Secretory cells contain a specific Rubisco, the small subunit of which belongs to a so-far-uncharacterized phylogenetic cluster and confers a modified pH-dependent activity.
Abstract
Ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) is the most abundant enzyme in plants and is responsible for CO2 fixation during photosynthesis. This enzyme is assembled from eight large subunits (RbcL) encoded by a single chloroplast gene and eight small subunits (RbcS) encoded by a nuclear gene family. Rubisco is primarily found in the chloroplasts of mesophyll (C3 plants), bundle-sheath (C4 plants), and guard cells. In certain species, photosynthesis also takes place in the secretory cells of glandular trichomes, which are epidermal outgrowths (hairs) involved in the secretion of specialized metabolites. However, photosynthesis and, in particular, Rubisco have not been characterized in trichomes. Here, we show that tobacco (Nicotiana tabacum) trichomes contain a specific Rubisco small subunit, NtRbcS-T, which belongs to an uncharacterized phylogenetic cluster (T). This cluster contains RbcS from at least 33 species, including monocots, many of which are known to possess glandular trichomes. Cluster T is distinct from the cluster M, which includes the abundant, functionally characterized RbcS isoforms expressed in mesophyll or bundle-sheath cells. Expression of NtRbcS-T in Chlamydomonas reinhardtii and purification of the full Rubisco complex showed that this isoform conferred higher Vmax and Km values as well as higher acidic pH-dependent activity than NtRbcS-M, an isoform expressed in the mesophyll. This observation was confirmed with trichome extracts. These data show that an ancient divergence allowed for the emergence of a so-far-uncharacterized RbcS cluster. We propose that secretory trichomes have a particular Rubisco uniquely adapted to secretory cells where CO2 is released by the active specialized metabolism.
Ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) is the most abundant enzyme in plants and probably on earth. It catalyzes the carboxylation of ribulose-1,5-biphosphate (RuBP) in chloroplasts and so is responsible for CO2 fixation during photosynthesis. The resulting products, two molecules of 3-phosphoglycerate, are then used to build carbohydrates. Rubisco is a complex made of eight copies of a large subunit (RbcL), which is encoded by a chloroplast gene, and eight copies of a small subunit (RbcS), which is encoded by a nuclear gene family (Spreitzer and Salvucci, 2002; Andersson and Backlund, 2008). Although the catalytic reaction is mostly controlled by the large subunit, the small subunit also influences the reaction. For instance, expressing higher plant RbcS subunits in a Chlamydomonas reinhardtii mutant that lacks rbcS genes showed that RbcS makes a significant contribution to the Rubisco activity (Genkov et al., 2010). Expressing an RbcS from sorghum, a C4 plant, in rice, a C3 plant, resulted in increased catalytic turnover of Rubisco (Ishikawa et al., 2011).
In the plant, Rubisco is found in photosynthetic tissues, i.e. mostly in the leaf mesophyll (C3 plants), bundle sheath (C4 plants), and guard cells. However, Rubisco is also found in more specialized cell types such as glandular trichomes (hairs) of certain species. Trichomes are expansions of the epidermis of most of the aerial tissues and can be glandular or nonglandular, depending on their secretion ability. Glandular trichomes comprise a stalk made of one or several cells topped by a head made of a single or several secretory cells. They function to synthesize and secrete specialized (also called secondary) metabolites, which are involved in the plant defense against abiotic (e.g. UV, light) as well as biotic (e.g. herbivores and pathogens) stress. Moreover, many of the metabolites synthesized in trichomes have fragrant and aromatic properties, which are exploited in food (e.g. thyme, basil, mint, hop) or in perfumery (e.g. lavender, sage). Some metabolites also have pharmacological properties such as artemisinin, an antimalaria agent secreted by Artemisia annua trichomes (Tissier, 2012; Glas et al., 2012; Lange and Turner, 2013; Lange, 2015; Wagner, 1991).
In some plants, trichomes contain chloroplasts that function to help provide the carbon and energy needs for specialized metabolite production (Keene and Wagner, 1985). In tobacco (Nicotiana tabacum), the trichome exudate (mainly made of diterpenes, acyl sugar esters) accounts for more than 10% of the dry weight of leaves (Wagner, 1991). In this species as well as in others, enzymes involved in the specialized metabolism have attracted much attention because of their role in the plant defense and their industrial or pharmacological interest (Lange and Turner, 2013; Tissier, 2012). On the contrary, the primary metabolism and, in particular, photosynthesis and the enzymes involved therein have barely been characterized, probably because they are thought to be similar, as in other photosynthetic cells.
In this report, we show that chloroplasts of glandular trichomes from tobacco contain a novel RbcS that is common among other trichome-producing plant species and whose sequence is phylogenetically distinct from the RbcS isoforms made in mesophyll cells. When transformed into an RbcS-lacking mutant of the green alga C. reinhardtii, both mesophyll and trichome RbcS isoforms from tobacco assembled with the algal RbcL to form functional RbcL8RbcS8 hybrid enzymes that restored photosynthetic growth. Differences in the pH-dependent activity of each hybrid Rubisco, as well as the native trichome and mesophyll Rubisco, suggest that the trichome-specific expression of the alternative RbcS may pose an example of evolutionary adaptation to the specialized metabolism of secretory cells.
RESULTS
Tobacco Trichomes Contain a Specific RbcS
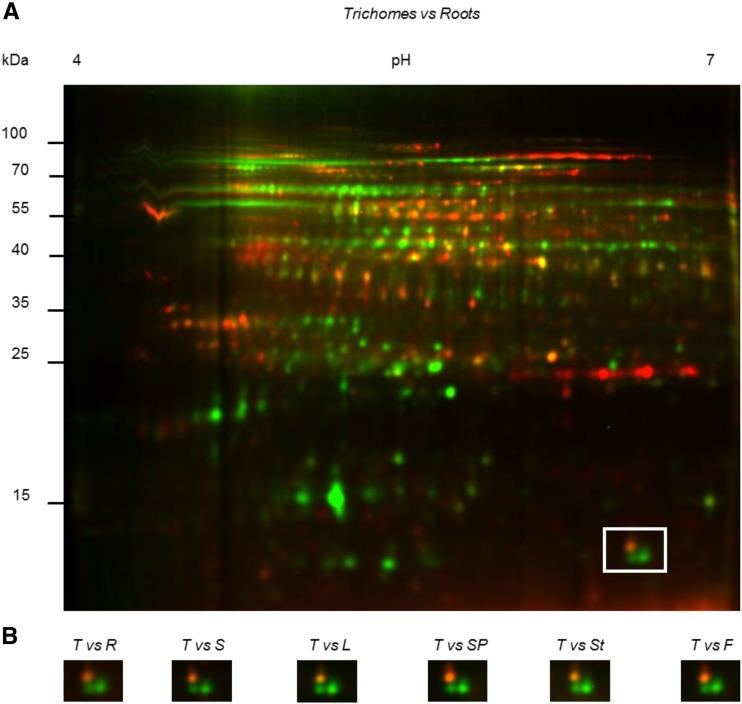
We were initially interested in finding proteins that are specifically expressed in photosynthetic glandular trichome cells of tobacco. This species contains two types of glandular trichomes, short ones and tall ones, only the latter being photosynthetic. Tall glandular trichome heads were purified by centrifugation on Percoll gradients, and their protein complement was compared to that of different plant tissues by two-dimensional difference gel electrophoresis (2D-DIGE). As an example, Figure 1A shows the comparison between trichomes and roots. Comparison between trichomes and other tissues can be found in Supplemental Figure S1.
Figure 1.
2D-DIGE of soluble proteins of tall glandular trichomes compared to other tissues. Proteins of tall glandular trichomes were compared to proteins from roots, germinating seeds, leaves, seed pods, stems, and flowers. Soluble proteins (20 µg) isolated from trichomes and other tissues were labeled with the Cy3 (green) and Cy5 (red) fluorescent dyes, respectively. A, Example of a full gel (trichomes versus roots). Other gels are shown in Supplemental Figure S1. B, The area delimited by a square in A is displayed for the indicated comparisons between trichomes and other tissues. Within this square, the right green spot was identified as NtRbcS-T (spot 42 in Supplemental Tables S1 and S2). T, Trichomes; R, roots; S, seeds; L, leaves; SP, seed pods; St, stems; F, flowers.
Spot analysis revealed 2516 protein spots, among which 118 exhibited increased expression levels (average ratio ≥2; t test and one-way ANOVA; P ≤ 0.005) in the tall glandular trichomes when compared to other tissues. The ratio was between 2 and 5 for 88 spots, and over 5-fold for 30 spots. The 49 most abundant spots were manually excised from a preparative gel of tall trichome soluble proteins (Supplemental Fig. S2), subjected to trypsin digestion, and the peptides analyzed by tandem mass spectrometry (MS/MS) analysis. A data search using the translated tobacco NCBI EST database yielded significant identifications (P < 0.05) for each spot selected. After BLAST searching, 47 could be functionally classified, and most were involved in specialized metabolism and plant defense (Supplemental Table S1).
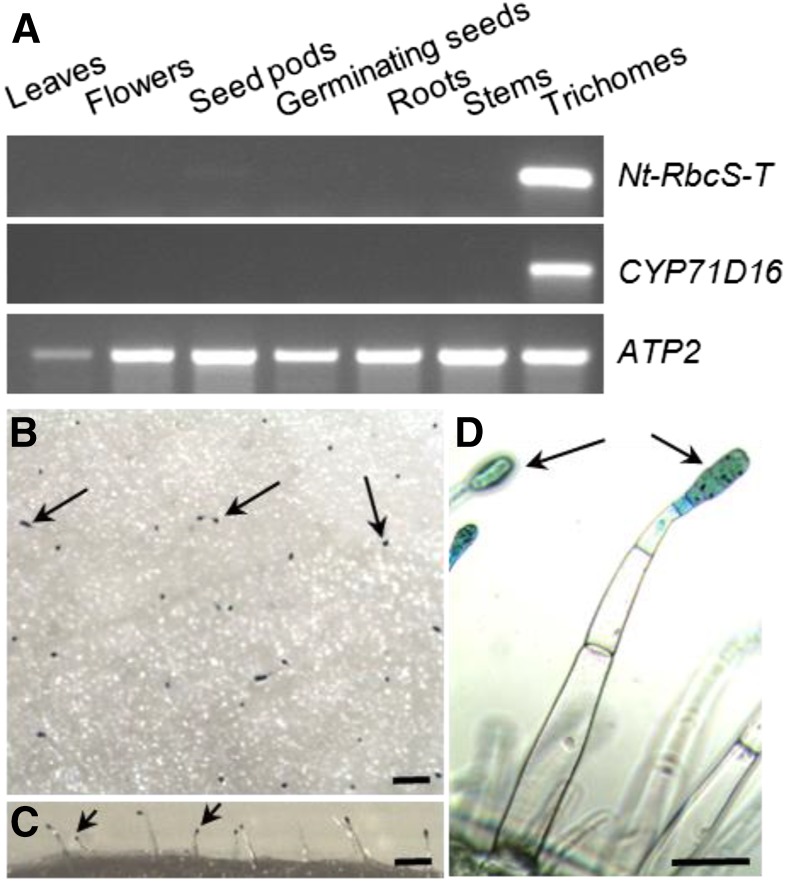
One of the proteins that was strongly enriched in the trichome sample (Fig. 1B; average enrichment ratio of 15.48) was a Rubisco small subunit (accession no. DV157962), from now on named NtRbcS-T. To confirm this observation, RNA was extracted from the same tissues as for the proteomic analysis, and reverse transcription-PCR was performed (Fig. 2A). CYP71D16, which has been demonstrated to be trichome specific (Wang et al., 2002), was used as a positive control, and ATP2, coding for the β-subunit of mitochondrial ATP synthase, was used as a loading control (Boutry and Chua, 1985). A strong NtRbcS-T signal was observed in the trichome sample. A faint signal was also observed for seed pods, but this was expected as trichomes were not removed from those tissues. For further confirmation, a 1.24-kb region encompassing the putative NbRbcS-T transcription promoter was retrieved from Nicotiana benthamiana and fused to the GUS reporter gene. Plants transformed with this construct showed GUS activity in the head cells of tall glandular trichomes, but not in the other leaf tissues (Fig. 2, B–D).
Figure 2.
Trichome-specific expression of NtRbcS-T. A, RNA was prepared from the indicated tissues and used for RT-PCR for the indicated transcripts. CYP71D16 is a marker of tall trichomes, and ATP2 was used as a loading control. For the leaf and stem samples, trichomes were removed before RNA extraction. B to D, Histochemical determination of pNbRbcS-T-GusVenus expression in tall glandular trichomes of tobacco leaves. Top (B), lateral (C), and detailed (D) views of the adaxial side of the leaf showing GUS activity in the head cells of glandular trichomes (examples are indicated with arrows). No other cell type shows GUS activity. Bars = 500 µm (B and C) and 100 µm (D).
NtRbcS-T Belongs to a Specific Phylogenetic Cluster
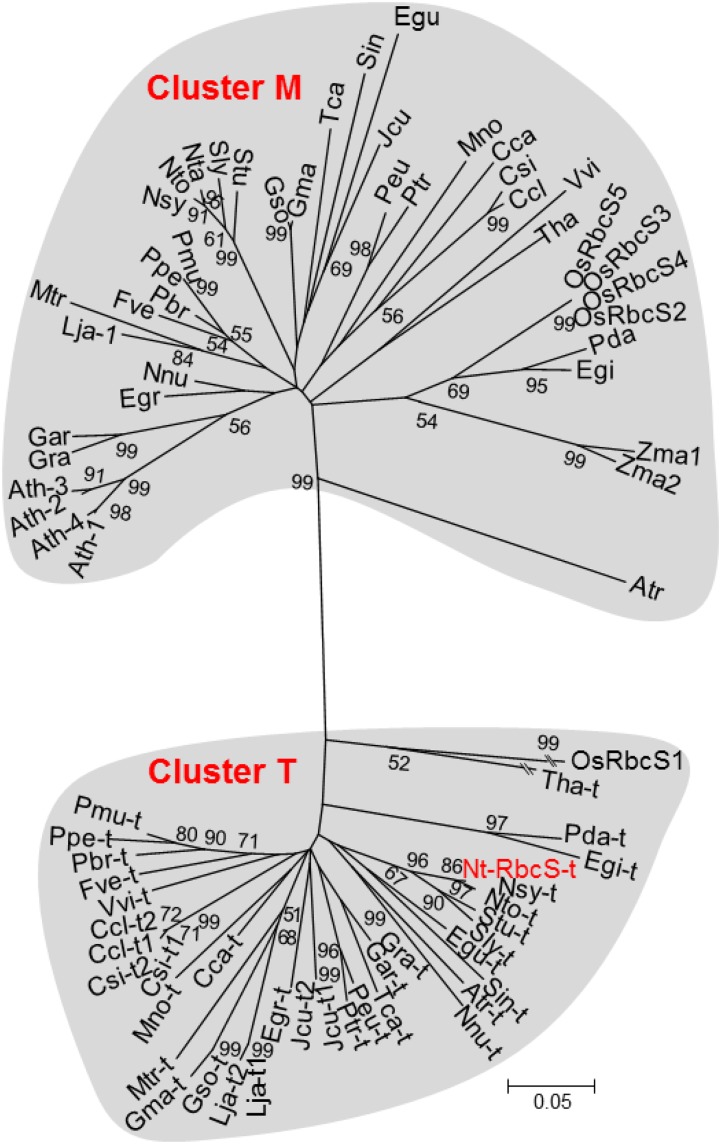
The mature NtRbcS-T amino acid sequence showed between 59.2% and 62.5% identity with the mature amino acid sequences of the recognized RbcS isoforms (e.g. Genbank accessions CAA26208.1 and AAP03874.1) produced in tobacco mesophyll cells. A BLAST homology search using NtRbcS-T identified 35 sequences from 31 C3 plant species with scores higher than 200 (Supplemental Table S2). The majority of these species are known to contain secretory organs such as glandular trichomes, colleters, nectaries, or secretory cavities. From each of these plant species, an additional RbcS sequence was included that represented an RbcS isoform produced in mesophyll cells. The isoform examined was that showing the highest sequence identity to the “trichome” RbcS isoform from the same species. Although the rice (Oryza sativa) Rubisco small subunit, OsRbcS1, displays a BLAST score lower than the 200 threshold, its sequence and other rice RbcS sequences were added to this analysis since OsRbcS1 also exhibits a specific expression pattern (Morita et al., 2014). Because no sequence from C4 plants was obtained through the BLAST analysis, the two RbcS sequences of maize (Zea mays), a C4 plant species, were also included. A phylogenetic analysis of these RbcS sequences identified two distinct branches (Fig. 3). One cluster, called T, comprised all of the “trichome” RbcS sequences, and the other cluster, called M, contained all of the mesophyll RbcS isoforms, including all four RbcS sequences from Arabidopsis (Arabidopsis thaliana), a species that lacks glandular trichomes. The two maize RbcS, which are expressed in bundle sheaths, were also localized in cluster M.
Figure 3.
Phylogenetic analysis of Rubisco small subunit sequences. The accession numbers and the abbreviations corresponding to the mature protein sequences used for this analysis are given in Supplemental Table S2. The analysis was performed as described in the “Materials and Methods” section. Bootstraps (1,000 iterations) were calculated by the nearest-neighbor-interchange method and values higher than 50% are indicated. Branch lengths are proportional to phylogenetic distances, except for Tha-t and OsRbcS1.
Trichome and Mesophyll Cells Have Electrophoretically Distinct Rubisco Complexes
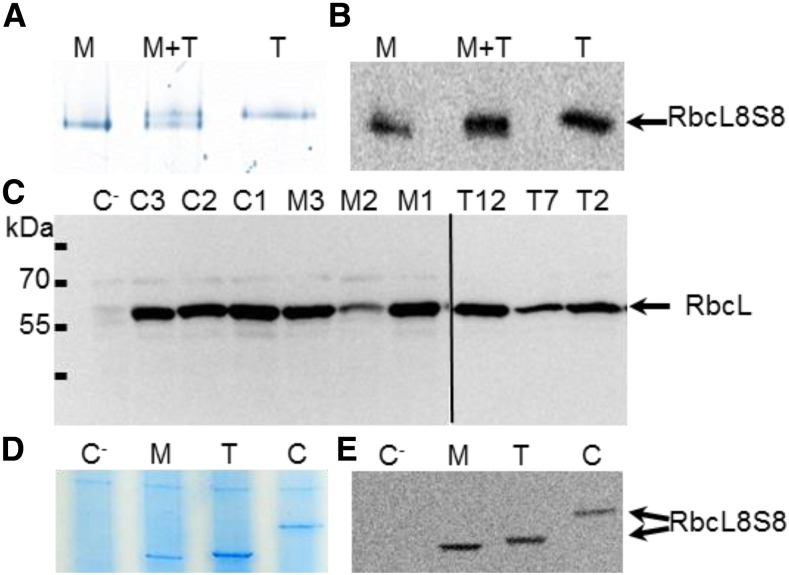
The RbcL is encoded by the chloroplast genome and is therefore expected to be the same in all photosynthetic cells. RbcS is thus the only part that can distinguish the Rubisco complex in different cell types. NtRbcS-T has a predicted pI (6.24) higher than that of the other tobacco small subunits (5.05–5.19). It is therefore possible that because of its small subunit, the whole Rubisco complex has a distinct electrophoretic mobility on native gel. Because of its hexadecameric organization and thus large size (∼540,000 D) as well as its abundance, the Rubisco complex is expected to run slowly and to be distinguished from the background of other proteins. A difference could not be observed by analyzing a whole-leaf extract because the contribution of the trichome Rubisco is too small relative to the total pool of leaf Rubisco. Protein extracts were thus obtained from isolated trichomes and from leaves cleared of trichomes and analyzed by electrophoresis. The leaf extract contains a majority of photosynthetic mesophyll cells, while in the trichome extract, the photosynthetic head cells of the tall glandular trichomes represent a minority compared to the cells that contain no or very few chloroplasts: stalk cells of the tall glandular trichomes, short glandular trichomes, as well as nonglandular trichomes. To take this difference into account, 10 times more protein was loaded for the trichome sample. Both colloidal blue staining of the gel (Fig. 4A) and western blotting (Fig. 4B) showed that the trichome Rubisco ran slower than the mesophyll enzyme. This was confirmed when both samples were mixed. We can therefore conclude that the trichome and mesophyll Rubisco complexes are electrophoretically homogenous and distinct.
Figure 4.
Electrophoretic analysis of leaf, trichome, and C. reinhardtii Rubisco. A, Native gel electrophoresis of soluble proteins from leaf tissues cleared of trichomes (M; 3 µg), from isolated trichomes (T; 30 µg). A mix of 1.5 µg of soluble proteins from leaf tissues cleared of trichomes and 15 µg of soluble proteins from isolated trichomes (M+T) was also analyzed in order to confirm that trichome and mesophyll Rubisco complexes are electrophoretically distinct. The gel was stained with colloidal blue. B, An electrophoresis performed as in A was followed by western blotting using an anti-RbcL antibody. C, Western-blotting analysis of RbcL in the C. reinhardtii lines. Soluble proteins (40 µg) from pSS1-CrRbcS1 (C1, C2, and C3), pSS1-NtRbcS-M (M1, M2, and M3), and pSS1-NtRbcS-T (T2, T7, and T12) transformants were analyzed by SDS-PAGE and western blotting using an anti-RbcL antibody. The recipient strain (C−) lacking RbcS genes was used as a negative control. D and E, Native gel electrophoresis of soluble proteins (40 µg protein) from C. reinhardtii transformants. Soluble extracts of the recipient strain lacking RbcS genes (C−), strains expressing pSS1-NtRbcS-M (M), pSS1-NtRbcS-T (T), or pSS1-CrRbcS1 (C) were electrophoresed on a native gel and either stained with Coomassie Blue (D) or subjected to western blotting using an anti-RbcL antibody (E).
The Trichome RbcS Complements the Absence of Endogenous RbcS in C. reinhardtii
The difficulty of collecting large amounts of trichomes prevented us from extensive biochemical characterization of the trichome Rubisco. The influence of trichome and mesophyll RbcS on Rubisco catalysis was compared by nucleus transformation in the algae C. reinhardtii. Codon-optimized genes (Supplemental Fig. S3) for NtRbcS-T and a mesophyll-expressed tobacco RbcS (CAA26208.1, termed NtRbcS-M) were separately cloned into the transforming plasmid pSS1-CrRbcS1 (Genkov et al., 2010; Supplemental Fig. S4). Each plasmid, including pSS1-CrRbcS1, was introduced into the nucleus of the Rubisco-deficient rbcSΔ-T60-3 C. reinhardtii mutant strain that lacks both rbcS alleles (Genkov et al., 2010; Wachter et al., 2013). Multiple transformed lines for each genotype were isolated, with those producing NtRbcS-T and NtRbcS-M showing restored growth under light conditions, thus demonstrating the capacity of both NtRbcS-T and NtRbcS-M to assemble with C. reinhardtii RbcL subunits to form a functional RbcL8RbcS8 hybrid Rubisco. However, their growth rate was reduced when compared with the line expressing a C. reinhardtii RbcS (Fig. 5). A similar observation was reported for the expression of Arabidopsis or sunflower RbcS in C. reinhardtii (Genkov et al., 2010). These authors suggested that this reduced growth resulted from the absence of the chloroplast pyrenoid, whose formation requires the C. reinhardtii RbcS.
Figure 5.
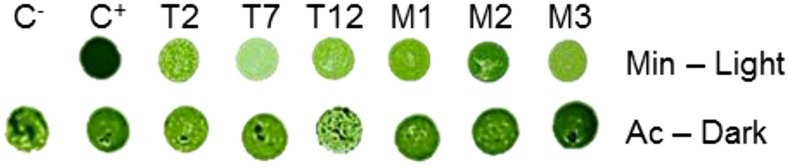
Tobacco RbcS-T and RbcS-M allow light-driven growth of C. reinhardtii. The recipient strain (C−) lacking the RbcS genes and the strains transformed with pSS1-CrRbcS1 (C+), pSS1-NtRbcS-T (T), or pSS1-NtRbcS-M (M) were spotted on either a minimal medium (Min) under light (75 µE m−2 s−1), or on an acetate medium (Ac) under dark conditions and grown for 11 d.
We confirmed the presence of Rubisco by western blotting using an antibody against the large subunit. The recipient strain lacking RbcS genes was used as a negative control, since this subunit is not detected although the gene is still present. Thus, the amount of RbcL observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) corresponds to the amount of Rubisco holoenzyme in vivo (Genkov and Spreitzer, 2009). RbcL was identified in all positive transformants (Fig. 4C). We then performed a native gel electrophoresis of a soluble extract of a clone obtained with each plasmid. Unlike the recipient C. reinhardtii strain, the three different transgenic lines produced distinct RbcL8RbcS8 enzymes with the hybrid complexes comprising C. reinhardtii RbcL and either NtRbcS-T or NtRbcS-M showing differences in mobility as seen in the plant extracts (Fig. 4D). The Rubisco complex with the C. reinhardtii small subunit had a lower mobility, in agreement with its higher pI (7.6) and higher Mw (16.25 kD). These differences were confirmed by western blotting (Fig. 4E).
The Trichome and Mesophyll RbcS Confer Different pH-Dependent Activity
Unlike the recipient strain used as a negative control, the three types of C. reinhardtii transgenic lines displayed Rubisco activity (data not shown). In order to determine if NtRbcS-T and NtRbcS-M confer different biochemical features to the Rubisco activity, this complex was purified by centrifugation on a Suc gradient (Supplemental Fig. S5). The Rubisco complex with NtRbcS-T displayed slightly higher Km (CO2) and Vmax of carboxylation than the complex with NtRbcS-M and reached values similar to those observed with CrRbcS1 (Table I). As O2 is a competitive inhibitor of CO2 fixation by Rubisco, the influence of NtRbcS-T and NtRbcS-M of the CO2/O2 competition of hybrid C. reinhardtii Rubisco was examined by comparing their relative carboxylase activities in assays equilibrated with no O2 (nitrogen bubbled) or high O2 under nonsaturating CO2 concentrations (2 mm NaHCO3; Table I). The difference in the activity between both conditions was similar, suggesting that both NtRbcS-T and NtRbcS-M have little or no impact on the CO2/O2 competition of the hybrid Rubisco at pH 8.0.
Table I. Kinetic properties of the carboxylase activity of Rubisco purified from the C. reinhardtii mutants expressing its own (CrRbcS1) small subunit as well as the mesophyll (NtRbcS-M) and the trichome (NtRbcS-T) small subunit from tobacco.
The means ± sd of three independent preparations are shown. For each parameter, asterisks indicate significant differences between enzymes according to a Kruskal-Wallis test (P < 0.05, n = 3) followed by a Tukey post-hoc test.
| Km (µm) | Vmax (µmol/min/mg) | Activity in the Presence of O2 (% of the Maximum Activity)a | |
|---|---|---|---|
| CrRbcS1 | 35.1 ± 2.9 | 1.84 ± 0.13 | 73.9 ± 3.5 |
| NtRbcS-M | 26.8 ± 1.6* | 1.60 ± 0.06* | 74.0 ± 3.0 |
| NtRbcS-T | 38.4 ± 5.1 | 1.96 ± 0.15 | 71.9 ± 3.9 |
Activity in the presence of O2 was calculated as a percentage of the maximum activity measured in the presence of N2 (see “Materials and Methods”).
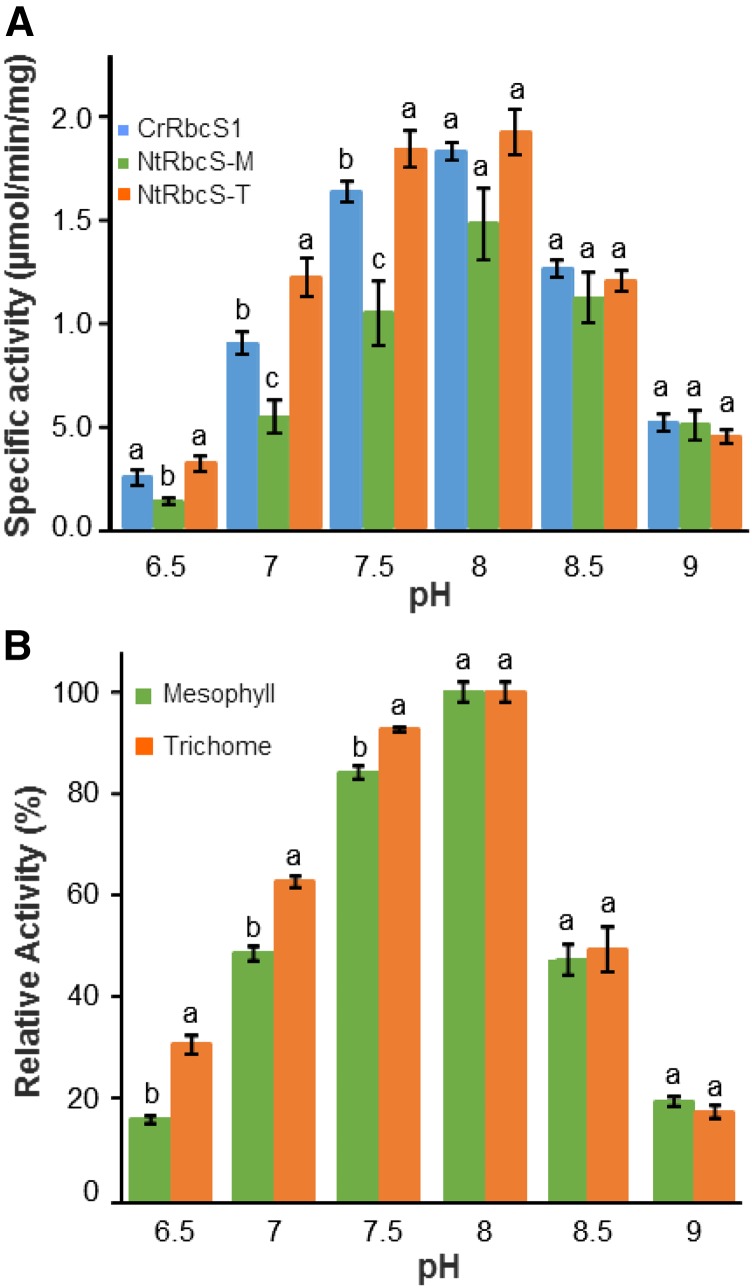
In the chloroplast, light irradiation and electron transport in the photosynthetic chain induce proton transport from the stroma to the thylakoid lumen. The resulting stroma alkalinization increases Rubisco activity (Ruuska et al., 2000). We investigated whether NtRbcS-T and NtRbcS-M confer different pH-dependent activities (Fig. 6A). Although all three enzymes had the same optimal pH (8.0), NtRbcS-T conferred a more acidic profile than NtRbcS-M and CrRbcS1.
Figure 6.
Comparison of Rubisco activity supported by NtRbcS-M and NtRbcS-T in C. reinhardtii and in tobacco. A, Specific activities of purified Rubisco from pSS1-CrRbcS1, pSS1-NtRbcS-M, or pSS1-NtRbcS-T C. reinhardtii transformants according to the pH values. Purified Rubisco (3 µg) was assayed for RuBP carboxylase activity in a buffer brought to the indicated pH. B, Relative activity of Rubisco from tobacco trichomes and leaves according to the pH. Soluble proteins from an extract from tobacco trichomes (30 µg) or leaf tissues clear of trichomes (15 µg) were assayed for RuBP carboxylase activity in a buffer brought to the indicated pH. The data represent the percentage of the maximal activity (at pH 8.0) for each sample. In A and B, the means ± the ses of four independent preparations are shown. Different letters indicate the significant differences between the transformants at a given pH according to a Kruskal-Wallis (A) or Mann-Whitney tests (B) followed by a Tukey post-hoc test (P < 0.05).
We sought to confirm with plant extracts the different behavior of Rubisco according to the pH. Purifying Rubisco from trichomes would require a huge amount of trichomes. We therefore directly measured the Rubisco activity in a soluble protein extract from isolated trichomes and from leaves cleared of trichomes. At the optimal pH (8.0), the RuBP carboxylase-specific activity was 0.41 ± 0.02 µmol min−1 mg−1 for the leaf extract, a value in the same range as that reported for a tobacco leaf extract (Bota et al., 2004). For the trichome extract, the activity was 0.078 ± 0.004 µmol min−1 mg−1. The 5-fold difference of carboxylase activity observed between the leaf and trichome extracts is in large part due to the dilution of the photosynthetic head cells of the long glandular trichomes by the stalk cells, the short trichomes, and the long nonglandular trichomes. This 5-fold difference in Rubisco activity is in agreement with the approximate 5-fold difference in Rubisco content between the trichome and leaf extracts as shown by western blotting (Supplemental Fig. S6).
We then compared the activity of the two extracts at different pH values. When reported with respect to the maximal activity at pH 8.0 (Fig. 6B), higher relative activity was observed at pH 6.5–7.5 for the trichome extract than for the leaf extract, which is thus in agreement with the data obtained with the purified Rubisco from the C. reinhardtii transgenic lines. For instance, at pH 6.5, the trichome relative activity was almost twice as high as that of the leaf extract.
DISCUSSION
Rubisco has been extensively studied in whole-leaf tissues in which mesophyll cells quantitatively prevail by large over glandular trichomes. No specific information is available concerning the Rubisco present in trichomes. This is probably due to the difficulty of isolating trichomes and the absence of clues indicating that trichome chloroplasts might differ from mesophyll chloroplasts regarding their basic metabolism. Here, we discovered that in tobacco, the Rubisco in trichomes has a specific small subunit, NtRbcS-T. This was supported by several pieces of evidence: proteomic and RT-PCR comparison of trichomes with other plant tissues, GUS gene reporter expression, and native gel electrophoresis.
The phylogenetic analysis showed a clear distinction between NtRbcS-T and the other tobacco RbcS. This was not just anecdotal since the 35 closest RbcS sequences (with a BLAST score higher than 200) representing 31 species all grouped together in the same cluster, cluster T (Fig. 3). However, the other RbcS sequences from the same species grouped in another cluster, cluster M. The RbcS-T sequences were predicted from annotated sequenced genomes, thus corresponding to proteins not yet functionally characterized, possibly because of their restricted expression, such is the case for the trichome-specific NtRbcS-T. On the other hand, cluster M includes all of the RbcS from Arabidopsis, which does not bear glandular trichomes. RbcS sequences corresponding to genes that are well expressed in mesophyll cells of C3 plants or in bundle-sheath cells of C4 plants belong to cluster M. For instance, this is the case for the RbcS of crystallized Rubisco (e.g. pea [Pisum sativum], tobacco, spinach [Spinacia oleracea], and rice) isolated from whole-leaf tissues. This double RbcS clustering is ancient, since it also concerns the monocots Elaeis guineensis (African oil palm) and Phoenix dactylifera (date palm), as well as Amborella trichopoda, which is the sole representative of a lineage in the angiosperms that appeared in parallel to the lineage leading to the current flowering plants (Amborella Genome Project, 2013). Since most of the RbcS-T were identified from genome sequences, it can be predicted that many more members will be identified as more plant genomes are sequenced. The observation that a T-type RbcS is absent in several monocot as well as dicot species suggests that independent losses of T-type RbcS genes occurred during evolution.
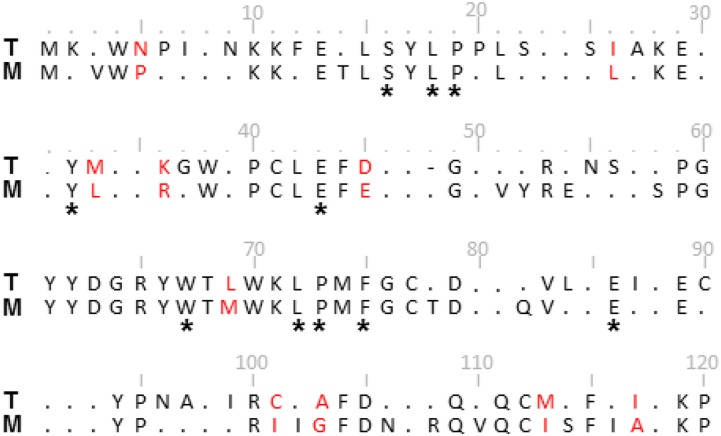
We aligned and generated a consensus sequence for NtRbcS-T and its closest sequences (T-consensus) and, independently, the RbcS sequences from the same species but belonging to the other cluster (M-consensus; Fig. 7; Supplemental Fig. S7). Comparison of the T- and M-consensus sequences shows almost the same number of conserved residues in cluster T (83) as in cluster M (79). The differences between the two consensuses are scattered all along the sequence. Ten amino acids differ between the two sequences, with major changes such as Pro-5 (M) into Asn (T), or Ile-101 (M) into Cys (T). Some positions that are poorly conserved in cluster M are highly conserved in cluster T, such as Asn-9, Ser-56, Ile-87, Cys-90, and Asn-96. A residue is deleted at position 48 of cluster T. Because the small subunit interacts with the large subunit, it may influence the catalytic efficiency and specificity of Rubisco (Spreitzer, 2003). Indeed, it has been demonstrated in several analyses of hybrid enzymes that small subunits can influence the catalytic efficiency and specificity of Rubisco (Spreitzer, 2003). We can thus hypothesize that NtRbcS-T induces a different Rubisco activity compared to NtRbcS-M. The C. reinhardtii expression system, used before to compare different plant RbcS (Genkov et al., 2010; Wachter et al., 2013), showed that NtRbcS-T and NtRbcS-M are both active. We then took advantage of the expression of NtRbcS-T and NtRbcS-M in C. reinhardtii in order to purify the Rubisco enzymes and compare their kinetic properties. Similar inhibition of the carboxylase activity was observed in the presence of saturating oxygen, suggesting that the CO2/O2 competition is the same for both enzymes. As previously shown by Wachter et al. (2013), the hybrid Rubisco made of NtRbcS-M displayed lower Vmax and Km than the wild-type C. reinhardtii Rubisco. However, NtRbcS-T conferred slightly but significantly higher Km and Vmax than NtRbcS-M (Table I), reaching values obtained with the C. reinhardtii RbcS. Higher catalytic turnover and lower affinity for CO2 are not only features of green algae Rubisco, but are also properties of Rubisco typically found in C4 plants (Sage, 2002). However, no close sequence from C4 plants was released from the BLAST analysis performed with NtRbcS-T as a query. In the phylogenetic tree, the two RbcS sequences of maize, a C4 plant, are included in cluster M. This observation indicates that the trichome- and C4-type Rubisco have independent origins.
Figure 7.
Alignment of the consensus sequences of the RbcS from clusters T and M. The consensus sequences were determined from the RbcS alignment shown in Supplemental Figure S7. Residues that differ between the two consensus sequences are in red. * indicates the RbcS residues conserved in greater than 95% of all known small subunit sequences (Spreitzer, 2003). Dots indicate less than 95% conservation within a cluster. Numbering is according to the RbcS-M alignment of Supplemental Figure S7. Note that the RbcS-T consensus sequence contains a deletion at position 48 (-).
Recently, an unusual rice Rubisco small subunit, OsRbcS1, genetically distant from other rice RbcS sequences, has been shown to be expressed in the leaf sheath, culm, anther, and root central cylinder (Morita et al., 2014). In the phylogenetic tree, OsRbcS1 is close to NtRbcS-T, being probably part of cluster T (Fig. 3). Expressing OsRbcS1 in the leaf blade of rice resulted in increased Rubisco catalytic turnover and lower affinity for CO2, as observed for the hybrid Rubisco made of NtRbcS-T and the C. reinhardtii RbcL. This result therefore indicates that RbcS sequences of cluster T might be sufficient to confer high catalytic activity to Rubisco.
Another interesting observation of this report is the different activity profile according to the pH between the hybrid RbcS-M and RbcS-T Rubisco enzymes (Fig. 6A). This was also the case when the Rubisco activity was directly compared between trichomes and leaf tissues cleared of trichomes (Fig. 6B). Although the profiles were slightly different in the C. reinhardtii and the native systems (this might be expected considering that the large subunit differs), a common observation was the relative higher activity observed at pH values below 8.0 for the trichome enzyme. These data thus demonstrate that NtRbcS-T is functionally different from NtRbcS-M.
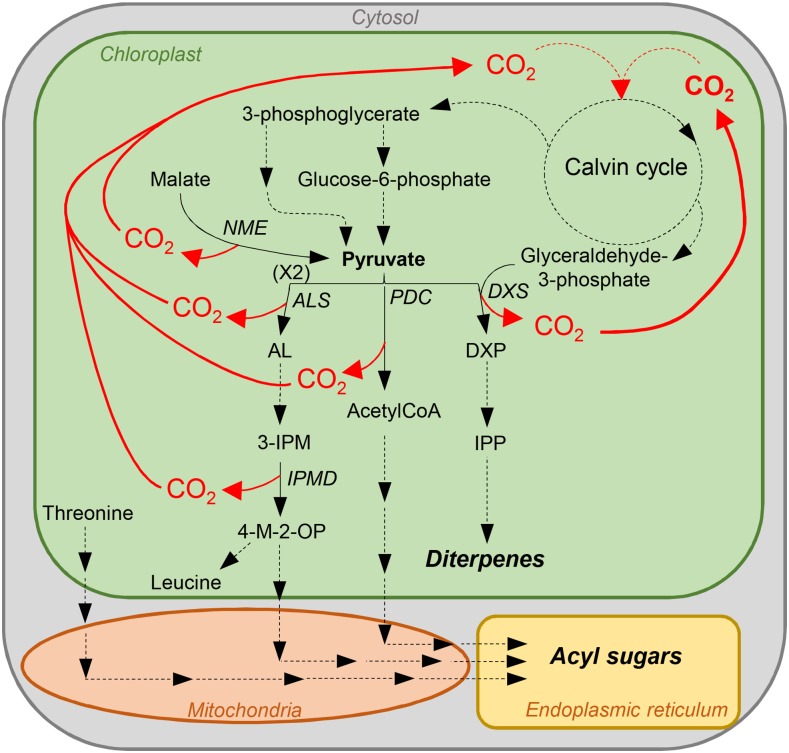
What might be the significance of a more active Rubisco preserving its activity at low pH? While the specialized metabolism has been the focus of trichome secretory cells, their overall primary and energy metabolism has barely been analyzed. CO2 is generated within these cells by the very active specialized metabolic pathways. Tobacco glandular trichomes indeed secrete large amounts of sugar esters and diterpenes (Wagner et al., 2004). In the plastid, the first step toward the C5 terpene precursor, isopentenyl pyrophosphate, combines pyruvate and glyceraldehyde-3-phosphate to give the C5 compound, 1-deoxy-d-xylulose 5-phosphate, as well as a CO2 molecule (Fig. 8). Glyceraldehyde-3-phosphate is a direct product of the Calvin cycle. Pyruvate might be generated together with CO2 from malate by the chloroplast NADP+-malic enzyme, which has been identified in proteomic analyses of tobacco trichomes (Van Cutsem et al., 2011; Sallets et al., 2014). Acyl acids of sugar esters are derived from Thr and the pyruvate pathways of branched-chain amino acid metabolism (Kandra et al., 1990). CO2 is produced by two enzymes: acetolactate synthase, which intervenes in the synthesis of branched-chain keto acid precursors, and 3-isopropylmalate dehydrogenase, which catalyzes the chemical reaction of 3-isopropylmalate into 4-methyl-2-oxopentanoate. Sugar esters are also acetylated via acetyl-coA produced together with CO2 from pyruvate by the pyruvate decarboxylase. These three enzymes have been identified in previous proteomic analyses of tobacco trichomes (Van Cutsem et al., 2011; Sallets et al., 2014). Thus, active terpene and sugar ester synthesis releases large amounts of CO2 in trichome cells, generating a CO2-rich environment similar to that found in algae or C4 plants and allowing for a Rubisco enzyme with lower affinity and higher catalytic activity. In addition, high CO2 production, since part of it is generated within the chloroplast stroma, might result in lowering the stroma pH (Savchenko et al., 2000). A Rubisco enzyme that keeps an activity at more acidic pH might thus be an asset under these conditions. Measuring the stroma pH in trichomes would be an important step to confirm this hypothesis. To decipher the physiological role of NtRbcS-T in CO2 fixation and the consequences on the trichome metabolism and the plant defense against biotic stress, genetic tools should be used. As trichomes are dispensable, silencing NtRbcS-T expression (e.g. by RNA interference or CRISPR-Cas9) would be an appropriate option.
Figure 8.
Model showing some of the CO2-generating metabolic pathways in tobacco glandular trichomes. NME, NADP+ malic enzyme; ALS, acetolactate synthase; AL, acetolactate; 3-IPM, 3-isopropylmalate; IPMD, 3-isopropylmalate dehydrogenase; 4-M-2-OP, 4-methyl-2-oxopentanoate; PDC, pyruvate decarboxylase; DXS, 1-deoxy-d-xylulose 5-phosphate synthase; DXP, 1-deoxy-d-xylulose 5-phosphate; IPP, isopentenyl pyrophosphate.
Except for NtRbcS-T described in this report and OsRbcS1 (Morita et al., 2014), the RbcS genes that belong to the cluster T have not been characterized so far, indicating that their expression is probably restricted to particular cell types. Nevertheless, their sequence conservation among distant species suggests that they are functional. It is tempting to hypothesize that the cluster T contains genes other than NtRbcS-T, which are expressed in glandular trichomes. Many of the 33 species represented in this cluster have glandular trichomes (Supplemental Table S2). Whether in all these cases glandular trichomes are photosynthetic is difficult to assess because this information is only mentioned in few cases. The obvious advantage of photosynthesis is that energy and carbon sources as well as oxygen are provided. However, leucoplasts are abundant in nonphotosynthetic secretory trichome cells and are very active in synthesizing specialized metabolites such as terpenoids. Considering the necessity mentioned above to recycle CO2 released during the synthesis of isoprenoids, a T-type Rubisco might still play the role of carbon recycling in the absence of photosynthesis. It would be therefore interesting to determine whether Rubisco and the other enzymes of the Calvin cycle are present in the leucoplasts.
We could also expect that in some cases, a T-type RbcS is expressed, not in trichomes, but in other cell types with chloroplasts or leucoplasts highly active in specialized metabolism. For instance, a T-type RbcS gene was found in Eucalyptus grandis, Citrus sinensis, and Citrus clementina. These species possess secretory cavities in the leaf and/or the fruit peel where essential oils (including terpenoids) are secreted from the surrounding specialized epithelial cells. Voo et al. (2012) compared transcripts from laser-dissected epithelial cells (which synthesize essential oils) and parenchyma cells (which do not synthesize oils) from grapefruit at two different developmental stages. An RbcS transcript (orange1.1g030738m) was 14.6 (earlier stage) and 7.7 (later stage) times more abundant in epithelial versus parenchyma cells. Actually, it turns out that this transcript corresponds to the RbcS gene from C. sinensis (KDO43856.1) listed in cluster T (Supplemental Table S2). This example, together with that of tobacco trichomes, supports a role for a T-type RbcS in secretory cells characterized by active specialized metabolism in the plastids. A direction for further research would consist of expressing a citrus or eucalyptus RbcS-T in C. reinhardtii and examining whether the resulting hybrid Rubisco recapitulates the kinetics and the pH profile obtained with NtRbcS-T.
In conclusion, we discovered that the photosynthetic glandular trichomes of tobacco contain a specific RbcS that is functional and confers different kinetics and different pH-dependent activity. We also showed that NtRbcS-T belongs to a distinct cluster that regroups RbcS from many other species. We propose that the T-type RbcS functions in a neutral or slightly acidic pH environment, which might be the case in specialized secretory cells where high CO2 concentration is found due to its release by the active specialized metabolism. This work also illustrates the originality of omics approaches to reveal genes that are expressed in more particular cell types and which code for variant enzymes that might be more adapted to particular metabolic environments.
MATERIALS AND METHODS
Plant Material
Tobacco (Nicotiana tabacum cv Petit Havana SR1; Maliga et al., 1973) was grown in a phytotron with a photoperiod of 16 h of light (300 µmol m−2 s−1) at 25°C and 8 h of darkness at 20°C. For analysis of root materials, plants were transferred to a hydroponic medium [1.25 mm Ca(NO)3, 1.25 mm MgSO4, 1.25 mm KNO3, 0.25 mm KH2PO4, 25 µm Fe(III)Na2EDTA, 12.5 µm H3BO3, 11.25 µm MnCl2, 9.5 µm ZnSO4, 0.75 µm CuSO4, 0.25 µm (NH4)6Mo7O24]. For analysis of the different plant tissues, stem, leaf, and root materials were taken just prior to the onset of flowering. Trichomes were removed from frozen leaves and stems as described by Van Cutsem et al. (2011). For analysis of seedlings, seeds were sterilized for 1 min in 70% ethanol and 5 min in 50% v/v commercial bleach. Seeds were vernalized at 4°C for 48 h and germinated in vitro on Murashige and Skoog medium (4.4 g/L MS salts [MP Biomedicals], pH 5.8 [KOH], 3% Suc, and 0.9% agar) at 25°C under a photoperiod of 16 h light (50 µmol photons m−2 s−1)/8 h darkness.
Tall Trichome Isolation for Proteomics Analysis
Tall trichomes were isolated as described by Sallets et al. (2014). In brief, trichomes were removed from leaf tissues by shaking in the presence of glass beads, and tall trichomes were then purified by centrifugation on a Percoll gradient.
2D-DIGE Analysis and Mass Spectrometry
Plant tissues were frozen in liquid nitrogen and ground with a mortar and pestle. Five hundred milligrams of each of the frozen samples were homogenized, and soluble proteins were obtained and precipitated by chloroform/methanol as previously described (Van Cutsem et al., 2011). Soluble protein samples of the different tissues were analyzed by 2D-DIGE (Champagne et al., 2012). For MS analysis, 500 µg of soluble proteins from tall trichomes were electrophoresed and stained with colloidal blue. Spots of interest were manually excised from the gel and digested with trypsin, and MALDI-MS/MS analysis was performed as previously described (Van Cutsem et al., 2011). The acquired MS/MS spectra were used to search the EST database of NCBI (July 5, 2012).
RNA Extraction and RT-PCR
Total RNA was isolated from trichomes, leaves, stems, flowers, seed pods, roots, and germinating seeds. Plant tissues were frozen in liquid nitrogen and ground with a mortar and pestle. Total RNA was extracted using a Sigma Spectrum Plant Total RNA Kit following the manufacturer’s protocol. Genomic DNA was removed using DNase I (Sigma-Aldrich). One microgram of total RNA was mixed with 2 µL of 40 µm oligo(dT), incubated for 5 min at 70°C, and then placed on ice for 5 min. Reverse transcription mixture (1 µL of RNase inhibitor [Promega], 1.5 µL of Moloney murine leukemia virus reverse transcriptase, 5 µL of 5× reaction buffer [Promega], 2.5 µL of 10 mm dNTPs [Promega], adjusted to 25 µL with water) was added, and the sample was incubated for 1 h at 42°C and for 5 min at 85°C, placed on ice for 5 min, and stored at −20°C. RT-PCR was performed for 35 cycles with denaturation for 30 s at 94°C, annealing for 30 s at 60°C, and extension for 1 min at 72°C, followed by a final extension of 7 min. The following primers were used: NtRbcS-T-F, 5′-ACATGATCCAAAAGGGTTGG-3′ and NtRbcS-T-R, 5′-CAAATGCCAAACAACGAATG-3′; CYP71D16-F, 5′-TGCTCAGTGCCAAAAATGTC-3′ and CYP71D16-R, 5′-ATTGAAGGCCTCCCTCTTTC-3′; ATP2-F, 5′-TCTTTGCTGGTGTTGGTCAA-3′ and ATP2-R, 5′-TGAGCTCATCCATACCCAAA-3′.
pNb-RbcS-T-GUSVENUS Fusion
The NbRbcS-T promoter region was identified by BLAST searching the draft genome of Nicotiana benthamiana in the Solgenomics database (http://solgenomics.net/) using the EST (GenBank accession number DV157962) sequence corresponding to NtRbcS-T. A 1240 bp fragment upstream of the start codon was amplified using the forward primer 5′-AAGCTTATTAGCATCAACCGGGTTAGC-3′ and the reverse primer 5′-GGTACCGTTCACCTTCACTTTAAGCTAC-3′ using N. benthamiana genomic DNA as a template. The PCR product was cloned into pGEM-T easy (Promega) and sequenced. The NbRbcS-T promoter was inserted in front of the GUSVENUS coding sequence in pAUX3131 (Navarre et al., 2011) using the HindIII-KpnI restriction sites. The fusion construct was excised using I-SceI and inserted into the pPZP-RCS2-nptII plant expression vector (Goderis et al., 2002), also cut with I-SceI. The construct was introduced into Agrobacterium tumefaciens LBA4404 virGN54D (van der Fits et al., 2000) for subsequent tobacco leaf disk transformation (Horsch et al., 1986).
Detection of GUS Expression
Histochemical staining of plant tissues for GUS activity was conducted as described (Bienert et al., 2012). Stained tissues were washed with 70% ethanol for chlorophyll extraction, and cleared trichomes were observed under a light microscope (Carl Zeiss MicroImaging) and photographed (Moticam 2300).
Chlamydomonas reinhardtii Strain, Plasmids, and Media
The C. reinhardtii strain used for the heterologous and homologous expression of tobacco (mesophyll and trichome) and C. reinhardtii RbcS genes was the cell-wall-less rbcSΔ-T60-3 strain (Genkov et al., 2010). This host strain lacks photosynthesis and requires acetate for growth due to the deletion of the 13-kb locus that contains the RbcS1 and RbcS2 gene family (Genkov et al., 2010). C. reinhardtii was cultured at 25°C in darkness on Tris-acetate phosphate medium (Harris, 1989). For selection of the transformants, C. reinhardtii was cultured in thymidine-5′-monophosphate medium (Harris, 1989) under light conditions (75 µE m−2 s−1). For solid medium, agar (Sigma-Aldrich) was added to the medium to a final concentration of 1.5% (w/v). Coding sequences for the mature small subunits of NtRbbcs-T and NtTbcS-M were synthesized (GenScript) with codons optimized for C. reinhardtii (Supplemental Fig. S3). These foreign RbcS coding sequences surrounded by their C. reinhardtii flanking sequences were digested with NcoI and BlpI and used to replace the NcoI-BlpI fragment in the CrRbcS1 gene of pSS1-ITP (Genkov et al., 2010) referred to here as pSS1-CrRbcS1. These new plasmids were named pSS1-NtRbcS-M and pSS1-NtRbcS-T. Transformation of cell-wall-less rbcSΔ-T60-3 cells was performed by electroporation (Shimogawara et al., 1998).
Soluble Protein Extraction from C. reinhardtii
Cells (250 mL culture) were collected by centrifugation at 4,000g for 10 min at room temperature (Beckman Coulter JLA10.500 rotor). After resuspension in 5 mL of extraction buffer (50 mm Bicine, pH 8.0 [NaOH], 10 mm NaHCO3, and 10 mm MgCl2), supplemented just before use with 10 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1 µg mL−1 each of protease inhibitors (leupeptin, aprotinin, antipain, pepstatine, chymostatin), the cells were sonicated (VibraCell 75022) at 4°C for 5 min. Cell debris were removed by two consecutive centrifugations at 3,000g (10 min) and 8,000g (15 min; Eppendorf 5430) at 4°C. After an additional centrifugation at 180,000g (Beckman-Coulter MLA80 rotor) for 25 min at 4°C, the soluble proteins in the supernatant were quantified.
Soluble Protein Extraction from Tobacco
Leaves were frozen in liquid nitrogen, and the trichomes were removed using a frozen painting brush. Trichomes and the trichome-free leaves were homogenized separately in 3 mL of extraction buffer described above using a Wheaton tissue grinder (5 mL capacity). The cell debris were removed by centrifugation at 3,000g (Eppendorf 5417C) at 4°C for 10 min. After an additional centrifugation at 180,000g (Beckman-Coulter TLA55 rotor) for 15 min at 4°C, the soluble proteins in the supernatant were quantified.
Rubisco Purification
Five milliliters of the soluble proteins from the C. reinhardtii cells were obtained as described above and supplemented with ammonium sulfate (60% saturation). After 2 h incubation, the samples were centrifuged at 4°C for 20 min at 10,000g. The pellet was solubilized in 1 mL of extraction buffer described above (without polyvinylpyrrolidone), and an additional centrifugation at 4°C for 15 min at 10,000g was performed in order to remove residual aggregates. One milliliter of proteins was then loaded on a 10 mL 10% to 30% Suc gradient prepared in the extraction buffer described above (without polyvinylpyrrolidone) and centrifuged at 250,000g for 17 h at 4°C (Beckman Coulter SW40 Ti swinging-bucket rotor). Fractions (0.5 mL) were collected and analyzed by SDS-PAGE and colloidal blue staining. Rubisco-containing fractions were pooled, and their Rubisco content was quantified by SDS-PAGE, using bovine serum albumin as a standard. The gel pictures were then analyzed using ImageJ software.
Rubisco Activity
The in vitro carboxylation activity of Rubisco was measured continuously in a buffered medium (50 mm Bicine, 50 mm Bis-Tris, and 50 mm HEPES) using an NADH-linked coupled enzyme system (Kubien et al., 2011). The RuBP (83895), NADH (N4505), ATP (A3377), phosphocreatine (P7936), and enzymes (creatine phosphokinase [C3755], carbonic anhydrase [C3134], glyceraldehyde-3-P dehydrogenase [G2267], 3-phosphoglyceric phosphokinase [7635], α-glycerophosphate dehydrogenase-triosephosphate isomerase [G1881]) were purchased from Sigma-Aldrich. Km and Vmax were determined at pH 8.0 using 0.3, 0.6, 0.9, 1.3, 2, 2.3, 4.3, 6.3, and 10.3 mm NaHCO3. The pH-dependent carboxylase activity was performed in the presence of 10 mm NaHCO3. The CO2/O2 competition of Rubisco was determined at pH 8.0 in the presence of 2 mm NaHCO3 and was calculated from the carboxylase activity measured in the presence of O2 as a percentage of the maximum activity measured in the presence of N2. The reaction medium (except for enzymes and NaHCO3) was bubbled for 20 min with the appropriate gas prior to measurements.
Electrophoretic Analysis
SDS-PAGE was performed according to Laemmli (1970) using iD PAGE Gel (Eurogentec). Native gel electrophoresis was performed using 4% to 20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) and the same buffers as in Laemmli (1970), except that SDS and dithiothreitol were omitted.
Antibodies and Western Blotting
Rabbit anti-RbcL (Rubisco large subunit, form I and form II; 1:4,000) and horseradish peroxidase-coupled anti-rabbit IgG (1:5,000) antibodies were purchased from Agrisera and Roche, respectively.
Sequence Alignment and Phylogenetic Tree Building
The RbcS amino acid sequences were aligned using the ClustalW Multiple alignment (Thompson et al., 1997) and imported into the Molecular Evolutionary Genetics Analysis package version 4 (Tamura et al., 2007). Phylogenetic analyses were conducted using the neighbor-joining method implemented in Molecular Evolutionary Genetics Analysis, (Saitou and Nei, 1987), with the pairwise deletion option for handling alignment gaps and with the equal input correction model with the heterogeneous pattern among lineages for distance computation. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The phylogenetic trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The accession numbers and the organisms corresponding to sequences used for this tree are listed in Supplemental Table S2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KU162868 (NtRbcS-T promoter sequence) and DV157962 (NtRbcS-T coding sequence).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. 2D-DIGE of soluble proteins of tall glandular trichomes compared to other tissues.
Supplemental Figure S2. Preparative 2-DE of soluble tall glandular trichome proteins.
Supplemental Figure S3. Synthetic coding sequences of NtRbcS-T and NtRbcS-M for expression in C. reinhardtii.
Supplemental Figure S4. Engineered plasmids containing the wild-type C. reinhardtii rbcS1 gene (pSS1-CrRbcS1) and the tobacco mesophyll- and trichome-expressed rbcS cDNA sequences (pSS1-NtRbcS-M and pSS1-NtRbcS-T).
Supplemental Figure S5. Electrophoretic analysis of the purified Rubisco from C. reinhardtii transformants.
Supplemental Figure S6. Comparison of the Rubisco amount in trichome and leaf samples of tobacco.
Supplemental Figure S7. Alignment of the RbcS amino acid sequences.
Supplemental Table S1. MS-MS identification of the 49 most abundant trichome-specific protein spots.
Supplemental Table S2. List of the protein sequences used for the phylogenetic analysis of the RbcS.
Supplementary Material
Acknowledgments
We thank Robert J. Spreitzer for helpful discussion and for the C. reinhardtii transformation system. We also thank Nadine Coosemans, Hervé Degand, Anne-Marie Faber, and Joseph Nader for excellent technical help.
Footnotes
This work was supported by the Interuniversity Poles of Attraction Program (Belgian State, Scientific, Technical, and Cultural Services), by the Belgian National Fund for Scientific Research, and by an ERA-CAPS grant. R.L. held a FRIA fellowship. M.P. held an EU Marie Skłodowska-Curie fellowship.
Articles can be viewed without a subscription.
References
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Andersson I, Backlund A (2008) Structure and function of Rubisco. Plant Physiol Biochem 46: 275–291 [DOI] [PubMed] [Google Scholar]
- Bienert MD, Delannoy M, Navarre C, Boutry M (2012) NtSCP1 from tobacco is an extracellular serine carboxypeptidase III that has an impact on cell elongation. Plant Physiol 158: 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota J, Medrano H, Flexas J (2004) Is photosynthesis limited by decreased Rubisco activity and RuBP content under progressive water stress? New Phytol 162: 671–681 [DOI] [PubMed] [Google Scholar]
- Boutry M, Chua NH (1985) A nuclear gene encoding the beta subunit of the mitochondrial ATP synthase in Nicotiana plumbaginifolia. EMBO J 4: 2159–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne A, Rischer H, Oksman-Caldentey KM, Boutry M (2012) In-depth proteome mining of cultured Catharanthus roseus cells identifies candidate proteins involved in the synthesis and transport of secondary metabolites. Proteomics 12: 3536–3547 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ (2010) Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: Engineered rbcS cDNA for expression in Chlamydomonas. J Biol Chem 285: 19833–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genkov T, Spreitzer RJ (2009) Highly conserved small subunit residues influence Rubisco large subunit catalysis. J Biol Chem 284: 30105–30112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas JJ, Schimmel BCJ, Alba JM, Escobar-Bravo R, Schuurink RC, Kant MR (2012) Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int J Mol Sci 13: 17077–17103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis IJWM, De Bolle MFC, François IEJA, Wouters PFJ, Broekaert WF, Cammue BPA (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol 50: 17–27 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego, CA [Google Scholar]
- Horsch RB, Klee HJ, Stachel S, Winans SC, Nester EW, Rogers SG, Fraley RT (1986) Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proc Natl Acad Sci USA 83: 2571–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H (2011) Functional incorporation of sorghum small subunit increases the catalytic turnover rrate of Rubisco in transgenic rice. Plant Physiol 156: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandra L, Severson R, Wagner GJ (1990) Modified branched-chain amino acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur J Biochem 188: 385–391 [DOI] [PubMed] [Google Scholar]
- Keene CK, Wagner GJ (1985) Direct demonstration of duvatrienediol biosynthesis in glandular heads of tobacco trichomes. Plant Physiol 79: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien DS, Brown CM, Kane HJ (2011) Quantifying the amount and activity of Rubisco in leaves. Methods Mol Biol 684: 349–362 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lange BM. (2015) The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu Rev Plant Biol 66: 139–159 [DOI] [PubMed] [Google Scholar]
- Lange BM, Turner GW (2013) Terpenoid biosynthesis in trichomes—current status and future opportunities. Plant Biotechnol J 11: 2–22 [DOI] [PubMed] [Google Scholar]
- Maliga P, Sz-Breznovits A, Márton L (1973) Streptomycin-resistant plants from callus culture of haploid tobacco. Nat New Biol 244: 29–30 [DOI] [PubMed] [Google Scholar]
- Morita K, Hatanaka T, Misoo S, Fukayama H (2014) Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of Rubisco in rice. Plant Physiol 164: 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre C, Sallets A, Gauthy E, Maîtrejean M, Magy B, Nader J, Pety de Thozée C, Batoko H, Boutry M (2011) Isolation of heat shock-induced Nicotiana tabacum transcription promoters and their potential as a tool for plant research and biotechnology. Transgenic Res 20: 799–810 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Andrews TJ, Badger MR, Price GD, von Caemmerer S (2000) The role of chloroplast electron transport and metabolites in modulating Rubisco activity in tobacco. Insights from transgenic plants with reduced amounts of cytochrome b/f complex or glyceraldehyde 3-phosphate dehydrogenase. Plant Physiol 122: 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. (2002) Variation in the kcat of Rubisco in C3 and C4 plants and some implications for photosynthetic performance at high and low temperature. J Exp Bot 53: 609–620 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sallets A, Beyaert M, Boutry M, Champagne A (2014) Comparative proteomics of short and tall glandular trichomes of Nicotiana tabacum reveals differential metabolic activities. J Proteome Res 13: 3386–3396 [DOI] [PubMed] [Google Scholar]
- Savchenko G, Wiese C, Neimanis S, Hedrich R, Heber U (2000) pH regulation in apoplastic and cytoplasmic cell compartments of leaves. Planta 211: 246–255 [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys 414: 141–149 [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Salvucci ME (2002) Rubisco: Structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53: 449–475 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tissier A. (2012) Glandular trichomes: What comes after expressed sequence tags? Plant J 70: 51–68 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Simonart G, Degand H, Faber AM, Morsomme P, Boutry M (2011) Gel-based and gel-free proteomic analysis of Nicotiana tabacum trichomes identifies proteins involved in secondary metabolism and in the (a)biotic stress response. Proteomics 11: 440–454 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Deakin EA, Hoge JHC, Memelink J (2000) The ternary transformation system: Constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol Biol 43: 495–502 [DOI] [PubMed] [Google Scholar]
- Voo SS, Grimes HD, Lange BM (2012) Assessing the biosynthetic capabilities of secretory glands in Citrus peel. Plant Physiol 159: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter RM, Salvucci ME, Carmo-Silva AE, Barta C, Genkov T, Spreitzer RJ (2013) Activation of interspecies-hybrid Rubisco enzymes to assess different models for the Rubisco-Rubisco activase interaction. Photosynth Res 117: 557–566 [DOI] [PubMed] [Google Scholar]
- Wagner GJ. (1991) Secreting glandular trichomes: More than just hairs. Plant Physiol 96: 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GJ, Wang E, Shepherd RW (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot 93: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Gan S, Wagner GJ (2002) Isolation and characterization of the CYP71D16 trichome-specific promoter from Nicotiana tabacum. J Exp Bot 53: 1891–1897 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In Bryson V, Vogel HJ, eds, Evolving Genes and Proteins. Academic Press, New York, pp 97–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.