The protein translocase SEC2 in the inner chloroplast envelope facilitates membrane integration of a subset of envelope proteins.
Abstract
Most chloroplast proteins are synthesized in the cytosol and imported into chloroplasts. Many imported proteins are further targeted to the thylakoid membrane and lumen by the SEC1, TAT, or SRP/ALB3 translocases. Others are targeted to the inner chloroplast envelope membrane by undescribed translocases. Recently, a second SEC system (SEC2) consisting of SCY2, SECE2, and SECA2 was found in the chloroplast envelope. Null mutants of SCY2 in Arabidopsis (Arabidopsis thaliana) exhibit a severe embryo-lethal phenotype. To investigate the function of the SEC2 system in plants, we used inducible RNA interference to knock down SCY2 in Arabidopsis. Seedlings cultured with inducer were chlorotic with aberrant chloroplasts and undeveloped thylakoids, indicating an essential role for SCY2 in chloroplast biogenesis beyond embryo development. In SCY2 down-regulated seedlings, several thylakoid membrane proteins, including SCY1, ALB3, and TATC, and inner envelope membrane proteins, including TIC40, TIC110, and FTSH12, were reduced substantially, suggesting that they may be SEC2 substrates. Additional insight was achieved by the in vitro reconstitution of protein integration into chloroplast membranes. The results show that SCY1 and ALB3 target directly to the thylakoid membrane and are likely independent of SEC2. FTSH12 was integrated into the envelope membrane in a coupled import-integration reaction that was impaired by the SECA inhibitor sodium azide. The stromal intermediate of TIC40 integrated into the envelope in a reaction that was largely inhibited when antibodies against epitope-tagged SCY2 or SECE2 were applied. These data demonstrate that the SEC2 translocase likely integrates a subset of inner envelope membrane proteins, such as FTSH12 and TIC40.
Chloroplasts of plants and algae evolved from an endosymbiotic cyanobacterium (Cavalier-Smith, 1987; Celedon and Cline, 2013). Initially, the endosymbiont contained ∼3,000 genes and synthesized and localized its proteins by means of protein translocases: that is, enzymes that translocate proteins across or into membrane bilayers. During evolution to the modern chloroplast, most endosymbiont genes were transferred to the host nucleus. Today, many of the ancestral proteins are returned to the chloroplast by synthesis in the cytosol, import into the organelle, and sorting to the six different chloroplast compartments: the outer envelope membrane, the inner envelope membrane, the inter envelope space, the aqueous stromal matrix, the internal thylakoid membrane, and the aqueous lumen it encloses (Cline and Dabney-Smith, 2008; Celedon and Cline, 2013).
Import into the chloroplast stroma is governed by a transit peptide appended to the N terminus of precursor proteins, which directs translocation through the TOC (translocon at the outer envelope membrane of chloroplasts) and TIC (translocon at the inner envelope membrane of chloroplasts) complexes (Cline and Dabney-Smith, 2008; Li and Chiu, 2010). Sorting from the stroma involves secondary sorting signals and, frequently, additional translocases (Celedon and Cline, 2013). In the case of many proteins destined for the thylakoid membrane and lumen, the imported proteins are directed from the stroma to thylakoids (Cline et al., 1992) by conserved ancestral translocases, such as the Signal Recognition Particle (SRP)/ALBINO3 (ALB3) integrase (Cline et al., 1992; Li et al., 1995; Moore et al., 2000), the chloroplast SEC1 translocase (Nakai et al., 1994; Yuan et al., 1994; Settles et al., 1997; Mori et al., 1999), and the chloroplast Twin Arginine Translocation (TAT) translocase (Mori et al., 1999), which are highly homologous to the SRP/YidC, Sec, and Tat systems of bacteria (Schuenemann et al., 1999; Cline and Dabney-Smith, 2008; Celedon and Cline, 2012, 2013). This organization of a general import translocase followed by an evolutionarily conserved destination translocase has been called conservative sorting (Hartl et al., 1986), a term that evokes the parsimonious use of ancient conserved systems that coevolved with their substrates more than one billion years ago.
Although much is known about the sorting of imported chloroplast proteins, there are many unknowns. For example, little is known about how multispanning membrane proteins, such as SCY1 (10 transmembrane domains [TMs]), TATC (six TMs), and ALB3 (five TMs), are integrated into thylakoid membranes. In addition, the integration of proteins of the inner envelope membrane proceeds by unknown mecahnisms, although pathways for some have been mapped. The two main pathways are called the stop-transfer pathway and the post-import integration pathway (Cline and Dabney-Smith, 2008; Li and Chiu, 2010; Viana et al., 2010). In the stop-transfer pathway the substrate’s TM stops its transfer at the TOC complex during plastid import (Froehlich and Keegstra, 2011). In the post-import pathway, proteins are initially imported across the TOC/TIC before insertion into the inner envelope membrane from the stromal side (Li and Schnell, 2006; Tripp et al., 2007; Vojta et al., 2007). This latter pathway suggests a conservative sorting mechanism and implies that a conserved traslocase is present in the inner envelope membrane. (For convenience, we followed the Arabidopsis [Arabidopsis thaliana] nomenclature and use uppercase to name general pathways [e.g. SEC and TAT] and Arabidopsis traslocase components [e.g. SEC1 and TATC] and designated proteins from other species with a species prefix [e.g. psALB3 and psSCY1].)
A conserved SEC translocase system called SEC2 was described recently in Arabidopsis (Skalitzky et al., 2011; Li et al., 2015). The identified components SECA2, SCY2, and SECE2 are distantly related to thylakoid SEC1 components (SECA1, SCY1, and SECE1), the bacterial SECA, SECY, and SECE (Skalitzky et al., 2011; Li et al., 2015), and the endoplasmic reticulum Sec61α and Sec61γ. SCY2 and SECE2 form a complex in the membrane (Li et al., 2015) and are localized primarily in the plastid envelope with possibly a small portion in the thylakoids (Skalitzky et al., 2011; Li et al., 2015). The SCY2/SECE2 complex is expected to serve as a protein-conducting channel similar to the orthologous components in other SEC systems. Substrates for SEC systems are targeted by N-terminal cleavable hydrophobic signal sequences or by N-terminal uncleaved TMs (termed signal anchors; Rapoport, 2007). Thus, SEC systems are versatile translocases that can fully translocate a preprotein across the membrane or integrate membrane proteins. Known SEC systems invariably transport proteins from the cytosol or cytosol equivalent compartment (cis; e.g. the plastid stroma) to the trans-compartment. In the case of plastid SEC2, the trans-compartment is the inter envelope space; for thylakoid SEC1, it is the thylakoid lumen. The SEC2 system is essential in Arabidopsis because null mutation of the gene for any SEC2 component results in embryo arrest at the globular stage (Skalitzky et al., 2011; Li et al., 2015). However, neither the translocase activity of SEC2 nor its substrates have been characterized.
In this study, two experimental approaches were used to identify SEC2 substrates. First, inducible RNA interference (RNAi) was employed to down-regulate SCY2 transcription during Arabidopsis seedling growth in order to assess the role of SCY2 in chloroplast biogenesis. Our results show that SCY2 deficiency results in drastically reduced accumulation of several plastid envelope and thylakoid membrane proteins. These proteins can be considered substrate candidates for SCY2. In a second approach, assays were developed for the integration of candidate proteins into isolated membranes, thereby allowing detailed analysis of the integration requirements and the use of inhibiting antibodies.
The results of these analyses suggest that, in chloroplasts, SCY1, ALB3, and probably TATC are integrated into thylakoids by an active integration mechanism that remains to be fully described. TIC40 and FTSH12 are integrated into the envelope with the expected characteristics of a SEC2-mediated mechanism. Specifically, antibodies to SCY2 and SECE2 inhibited TIC40 integration. Sodium azide, a potent inhibitor of SECA proteins, inhibited FTSH12 integration. This study provides, to our knowledge, an initial identification of substrates of the SEC2 system and, importantly, describes methods to achieve the in vitro integration of membrane proteins essential for envelope and thylakoid biogenesis.
RESULTS
Inducible Down-Regulation of SCY2 in Arabidopsis
In order to bypass the embryo development defect and investigate potential functions of SCY2 after germination, Arabidopsis was transformed with a hairpin SCY2-RNAi construct under the control of an estrogen-inducible promoter in vector pMDC7 (Curtis and Grossniklaus, 2003). The sequence used for the hairpin was from the second exon of SCY2 (Fig. 1A), which lacks homology to other sequences in the Arabidopsis genome and, in particular, SCY1. Plants transformed with empty vector (pMDC7) and plants transformed with a SCY1-RNAi construct served as the negative and positive controls, respectively (Fig. 1A). Multiple lines containing RNAi constructs were isolated and confirmed through PCR screening.
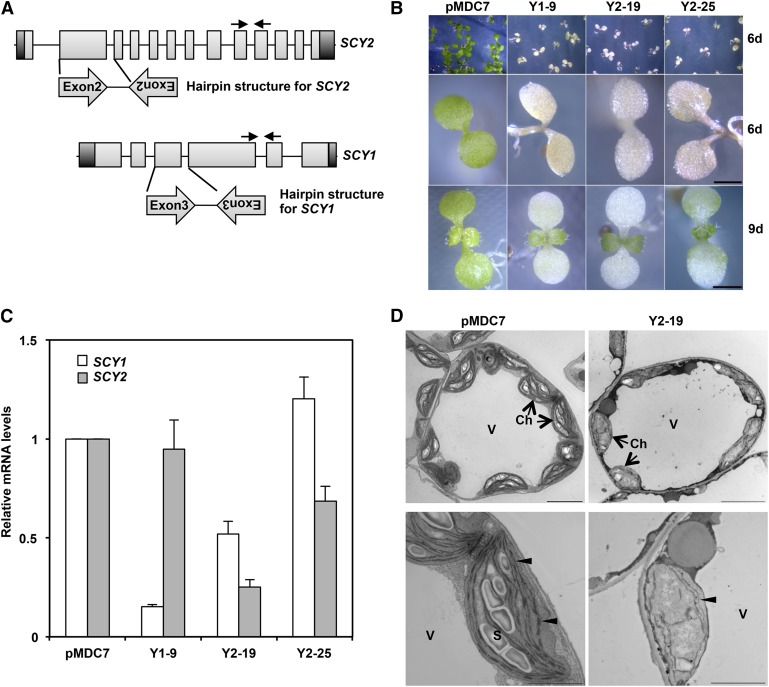
Figure 1.
RNAi-mediated silencing of SCY2 and SCY1 results in chlorotic cotyledons in Arabidopsis. A, Scheme of inducible RNAi constructs used to silence SCY2 or SCY1 in Arabidopsis. The hairpin structures were generated from the most diverged regions between SCY2 (the second exon and intron) and SCY1 (the third exon and intron) and cloned downstream of the estrogen-inducible promoter XEV in the pMDC7 vector. Black arrows mark the positions of primers used in quantitative real-time (qRT)-PCR to quantify SCY2 and SCY1 transcripts. B, Phenotypes of 6-d-old (top and middle rows) and 9-d-old (bottom row) seedlings germinated on MS medium containing 15 μg mL−1 hygromycin and 20 μm estrogen. pMDC7, Empty vector; Y1-9, SCY1-RNAi; Y2-19 and Y2-25, two lines of SCY2-RNAi. Bars = 1 mm. C, Relative mRNA levels of SCY1 (white bars) and SCY2 (gray bars) in 6-d-old seedlings germinated on MS medium containing 20 μm estrogen as determined by qRT-PCR analysis. Data represent mean values of at least four biological replicates and are normalized to the control (pMDC7). Error bars indicate se. D, Electron micrograph images of cotyledon mesophyll cells of 6-d-old seedlings germinated on MS medium with 20 μm estrogen. The top row shows overviews of mesophyll cells from the empty vector control (pMDC7) or SCY2-RNAi (Y2-19) cotyledons, and the bottom row shows higher magnification views of individual plastids from the same cells. The number of starch grains (S) is reduced significantly, and thylakoids (arrowheads) are undeveloped in SCY2-RNAi plastids. Ch, Chloroplast; V, vacuole. Bars = 5 μm (top row) and 2 μm (bottom row).
T3 seeds bearing empty vector or hairpin constructs were germinated on Murashige and Skoog (MS) medium containing 20 μm β-estrogen. The cotyledons and hypocotyls of SCY2-RNAi lines were white and, in some cases, purple, due to the accumulation of anthocyanin (Fig. 1B). Induced SCY1-RNAi lines had white to yellow cotyledons without anthocyanin accumulation (Fig. 1B). Plants transformed with empty vector had green seedlings with or without induction (Fig. 1B). When germinated on medium without estrogen, all transgenic plants were indistinguishable from wild-type plants (data not shown). Plastids in the cotyledons of treated SCY2-RNAi seedlings had irregular morphologies, limited thylakoid development, and virtually no starch grains (Fig. 1D). Appressed grana regions were absent in the internal membranes, consistent with the reduction in chlorophyll (Chl). These observations indicated that SCY2 silencing causes global defects in chloroplast and thylakoid development in Arabidopsis.
Phenotypic changes correlated with the extent of silencing in different lines, as revealed by qRT-PCR. In the line with the most severe phenotype (Y2-19), the SCY2 transcript level was 25% of that in control seedlings, whereas in the more moderate Y2-25 line, SCY2 transcripts accumulated at ∼68% of the control (Fig. 1C). In the severe SCY1-RNA1 line Y1-9, the SCY1 transcript level was 15% of the control (Fig. 1C). Although the SCY2 transcript level was not affected in SCY1-RNAi lines, the SCY1 transcript level was reduced in the SCY2-RNAi line Y2-19. The phenotypic effects for SCY2-RNAi and SCY1-RNAi were lost at the first pair of true leaves, although they could be extended to the first true leaf by repeated application of estrogen either by application to seedlings or by liquid culture in the presence of β-estrogen. This appears to be due to a breakdown of RNAi, as SCY1 null mutants are stably and uniformly chlorotic (Martin et al., 2009; Skalitzky et al., 2011). qRT-PCR analysis confirmed that both SCY1 and SCY2 transcripts had returned to wild-type levels in the green true leaves (Martin, 2010).
Immunoblot analysis of tissue extracts from treated SCY2-RNAi seedlings showed reduced accumulation of the envelope proteins TIC110, TIC40, and FTSH12 and the thylakoid proteins SCY1, TATC, and ALB3 compared with empty vector control plants (Fig. 2, A and B). The levels of SRP/ALB3 and TAT substrates, light-harvesting complex protein (LHCP) and the 23-kD oxygen-evolving complex protein (OE23), respectively, also were reduced severely (Supplemental Fig. S1). Only slight reductions of the outer envelope protein TOC75 were observed. In contrast, the abundance of most of these proteins, with the exception of SCY1, was unaffected in SCY1-RNAi seedlings, consistent in part with previous results (Martin et al., 2009). These results suggest that the loss of SCY2 creates a different and more global effect on chloroplast biogenesis in developing seedlings than loss of SCY1.
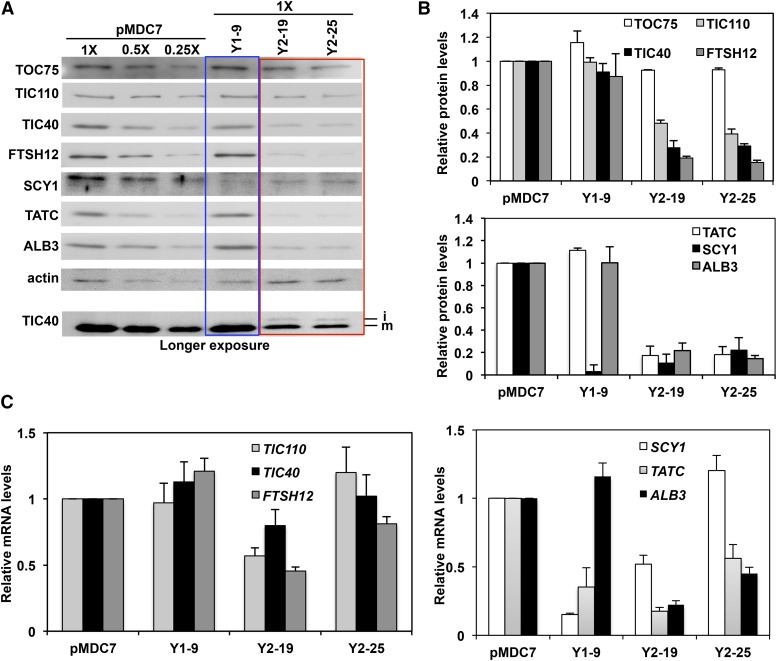
Figure 2.
Several membrane proteins were deficient in SCY2-RNAi plants. A, Total protein extracts from 6-d-old estrogen-induced seedlings were subjected to SDS-PAGE and immunoblotting with various antibodies. Actin was used as the internal control for equal protein loading. For better comparison, samples from empty vector-transformed plants were diluted to 0.5- and 0.25-fold. At the bottom, the blot for TIC40 was exposed to film for a longer time to show iTIC40 accumulation in SCY2-RNAi plants. i, iTIC40; m, mTIC40. B, Bar graph of the relative amounts of immunoblot signals of the proteins from A. Each bar presents the mean value of three biological replicates normalized to control plants (pMDC7). Error bars indicate se. C, Relative mRNA levels of TIC110, TIC40, and FTSH12 (left chart) and SCY1, TATC, and ALB3 (right chart) in 6-d-old seedlings germinated on MS medium containing 20 μm estrogen as determined by qRT-PCR analysis. Data represent mean values of at least four biological replicates normalized to control plants (pMDC7). Error bars indicate se.
Direct or indirect effects might cause a reduced accumulation of proteins in SCY2 down-regulated plants. For example, a block in membrane integration could lead to the accumulation of the integration substrate and its subsequent degradation. This appears to be the case for TIC40, because a small amount of TIC40 intermediate (iTIC40) is visible in long exposures of immunoblots of SCY2-RNAi lines (Fig. 2A, bottom). This might reflect a very active degradation process, as seen for other proteins (Bennett, 1981; Cline et al., 1989; Viitanen et al., 1990). Reduced protein accumulation also might be due to the down-regulation of transcription (retrograde signaling; Nott et al., 2006). Transcript quantification by qRT-PCR showed a reduction in transcripts for many of the proteins affected in SCY2-RNAi lines (Fig. 2C), with TIC110, TIC40, and FTSH12 least affected and TATC and ALB3 most affected. In order to determine which proteins are SEC2 substrates and which proteins are affected primarily by secondary effects, a different and more direct approach was necessary.
In Vitro Reconstitution of Membrane Integration to Assess the Utilization of SEC2 for Candidate Substrate Proteins
In vitro chloroplast protein import and protein integration into isolated membranes were attempted as a more direct assessment of the role of the SEC2 system in the integration of candidate proteins. For this approach, we used pea (Pisum sativum) chloroplasts because of their ease of isolation and robust protein trafficking activities and verified the results with Arabidopsis chloroplasts where possible. As preliminaries, we first developed a chloroplast lysis method in which the resulting inner envelope membrane vesicles and the thylakoid membrane would present their stromal face to added reagents. Several lysis methods were employed that included hypotonic lysis as well as mechanical or freeze-thaw lysis under hypertonic conditions. All of these methods produced the desired inside-out inner envelope membranes and right-side-out thylakoids, as evidenced by the susceptibility of TIC110 and TATC to added protease and the corresponding protection of TOC75 and OE23 from the protease (Supplemental Fig. S2). The highest yield of such vesicles was obtained with chloroplasts hypotonically lysed with 10 mm HEPES-KOH, pH 8; this lysate was used in our assays.
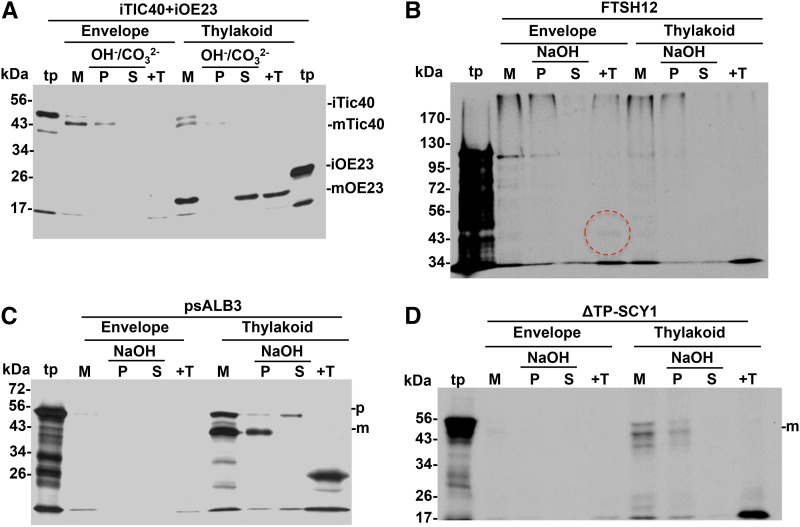
Second, it was important to establish criteria to assess the correct integration of candidate proteins into the membrane. Resistance to exaction with 0.2 m Na2CO3, pH 11.5, or the more rigorous 0.1 m NaOH, is an accepted method to determine if proteins are integrally inserted into the membrane bilayer (Steck and Yu, 1973; Cline, 1986). The 0.1 m NaOH treatment has the added benefit of opening membrane vesicles such that peripheral proteins on both sides of the bilayer are exposed to extractant. Analysis of endogenous SCY1, ALB3, and TATC from pea and FTSH12 from Arabidopsis (Supplemental Fig. S3A; Martin et al., 2009) as well as imported proteins showed that their integrated forms are resistant to 0.1 m NaOH extraction (Supplemental Fig. S3, B–D). Due to a better production of the full-length protein, we used a pea ALB3 (psALB3) cDNA clone to generate in vitro translated ALB3 in our assays. The imported/integrated forms of TATC (Mori et al., 2001; Martin et al., 2009), psALB3, SCY1, and FTSH12, also yield protected products when the membrane was treated with the broad specificity protease thermolysin (Supplemental Fig. S3, B–D). In our hands, psALB3 produced an ∼26-kD degradation product, consistent with TM2 to TM4 and intervening loops being protected. SCY1 produced a major band at 20 to 24 kD, which likely represents protected TM1 to TM6 (SCY1 has 10 TMs). FTSH12 produced a 43-kD band and, frequently, a 39-kD band. The 43-kD band diminished with concomitant increase of the 39-kD band in a protease concentration series (Supplemental Fig. S4). This is consistent with the protection of FTSH12’s two TMs and intervening loop and strongly suggests that the large FTSH12 C-terminal protease and ATPase domains are exposed to the stroma. We routinely relied on protease-protected bands as the best evidence for integration.
In contrast to these results, TIC40 and TIC110 were extracted by 0.1 m NaOH but resistant to extraction by 0.2 m Na2CO3 (Supplemental Fig. S5A). However, in our hands, Na2CO3 was not sufficiently rigorous to discriminate between membrane-associated and membrane-integrated TIC40 and TIC110 because it did not extract bound precursor proteins (Supplemental Fig. S5B). The second measure of TIC40 integration is processing of its N-terminal propeptide, which occurs upon insertion into the inner envelope. We found that a mixture of 30% 0.1 m NaOH and 70% 0.2 m Na2CO3 (OH−/CO32−) extracted most bound precursor protein as well as iTIC40 but a minimal amount of the mature protein mTIC40 (Supplemental Fig. S5B). Therefore, this mixture was routinely used for alkaline extraction of TIC40. Ultimately, we relied most heavily on the amount of mTIC40 as evidence of TIC40 integration. We have not identified conditions that sufficiently discriminated bound from integrated TIC110, and unfortunately, the integrated protein is the same size as the nonintegrated protein. As a result, studies of TIC110 were not pursued.
Sensitivity of Imported Proteins to Sodium Azide and Ionophores
Certain inhibitor treatments of chloroplasts can diagnostically impair only the membrane translocation step. Sodium azide can inhibit Sec-dependent protein transport by impairing SECA, the translocation motor protein. For example, sodium azide inhibits the thylakoid localization of the SECA1/SCY1/SECE1-dependent plastocyanin and the 33-kD oxygen-evolving complex protein OE33 (Yuan et al., 1994) without affecting their import into the stroma. In the SEC2 system, SECA2, a stroma-localized homolog to SECA1 and Escherichia coli SECA (Skalitzky et al., 2011), could share a similar sensitivity to sodium azide.
Inclusion of sodium azide during light-plus ATP-meditated chloroplast protein import strongly inhibited FTSH12 integration, as evidenced by NaOH extraction of the imported (and processed) mFTSH12 (Fig. 3B), whereas there was little effect on SCY1 integration (Fig. 3C) and a marginal effect on TIC40 (Fig. 3A) and psALB3 (Fig. 3D) integration. A previous analysis showed that TATC integration is unaffected by sodium azide (Martin et al., 2009). These results support the conclusion that FTSH12 employs a SECA-like protein for integration into the inner envelope membrane.
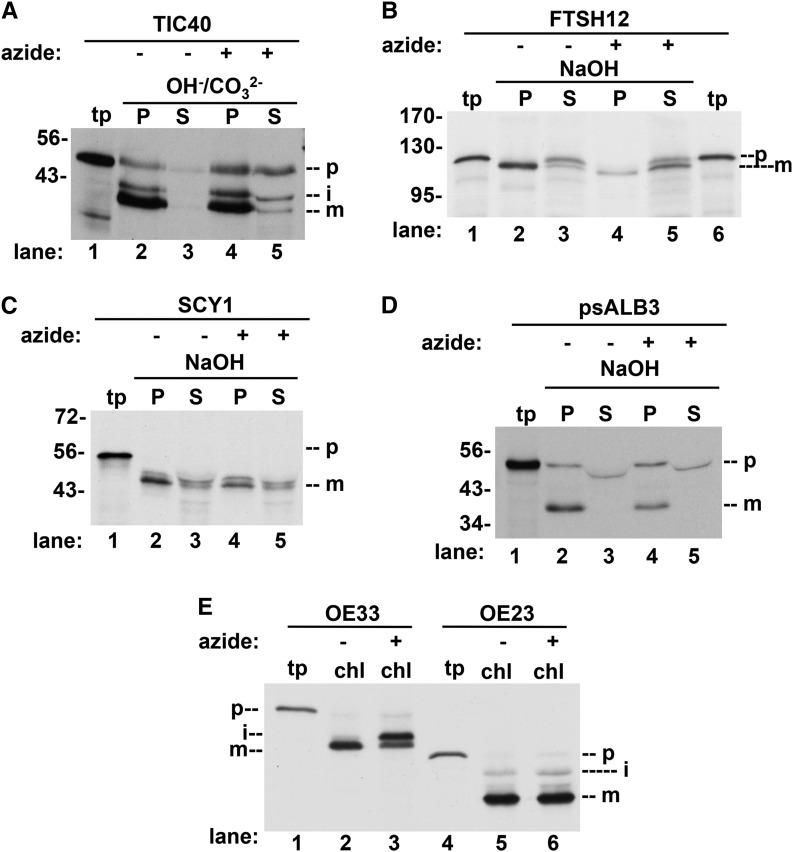
Figure 3.
Sodium azide impaired the membrane integration of FTSH12 during chloroplast protein import in vitro. A to D, In vitro translated precursor proteins TIC40 (A), FTSH12 (B), SCY1 (C), and psALB3 (D) were incubated with pea chloroplasts and 2.5 mm ATP with or without pretreatment with 7 mm sodium azide. After 20 min in the light at 25°C, recovered chloroplasts were extracted with 0.2 m Na2CO3, 0.1 m NaOH, or OH−/CO32− as shown above the gels. The pellet (P) and supernatant (S) samples from extraction were loaded at the same stoichiometry. E, OE33 and OE23 were used as positive and negative controls for sodium azide treatment. Here, recovered chloroplasts were analyzed directly, as impairment of thylakoid transport is apparent by the accumulation of the stromal intermediates. Note that the FTSH12 translation product (tp) is loaded next to the extract of the +azide sample to emphasize that mature FTSH12 is extracted with 0.1 m NaOH (B, compare lanes 5 and 6). i, Intermediate protein; m, mature protein; p, precursor protein.
Ionophores that dissipate the proton motive force (PMF) impair the thylakoid localization of some membrane and lumenal proteins but do not impair import into the organelle if ATP is provided in the assay. Ionophores had little effect on the localization of envelope proteins, as assessed by chloroplast subfractionation and alkaline extraction (Fig. 4, A and B), but seriously impaired the localization of SCY1 and psALB3 (Fig. 4, C and D), resulting in significant accumulation of the imported proteins in the soluble stromal fraction (lane 8, compare with lane 3) and increased alkaline extraction of the membrane-associated proteins. Previous work showed that TATC integration is unaffected by ionophores (Martin et al., 2009). These results indicate that the PMF is involved in SCY1 and ALB3 integration but may not contribute to TIC40 and FTSH12 integration.
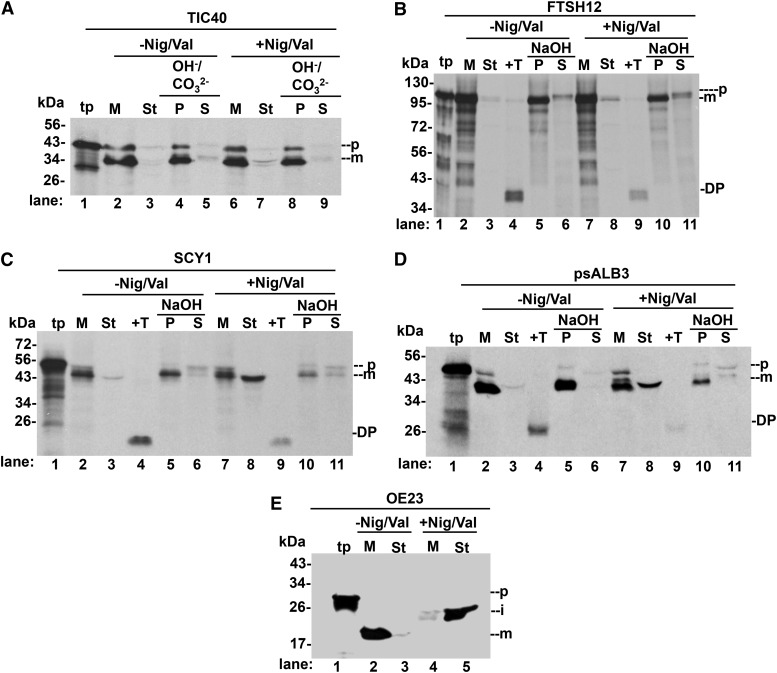
Figure 4.
Ionophores impaired the membrane integration of psALB3 and SCY1 but had no effect on TIC40 and FTSH12. Chloroplasts treated without or with 1 μm nigericin and 2 μm valinomycin were incubated with in vitro translated precursor proteins and 5 mm ATP in import buffer (IB) for 20 min in light at 25°C. Intact chloroplasts were lysed and centrifuged to separate SE (St) from membranes (M). Membranes were further extracted with 0.2 m Na2CO3, 0.1 m NaOH, or OH−/CO32− as shown above the gels. An equivalent aliquot of membranes from the SCY1, psALB3, and FTSH12 assays was treated with thermolysin (+T). OE23 localization (i.e. accumulation of iOE23) served as the positive control for the effect of ionophores. DP, Degradation product; i, intermediate protein; m, mature protein; p, precursor protein; P, pellet; S, supernatant; tp, translation product.
Reconstituted Integration into Organelle-Free Membranes
A more direct approach for discriminating SEC2-dependent substrates from SEC2-independent substrates was to reconstitute their integration in organelle-free assays and assess both the requirements for integration and the response to translocase inhibitors. Our basic assay was to include freshly prepared lysate, freshly prepared 10× concentrated stromal extract (SE), and Mg-ATP and to incubate this mixture with freshly prepared in vitro translated precursor protein in the light at 25°C. The substrates were iTIC40, mTATC from pea (psmTATC), pre-ALB3, and SCY1 and FTSH12 that were N-terminally truncated to remove the first ∼10 residues of the transit peptide. This prevented protein import into any chloroplasts that survived the lysis procedure, although that proved not to be a problem with hypotonic lysis. With this assay, we obtained significant integration of TIC40, SCY1, and both pea and Arabidopsis ALB3. Supplemental Figure S3 shows a comparison of import versus integration for psALB3 (B) and SCY1 (C). Integration occurred as assessed by both alkaline extraction and partial protease protection. We were only able to obtain very small amounts of integration with FTSH12 (Supplemental Fig. S3D) and TATC (data not shown).
Integration of Thylakoid Protein Substrates
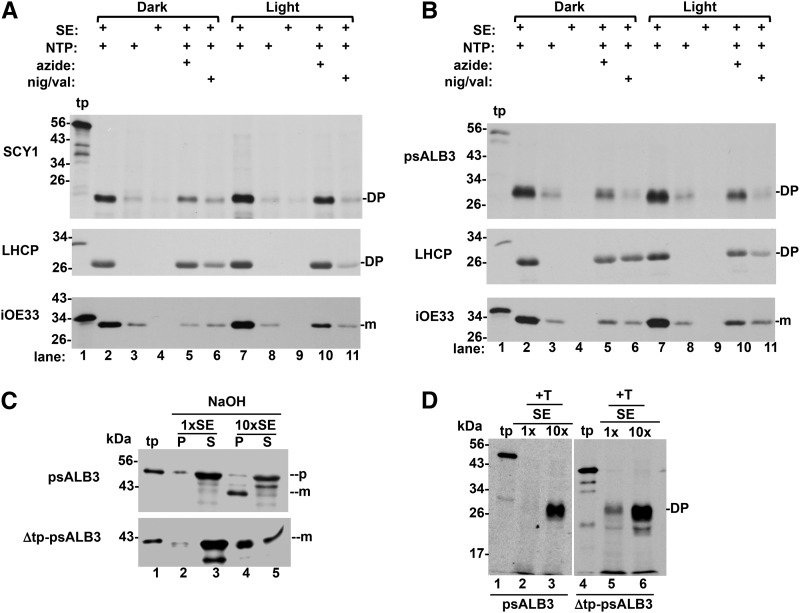
The requirements for SCY1 and ALB3 integration were assayed by either selectively omitting assay components or by including selective inhibitors (Fig. 5). Assays were conducted in the light or darkness. Both SCY1 (Fig. 5A) and ALB3 (Fig. 5B) required SE (lanes 3 and 8), NTPs and/or NDPs (lanes 4 and 9), and were inhibited by ionophores that dissipate the PMF (lanes 6 and 11). The effects of azide in dark versus light are informative. The thylakoid PMF can be established either by photo electron transport in light or by ATP synthase (CF1/CF0) proton pumping via ATP hydrolysis. Although azide inhibits SECA, it also inhibits ATP synthase (Murataliev et al., 1991). Thus, the inhibition by azide in dark but not in light (Fig. 5, A and B; compare lanes 5 and 10) confirms that the dark integration of SCY1 and ALB3 depends on PMF generated by ATP synthase rather than on SECA. This is seen in the integration of LHCP, which requires the PMF but not SECA, and OE33, which requires the PMF and employs SECA. More importantly, this differential effect of azide strongly suggests that the thylakoid membrane, where the ATP synthase resides, is the target membrane for both SCY1 and ALB3 integration.
Figure 5.
SCY1 and ALB3 integration into membranes requires stromal factors, PMF and NTP. A and B, Requirements for the integration of SCY1 and psALB3 into membranes were determined with specific inhibitors or by selective removal of components from the complete assay mixture, which consisted of lysate or an equivalent amount of membranes (i.e. assays completely lacking SE; lanes 3 and 8), 10× SE, Mg-ATP, in vitro translated precursor proteins, and IB (see “Materials and Methods”). Assays lacking ATP were additionally treated with apyrase (10 units per assay) before the addition of apyrase-treated translation products (tp; see “Materials and Methods”). Where noted above the gels, assays received sodium azide to 10 mm final concentration or nigericin/valinomycin (1 μm/2 μm) before initiation of the assay. Assays were at 25°C in darkness or light for 50 min. Membranes were recovered by centrifugation and treated with thermolysin (see “Materials and Methods”) before analysis by SDS-PAGE/fluorography. Substrates used were in vitro translated Δtp-SCY1, pre-psALB3, pre-LHCP, and iOE33. iOE33 and LHCP assays served as controls for these assays. iOE33 transport requires SE (SECA1) and is inhibited by azide and ionophores. LHCP requires SE (SRP) and is inhibited by ionophores but not sodium azide. C, In vitro translated pre-psALB3 and an N-terminally truncated psALB3(Δtp-psALB3) lacking the first 60 amino acids were used in integration assays with 1× SE or a 10× excess of SE. Membranes recovered from assays were subjected to extraction with 0.1 m NaOH. D, Membranes recovered from the same assays as in C were treated with thermolysin (+T) to produce the diagnostic 26-kD protease-protected degradation product (DP). m, Mature protein; p, precursor protein; P, pellet; S, supernatant.
As is apparent in the ALB3 assays in Supplemental Figure S3B, right, lane 2, the full-length precursor was added to the assay, but only mALB3 was found integrally associated with the membrane. This raised the possibility that the stroma requirement is for enzymatic cleavage of the transit peptide before integration. We tested this by creating an N-terminally truncated version (Δtp-psALB3) lacking the first 60 residues of pre-ALB3; the resulting in vitro translated Δtp-psALB3 migrated at a similar molecular mass to mALB3 by SDS-PAGE, indicating that its transit peptide was fully deleted (Fig. 5C, compare lanes 1 and 4 for psALB3 and Δtp-psALB3). Integration of the full-length and the truncated psALB3 was strongly stimulated by SE, as judged by either 0.1 m NaOH extraction (Fig. 5C) or protease protection (Fig. 5D), suggesting that some other stromal factor is involved in ALB3 integration.
Integration of TIC40
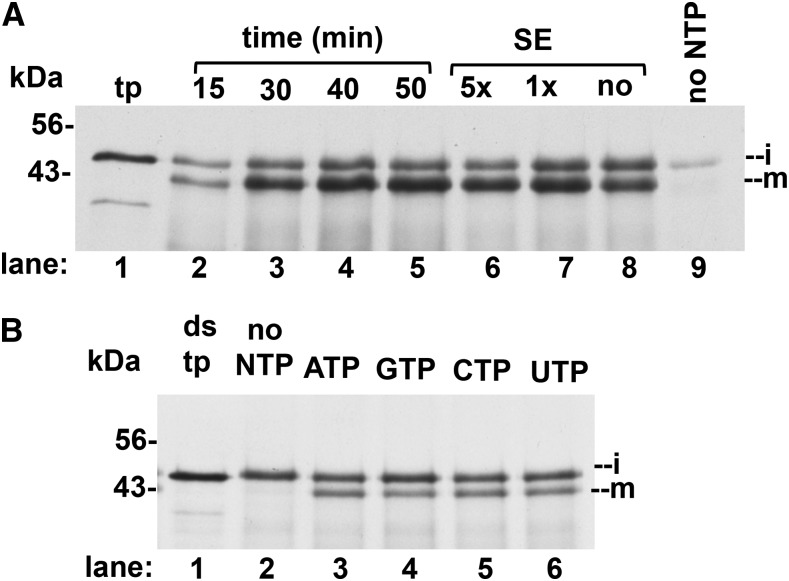
The above reaction conditions were used to assay for iTIC40 integration. This was modeled on the assay described by Li and Schnell (2006) except that we used freshly prepared chloroplast lysate (rather than frozen envelope membranes) and more stringent alkaline extraction conditions. We found that integration was maximal at 40 min (Fig. 6A, lanes 2–5), did not require SE (lanes 6–8), in agreement with Li and Schnell (2006), but did require NTPs/NDPs (lane 9). In order to examine this requirement more carefully, we used washed membranes and translation extract that was twice desalted. Without any addition, integration was undetectable (Fig. 6B, lane 2). However, unexpectedly, activity was obtained with ATP, GTP, CTP, and UTP (Fig. 6B, lanes 3–6). Titration of each NTP showed that ATP, CTP, and UTP were similarly effective; GTP was slightly less effective (Y. Li and K. Cline, unpublished data). The significance of this requirement is unclear, because TIC40 integration into Arabidopsis membranes shows much less dependence on NTPs (data not shown).
Figure 6.
Characteristics of TIC40 integration. A, Complete assays contained in vitro translated iTIC40, lysate or membranes obtained from lysate, varying amounts of SE, and 2.5 mm ATP or apyrase-treated lysate (lane 9) as described in Figure 5 and “Materials and Methods.” Time-course samples contained 10× SE. Assays were for 50 min or the times indicated at 25°C. Membranes were pelleted and extracted with OH−/CO32− (see “Materials and Methods”). Only the alkaline-resistant pellets are shown. B, Assays contained membrane obtained from freshly prepared lysate by centrifugation followed by washing with IB, in vitro translated iTIC40 desalted twice through 1-mL Sephadex G25 spin columns, and either IB (lane 2) or 1 mm NTP (lanes 3–6). The HPLC-grade NTPs were purchased from MilliporeSigma. Before use, each NTP stock received a 1.5× molar amount of MgCl2. Assays were for 40 min at 25°C in darkness or light and were extracted with the alkaline mixture as in A. There was no apparent difference in the results from the dark assays versus the light assays. Assays conducted in light are shown. i, Intermediate protein; m, mature protein; tp, translation product; ds, desalted.
FTSH12 Integrates into Chloroplast Membranes in a Binding/Chase Assay
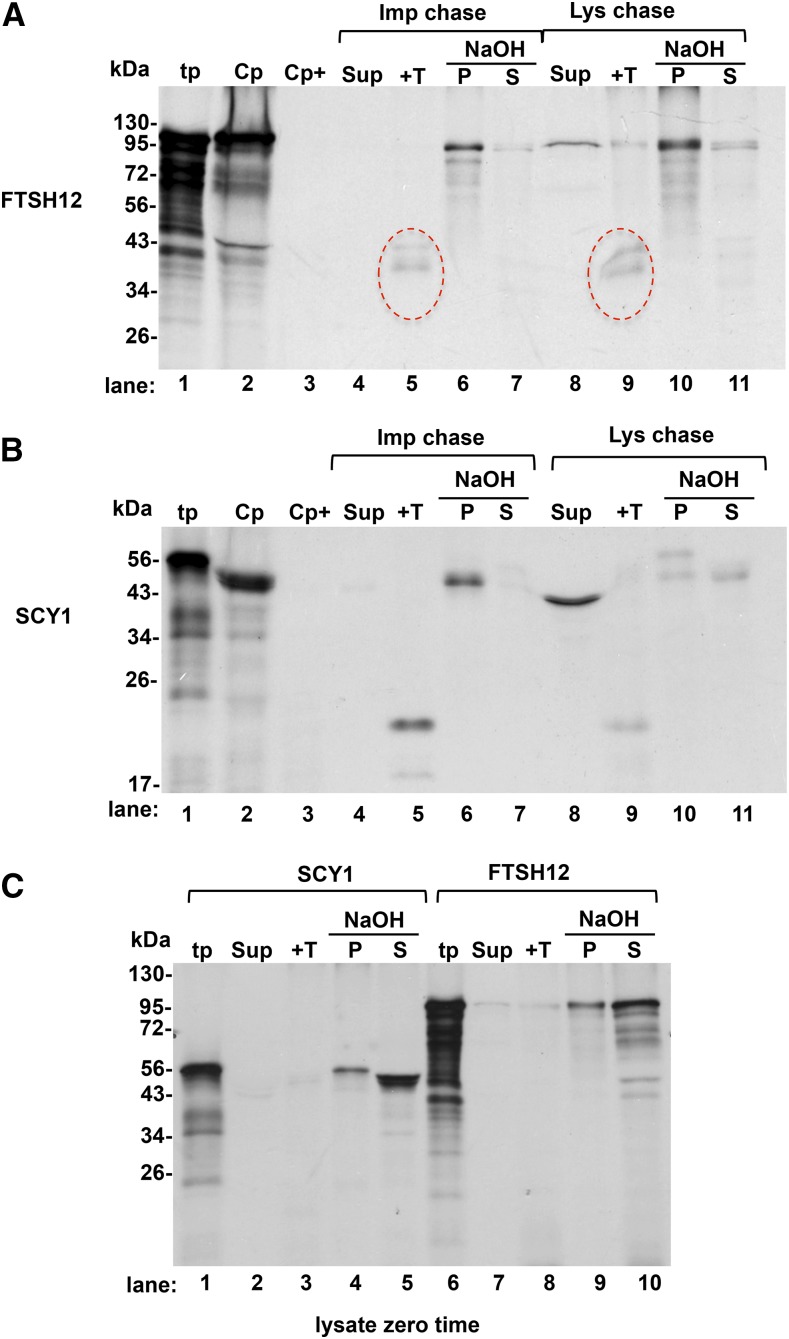
One possibility for the very low level of FTSH12 integration in the lysate assay is that integration is coupled to its import across TOC-TIC. Such a coupled mechanism was observed for TIC40 in a time course of import (Li and Schnell, 2006). An experiment to test this idea was to conduct a bind-and-chase reaction, in which the binding step was one termed early intermediate of protein import (Schnell et al., 1994) and the chase and integration reaction were conducted with lysate obtained from the binding chloroplasts. This is shown in Figure 7 for both FTSH12 and SCY1. The chloroplasts obtained from the binding reaction (Fig. 7, A and B, lane 2) have a substantial amount of associated precursor, which is degraded by thermolysin added to the chloroplasts (lane 3), demonstrating that a considerable portion of the precursor protein chain is exposed to the surface of the chloroplasts. If the binding chloroplasts are subjected to a chase reaction with 5 mm ATP, the majority of the recovered substrate in the membrane fraction is alkaline resistant (Fig. 7, A and B, lanes 6 and 7) and produces the diagnostic protease-protected products (lane 5). A similar integration occurred for FTSH12 when the precursor-bound chloroplasts were first lysed and then incubated with 10× SE and ATP (Fig. 7A, lanes 8–11). Comparable amounts of the protease degradation product (Fig. 7A, lane 9 compared with lane 5) and alkaline-resistant protein (Fig. 7A, lane 10 compared with lane 6) were obtained. Some FTSH12 was recovered in the soluble fraction (Fig. 7A, lane 8), presumably protein that was imported across TOC-TIC but failed to engage the machinery of the inner envelope.
Figure 7.
FTSH12 is membrane integrated during a coupled import-integration bind-and-chase assay. The full precursors pre-FTSH12 and pre-SCY1 were translated in a homemade wheat germ system (see “Materials and Methods”). Translation products (tp) were diluted 15-fold in the final assay mixture to an estimated 0.1 mm ATP, which contained intact chloroplasts (Cp) at 0.33 mg mL−1 Chl. Precursor binding to chloroplasts (i.e. to the early intermediate stage; Schnell et al., 1994) was for 15 min at 25°C in darkness. Chloroplasts were recovered on Percoll cushions with (A and B, lane 3) or without (A and B, lane 2) thermolysin pretreatment. An aliquot of the untreated chloroplasts was subjected to a chase assay with 3 mm ATP (A and B, lanes 4–7). The remaining chloroplasts were lysed in 10 mm HEPES/KOH and adjusted to IB. A portion of the lysate was saved as a zero time control (C). The remainder was combined with ATP and 10× SE to 5 mm and 2.5× SE, respectively. Chase assays were for 30 min at 25°C in the light. Following the chase assays, the chloroplasts from the import chase were lysed, then this lysate as well as the lysate zero time and lysate chase were subfractionated and analyzed as follows: lysates were centrifuged to obtain a membrane fraction and a supernatant (Sup). One aliquot of membranes was treated with thermolysin (+T; lanes 5 and 9 in A and B and lanes 3 and 8 in C) and the other was extracted with 0.1 m NaOH as described (see “Materials and Methods”) into NaOH-resistant pellet (P) and NaOH-extracted supernatant (S). All gel-loaded samples were stoichiometrically equivalent. Red circle, degradation products from thermolysin digestion.
Figure 7C shows an analysis of the lysates obtained from the binding chloroplasts at time zero. Nearly all of the FTSH12 and SCY1 was membrane bound, largely extracted by alkali (lanes 4/5 and 9/10), and did not produce the diagnostic protease-protected bands (lanes 3 and 8). In fact, there was very little material in protease-treated samples. This suggests that a significant amount of the precursor protein was exposed to the stromal side of the membrane in the chloroplasts obtained from the binding reaction.
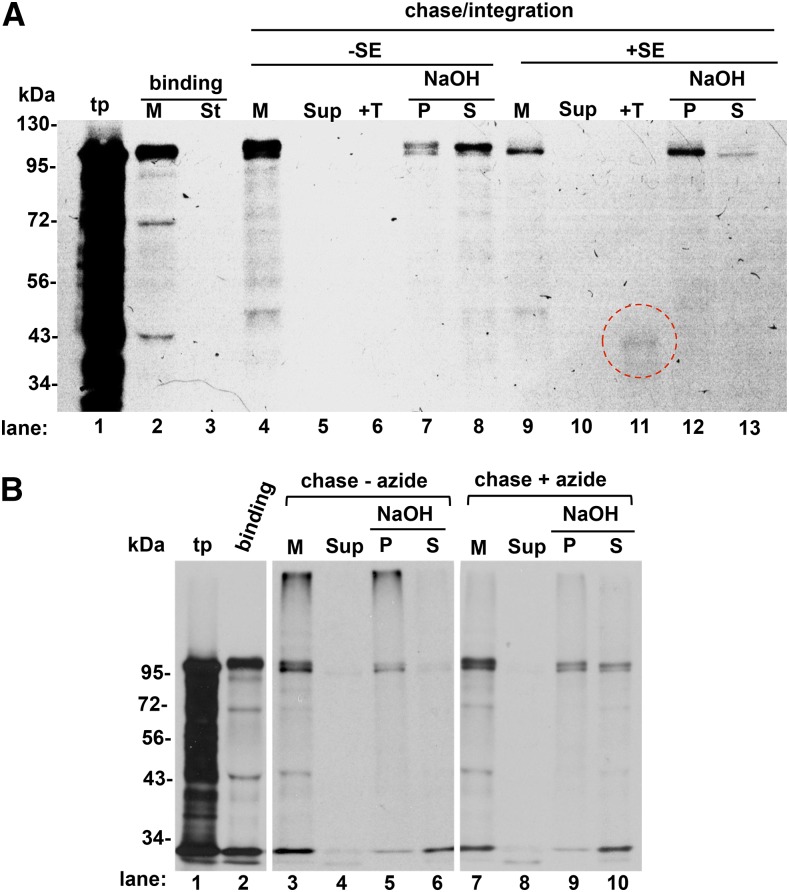
Membrane integration of FTSH12 in the chase assays was time dependent, with more integrated at 30 min than at 10 min (Supplemental Fig. S6). More importantly, chase integration was dependent on SE (Fig. 8A) and was inhibited by sodium azide (Fig. 8B). It is not possible to say whether SE was required for progression through TOC/TIC or for inner envelope integration. However, because azide does not interfere with protein import, its inhibitory effects suggest the involvement of a SECA protein in the integration mechanism.
Figure 8.
Membrane integration of FTSH12 in a bind-and-chase assay requires SE and is inhibited by sodium azide. A, Intact pea chloroplasts were incubated with in vitro translated pre-FTSH12 on ice in the dark for 15 min as described in “Materials and Methods.” Chloroplasts then were pelleted at 1,000g for 5 min at 2°C, lysed at 2 mg mL−1 Chl on ice with 10 mm HEPES-KOH, and then adjusted to 1 mg mL−1 Chl with 2× IB. Membranes (M; lane 2) were separated from stroma (St; lane 3) by centrifugation. Membranes were suspended in IB (lanes 4–8) or SE (lanes 9–13), mixed with ATP to 5 mm, and incubated at 25°C for 50 min in 120 μE m−2 s−1 white light. Membranes (lanes 4 and 9) were separated from supernatant (Sup; lanes 5 and 10) and then treated with thermolysin (+T; lanes 6 and 11) or extracted with 0.1 m NaOH to a resistant pellet (P; lanes 7 and 12) or extracted supernatant (S; lanes 8 and 13) as described in “Materials and Methods.” B, A similar binding/chase-integration assay was conducted as described in A, except that 7 mm sodium azide was added to the mixture of membranes and 10× SE and incubated for 10 min on ice before the chase step. Chase reactions were started with 5 mm ATP at 25°C in the light and continued for 50 min. Reactions were separated into supernatant (lanes 4 and 8) and membranes (lanes 3 and 7). Membranes were extracted with NaOH (lanes 5, 6, 9, and 10) as in A. tp, Translation product. Red circle, degradation products from thermolysin digestion.
The lysate chase of SCY1 was very different (Fig. 7B). Most of the mature SCY1 was found in the soluble fraction (lane 8), and less of the membrane-associated SCY1 was alkaline resistant (lanes 10 and 11) or protease resistant (lane 9). A likely explanation for the behavior of SCY1 in this assay is that SCY1 integration is not coupled to protein import and that lysis dramatically increases the physical distance between envelope and thylakoids compared with that in chloroplasts. A similar result was obtained for a bind-and-chase reaction with TATC, in which most was recovered in the soluble fraction (Y. Li and K. Cline, unpublished data).
The Envelope Is the Target Membrane for TIC40 and FTSH12 Integration; the Thylakoid Is the Target Membrane for SCY1 and ALB3 Membranes
Our assay employed chloroplast lysate and unfractionated chloroplast membranes to minimize manipulations that invariably result in reduced integration activity. Thus, it was important to verify target membranes for the different substrates. After organelle-free integration reactions, recovered membranes were sonicated briefly and then separated by Suc gradient centrifugation into envelope and thylakoid-enriched membranes (see “Materials and Methods”). Membranes were treated with thermolysin or extracted with NaOH or OH−/CO32−. As shown in Figure 9, both mTIC40 and mFTSH12 are localized mainly in the envelope fraction, whereas mALB3 and mSCY1 are localized mainly in the thylakoid fraction, as expected.
Figure 9.
Fractionation of membranes from in vitro integration and binding/lysate chase assays. Membranes (M) recovered from in vitro integration assays (as in Figs. 5 and 6) and a lysate chase assay (as in Fig. 7) were further fractionated into envelope and thylakoid fractions by Suc gradient centrifugation (see “Materials and Methods”). Each fraction was extracted with 0.1 m NaOH or OH−/CO32−, separated into pellet (P) and supernatant (S), or treated with thermolysin (+T). Samples were analyzed with SDS-PAGE and fluorography. In A, membranes assayed with iTIC40 and iOE23 were mixed together before fractionation; here, mOE23 served as a positive control for thylakoid localization. The red circle in B marks the protease-protected FTSH12 degradation fragments. m, Mature protein; p, precursor protein; tp, translation product.
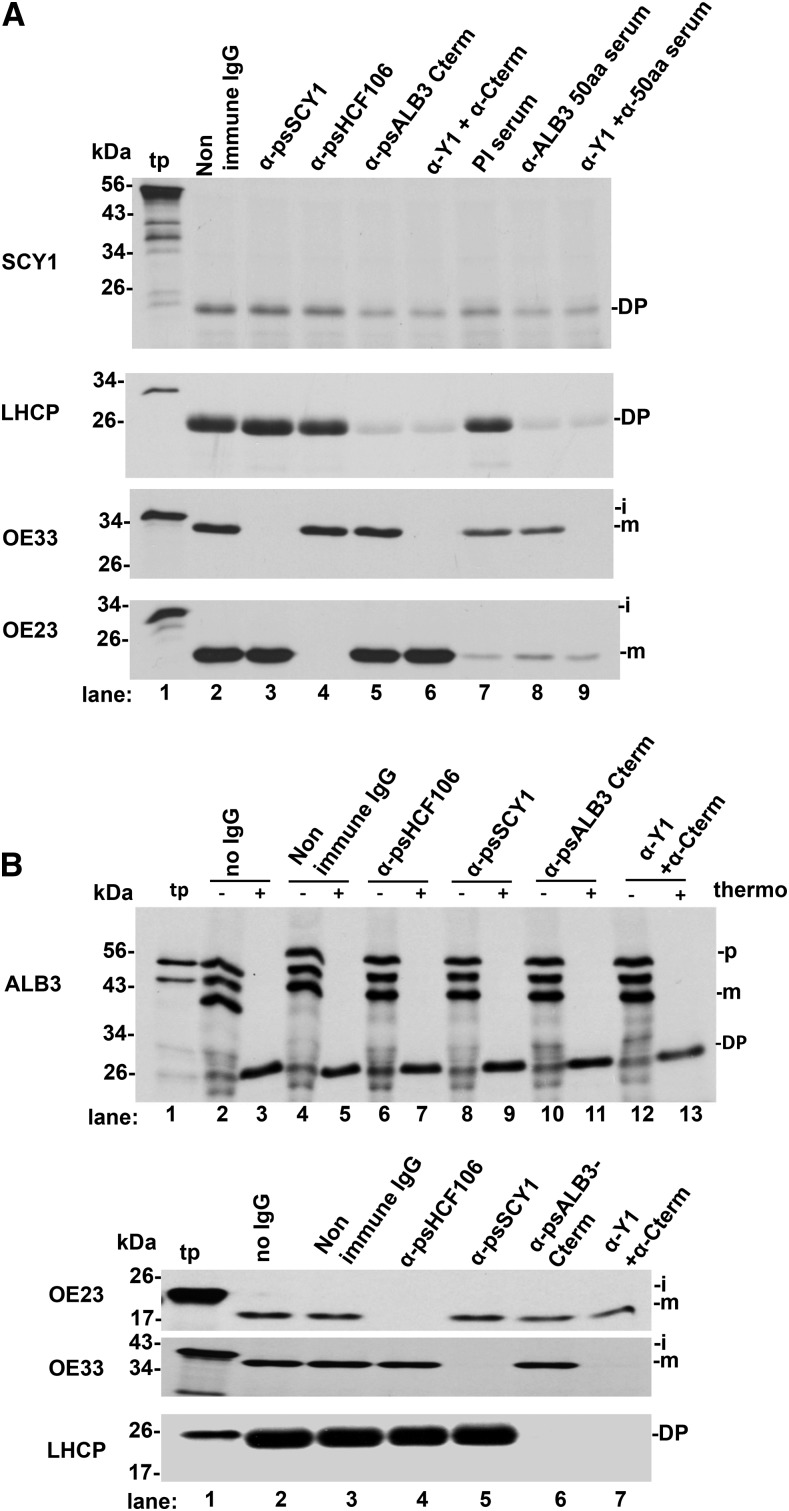
ALB3 Integration Was Not Inhibited by Any Thylakoid Translocase Antibodies; SCY1 Integration Was Reduced Significantly by Anti-ALB3
Because ALB3 and SCY1 integration appears to occur directly into thylakoids, their integration was examined by assays in which antibodies against key components of the pea thylakoid translocases inhibit translocation on those pathways. Anti-SCY1 (SEC1), anti-HCF106 (TAT), and anti-ALB3 C terminus (SRP/ALB3 pathway) IgGs were prebound to membranes, and the membranes were assayed for integration. In the case of SCY1 integration, antibodies to SCY1 or HCF106 had no effect (Fig. 10A), but antibodies to the ALB3 C terminus had a reproducible reduction of integration. The C terminus is involved in the docking the SRP; thus, this antibody is very effective in inhibiting LHCP integration. However, an antibody to an ALB3 50-amino acid peptide (not involved in docking) had the same partial inhibition on SCY1 integration. Quantification of the fluorogram in Figure 10A showed that anti-ALB3 reduced SCY1 integration to ∼40% to 50% of the control (Supplemental Fig. S7). It has been reported that E. coli TATC can be integrated either by the Sec translocase or YidC (ALB3 ortholog; Welte et al., 2012). However, we found no enhancement of inhibition on SCY1 integration by including anti-SCY1 IgGs with anti-ALB3 (Fig. 10A, lanes 6 and 9). Using essentially the same methodology for ALB3 integration, we found no effect for any of the antibodies or the combination of SCY1 and ALB3 antibodies on ALB3 integration (Fig. 10B). The assays LHCP, OE33, and OE23 are controls for the specificity and efficacy of the antibodies (i.e. nearly complete inhibition by antibodies to their respective pathways).
Figure 10.
ALB3 integration was not inhibited by any thylakoid translocase antibodies; SCY1 integration was reduced significantly by anti-ALB3. Membranes recovered from a chloroplast lysate were incubated at 0.33 mg mL−1 Chl with IgGs at 0.8 μg mL−1 or anti-ALB3 50 aa serum and its prebleed at 100 μL of serum in import buffer containing 5 mm MgCl2 (IBM) and 0.7% bovine serum albumin for 1 h on ice. Membranes were pelleted, washed, and resuspended in IBM. Treated membranes were assayed with 10× SE, 5 mm Mg-ATP, and in vitro translated precursor proteins Δtp-SCY1, pre-LHCP (in A), mLHCP (in B), iOE33, iOE23, and pre-ALB3 as designated to the left of the gels for 50 min in the light. Membranes were recovered, treated with thermolysin, and analyzed with SDS-PAGE and fluorography. A, LHCP served as a positive control for the SRP/ALB3 pathway, OE33 for the SEC1 pathway, and OE23 for the TAT pathway. Note that serum has a severe nonspecific inhibitory effect on the TAT pathway (lanes 7–9). B, ALB3 assays were conducted similarly, except that mLHCP was used in control experiments and aliquots of untreated assay membranes were loaded adjacent to thermolysin-treated membranes. DP, Degradation product; i, intermediate protein; m, mature protein; p, precursor protein.
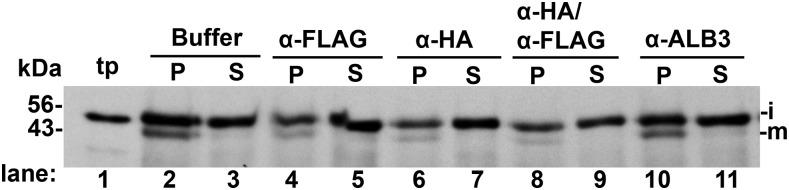
TIC40 Integration Was Largely Inhibited by Antibodies against Epitope-Tagged SCY2/SECE2
The potential involvement of the SEC2 system in TIC40 integration was tested in lysate obtained from recombinant Arabidopsis, in which wild-type SCY2 and SECE2 were replaced with SCY2-HA and FLAG-SECE2, respectively (Li et al., 2015). TIC40 integration was substantially inhibited by α-FLAG IgG and α-HA IgG individually (Fig. 11, lanes 4–7) and reduced further by a combination of both antibodies (lanes 8 and 9). Quantification showed reductions (Supplemental Fig. S8) to ∼40% of control for α-FLAG, to ∼30% for α-HA, and to ∼10% for the combination. TIC40 integration was not impaired by the antibody buffer (Fig. 11, lanes 2 and 3) or by the irrelevant antibody α-ALB3 (lanes 10 and 11). This constitutes one line of evidence that TIC40 is a substrate of the SEC2 system. At present, it is unknown if SECA2 participates in the integration reaction, considering the small effect of sodium azide during protein import (Fig. 3).
Figure 11.
The membrane integration of TIC40 depends on SCY2/SECE2. Arabidopsis chloroplast lysate (scy2 sece2 double mutant complemented with SCY2-HA/FLAG-SECE2; Li et al., 2015) was mixed with anti-FLAG, anti-HA, or unrelated anti-ALB3 IgG for 1 h before being assayed with in vitro translated iTIC40. The buffer in commercial anti-FLAG solution was exchanged to 20 mm HEPES-KOH (see “Materials and Methods”). Both the buffer-exchanged IgG and the original buffer were tested in integration assays, which were for 50 min at 25°C in the light (120 μE m−2 s−1). Membranes were then recovered and extracted with OH−/CO32− and separated into alkaline-resistant pellet (P) and alkaline-extracted supernatant (S) samples (see “Materials and Methods”) and were analyzed with SDS-PAGE and fluorography. i, iTIC40; m, mature TIC40; tp, translation product.
We attempted anti-SCY2/SECE2 inhibition of the chase of FTSH12 into the membrane, but this was without effect. There is a likely explanation for this result (see “Discussion”).
DISCUSSION
In this study, we investigated the possibility that the plastid SEC2 system is part of a conservative sorting pathway for proteins that are initially imported into the stroma by the envelope TOC/TIC translocase and then integrated into the envelope. In this type of sequential arrangement of translocases, mutations in components of the TOC/TIC translocase can have global effects on downstream events (e.g. the development of the thylakoids). For example, mutations in TOC159 and TIC56 (Köhler et al., 2015) result in chlorotic seedlings and plastids with reduced and disorganized internal membranes. Mutations in components of a secondary destination translocase (e.g. the thylakoid translocases; Li et al., 1995; Voelker and Barkan, 1995; Hutin et al., 2002; Martin et al., 2009) generally have effects limited to that compartment. Although SEC2 can be considered a destination translocase, reduced SCY2 expression through inducible RNAi exhibited global effects (i.e. loss of thylakoid membrane amount, structure, and pigment) and severely reduced amounts of the thylakoid translocase proteins SCY1, TATC, and ALB3 as well as the envelope proteins TIC40, TIC110, and FTSH12 (Figs. 1 and 2). All of these proteins could tentatively be considered substrates of SEC2. Even the thylakoid translocase components needed to be seriously considered, because they are multispanning membrane proteins that appear not to be localized by the known thylakoid pathways (Kroll et al., 2001; Asakura et al., 2004; Martin et al., 2009) and because inner envelope invaginations are thought to produce thylakoids via membrane flow during chloroplast development (Hoober et al., 1991; Morré et al., 1991; Kroll et al., 2001).
The in vitro import and integration studies conducted here were essential for sorting out the primary from the secondary effects of a disabled SEC2 system. They indicate that, in developed or mostly developed chloroplasts, SCY1 and ALB3 are imported into the stroma first and then integrated directly into thylakoid membranes. A similar pathway was demonstrated previously for TATC (Martin et al., 2009). The thylakoid integration of SCY1 and ALB3 requires PMF, stroma components, and NTPs, indicating the involvement of active integration machineries. The identity of the machinery is uncertain at this stage. Antibody inhibition experiments suggest that ALB3 integration involves none of the known thylakoid pathways and that SCY1 integration involves ALB3 (Fig. 10). However, that anti-ALB3 treatment with two different antibodies (to different regions of ALB3) reduced SCY1 integration to 40% to 50% of the control, whereas it virtually eliminated LHCP integration, suggesting an important but nonessential role for ALB3 in SCY1 integration (Supplemental Fig. S7). Integration of LHCP family members involves targeting by the SRP54 and SRP43 subunits and the FTSY receptor in addition to ALB3 (Figs. 5 and 10; DeLille et al., 2000; Henry, 2010; Richter et al., 2010). SRP54 binds LHCP TM2 or TM3 and SRP43 binds an 18-residue sequence (L18) in the loop between the two TMs (DeLille et al., 2000). We noticed that SCY1 has a partial L18 consensus amino acid proximal to TM1. However, we did not detect SRP interaction, nor could we replace SE in an SCY1 integration assay with recombinant SRP components (K. Cline, unpublished data).
One caveat to the antibody experiments is that inhibition by anti-SCY1 (directed to the C-terminal tail) has only been demonstrated for substrates that employ SECA1 in addition to SCY1/SECE1 (Mori et al., 1999). As SECA appears not to be required for SCY1 or ALB3 integration, the possible involvement of SCY1/SECE1 needs to be examined more thoroughly. However, it should be noted that antibody to the C-terminal tail of SCY2 (Fig. 11) was very effective for inhibiting the SECA2-independent integration of TIC40 and that E. coli SECY integration does not require preexisting SECY (Swidersky et al., 1992).
The biochemical results for SCY1 and ALB3 integration presented here largely agree with genetic experiments conducted with translocase mutants. TATC accumulated normally in alb3, ftsy, or scy1 mutants in Arabidopsis (Asakura et al., 2008; Martin et al., 2009). SCY1 accumulated normally in alb3 mutants in Arabidopsis (Asakura et al., 2004; Martin et al., 2009) and in ftsy mutants in Arabidopsis and maize (Zea mays; Asakura et al., 2004, 2008). ALB3 accumulated normally in ftsy mutants in Arabidopsis and maize. On the other hand, ALB3 was reduced significantly in a scy1 null mutant in Arabidopsis (Martin et al., 2009). Clearly, additional experiments are required to sort out the possible involvement of thylakoid translocases in SCY1 and ALB3 integration. And it is still possible (although unlikely) that the minor amount of SCY2 in thylakoids (Skalitzky et al., 2011) is involved in their integration. The assays developed here should allow a thorough analysis of SCY1 and ALB3 integration, clarifying the role of known as well as cryptic translocases.
We obtained reliable integration for two envelope proteins affected by SCY2 down-regulation: TIC40 and FTSH12. The integration of iTIC40 into envelope membranes was reported previously (Li and Schnell, 2006). Our results agree that stroma components are not required for integration. However, in our hands, integration of iTIC40 into both pea and Arabidopsis chloroplast membranes proceeded with faster kinetics (Fig. 6). Differences between assay conditions may contribute to this difference. Whereas Li and Schnell (2006) used frozen inner envelope membrane preparations and stromal iTIC40 recovered from lysed chloroplasts, our assay employed freshly prepared chloroplast lysates or total membranes obtained from the lysates and in vitro translated iTIC40, minimizing the reductions of activity that frequently result from manipulations and freezing. In addition, we also observed an NTP requirement for iTIC40 integration into pea membranes. This is somewhat difficult to understand mechanistically because ATP, GTP, UTP, and CTP all produced comparable stimulation of integration. The NTP requirement needs to be investigated rigorously, especially as Vojta et al. (2007) reported an ATP requirement for TIC110 insertion during a chloroplast import assay. Nevertheless, all of these studies consistently point to integration machinery involved in TIC40 integration.
Inhibition of iTIC40 integration by antibodies to FLAG-SECE2 and/or SCY2-HA provided the most definitive evidence for the involvement of integration machinery. To our knowledge, this is the first direct evidence of a substrate that requires the SEC2 system. For SEC2-mediated insertion of TIC40, the amino-proximal end of the TM would need to insert first. This occurs in other Sec systems if there are more positively charged residues on the carboxyl end than on the amino end flanking the TM (Rychkova and Warshel, 2013). This is called the positive inside rule (von Heijne, 1989). TIC40 obeys this rule, lacking charged residues amino proximal to the TM but having a conserved twin Lys carboxyl proximal to the TM. It is clear that SECA2 is not essential for TIC40 integration (Fig. 6). However, partial inhibition by sodium azide in a chloroplast import assay suggests the possibility of SECA2 as an accessory factor. Chiu and Li (2008) reported that TIC40 may be involved in its own integration as well as in the integration of TIC110 and TIC21. This suggests the possibility that TIC40 also may be an accessory factor for SEC2 operation.
Our results implicating SEC2 in FTSH12 integration are less definitive. FTSH proteins are quality control proteases of prokaryotic origin that generally contain two TMs, separated by an intervening loop, followed by a large globular catalytic carboxyl-proximal domain exposed to the cytosol or cytosol-equivalent compartment (e.g. the plastid stroma; Ito and Akiyama, 2005). There are some exceptions to this topology and number of TMs (Arnold and Langer, 2002; Rodrigues et al., 2011). However, protease-protection studies (Supplemental Fig. S3) indicate that FTSH12 does indeed have two TMs, a loop facing the interenvelope space, and a stromal 550-residue catalytic domain. There are several conceivable ways of envelope membrane integration that could yield this topology. However, the most straightforward is for the entire FTSH12 polypeptide to be imported to the stromal side of the envelope followed by integration of the two TMs and loop by a stroma-facing translocase, such as the SEC2 translocase. The fact that a significant amount of soluble FTSH12 is produced during lysate chase from prebound chloroplasts is consistent with this model, as is the nearly complete inhibition of integration by the SECA inhibitor sodium azide either during protein import or during the chase of lysates from prebound preFTSH12 (Figs. 3 and 8). SECA (presumably SECA2) is expected to be essential to transport the long loop (∼280 residues) across the membrane, based on studies in bacteria and also on the thylakoid FTSH5 (Deitermann et al., 2005; Rodrigues et al., 2011).
Two aspects of this process are still not understood. First is that only a very minor amount of integration was obtained for in vitro translated FTSH12 added directly to the chloroplast lysate assay. Second, antibodies to SCY2 and/or SECE2 did not inhibit integration during the FTSH12 chase assay. One possible explanation for both observations is that FTSH12 may need to initiate integration while the polypeptide chain is still being imported across the envelope. Such a coupled import-integration process has been observed previously for TIC40 (Li and Schnell, 2006), and FTSH12 is a much longer polypeptide. If FTSH12 initiated integration during the binding step, SECA2 might mask both the SCY2-HA and FLAG-SECE2 epitopes for antibody binding. Bacterial SECA and SECE binding are concentrated toward the C-terminal regions of SCY, with a particularly strong interaction on the most C-terminal cytoplasmic loop and C-terminal SCY tail (i.e. the location of the HA epitope on SCY2; Mori and Ito, 2006). Because of the very large size of SECA (SECA2), it is likely that if SECA2 is engaged with SCY2/SECE2, it could occlude the epitope regions from antibody access.
In summary, two approaches were used to identify substrates for the plastid envelope-located SEC2 system. Inducible RNAi of SCY2 influenced both the envelope and thylakoid, reducing the accumulation of thylakoid translocase components and inner envelope membrane proteins. In vitro protein import and membrane integration studies with mostly developed chloroplasts indicated that the RNAi effect on envelope proteins is a primary effect, because the SEC2 translocase system is involved in the post-import integration of proteins such as TIC40 and FTSH12, whereas the RNAi effect on thylakoid translocase components is likely due to secondary effects, either the down-regulation of transcription or the impairment of the TIC protein import apparatus in the envelope, or both. Clearly, these initial results will foster more detailed analyses to understand the integration process of chloroplast membrane proteins.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized, stratified at 4°C for 48 h, and grown on 0.8% (w/v) agar plates containing MS basal medium with vitamins (PhytoTechnology Laboratories), 50 mm MES-KOH, pH 5.7, and 1% (w/v) Suc under a 14-h-light (100 μE m−2 s−1)/10-h-dark cycle at 23°C. For inducible RNAi plants, seeds were germinated on MS medium containing 15 μg mL−1 hygromycin and 20 μm β-estradiol (MilliporeSigma) and grown under the same conditions as wild-type plants.
Hairpin Construction for Inducible RNAi of SCY1 and SCY2
A DNA fragment containing the second exon and the following intron of SCY2 was amplified directly from Arabidopsis genomic DNA by PCR and assembled into the hairpin structure with the inverted second exon fragment. To generate the SCY1 hairpin fragment, the third exon and the following intron with the inverted third exon were used. Both hairpin fragments were cloned into the estrogen-inducible binary vector, pMDC7 (Curtis and Grossniklaus, 2003), through the Gateway system (Invitrogen). All primer sets that were used to synthesize the RNAi constructs are listed in Supplemental Table S1.
Generation of Transgenic Plants and Analysis of RNAi Plants
Each RNAi construct was introduced into Arabidopsis (Columbia-0) by Agrobacterium tumefaciens-mediated floral dip transformation (Zhang et al., 2006). T1 lines carrying estrogen-inducible constructs or the empty vector pMDC7 were initially selected on medium containing 30 μg mL−1 hygromycin. Transformants were confirmed by PCR amplification from plant DNA using the vector primers and gene-specific primers listed in Supplemental Table S1. A total of 19 SCY1-RNAi lines and 44 SCY2-RNAi lines were identified, and lines that showed strong and stable phenotypes following estrogen treatment were selected. T3 plants germinated on selection medium containing 20 μm β-estradiol and 15 μg mL−1 hygromycin were used for further protein and transcriptional analysis.
qRT-PCR
Total RNA from 6-d-old Arabidopsis seedlings germinated on MS selection medium was extracted using the Qiagen Plant RNeasy miniprep kit. Three micrograms of total RNA was reverse transcribed with oligo(dT)22 and SuperScript II reverse transcriptase (Thermo Fisher) in 20-μL reactions according to the manufacturer’s instructions. SYBR Green-based quantitative PCR was performed in triplicate and quantified in the StepOnePlus real-time system (Thermo Fisher). Each 10-μL reaction contained 0.1 µL of cDNA, 0.2 µm primers, and 5 μL of SYBR Green select master mix (ABI, Life Technologies). Primers used in this study are listed in Supplemental Table S1. Expression was normalized against UBIQUITIN10 (AT4G05320; Czechowski et al., 2004) as the internal control. The melting curve was examined for each reaction to ensure that no primer dimers or nonspecific PCR products were present.
Arabidopsis Total Protein Extraction
Total proteins were extracted from 50 to 100 mg of 6-d-old Arabidopsis seedlings grown on MS medium by freezing in liquid nitrogen and homogenizing with micropestles in 0.5 to 1 mL of extraction buffer (125 mm Tris-HCl, pH 8, 1% SDS, 5 mm EDTA, 1 mm DTT, 1× plant protease inhibitor cocktail [Roche], and 2 mm PMSF). Samples were centrifuged at 14,000g for 10 min at 4°C. Protein concentrations of supernatants were determined with bicinchoninic acid reagent (Thermo Fisher). Equal protein amounts were mixed with 1 volume of 2× SDS-PAGE sample buffer (SSB) plus urea (0.1 m Tris-HCl, pH 6.8, 5% SDS, 5 mm EDTA, 0.1% bromophenol blue, 0.2 m DTT, and 8 m urea) and incubated at 37°C for 20 min before being subjected to SDS-PAGE.
Quantifying Immunoblot Signals
Proteins on SDS-PAGE gels were electroblotted to PVDF membranes in transfer buffer (25 mm Tris, 192 mm Gly, and 20% methanol). Blots were blocked with TBST (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Tween 20) containing 5% nonfat dry milk for 20 min at room temperature and probed with primary antibodies diluted in TBST with 0.5% dry milk for 2 h at room temperature. Antibody dilutions were as follows: anti-Tic110 (1:10,000; Inaba et al., 2005), anti-Tic40 (1:10,000; Chou et al., 2003), anti-TOC75 (1:5,000), anti-SCY1 (1:5,000; Schuenemann et al., 1999), anti-ALB3-50 aa (1:5,000; Moore et al., 2000), anti-FTSH12 (1:2,000; Dr. Masato Nakai, Osaka University), anti-TATC (1:10,000; Martin et al., 2009), and mouse anti-actin IgG (1:1,000; Pan Ab-5 [Thermo Fisher]). After washing in TBST for 3 × 10 min, the blot was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000; Bio-Rad) or horseradish peroxidase-conjugated goat anti-mouse IgG (1:5,000; Bio-Rad) for 1 h at room temperature, blots were washed in TBST for 3 × 10 min and processed for enhance chemiluminescence detection (GE Healthcare) and exposure to Fuji medical x-ray film (Super RX-N).
Immunoblot signals were quantified from scanned x-ray films by densitometric analysis of bands with Quantity One software (Bio-Rad). The actin band was used to normalize the amount of proteins loaded from each sample. The relative abundance of proteins in mutant versus wild-type plant extracts was calculated based on intensities of their immunoblot signals.
Construction of Plasmids for in Vitro Transcription
Full-length cDNAs of TIC110 and TIC40 were amplified from pET21d-AtTic110 and pET21d-AtTic40-6xHis provided by Dr. Danny Schnell (Michigan State University) with primer sets Tic110-1F/Tic110-His-R and Tic40-1F/Tic40-1R (Supplemental Table S1) and cloned into pGEM-4Z at KpnI/XbaI and EcoRI/KpnI sites, respectively, in which the transcription was driven by the Sp6 promoter. The full-length cDNA of FTSH12 was amplified from cpDEST14-atFTSH12 (from Dr. John Froehlich, Michigan State University) and cloned into pGEM-4Z vector at EcoRI/SalI sites. Other full-length precursor clones used for in vitro transcription, including SCY1, SCY2, ALB3, pea (Pisum sativum) ALB3, pea TATC, pea LHCP, pea OE23, and wheat (Triticum aestivum) OE33 and iOE33, were described previously (Cline et al., 1989, 1993; Moore et al., 2000; Mori et al., 2001; Martin et al., 2009; Skalitzky et al., 2011; Li et al., 2015).
A cDNA fragment encoding the intermediate form of TIC40 (iTIC40; Chiu and Li, 2008) was generated through PCR amplification with primers iTic40Xba5′ and Tic40-1R (Supplemental Table S1) from pGEM-4Z-Tic40 and cloned into pGEM-4Z at XbaI/KpnI sites. To generate ΔTP-SCY1 and ΔTP-FTSH12 (transit peptide deletion), the coding sequences of SCY1 and FTSH12 lacking their first 10 amino acids were PCR amplified with the primers listed in Supplemental Table S1 and cloned into pGEM-4Z vector. ΔTP-psALB3, lacking the first 60 amino acids, was PCR amplified from pGEM-4Z-psALB3 and cloned into pGEM-4Z at EcoRI/SaII sites. The translation initiation codon ATG was added in all truncated gene fragments through PCR amplification.
Isolation of Chloroplasts, Lysis, and Preparation of Membranes and SE
Intact chloroplasts were isolated from 9- to 10-d-old pea seedlings as described previously (Cline, 1986). Arabidopsis chloroplasts were prepared as described (Li et al., 2015). Chloroplasts were resuspended in IB (50 mm HEPES-KOH, pH 8, and 0.33 m sorbitol) at 1 mg mL−1 Chl. Chloroplasts were lysed by suspending chloroplast pellets in 10 mm HEPES-KOH, pH 8, at 1 to 2 mg mL−1 Chl for 5 to 10 min on ice, followed by the addition of 1 volume of 2× IB. Membranes were separated from SE by centrifuging lysates at 20,000g for 20 min at 4°C, and membrane pellets were resuspended in IB. For the preparation of 10× SE, lysates at 1 mg mL−1 Chl were centrifuged at 100,000g for 20 min at 2°C. The supernatant was then concentrated 10-fold with a Centrifugal Filter Unit (Millipore Ultracel-30K) at 3,800g for 30 min at 4°C.
In Vitro Assay for Protein Import into Chloroplasts
Capped mRNAs for precursor proteins were transcribed in vitro with SP6 polymerase (Promega) and translated in vitro in the presence of [3H]Leu and the full complement of unlabeled amino acids lacking Leu with a homemade wheat germ translation system (Cline, 1986). Translations were diluted with 1 volume of 2× IB containing 60 mm unlabeled Leu before use in assays. Translation reactions in assays lacking ATP were treated with apyrase at 6 units per 100 μL of undiluted translation product for at least 15 min on ice prior to use. Diluted translation products were incubated with intact chloroplasts (0.33 mg mL−1 Chl) and 5 mm Mg-ATP (MilliporeSigma) for 20 to 50 min in a 25°C water bath supplied with 120 μE m−2 s−1 white light. After import, intact chloroplasts were reisolated by centrifugation through 35% Percoll in IB and washed with 1 mL of IB. Alternatively, chloroplasts were treated with thermolysin at 0.1 mg mL−1 for 30 min at 4°C. Reactions were terminated with IB containing EDTA to 10 mm final concentration, and intact chloroplasts were recovered by centrifugation through a 35% Percoll cushion containing 5 mm EDTA and washed with IB containing 5 mm EDTA. Import assays conducted in the presence of inhibitors received sodium azide (7 mm final concentration) or a combination of nigericin and valinomycin (1 and 2 μm final concentrations, respectively) for 10 min on ice before initiation of the assay.
Reconstituted Membrane Integration in Chloroplast Lysates or Isolated Chloroplast Membranes
Assays contained adjusted translation products, chloroplast lysates, or total membranes at 0.17 mg mL−1 Chl, SE at a stoichiometric ratio of 10 with respect to Chl, and 2.5 mm Mg-ATP in IB. Assays were generally conducted for 50 min at 25°C either in 120 μE m−2 s−1 white light or in darkness as stipulated in the figure legends. Membranes were then centrifuged at 20,000g for 20 min. An aliquot of membranes was resuspended in IB and treated with thermolysin as above. As stated in the figure legends, another aliquot was extracted with 150 to 300 μL of 0.1 m NaOH, 0.2 m Na2CO3, or OH−/CO32− and incubated for 30 min on ice, then centrifuged at 100,000g for 20 min at 2°C. The pellet was saved as alkaline-resistant membranes; the proteins of the supernatant were precipitated with 10% (w/v) TCA on ice for 30 min, followed by centrifugation in a microfuge for 30 min at 4°C. The resulting pellet was washed with 1 mL of ice-cold acetone, centrifuged for 10 min at 4°C, and air dried. Both membrane pellet and extracted supernatant pellet were dissolved in stoichiometrically equivalent volumes in SSB plus urea at room temperature. The alkaline extraction protocol was essentially as described by Chu and Li (2011).
Coupled Binding and Chase Integration Assay
The full-length precursor proteins were translated in a homemade wheat germ system (Cline, 1986), which contains 1.5 mm ATP plus an ATP regenerating system. Translation products were subsequently diluted 15-fold in the final assay mixture to 0.1 mm ATP, which contained intact chloroplasts at 0.33 mg mL−1 Chl. Precursor binding to chloroplasts (i.e. to the early intermediate stage; Schnell et al., 1994) was for 15 min at 25°C in darkness or as stated in the figure legends. Chloroplasts were recovered on Percoll cushions, washed in IB, lysed in 10 mm HEPES/KOH, and adjusted to IB. The lysate was combined with ATP to 5 mm and 10× SE to 2.5×. Chase assays were for 30 min at 25°C in the light or as designated in the figure legends. Lysate chase reactions were centrifuged at 20,000g for 20 min at 4°C to obtain a membrane fraction and a supernatant. Membranes were resuspended in a small amount of IB and divided into two aliquots; one aliquot was treated with thermolysin as described above, and the other was extracted with a 6-fold excess of 0.1 m NaOH as described above into an NaOH-resistant pellet and an NaOH-extracted supernatant. All samples were dissolved in SSB plus urea and gel loaded in stoichiometrically equivalent amounts.
Antibody Inhibition Assay
For the inhibition of pea thylakoid translocases, IgGs against psSCY1 (SEC1 pathway; Mori et al., 1999), psALB3-C terminus (SRP/ALB3 pathway; Mori et al., 2001), and psHCF106 (TAT pathway; Mori et al., 2001) were incubated at 0.8 μg mL−1 with membranes from a chloroplast lysate at 0.33 mg mL−1 Chl in one-third-strength IBM and 0.7% bovine serum albumin for 1 h on ice. In the case of α-ALB3 50 aa and its prebleed, 100 μL mL−1 serum was used instead of IgG (Moore et al., 2000). Membranes were then washed and resuspended in IBM. For combinations of antibodies, each antibody was used at 0.8 μg mL−1 or serum at 100 μL mL−1. Treated membranes were assayed with 10× SE, 2.5 mm Mg-ATP, and in vitro translated precursor proteins in an integration assay for 50 min in light as described above. Membranes were then treated with thermolysin.
For antibody inhibition of the SCY2/SEC2 translocase, chloroplast total lysate was prepared from a scy2 sec2 double mutant Arabidopsis line that had been rescued with FLAG-SEC2 and SCY2-HA (Li et al., 2015). Each 50 μL of lysate was incubated with 10 μg of anti-HA IgG (Roche; 3F10), 10 μg of anti-FLAG IgG (Sigma-Aldrich; F7425), or a mixture of the two on ice for 1 h. The anti-FLAG IgG was supplied in phosphate buffer plus sodium azide. Before use, 100 μL of anti-FLAG IgG solution buffer was exchanged by three rounds of concentration to ∼10 μL with Microcon Centrifugal Filter Device YM-30 (EMD Millipore) followed by the addition of 100 μL of 20 mm HEPES-KOH, pH 7.5. Both the buffer-exchanged antibody and the original buffer that the antibody was supplied in were tested for effects in the iTIC40 integration assay.
Subfractionation of Envelope and Thylakoid Membranes
Total membranes were pelleted at 20,000g for 20 min at 4°C after reconstitution or binding-chase integration assays, resuspended in 0.3 m Suc in 10 mm Tricine-KOH, pH 8, containing 2 mm EDTA (TE), equivalent to 1 mg mL−1 Chl, and sonicated with Microson XL 2000 (Misonmix) at 3 W of output power for 10 s on ice. After this treatment, no intact thylakoid networks were visible with the microscope. Samples (300 μL) were loaded on step Suc gradients made up with 0.3 mL of 1.3 m, 1.2 mL of 1.2 m, and 0.3 mL of 1.1 m Suc in TE buffer in a precooled ultracentrifuge tube (Beckman; 347356) and centrifuged at 45,000g for 5 h at 2°C with a Beckman TLS55 rotor. The interface between the 0.3 m and 1.1 m Suc layers (∼600 μL) was collected, diluted with 900 μL of TE buffer, and centrifuged at 100,000g for 20 min at 2°C. The resulting pellet was used as the total envelope fraction. The 1.3 m Suc layer and the pellet (∼300 μL) were diluted with 1 mL of TE buffer and centrifuged at 100,000g for 20 min at 2°C. The pellet was saved as thylakoids. Each total envelope or thylakoid fraction was resuspended in 50 μL of SSB plus urea, incubated at 37°C for 20 min, and subjected to SDS-PAGE and fluorography analysis. For alkaline or protease treatment, equivalent amounts of total envelope or thylakoid fractions were subjected to 0.1 m NaOH or thermolysin treatment as described above.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The levels of substrate proteins of SRP/ALB3 and TATC were reduced in SCY2-RNAi plants.
Supplemental Figure S2. Chloroplast lysis methods produce inside-out inner envelope vesicles and right-side-out thylakoid vesicles.
Supplemental Figure S3. Properly integrated ALB3, SCY1, and FTSH12 are resistant to 0.1 m NaOH extraction and produce diagnostic protease-protected degradation products.
Supplemental Figure S4. Properly integrated FTSH12 produces a protease-resistant degradation product of ∼39 kD that likely represents its two TMs and intervening loop exposed to the interenvelope space.
Supplemental Figure S5. A mixture of 30% NaOH and 70% Na2CO3 can be used for the diagnosis of TIC40 integration.
Supplemental Figure S6. The membrane integration of FTSH12 is time dependent.
Supplemental Figure S7. Quantitative analysis of integrated SCY1 and ALB3 in antibody inhibition assays.
Supplemental Figure S8. Quantitation of membrane-integrated mTIC40 in antibody inhibition assays.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Michael McCaffery for excellent technical support in chloroplast isolation and DNA construct preparation, Dr. John Froehlich for the Arabidopsis FTSH12 cDNA clone, Dr. Danny Schnell for Arabidopsis TIC110 and TIC40 cDNA clones, Dr. Hsou-min Li and Dr. Danny Schnell for antibodies against TOC75, TIC40, and TIC110, Dr. Danja Schuenemann for antibodies against SCY1, Dr. Nakai Masato for serum against FTSH12, and Dr. Ralph Henry for anti-ALB3-50 aa serum and E. coli-expressed cpFTSY, cpSRP54, and cpSRP43 proteins.
Glossary
- TM
transmembrane domain
- RNAi
RNA interference
- MS
Murashige and Skoog
- qRT
quantitative real-time
- PMF
proton motive force
- SE
stromal extract
- IB
import buffer
- Chl
chlorophyll
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. MCB 1158110 to K.C. and grant no. MCB 1158173 to D.E.F.).
Articles can be viewed without a subscription.
References
- Arnold I, Langer T (2002) Membrane protein degradation by AAA proteases in mitochondria. Biochim Biophys Acta 1592: 89–96 [DOI] [PubMed] [Google Scholar]
- Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, Terashima I, Barkan A, Nakai M (2004) Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Kikuchi S, Nakai M (2008) Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J 56: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Bennett J. (1981) Biosynthesis of the light-harvesting chlorophyll a/b protein: polypeptide turnover in darkness. Eur J Biochem 118: 61–70 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (1987) The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann N Y Acad Sci 503: 55–71 [DOI] [PubMed] [Google Scholar]
- Celedon JM, Cline K (2012) Stoichiometry for binding and transport by the twin arginine translocation system. J Cell Biol 197: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon JM, Cline K (2013) Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim Biophys Acta 1833: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CC, Li HM (2008) Tic40 is important for reinsertion of proteins from the chloroplast stroma into the inner membrane. Plant J 56: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Li HM (2011) Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol Biol 774: 339–350 [DOI] [PubMed] [Google Scholar]
- Cline K. (1986) Import of proteins into chloroplasts: membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J Biol Chem 261: 14804–14810 [PubMed] [Google Scholar]
- Cline K, Dabney-Smith C (2008) Plastid protein import and sorting: different paths to the same compartments. Curr Opin Plant Biol 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Ettinger WF, Theg SM (1992) Protein-specific energy requirements for protein transport across or into thylakoid membranes: two lumenal proteins are transported in the absence of ATP. J Biol Chem 267: 2688–2696 [PubMed] [Google Scholar]
- Cline K, Fulsom DR, Viitanen PV (1989) An imported thylakoid protein accumulates in the stroma when insertion into thylakoids is inhibited. J Biol Chem 264: 14225–14232 [PubMed] [Google Scholar]
- Cline K, Henry R, Li C, Yuan J (1993) Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J 12: 4105–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Deitermann S, Sprie GS, Koch HG (2005) A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J Biol Chem 280: 39077–39085 [DOI] [PubMed] [Google Scholar]
- DeLille J, Peterson EC, Johnson T, Moore M, Kight A, Henry R (2000) A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc Natl Acad Sci USA 97: 1926–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JE, Keegstra K (2011) The role of the transmembrane domain in determining the targeting of membrane proteins to either the inner envelope or thylakoid membrane. Plant J 68: 844–856 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Schmidt B, Wachter E, Weiss H, Neupert W (1986) Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell 47: 939–951 [DOI] [PubMed] [Google Scholar]
- Henry RL. (2010) SRP: adapting to life in the chloroplast. Nat Struct Mol Biol 17: 676–677 [DOI] [PubMed] [Google Scholar]
- Hoober JK, Boyd CO, Paavola LG (1991) Origin of thylakoid membranes in Chlamydomonas reinhardtii y-1 at 38°C. Plant Physiol 96: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Havaux M, Carde JP, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L (2002) Double mutation cpSRP43−/cpSRP54− is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J 29: 531–543 [DOI] [PubMed] [Google Scholar]
- Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell DJ (2005) Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17: 1482–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Akiyama Y (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–31 [DOI] [PubMed] [Google Scholar]
- Köhler D, Montandon C, Hause G, Majovsky P, Kessler F, Baginsky S, Agne B (2015) Characterization of chloroplast protein import without Tic56, a component of the 1-megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol 167: 972–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chiu CC (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Li M, Schnell DJ (2006) Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J Cell Biol 175: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Henry R, Yuan J, Cline K, Hoffman NE (1995) A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA 92: 3789–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Singhal R, Taylor IW, McMinn PH, Chua XY, Cline K, Fernandez DE (2015) The Sec2 translocase of the chloroplast inner envelope contains a unique and dedicated SECE2 component. Plant J 84: 647–658 [DOI] [PubMed] [Google Scholar]
- Martin JR (2010) Biogenesis of chloroplast thylakoid translocase subunit cpTatC. PhD thesis. University of Florida, Gainesville, FL [Google Scholar]
- Martin JR, Harwood JH, McCaffery M, Fernandez DE, Cline K (2009) Localization and integration of thylakoid protein translocase subunit cpTatC. Plant J 58: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Harrison MS, Peterson EC, Henry R (2000) Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J Biol Chem 275: 1529–1532 [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K (2006) Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA 103: 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Cline K (2001) Chloroplast TatC plays a direct role in thylakoid (Delta)pH-dependent protein transport. FEBS Lett 501: 65–68 [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K (1999) Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol 146: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré DJ, Selldén G, Sundqvist C, Sandelius AS (1991) Stromal low temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiol 97: 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murataliev MB, Milgrom YM, Boyer PD (1991) Characteristics of the combination of inhibitory Mg2+ and azide with the F1 ATPase from chloroplasts. Biochemistry 30: 8305–8310 [DOI] [PubMed] [Google Scholar]
- Nakai M, Goto A, Nohara T, Sugita D, Endo T (1994) Identification of the SecA protein homolog in pea chloroplasts and its possible involvement in thylakoidal protein transport. J Biol Chem 269: 31338–31341 [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Rapoport TA. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Richter CV, Bals T, Schünemann D (2010) Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur J Cell Biol 89: 965–973 [DOI] [PubMed] [Google Scholar]
- Rodrigues RA, Silva-Filho MC, Cline K (2011) FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J 65: 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychkova A, Warshel A (2013) Exploring the nature of the translocon-assisted protein insertion. Proc Natl Acad Sci USA 110: 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G (1994) Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Schuenemann D, Amin P, Hartmann E, Hoffman NE (1999) Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex. J Biol Chem 274: 12177–12182 [DOI] [PubMed] [Google Scholar]
- Settles AM, Yonetani A, Baron A, Bush DR, Cline K, Martienssen R (1997) Sec-independent protein translocation by the maize Hcf106 protein. Science 278: 1467–1470 [DOI] [PubMed] [Google Scholar]
- Skalitzky CA, Martin JR, Harwood JH, Beirne JJ, Adamczyk BJ, Heck GR, Cline K, Fernandez DE (2011) Plastids contain a second Sec translocase system with essential functions. Plant Physiol 155: 354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck TL, Yu J (1973) Selective solubilization of proteins from red blood cell membranes by protein perturbants. J Supramol Struct 1: 220–232 [DOI] [PubMed] [Google Scholar]
- Swidersky UE, Rienhöfer-Schweer A, Werner PK, Ernst F, Benson SA, Hoffschulte HK, Müller M (1992) Biochemical analysis of the biogenesis and function of the Escherichia coli export factor SecY. Eur J Biochem 207: 803–811 [DOI] [PubMed] [Google Scholar]
- Tripp J, Inoue K, Keegstra K, Froehlich JE (2007) A novel serine/proline-rich domain in combination with a transmembrane domain is required for the insertion of AtTic40 into the inner envelope membrane of chloroplasts. Plant J 52: 824–838 [DOI] [PubMed] [Google Scholar]
- Viana AA, Li M, Schnell DJ (2010) Determinants for stop-transfer and post-import pathways for protein targeting to the chloroplast inner envelope membrane. J Biol Chem 285: 12948–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitanen PV, Lubben TH, Reed J, Goloubinoff P, O’Keefe DP, Lorimer GH (1990) Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry 29: 5665–5671 [DOI] [PubMed] [Google Scholar]
- Voelker R, Barkan A (1995) Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J 14: 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta L, Soll J, Bölter B (2007) Requirements for a conservative protein translocation pathway in chloroplasts. FEBS Lett 581: 2621–2624 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341: 456–458 [DOI] [PubMed] [Google Scholar]
- Welte T, Kudva R, Kuhn P, Sturm L, Braig D, Müller M, Warscheid B, Drepper F, Koch HG (2012) Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol Biol Cell 23: 464–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Henry R, McCaffery M, Cline K (1994) SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science 266: 796–798 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.