to pull down AtToc159 bound to prRBCS-GST or GST. The beads were washed with 1 mL of wash buffer (50 mm HEPES-NaOH, pH 8, 5 mm MgCl2, and 150 mm NaCl) three times. For prFd-protAHis, 75 pmol of prFd-protAHis or FeBABE-conjugated prFd-protAHis was incubated with 75 pmol of purified wild-type AtToc159G in 100 μL of binding buffer at 4°C for 2 h. After incubation, 10 μL of IgG Sepharose (GE Healthcare) was added to the reactions and further incubated at 4°C for another 30 min. The IgG Sepharose was pelleted to pull down the AtToc159G bound to prFd-protAHis or Fe-BABE-conjugated prFd-protAHis. The beads were washed with 500 μL of wash buffer three times.
FeBABE cleavage was conducted as described previously (Chen and Hahn, 2003). The beads with FeBABE-conjugated preproteins and bound AtToc159G were resuspended in 20 μL of 50 mm HEPES-KOH, pH 8, and 2.5 μL of 100% glycerol was then added to scavenge diffusible hydroxyl radicals during the subsequent cleavage reaction. The cleavage reaction was initiated by the sequential addition of 3.2 μL of 40 mm sodium ascorbate and 2.5 μL of freshly prepared initiating reagent (6 μL of 30% hydrogen peroxide added to 994 μL of 50 mm HEPES-KOH, pH 8, and 10 mm EDTA). The reactions were incubated at room temperature for 5 min and then quenched by adding 8 μL of 4× NuPAGE LDS sample buffer (Invitrogen). The samples were analyzed by SDS-PAGE, and the cleaved fragments were visualized by immunoblotting using the anti-AtToc159G or anti-His6 antibody.
Site-Directed Mutagenesis
The four endogenous Cys residues in AtToc159G were mutated to Ala, and other desired positions were further mutated to Cys to generate single-Cys AtToc159G mutants using the QuikChange Site-Directed Mutagenesis Kit (Agilent). For prFd-protAHis(-37S/C), the five endogenous Cys residues in prFd-protAHis were mutated to Ala, and the Ser at the −37 position in the transit peptide was further mutated to Cys. Five evenly spaced positions in the prFd transit peptide were initially chosen and mutated to Cys. In pilot experiments, prFd-protAHis(-37S/C) showed the best cross-linking to AtToc159G.
Homology Modeling of AtToc159G
Residues 727 to 1,092 of AtToc159G (i.e. covering the entire predicted GTPase domain) were initially used to generate a monomer model of AtToc159G on the Phyre2 sever (Kelley et al., 2015) using pea Toc34G (Protein Data Bank no. 1h65) as a template. Two regions could not be aligned to Toc34G, the N-terminal portion of AtToc159G from residues 727 to 834 and the C-terminal portion from residues 1,062 to 1,092, so no model for these two regions could be generated. A dimer structure for AtToc159G was obtained by further superimposing the AtToc159G monomer structure (residues 835–1,061) or the AtToc159G(A864C) mutant onto the peaToc34G dimer structure with energy minimization by the CNS program (Brünger et al., 1998; Brunger, 2007).
In Vitro Cross-Linking Assays Using BMH
[35S]Met-labeled wild-type or mutant AtToc159G were in vitro transcribed/translated using a TNT-coupled rabbit reticulocyte lysate system and SP6 RNA polymerase (Promega). One microliter of [35S]Met-labeled wild-type or mutant AtToc159G was incubated with 100 pmol of purified recombinant preproteins at room temperature for 20 min. BMH was added to the reaction to a final concentration of 0.2 mm and incubated for another 20 min. Cross-linking reactions were terminated by adding 6 μL of 4× NuPAGE LDS sample buffer (Invitrogen). The samples were analyzed by SDS-PAGE and fluorography.
Generation of AtToc159GM Transgenic Plants
A cDNA fragment encoding AtToc159GM (residues 727–1,503) was amplified by PCR using the forward primer B1-Toc86 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGACATCTCAGGATGGCACGAAGC-3′) and the reverse primer Toc86-B2R (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAGTACATGCTGTACTTGTC-3′). The PCR product was cloned into the pDONR vector (Earley et al., 2006) by performing the BP recombination reaction (Invitrogen), creating the plasmid pDONR-AtToc159GM. The Ala-864 and Arg-894 of AtToc159GM in pDONR-AtToc159GM were mutated to Cys separately using the QuikChange Site-Directed Mutagenesis Kit (Agilent), and the sequences were confirmed by sequencing. The three entry clones carrying wild-type AtToc159GM, AtToc159GM(A864C), and AtToc159GM(R894C) were then cloned into the pEarleyGate 202 vector (Earley et al., 2006) to generate the plant transformation clones by performing the LR recombination reaction. The three plant transformation clones were then transformed into Agrobacterium tumefaciens GV3101, and the transformed agrobacteria were used to transform plants heterozygous for the ppi2 mutation (CS11072; Bauer et al., 2000) using the floral dip method (Clough and Bent, 1998). T1 seeds were screened on MS medium containing 0.05 mm Basta (phosphinotricin) to select for plants containing the transgene encoding the introduced AtToc159GM and 50 μg mL−1 kanamycin to select for the ppi2 mutation. T2 seeds from individual T1 plants resistant to Basta and kanamycin were again sown on MS medium containing Basta and kanamycin. Basta- and kanamycin-resistant T2 plants were genotyped to identify plants homozygous for the ppi2 mutation, and their seeds were harvested separately. T3 plants homozygous for the ppi2 mutation and stably expressing the introduced AtToc159GM were used for the chloroplast isolation and BMH cross-linking experiments. Primers used for genotyping by PCR were as follows: for the endogenous wild-type AtToc159, 4T 4-3 (5′-TCGAATTCATGGACTCAAAGTCGGTTACTCCA-3′) and 4T 4-6 (5′-CCTTCTTCTGCATCATCGACTATG-3′); for the ppi2 mutation, 4T 4-3 and LB102A (5′-GATGCAATCGATATCAGCCAATTTTAGAC-3′); and for the AtToc159GM transgenes, AtToc159G 271F (5′-CACCCCATTGTGTTATGATGTGTTT-3′) and M13 forward primer (5′-GTAAAAACGACGGCCAG-3′). The ppi2 mutant, the nontransformed wild-type control, and the transgenic lines are all in the Wassilewskija ecotype.
BMH Cross-Linking of [35S]Met-prRBCS Bound on Chloroplasts and Immunoprecipitation
Isolated chloroplasts of the transgenic plants were incubated in the dark on ice for 30 min to deplete internal ATP. In vitro-translated [35S]Met-labeled prRBCS and prRBCS(-1C/A) were synthesized using TNT Coupled Wheat Germ Extract (Promega). For the binding assays, [35S]Met-labeled prRBCS or prRBCS(-1C/A) was gel filtrated to remove free nucleotides from the TNT wheat germ extract and then incubated with ATP-depleted chloroplasts in the presence of 100 µm ATP in the dark at room temperature for 15 min. After the binding, intact chloroplasts were reisolated through a 40% Percoll cushion and resuspended in import buffer, and BMH was added to a final concentration of 0.5 mm. The reaction was incubated at 4°C for 30 min and then quenched by adding 4× NuPAGE sample buffer. The samples were analyzed by SDS-PAGE and fluorography. If the samples were to be used for immunoprecipitation, after BMH cross-linking, DTT was added to the reaction to a final concentration of 50 mm and further incubated at 4°C for 15 min to quench the BMH. Chloroplast solubilization with 1% LDS and subsequent immunoprecipitation were performed as described previously (Huang et al., 2016). The antibodies used for immunoprecipitation were against the AtToc75 POTRA domain (Chen et al., 2016) and against the AtToc159 M domain (Tu et al., 2004), both of which have been described previously.
GTPase Activity Assay
Two microliters of [γ-32P]GTP was mixed with 2 μg of purified wild-type or single-Cys mutant AtToc159G in a final volume of 10 μL containing 50 mm HEPES-NaOH, pH 7.5, 2 mm MgCl2, and 0.2 mm cold GTP and incubated at 30°C for 30 min. The GTPase reactions were terminated by adding 1 μL of 0.5 m EDTA, pH 8. The samples were loaded onto thin-layer chromatography plates (Sigma-Aldrich; Z122882) and developed with 1 m formic acid and 0.5 m LiCl for 15 min to separate the free phosphates and GTP. The thin-layer chromatography plates were analyzed by autoradiography and quantified by a phosphorimager (Typhoon; FLA 9000).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/TAIR/UniProtKB data libraries under accession numbers At4g02510 (Arabidopsis Toc159), AAF75761 (pea Toc159), XP_009769991 (tobacco [Nicotiana tabacum] Toc159), NP_001147969 (maize [Zea mays] Toc159), XP_015639221 (rice [Oryza sativa] Toc159), XP_004152365 (cucumber [Cucumis sativus] Toc159), XP_002312975 (Populus spp. Toc159), and Q41009 (pea Toc34).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GTPase activities of wild-type and single-Cys mutant AtToc159G.
Supplemental Figure S2. The rat anti-AtToc159G antibody.
Supplemental Figure S3. The cross-linked adducts contain the preprotein added.
Supplementary Material
Acknowledgments
We thank Dr. Hung-Ta Chen for assistance in performing FeBABE cleavage analysis, Chia-Yun Chang for assistance in generating the Arabidopsis transgenic plants, and the Institute of Molecular Biology English Editing Core for English editing.
Glossary
- BMH
bismaleimidohexane
- LDS
lithium dodecyl sulfate
- MS
Murashige and Skoog
Footnotes
This work was supported by the Ministry of Science and Technology, Taiwan (grant no. MOST 105-2321-B-001-011 to H.-m.L.), and Academia Sinica of Taiwan.
Articles can be viewed without a subscription.
References
- Agne B, Infanger S, Wang F, Hofstetter V, Rahim G, Martin M, Lee DW, Hwang I, Schnell D, Kessler F (2009) A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J Biol Chem 284: 8670–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 [DOI] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 [DOI] [PubMed] [Google Scholar]
- Brunger AT. (2007) Version 1.2 of the Crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chen HT, Hahn S (2003) Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol Cell 12: 437–447 [DOI] [PubMed] [Google Scholar]
- Chen K, Chen X, Schnell DJ (2000) Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol 122: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Chen LJ, Li HM (2016) Polypeptide transport-associated domains of the Toc75 channel protein are located in the intermembrane space of chloroplasts. Plant Physiol 172: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Chardin P (1999) GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci 24: 306–311 [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309 [DOI] [PubMed] [Google Scholar]
- Chu CC, Li HM (2011) Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol Biol 774: 339–350 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Datwyler SA, Meares CF (2000) Protein-protein interactions mapped by artificial proteases: where sigma factors bind to RNA polymerase. Trends Biochem Sci 25: 408–414 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J (1994) A receptor component of the chloroplast protein translocation machinery. Science 266: 1989–1992 [DOI] [PubMed] [Google Scholar]
- Holbrook K, Subramanian C, Chotewutmontri P, Reddick LE, Wright S, Zhang H, Moncrief L, Bruce BD (2016) Functional analysis of semi-conserved transit peptide motifs and mechanistic implications in precursor targeting and recognition. Mol Plant 9: 1286–1301 [DOI] [PubMed] [Google Scholar]
- Huang PK, Chan PT, Su PH, Chen LJ, Li HM (2016) Chloroplast Hsp93 directly binds to transit peptides at an early stage of the preprotein import process. Plant Physiol 170: 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelic M, Sveshnikova N, Motzkus M, Hörth P, Soll J, Schleiff E (2002) The chloroplast import receptor Toc34 functions as preprotein-regulated GTPase. Biol Chem 383: 1875–1883 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ (1994) Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Koenig P, Oreb M, Höfle A, Kaltofen S, Rippe K, Sinning I, Schleiff E, Tews I (2008a) The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures. Structure 16: 585–596 [DOI] [PubMed] [Google Scholar]
- Koenig P, Oreb M, Rippe K, Muhle-Goll C, Sinning I, Schleiff E, Tews I (2008b) On the significance of Toc-GTPase homodimers. J Biol Chem 283: 23104–23112 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ (1997) Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol 139: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lee J, Wang F, Schnell DJ (2009) Toc receptor dimerization participates in the initiation of membrane translocation during protein import into chloroplasts. J Biol Chem 284: 31130–31141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chiu CC (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Li HM, Kesavulu MM, Su PH, Yeh YH, Hsiao CD (2007) Toc GTPases. J Biomed Sci 14: 505–508 [DOI] [PubMed] [Google Scholar]
- Lubben TH, Keegstra K (1986) Efficient in vitro import of a cytosolic heat shock protein into pea chloroplasts. Proc Natl Acad Sci USA 83: 5502–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumme C, Altan-Martin H, Dastvan R, Sommer MS, Oreb M, Schuetz D, Hellenkamp B, Mirus O, Kretschmer J, Lyubenova S, et al. (2014) Nucleotides and substrates trigger the dynamics of the Toc34 GTPase homodimer involved in chloroplast preprotein translocation. Structure 22: 526–538 [DOI] [PubMed] [Google Scholar]
- Ma Y, Kouranov A, LaSala SE, Schnell DJ (1996) Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J Cell Biol 134: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreb M, Höfle A, Koenig P, Sommer MS, Sinning I, Wang F, Tews I, Schnell DJ, Schleiff E (2011) Substrate binding disrupts dimerization and induces nucleotide exchange of the chloroplast GTPase Toc33. Biochem J 436: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila YD, Richardson LG, Schnell DJ (2015) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol 427: 1038–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Keegstra K (1994) Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Li HM, Keegstra K (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Reddick LE, Vaughn MD, Wright SJ, Campbell IM, Bruce BD (2007) In vitro comparative kinetic analysis of the chloroplast Toc GTPases. J Biol Chem 282: 11410–11426 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol 7: 303–304 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G (1994) Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2013) The chloroplast protein import system: from algae to trees. Biochim Biophys Acta 1833: 314–331 [DOI] [PubMed] [Google Scholar]
- Smith MD, Hiltbrunner A, Kessler F, Schnell DJ (2002) The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J Cell Biol 159: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Rounds CM, Wang F, Chen K, Afitlhile M, Schnell DJ (2004) atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J Cell Biol 165: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YJ, Forouhar F, Li HM, Tu SL, Yeh YH, Kao S, Shr HL, Chou CC, Chen C, Hsiao CD (2002) Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nat Struct Biol 9: 95–100 [DOI] [PubMed] [Google Scholar]
- Tu SL, Chen LJ, Smith MD, Su YS, Schnell DJ, Li HM (2004) Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Agne B, Kessler F, Schnell DJ (2008) The role of GTP binding and hydrolysis at the atToc159 preprotein receptor during protein import into chloroplasts. J Cell Biol 183: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem 80: 943–971 [DOI] [PubMed] [Google Scholar]
- Yeh YH, Kesavulu MM, Li HM, Wu SZ, Sun YJ, Konozy EH, Hsiao CD (2007) Dimerization is important for the GTPase activity of chloroplast translocon components atToc33 and psToc159. J Biol Chem 282: 13845–13853 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.